Abstract
Objective:
Cocaine use disorders (CUDs) represent a major public health problem in many countries. To better understand the interaction between the environmental modulations and phenotype, the aim of the present study was to investigate the DNA methylation pattern of CUD patients, who had concomitant cocaine and crack dependence, and healthy controls.
Methods:
We studied DNA methylation profiles in the peripheral blood of 23 CUD patients and 24 healthy control subjects using the Illumina Infinium HumanMethylation450 BeadChip arrays.
Results:
Comparison between CUD patients and controls revealed 186 differentially methylated positions (DMPs; adjusted p-value [adjP] < 10-5) related to 152 genes, with a subset of CpGs confirmed by pyrosequencing. DNA methylation patterns discriminated CUD patients and control groups. A gene network approach showed that the EHMT1, EHMT2, MAPK1, MAPK3, MAP2K1, and HDAC5 genes, which are involved in transcription and chromatin regulation cellular signaling pathways, were also associated with cocaine dependence.
Conclusion:
The investigation of DNA methylation patterns may contribute to a better understanding of the biological mechanisms involved in CUD.
Keywords: Cocaine, crack cocaine, DNA methylation, drug dependence, epigenetics
Introduction
Cocaine is an alkaloid extracted from Erythroxylon coca and Erythroxylon novogranatense, two plant species that grow mainly in South America and whose use has been reported for over 5,000 years.1 However, over the last 30 years, abuse of and dependence on cocaine and its smoked form, known as crack, emerged as a major public health problem in many countries.2 Brazil is one of the greatest consumer markets for cocaine worldwide (3.2 million people), and the world’s largest consumer of crack (1.8 million people).3,4
Crack has become a drug of choice due to its low price and more intense effects compared to other drugs; its onset of action is immediate and it provides higher cocaine concentration levels (40-85%) compared to other routes, such as snorted, injected, and inhaled.5,6 Since crack and other similar products simply represent different formulations of the same substance (cocaine), they exert similar effects in the body, with differences arising from the route of administration. The main effects of cocaine and crack in the brain involve modulation of neurotransmitters from the mesolimbic-mesocortical system by blocking dopamine reuptake in the synaptic cleft. This results in euphoria and a feeling of wellbeing, which may lead the user to repeat the experience and, therefore, increase vulnerability to drug dependence.1,6
Drug dependence behavior results from complex interactions between genetic and environmental factors, and the study of epigenetics has been suggested as a potential method of better understanding these mechanisms.7 Epigenetics modulate gene expression throughout life in response to environmental exposures, such as drug consumption. Different epigenetic approaches have been used to elucidate this interaction, such as DNA methylation and histone modifications, which are associated with synaptic plasticity and memory formation, as well as with behavioral responses to drugs of abuse.8,9 DNA methylation machinery in brain regions related to reward undergo changes after cocaine exposure. These include the enzymes that catalyze DNA methylation (DNMTs) and demethylation (TETs), both able to induce stable changes in neuronal function and behavior, which are directly linked to chromatin remodeling and epigenetic modulation.9,10
Changes in genome-wide DNA methylation have been explored successfully in other psychiatric disorders, such as bipolar disorder, schizophrenia, major depression, and posttraumatic stress disorder,11,12 as well as to evaluate the correspondence between DNA methylation in brain tissue and accessible peripheral tissues (e.g., blood).13,14 Within this context, we researched DNA methylation patterns in patients with cocaine use disorder (CUD) and controls, aiming to explore this biological mechanism potentially involved with drug dependence by in silico analyses.
Methods
Sample groups
Samples were selected from the DNA Bank of the Departamento e Instituto de Psiquiatria, Faculdade de Medicina da Universidade de São Paulo (FMUSP), São Paulo, Brazil. Genomic DNA extracted from peripheral blood was selected from 23 CUD patients (originally, 24 patients were selected, but one of them was identified as being 47, XXY during the analysis and therefore excluded from the study) and 24 healthy controls, all males ranging between 23 and 29 years of age. Cases were recruited and evaluated as inpatients and outpatients from seven drug-dependence treatment clinics in São Paulo. Individuals with another psychiatric diagnosis (such as psychosis) or any chronic organic illness, such as diabetes or other metabolic disorders, were excluded. All current cocaine users then took part in a structured interview to collect data on sociodemographic characteristics, sexual behaviors, and a drug-use profile. Interviews were conducted by three researchers (experienced psychiatrists or psychologists) who had been trained to use the questionnaire. All cases met ICD-10 criteria for diagnosis of cocaine dependence and had 8-to-10-year histories of concomitant cocaine and crack dependence as well as use/abuse of other drugs, such as cannabis, solvents, heroin, and tobacco. The control group consisted of volunteer blood donors who completed a short questionnaire designed to screen for contagious diseases and use of any kind of drug. Volunteers with a history of abuse or recent contact with any potential drugs of abuse, contagious diseases, or who had any known current psychiatric condition or lifetime history of psychiatric hospitalization were not selected for this study (Table 1). A detailed description of the above groups can be found elsewhere.15,16 The study was approved by the local research ethics committee (protocol 143/13) and conducted in accordance with the Declaration of Helsinki.
Table 1. Sample sociodemographic and drug-use characteristics.
| Cases (n=23) | Controls (n=24) | p-value* | |
|---|---|---|---|
| Age (years) | |||
| 23 | 7 (30.4) | 7 (29.2) | 0.9999 |
| 24 | 4 (17.4) | 4 (16.7) | |
| 25 | 2 (8.7) | 2 (8.3) | |
| 26 | 1 (4.3) | 1 (4.2) | |
| 27 | 2 (8.7) | 3 (12.5) | |
| 28 | 3 (13.0) | 3 (12.5) | |
| 29 | 4 (17.4) | 4 (16.7) | |
| Ethnicity | |||
| White | 13 (56.5) | 13 (54.2) | 0.8710 |
| African | 10 (43.5) | 11 (45.8) | |
| Education | |||
| Primary education or less | 19 (82.6) | 10 (41.7) | 0.0039 |
| Secondary education | 4 (17.4) | 14 (58.3) | |
| Other drugs used | |||
| Tobacco | 21 (91.3) | - | - |
| Cannabis | 23 (100) | - | - |
| Solvents | 6 (26.1) | - | - |
| Heroin | 1 (4.3) | - | - |
Data presented as n (%).
Chi-square test (99% confidence interval).
Genome-wide DNA methylation
Genomic DNA was extracted from peripheral blood using the “salting out” method. A total of 500 ng of DNA was bisulfite-converted using the EZ DNA methylation kit (Zymo Research, Irvine, California, USA), following the manufacturer’s instructions. Converted DNA was denatured, neutralized, and amplified by isothermal polymerase chain reaction (PCR) for 16 hours (16 cycles: 95 °C, 30 seconds; 50 °C, 60 minutes; 4 °C, 10 minutes). DNA was hybridized using the Illumina Infinium HumanMethylation450 BeadChip arrays (450K) (Illumina Inc, San Diego, California, USA), following the Illumina Infinium HD methylation protocol. Arrays were then scanned with the iScan Microarray Scanner (Illumina), and IDAT files were extracted using GenomeStudio software (version 1.8). Data were submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) (GSE77056).
DNA methylation analyses
Methylation data was analyzed in ChAMP (Chip Methylation Analysis Pipeline)17 and RnBeads18 packages, both available in Bioconductor (https://www.bioconductor.org/). ChAMP quality control steps used a threshold of p > 0.01 to remove 1,261 probes with poor intensity detection values. Next, we removed 281 probes represented by less than three beads and 37,301 probes located in single nucleotide polymorphisms (SNPs) (dbSNP database) or that aligned at multiple locations.19 These pre-processing steps left 446,669 probes to be analyzed in 47 samples. To account for the different contributions of DNA methylation levels due to the heterogeneity underlying the cell composition of the blood, we applied cellular correction using the estimateCellCounts function, which inferred the distribution of the white blood cells (CD8+ and CD4+ T cells, B cells, monocytes, granulocytes, and natural killer cells).20,21 The beta-mixture quantile (BMIQ) normalization method was used to adjust the intensity distribution of the Infinium II probes to fit that of the Infinium I probes.22
For RnBeads, the Greedycut algorithm removed unreliable measurements for 813 probes (p > 0.05), probes that were associated with SNPs (4,713 probes), as well as those located in specific contexts (non-CpG positions, 3,140 probes). The background was corrected with the noob method, which is based on a normal-exponential convolution using out-of-band probes,23 resulting in 476,911 probes in 47 samples. Signal intensities from type I and II probes were normalized using the SWAN method, which adjusts intensities based on a quantile approach.24
Statistical analysis
Comparison between cases and controls using the ChAMP and RnBeads packages was applied to M-values (logit of B-values) employing an empirical bayesian framework linear model from limma.25,26 Positions with an adjusted p-value (adjP) < 10-5 as given by the Benjamini & Hochberg method, presenting the same methylation direction, were considered as differentially methylated positions (DMPs). We also used RnBeads to identify methylation differences at predefined genomic regions; to do so, we combined the uncorrected CpG-specific p-values within a given region using an extension of Fisher’s method that resulted in a single aggregated p-value for each region, also adjusted for multiple testing correction using the false discovery rate (FDR).18 We considered differentially methylated regions (DMRs) as those presenting an adjP of < 0.05.
To verify the methylation pattern in cases and controls, a hierarchical cluster analysis was applied to the M-values from DMPs using Ward’s method, Euclidean distance with 1,000 resampling, and z-score normalization as parameters, as implemented by the fpc package.27
Pyrosequencing
Gene-specific CpG methylation was evaluated by pyrosequencing. Primers were designed using PyroMark Assay Design SW 2.0 software (Qiagen, Hilden, Germany) to amplify the bisulfite-treated DNA of six DMPs located in the C11orf58 (cg10633981), HDAC5 (cg11049075), MAP2K1 (cg24449302), MPV17L (cg09844907), LOC401431 (cg17479131) genes and in an intergenic region (IGR) (cg19933985). Primers and PCR conditions are shown in Table 2. PCR was performed using the PyroMark PCR Kit (Qiagen) and PyroMark Q24 system (Qiagen), following the manufacturer’s instructions. For each assay, the software included at least one control dispensation to adjust signal over background noise and verify the efficiency of bisulfite conversion. Methylated and unmethylated bisulfite-converted human control DNA (EpiTect PCR control DNA, Qiagen) was used as positive and negative controls.
Table 2. Pyrosequencing PCR primer sequences and conditions.
| ProbeID | Gene | Primers | Tm (°C) |
|---|---|---|---|
| cg10633981 | C11orf58 | Forward: 5′-TAGAAATAATTGGTGAGGGTAGGA-3′ | 58 |
| Reverse: 5′-ACTTCCTAATCTATCAATTCAATATCATAA-3′ – Biotin | 58 | ||
| Sequencing: 5′-AGGGTTGGGAATTTT-3′ | 44 | ||
| cg19933985 | IGR | Forward: 5′-TTGGAATTTAGGTGAAGATGTT-3′ – Biotin | 56 |
| Reverse: 5′-CATTAACTAAACCATCATTATACAACC-3′ | 58 | ||
| Sequencing: 5′-ACCATCATTATACAACCT-3′ | 44 | ||
| cg11049075 | HDAC5 | Forward: 5′-TTTTTTAAAGGGATGTGAGTAGGAAGAATT-3′ | 61 |
| Reverse: 5′-CCCAAATAACTACAAAAACAAAAATTTACT-3′ – Biotin | 59 | ||
| Sequencing: 5′-ATTTTTAAATATATAATTAAGATGT-3′ | 38 | ||
| cg24449302 | MAP2K1 | Forward: 5′-GGTATTTGGTTTGGAGAGTTTAT-3′ | 57 |
| Reverse: 5′-CTTTCCACACTCCCACTC-3′ – Biotin | 57 | ||
| Sequencing: 5′-GGATTTAGTTTTTAGGTAGT-3′ | 39 | ||
| cg09844907 | MPV17L | Forward: 5′-GGGTTTTTTTAGTTTTTAGTAGTTATTGT-3′ | 57 |
| Reverse: 5′-CCTTCTACCTCTATAACTTTACCTATTCT-3′ – Biotin | 60 | ||
| Sequencing: 5′-TGTATTTGTTTTTTTTAGAAGTTT-3′ | 44 | ||
| cg17479131 | LOC401431 | Forward: 5′-GAATTAGAAAGTATATGATGGAGGTTTTAG-3′ | 58 |
| Reverse: 5′-ATATAAACCTCTTAAAACAAACTACACT-3′ – Biotin | 57 | ||
| Sequencing: 5′-AAGAGTATATATGAGTTGATATAG-3′ | 44 |
Tm = temperature (annealing).
For each CpG position, methylation percentages provided by pyrosequencing were calculated as the ratio of C to C + T, as implemented in the PyroMark Q24 2.0.7 software (Qiagen) and normalized by positive controls. Mann-Whitney U tests were applied to identify differences between case and control groups, considering significant those CpG positions with p < 0.05. Correlations between genome-wide and pyrosequencing data were analyzed with linear regression using GraphPad Prism version 7.03 (GraphPad for Science, San Diego, CA, USA).
Network analysis
Genes associated with DMPs were submitted to WebGestalt28 to identify enriched protein-protein interaction (PPI) modules using the Biological General Repository for Interaction Datasets29 (BioGRID), considering significant modules with adjP < 0.05 as given by the Benjamini-Hochberg test. The random walk method and Gene Ontology (GO) enrichment analysis based on the retrieved sub-network (FDR < 0.05) were used to identify hyperrepresented PPI modules.28 We also used MetaCore™ software with standard parameters (https://portal.genego.com/) (p < 0.05, hypergeometric distribution; z score > 45) to further explore enriched functional annotation of DMPs in a gene network context.
Results
Identification of changes in DNA methylation in the peripheral blood of cocaine use disorder (CUD) patients
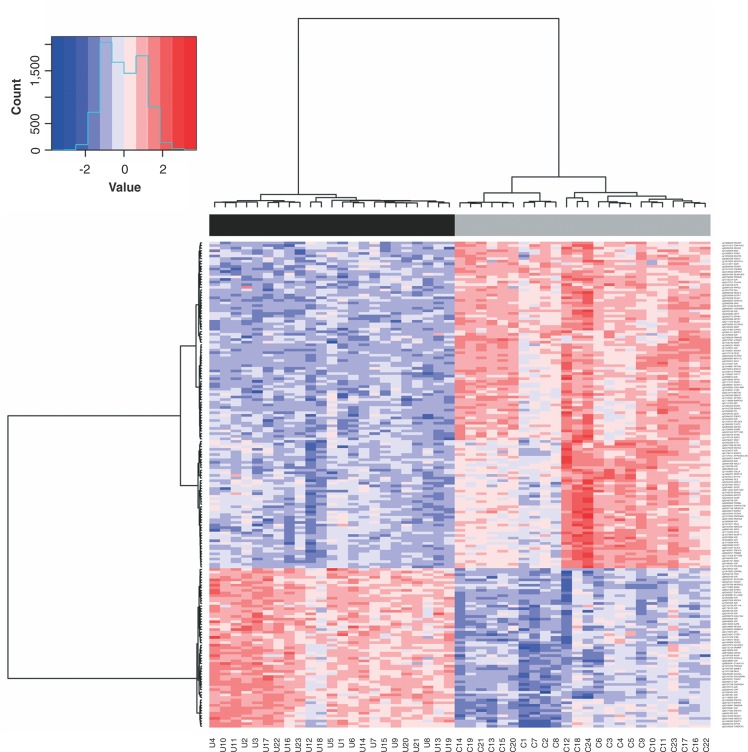
Genome-wide DNA methylation differences between 23 CUD patients and 24 healthy controls were identified using the ChAMP & RnBeads package analysis. ChAMP revealed 249 DMPs, with 108 hypermethylated and 141 hypomethylated (adjP < 10-5), and RnBeads showed 361 DMPs, with 69 hypermethylated and 292 hypomethylated (adjP < 10-5). The overlap between both methods resulted in 186 DMPs related to 152 genes, all in the same direction, with 61 hypermethylated and 125 hypomethylated CpG positions. According to the distribution of the positions in relation to their genomic location and CpG characterization, most of the DMPs are located in promoter regions (5′UTR, TSS200, and TSS1500) (82-44%) and in a CpG island (83-45%) (Table S1 (1MB, pdf) , available as online-only supplementary material). Thus, we verified the existence of DMRs, identifying 118 DMRs: four located in CpG islands, 48 in promoters, 37 in gene bodies, and 29 in genome-wide tiling regions (Table S2 (1MB, pdf) , available as online-only supplementary material). Five genes were common to DMP and DMR analyses: KLHL9, WDR61, OSMR, SNORD94, and ARF4. A hierarchical clustering analysis applied to M-values showed that methylation patterns from the 186 DMPs discriminated two groups of samples (cluster stability [boot mean]), which completely separated CUD patients from healthy controls (Figure 1).
Figure 1. Hierarchical clustering based on the methylation patterns of 186 differentially methylated positions (DMPs) between patients with cocaine use disorder (CUD) and controls (Table S1 (1MB, pdf) ). Top: Complete separation between CUD and control groups; Bottom: Sample name. DNA methylation level is represented by color intensity in the users in relation to controls. C = controls; U = CUD patients.
Correlations between genome-wide and pyrosequencing data
To confirm the methylation levels of selected DMPs, pyrosequencing was carried out for six CpG positions: five located next to genes and one in the IGR. Four CpGs were chosen, as they presented the highest methylation differences: cg10633981 (C11orf58), cg09844907 (MPV17L), cg17479131 (LOC401431), and cg19933985 (IGR), as well as two additional CpGs located at genes previously related to cocaine dependence (cg11049075 – HDAC5, and cg24449302 – MAP2K1).30,31 Methylation levels of three CpG positions were correlated between B-values and pyrosequencing data (HDAC5, MAP2K1, and cg19933985) (Table 3).
Table 3. Correlation between genome-wide and pyrosequencing methylation data.
| Δβ | Pyrosequencing | Mann-Whitney U test | Linear regression | ||||
|---|---|---|---|---|---|---|---|
| ProbeID | Gene | ChAMP | RnBeads | Log2(meancontrol/meancase) | p-value | R2 | p-value |
| cg10633981 | C11orf58 | -0.167 | -0.166 | 0.043* | < 0.0001 | 0.319 | < 0.0001 |
| cg19933985 | IGR | -0.145 | -0.148 | -0.072 | < 0.0001 | 0.675 | < 0.0001 |
| cg11049075 | HDAC5 | -0.090 | -0.099 | -0.040 | 0.0004 | 0.130 | 0.016 |
| cg24449302 | MAP2K1 | 0.063 | 0.088 | 0.178 | 0.0001 | 0.277 | 0.0002 |
| cg17479131 | LOC401431 | 0.145 | 0.133 | -0.352* | 0.002 | 0.021 | 0.3433 |
| cg09844907 | MPV17L | 0.048 | 0.140 | 0.095 | 0.5651 | 0.045 | 0.1731 |
Discordant.
Characterization of genes associated with differentially methylated CpG positions
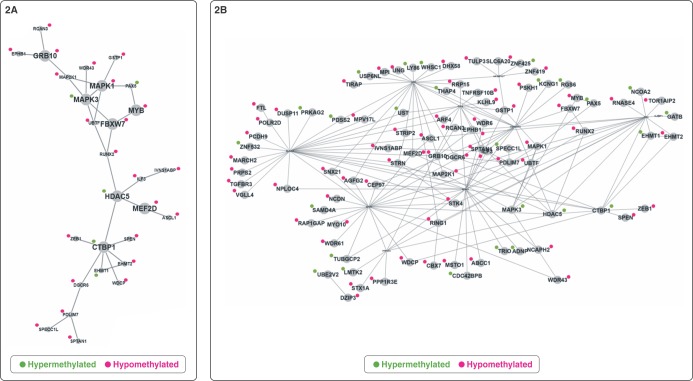
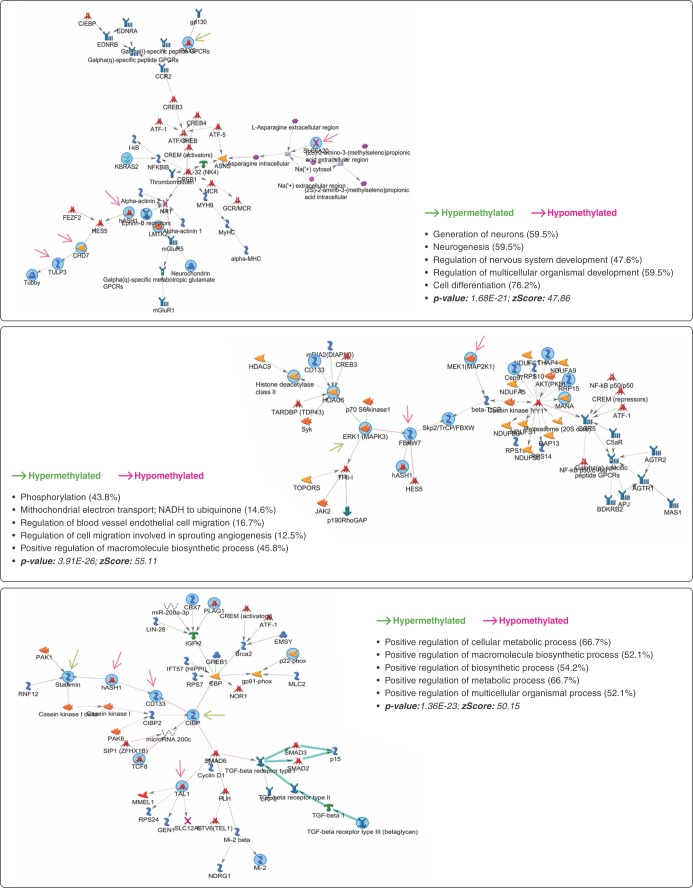
The 152 genes related to DMPs were submitted to PPI networks modules enrichment analysis, which identified a module of genes involved with G1/S transition of the mitotic cell cycle, histone deacetylase complex, and RNA polymerase II core promoter proximal region sequence-specific DNA binding (C = 1329; O = 29; E = 10.48; R = 2.77; p = 3.35E-07; FDR = 2.91E-04). To better understand the importance of the biological process associated with these genes, 137 out of the 152 were mapped on BIOGRID, with the network prioritization showing 28 genes directly connected (Figure 2A). We noted that, considering gene methylation status, the distribution was not random (p = 0.087): 23 genes were hypomethylated and five were hypermethylated in CUD patients compared to controls. There was enrichment for 138 GO biological process categories (Table S3 (1MB, pdf) , available as online-only supplementary material). Ninety-one genes were used as seeds in the expanded sub-network composed of 101 genes (Figure 2B). Using the expanded network, 59 GO biological process categories were enriched (Table S3 (1MB, pdf) ). Both prioritization and expanded networks presented biological processes related to neurogenesis, histone modification, regulation of signal transduction, regulation of transcription, and regulation of the ERK1/ERK2 cascade and MAPK pathway. In addition, enriched networks identified by MetaCore™ contained genes mainly involved with cell differentiation (p = 1.68E-21; z score: 47.86), macromolecule biosynthesis (p = 3.91E-26; z score: 55.11), and metabolic process (p-value: 1.36E-23; z score: 50.5). CHD7, SLC6A20, ASCL1, PAX5, and TULP3 are also related to neurogenesis and regulation of nervous system development (Figure 3). The 28 genes associated with the 48 DMRs located in promoter regions were enriched mainly for the PI3K-Akt signaling pathway (C = 341; O = 4; E = 0.38; R = 10.52; p = 3.01E-04; FDR = 2.62E-02).
Figure 2. Enriched expanded sub-networks from Biological General Repository for Interaction Datasets (BioGRID). A) Network prioritization showing 28 of the 152 genes related to differentially methylated positions (DMPs); B) Network expansion with the 91 genes selected as seed. Green and red denote gene methylation status.
Figure 3. Enrichment analysis report from the 152 genes using MetaCore™ software. Three networks were identified by the GeneGO Metacore enrichment analysis. Green and red denote seed gene methylation status.
Discussion
Chronic exposure to cocaine and crack interferes with the brain reward system by several mechanisms, including disruption of DNA methylation dynamics.10 We explored changes in DNA methylation in the peripheral blood of CUD patients, aiming to understand the potential contribution of this mechanism to this disorder. Hierarchical clustering based on the methylation patterns of DNA extracted from peripheral blood discriminated cases from controls. Pyrosequencing analysis confirmed the methylation levels identified on the genome-wide DNA methylation in three out of six (50%) genes analyzed. Our results showed that most CpGs were hypomethylated in CUD patients compared to controls. A similar pattern was previously observed in the nucleus accumbens (NAc) in rodent models, with global DNA hypomethylation after cocaine administration.32 It is important to note that, among the 28 most connected genes, 23 (82.1%) were hypomethylated and only five (17.9%) were hypermethylated; however, two CpGs (CBPT1, MAPK3) were located in the gene body, and one CpG (HDAC5) was located in the 5′UTR, within the shore region.
Cocaine use results in long-term adaptive effects in the central nervous system by changing neuronal structure and function. This neural activity may modulate DNA methylation patterns.8,33 Wright et al. observed upregulation of DNMT3A/B after cocaine administration and drug-seeking behaviors in rodents, thus connecting (at least in part) cocaine use to the DNA methylation process.32 Here, enrichment of the network resulted mainly in biological pathways already known to be related to cocaine use, such as transcription and chromatin regulation, histone activity, and cell cycle (neurogenesis and neuronal differentiation). These findings are consistent with the literature, but here we highlight DNA methylation as one of the mechanisms involved in these biological pathways.
Regarding the genes related to chromatin regulation and histone activity pathways, EHMT1 and EHMT2 encode histone methyltransferases involved in transcriptional repression and, in the mouse brain, are associated with prolonged cocaine exposure.34,35 HDAC5 encodes a histone deacetylase that alters the chromosome structure, affecting the access of transcription factors to DNA. It maintains brain homeostasis, which is disrupted in several disease conditions.33,36 Animal-model studies have reported an association of HDAC5 with adaptations to cocaine exposure, due its involvement in behavioral transition from drug experimentation to compulsive drug use.30,33 MEF2 is a possible mediator of HDAC5 function, as it reduces cocaine reward sensitivity after repeated cocaine exposures,30 and MEF2D, a survival-related transcription factor,37 is connected to HDAC5 in gene-gene interaction analysis.
The extracellular signal-related kinase (ERK) pathway was also enriched in the network analysis, represented by CTBP1, TAL1, and FBXW7. ERK pathway activity mediates changes in neuronal activities as a consequence of cocaine use, which could lead to epigenomic changes.10 CTBP1 and TAL1 are related to cellular and metabolic processes. CTBP1 is downregulated in cerebellar granule neurons that induces apoptosis,38 whereas TAL1 is a transcription factor necessary for the specification of the blood formation during development,39 with a role in the MEK/ERK pathway.40 FBXW7, a gene involved in cell proliferation and differentiation, is also involved in the regulation of ERK1/2 MAP kinase signaling.41
We also identified methylation changes in CpG positions from MAPK1, MAPK3, MAP2K1, and OSMR, all involved in the MAPK pathway. This pathway is activated by increases in extracellular dopamine in the striatum in response to cocaine abuse, and plays an important role in gene regulation by direct activation of transcription factors and chromatin remodeling.42 These data corroborate our analysis, as we identified hypomethylation in the promoter region of MAPK1, which might increase expression in drug-dependent individuals. MAPK3 showed a hypermethylated CpG in the gene and higher expression in dorsolateral prefrontal cortex.31 MAP2K1 had a hypomethylated CpG located in the CpG island at the gene promoter, suggesting that this CpG could be associated with the higher expression levels found in the prefrontal cortex of cocaine and crack dependents.31 Moreover, STMN1 and UBE2V2 presented CpG hypermethylation in the gene body and in the gene promoter, respectively, and are known to be downregulated in the hippocampus of cocaine dependents. HIPK3 and LMTK2 reported hypomethylation in a CpG located in the CpG island and gene body, respectively, and are upregulated in cocaine dependents.43
Comparing our data with those of other DNA methylation studies that evaluated use/abuse of other substances, such as tobacco, we found similarities in 30 differentially methylated genes (adjP < 10-5): STX1A, ATP11A, ZNRF3, MYO10, UNG, PCDH9, TGFBR3, CARS2, CTBP1, PSMB8, and SNX21 from Zeilinger et al.44; PDLIM7, ATP11A, ADCK4, MPI, CARS2, VWF, RCAN3, PSMB8, PRKAG2, PHF17, SDK1, CBX7, PCDH9, CDC42BPB, ABCC1, and PAX5 from Joehanes et al.45; and PCDH9, TRIO, and ATP11A from Wilson et al.46 Among these genes, we highlight CTBP1 (described above), PSMB8, and PCDH9. PSMB8 is a gene involved in the ERK1/2 and PI3K/AKT signaling pathways,47 which can be activated by cocaine intake and have enhanced activity in reward-related brain areas.48 PCDH9, which is a gene found in all of the aforementioned studies, has a role in cell adhesion and recognition and is associated with protective effects against cocaine-induced cytotoxicity in the mouse brain.49
We identified two important genes by both DMP and DMR analyses: OSMR, involved in the MAPK pathway as described above50; and ARF4, involved in protein activity and required for mammalian development. Both have been related to cocaine exposure response in the NAc of rats.51 Four genes associated to 48 DMRs located in promoters were enriched for Pi3K signaling pathway: PIK3CB, CCNE2, OSMR, and CDKN1B. Of these, CDKN1B has been previously reported as involved with gene regulation in the NAc after chronic cocaine exposure.51,52 Comparison of genes associated with 48 DMRs located in promoters with tobacco DNA methylation studies showed overlap for three genes: EHD1, PITPNB,44 and NRARP.45
Altogether, genes related to the ERK1/2 cascade, MAP kinase pathway, Pi3K signaling pathway, neurogenesis, and neuronal differentiation – already known to be related to cocaine and crack dependence – were found to be differentially methylated in peripheral blood. Moreover, the gene network analysis also suggested that these genes could be involved with the underlying pathology of CUD.
This study has some methodological limitations. Drug dependence classification was recently updated in the newest version of the DSM (DSM-5). Unlike in the DSM-IV and ICD system (ICD-10 and its updated version, ICD-11), which assume different diagnostic criteria for substance abuse and substance dependence, the DSM-5 includes both substance dependence and substance abuse in a dimensional classification spectrum.53 Although cases had to meet the ICD-10 criteria for cocaine and crack dependence to be included in this study, we used the current nomenclature proposed by DSM-5, i.e., cocaine use disorder (CUD), throughout the paper. Cases also used and abused other substances (e.g., tobacco, cannabis, solvents, and heroin); however, cocaine/crack was the only drug abused by all participants in the CUD group. Although we controlled the methylation analysis for associations with traits and co-variables, there is a possibility that substance comorbidities among the CUD group interfered with methylation evaluation, because of the difficulty of controlling these variables since controls did not use any substance. Another limitation is the small sample size; to mitigate this, our sample selection criteria (including years of dependence, age group, only males) sought to ensure the best possible homogeneity in each group, reducing biases and individual interferences. Considering these limitations, our findings should be explored in other groups, such as women, other age ranges, and other substances, with proper controls for these are properly controlled in the analysis.
In conclusion, the present study found significant differences in peripheral-blood DNA methylation patterns between CUD patients and controls, suggesting this approach is relevant and can contribute to a better understanding of the biological mechanisms involved in CUD. In addition, our data suggest that DNA methylation changes in the peripheral blood may be used to identify the CUD vulnerability phenotype. However, future investigations using this approach in independent samples are needed to explore these findings, both in peripheral tissues and, particularly, in brain regions involved with CUD.
Disclosure
The authors report no conflicts of interest.
Acknowledgements
The authors thank Bianca Lisboa and Thaís Chile for technical assistance. Funding was provided by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; grant 2013/00659-2) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES; PROEX 1229245 to CC). MM and ACT were supported by FAPESP grants 2015/06281-7 and 2014/00041-1, respectively.
Footnotes
How to cite this article: Camilo C, Maschietto M, Vieira HC, Tahira AC, Gouveia GR, Feio dos Santos AC, et al. Genome-wide DNA methylation profile in the peripheral blood of cocaine and crack dependents. Braz J Psychiatry. 2019;41:485-493. http://dx.doi.org/10.1590/1516-4446-2018-0092
References
- 1.Chasin AAdM, Silva ESd, Carvalho VM. Estimulantes do sistema nervoso central. In: Oga S, Camargo MMdA, Batistuzzo JAdO, editors. Fundamentos de toxicologia 3a ed. São Paulo: Atheneu; 2008. pp. 353–74. [Google Scholar]
- 2.Associação Brasileira de Psiquiatria Abuse and addiction: crack. Rev Assoc Med Bras. 2012;58:138–40. [PubMed] [Google Scholar]
- 3.Abdalla RR, Madruga CS, Ribeiro M, Pinsky I, Caetano R, Laranjeira R. Prevalence of cocaine use in Brazil: data from the II Brazilian national alcohol and drugs survey (BNADS). Addict Behav. 2014;39:297–301. doi: 10.1016/j.addbeh.2013.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Escobar M, Scherer JN, Soares CM, Guimarães LS, Hagen ME, von Diemen L, et al. Active Brazilian crack cocaine users: nutritional, anthropometric, and drug use profiles. Braz J Psychiatry. 2018;40:354–60. doi: 10.1590/1516-4446-2017-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn J, Laranjeira RR. Transitions in the route of cocaine administration--characteristics, direction and associated variables. Addiction. 1999;94:813–24. doi: 10.1046/j.1360-0443.1999.9468135.x. [DOI] [PubMed] [Google Scholar]
- 6.Lizasoain I, Moro MA, Lorenzo P. Cocaína: aspectos farmacológicos. Adicciones. 2002;14:57–64. [Google Scholar]
- 7.Wong CC, Mill J, Fernandes C. Drugs and addiction: an introduction to epigenetics. Addiction. 2011;106:480–9. doi: 10.1111/j.1360-0443.2010.03321.x. [DOI] [PubMed] [Google Scholar]
- 8.McQuown SC, Wood MA. Epigenetic regulation in substance use disorders. Curr Psychiatry Rep. 2010;12:145–53. doi: 10.1007/s11920-010-0099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alaghband Y, Bredy TW, Wood MA. The role of active DNA demethylation and Tet enzyme function in memory formation and cocaine action. Neurosci Lett. 2016;625:40–6. doi: 10.1016/j.neulet.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaillancourt K, Ernst C, Mash D, Turecki G. DNA methylation dynamics and cocaine in the brain: progress and prospects. Genes (Basel). 2017;8:E138. doi: 10.3390/genes8050138. doi: http:/10.3390/genes8050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Zhang C, Cheng L, Reilly JL, Bishop JR, Sweeney JA, et al. Correlation between DNA methylation and gene expression in the brains of patients with bipolar disorder and schizophrenia. Bipolar Disord. 2014;16:790–9. doi: 10.1111/bdi.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuan PF, Waszczuk MA, Kotov R, Marsit CJ, Guffanti G, Gonzalez A, et al. An epigenome-wide DNA methylation study of PTSD and depression in World Trade Center responders. Transl Psychiatry. 2017;7:e1158. doi: 10.1038/tp.2017.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walton E, Hass J, Liu J, Roffman JL, Bernardoni F, Roessner V, et al. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr Bull. 2016;42:406–14. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannon E, Lunnon K, Schalkwyk L, Mill J. Interindividual methylomic variation across blood, cortex, and cerebellum: implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics. 2015;10:1024–32. doi: 10.1080/15592294.2015.1100786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guindalini C, Howard M, Haddley K, Laranjeira R, Collier D, Ammar N, et al. A dopamine transporter gene functional variant associated with cocaine abuse in a Brazilian sample. Proc Natl Acad Sci U S A. 2006;103:4552–7. doi: 10.1073/pnas.0504789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guindalini C, Vallada H, Breen G, Laranjeira R. Concurrent crack and powder cocaine users from Sao Paulo: do they represent a different group? BMC Public Health. 2006;6:10. doi: 10.1186/1471-2458-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, et al. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2014;30:428–30. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods. 2014;11:1138–40. doi: 10.1038/nmeth.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordlund J, Bäcklin CL, Wahlberg P, Busche S, Berglund EC, Eloranta ML, et al. Genome-wide signatures of differential DNA methylation in pediatric acute lymphoblastic leukemia. Genome Biol. 2013;14:r105. doi: 10.1186/gb-2013-14-9-r105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30:1431–9. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–96. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Triche TJ, Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013;41:e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maksimovic J, Gordon L, Oshlack A. SWAN: subset-quantile within array normalization for illumina infinium Humanmethylation 450 BeadChips. Genome Biol. 2012;13:R44. doi: 10.1186/gb-2012-13-6-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hennig C. Flexible procedures for clustering: R package version 2.1-10 [Internet]. 2018. [cited 2018 Nov 7]. cran.r-project.org/web/packages/fpc/fpc.pdf. [Google Scholar]
- 28.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–7. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatr-Aryamontri A, Oughtred R, Boucher L, Rust J, Chang C, Kolas NK, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–79. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi M, Carreira MB, Smith LN, Zirlin BC, Neve RL, Cowan CW. Histone deacetylase 5 limits cocaine reward through cAMP-induced nuclear import. Neuron. 2012;73:108–20. doi: 10.1016/j.neuron.2011.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehrmann E, Oyler J, Vawter MP, Hyde TM, Kolachana B, Kleinman JE, et al. Transcriptional profiling in the human prefrontal cortex: evidence for two activational states associated with cocaine abuse. Pharmacogenomics J. 2003;3:27–40. doi: 10.1038/sj.tpj.6500146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright KN, Hollis F, Duclot F, Dossat AM, Strong CE, Francis TC, et al. Methyl supplementation attenuates cocaine-seeking behaviors and cocaine-induced c-Fos activation in a DNA methylation-dependent manner. J Neurosci. 2015;35:8948–58. doi: 10.1523/JNEUROSCI.5227-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ajonijebu DC, Abboussi O, Russell VA, Mabandla MV, Daniels WM. Epigenetics: a link between addiction and social environment. Cell Mol Life Sci. 2017;74:2735–47. doi: 10.1007/s00018-017-2493-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Covington HE, 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–70. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maze I, Chaudhury D, Dietz DM, Von Schimmelmann M, Kennedy PJ, Lobo MK, et al. G9a influences neuronal subtype specification in striatum. Nat Neurosci. 2014;17:533–9. doi: 10.1038/nn.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–29. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 37.Gao L, She H, Li W, Zeng J, Zhu J, Jones DP, et al. Oxidation of survival factor MEF2D in neuronal death and Parkinson's disease. Antioxid Redox Signal. 2014;20:2936–48. doi: 10.1089/ars.2013.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stankiewicz TR, Schroeder EK, Kelsey NA, Bouchard RJ, Linseman DA. C-terminal binding proteins are essential pro-survival factors that undergo caspase-dependent downregulation during neuronal apoptosis. Mol Cell Neurosci. 2013;56:322–32. doi: 10.1016/j.mcn.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porcher C, Chagraoui H, Kristiansen MS. SCL/TAL1: a multifaceted regulator from blood development to disease. Blood. 2017;129:2051–60. doi: 10.1182/blood-2016-12-754051. [DOI] [PubMed] [Google Scholar]
- 40.Zhou RQ, Wu JH, Gong YP, Guo Y, Xing HY. Transcription factor SCL/TAL1 mediates the phosphorylation of MEK/ERK pathway in umbilical cord blood CD34+ stem cells during hematopoietic differentiation. Blood Cells Mol Dis. 2014;53:39–46. doi: 10.1016/j.bcmd.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Duhamel S, Girondel C, Dorn JF, Tanguay PL, Voisin L, Smits R, et al. Deregulated ERK1/2 MAP kinase signaling promotes aneuploidy by a Fbxw7β-Aurora A pathway. Cell Cycle. 2016;15:1631–42. doi: 10.1080/15384101.2016.1183851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brami-Cherrier K, Roze E, Girault JA, Betuing S, Caboche J. Role of the ERK/MSK1 signalling pathway in chromatin remodelling and brain responses to drugs of abuse. J Neurochem. 2009;108:1323–35. doi: 10.1111/j.1471-4159.2009.05879.x. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Z, Yuan Q, Mash DC, Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci U S A. 2011;108:6626–31. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet. 2016;9:436–47. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson R, Wahl S, Pfeiffer L, Ward-Caviness CK, Kunze S, Kretschmer A, et al. The dynamics of smoking-related disturbed methylation: a two time-point study of methylation change in smokers, non-smokers and former smokers. BMC Genomics. 2017;18:805. doi: 10.1186/s12864-017-4198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang BY, Song JW, Sun HZ, Xing JC, Yang ZH, Wei CY, et al. PSMB8 regulates glioma cell migration, proliferation, and apoptosis through modulating ERK1/2 and PI3K/AKT signaling pathways. Biomed Pharmacother. 2018;100:205–12. doi: 10.1016/j.biopha.2018.01.170. [DOI] [PubMed] [Google Scholar]
- 48.Corominas M, Roncero C, Ribases M, Castells X, Casas M. Brain-derived neurotrophic factor and its intracellular signaling pathways in cocaine addiction. Neuropsychobiology. 2007;55:2–13. doi: 10.1159/000103570. [DOI] [PubMed] [Google Scholar]
- 49.Hayase T, Yamamoto Y, Yamamoto K, Muso E, Shiota K. Microarray profile analysis of toxic cocaine-induced alterations in the expression of mouse brain gene sequences: a possible ‘protective’ effect of buprenorphine. J Appl Toxicol. 2004;24:15–20. doi: 10.1002/jat.939. [DOI] [PubMed] [Google Scholar]
- 50.Kurosawa T, Yamada A, Takami M, Suzuki D, Saito Y, Hiranuma K, et al. Expression of nephronectin is inhibited by oncostatin M via both JAK/STAT and MAPK pathways. FEBS Open Bio. 2015;5:303–7. doi: 10.1016/j.fob.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lichti CF, Fan X, English RD, Zhang Y, Li D, Kong F, et al. Environmental enrichment alters protein expression as well as the proteomic response to cocaine in rat nucleus accumbens. Front Behav Neurosci. 2014;8:246. doi: 10.3389/fnbeh.2014.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferguson D, Shao N, Heller E, Feng J, Neve R, Kim HD, et al. SIRT1-FOXO3a regulate cocaine actions in the nucleus accumbens. J Neurosci. 2015;35:3100–11. doi: 10.1523/JNEUROSCI.4012-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saunders JB. Substance use and addictive disorders in DSM-5 and ICD-10 and the draft ICD-11. Curr Opin Psychiatry. 2017;30:227–37. doi: 10.1097/YCO.0000000000000332. [DOI] [PubMed] [Google Scholar]