Abstract
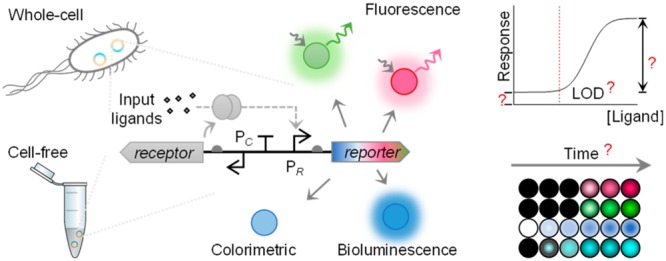
Whole-cell and cell-free transcription-translation biosensors have recently become favorable alternatives to conventional detection methods, as they are cost-effective, environmental friendly, and easy to use. Importantly, the biological responses from the biosensors need to be converted into a physicochemical signal for easy detection, and a variety of genetic reporters have been employed for this purpose. Reporter gene selection is vital to a sensor performance and application success. However, it was largely based on trial and error with very few systematic side-by-side investigations reported. To address this bottleneck, here we compared eight reporters from three reporter categories, i.e., fluorescent (gfpmut3, deGFP, mCherry, mScarlet-I), colorimetric (lacZ), and bioluminescent (luxCDABE from Aliivibrio fischeri and Photorhabdus luminescens, NanoLuc) reporters, under the control of two representative biosensors for mercury- and quorum-sensing molecules. Both whole-cell and cell-free formats were investigated to assess key sensing features including limit of detection (LOD), input and output dynamic ranges, response time, and output visibility. For both whole-cell biosensors, the lowest detectable concentration of analytes and the fastest responses were achieved with NanoLuc. Notably, we developed, to date, the most sensitive whole-cell mercury biosensor using NanoLuc as reporter, with an LOD ≤ 50.0 fM HgCl2 30 min postinduction. For cell-free biosensors, overall, NanoLuc and deGFP led to shorter response time and lower LOD than the others. This comprehensive profile of diverse reporters in a single setting provides a new important benchmark for reporter selection, aiding the rapid development of whole-cell and cell-free biosensors for various applications in the environment and health.
Whole-cell biosensors are cells that detect and report a target or condition of interest.1−4 Due to being renewable, environmental friendly, and cost-effective, they have drawn increasing attention as viable alternatives to electronic or chemical sensors over the last three decades.2,3 Notably, in the rising era of synthetic biology, a growing number of engineered whole-cell biosensors have been researched for a broad range of applications, such as environmental assessment,3,5−7 clinical diagnosis8,9 and biotherapy,10,11 controlled bioprocessing,12,13 mineral surveying,14 and landmine clearing.15
Meanwhile the cell-free transcription-translation (TX-TL) system is becoming a favorable technology for in vitro synthetic biology study due to its capability of flexibility, stability, portability, and fast prototyping as well as creating a minimal cellular environment.16−19 Because the cell-free system (CFS) contains no cells but only the basic biological machineries and energy sources required for TX-TL,20 it has been proposed as a feasible solution to circumvent the biosafety issues associated with whole-cell biosensing.19 Hence, a number of cell-free biosensors have been developed recently to detect heavy metals,21 antibiotics,22 and pathogens.23,24
Both whole-cell and cell-free biosensors share a similar architecture comprising a sensing module, a computing module, and an output actuating module.1−3 Many reporter proteins that produce light,8,25,26 fluorescence,5,15 colors,23,27 electrons,28 or gas vesicles29 can be used as genetic reporters in the output module.
Like other genetic devices or gene expression studies,17,30 many biosensors were first built with fluorescent reporters as the output actuator,3,5,15,24 thus simplifying their characterization in laboratory settings. Fluorescent proteins are relatively stable and take a short time to mature, and their light emission can be readily measured by a fluorimeter under specific light excitation. In addition, they can be used to study sensor cells at single cell level by fluorescence microscopy or fluorescence activated cell sorting.
Colorimetric reporters are often used to allow direct visualization of sensor output by the naked eye, which can drive down the operating costs of readout machines. As the first enzyme used to produce colorimetric output in engineered biosensors,31 β-galactosidase (i.e., LacZ) from Escherichia coli lac operon is the most popular enzyme used for both whole-cell and cell-free biosensors.19,21,23 The LacZ-catalyzed hydrolysis is fast,19 but many bacterial strains contain an intact lac operon, which will increase the background of the colorimetric output.
Bioluminescent reporters are also based on biochemical reactions which produce light without the need of an excitation light source. Among them, bacterial (LuxCDABE or LuxAB)8,32 and firefly (LucFF)33 luciferases are often used in whole-cell or cell-free biosensors. Owing to its high luminescent activity and small size (19 kDa), the recently engineered NanoLuc luciferase has become a favored bioluminescent reporter for whole-cell biosensors.25,34 Many of these bioluminescent biosensors have been coupled with portable devices for field testing or on-site diagnosis.8,25,35 However, unless the whole luciferase cassette is present, an external substrate such as d-luciferin, coelenterazine, or furimazine is required, limiting their applications for continuous monitoring.
Although a variety of genetic reporters have been thoroughly studied, their selections for biosensor engineering were more based on one’s experience rather than systematic side-by-side investigations. A few prior studies have been carried out to compare a couple of reporter categories; however, they were either not from the view of biosensor applications or not in directly comparable settings.7,36−38 To facilitate biosensor engineering, it is of great importance to compare different reporter categories systemically while evaluating their contributions to sensing performance. To this end, here we characterized and compared three widely used reporter categories, i.e., fluorescent, colorimetric, and bioluminescent reporters, under two representative biosensors of mercury- and quorum-sensing molecule within two different sensor settings, i.e., whole-cell and cell-free contexts. We investigated their properties in terms of contributions to analytical performance and key sensing features including limit of detection (LOD), input and output dynamic ranges, response time, and output visibility. Such a comprehensive profile provides a new benchmark reference for reporter gene selection, which will aid the rapid development of whole-cell and cell-free biosensors for various applications.
Experimental Section
Strains, Chemicals, and Reagents
Plasmid cloning and in vivo genetic circuit characterization were all performed in E. coli TOP10. Cells were cultured in Lysogeny broth (LB) medium (10 g L–1 peptone, 5 g L–1 NaCl, 5 g L–1 yeast extract), with appropriate antibiotics. The antibiotic concentrations used were 50 μg mL–1 for both kanamycin and ampicillin (for low copy number plasmid) or 100 μg mL–1 for ampicillin (for high copy number plasmid). Antibiotics and inducers (i.e., mercury(II) chloride (HgCl2) and N-(β-ketocaproyl)-l-homoserine lactone (3OC6HSL)) were analytical grade and purchased from Sigma-Aldrich. They were dissolved in ddH2O or nuclease free H2O (W4502, Sigma-Aldrich) and were then filtered using 0.22 μm syringe filters (SLGP033RS, Millipore).
LacZ substrate 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-gal) (MB1001, Melford) was dissolved in dimethyl sulfoxide (DMSO) (D8418, Sigma-Aldrich) to make 2% or 5% (w/v) stock solutions. Substrate furimazine for NanoLuc luciferase was from Nano-Glo Luciferase Assay System (N1110, Promega).
Plasmid Circuit Construction
Standard molecular biology techniques were used to construct plasmids containing mercury- and quorum-sensing molecule-responsive genetic circuits. All plasmids were constructed via BioBrick assembly39 and standard PCR. BioBrick vectors pSB1A3, pSB4A3, and pSB3K3 were used for plasmids cloning, and pSB3K3 was used for sensor circuit characterization (http://biobricks.org). Plasmid maps and detailed configurations are provided in relevant figures and summarized in Figure S1. Sequence details and sources of relevant parts are listed in Table S1. All primers used in this study, listed in Table S2, were purchased from Sigma-Aldrich. All plasmids constructed in this study have been confirmed by Sanger sequencing (Source BioScience). Detailed plasmid construction procedures are described in Supporting Information.
Reporter Gene Expression Assay in Vivo
For in vivo reporter gene characterization, engineered E. coli with constructed sensor plasmids were induced with HgCl2 or 3OC6HSL, and the reporter signals were measured using a plate reader (BMG FLUOstar) post 30, 60, 90,120, 180, 240, 300, and 360 min induction and incubation. NanoLuc-conducted bioluminescent and LacZ-conducted colorimetric measurements were acquired in lysing and nonlysing conditions using the same concentration of substrates (X-gal = 0.04 mg mL–1 from 2% X-gal stock solution and 0.2 μL of furimazine stock solution per 200 μL of culture). NanoLuc-derived bioluminescent kinetics were measured for 30 min after substrate addition, and the highest signal was chosen for data analysis. Colorimetric signal measurement was preceded by 30 min incubation at 37 °C, with orbital shaking at 300 rpm in the plate reader. To determine the cell density, absorbance (A600) was also read prior to each reporter measurement. For the lux operon reporter, the bioluminescent signal was measured immediately after absorbance measurement without the addition of substrates. Unless indicated otherwise, each reporter within different sensors was tested with three biological replicates. All the data shown are mean values with standard deviation as error bars. Detailed experimental procedures, data analysis, and instrument settings for reporter measurements and visualization are described in Supporting Information.
Reporter Gene Expression Assay in Vitro
The cell-free reactions were performed using E. coli S30 Extract System for Circular DNA (L1020, Promega) according to the manufacturer’s protocol. Each reaction contained 40% (v/v) S30 premix, 10% (v/v) amino acid mix, 30% (v/v) S30 extract, and 2% (v/v) inducer. The remaining 18% (v/v) contained DNA template with or without substrates. A 9.6 nM DNA template was used for each reporter within the mercury-sensing circuit and for the negative control (reaction with empty pSB3K3). A 9.2 nM DNA template was used for each reporter within the quorum-sensing molecule-responsive circuit. Fluorescent and colorimetric reporters within each sensor were measured continuously using BMG FLUOstar plate reader after induction. For LacZ reporter characterization, 5% X-gal was supplied into cell-free mixture before incubation (with 0.042% X-gal as the final concentration). For NanoLuc reporter characterization, 0.5 μL of furimazine stock solution was added to each well of the cell-free mixture after 20, 40, 60, 120, 180, and 240 min incubation. Unless indicated otherwise, all the reporters within different sensors were tested in two independent experiments and each with three technical replicates. All the data shown are mean values with standard deviation as error bars. Detailed experimental procedures, data analysis, and instrument settings for reporter measurements are described in Supporting Information. Calculation of sensor detection limit, mathematical modeling, and data fitting for both whole-cell and cell-free biosensors are described in Supporting Information.
Results and Discussion
Design and Standardization of Reporter Expression Characterization for Biosensors
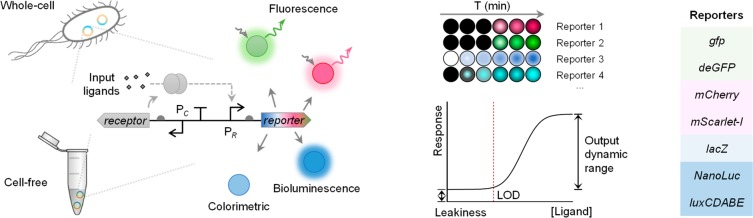
We characterized eight different genetic reporters to compare their main advantages and limitations contributing to the sensing performance of both whole-cell and cell-free biosensors. Fluorescent reporter genes gfp (gfpmut3), deGFP, mCherry, and mScarlet-I, luciferase genes NanoLuc and lux operons from Aliivibrio fischeri and Photorhabdus luminescens, and colorimetric output gene lacZ have been selected and profiled within the same biosensor settings both in vivo and in vitro (Figure 1).
Figure 1.
Comprehensive profiling of diverse genetic reporters in whole-cell and cell-free expression biosensor systems. Three categories of genetic reporters are selected to compare their profiles when applied in whole-cell and cell-free biosensors: (1) green fluorescent reporters (i.e., gfp and deGFP) and red fluorescent reporters (i.e., mCherry and mScarlet-I), (2) colorimetric reporter (i.e., lacZ), and (3) bioluminescent reporters (i.e., NanoLuc and lux operons from A. fischeri and P. luminescens). Response time, limit of detection (LOD), output dynamic range, and basal expression (leakiness) are tested for those reporters within both the mercury- and quorum-sensing molecule-responsive sensors.
Two biosensing systems for the mercury ion and 3OC6HSL quorum-sensing molecule have been designed and standardized to characterize and compare the selected reporters (Figure S1). The medium copy number plasmid pSB3K3 (10–12 copies per cell40) was used for characterizing all reporters to limit the metabolic burden. The output actuating modules coupled with selected reporters were placed in opposite directions to the sensing modules to prevent potential transcriptional read-through to the reporter from the sensing module. All reporters within the two sensing systems were tested under the same condition either in E. coli TOP10 or in E. coli S30 CFS. LOD, input and output dynamic ranges, response time, and output visibility were profiled for each reporter.
In Vivo Characterization and Comparison of Genetic Reporters
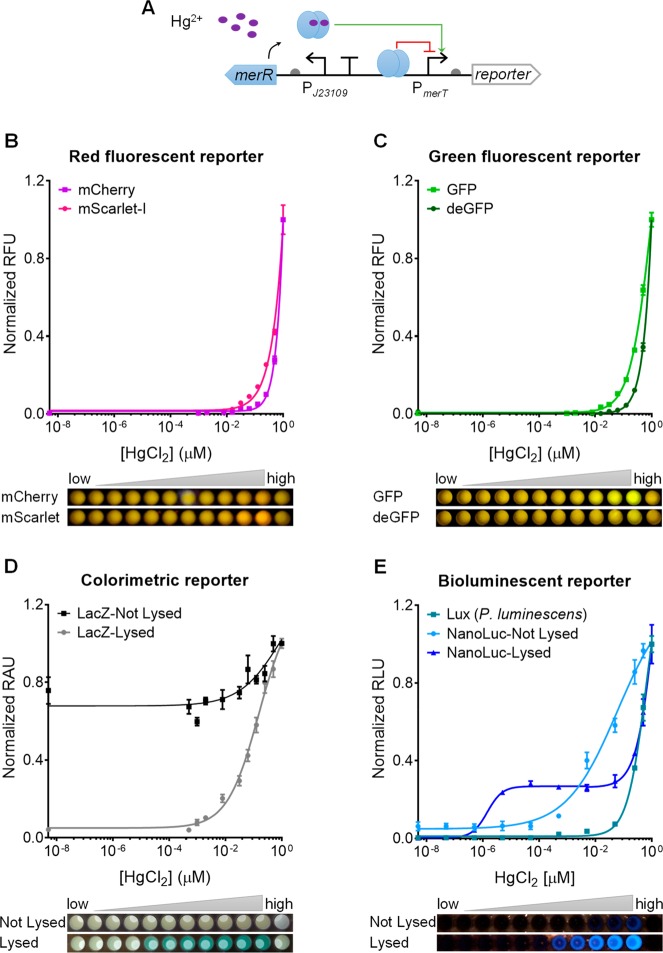
We first tested all chosen genetic reporters within a sensitive mercury sensor (i.e., J23109-merR-PmerT).41 This sensor has a constitutive promoter (J23109) that drives the expression of the mercury receptor MerR, which would derepress its cognate promoter PmerT upon mercury (Hg2+) binding and trigger the expression of the downstream reporter gene (Figure 2A).42 Cell phone images, induction fold, and dose–response curves were obtained postinduction of mercury (HgCl2) at various concentrations and different incubation times (Figure 2B–E, Figures S2–S4).
Figure 2.
Characterization of diverse genetic reporters within a mercury-responsive whole-cell biosensor. (A) Schematic of a mercury-responsive sensor module (J23109-merR-PmerT) coupled to a genetic reporter. (B–E) Dose–response curves and cell phone images of the mercury sensor with red fluorescent reporters mCherry and mScarlet-I (B), green fluorescent reporters GFP and deGFP (C), colorimetric reporter LacZ with the cells lysed or not lysed (D), and bioluminescent reporters NanoLuc with cells lysed or not lysed and LuxCDABE from P. luminescens (E). The last well in each cell phone image shows the reporter-free negative control cultures. Data were collected 360 min postinduction for B and C, and 90 min postinduction for D and E. Values are mean ± SD (n = 3 biologically independent experiments).
Comparing among the red fluorescent proteins (Figure 2B), we reported an induction fold over the control of 59.2 and 14.6 for mScarlet-I and mCherry, respectively (Figure S3), and LOD of 15.63 nM mercury for mCherry and 7.81 nM mercury for mScarlet-I (Figure S4), suggesting that mScarlet-I performs better than mCherry as a reporter. We deem this could be due to faster maturation and higher brightness of mScarlet than mCherry.43 Comparing GFP and deGFP (Figure 2C), we reported that GFP had a higher fold of induction (51.3 vs 17.3), lower LOD (7.81 nM vs 125.00 nM of HgCl2, Figure S4), and faster response (Figures S2,S3).
Both colorimetric reporter LacZ and bioluminescent reporter NanoLuc under the mercury sensor were monitored with lysed and nonlysed cells (see Experimental Section). Both reporters’ performance was improved in cell lysing conditions in terms of response time, LOD, output dynamic range, and output visibilities (Figure 2D,E, Figures S2–S4), suggesting that the cell membrane could limit diffusion and transport of the substrates. For LacZ, the best LOD (0.49 nM of HgCl2) with cell lysis was achieved 60 min postinduction, while the best LOD without cell lysis was 7.81 nM HgCl2 (16-fold higher) after 6 h induction (Figure S4). For NanoLuc, the best LOD (5.00 × 10–5 nM HgCl2) with cell lysis was obtained 30 min postinduction, while the best LOD without cell lysis was three orders higher (0.05 nM HgCl2) and was observed 3 h postinduction and postincubation.
As concerns lux operons from A. fischeri and P. luminescens, only the lux operon from P. luminescens showed notable output upon mercury induction (Figure 2E, Figure S6). In addition, the P. luminescens luciferase contributed to a lower LOD (5.00 × 10–4 nM of HgCl2) at early stage after induction (30–90 min), and higher output dynamic range at intermediate stage (90–120 min) (Figures S4 and S6). However, its overall performance was not comparable to that of NanoLuc with cell lysis. Compared to the NanoLuc without cell lysis, it remains a good bioluminescent reporter due to no requirement of adding substrates and lysing cells. Similar results were observed when testing the two operons under the quorum-sensing molecule sensor (Figures S6 and S7). Previous studies have shown that the luciferase from A. fischeri was thermolabile, with enzyme denaturation occurring above 30 °C, while the luciferase from P. luminescens was thermostable.44 As all the experiments were performed at 37 °C, the activity of the luciferase from A. fischeri may have been inhibited due to denaturation. This was confirmed by characterizing the luciferase reporter under different temperatures, where the A. fischeri luciferase showed better activity at lower temperature in LB (Figure S7). Different media (i.e., M9 with glycerol or glucose as carbon source) were also tested, indicating that salt and glucose levels could affect the two luciferases’ activities (Figure S7).45
Pros and Cons of Different Reporter Categories Acting in Vivo
mScarlet-I, GFP, LacZ (with cell lysis), and NanoLuc (with cell lysis) were selected for further analyses and investigation, as they showed superior performance in response time, LOD, and output dynamic range when compared to other reporters within their own categories.
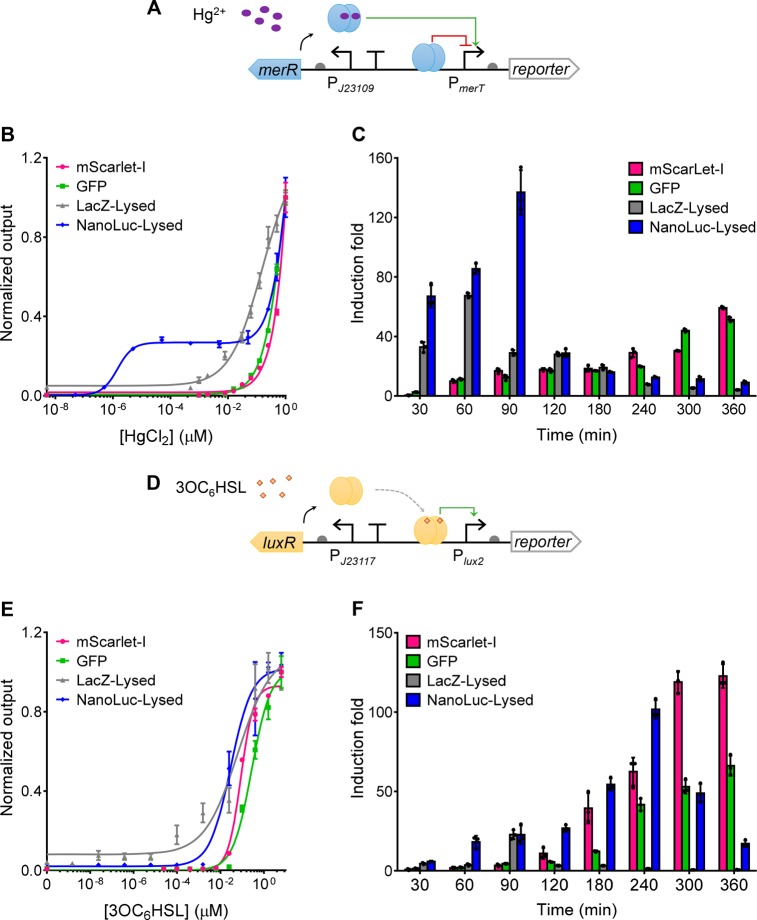
We first compared the four reporters under the mercury sensor (Figure 3A). The best dose–responses for each reporter and the cognate fold of induction were analyzed (Figure 3B,C). The sensors with mScarlet-I and GFP reporter showed similar LOD and induction fold, both of which were improved with longer incubation time (Figure 3C, Figure S4). However, we observed high background level, especially for green fluorescence, due to autofluorescence from bacterial cells and LB medium, limiting their use for direct visualization (Figure S2). In contrast, LacZ and NanoLuc showed very low background, facilitating their direct visualization by the naked eye. Moreover, they contributed to faster response (30 min for LacZ and NanoLuc vs 60 min for GFP and mScarlet-I), much lower LOD (16-fold lower for LacZ and 5 orders of magnitude lower for NanoLuc) and broader input dynamic ranges than the fluorescent reporters, which indicates the fast enzymatic reaction-based reporters are preferable for sensors requiring a short and sensitive response. This observation is consistent with previous studies.7,36,38 Nevertheless, both reporters with cell lysis are only suitable for disposable sensors with single-time-point readout. In addition, short time incubation will be required to obtain the best LOD and output dynamic range (Figure 3C, Figure S4) due to background activity induced by the sensor’s leakiness, which is more sensitive toward amplified enzymatic reactions than fluorescent reporters (Figure S2).
Figure 3.
Comparing diverse genetic reporters within mercury and quorum-sensing molecule whole-cell biosensors. (A, D) Schematics showing the mercury- or quorum-sensing molecule sensor module (J23109-merR-PmerT or J23117-luxR-Plux2) coupled to an output genetic reporter. (B, E) Dose–response of the mercury- and quorum-sensing molecule sensors with different reporters. For mScarlet-I and GFP, data were collected 360 min postinduction and postincubation. For LacZ and NanoLuc, data were collected 90 min postinduction. Dose–responses for the sensors at different time points are shown in Figures S4 and S5, and the relevant cell phone images are shown in Figures S2 and S8. (C, F) Fold of induction over time of the mercury- or quorum-sensing molecule sensors with different reporters responding to 0.1 μM HgCl2 (C) and 0.04 μM 3OC6HSL (F) respectively. Induction fold was calculated using the output with induction divided by the output without induction. Values are mean ± SD (n = 3 biologically independent experiments).
To test the generality of the aforementioned reporters’ performance, we next characterized the same set of reporters under a different sensing system, i.e., a quorum-sensing molecule (3OC6HSL)-responsive sensor (J23117-luxR-Plux2, Figure 3D–F).46 In this sensing system, a constitutive promoter (J23117) drives the expression of the 3OC6HSL-responsive LuxR receptor which activates its cognate promoter Plux2 when bound to 3OC6HSL (Figure 3D).46 Similar to the performance of reporters under the mercury sensor, GFP and mScarlet-I under the quorum-sensing molecule sensor showed similar dose–response curves and LOD, and both their LOD and induction fold were improved with longer incubation time (Figure 3E,F, Figure S5). Similarly, LacZ and NanoLuc showed much lower LOD (3–4 orders of magnitude lower than the fluorescent reporters) and faster responses (30 min for LacZ and NanoLuc vs 60 min for GFP and mScarlet-I) than the fluorescent reporters. Notably, NanoLuc provided the lowest LOD (3.81 × 10–4 nM of 3OC6HSL) among all reporters characterized (Figure S5). Similar high background leakiness and decreasing induction fold across time were observed under the quorum-sensing molecule sensor (Figure 3F, Figure S8). In contrast to reporters under the mercury sensor, the best induction folds of LacZ and NanoLuc under the quorum-sensing molecule sensor were observed at longer incubation time. However, it is worth noting that the induction levels for the two types of sensors are not comparable and their output kinetics are different.
Interestingly, we observed a biphasic dose–response curve for the mercury sensor particularly with the NanoLuc reporter with cell lysis (Figures 2E,3B). Such a response curve was not observed for the quorum-sensing molecule sensor, suggesting the biphasic dose–response is largely due to the intrinsic sensing behavior of the mercury-sensing system. Unlike the transcriptional activator LuxR, MerR is a repressor-activator.42 Moreover, previous studies suggested that with only one Hg2+ binding to the MerR homodimer, the MerR could activate transcription but at a moderate rate, while the MerR homodimer bound with two Hg2+ could fully activate the transcription.42 Therefore, the mercury sensor may respond to low mercury induction at a moderate rate while the response may be significantly increased in the presence of high mercury. This may explain why the biphasic dose–response curve occurred for the mercury sensor, particularly using NanoLuc reporter with cell lysis (Figures 2E and 3B). If the biphasic dose–response curve is not preferred, the data can be collected at an earlier time point postinduction and postincubation to eliminate such response behavior while maintaining similar high sensitivity (Figure S4A).
Characterization and Comparison of Genetic Reporters in TX-TL CFS
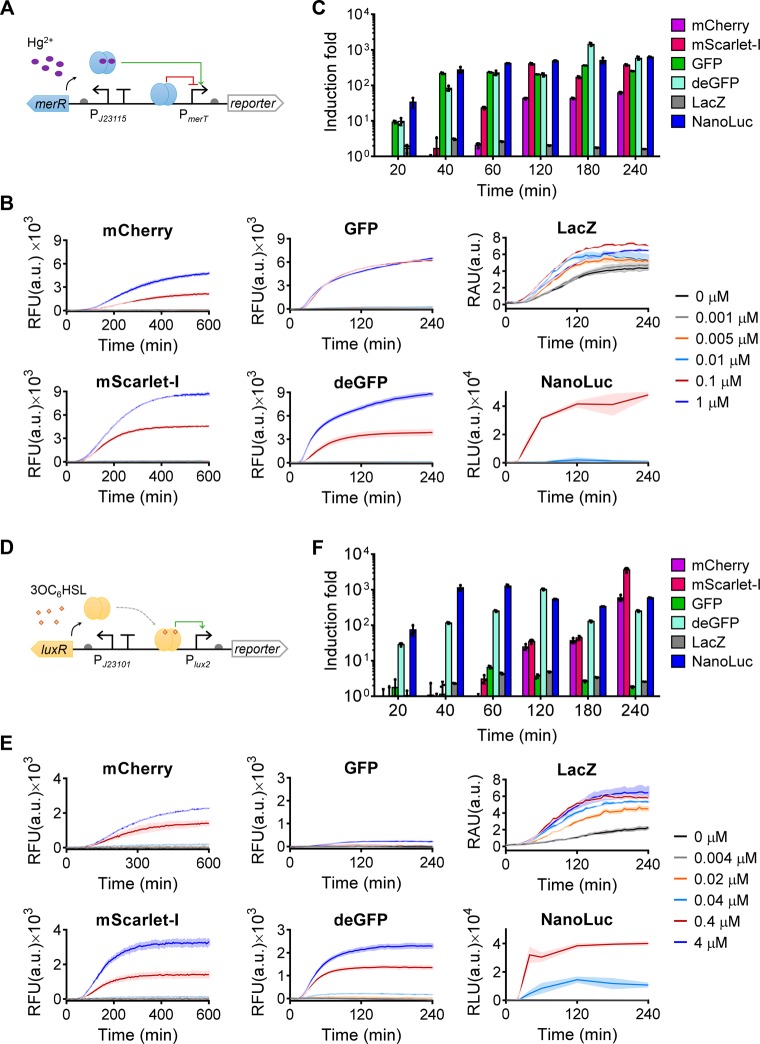
To provide a more comprehensive profiling of the different genetic reporters for biosensing, we compared the same reporters in an in vitro TX-TL CFS using cell-free biosensors. The CFS can be either based on crude cell extract47 or a system of purified recombinant elements (PURE) necessary for transcription-translation.48 The former is cheaper, easier to produce and more widely used in the field and therefore was selected for the reporter characterization in this study. Mercury- and quorum-sensing molecule sensors were also used for the in vitro test to provide a comparable context to their in vivo performance (Figure 4). To generate the same sensing activities for each sensor with different genetic reporters in the CFS, the same molar concentration of the sensory plasmids for each sensor was tested. Time-course response curves (Figure 4B,E), fold of induction (Figure 4C,F) and dose–responses (Figures S9 and S10) were analyzed for both sensors of different reporters. The experiments have been repeated at least twice independently (Figure S11).
Figure 4.
Profiling diverse genetic reporters within cell-free biosensors. (A, D) Schematics showing the mercury- or quorum-sensing molecule sensor (J23115-merR-PmerT or J23101-luxR-Plux2) coupled to diverse genetic reporters. (B, E) Dynamic output responses of the sensors responding to varying concentrations of HgCl2 (B) or 3OC6HSL (E). (C, F) Fold of induction over time of the mercury- or quorum-sensing molecule cell-free sensors of different reporters responding to 0.1 μM HgCl2 (C) and 0.4 μM 3OC6HSL (F), respectively. Induction fold was calculated using the sensor output with induction divided by the sensor output without induction. Values are mean ± SD (n = 3 technical replicates). a.u., arbitrary units.
Overall, the sensors with green fluorescent reporters or enzymatic reporters responded faster (20 min) and were more sensitive than the sensors with red fluorescent reporters (60 min) (Figure 4C,F, Figures S9, S10, and S11C,F). This meets our expectation, as the green fluorescent reporters generally mature faster than the red fluorescent reporters,49 and the enzymatic reactions are usually more sensitive and can amplify the sensor’s output signals. However, additional substrates are required for the enzymatic reaction, which is costly and unstable, whereas the high autofluorescence from the cell-free reagent could affect the measurement of the green fluorescent reporters. The LacZ substrate X-gal is more stable and cheaper than the NanoLuc substrate furimazine and can be added into the cell-free mixture at the beginning of induction. Nevertheless, cell-free sensors with LacZ reporter have low induction fold due to the sensors’ high background caused by leakiness and hence low output dynamic range of the cognate color change. In addition, the commercial CFS we used was made from a lacZ+ cell strain and hence already contains some background level of LacZ, thus increasing the background leakiness and reducing the output dynamic range. Using a LacZ-free CFS or a more sensitive LacZ substrate may improve this reporter’s performance in vitro.
Surprisingly, both green fluorescent reporters and NanoLuc provided the best LOD among all the reporters (Figures S9 and S10) but with a shorter incubation time for the latter (Figure S10B). For the mercury sensor, the GFP reporter contributed to an LOD < 1.0 × 10–3 μM mercury, making it the most sensitive one among all cell-free mercury sensors constructed to date. For the quorum-sensing molecule sensor, both deGFP (4 h) and NanoLuc (40 min) contributed to an LOD < 4.0 × 10–3 μM 3OC6HSL, 5–10 times lower than the rest of reporters.
When each reporter category was compared, mScarlet-I was superior to mCherry in terms of response time and induction fold (Figure 4C,F, Figure S11C,F), similar to their performance in vivo. However, the comparison of GFP and deGFP was less conclusive. The fluorescence output of deGFP was much higher than that of GFP under the quorum-sensing molecule sensor (Figure 4E, Figure S11E) while it was only true for the mercury sensor under high mercury induction levels (Figure 4B, Figure S11B). deGFP was designed to be more translatable in CFS than its original eGFP,50 but its performance against GFPmut3 (i.e., the GFP we tested here) has not been studied previously. Because deGFP worked well for both mercury- and quorum-sensing molecule sensors, it may be a more reliable reporter for regular cell-free biosensors. Further investigation would aid an in-depth comparison of their performance as a reporter, for example, by measuring the reporters’ fluorescent intensity, maturation, and transcription and translation efficiency in different genetic contexts.
Conclusions
In summary, we provided a comprehensive profiling of eight different genetic reporters from three commonly used reporter categories (i.e., fluorescent, colorimetric, and bioluminescent detection) within two representative sensor systems both in vivo and in vitro. The selected reporters have been frequently used in either whole-cell or cell-free biosensors but barely compared systematically in terms of their contributions to sensing features, limiting the biosensors’ development and applications. NanoLuc luciferase is a noticeable reporter due to its small size and high luminescent activity. Our study for the first time showed its characteristics in bacterial whole-cell biosensors and cell-free biosensors and indicated its superior reporting performance in both sensing systems. Overall, we reported that enzymatic reporters (especially bioluminescent reporter NanoLuc) provided the fastest response and lowest LOD in vivo. Both green fluorescent reporters and the enzymatic reporters contributed to the fastest response and lowest LOD in vitro. Considering the drastic differences and wide representation of the two sensor systems tested, similar conclusions obtained from the two sensor systems indicate the generality of our findings regarding the performance of these different genetic reporters. Therefore, these results can be reasonably applied to and inform the development of biosensors for other targets. However, the choice of sensor reporters also need to take into account several other important factors pertinent to their real world application requirements, such as the background signal level (green fluorescent reporter has the highest background activity), cost, and stability of the substrates for enzymatic reporters, whether an end-point data acquisition is sufficient or a continuous monitoring is preferred, and whether the cells can be lysed or not. This study provides new important benchmark for biosensor reporter gene selection, which will aid the rapid development of different whole-cell and cell-free biosensors for a variety of applications in the environment and health.
Acknowledgments
We thank Shimshon Belkin (Hebrew University of Jerusalem) for providing the plasmids comprising the lux operons. This work was supported by UK BBSRC grant [BB/N007212/1], Leverhulme Trust [RPG-2015-445], UKRI Future Leaders Fellowship [MR/S018875/1], Wellcome Trust Seed Award in Science [202078/Z/16/Z], and EPSRC/BBSRC Global Challenges Research Fund Awards to B.W. A.L., E.M., and A.R. were supported by the NATO Science for Peace and Security Programme under grant no. 985042.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b04444.
Experimental procedures, plasmid maps, cell phone images, dose–response curves over time for in vivo and in vitro analyses, list of LOD, lux operon characterization, independent replicates of in vitro assay, list of genetic parts and sequences, list of oligonucleotides, and list of abbreviations (PDF)
Author Contributions
X.W. and A.L. contributed equally to this work. B.W. conceived and led the study. B.W., A.L., and X. W. designed the experiments. X.W., A.L., E.M., and A.R. designed and optimized the bioluminescent assays. X.W. and A.L. performed all the experiments and data analysis. All authors took part in the interpretation of results and preparation of materials for the manuscript. X.W., A.L., and B.W. wrote the manuscript with inputs from all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Wang B.; Buck M. Trends Microbiol. 2012, 20 (8), 376–384. 10.1016/j.tim.2012.05.001. [DOI] [PubMed] [Google Scholar]
- van der Meer J. R.; Belkin S. Nat. Rev. Microbiol. 2010, 8 (7), 511–522. 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- Kim H. J.; Jeong H.; Lee S. J. Anal. Bioanal. Chem. 2018, 410 (4), 1191–1203. 10.1007/s00216-017-0751-6. [DOI] [PubMed] [Google Scholar]
- Hicks M.; Bachmann T. T.; Wang B. ChemPhysChem 2019, 20, 1–14. 10.1002/cphc.201900739. [DOI] [Google Scholar]
- Wang B.; Barahona M.; Buck M. Biosens. Bioelectron. 2013, 40 (1), 368–376. 10.1016/j.bios.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Feng Y.; Ryser M. D.; Zhu K.; Herschlag G.; Cao C.; Marusak K.; Zauscher S.; You L. Nat. Biotechnol. 2017, 35 (11), 1087–1093. 10.1038/nbt.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker J.; Balluch D.; Gsell M.; Harms H.; Feliciano J.; Daunert S.; Malik K. A.; van der Meer J. R. Environ. Sci. Technol. 2003, 37 (20), 4743–4750. 10.1021/es034258b. [DOI] [PubMed] [Google Scholar]
- Mimee M.; Nadeau P.; Hayward A.; Carim S.; Flanagan S.; Jerger L.; Collins J.; McDonnell S.; Swartwout R.; Citorik R. J.; et al. Science 2018, 360 (6391), 915–918. 10.1126/science.aas9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet A.; Endy D.; Renard E.; Molina F.; Bonnet J. Sci. Transl. Med. 2015, 7 (289), 289ra83. 10.1126/scitranslmed.aaa3601. [DOI] [PubMed] [Google Scholar]
- Ho C. L.; Tan H. Q.; Chua K. J.; Kang A.; Lim K. H.; Ling K. L.; Yew W. S.; Lee Y. S.; Thiery J. P.; Chang M. W. Nat. Biomed. Eng. 2018, 2 (1), 27–37. 10.1038/s41551-017-0181-y. [DOI] [PubMed] [Google Scholar]
- Chen J. X.; Steel H.; Wu Y. H.; Wang Y.; Xu J.; Rampley C. P. N.; Thompson I. P.; Papachristodoulou A.; Huang W. E. Appl. Environ. Microbiol. 2019, 85 (6), e02959. 10.1128/AEM.02959-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Carothers J. M.; Keasling J. D. Nat. Biotechnol. 2012, 30 (4), 354–359. 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- Dekker L.; Polizzi K. M. Curr. Opin. Chem. Biol. 2017, 40, 31–36. 10.1016/j.cbpa.2017.05.014. [DOI] [PubMed] [Google Scholar]
- Cerminati S.; Soncini F. C.; Checa S. K. Biotechnol. Bioeng. 2011, 108 (11), 2553–2560. 10.1002/bit.23213. [DOI] [PubMed] [Google Scholar]
- Belkin S.; Yagur-Kroll S.; Kabessa Y.; Korouma V.; Septon T.; Anati Y.; Zohar-Perez C.; Rabinovitz Z.; Nussinovitch A.; Agranat A. J. Nat. Biotechnol. 2017, 35 (4), 308–310. 10.1038/nbt.3791. [DOI] [PubMed] [Google Scholar]
- Garamella J.; Marshall R.; Rustad M.; Noireaux V. ACS Synth. Biol. 2016, 5 (4), 344–355. 10.1021/acssynbio.5b00296. [DOI] [PubMed] [Google Scholar]
- Niederholtmeyer H.; Sun Z. Z.; Hori Y.; Yeung E.; Verpoorte A.; Murray R. M.; Maerkl S. J. eLife 2015, 4, e09771. 10.7554/eLife.09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampley C. P. N.; Davison P. A.; Qian P.; Preston G. M.; Hunter C. N.; Thompson I. P.; Wu L. J.; Huang W. E. Sci. Rep. 2017, 7 (1), 7261. 10.1038/s41598-017-07391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee K.; Green A. a; Ferrante T.; Cameron D. E.; Daleykeyser A.; Yin P.; Collins J. J. Cell 2014, 159 (4), 940–954. 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J. G.; Stark J. C.; Jewett M. C. Cold Spring Harbor Perspect. Biol. 2016, 8 (12), a023853. 10.1101/cshperspect.a023853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didovyk A.; Tonooka T.; Tsimring L.; Hasty J. ACS Synth. Biol. 2017, 6 (12), 2198–2208. 10.1021/acssynbio.7b00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyen T. T. M.; Matsuura H.; Ujiie K.; Muraoka M.; Harada K.; Hirata K. J. Biosci. Bioeng. 2017, 123 (1), 96–100. 10.1016/j.jbiosc.2016.07.015. [DOI] [PubMed] [Google Scholar]
- Pardee K.; Green A. A.; Takahashi M. K.; Braff D.; Lambert G.; Lee J. W.; Ferrante T.; Ma D.; Donghia N.; Fan M.; et al. Cell 2016, 165 (5), 1255–1266. 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- Wen K. Y.; Cameron L.; Chappell J.; Jensen K.; Bell D. J.; Kelwick R.; Kopniczky M.; Davies J. C.; Filloux A.; Freemont P. S. ACS Synth. Biol. 2017, 6 (12), 2293–2301. 10.1021/acssynbio.7b00219. [DOI] [PubMed] [Google Scholar]
- Lopreside A.; Calabretta M. M.; Montali L.; Ferri M.; Tassoni A.; Branchini B. R.; Southworth T.; D’Elia M.; Roda A.; Michelini E. Anal. Bioanal. Chem. 2019, 411 (19), 4937–4949. 10.1007/s00216-019-01805-2. [DOI] [PubMed] [Google Scholar]
- Michelini E.; Calabretta M. M.; Cevenini L.; Lopreside A.; Southworth T.; Fontaine D. M.; Simoni P.; Branchini B. R.; Roda A. Biosens. Bioelectron. 2019, 123, 269–277. 10.1016/j.bios.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Watstein D. M.; Styczynski M. P. ACS Synth. Biol. 2018, 7 (1), 267–275. 10.1021/acssynbio.7b00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster D. P.; TerAvest M. a; Doud D. F. R.; Chakravorty A.; Holmes E. C.; Radens C. M.; Sureka S.; Gralnick J. a; Angenent L. T. Biosens. Bioelectron. 2014, 62, 320–324. 10.1016/j.bios.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Bourdeau R. W.; Lee-Gosselin A.; Lakshmanan A.; Farhadi A.; Kumar S. R.; Nety S. P.; Shapiro M. G. Nature 2018, 553 (7686), 86–90. 10.1038/nature25021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.; Kitney R. I.; Joly N.; Buck M. Nat. Commun. 2011, 2 (1), 508. 10.1038/ncomms1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillardet P.; Huisman O.; D’Ari R.; Hofnung M. Proc. Natl. Acad. Sci. U. S. A. 1982, 79 (19), 5971–5975. 10.1073/pnas.79.19.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J. M. H.; DiGrazia P. M.; Applegate B.; Burlage R.; Sanseverino J.; Dunbar P.; Larimer F.; Sayler G. S. Science 1990, 249 (4970), 778–781. 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- Pellinen T.; Huovinen T.; Karp M. Anal. Biochem. 2004, 330 (1), 52–57. 10.1016/j.ab.2004.03.064. [DOI] [PubMed] [Google Scholar]
- Eggers C. T.; Valley M. P.; Klaubert D. H.; Unch J.; Encell L. P.; Wood K. V.; Hall M. P.; Benink H. A.; Otto P.; Zimmerman K.; et al. ACS Chem. Biol. 2012, 7 (11), 1848–1857. 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried K.; Endes C.; Bhuiyan A. F. M. K.; Kuppardt A.; Mattusch J.; van der Meer J. R.; Chatzinotas A.; Harms H. Environ. Sci. Technol. 2012, 46 (6), 3281–3287. 10.1021/es203511k. [DOI] [PubMed] [Google Scholar]
- Huang C. W.; Yang S. H.; Sun M. W.; Liao V. H. C. Environ. Sci. Pollut. Res. 2015, 22 (13), 10206–10213. 10.1007/s11356-015-4216-1. [DOI] [PubMed] [Google Scholar]
- Garcia H. G.; Lee H. J.; Boedicker J. Q.; Phillips R. Biophys. J. 2011, 101 (3), 535–544. 10.1016/j.bpj.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkila K.; Maksimow M.; Karp M.; Virta M. Anal. Biochem. 2002, 301 (2), 235–242. 10.1006/abio.2001.5517. [DOI] [PubMed] [Google Scholar]
- Shetty R. P.; Endy D.; Knight T. F. J. Biol. Eng. 2008, 2, 5. 10.1186/1754-1611-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Schumacher J.; Wan X.; Lou C.; Wang B. ACS Synth. Biol. 2018, 7 (2), 553–564. 10.1021/acssynbio.7b00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X.; Volpetti F.; Petrova E.; French C.; Maerkl S. J.; Wang B. Nat. Chem. Biol. 2019, 15 (5), 540–548. 10.1038/s41589-019-0244-3. [DOI] [PubMed] [Google Scholar]
- Chang C. C.; Lin L. Y.; Zou X. W.; Huang C. C.; Chan N. L. Nucleic Acids Res. 2015, 43 (15), 7612–7623. 10.1093/nar/gkv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels D. S.; Haarbosch L.; Van Weeren L.; Postma M.; Wiese K. E.; Mastop M.; Aumonier S.; Gotthard G.; Royant A.; Hink M. A.; et al. Nat. Methods 2017, 14 (1), 53–56. 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- Manukhov I. V.; Eroshnikov G. E.; Vyssokikh M. Y.; Zavilgelsky G. B. FEBS Lett. 1999, 448 (2–3), 265–268. 10.1016/S0014-5793(99)00384-1. [DOI] [PubMed] [Google Scholar]
- Waters P.; Lloyd D. Microbiology 1985, 131 (11), 2865–2869. 10.1099/00221287-131-11-2865. [DOI] [Google Scholar]
- Wang B.; Barahona M.; Buck M. Nucleic Acids Res. 2015, 43 (3), 1955–1964. 10.1093/nar/gku1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. M.; Kigawa T.; Choi C. Y.; Yokoyama S. Eur. J. Biochem. 1996, 239 (3), 881–886. 10.1111/j.1432-1033.1996.0881u.x. [DOI] [PubMed] [Google Scholar]
- Shimizu Y.; Inoue A.; Tomari Y.; Suzuki T.; Yokogawa T.; Nishikawa K.; Ueda T. Nat. Biotechnol. 2001, 19 (8), 751–755. 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- Balleza E.; Kim J. M.; Cluzel P. Nat. Methods 2018, 15 (1), 47–51. 10.1038/nmeth.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.; Noireaux V. J. Biol. Eng. 2010, 4, 8. 10.1186/1754-1611-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.