Short abstract
What's already known about this topic?
Incidental diagnoses of an occult maternal malignancy have been reported upon aberrant routine noninvasive prenatal testing (NIPT).
The presence of tumor‐derived cell‐free DNA in the maternal circulation can skew the NIPT profile.
What does this study add?
Pregnant women with a confirmed neoplastic disease should not have NIPT testing for fetal aneuploidy screening since NIPT results cannot accurately be applied to assess the fetal chromosomal constitution in this condition.
What's already known about this topic?
Incidental diagnoses of an occult maternal malignancy have been reported upon aberrant routine noninvasive prenatal testing (NIPT).
The presence of tumor‐derived cell‐free DNA in the maternal circulation can skew the NIPT profile.
What does this study add?
Pregnant women with a confirmed neoplastic disease should not have NIPT testing for fetal aneuploidy screening since NIPT results cannot accurately be applied to assess the fetal chromosomal constitution in this condition.
Noninvasive prenatal testing (NIPT), using massively parallel sequencing of plasma cell‐free DNA (cfDNA), has been adopted worldwide for prenatal screening of common fetal aneuploidies1. It is based on the analysis of fetal cfDNA fragments, derived from the placenta and freely circulating in the maternal bloodstream. Two basic sequencing approaches are currently in use to analyze circulating fetal cfDNA, namely, random (whole‐genome) and targeted sequencing, being outlined in Bianchi and Chiu1. In the genome‐wide method, chromosomal ratios are calculated based on the number of sequencing reads of the chromosome of interest (eg, chromosome 21 in the case of Down syndrome) relative to the reads of a reference chromosome in a set of normal (diploid) samples. From these ratios, one z‐score per chromosome is calculated to determine fetal aneuploidy. A z‐score of three is commonly used as a risk threshold above which a trisomy might be suspected. Because the fraction of placenta‐derived “fetal” cfDNA exists against a high background of maternal plasma cfDNA, NIPT profiling not only examines fetal but also maternal cfDNA, implying that maternal chromosomal abnormalities can be detected as well2. Since the introduction of NIPT in prenatal diagnostics, incidental findings of an occult maternal malignancy following a “false‐positive” NIPT test have been reported repeatedly. Common cancer types encountered in pregnancy (such as breast cancer, lymphoma, and leukemia) and also other cancers (like ovarian cancer, multiple myeloma, digestive cancers, malignant melanoma, or sarcomas) and benign tumors (uterine leiomyomas) have been accidentally identified upon aberrant NIPT testing (previous work 3, 4, 5, 6, 7, 8, 9, 10 and unpublished results). From these cases, it is now appreciated that the presence of tumor‐derived cfDNA can skew the NIPT profile and confound its interpretation. Three particular scenarios might be encountered. Firstly, when the observed imbalances are incompatible with fetal development, a maternal malignancy might be invoked. In a second scenario, where such imbalances are compatible with fetal development, a false positive prenatal diagnosis could be made.10. This is illustrated in Table 1, representing NIPT data from a series of 26 pregnant cases that had a known diagnosis of breast cancer (n = 24), colon cancer (n = 1), or lymphoma (n = 1), prior to participating to a research study in which genome‐wide NIPT testing in this cancer‐in‐pregnancy setting was evaluated. In six out of the 26 cases, an aberrant NIPT output with chromosome‐wide z‐scores higher than three for chromosomes 21, 18, and/or 13 was observed, suggesting a fetal trisomy for (one of) the respective chromosomes. However, upon low‐pass sequencing of tumor biopsy specimens of these women, it was clear that the observed gains of chromosomes 21, 18, and 13 in cfDNA were derived from tumor DNA. This resulted in false positive scores of 15.4%, 15.4%, and 19.2% for trisomy 21, 18, and 13, respectively, in this study group of pregnant cancer patients. Figure 1 visualizes the NIPT output for one of these six cases, i.e. a woman that was diagnosed with a stage II, triple negative breast cancer when being 8 weeks pregnant. When limiting the analyses to the commonly tested fetal chromosomes, z‐scores higher than 3 were observed for chromosomes 21, 18, and 13. A genome‐wide inspection however showed the presence of chromosomal imbalances in almost all 22 autosomes. Upon comparison with the copy number profile of matched tumor biopsy DNA, the (sub)chromosomal CNAs and aneuploidies observed in cfDNA were shown to originate from tumor DNA. This woman gave birth to a baby boy with a normal neonatal outcome. Finally, also an NIPT outcome with an apparently normal result (for the investigated fetal chromosomes) cannot accurately be applied to assess the fetal chromosomal constitution as (a) z‐scores of particular fetal chromosomes or chromosomal fragments might be skewed due to excessive presentation of highly amplified tumoral chromosomes or chromosome arms or (b) chromosomal amplifications and deletions in the fetal and tumoral cfDNA may cancel each other out resulting in a neutral z‐score for a particular chromosome. In our study cohort of pregnant cancer patients, five women had a negative z‐score (z ≤ −3) for chromosomes 21, 18, 13 or a combination of these chromosomes. Except for one case, all observed aneuploidies in cfDNA were shown to reflect true monosomies in the tumor DNA (Table 1). All these five women gave birth to a child with no congenital malformations. If, however, one of these children would have been affected by a true fetal trisomy (characterized by a z‐score ≥ 3), then the monosomies in the tumor DNA would have neutralized the final z‐score for the respective chromosomes, resulting in a false negative NIPT output. The theoretical risk of such a false negative NIPT score in our patient cohort ranged from 7.7% to 15.4% for chromosomes 21, 18, and 13 (Table 1).
Table 1.
Risk of false positive and false negative NIPT scores for chromosomes 21, 18, and 13 in a cohort of pregnant women with a known maternal malignancy (n = 26)
| NIPT Profile in Plasma cfDNA | Copy Number Profile in Tumor DNAa | |||||
|---|---|---|---|---|---|---|
| chr21 | chr18 | chr13 | chr21 | chr18 | chr13 | |
| Number of cases with normal z > −3 and z < 3 | 22 | 22 | 20 | na | na | na |
| Number of cases with z ≥ 3 | 2 | 2 | 2 | 2/2 | 2/2 | 2/2 |
| Number of cases with z ≤ −3 | 2 | 2 | 4 | 2/2 | 2/2 | 3/4 |
| Percentage of false positive NIPT scores (%) | 15.4 | 15.4 | 19.2 | |||
| Theoretical risk of false negative NIPT scores (%) | 7.7 | 7.7 | 15.4 | |||
Abbreviations: cfDNA, cell‐free DNA; chr, chromosome; na, not applicable; z, z‐score.
Low‐pass sequencing (0,1 × coverage) of matched tumor biopsy DNA.
Figure 1.
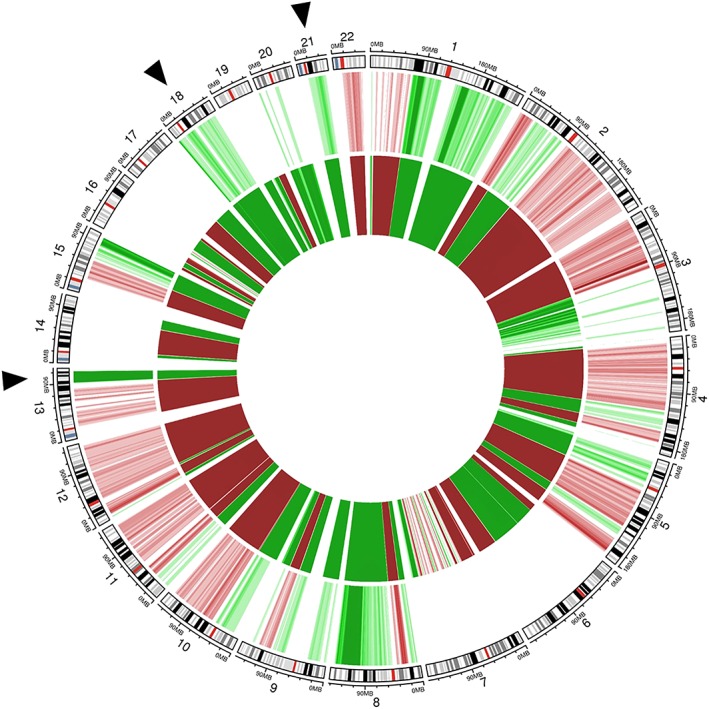
Circos plot showing chromosomal anomalies detectable in plasma cell‐free (cfDNA) and tumor DNA of a pregnant women being 8 weeks pregnant and with a known breast cancer diagnosis. The genomic representation profile of the autosomal chromosomes is shown in clockwise order, aligned with chromosomal ideograms (outer circle). Chromosomal anomalies with a chromosomal z‐score ≥ 3 (suggesting gain) are indicated in green; those with a z‐score ≤ −3 (suggesting loss) are shown in red. Color grades are used to indicate four z‐score intervals of length 1.5 ranging from 3 (−3) to 9 (−9). The fifth darkest color is reserved for values greater than 9 or less than −9. The middle circle depicts the genome‐wide NIPT profile in plasma cfDNA with elevated z‐scores for chromosomes 21, 18, and 13 (indicated by black arrows). Upon a genome‐wide view, (sub)chromosomal imbalances across multiple autosomal chromosomes can be observed. The inner circle shows the copy number profile of matched tumor DNA extracted from formalin‐fixed paraffin‐embedded tumor biopsy material (whole‐genome low‐pass sequencing, 0,1 × coverage). Comparison of both profiles reveals that the (sub)chromosomal CNAs and aneuploidies observed in plasma cfDNA are derived from tumor DNA. Details about the NIPT data analysis pipeline can be found elsewhere 11. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Together, these examples illustrate that the presence of tumor‐derived cfDNA can induce an aberrant NIPT result masking the fetal chromosomal profile. Therefore, we here advocate excluding pregnant women with a confirmed neoplastic disease from NIPT for fetal aneuploidy screening. Particular difficulties might arise with targeted NIPT assays, where information about genome‐wide distribution of cfDNA fragments is lacking to aid in the interpretation of deviating results of chromosomes 21, 18, and/or 13. However, even with full genome information, correct interpretation of the fetal genetic constitution might be disturbed, as shown above. Hence, NIPT testing as a screening tool for fetal aneuploidies is contraindicated in cases with a known neoplastic disease. With future novel algorithms taking into account the origin of cfDNA, advanced approaches to measure fetal fraction and improved algorithms for aneuploidy detection, it may well become possible to identify and exclude analysis of tumor‐derived cfDNA and avoid misdiagnoses. Until that time, we argue that pregnant cancer patients should be offered a detailed structural anomaly screening by ultrasound and an amniocentesis for karyotyping if certainty on chromosomal abnormalities is desired. Although not offered anymore in some centers 12, a combined first‐trimester screening can be performed to screen for trisomy 21, 13, and 18 in case of a cancer diagnosis before 14 weeks.
DATA SHARING STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
ETHICS APPROVAL
Plasma samples for NIPT testing were collected between August 2014 and November 2018. The study was approved by the ethics committee of University Hospitals Leuven (S/57197). Written informed consent was obtained from all participants.
CONFLICTS OF INTEREST
The authors have declared no conflicts of interest.
FUNDING SOURCES
This work was supported by a research grant from Research Fund Flanders (FWO‐Vlaanderen) (G080217N to FA and JRV).
ACKNOWLEDGMENTS
We thank H. Wildiers, S. Hatse, P. Neven, G. Floris, T. Tousseyn, and E. Cardonick for providing cell‐free DNA samples or matching tumor tissue biopsy for the pregnant women that participated in this study; B. Van den Bosch for DNA extractions from tumor tissue; and M. Neofytou and D. Villela for preparing sequencing libraries.
Lenaerts L, Van Calsteren K, Che H, Vermeesch JR, Amant F. Pregnant women with confirmed neoplasms should not have noninvasive prenatal testing. Prenatal Diagnosis. 2019;39:1162–1165. 10.1002/pd.5544
Joris Robert Vermeesch and Frédéric Amant contributed equally to this study.
REFERENCES
- 1. Bianchi DW, Chiu RWK. Sequencing of circulating cell‐free DNA during pregnancy. N Engl J Med. 2018;379(5):464‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brison N, Van Den Bogaert K, Dehaspe L, et al. Accuracy and clinical value of maternal incidental findings during noninvasive prenatal testing for fetal aneuploidies. Genet Med. 2017;19(3):306‐313. [DOI] [PubMed] [Google Scholar]
- 3. Amant F, Verheecke M, Wlodarska I, et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol. 2015;1(6):814‐819. [DOI] [PubMed] [Google Scholar]
- 4. Vandenberghe P, Wlodarska I, Tousseyn T, et al. Non‐invasive detection of genomic imbalances in Hodgkin/Reed‐Sternberg cells in early and advanced stage Hodgkin's lymphoma by sequencing of circulating cell‐free DNA: a technical proof‐of‐principle study. Lancet Haematol. 2015;2(2):e55‐e65. [DOI] [PubMed] [Google Scholar]
- 5. Dharajiya NG, Grosu DS, Farkas DH, et al. Incidental detection of maternal neoplasia in noninvasive prenatal testing. Clin Chem. 2018;64(2):329‐335. [DOI] [PubMed] [Google Scholar]
- 6. Bianchi DW, Chudova D, Sehnert AJ, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA. 2015;314(2):162‐169. [DOI] [PubMed] [Google Scholar]
- 7. Ji X, Li J, Huang Y, et al. Identifying occult maternal malignancies from 1.93 million pregnant women undergoing noninvasive prenatal screening tests. Genet Med. 2019;1;1‐10. 10.1038/s41436-019-0510-5 [DOI] [PubMed] [Google Scholar]
- 8. Janssens K, Deiteren K, Verlinden A, et al. Detection of a case of chronic myeloid leukaemia with deletions at the t(9;22) translocation breakpoints by a genome‐wide non‐invasive prenatal test. Prenat Diagn. 2016;36(8):760‐765. [DOI] [PubMed] [Google Scholar]
- 9. Imbert‐Bouteille M, Chiesa J, Gaillard JB, et al. An incidental finding of maternal multiple myeloma by non invasive prenatal testing. Prenat Diagn. 2017;37(12):1257‐1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osborne CM, Hardisty E, Devers P, et al. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenat Diagn. 2013;33(6):609‐611. [DOI] [PubMed] [Google Scholar]
- 11. Bayindir B, Dehaspe L, Brison N, et al. Noninvasive prenatal testing using a novel analysis pipeline to screen for all autosomal fetal aneuploidies improves pregnancy management. Eur J Hum Genet: EJHG. 2015;23(10):1286‐1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Elslande J, Brison N, Vermeesch JR, et al. The sudden death of the combined first trimester aneuploidy screening, a single centre experience in Belgium. Clin Chem Lab Med. 2019;XXX 10.1515/cclm-2019-0231 [DOI] [PubMed] [Google Scholar]


