Summary
Accumulation of lactate in the tissue microenvironment is a feature of both inflammatory disease and cancer. Here, we assess the response of immune cells to lactate in the context of chronic inflammation. We report that lactate accumulation in the inflamed tissue contributes to the upregulation of the lactate transporter SLC5A12 by human CD4+ T cells. SLC5A12-mediated lactate uptake into CD4+ T cells induces a reshaping of their effector phenotype, resulting in increased IL17 production via nuclear PKM2/STAT3 and enhanced fatty acid synthesis. It also leads to CD4+ T cell retention in the inflamed tissue as a consequence of reduced glycolysis and enhanced fatty acid synthesis. Furthermore, antibody-mediated blockade of SLC5A12 ameliorates the disease severity in a murine model of arthritis. Finally, we propose that lactate/SLC5A12-induced metabolic reprogramming is a distinctive feature of lymphoid synovitis in rheumatoid arthritis patients and a potential therapeutic target in chronic inflammatory disorders.
Keywords: lactate, lactate transporter, signaling, inflammation, immunometabolism, T cell, translational research, cytokines, metabolic crosstalk
Graphical Abstract
Highlights
-
•
Lactate induces the expression of SLC5A12 on human CD4+ T cells in the inflamed tissue
-
•
Lactate promotes CD4+ T cell IL17 production via PKM2/STAT3 signaling and FA synthesis
-
•
Lactate inhibits CD4+ T cell motility via increased FA synthesis and reduced glycolysis
-
•
SLC5A12 blockade ameliorates the disease severity in a murine model of arthritis
With buildup in the inflamed tissue, lactate can exacerbate the inflammatory response. Pucino et al. report that lactate induces expression of its own transporter, SLC5A12, driving lactate uptake into CD4+ T cells and resulting in increased IL17 production via PKM2/STAT3 signaling and enhanced fatty acid synthesis. They provide evidence that targeting SLC5A12 may ameliorate chronic inflammatory disorders.
Context and Significance
Lactate is one of the most enriched by-products of cellular metabolism in inflamed tissues. Its accumulation leads to the exacerbation of the inflammatory response in chronic inflammatory disorders like rheumatoid arthritis. Researchers in Birmingham, UK, and their collaborators have used human cells and tissues to investigate the processes by which lactate drives inflammation. Here, they report on a mechanism that involves a transporter known as SLC5A12, which brings lactate into the cell. This transporter is expressed by a specific subset of lymphocytes, the CD4+ T cells, often infiltrating the inflamed tissues. They discover that SLC5A12-driven lactate uptake leads to a stepwise reprogramming of cellular metabolism, which supports a pro-inflammatory response by CD4+ T cells. Targeting SLC5A12 pharmacologically with a monoclonal antibody shows promising results in a pre-clinical model of rheumatoid arthritis characterized by the infiltration of CD4+ T cells.
Introduction
The recent discovery of the fundamental role of metabolism in immune cell biology is contributing immensely to our understanding of immune cell regulation (Buck et al., 2016, O'Neill et al., 2016).
So far, most studies have focused on the role of metabolic pathways in the establishment of the immune response. More recently, novel signaling functions of metabolic intermediates in the regulation of immunity, including the small metabolites lactate, acetyl-CoA, succinate, itaconate, and others have been revealed. The roles of metabolite signaling stretch from regulation of cytokine production via effects on the cellular redox state, to interactions with transcription factors binding to specific cytokine promoter elements, to modulating the activity of transmembrane ion channels, and interference with cell migration and differentiation. Hence, the signaling functions of metabolites extend beyond self-regulatory roles and include cell-to-cell communication and sensing of micro-environmental conditions, i.e., within the inflammatory microenvironment, to elicit stress responses and cellular adaptation (Haas et al., 2015, Haas et al., 2016, Tannahill et al., 2013).
Although known for its role in the muscle-liver Cori cycle and neuron-astrocyte shuttle, lactate has mainly been seen as a by-product of metabolism or as a biomarker in critical care at best rather than a bioactive molecule, and its functional effects have thus been neglected for long time. Far from being inert, lactate accumulation in the disease microenvironment has major effects on tissue-resident and infiltrating immune cells. Recently reported outcomes include tumor escape from immune surveillance mechanisms via reshaping of macrophage and effector T cell functions to immune-suppressive and tumor promoting regulatory T cells and tumor-associated macrophages (Angelin et al., 2017, Brand et al., 2016, Colegio et al., 2014, Reina-Campos et al., 2017). In contrast, accumulation of lactate in the tissue microenvironment in the course of inflammatory disorders serves as an amplifier of inflammation (Haas et al., 2015, Haas et al., 2016, Weyand et al., 2017). Lactate has recently been shown to be a major source of carbons for the tricarboxylic acid cycle (TCA), surprisingly even in excess of glucose, both in normal and cancerous tissues (Faubert et al., 2017, Hui et al., 2017). However, whether this contributes to its signaling properties is not well understood.
The physiological lactate concentration in blood and healthy tissues is approximately 1.5–3 mM, but it can rise to 10–40 mM at inflamed tissues as shown in tumor microenvironments and arthritic joints as well as atherosclerotic plaques and adipose tissue in obese individuals. Elevated levels of lactate have also been reported in the serum of multiple sclerosis and Sjögren’s syndrome patients, in the latter correlating with fatigue and exercise intolerance (Pucino et al., 2017, Amorini et al., 2014). Lactate is mainly produced in the cytoplasm during hypoxia or as a consequence of aerobic glycolysis in proliferating cells, and it is then secreted through the plasma membrane. This transport is dependent on solute carrier transporters that perform proton-lactate symport (i.e., MCT1-4) or sodium-dependent transport (i.e., SLC5A8 and SLC5A12). Indeed, only MCT1 (also known as SLC16A1, Km 4.5) and MCT4 (also known as SLC16A3, Km 28) have a high specificity for lactate and broad tissue expression. Sodium-coupled lactate transport is carried out by the high-affinity transporter SLC5A8 or the low-affinity transporter SLC5A12, which have been initially reported for their expression in kidney (Srinivas et al., 2005, Gopal et al., 2007). More recently, we have reported the expression of SLC5A12 by CD4+ T cells (Haas et al., 2015). Even though some transporters facilitate extrusion (e.g., MCT4) and others influx (e.g., MCT1) of lactate, the main factor determining the transport direction is the lactate gradient, facilitating lactate import when extracellular lactate is high, such as in inflamed tissues (Halestrap and Wilson, 2012, Srinivas et al., 2005).
For more than 50 years, the inflamed joint has been recognized as a site of low glucose and high lactate concentrations (Goetzl et al., 1971, Treuhaft and MCCarty, 1971), reflective of the intense cellular turnover in the rheumatoid pannus. Synovial fibroblasts adopt an anaerobic glycolysis type of metabolism, producing and secreting high amounts of lactate in the microenvironment. There, lactate contributes to the regulation of the functions of surrounding cells (i.e., plasticity), including infiltrating immune cells (Fujii et al., 2015).
Rheumatoid arthritis (RA) is characterized by three distinctive histological pattern of synovitis (i.e., pathotypes). In 40% of patients, the inflammatory infiltrate is constituted mainly by monocyte and/or macrophages in the synovial sublining, i.e., myeloid pathotype. A rarer subset, ∼20% of RA patients, is characterized by a prevalent fibroid signature, i.e., fibroid and pauci-immune pathotype. In the remaining 40% of RA patients, immune cells can be found spatially grouped in follicular structures, which can acquire features of secondary lymphoid organs (SLO) with high T and B cell infiltration and segregation, i.e., lymphoid pathotype. These structures, which develop in the inflammatory tissue, are called ectopic lymphoid-like structures (ELS). The recognition that ELS may play a key pathogenic role in autoimmunity and may be exploited as potential biomarker for disease evolution and response to therapy is gaining attention (Pitzalis et al., 2013, Pitzalis et al., 2014, Bombardieri et al., 2017). Our group has recently found that SLC5A12 is highly expressed in human RA synovial tissues. Strikingly, its levels significantly increased in correlation with the RA synovial tissue T cell score and with the formation of ELS which are rich in IL17 (Haas et al., 2015, Peters et al., 2011, Jones et al., 2015, Jones and Jones, 2016), thus suggesting a possible role of lactate/SLC5A12-induced metabolic signaling in promoting chronic inflammation in RA.
Here, we explored the response of CD4+ T cells to lactate in the context of the tissue microenvironment in inflammatory disorders. We identified several mechanistic steps leading from the influx of lactate into CD4+ T cells to the plastic reshaping of their effector functions and their induced exacerbation of the inflammatory response.
Results
The Expression of the Lactate Transporter SLC5A12 by Immune Cells Is Regulated by Activating and Inflammatory Stimuli
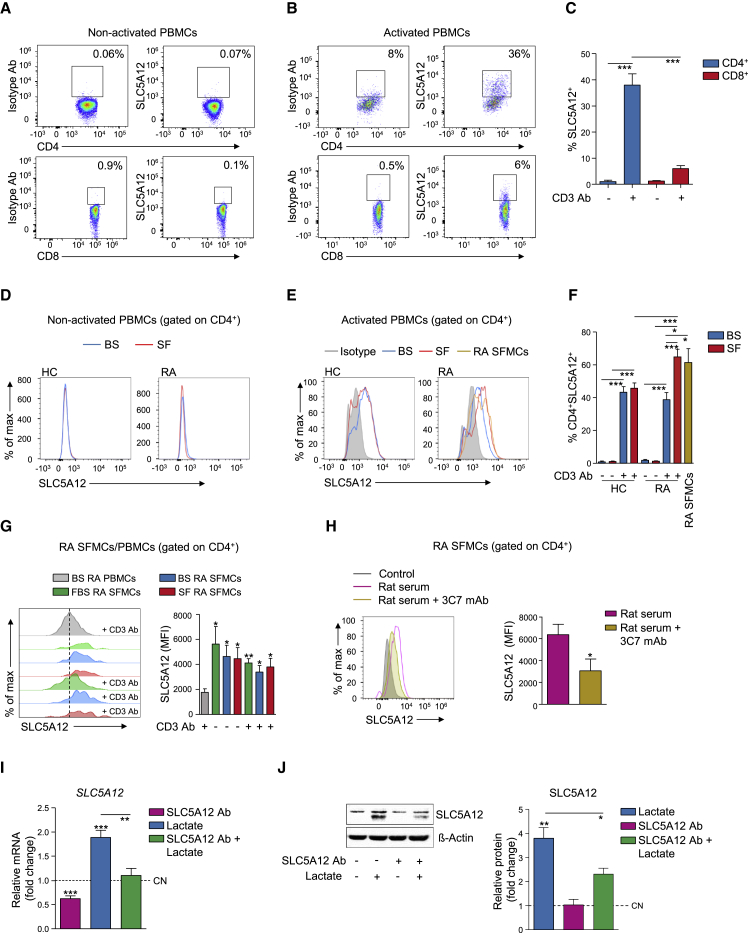
Lactate modulates CD4+ T cell migratory abilities and cytokine production via the sodium-coupled lactate transporter SLC5A12, which is selectively expressed by CD4+ but not by CD8+ T cells, at least in the murine system (Haas et al., 2015). To assess whether the expression of SLC5A12 may be regulated by activating stimuli, peripheral blood mononuclear cells (PBMCs) from healthy control (HC) subjects were activated for 48 h with anti-CD3 monoclonal antibody (mAb) or left untreated. Activation led to upregulation of SLC5A12 by peripheral CD4+ while CD8+ T cells were mostly negative (Figures 1A–1C). In the same experiment, activation led to upregulation of SLC5A12 also by peripheral CD14+ monocytes and to a less extent by CD19+ B cells (Figures S1A–S1C), conceivably via signals initiated by T cell interactions with antigen presenting cells upon CD3 stimulation in the context of the PBMCs. In comparison, the same immune cell subsets were negative for SLC5A12 or expressed it at low levels in untreated PBMCs (Figures 1A–1C and S1A–S1C). In additional experiments we carried out, we observed that SLC5A12 was already upregulated by CD4+ T cells at the 12-h activation time point of HC PBMCs and that indeed this time point may have the peak of SLC5A12 expression on the cell membrane. Indeed, SLC5A12 expression on the membrane was reduced at the subsequent 24- and 48-h time points (Figure S1D).
Figure 1.
SLC5A12 Expression by CD4+ T Cells Is Regulated by Activating and Inflammatory Stimuli
(A–C) Representative flow cytometry plots of SLC5A12 expression by CD4+ or CD8+ T cells from non-activated (n = 3; A) or anti-CD3 mAb-activated (n = 6; B) HC PBMCs. Quantification shown in (C).
(D–F) Representative flow cytometry histograms (D and E) and quantification (F) of SLC5A12 expression by CD4+ T cells from non-activated HC (n = 4) and RA (n = 4; D and F), or anti-CD3 mAb-activated HC (n = 4) and RA (n = 5; E and F) PBMCs. CD4+ T cells from non-activated RA SFMCs (n = 8; E and F) were also analyzed. Briefly, PBMCs were cultured in RPMI medium supplemented with 5% RA or HC autologous blood serum (BS), or 5% RA synovial fluid (SF); SFMCs were cultured in RPMI medium supplemented with 5% autologous SF.
(G) Representative flow cytometry histograms (left) and quantification (right) of SLC5A12 expression by CD4+ T cells from non-activated or anti-CD3 mAb-activated RA SFMCs. Briefly, cells were cultured in RPMI medium supplemented with 5% FBS (n = 3), 5% autologous BS (n = 8) or 5% autologous RA SF (n = 8). Activated RA PBMCs cultured in 5% BS RPMI (n = 5) were used as controls (H). MFI, mean fluorescent intensity.
(H) Representative flow cytometry histograms (left) and quantification (right) of SLC5A12 expression by CD4+ T cells from RA SFMCs (n = 5) incubated with 3C7 mAb or control rat sera.
(I and J) SLC5A12 mRNA (n = 5; I) and protein (representative western blots [left] and densitometric quantification [right; n = 3]; J) expression by CD4+ T cells isolated from HC PBMCs, then activated with anti-CD3 and anti-CD28 mAb in the presence of sodium lactate (10 mM) and/or SLC5A12 Ab, or left untreated. Lactate-untreated CD4+ T cells (CN, dotted line) set to 1.
One-way ANOVA (C and F) or two-tailed Student’s t test (G–J). Data expressed as mean ± SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001. See also Figures S1, S2, and S3.
Next, we activated PBMCs from HC or RA subjects for 48 h with anti-CD3 mAb or left them untreated and then compared the expression of SLC5A12 by CD4+ T cells. We found that peripheral RA CD4+ T cells became SLC5A12+ only upon T cell receptor (TCR) engagement, much like their HC counterparts (Figures 1D–1F). We analyzed the phenotype of CD4+SLC5A12+ T cells in further depth in Figure 3.
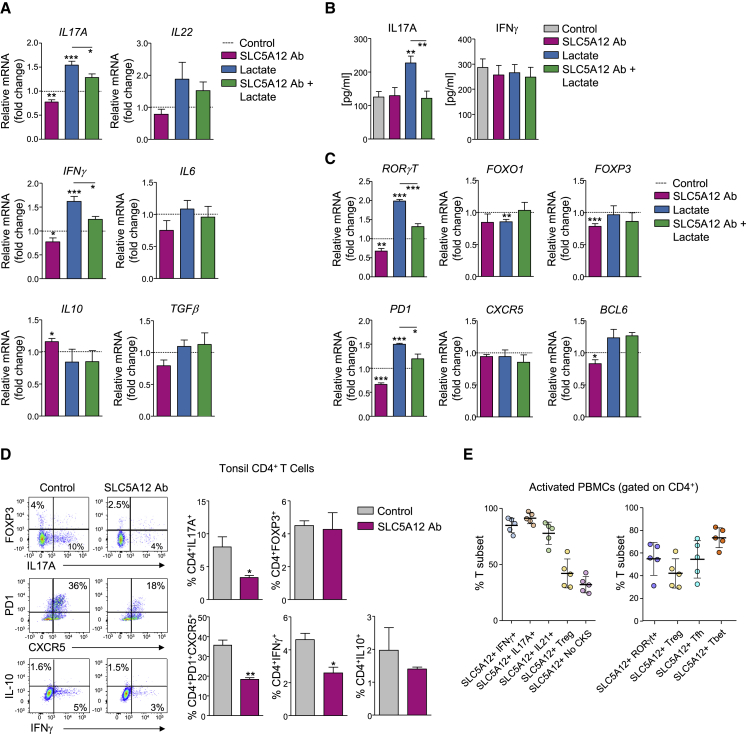
Figure 3.
Lactate Shapes the Effector Phenotype of CD4+ T Cells at the Site of Inflammation via SLC5A12
(A) Relative mRNA expression levels of IL17A, IL22, IFNγ, IL6, IL10, and TGFβ as assessed by qRT-PCR in tonsil CD4+ T cells treated with sodium lactate (10 mM) and/or SLC5A12 Ab or left untreated (n = 5). Levels of mRNA of each cytokine expressed by lactate-untreated CD4+ T cells were set to 1 (CN, dotted line).
(B) IL-17A and IFNγ ELISAs from supernatants of tonsil CD4+ T cells treated as in (A), (n = 5, each in duplicate).
(C) Relative mRNA expression levels of RORγT, FOXO1, FOXP3, PD1, CXCR5, and BCL6 as assessed by qRT-PCR in tonsil CD4+ T cells treated as in (A), (n = 5). Levels of mRNA of each cytokine expressed by lactate-untreated CD4+ T cells set to 1 (CN, dotted line).
(D) Representative flow cytometry plots of CD4+IL17+, CD4+FOXP3+, CD4+PD1+CXCR5+, CD4+IFNγ+, and CD4+IL10+ tonsil CD4+ T cells incubated in the presence or absence of SLC5A12 Ab (left; n = 3). Quantification bar charts (right).
(E) Percentage of IFNγ+, IL17A+, IL21+, Treg (CD25+Foxp3+), and cytokine-negative (Neg CKS; left) or RORγt+, Treg (CD25+Foxp3+), Tfh (CXCR5+PD-1+ICOS+), and Tbet+ (right) CD4+SLC5A12+ T cell subsets in 48-h activated human HC PBMCs (n = 5).
Two-tailed Student’s t test. Data expressed as mean ± SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001.
To test whether inflammatory cues, in addition to activating stimuli, may also contribute to the expression of SLC5A12 by CD4+ T cells, we cultured HC or RA PBMCs in medium supplemented with 5% HC or RA autologous blood serum (BS), respectively, or with 5% RA synovial fluid (SF). The percentage of CD4+SLC5A12+ T cells was very low in both non-activated HC and RA PBMCs cultured in medium containing autologous BS or RA SF (Figures 1D and 1F). Anti-CD3 mAb-mediated activation led to upregulation of SLC5A12 by CD4+ T cells; however, no difference was observed in the percentage of CD4+SLC5A12+ T cells from HC and RA PBMCs activated in medium containing autologous BS (Figures 1E and 1F). In contrast, anti-CD3 mAb-mediated activation of RA but not of HC PBMCs in the presence of 5% RA SF led to a robust further upregulation of SLC5A12 by CD4+ T cells as compared to HC and RA CD4+ T cells from PBMCs activated in the presence of BS (Figures 1E and 1F). Importantly, we observed that SLC5A12 expression levels by CD4+ T cells from RA PBMCs activated in the presence of RA SF were comparable to those expressed by CD4+ T cells in synovial fluid mononuclear cells (SFMCs) from RA joints in the absence of any ex vivo stimulation (Figures 1E and 1F). We also found that CD4+ T cells from RA SFMCs presented high levels of SLC5A12 irrespective of any activating or inflammatory stimuli we used ex vivo, as compared to CD4+ T cells from RA PBMCs activated in the presence of autologous BS, suggesting they have already experienced maximal levels of SLC5A12 inducing factors, i.e., antigen and exposure to inflammatory cues (Figure 1G). As for RA SFMCs, when we analyzed mononuclear cells (MCs) from inflamed tonsils excised from patients subjected to tonsillectomy, CD4+ T cells were SLC5A12+, independent of any activating stimuli we used ex vivo (Figures S2A–S2C and S2G). Likewise, analysis of CD14+ and CD19+ cells by fluorescence-activated cell sorting (FACS) or CD68+ and CD20+ cells by fluorescence microscopy in the same samples revealed that they were SLC5A12+, independent of any activating stimuli we used ex vivo (Figures S2D–S2G). In contrast, CD8+ T cells were mostly negative for SLC5A12 (Figures S2A–S2C and S2G), which was consistent with data in Figures 1A–1C.
We then wondered whether lactate may contribute to the regulation of the expression of SLC5A12. We generated mAbs targeting SLC5A12 by immunization of rats with a peptide comprising the predicted main extracellular loop of SLC5A12 (Gopal et al., 2007), with the aim of inhibiting the carrier function of the transporter. Out of ∼400-screened clones, we selected 3C7 for its ability to specifically recognize SLC5A12 (Figure S3). Treatment of RA SFMCs with 3C7 mAb led to reduced expression of the transporter itself by CD4+ T cells (Figure 1H). Furthermore, incubation of anti-CD3 and anti-CD28 mAb-activated peripheral CD4+ T cells—isolated from HC PBMCs with magnetic bead-based negative selection prior to activation—with 10 mM sodium lactate, a concentration similar to what is measured in RA SF (Haas et al., 2015), contributed to the induction of SLC5A12 expression both at mRNA and protein level. Pre-incubation with a blocking anti-SLC5A12 polyclonal antibody (SLC5A12 Ab; Haas et al., 2015) prevented lactate-induced upregulation of SLC5A12 (Figures 1I and 1J). These data suggested that accumulation of extracellular lactate, such as at sites of inflammation, contributes to the upregulation of the lactate transporter SLC5A12 by activated CD4+ T cells.
SLC5A12 Facilitates Lactate Uptake and Oxidation by TCA Cycle in Activated CD4+ T Cells
Having observed an increase in the expression levels of the lactate transporter SLC5A12 in response to activating and inflammatory stimuli in CD4+ T cells (Figure 1) and knowing that these cells are mostly MCT1− (Haas et al., 2015), we wondered whether SLC5A12 may serve as the main carrier of lactate into activated CD4+ T cells that have reached the inflamed tissue and hence are exposed to a high concentration of extracellular lactate.
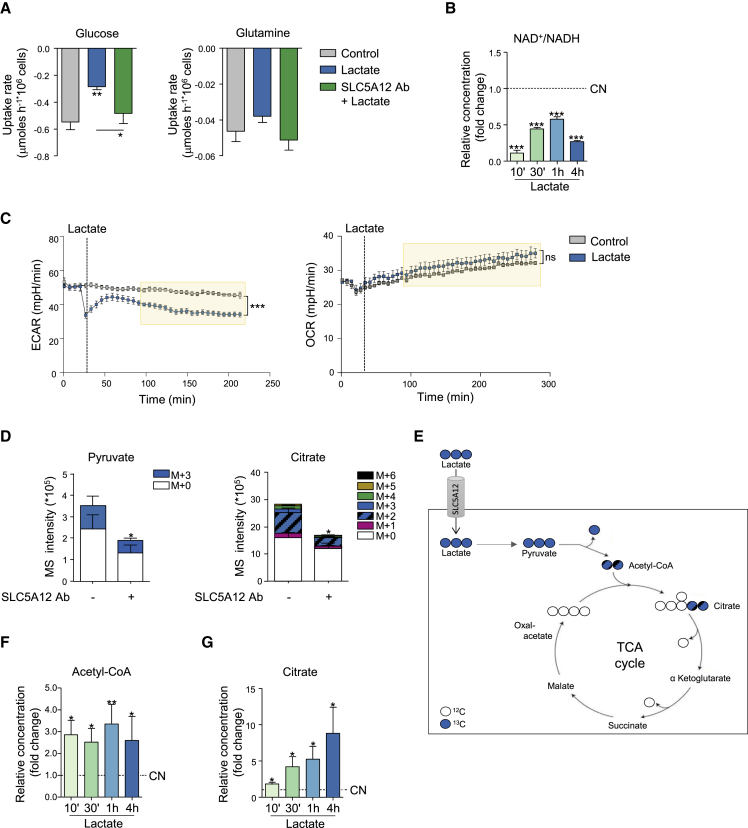
Indeed, exposure to lactate caused a decrease in glucose uptake by activated CD4+ T cells, which was reversed by incubation of cells with SLC5A12 Ab (Figure 2A). No significant change in glutamine uptake was observed in the same experiment indicating a specific block in glycolysis (Figure 2A). NAD+ is a key co-factor of the sixth reaction of the glycolytic cascade catalyzed by glyceraldehyde 3-phosphate dehydrogenase. As a consequence of this reaction, NAD+ is reduced to NADH, which acts as an inhibitory feedback on glycolysis. NADH can be re-oxidized to NAD+ via the lactate dehydrogenase reaction converting pyruvate to lactate. This reaction is important to maintain a steady flux of glycolysis. However, lactate dehydrogenase can perform the reverse reaction when cells are exposed to high levels of extracellular lactate as it happens at the site of inflammation, with reduction of NAD+ to NADH and consequent inhibitory feedback on glycolysis. Indeed, upon exposure to lactate we observed a drop in the NAD+/NADH ratio in activated CD4+ T cells, indicating a relative increase in intracellular NADH (Figure 2B). Data in Figures 2A and 2B were consistent with the reduced rate of glycolysis (ECAR) we observed in the presence of lactate (Figure 2C, left), while the oxygen consumption rate (OCR) in the mitochondria was not affected by lactate (Figure 2C, right).
Figure 2.
Lactate Uptake by CD4+ T Cells Impacts Intracellular Utilization of Central Carbon Metabolic Pathways
(A) Glucose and glutamine uptake rates for CD4+ T cells isolated from HC PBMCs, then activated with anti-CD3 and anti-CD28 mAbs for 24 h followed by further 48-h culture with lactate alone or in the presence of SLC5A12 Ab, or left untreated, in medium containing low glucose (5 mM) and 5% FBS (n = 3, each in duplicate).
(B) NAD+ and NADH intracellular levels in CD4+ T cells (n = 2) treated with sodium lactate (10 mM) for the indicated time points after 72-h activation and shown as NAD+/NADH ratio. Lactate-untreated CD4+ T cells (CN, dotted line) set to 1.
(C) Seahorse measurements of extracellular acidification (left) and oxygen consumption (right) rates (ECAR and OCR, respectively) by 12-h-activated CD4+ T cells (n = 3, technical replicates). 1 h prior to the experiment, cells were seeded in a 96-well microplate in XF Assay medium in the presence of 10 mM of glucose. Sodium lactate (10 mM) or PBS was injected during measurement. Data representative of n = 2 independent experiments.
(D and E) 13C tracing of [U13C]-lactate into pyruvate and citrate. Activated CD4+ T cells were incubated for 48 h with [U13C]-lactate in the presence or absence of SLC5A12 Ab in medium containing low glucose (5 mM) and 5% FBS (n = 2, each in duplicate). Polar metabolites were extracted, analyzed by LC-MS and peak areas of mass isotopologues normalized to cell number are represented.
(F and G) Acetyl-CoA (F) and citrate (G) intracellular levels in CD4+ T cells (n = 3) treated with sodium lactate (10 mM) for the indicated time points after 72-h activation. Lactate-untreated CD4+ T cells (CN, dotted line) set to 1.
Two tailed Student’s t test. Data expressed as mean ± SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001.
We reasoned that these findings might be explained by an uptake of lactate by activated CD4+ T cells when they are in an inflamed, lactate-rich tissue. Pyruvate may then enter the TCA cycle, but since we did not observe an increase in mitochondrial OCR, we wondered what the fates of the lactate-derived carbons were. To test our rationale in a direct fashion, we performed mass spectrometry-based tracer analysis of [U13C]-lactate. Specifically, we activated CD4+ T cells and then incubated them with [U13C]-lactate in the presence or absence of SLC5A12 Ab, similarly to Figure 2A. We then extracted intracellular metabolites and performed mass spectrometry analysis. We found that a significant proportion of 13C-carbons from [U13C]-lactate were incorporated in to pyruvate (M+3) and citrate (M+2), and that this effect was reduced by incubation with SLC5A12 Ab (Figures 2D and 2E). Additionally, the M0 (unlabeled isotope) in each of these metabolites is also reduced: as the antibody blocks the lactate transporter SLC5A12, influx of 13C-lactate but also of glucose-derived 12C-lactate is reduced as a consequence of SLC5A12 “re-importing” some lactate previously secreted by the cell. Consistent with these data, we found an increase in citrate and acetyl-CoA levels in activated CD4+ T cells exposed to lactate at different time points (Figures 2F and 2G).
Altogether, these data suggested that when exposed to high levels of lactate, such as those found in an inflamed tissue, activated CD4+ T cells could take up lactate via the specific carrier SLC5A12. This causes glycolysis to slow down (through NAD) and in turn leads to more carbons going into the TCA cycle. Therefore, we hypothesized that the carbon flux we observed may be required to replenish intermediates of the TCA cycle that feed biosynthetic processes.
Lactate Shapes the Effector Phenotype of CD4+ T Cells at the Site of Inflammation via SLC5A12
We then tested the effects of exposure to inflamed tissue levels of lactate in the presence or absence of SLC5A12 Ab on the effector phenotype of anti-CD3 and anti-CD28 mAb-activated CD4+ T cells that were isolated from inflamed tonsils. We observed an upregulation of IL17A and IFNγ mRNAs in response to lactate, which was reversed by incubation with SLC5A12 Ab. IL17-family member IL22 also showed a tendency to upregulation in response to lactate. We did not observe any significant modulation in other cytokines (i.e., inflammatory IL6 or immunosuppressive IL10 and TGFβ; Figure 3A). However, the observed upregulations at the mRNA level resulted in only IL17A but not IFNγ upregulation at the protein level upon treatment with lactate and again this response was abolished by incubation with SLC5A12 Ab (Figure 3B).
Supporting the findings of IL17 upregulation, also the mRNA of RORγT, the signature transcription factor of the Th17 T cell subset, was elevated as a consequence of exposure to lactate and again this response was abolished by incubation with SLC5A12 Ab (Figure 3C). Interestingly, the expression of the transcription factor FOXO1, which limits the differentiation of CD4+ T cells into the Th17 subset and the consequent production of IL17 (Ouyang et al., 2009), was reduced by treatment with lactate, even though incubation with SLC5A12 Ab did not have any effects on its expression (Figure 3C). Consistent with data in Figure 3A, expression of FOXP3, the signature transcription factor of regulatory T (Treg) cells producing TGFβ and IL10 was not impacted by lactate treatment (Figure 3C). Furthermore, we observed a lactate-dependent regulation of PD1 but not of the transcription factor BCL6 or the chemokine receptor CXCR5 (Figure 3C).
To gain direct insights on the impact of lactate on the effector phenotype of CD4+ T cells at the site of inflammation, we conducted intracellular staining of CD4+IL17+ (Th17), CD4+IFNγ+ (Th1), CD4+PD1+CXCR5+ (Tfh), and CD4+FOXP3+ or CD4+IL10+ (Treg) subsets from activated MCs from inflamed tonsils (Figure 3D). Incubation with SLC5A12 Ab resulted in a reduction in the Th17 and Tfh T cell subsets with a less pronounced reduction in the Th1 and no modulation of the Treg subsets (Figure 3D). We further characterized the phenotype of human CD4+SLC5A12+ T cells from 48-h activated HC PBMCs and observed that RORγt+, CXCR5+PD-1+ICOS+ (Tfh), and Tbet+ are the CD4+ T cell subsets that are most positive for SLC5A12 while CD25+Foxp3+ (Treg) CD4+ T cells are less frequently positive for SLC5A12 (Figure 3E, right). Furthermore, we observed that IFNγ+ (signature cytokine of Tbet+), IL17A+ (signature cytokine of RORγt+), and IL21+ (signature cytokine of Tfh) CD4+ T cells are mostly positive for SLC5A12, while Treg and CD4+ T cells that do not produce cytokines (Neg CKS) are much less frequently positive for SLC5A12 (Figure 3E, left).
Lactate Induces IL17 Expression via Nuclear PKM2- and Fatty Acid Synthesis-Mediated STAT3 Phosphorylation
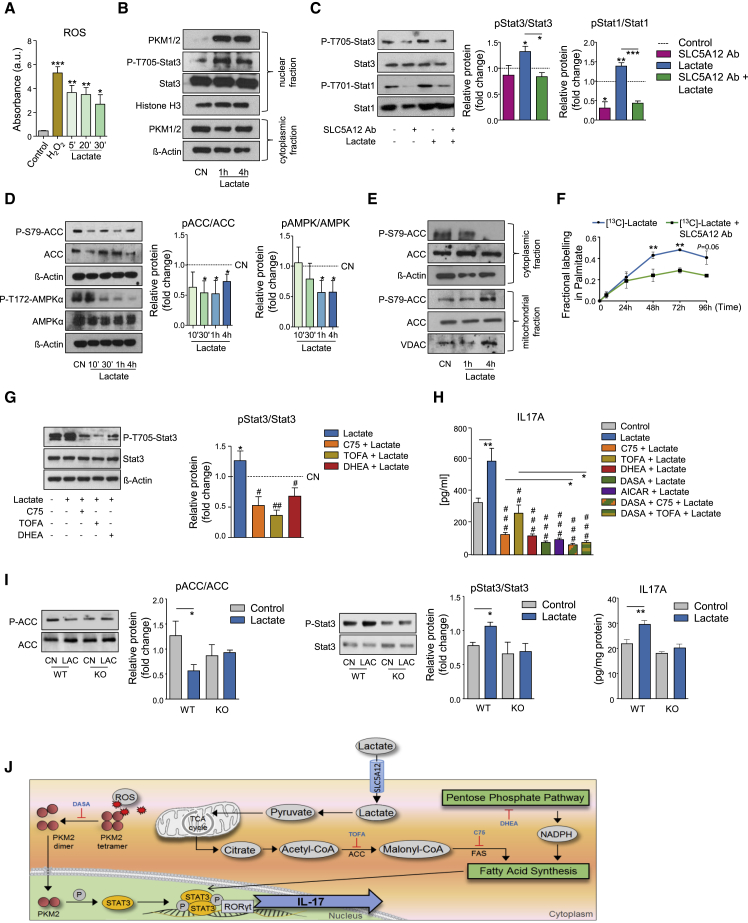
We next asked how lactate may promote IL17 expression and whether the lactate uptake via SLC5A12 and its induced increase of citrate may play a role in this response of CD4+ T cells. Exposure of activated CD4+ T cells to inflamed tissue levels of lactate caused a rapid, marked elevation of intracellular reactive oxygen species (ROS; Figure 4A). The glycolytic enzyme pyruvate kinase M2 (PKM2) functions as a homo-tetramer in the cytosol converting phosphoenolpyruvate to pyruvate in the last reaction of glycolysis. ROS can promote the oxidation and subsequent dimerization of PKM2. Dimers of PKM2 localize in the nucleus where they phosphorylate transcription factors, including signal transducer and activator of transcription 3 (STAT3), a known transcriptional regulator of IL17 (Shirai et al., 2016, Yang et al., 2007). Indeed, we found that lactate promoted the translocation of PKM2 in the nucleus and the phosphorylation of STAT3 (Figure 4B). Activation of STAT3 occurred as early as 1 h after cell treatment with lactate (Figure 4B) and could still be observed at 12 h (Figure 4C). STAT1, another STAT family member implicated in Th17 differentiation (Peters et al., 2015), was also phosphorylated at the same time point (Figure 4C). Phosphorylation of STAT1/3 returned to basal levels upon incubation with SLC5A12 Ab (Figure 4C).
Figure 4.
Lactate Induces IL17 via Nuclear PKM2- and FAS-Mediated STAT3 Phosphorylation
CD4+ T cells were isolated from HC PBMCs and activated with anti-CD3 and anti-CD28 mAb.
(A) ROS levels in CD4+ T cells (n = 3) treated with sodium lactate (10 mM) for the indicated time points after 72-h activation. PBS and H2O2 were used as negative and positive control, respectively.
(B) Representative western blots showing nuclear PKM1/2, P-STAT3, STAT3, and cytosolic PKM1/2 in activated CD4+ T cells treated with sodium lactate (10 mM) for the indicated time points or left untreated (CN). Histone H3 and β-actin were used as controls for nuclear and cytosolic fraction, respectively. Data representative of n = 3 independent experiments.
(C) Representative western blots (left) and densitometric quantification (right; n = 3) of P-STAT3, STAT3, P-STAT1, and STAT1 expression by activated CD4+ T cells treated with sodium lactate (10 mM) and/or SLC5A12 Ab, or left untreated. Untreated CD4+ T cells (CN, dotted line) set to 1.
(D) Representative western blots (left) and densitometric quantification (right; n = 3) of P-ACC, ACC, P-AMPK, and AMPK expression by activated CD4+ T cells treated with sodium lactate (10 mM) for the indicated time points or left untreated (CN). Untreated CD4+ T cells (CN, dotted line) set to 1.
(E) Representative western blots showing cytosolic and mitochondrial P-ACC and ACC in activated CD4+ T cells treated with sodium lactate (10 mM) for the indicated time points or left untreated (CN). β-actin and VDAC were used as controls for cytosolic and mitochondrial fraction, respectively. Data representative of n = 2 independent experiments.
(F) Mass spectrometry carbon tracer analysis of palmitate in 48-h [U13C]-lactate-fed activated CD4+ T cells treated as in Figure 2D (n = 4, time points 0, 24, and 48 h; n = 2, time points 72 and 96 h).
(G) Representative western blots (left) and densitometric quantification (right; n = 2) of P-STAT3 and STAT3 expression by activated CD4+ T cells treated with sodium lactate (10 mM) alone or in combination with C75 (10 μM), TOFA (20 μM), and DHEA (20 μM) or left untreated (CN). Untreated CD4+ T cells (CN, dotted line) set to 1.
(H) IL-17A and IFNγ ELISAs from supernatants of activated CD4+ T cells treated with sodium lactate (10 mM) alone or in combination with C75 (10 μM), TOFA (20 μM), DHEA (20 μM), DASA (20 μM), AICAR (1 mM), or left untreated (n = 5, each in duplicate; for lactate + DASA + C75 or lactate + DASA + TOFA, n = 2, each in duplicate).
(I) Representative western blots (left) and densitometric quantifications (right; n = 3) of P-ACC, ACC, P-STAT3, and STAT3 expression by activated CD4+ T cells from Slc5a12 WT or KO mice, treated with sodium lactate (10 mM) or left untreated (CN). Also, IL-17A ELISA from supernatants of activated CD4+ T cells from Slc5a12 WT or KO mice, treated with sodium lactate (10 mM) or left untreated (n = 3, each in duplicate).
Two-tailed Student’s t test (A), (C), (F), (H), and (I) or one-way ANOVA (G). Data expressed as mean ± SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ###p ≤ 0.001 versus lactate (H).
(J) Schematic depicting the described findings: lactate modulates IL17 expression by activating two pathways, PKM2 translocation into the nucleus and FAS induction, converging on STAT3-induced transcription of IL17. See also Figure S4.
De novo fatty acid synthesis (FAS) is another biological process that has been implicated in the differentiation of the Th17 T cell subset (Berod et al., 2014). We observed that activated CD4+ T cells take up lactate and consequently increase the intracellular pool of citrate and acetyl-CoA (Figures 2D–2G), which are the substrates of FAS. We therefore asked whether exposure to lactate may induce FAS in these cells, by assessing the activation levels of acetyl-CoA-Carboxylase (ACC) and 5′-AMP activated protein kinase (AMPK), two key enzymes in the regulation of fatty acid metabolism. We found that exposure to lactate caused a decrease in phosphorylated ACC at Serine 79 indicating increased ACC enzymatic activity (Figure 4D). Consistently, we also detected a decrease in phosphorylated AMPKα at Threonine 172 (Figure 4D), indicating reduced AMPK enzymatic activity. ACC exists in humans and other mammals as two isoforms, ACC1 and ACC2. Whereas ACC1 is present in the cytosol and initiates de novo synthesis of fatty acids by converting acetyl-CoA to malonyl-CoA (Chirala and Wakil, 2004), ACC2 is associated with the outer mitochondrial membrane and is a key enzyme in the oxidation of fatty acids (FAO; Abu-Elheiga et al., 2000, Abu-Elheiga et al., 2001). We found a marked decrease in phosphorylated ACC in the cytosol of activated, lactate-treated CD4+ T cells but no major change in phosphorylated ACC in the mitochondria (Figure 4E).
To test any effects of lactate on FAO in a more direct fashion, we measured OCR in activated CD4+ T cells cultured in 2.5 mM glucose and 1 μM BSA-palmitate that served as a substrate for FAO, or BSA alone. BSA-palmitate raised OCR as compared to BSA alone, but lactate did not affect either conditions. Addition of etomoxir, an inhibitor of the key enzyme carnitine palmitoyltransferase-1 (CPT-1) in the initiation of FAO, reduced BSA-palmitate OCR to the levels observed in the BSA alone control and again this effect was not affected by lactate (Figure S4A).
To test whether lactate feeds FAS, we incubated activated CD4+ T cells with [U13C]-lactate and traced 13C labeling in palmitate. Lactate 13Cs labeled nearly 50% of newly synthesized palmitate. This effect was inhibited in cells that were incubated with SLC5A12 Ab, indicating incorporation of lactate-derived carbons in palmitate backbone (Figure 4F). An induction of FAS by lactate was also confirmed by an increment in total free fatty acid (FFA) cellular content (Figure S4B).
Given the importance of both STAT3 and FAS in the differentiation of the Th17 T cell subset (Shirai et al., 2016, Yang et al., 2007, Berod et al., 2014, Shi et al., 2011), we asked whether lactate may modulate the expression of IL17 via either or both pathways. We treated activated CD4+ T cells with 5-(tetradecyloxy)-2-furoic acid (TOFA), a competitive inhibitor of ACC (Berod et al., 2014), 4-methylene-2-octyl-5-oxotetrahydrofuran-3-carboxylic acid (C75), a FAS inhibitor (Shen et al., 2017) and dehydroepiandrosterone (DHEA), an inhibitor of glucose-6-phosphate dehydrogenase (G6PDH, Raineri and Levy, 1970; Gordon et al., 1995), a key step in the pentose phosphate pathway (PPP) providing NADPH equivalents for FAS (see also Figures 6C–6E). As expected, all three inhibitors increased phosphorylated ACC levels (Figure S4C), indicating an inhibitory effect on FAS. DHEA also increased phosphorylated AMPK (Figure S4C), which is suggestive of a switch toward FAO. With these three compounds, we then tested the impact of lactate-induced FAS on STAT3 activation and IL17 production. All three compounds reduced lactate-induced phosphorylation of STAT3 (Figure 4G) and expression of IL17A (Figure 4H). Also, the AMPK activator aminoimidazole-4-carboxamide 1-β-D-ribofuranoside (AICAR; Corton et al., 1995) and the potent and selective PKM2 activator N,N’-diarylsulfonamide (DASA), which stabilizes cytosolic PKM2 homo-tetramers and prevents PKM2 dimers translocation into the nucleus (Anastasiou et al., 2011), markedly reduced the expression of IL17A (Figure 4H). Interestingly, a co-treatment with DASA and C75 or TOFA resulted in an additional reduction of lactate-induced IL17A expression as compared to each compound alone (Figure 4H).
Figure 6.
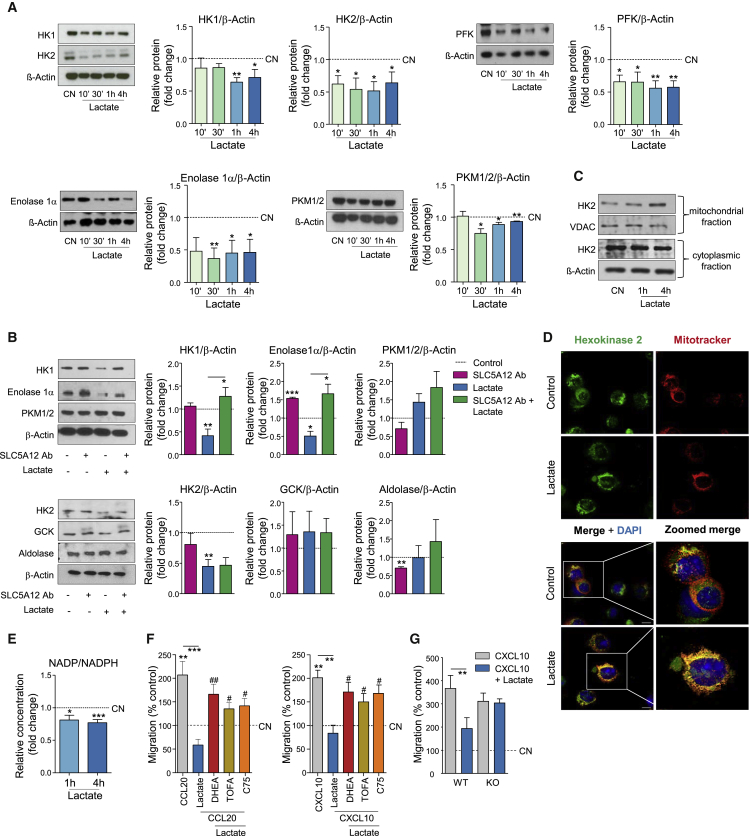
Lactate Reduces the Motility of CD4+ T Cells via Reduced Glycolysis and Enhanced FAS
(A) Representative western blots (left) and densitometric quantification (right; n = 3) of HK1, HK2, PFK, enolase 1α, and PKM1/2 expression by activated CD4+ T cells treated with sodium lactate (10 mM), or left untreated. Untreated CD4+ T cells (CN, dotted line) set to 1.
(B) Representative western blots (left) and densitometric quantification (right; n = 3) of HK1, HK2, enolase 1α, PKM1/2, GCK, and aldolase expression by activated CD4+ T cells treated with sodium lactate (10 mM) and/or SLC5A12 Ab, or left untreated. Untreated CD4+ T cells (CN, dotted line) set to 1.
(C) Representative western blots showing mitochondrial and cytosolic HK2 in activated CD4+ T cells treated with sodium lactate (10 mM) for the indicated time points or left untreated (CN). VDAC and β-actin were used as controls for mitochondrial and cytosolic fraction, respectively. Data representative of n = 2 independent experiments.
(D) Representative immunofluorescence images of untreated and lactate-treated CD4+ T cells. Co-staining for hexokinase 2 (green), MitoTracker (red), and DAPI (blue). Scale bar, 10 μm.
(E) NADP+ and NADPH intracellular levels in CD4+ T cells (n = 5) treated with sodium lactate (10 mM) for the indicated time points after 72-h activation and shown as NADP+/NADPH ratio. Lactate-untreated CD4+ T cells (CN, dotted line) set to 1.
(F) In vitro chemokinesis of activated CD4+ T cells in response to CCL20 (500 ng/mL; n = 4) or CXCL10 (300 ng/mL; n = 3) in the presence of sodium lactate (10 mM) with or without the metabolic drugs C75 (10 μM), TOFA (20 μM), and DHEA (20 μM). Untreated CD4+ T cells (w/o CXCL10, dotted line) were set to 100.
(G) In vitro chemokinesis of activated CD4+ T cells (n = 4) from Slc5a12 WT or KO mice in response to CXCL10 (300 ng/mL; 4 h) in the presence of sodium lactate (10 mM). Untreated CD4+ T cells (CN, dotted line) were set to 100.
Two-tailed Student’s t test (A), (B), and (E) or one-way ANOVA (F) and (G). Data expressed as mean ± SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; #p ≤ 0.05 versus lactate + chemokine. See also Figure S6.
To further support our findings, we took advantage of a new Crispr-Cas9 Slc5a12 KO mouse. We show that in response to lactate both ACC and STAT3 phosphorylation as well as IL17 production are impaired (Figure 4I).
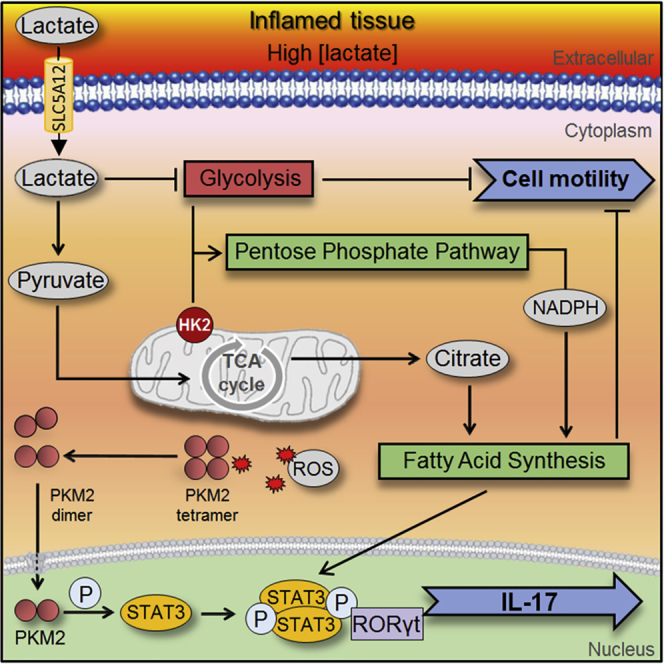
Taken together these data indicate that lactate modulates IL17 expression by activating two pathways, PKM2 translocation into the nucleus and enhanced FAS, converging on STAT3-induced transcription of IL17 (Figure 4I).
Reduced Glycolysis and Enhanced FAS Are the Mechanisms through Which Lactate Induces CD4+ T Cell Retention in the Inflamed Tissue
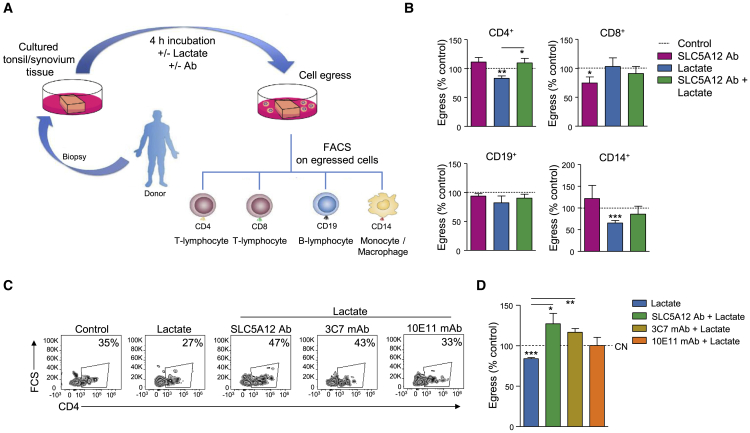
Building upon our previous findings that inflamed tissue levels of lactate induce a “stop migration signal” in activated CD4+ T cells (Haas et al., 2015, Pucino et al., 2017), we assessed the impact of interfering with SLC5A12 function—via the use of both SLC5A12 Ab and the mAbs we generated—on human CD4+ T cell migration in vitro and ability to egress from the inflamed tissue ex vivo. We started by culturing equal size tissue sections from juxtaposing areas of tonsil biopsies—isolated from patients who had been subjected to tonsillectomy—in the presence or absence of lactate and/or SLC5A12 Ab. We then assessed by flow cytometry the type and number of immune cells released in the culture media in each condition (Figure 5A). Lactate reduced the egress of CD4+ T cells as compared to the control condition and this effect was reversed by SLC5A12 Ab. Lactate also reduced the egress of CD14+ cells but treatment with SLC5A12 Ab did not reverse this effect, indicating SLC5A12 is not a dominant lactate transporter in this subset. No significant effects were observed by treatments with lactate and lactate plus SLC5A12 Ab in CD8+ or CD19+ cells (Figure 5B).
Figure 5.
SLC5A12 Blockade Promotes the Egress of CD4+ T Cell from the Inflamed Tissue
(A) Organ culture schematic describing the analysis performed to assess the egress of mononuclear cells (MCs) from the inflamed tissue.
(B) Analysis of MCs (CD4+, CD8+, CD19+, and CD14+) egress from tonsil tissues (n = 3, each in duplicate) cultured with sodium lactate (10 mM) and/or SLC5A12 Ab, or left untreated. Untreated MCs (CN, dotted line) set to 100.
(C and D) Representative flow cytometry plots (D) and quantification (E) of egressed CD4+ T cells from RA synovial tissues (n = 3) cultured with sodium lactate (10 mM) and/or SLC5A12 Ab, 3C7 mAb, 10E11 mAb, or left untreated. Untreated MCs (CN, dotted line) set to 100.
Two-tailed Student’s t test. Data expressed as mean ± SEM. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001. See also Figure S5.
We then sought to assess whether a similar effect could be observed in synovial tissue biopsies from patients who had been subjected to joint replacement. Concurrently, we also assessed whether the SLC5A12 mAbs we generated were able to reverse lactate-induced block on CD4+ T cell migratory abilities. We treated activated CD4+ T cells with lactate or left them untreated in the presence or absence of SLC5A12 Ab or seven SLC5A12 mAb clones. We then assessed cell chemokinesis in response to the chemokine CXCL10 in trans-wells. As expected, SLC5A12 Ab reversed the “stop migration signal” induced by lactate (Figure S5A; Haas et al., 2015). In addition, the mAb clones 3C7 and 9G7 were able to consistently reverse the “stop migration signal” induced by lactate (Figure S5A). Furthermore, as shown in Figure S3, 3C7 mAb was able to specifically recognize SLC5A12. We therefore tested 3C7 mAb alongside SLC5A12 Ab in the ex vivo egress model in synovial tissues. Again, SLC5A12 Ab was able to reverse the lactate-mediated retention of CD4+ T cell in the tissue and a similar effect was obtained with 3C7 mAb (Figures 5C and 5D).
Next, we asked how the metabolic adaptation of activated CD4+ T cells to inflamed tissue levels of lactate impacted on their response to migratory stimuli. Glycolysis is required for the motility of activated, murine CD4+ T cells (Haas et al., 2015), and FAS supports invasiveness of inflamed tissues by peripheral CD4+ T cells (Shen et al., 2017).
We first analyzed the effect of exposure of activated, human CD4+ T cells to lactate on several glycolytic enzymes in a time-course experiment. We observed reduced levels of hexokinase 1 (HK1), HK2, phosphofructokinase (PFK), enolase1α and PKM1/2 (Figure 6A), indicating reduced rates of glycolysis, consistent with data in Figures 2A–2C. However, lactate-induced downregulation of HK1 and enolase1α but not of HK2 and PKM1/2 was impeded by cell incubation with SLC5A12 Ab (Figure 6B). These data suggest specific checkpoints of lactate-mediated control of glycolysis, in addition to the observed increased reduction of NAD+ to NADH, as shown in Figure 2B. Based on our observation that lactate caused an increase in the intracellular pools of citrate and acetyl-CoA, we investigated whether lactate may induce post-translational acetylation of cytosolic proteins in a 12-h time course but we did not observe any major changes (Figure S6A). This potential checkpoint control will require further investigations.
Intracellular localization of HK2 was suggested to serve as a checkpoint channeling intracellular metabolic fluxes. While cytosolic HK2 mediates glycolysis, VDAC-dependent binding of HK2 to the outer membrane of mitochondria promotes cell survival (Anderson et al., 2017, Woldetsadik et al., 2017, Mathupala et al., 2009, Anflous-Pharayra et al., 2007). Indeed, mitochondrial HK2 may favor glucose-6-phosphate entry in the PPP producing NADPH equivalents and anabolic intermediates (Cheung et al., 2012). We found an increase in mitochondrial HK2 after 4-h lactate treatment as compared to cells left untreated (Figure 6C). This observation was supported by confocal microscopy data showing co-localization of HK2 with mitochondria upon 4 h treatment with lactate (Figure 6D). We also found a reduction in the NADP+/NADPH ratio at 1 h and 4 h after cell treatment with lactate (Figure 6E), consistent with an increased shunt of glucose-6-phosphate into PPP and with the observed induction of FAS. The observed inhibition of PKM2 may also contribute to divert glucose into the PPP and thereby generate NADPH (Le Goffe et al., 2002). Therefore, we asked whether induction of FAS by lactate might play a role in the entrapment of CD4+ T cells in the inflamed tissue. Activated CD4+ T cells were treated with lactate in the presence of DHEA, TOFA, and C75 or left untreated and then subjected to chemokinesis in response to either CCL20 or CXCL10. All compounds blocking FAS at different key steps released CD4+ T cells from lactate-induced “stop migration signal” (Figure 6F). Again, to further support our findings, we took advantage of the Slc5a12 KO mouse and found that in response to lactate migration response to CXCL10 was impaired (Figure 6G).
Overall, our findings indicate that lactate-induced inhibition of CD4+ T cell response to migratory stimuli and retention in the inflamed tissue is due to a metabolic adaptation to local levels of lactate that entails reduced glycolysis and translocation of HK2 to the outer membrane of mitochondria, with metabolic fluxes diverted into NADPH-dependent de novo FAS (Figure S6B).
Lactate/SLC5A12-Induced Metabolic Reprogramming Is Operational in the CD4+ T Cell Infiltrated RA Synovium
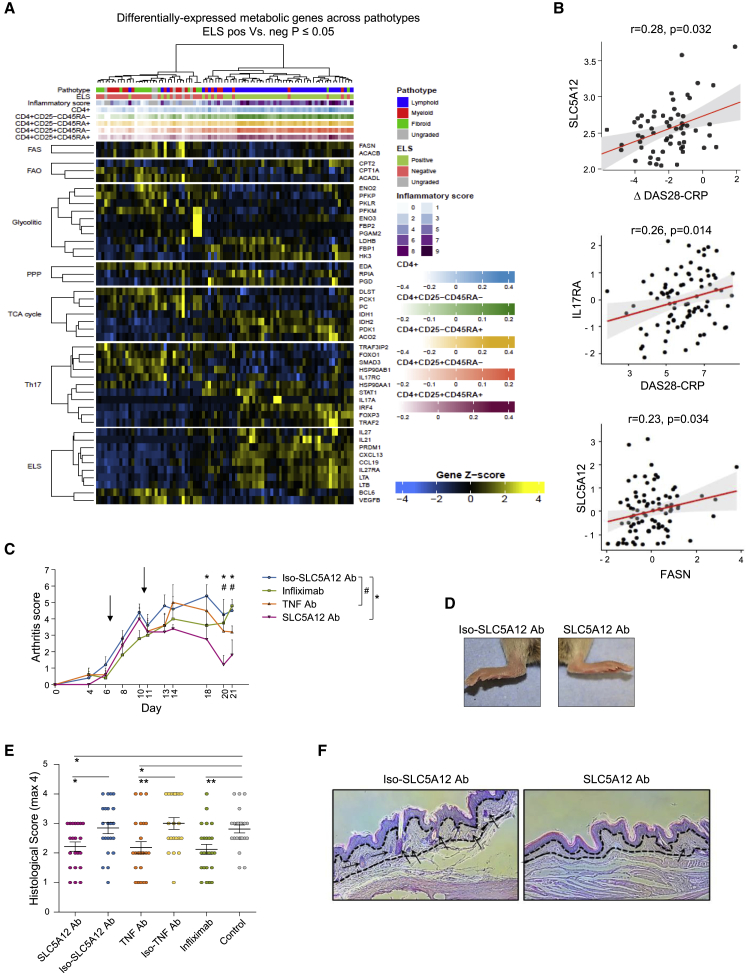
To examine the lactate/SLC5A12-induced metabolic signaling network in the clinical settings of RA, we took advantage of the pathobiology of early arthritis cohort (PEAC). This is a cohort of adults over the age of 18 manifesting early symptomatic inflammatory arthritis (<12 months) and who are naive to treatment with conventional or biologic disease-modifying anti-rheumatic drugs (DMARDs; http://www.peac-mrc.mds.qmul.ac.uk/index.php). Synovial biopsies collected for this cohort were classified according to histological pattern of synovitis (i.e., lymphoid, myeloid, or fibroid; Pitzalis et al., 2013, Cañete et al., 2009) and presence of ELSs. These are organized aggregates of T and B cells that develop at sites of chronic inflammation and are associated with more severe disease course and autoimmune responses as well as reduced response to therapy (Pitzalis et al., 2013, Cañete et al., 2009). ELSs are rich in CD4+IL17A+ cells that play a pivotal role in ELS formation and maintenance (Jones and Jones, 2016). Synovial biopsies were also classified by histological analysis according to inflammatory score (Krenn score; Krenn et al., 2002, Pitzalis et al., 2013) and expression of cell-lineage CD4+ T cell gene modules. As expected, the synovial biopsies with a lymphoid pathotype were also ELS positive and showed the highest inflammatory score and degree of infiltration by CD4+ T cells (Figure 7A).
Figure 7.
SLC5A12 Expression Correlates with RA Disease Activity and Its Blockade Improves Clinical Scores in a Murine Model of (CD4+ T Cells Joint-Enriched) Arthritis
(A) Heatmap showing RNA-sequencing expression of groups of metabolic genes differentially expressed (FDR < 0.05) between synovial biopsies (n = 87) from early rheumatoid arthritis. Synovial biopsies were classified as positive or negative for ectopic lymphoid structures (ELS) by histological analysis. Upper tracks show synovial histology inflammatory score (Krenn score), expression level of cell-lineage CD4+ T cell gene modules, ELS histology grouping and overall histology pathotype (lymphoid, myeloid or fibroid).
(B) Synovium SLC5A12 transcript positively correlates with delta disease activity score (Δ DAS28-CRP) calculated as the difference between DAS28-CRP at baseline and DAS28-CRP at 6 months and FASN transcript; also shown is the positive correlation between the inflammatory score DAS28-CRP with IL17RA transcript (n = 87). Correlation analyses performed using Spearman's correlation coefficients.
(C–F) Arthritis score in mice treated with the indicated antibodies versus controls. Arrows indicate days at which antibodies were injected. A score of 0 indicates no clinical signs of arthritis; a score of 1 for each of the toes, pad, and ankle indicates swelling and redness. Maximum score for each paw is 7 (n = 6 per group; C). Representative images of the paws at day 21 post-immunization showing the effects (i.e., swelling and redness) of treatment with SLC5A12 Ab as compared to the SLC5A12 isotype control antibody (D). Histological score in pads of mice subjected to different treatments, as shown (each dot corresponds to the assessment of an H&E slide acquired from the representative group, n = 19–24 slides/group; E). IHC with haematoxylin counterstain showing immune infiltrate (arrows) in the pad of mice treated with SLC5A12 Ab as compared to isotype control antibody-treated mice (F). Two-tailed Student’s t test (C) and (E). Data represent mean ± SD. ∗p ≤ 0.05; ∗∗p ≤ 0.01; #p ≤ 0.05. See also Figure S7.
In this cohort we analyzed the expression of groups of metabolic genes on synovial biopsies by RNA sequencing (n = 87). In the lymphoid pathotype, we found evidence of expected patterns of Th17 differentiation genes, i.e., reduced FOXO1 and increased IL17A, as well as of ELS genes (Figure 7A). When we analyzed the metabolic genes, we found a downregulation of glycolytic genes concurrent with an upregulation of PPP and TCA cycle genes in the lymphoid pathotype as compared to the other pathotypes (Figure 7A). Furthermore, we found a positive correlation between synovial SLC5A12 expression and disease activity measured as ΔDAS28-CRP (Figure 7B). We also found that DAS28-CRP correlates with IL17RA (Figure 7B) as well as CXCL13, LTB, and FOXO1 (Figure S7A). Furthermore, some key Th17 and metabolic genes described in our study correlate, and in particular, FASN with SLC5A12 (Figure 7B) as well as FASN with IL17RA, FASN with STAT3, and ACACA with STAT3 (Figure S7B).
Overall, our data support a role for lactate/SLC5A12-induced metabolic reprogramming in CD4+ T cells as a distinctive mechanism operational in the RA subset characterized by CD4+ T cell infiltration. In further support, we used a well-established murine model of arthritis where the disease is induced by subcutaneous injection of human glucose 6-phosphate isomerase and the inflammatory infiltrate into the joints is rich in T cells (Schubert et al., 2004, Bruns et al., 2009, Iwanami et al., 2008), hence resembling the human lymphoid RA. SLC5A12 Ab treatment reduced both clinical and histological scores of arthritis as compared to the isotype control and showed a trend effect superior to anti-TNF treatment (Figures 7C–7F)
Discussion
Historically, lactate has been considered a waste product or at best a biomarker in critical care. Yet in the past decades, it was already shown to be a major substrate for oxidative phosphorylation (OXPHOS) in neurons, for gluconeogenesis in the Cori cycle and for the synthesis of glycogen in the skeletal muscle (Pellerin and Magistretti, 1994, Magistretti and Allaman, 2018, Cornell et al., 1973). Recent evidence further supports lactate as a major carbon source for cellular metabolism both in normal and cancerous tissues. Infusion of [U13C]-lactate in fed and fasted mice, revealed extensive labeling of TCA cycle intermediates in all tissues (Hui et al., 2017, Faubert et al., 2017). Strikingly, in lung and pancreatic tumors, the contribution of lactate to the TCA cycle was greater than that of glucose (Faubert et al., 2017).
Here, we showed that lactate accumulation in the inflamed tissue contributes, together with activating and inflammatory stimuli, to the upregulation of the sodium-coupled lactate transporter SLC5A12 on human CD4+ T cells. SLC5A12 is already upregulated by CD4+ T cells at the 12-h activation time point of HC PBMCs and indeed this time point may have the peak of SLC5A12 expression on the cell membrane. Indeed, SLC5A12 expression on the membrane is reduced at the subsequent 24- and 48-h time points. These data suggest that a longer kinetic may not be required. Yet, the 48-h time point in most of our experiments is justified by the fact that we need to allow time for cells to respond functionally to lactate.
SLC5A12-mediated lactate uptake by human CD4+ T cells initiated an anabolic response leading to de novo FAS and involved the translocation of PKM2 in the nucleus. Both mechanisms contributed to the activation of downstream STAT3 transcription factors. Such integration between metabolism and signaling modules led to the plastic reshaping of the CD4+ T effector phenotype within the inflamed tissue.
Our data are in line with Yabu et al. who showed that lactic acid enhances the production of IL23/IL17 by CD4+ T cells, acting as a pro-inflammatory signal (Yabu et al., 2011). However, the mechanisms were not known. In line with lactate being a major fuel for the TCA cycle (Hui et al., 2017, Faubert et al., 2017), we observed an increase in citrate and acetyl-CoA levels upon exposure of activated CD4+ T cells to lactate at concentrations as in the chronic inflamed site. Acetyl-CoA is produced by the breakdown of both glucose (by glycolysis) and fatty acids (by FAO). It then enters the TCA cycle in the mitochondrion and forms citrate by reacting with oxaloacetate. Citrate can be exported to the cytosol, where it may be converted back to acetyl-CoA and then serve as a substrate for FAS through carboxylation in to malonyl-CoA by ACC, the first committed step in the synthesis of fatty acids. Indeed, after lactate treatment, we detected activation of ACC, which in line with our data has been shown to be indispensable for IL17 production (Berod et al., 2014, Endo et al., 2015).
Our in vitro data were further extended to disease pathobiology by RNA sequencing analysis showing a metabolic dysregulation within the synovium of the subset of RA patients, which is characterized by CD4+ T cell infiltration (∼40% of total patients). In particular, these patients displayed a low expression of glycolysis and increased levels of TCA cycle and PPP related genes, in line with data published by the Weyand’s group on the peripheral RA CD4+ T cells (Yang et al., 2013, Yang et al., 2016, Shen et al., 2017).
In contrast to its splice variant PKM1, which is constitutively expressed in most adult tissues, PKM2 is allosterically activated in a feed-forward regulatory loop by an upstream glycolytic metabolite, fructose-1,6-bisphosphate (FBP), and is susceptible to inhibition by growth factor signaling through interaction with phospho-tyrosine containing proteins. These properties of PKM2 allow proliferating cells to divert glucose into anabolic pathways emanating from glycolysis in order to meet the increased biosynthetic demands of proliferation. Association of PKM2 subunits into homo-tetramers is required for optimal enzymatic activity (Eigenbrodt et al., 1992). Both reduced FBP and increased ROS cause decreased PKM2 activity (Anastasiou et al., 2011). Incidentally, lactate uptake causes both these effects. PKM2 inhibition is able to divert glucose into the PPP and thereby generate NADPH (Le Goffe et al., 2002), which in turn provides reducing equivalents for detoxification of ROS by increasing reduced glutathione (GSH) and hence allowing cells to withstand oxidative stress. The small-molecule PKM2 activator DASA-10 (NCGC00181061, a substituted N,N′-diarylsulfonamide) prevented inhibition of PKM2 by H2O2 (Anastasiou et al., 2011). PKM2 has been investigated in atherosclerotic coronary artery disease (CAD) and RA patient-derived macrophages. Here, the increased glucose uptake and glycolytic flux due to inflammation fuel the generation of mitochondrial ROS, which in turn promote the destabilization of the PKM2 tetramer, favoring its dimerization and subsequent nuclear translocation. Nuclear PKM2 functions as a protein kinase that phosphorylates the transcription factor STAT3, thus boosting IL6 and IL1β production (Shirai et al., 2016, Weyand et al., 2017).
In line with these findings, we observed an increase of PKM2 nuclear translocation with concomitant enhanced STAT3 phosphorylation upon treatment of activated CD4+ T cells with lactate. STAT3 is implicated in Th17 differentiation (Yang et al., 2007) and interestingly, the inhibition of PKM2 nuclear translocation with DASA was able to reduce lactate-mediated IL17 production.
Similarly, lactate dehydrogenase A (LDHA) enzymatic activity was recently shown to be necessary to sustain IFNγ production by CD4+ T cells via induced aerobic glycolysis (Peng et al., 2016). Genetic deletion of LDHA in CD4+ T cells significantly reduced glucose consumption and promoted a shift toward oxidative metabolism, as well as the reduction of IFNγ expression. The decrease in IFNγ transcripts was due to reduced histone acetylation. Acetyl-CoA serves as a substrate for lysine acetyltransferases (KATs), which catalyze the transfer of acetyl groups to the epsilon-amino groups of histone lysines and many other proteins. Fluctuations in the concentration of acetyl-CoA, reflecting the metabolic state of the cell, are translated into dynamic protein acetylation that regulates a variety of cell functions, including transcription, replication, DNA repair, cell cycle progression, and aging (Shahbazian and Grunstein, 2007). In the absence of LDHA, the increase in the rate of the TCA cycle necessary to compensate the drop in glycolysis did not allow the export of acetyl-CoA from the mitochondria to the cytosol, thus reducing the pool of acetyl groups available for histone acetylation. These data demonstrate the regulation of INF-γ production in Th1 cells by lactate metabolism through a fine-tuned epigenetic mechanism of histone acetylation coupled to cellular metabolism (Peng et al., 2016).
Furthermore, N-Terminal acetylation of cellular proteins initiates specific protein degradation processes (Hwang et al., 2010). We did not observe increased lysine acetylation of cytosolic proteins upon cell exposure to lactate, suggesting this may not be the mechanism responsible for the reduced expression of glycolytic enzymes we observed in the presence of lactate, which may instead be due to reduced pathway usage consequent to NADH build up in the cell, as we have shown in line with literature. Moreover, citrate is a known inhibitor of glycolysis and its accumulation might have been responsible for the inhibition of glycolysis observed (Newsholme et al., 1977).
In contrast, the immune cell response to lactate in the tumor microenvironment is quite different from that seen in the context of chronic inflammation. Recent studies have reported the ability of tumor-derived lactate to suppress the immune response against the tumor itself, thus creating an environment permissive to the tumor growth (Brand et al., 2016, Angelin et al., 2017, Colegio et al., 2014).
Lactate accumulation in the inflamed tissue also caused CD4+ T cell retention at the site of inflammation, as a consequence of impaired cell motility caused by reduced glycolysis and enhanced FAS. In addition to a reduced glucose flux through glycolysis, several glycolytic enzymes, including HK, were decreased upon cell exposure to lactate in a time-dependent manner. HK catalyzes the first committed step of glucose metabolism. Glucose entering the cell through glucose transporters (GLUTs) is phosphorylated by HK to produce G6P. The two most common isoforms, HK1 and HK2, have overlapping tissue expression, but different subcellular distributions, with HK1 associated mainly with mitochondria and HK2 shuttling between mitochondrial and cytoplasmic compartments. HK2 binds to the voltage-dependent anion channel (VDAC), an outer mitochondrial membrane protein, which interacts with the adenine nucleotide translocase (ANT), forming a contact site between the outer and inner mitochondrial membranes (Fiek et al., 1982, Vyssokikh and Brdiczka, 2003).
Accordingly, we found increased mitochondrial localization of HK2 upon lactate treatment. We also found a reduction of NADP+/NADPH ratio, suggesting a shunt toward PPP and anabolic metabolism. Moreover, mitochondria-associated HK2 has a pro-survival function via antagonizing apoptotic Bcl-2 family proteins and thereby protects cells from apoptosis (Pastorino et al., 2002). This may be a mechanism that allows CD4+ T cell survival in lactate-rich environments.
Our data fit with RA naive CD4+ T cells displaying low basal glycolysis due to a deficiency in 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3). This caused a shunt of glucose-6-phosphate (G6P) toward the pentose phosphate pathway (PPP) with generation of NADPH equivalents and altered activation of ataxia telangiectasia mutated (ATM), a key enzyme in the control of cell cycle. Such alterations resulted in a high capacity of proliferation by RA CD4+ T cells and a switch to pro-inflammatory subsets such as Th1 and Th17 and chronic inflammation (Yang et al., 2013, Yang et al., 2016).
ATPlow pyruvatelow NADPHhigh RA CD4+ T cells also displayed increased FAS and consequent deposition of cytoplasmic lipid droplets. This resulted in the upregulation of the podosome scaffolding protein TKS5. TKS5hi RA CD4+ T cells spontaneously formed actin- and cortactin-rich membrane ruffles, which empowered them to penetrate into non-lymphoid tissue and establish inflammatory infiltrates. All these effects were abolished by FAS inhibition (Shen et al., 2017).
Building on these results, our data suggest that FAS is not only responsible for increased infiltration of the inflamed site by CD4+ T cells, but also for their retention in the site, together with reduced glycolysis, once they have reached it. De novo FAS is required for Th17 differentiation and pharmacologic inhibition of acetyl-CoA carboxylase, a key enzyme in FAS, was able to delay the disease and to reduce the severity of experimental autoimmune encephalomyelitis (EAE), (Berod et al., 2014). Hence, inhibiting FAS and/or promoting glycolysis may support resolution of inflammation.
If RA CD4+ T cells display a deficit in glycolysis, synovial RA fibroblasts are highly glycolytic. Indeed, glucose deprivation or glycolytic inhibitors such as 2-deoxy-D-glucose (2-DG), bromopyruvate (BrPa) and 3-(3-Pyridinyl)-1-(4-pyridinyl)-2-propen-1-one, impaired cytokine secretion, proliferation, and migration by synovial fibroblasts as well as disease severity in a mouse model of arthritis (Garcia-Carbonell et al., 2016). In line with this evidence, the inducible isoform of hexokinase, HK2, which catalyzes the phosphorylation of glucose to G6P—the first committed step in glucose metabolism—was found highly expressed by RA as compared to OA synovial fibroblasts. Interestingly, after HK2 silencing, RA fibroblasts were less invasive while the overexpression of HK2 enhanced the levels of MMP, IL6, and IL8 other than their migratory capabilities (Bustamante et al., 2018). Altered metabolism in RA fibroblasts has also been associated with the hypoxic microenvironment typical of the inflamed sites. Specifically, it was found that hypoxia induced a downregulation of mitochondrial respiration and an increase of glycolysis in RA fibroblast, which in turn promoted synovial invasive mechanisms (Biniecka et al., 2014). Hence, it is tempting to speculate that synovial fibroblasts produce large amounts of lactate in the arthritic synovium, which infiltrating CD4+ T cells have to face and adapt to. Here, we have described how they adapt to such condition.
Interestingly, the response to lactate by CD4+ T cells is mediated by a specific sodium lactate transporter, SLC5A12, which seems to be a major lactate transporter in these cells, unlike other immune cells, i.e., macrophages. We previously described in Haas et al. (2015) that CD4+ T cells express SLC5A12 but not SLC16A1 (MCT1). Here, we show that when we interfere with the carrier function of SLC5A12 in CD4+ T cells we see an impairment of some of their effector functions, namely migration and IL17 production. In comparisons, we do not see any effects of blocking SLC5A12 in macrophages—although they do express it—when we assess their migratory response to lactate (Figure 5B). This suggests that SLC5A12 may not be a major lactate transporter in macrophages and/or they may be able to use others, unlike what we observe in CD4+ T cells.
A selective expression of lactate transporters by immune cells may orchestrate their spatial distribution inside the inflamed tissue as well as affect their functional response (Ene-Obong et al., 2013, Haworth et al., 2008, Olloquequi et al., 2010). Thus, modulating selective T cell subsets via targeting specific lactate transporters may provide novel therapeutic tools to reduce inflammation as well as contributing to a better understanding of the pathogenesis of chronic inflammation. In this respect, our findings show that SLC5A12 does not particularly discriminate among CD4+ T cell subsets, which is what we would expect since we observe ∼40% SLC5A12+ CD4 T cells while only a small fraction of activated CD4+ T cells are normally IL-17 producers. It remains our message that lactate promotes IL-17 responses via metabolic rewiring downstream of SLC5A12-mediated lactate uptake and we have elucidated the mechanisms for this response. It also remains to be assessed whether lactate may modulate Th1 and/or Tfh responses.
Altogether this evidence supports a role for lactate as a major signaling molecule in its own right, able to operate the plastic shift of the immune response within the diseased site, whether in the tumor or the chronic inflammatory environment. In the site of inflammation, such as the inflamed synovium in RA, lactate seems to act as an amplifier of inflammation leading to the entrapment of CD4+ T cells and stimulation of inflammatory cytokines. In further support to the signaling activity of lactate, a receptor of lactate, GPR81, has been identified and involved in the regulation of lipolysis (Liu et al., 2009) and cancer cell survival (Roland et al., 2014). Lactate binding to GPR81 resulted in the upregulation of PD-L1, causing the suppression of the effector function of T cells in co-culture experiments (Feng et al., 2017).
Th17 CD4+ T cells play an important role in RA, multiple sclerosis, and psoriasis (Patel and Kuchroo, 2015). Rising levels of circulating Th17 cells and IL17 were observed in patients with an inadequate response to anti-TNF-α therapy (Chen et al., 2011). Despite several studies revealed the importance of IL17 in the pathogenesis of RA, clinical trials with IL17 blocking agents in RA have not reached striking results so far (Kugyelka et al., 2016), thus other targets are needed. This can be in part due to the heterogeneity of RA in terms of pathogenesis and histological patterns of synovitis (Pitzalis et al., 2013). IL17 and IL17R family members show a high variability in the expression in individual patients (Van Baarsen et al., 2014). Therefore, it is not surprising that the blockade of IL17A or its receptor with monoclonal antibodies did not lead to complete disease remission so far. Currently it is more likely that IL17 targeting agents could be used to complement and/or augment current therapies (Kugyelka et al., 2016). Moreover, IL17-signaling cascade is a complex system. Indeed, it consists of 6 members with 5 known receptors, thus widening the frontier for the development of new blocking and/or modifying agents, which might offer exciting new treatments in autoimmunity. For this reason, targeting the IL17 axis at different levels (i.e., Th17 differentiation and signaling), including blocking SLC5A12, may provide new therapeutic avenues for Th17-mediated inflammatory disorders.
Limitations of Study
Immunization with human recombinant G6PI or with the immune-dominant peptide G6PI325-339 was shown to induce an inflammatory polyarthritis which can be modulated by therapeutic interventions aimed at targeting T cell function (i.e., by inducing a switch in the balance between Th1/Th17 cells and Tregs; Schubert et al., 2004, Yoshida et al., 2016, Hirota et al., 2017). Based on these observations and our findings of a dominant effect of SLC5A12 on CD4+ T cells, we chose this experimental model of arthritis to test the effect of blocking SLC5A12. Nevertheless, a caveat to our findings is that the beneficial effects of targeting SLC5A12 may be due at least in part to its effects of macrophages and/or B cells. This could be further tested in other models of arthritis, such as collagen-induced arthritis. Anyhow, a previous demonstration of the functional relevance of SLC5A12 blockade was provided by us, using a zymosan model of peritonitis (Haas et al., 2015).
Furthermore, intra-articular injections are required in our model currently to achieve high enough concentration of the antibody within the joint as the effect of anti-SLC5A12 is elicited at the site of inflammation where high levels of lactate are present. This procedure would not lead to a biological effect systemically because a single dose of the antibody at the concentration used would be too low to achieve sufficient blood concentrations to enter efficiently other joints and/or organs. The same can be said for the control antibodies. Thus, we would not expect any systemic effect clinically. Additionally, we used the contralateral joint as control as this is routinely used in experimental arthritis, offering the advantage of minimizing the inherent variability in the severity of arthritis observed among different animals and allows direct comparison of data (Chen et al., 2015, Min et al., 2013, Mor-Vaknin et al., 2017). Regardless of these caveats to our current study, to advance in the pre-clinical space, our next steps will be to generate and characterize recombinant murine and humanized mAbs targeting SLC5A12, which we will then administer systemically
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Acetylated-Lysine Antibody | Cell Signaling Technology | Cat#9441; RRID:AB_331805 |
| Acetyl-CoA Carboxylase Antibody | Cell Signaling Technology | Cat#3662; RRID:AB_2219400 |
| Aldolase A Antibody | Cell Signaling Technology | Cat#3188; RRID:AB_2226674 |
| AMPKα (D63G4) Rabbit mAb | Cell Signaling Technology | Cat#5832; RRID:AB_10624867 |
| CD14 mouse antibody (clone 63D3) | BioLegend | Cat#367123; RRID:AB_2716228 |
| CD14 mouse antibody (clone M5E2) | BioLegend | Cat301804; RRID:AB_314186 |
| CD19 mouse antibody (clone HIB19) | BioLegend | Cat#302241; RRID:AB_2561381 |
| CD19 mouse antibody (clone SJ25C1) | BioLegend | N/A |
| CD20 (CD20cy) mouse antibody (clone L26) | Dako | GA60461-2 |
| CD25 mouse antibody (clone BC96) | BioLegend | Cat#302607; RRID:AB_314277 |
| CD3 monoclonal antibody (OKT3) | ThermoFisher Scientific | MA1-10175; RRID:AB_11157849 |
| CD4 mouse antibody (clone 4B12) | Dako | M731001-2 |
| CD4 mouse antibody (clone RPA-T4) | BioLegend | Cat#300558; RRID:AB_2564393 |
| CD4 rat antibody (clone A161A1) | BioLegend | Cat#357411; RRID:AB_2565663 |
| CD68 mouse antibody (clone PG-M1) | Dako | M087601-2 |
| CD8 mouse antibody (clone C8/144B) | Dako | M710301-2 |
| CD8 mouse antibody (clone HIT8a) | BioLegend | Cat#300930; RRID:AB_2629639 |
| CD8 mouse antibody (clone SK1) | BioLegend | Cat#344713; RRID:AB_2044005 |
| CXCR5 mouse antibody (clone J252D4) | BioLegend | Cat#356929; RRID:AB_2566226 |
| Dynabeads Human T-Activator CD3/CD28 | ThermoFisher Scientific | Cat#11161D |
| Enolase-1 Antibody | Cell Signaling Technology | Cat#3810; RRID:AB_2246524 |
| Foxp3 mouse antibody (clone 206D) | BioLegend | Cat#320108; RRID:AB_2140838 |
| GCK Antibody | Cell Signaling Technology | Cat#3782; RRID:AB_2140838 |
| Hexokinase I (C35C4) Rabbit mAb | Cell Signaling Technology | Cat#2024; RRID:AB_2116996 |
| Hexokinase II (C64G5) Rabbit mAb | Cell Signaling Technology | Cat#2867; RRID:AB_2232946 |
| Histone H3 Antibody | Cell Signaling Technology | Cat#9715; RRID:AB_331563 |
| ICOS mouse antibody (clone C398.4A) | BioLegend | Cat#313519 |
| IFNγ mouse antibody (clone 4S.B3) | BioLegend | Cat#502541 |
| IL10 mouse antibody (clone JES3-19F1) | BioLegend | Cat#506804 |
| IL17A mouse antibody (clone BL168) | BioLegend | Cat#512328 |
| IL21 mouse antibody (clone 3A3-N2) | BioLegend | Cat#513006 |
| MitoTracker™ Deep Red FM | ThermoFisher Scientific | M22426 |
| PD1 mouse antibody (clone EH12.2H7) | BioLegend | Cat#329913 |
| Phospho-Acetyl-CoA Carboxylase (Ser79) (D7D11) Rabbit mAb | Cell Signaling Technology | Cat#11818; RRID:AB_2687505 |
| Phospho-AMPKα (Thr172) (40H9) Rabbit mAb | Cell Signaling Technology | Cat#2535 |
| Phospho-Stat1 (Tyr701) (58D6) Rabbit mAb | Cell Signaling Technology | Cat#9167; RRID:AB_10859888 |
| Phospho-Stat3 (Tyr705) (D3A7) XP® Rabbit mAb | Cell Signaling Technology | Cat#9145; RRID:AB_2491009 |
| PKM1/2 (C103A3) Rabbit mAb | Cell Signaling Technology | Cat#3190; RRID:AB_2163695 |
| Rabbit polyclonal anti-PFK | Novus Biologicals | NEP1-37473 |
| RORγt mouse antibody (clone Q21-559) | BD Biosciences | Cat#563424 |
| SLC5A12 Antibody | Atlas Antibodies | HPA060904 |
| SLC5A12 Antibody | Abcam | Ab107749; RRID:AB_10864245 |
| SLC5A12 Antibody | Novus Biologicals | NBP2-49322 |
| SLC5A12 monoclonal (clone 10E11) | Aldevron GmbH | N/A |
| SLC5A12 monoclonal (clone 3C7) | Aldevron GmbH | N/A |
| SLC5A12 monoclonal (clone 4G2) | Aldevron GmbH | N/A |
| SLC5A12 monoclonal (clone 6E1) | Aldevron GmbH | N/A |
| SLC5A12 monoclonal (clone 7C1) | Aldevron GmbH | N/A |
| SLC5A12 monoclonal (clone 9G4) | Aldevron GmbH | N/A |
| SLC5A12 monoclonal (clone 9G7) | Aldevron GmbH | N/A |
| Stat1 (D1K9Y) Rabbit mAb | Cell Signaling Technology | Cat#14994; RRID:AB_2737027 |
| Stat3 (D3Z2G) Rabbit mAb | Cell Signaling Technology | Cat#12640; RRID:AB_2629499 |
| Tbet mouse antibody (clone 4B10) | BioLegend | Cat#644807 |
| VDAC (D73D12) Rabbit mAb | Cell Signaling Technology | Cat#4661; RRID:AB_10557420 |
| β-Actin (13E5) Rabbit mAb | Cell Signaling Technology | Cat#4970 |
| Biological Samples | ||
| Blood obtained from healthy anonymous adult donors | N/A | N/A |
| Mouse paw tissue | N/A | N/A |
| Synovial fluids obtained from rheumatoid arthritis patients | N/A | N/A |
| Synovial tissues obtained from rheumatoid arthritis patients | N/A | N/A |
| Tonsils from subjects undergoing tonsillectomy | N/A | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AICAR | Merck | Cat#A9978 |
| C75 | Santa Cruz Biotechnology | SC-202511 |
| DASA | Merck | Cat#550602 |
| DHEA | Cayman | Cat#15728 |
| Etomoxir | Merck | Cat#236020 |
| FCCP | Merck | Cat#C2920 |
| Histopaque 1077 | Sigma-Aldrich | Cat#10771 |
| Leukocyte Activation Cocktail | BD Biosciences | Cat#550583 |
| Lymphoprep | Stemcell Technologies | Cat#07801 |
| Oligomycin | Merck | Cat#495455 |
| Palmitate | Sigma-Aldrich | P9767 |
| Sodium L-lactate | Sigma-Aldrich | Cat#71718 |
| TOFA | Santa Cruz Biotechnology | SC-200653 |
| Critical Commercial Assays | ||
| Citrate Colorimetric/Fluorimetric Assay Kit | BioVision | Cat#K655-100 |
| EasySep™ Human CD4+ T Cell Isolation Kit | STEMCELL Technologies | Cat#17952 |
| EasySep™ Mouse CD4+ T Cell Isolation Kit | STEMCELL Technologies | Cat#19852 |
| Free Fatty Acid Assay Kit – Quantification | abcam | Cat#ab65341 |
| H2DCFDA | ThermoFisher Scientific | Cat#D399 |
| Human IFN gamma ELISA Ready-SET-Go!™ Kit | fisherscientific | Cat#15541107 |
| Human IL-17A (homodimer) ELISA Ready-SET-Go!™ Kit | fisherscientific | Cat#15501077 |
| Mitochondria Isolation Kit for Cultured Cells | ThermoFisher Scientific | Cat#89874 |
| NAD+/NADH Quantification Colorimetric Kit | BioVision | Cat#K337-100 |
| NADP/NADPH Assay Kit | Abcam | Cat#ab65349 |
| Nuclear Extraction Kit | abcam | Cat#ab113474 |
| PicoProbeTM Acetyl CoA Fluorimetric Assay Kit | BioVision | Cat#K317-100 |
| Zombie NIR™ Fixable Viability Kit | BioLegend | Cat#423105 |
| Deposited Data | ||
| Raw data files for RNA sequencing | ArrayExpress | https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6141 |
| Experimental Models: Cell Lines | ||
| Human: MCs from tonsils (primary) | N/A | N/A |
| Human: PBMCs from healthy controls (primary) | N/A | N/A |
| Human: PBMCs from rheumatoid arthritis patients (primary) | N/A | N/A |
| Human: SFMCs from rheumatoid arthritis patients (primary) | N/A | N/A |
| Mouse: CD4+ T cells from Slc5a12 WT or KO mice (primary) | N/A | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: SLC5A12 KO (Allele: Slc5a12em1(IMPC)Wtsi) | Sanger Institute | N/A |
| Oligonucleotides | ||
| BCL6 forward primer: 5’- CGAATCCACACAGGAGAGAAA-3’ |
Invitrogen | 347017 U4335 (B08) |
| BCL6 reverse primer: 5’- ACGCGGTATTGCACCTTG-3’ |
Invitrogen | 347017 U4335 (B09) |
| CXCR5 forward primer: 5’- GCTAACGCTGGAAATGGA-3’ |
Invitrogen | 347017 U4335 (C04) |
| CXCR5 reverse primer: 5’- GCAGGGCAGAGATGATTT-3’ |
Invitrogen | 347017 U4335 (C05) |
| Foxo1 forward primer: 5’- AGGGTTAGTGAGCAGGTTACAC-3’ |
Invitrogen | 347017 U4335 (C02) |
| Foxo1 reverse primer: 5’- TGCTGCCAAGTCTGACGAAA-3’ |
Invitrogen | 347017 U4335 (C03) |
| Foxp3 forward primer: 5’- CTGACCAAGGCTTCATCTGTG-3’ |
Invitrogen | 347017 U4335 (A06) |
| Foxp3 reverse primer: 5’- ACTCTGGGAATGTGCTGTTTC-3’ |
Invitrogen | 347017 U4335 (A07) |
| GAPDH forward primer: 5’- TCCTCTGACTTCAACAGCGA-3’ |
Invitrogen | 347017 U4335 (B02) |
| GAPDH reverse primer: 5’- GGGTCTTACTCCTTGGAGGC-3’ |
Invitrogen | 347017 U4335 (B03) |
| IFNγ forward primer: 5’- GGCATTTTGAAGAATTGGAAAG-3’ |
Invitrogen | 347017 U4335 (B04) |
| IFNγ reverse primer: 5’- TTTGGATGCTCTGGTCATCTT-3’ |
Invitrogen | 347017 U4335 (B05) |
| IL10 forward primer: 5’- ACCTGCCTAACATGCTTCGAG-3’ |
Invitrogen | 347017 U4335 (B10) |
| IL10 reverse primer: 5’- CCAGCTGATCCTTCATTTGAAAG-3’ |
Invitrogen | 347017 U4335 (B11) |
| IL17A forward primer: 5’- TGTCCACCATGTGGCCTAAGAG-3’ |
Invitrogen | 347017 U4335 (A08) |
| IL17A reverse primer: 5’- GTCCGAAATGAGGCTGTCTTTGA-3’ |
Invitrogen | 347017 U4335 (A09) |
| IL22 forward primer: 5’- TCCAGAGGAATGTGCAAAAG-3’ |
Invitrogen | 347017 U4335 (D07) |
| IL22 reverse primer: 5’- ACAGCAAATCCAGTTCTCCAA-3’ |
Invitrogen | 347017 U4335 (D08) |
| IL6 forward primer: 5’- AGTGAGGAACAAGCCAGAGC-3’ |
Invitrogen | 347017 U4335 (E07) |
| IL6 reverse primer: 5’- GTCAGGGGTGGTTATTGCAT-3’ |
Invitrogen | 347017 U4335 (E08) |
| PD1 forward primer: 5’- ACCTGGGTGTTGGGAGGGCA-3’ |
Invitrogen | 347017 U4335 (B12) |
| PD1 reverse primer: 5’- GGAGTGGATAGGCCACGGCG-3’ |
Invitrogen | 347017 U4335 (C01) |
| RORγt forward primer: 5’- CCTGGGCTCCTCGCCTGACC-3’ |
Invitrogen | 347017 U4335 (A04) |
| RORγt reverse primer: 5’- TCTCTCTGCCCTCAGCCTTGCC-3’ |
Invitrogen | 347017 U4335 (A05) |
| SLC5A12 forward primer: 5’- GTGTGCTGTCTTCTCTGGCT-3’ |
Eurofins MWG Operon | H680 31-3128-11/12 |
| SLC5A12 reverse primer: 5’- GCCACAAAAAGTCCTGGCAG-3’ |
Eurofins MWG Operon | H680 31-3128-12/12 |
| TGFβ forward primer: 5’- AGCGACTCGCCAGAGTGGTTA-3’ |
Invitrogen | 347017 U4335 (A10) |
| TGFβ reverse primer: 5’- GCAGTGTGTTATCCCTGCTGTCA-3’ |
Invitrogen | 347017 U4335 (A11) |
| Β-Actin forward primer: 5’- AGTTGCGTTACACCCTTTCTTG-3’ |
Invitrogen | 347017 U4335 (A12) |
| Β-Actin reverse primer: 5’- TCACCTTCACCGTTCCAGTTT-3’ |
Invitrogen | 347017 U4335 (B01) |
| Software and Algorithms | ||
| GraphPad Prism 7 | GraphPad Software, Inc | http://www.graphpad.com/scientific-software/prism/ |
| FlowJo 7.6.5 | Tree Star | www.flowjo.com |
| ImageJ | ImageJ | https://imagej.nih.gov/ij/ |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Claudio Mauro (c.mauro@bham.ac.uk). Mouse lines used in this study are available at INFRAFRONTIER/EMMA (www.infrafrontier.eu), (Allele: Slc5a12em1(IMPC)Wtsi). There are restrictions to the availability of SLC5A12 monoclonal antibodies due to intellectual property and ongoing patenting.
Experimental Model and Subject Details
Patient Samples
Blood, synovial fluids and synovial tissues were obtained from the same cohort of rheumatoid arthritis (RA) patients diagnosed according to the revised American College of Rheumatology (ACR) criteria (Aletaha et al., 2010). Demographic and clinical characteristics of the study cohort are presented in Table S1.
Healthy individuals, age and sex matched with RA patients (Table S1), were recruited through NHS Blood and Transplant service. Individuals with cancer, infections or other inflammatory comorbidities were excluded. Written informed consent was obtained by all participants according to ethical approval from National Research Ethics Service Committee London (LREC07/Q0605/129).
For RNA sequencing analysis, mRNA was extracted from synovial tissue samples obtained by ultrasound-guided biopsy from patients with early active RA (< 12 months' time) who were naïve-to-treatment with disease-modifying anti-rheumatic drugs (DMARDs) (n = 87). Patients were enrolled in the Pathobiology of Early Arthritis Cohort (PEAC, details at http://www.peac-mrc.mds.qmul.ac.uk/index.php) at the Centre for Experimental Medicine and Rheumatology at Queen Mary University of London (REC 05/Q0703/198, London, UK), as previously described (Kelly et al., 2015, Humby et al., 2019, Lewis et al., 2019). All patients belonging to this cohort underwent baseline ultrasound-guided biopsy on the most inflamed accessible joint. Afterward, patients started treatment with conventional DMARDs (methotrexate, leflunomide and/or sulphasalazine and/or hydroxychloroquine and/or corticosteroids). The response to treatment was evaluated at six months according to DAS28-CRP (Fransen, and van Riel, 2009).
Cell Isolation and Culture
Peripheral blood mononuclear cells (PBMCs) from healthy controls (HC) and paired PBMCs and synovial fluid mononuclear cells (SFMCs) were obtained from RA patients. PBMCs and SFMCs were isolated by density gradient centrifugation [lymphoprep (StemCell Technologies) and histopaque 1077 (Sigma-Aldrich), respectively]. Cells (2 × 106/mL) were cultured in 48-well plates (37°C, 5% CO2) in medium (RPMI 1640 - ThermoFisher) supplemented with 10% FBS (ThermoFisher) or 10% autologous blood serum or 10% RA synovial fluid and activated with 0.2μg/ml anti-CD3 monoclonal antibody (CD3 mAb, EBioscience) or left non-activated, according to well-established protocols (Pucino et al., 2016, Landegren et al., 1984, Van Wauwe et al., 1980).
For the experiments with human tonsils, tissues were mashed through a cell strainer and mononuclear cells (MCs) were isolated. Cells were cultured (2 × 106/ml, 37°C, 5% CO2) in RPMI 1640 plus 10% FBS and activated with 0.2μg/ml anti-CD3 mAb (Thermo Fisher Scientific) or left non-activated.
All samples from HC (PBMCs and tonsils) were gender- and age- matched with samples (SFMCs, PBMCs) from RA patients (for details see Table S1).
CD4+ T cells were purified from human PBMCs or MCs, or from spleen and lymph nodes of Slc5a12 WT or KO female mice by negative selection using a magnetic cell separation (EasySep, Stem Cell Technology), cultured in RPMI 1640 plus 10% FBS (37°C, 5% CO2) and then stimulated for 3 days in the presence of anti-CD3 and anti-CD28 mAbs coated Dynabeads (0.1 beads per cell; Thermo Fisher Scientific).
Mouse Models
We thank the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP) and its funders as well as INFRAFRONTIER/EMMA (www.infrafrontier.eu), for accepting our gene nomination for Slc5a12, and for generating and providing the Crispr/Cas9 mutant mouse line (Allele: Slc5a12em1(IMPC)Wtsi) in the C57BL/6N background (White et al., 2013, Skarnes et al., 2011, Bradley et al., 2012, Pettitt et al., 2009). All procedures were consented by the UK Home Office and animals were sacrificed following an accepted Schedule 1 method.
DBA/1 female mice purchased from Charles River were immunized s.c. with 20μg human hG6PI synthetic peptide (hG6PI325-339; ThermoFisher Scientific) in CFA (Sigma-Aldrich). The indicated amount of peptide was mixed with CFA in a 1:1 ratio (v/v) and emulsified by sonication. For induction of arthritis, 100μl of the emulsion was injected subcutaneously at the base of the tail. At day 7 – a time point at the onset of the disease – and 11 post-induction, mice were left untreated or treated with intra-articular injection of 20μl of 0.1mg/ml antibody into the rear paws; Infliximab (Remicade, Janssen Biologics), anti-TNF (TN3-19.12, BD Bioscience), anti-SLC5A12 (Atlas Antibodies), Iso-TNF (BD biosciences) and Iso-SLC5A12 (Atlas Antibodies). Specific antibody and the corresponding isotype control were injected into the right or left back paw of the same mouse. The development of disease was monitored daily by visually assessing the arthritis score. A score of 0 indicates no clinical signs of arthritis; a score of 1 for each of the fingers, pad and ankle indicates swelling and redness. Maximum score for each paw is 7. A trained observer who was blinded to the immunization status of the mice performed the scoring.
All mice were group-housed and maintained under SPF health/immune status in individually ventilated cages with standard enrichment. Mice were housed in a temperature (24°C) and humidity-controlled room on a 12 h light/dark cycle (lights on 7:00) with ad libitum access to water and food.
Method Details
Flow Cytometry
In order to assess SLC5A12 expression on different immune cell types, we stained PBMCs or tonsil MCs or RA SFMCs with Live/Dead (ZOMBIE/NIR, fixable viability dye, 1:1000, BioLegend) for 15 minutes at room temperature protected from light to allow detection and exclusion of dead cells from the analysis. Without washing, cells were then stained with BV711–labelled anti–CD4 (clone RPA-T4, 1:100), PE/Dazzle–labelled anti–CD8 (clone HIT8a, 3:1000), FITC–labelled anti–CD14 (clone 63D3, 1:100), PeCy7–labelled anti–CD19 (clone HIB19, 1:100). Rabbit anti-SLC5A12 unconjugated primary antibody (4:1000, HPA060904 – Atlas Antibodies) was added after fixing and permeabilizing cells (fixation-permeabilization buffer; eBioscience) for 30 minutes followed by Alexa Fluor-555 goat anti-rabbit (1:1000, Invitrogen) secondary antibody. Polyclonal rabbit IgG (DAKO) was used as isotype control.
To assess SLC5A12 expression on T cell subsets identified on the basis of transcription factors expression or cytokines production, healthy donors PBMCs were activated for 48h and incubated with Leucocyte Activating cocktails (BD) for the last 3 hours. Cells were first stained for Live/Dead Zombie for 15 minutes at room temperature protected from light, washed and incubated for 10 minutes with human Fc TruStain FcX (BioLegend, 5:100) to block Fc receptors (CD16, CD32, CD64) and avoid non-specific binding. Surface antigens were stained with BV510-labelled anti-CD14 (clone 63D3, 1:100), BV510-labelled anti-CD19 (clone HIB19, 1:100), PE/Dazzle 594-labelled anti-CD4 (clone A161A1, 1:100), APC Cy7-labelled anti-CD8 (clone SK1, 1:100), BV605-labelled anti-CXCR5 (J252D4, 5:100), PECy7-labelled anti-ICOS (clone C398.4A, 1:100), PerCpCy5.5-labelled anti-PD1 (clone EH12.2H7, 1:100), PECy5-labelled anti-CD25 (clone BC96, 1:100) (BioLegend). Cells were then fixed, permeabilized (fixation-permeabilization buffer; eBioscience) and stained first with Pacific Blue-labelled anti-Tbet (clone 4B10, 2:100), PE-labelled anti-Foxp3 (clone 206D, 2.5:100), BV711-labelled anti-IL17A (clone BL168, 5:100), BV785-labelled anti-IFNγ (clone 4S.B3, 3:100), Alexa647-labelled anti-IL21 (clone 3A3-N2, 3:100) (BioLegend) and BV650-labelled anti-RORγt (clone Q21-559, 2:100) (BD Biosciences). Rabbit anti-SLC5A12 unconjugated primary antibody (0.4:100, HPA060904 – Atlas Antibodies) was then added for 30 minutes followed by Alexa Fluor-555 goat anti-rabbit (1:1000, Invitrogen) secondary antibody.
Cells were acquired using a LSR Fortessa II (BD Biosciences) flow cytometer and analyzed with FlowJo version 7.6.5 software. All monoclonal antibodies were from Biolegend.
Immunofluorescence, Immunohistochemistry, and Confocal Microscopy
For SLC5A12 single and double (with CD4, CD8, CD20 or CD68) immunofluorescence, after antigen retrieval (S2367, Dako; 45 minutes) and block of non-specific binding (1 hour hour), paraffin-embedded tonsil tissue sections were incubated for 1 hour hour with anti-SLC5A12 Ab (1:50, Novus Biologicals) and then overnight at 4°C with anti-CD4, anti-CD8, anti-CD20 or anti-CD68 (1:50, Dako). The following day, slides were washed in PBS and incubated with fluorochrome-conjugated secondary antibodies (1:300, Invitrogen, Eugene, Oregon, USA). The slides were then washed in PBS for up to 5 minutes, mounted in fluorescence mounting medium (DakoCytomation) containing 1 μg/ml DAPI, and examined by Olympus IX81 fluorescence microscope. The list of primary and secondary antibodies is shown in Key Resources Table.
For the intracellular detection of HK2, human CD4+ T cells cultured on glass coverslips were incubated for 5 minutes with 300 nM mitotracker deep red FM (ThermoFisher Scientific) at 37°C in 5% CO2. After the incubation period, cells were washed twice and fixed/permeabilized in permeabilization/fixation buffer (Bioscience) overnight at 4°C. After washing with PBS, cells were incubated with the primary antibodies against anti-HK2 (dilution 1:200, Cell Signaling Technology) for 1 hour at room temperature followed by 30 minutes incubation with 1:200 secondary Alexa Fluor 555 conjugated goat anti-rabbit (Invitrogen). Alexa Fluor 488 phalloidin (ThermoFisher) for the actin staining was also added at this stage. One million cells resuspended in 100 μl PBS were counterstained with DAPI to detect nuclei, spun in the cytospin (250 rpm for 5 minutes) to allow the attachment to the coverslips and then mounted for microscopy. All images were acquired using a confocal microscope LSM880 (Zeiss).
FFPE tissue blocks from murine paws were sectioned at a thickness of 3 μm using a Leica RM1235 microtome. Sections were mounted on to Superfrost plus (+) slides and left to dry in a slide rack at room temperature for 30 minutes. Slides were heated (60°C) for a minimum of 30 minutes prior to staining. Tissue sections were stained with haematoxylin and eosin (H&E), mounted with DePex and left to dry overnight. Slides were imaged using an Olympus BX61 microscope for initial grading. 0 = normal; 1 = minimal infiltration of inflammatory cells in synovium and periarticular tissue of affected joints; 2 = mild infiltration. 3 = moderate infiltration with moderate oedema. If referring to paws, restricted to affected joints; 4 = Marked infiltration affecting most area with marked oedema.
Chemokinesis Assays and Tissue Organ Culture
For the analysis of egressed MCs, equal size tissue sections from juxtaposing areas of tonsil or synovium biopsies were seeded in 48-well-plates in RPMI 1640 supplemented with 10% FBS and treated as indicated in figure. After 4 hours of tissue culture, the supernatants containing egressed cells were collected, followed by staining MCs for CD4, CD8, CD19 or CD14 and counting by FACS the percentage of events for each cell type. Data were then expressed as percentage fold change as compared to the respective controls.
Metabolic Profiling
Real-time measurements of extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were performed with a Seahorse XF96 Extracellular Flux Analyser (Agilent). Briefly, CD4+ T cells were grown in RPMI medium supplemented with 10% FBS. One hour before the experiment, 3 x 105 CD4+ T cells were seeded in a 96-well microplate in XF Assay medium (Dulbecco’s Modified Eagle’s Medium, DMEM) in the presence of 10 mM glucose. Sodium lactate or PBS were injected during measurement. Fatty acid oxidation was analyzed by measuring OCR in the presence of palmitate. Briefly, 2.5 x 105 CD4+ T cells were seeded in a 96-well microplate in XF Assay Modified DMEM containing 2.5mM glucose. Fifteen minutes before the assay, control cells were treated with 40μM etomoxir to block CPT-1a. Just before starting the measurements, cells were treated with 167μM BSA-palmitate or BSA alone (Sigma-Aldrich). During the assay 1μM oligomycin and 1μM FCCP were injected. All data were analysed using XF software.
Intracellular metabolites content was determined by using dedicated quantification kits [NAD+/NADH Quantification Colorimetric Kit, Citrate Colorimetric/Fluorimetric Assay Kit (BioVision), Acetyl CoA Fluorimetric Assay Kit (BioVision) and NADP+/NADPH Colorimetric Assay Kit (Abcam)], according to the manufacturer’s instructions.
Reactive oxygen species (ROS) were measured using the fluorescent probe carboxy-H2DCFDA (ThermoFisher Scientific). FFAs were measured with Free Fatty Acid Colorimetric/Fluorometric Assay Kit (Abcam).
Metabolomics and Stable Isotope Tracing
Following isolation, CD4+ T cells were initially activated in media with anti-CD3 and anti-CD28 mAbs for 24 hours followed by further 48 hours culture with lactate alone or in the presence of SLC5A12 Ab in medium containing low glucose (5mM) and 5% FBS. Spent medium was collected and processed for metabolite extraction as described (Mackay et al., 2015). Briefly, medium from each condition was diluted 50 fold with cold extraction solvent consisting of 50% methanol, 30% acetonitrile and 20% water. Polar metabolites were extracted by vortexing the tubes for 10 minutes followed by centrifugation at 16,000 × g for 10 minutes at 4°C. The supernatants were transferred to glass vials and analysed by LC-MS as described (Mackay et al., 2015). Glucose and glutamine concentrations in the spent medium were quantified using external calibration curves generated by spiking in different concentrations of 13C-glucose and 13C-glutamine in the medium and extraction into extraction solvent. Peak areas from the samples were extrapolated to the standard curve peak areas and absolute concentrations of glucose and glutamine were obtained. Uptake rates of metabolites (glucose and glutamine) were calculated as difference in concentrations normalized to the area under the growth curve of cells.
For lactate tracing into polar metabolites, CD4+ T cells were activated for 24 hours in low glucose medium in a 6-well cell culture plate. The medium was then replaced with fresh medium containing 10mM [U13C]-lactate (Sigma-Aldrich) with or without SLC5A12 Ab and cells were cultured for additional 48 hours. Medium was removed by centrifugation and metabolites were extracted from the cell pellets with cold extraction solvent (50% methanol, 30% acetonitrile and 20% water). The LC-MS parameters for data acquisition were kept the same as described (Mackay et al., 2015).
For lactate tracing into palmitate, CD4+ T cells were activated for 24 hours in low glucose medium (5mM) in a 6-well cell culture plate. The medium was then replaced with fresh medium containing 10mM [U13C]-lactate (Sigma-Aldrich) with or without SLC5A12 Ab for 0, 24, 48, 72 and 96 hours. At the end of each time point, medium was aspirated and cell pellets were treated with 750μl of 1:1 cold PBS: methanol and total lipid fraction was extracted in 500μl of chloroform. This extract was dried under inert nitrogen, reconstituted in 90 μl chloroform and total fatty acids were derivatized using the transesterification reagent MethPrep II (Thermofisher Scientific, UK) before analysis by GC-MS as described (Tumanov et al., 2015).
For cytokine detection, in the final 4 hours of culture, cells were treated with 50ng/ml PMA and 500ng/ml ionomycin and 1:1.000 brefeldin A (Sigma-Aldrich), followed by surface staining of CD4 (RPA-T4, Biolegend).
To assess the impact of SLC5A12 blockade on cytokine production and CD4+ T cell subsets, we stained activated tonsil MCs, treated with or without SLC5A12 Ab (48 hours), with BV711–labelled anti–CD4 (clone RPA-T4, 1:100), BV421–labelled anti–PD1 (clone EH12.2H7, 1:100) and PerCP/Cyanine5.5 anti-human CXCR5 (clone J252D4, 5:100). Thereafter, we washed, fixed, and permeabilized cells (fixation-permeabilization buffer; eBioscience) and stained with BV450-labelled anti-IL17A (clone BL168, 5:100), AF488-labelled anti-FOXP3 (clone 206D, 2.5:100), PE-labelled anti-IL10 (clone JES3-19F1, 5:100), FITC-labelled IFNγγ (clone 4S.B3, 3:100). Intracellular staining was assessed by flow cytometry using a LSR Fortessa II (BD Biosciences) and FlowJo version 7.6.5 software. All monoclonal antibodies were from Biolegend.
Chemokinesis assays were performed in 5μm trans-well inlays (Corning). One hour before the assay, CD4+ T cells purified from human PBMCs were incubated with sodium lactate (10mM) and pre-treated (1 hour hour) with SLC5A12 polyclonal Ab (2.5 ug/ml, Atlas antibodies) or monoclonal mAbs (dilution 1: 50; SLC5A12 mAb clones: 3C7, 4G2, 6E1, 7C1, 9G4, 9G7 and 10E11) purified from sera of rat immunized with SLC5A12 recombinant peptide or left untreated. In some experiments sodium lactate treated cells were pre-treated (2 hours) with metabolic drugs: C75 (10uM), TOFA (20uM), DHEA (20uM), DASA (20uM), AICAR (1mM) or left untreated. For experiments with CD4+ T cells from spleen and lymph nodes of Slc5a12 WT or KO female mice, cells were incubated with sodium lactate (10mM) or left untreated. In all the assays 3 x 105 lymphocytes suspended in migration medium (RPMI 2% FCS) were seeded in the upper trans-well chamber; CXCL10 (300 ng/ml) or CCL20 (500 ng/ml) chemokines were added to the lower chamber. Migrated T cells were counted in cell counting chambers 4 hours after seeding, and then the percentage of migrated cells was calculated.
Molecular Signaling and Western Blot Analyses
For western blot analyses, proteins were extracted by lysing anti-CD3/CD28 mAbs activated CD4+ T cells (2-4 x 106 per condition) in RIPA lysis buffer (65mM Tris-HCl, pH 7.5, 150mM NaCl, 1mM EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail tablets (#04693132001, Roche). Equivalent amounts of protein (30μg), as determined by standard Bradford assay (Bio-Rad), were loaded, separated by SDS-PAGE and transferred to polyvinyildene difluoride membranes using a transfer apparatus according to the manufacturer’s protocols (Bio-Rad). After incubation with 5% non-fat milk in TBST (10mM Tris pH 8.0, 150mM NaCl, 0.5% Tween 20) for 60 minutes, membranes were washed twice with TBST and incubated overnight at 4°C with a 1:1000 dilution of primary antibodies against pSTAT3, STAT3, pSTAT1, STAT1, PKM1/2, pACC, ACC, pAMPK, AMPK, HK1, HK2, enolase1α, GCK, Aldolase, Acetyl lysine, Histone H3, VDAC, β-Actin (Cell Signalling Technology), PFK (Novus Biologicals), SLC5A12 (Abcam) Membranes were then incubated for 1 hour hour at room temperature with horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies (1:2000). Blots were washed twice with TBST and developed with the ECL system (Amersham Biosciences) according to the manufacturer’s protocols. Density of bands was calculated with ImageJ software.
Nuclear, mitochondrial and cytosolic fractions were extracted by using Nuclear Extraction Kit (Abcam) and Mitochondria Isolation Kit for Cultured Cells (Thermo Scientific) according to the manufacturer's instructions.
RNA Isolation, Reverse Transcription, and qRT-PCR
Total RNA was isolated from 1 x 106 CD4+ T cells or 10mg RA synovial tissue using RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions and assessed for quality and quantity using absorption ratios of 260/280 nm and 260/230 nm. Cells were lysed in RLT lysis buffer and nucleic acids were precipitated with 70% ethanol and RNA bound to spin columns. Following several washing steps, RNA was eluted in dH2O. The isolated RNA was reverse transcribed to complementary DNA (cDNA) using commercially available kits according to the manufacturer’s instructions (Applied Biosystems). Briefly, 1 μg of total RNA was mixed with buffer, deoxy-nucleotides (dNTPs) and reverse transcriptase and incubated for 2 hours at 30°C, followed by a 5 minutes heat inactivation step at 85°C. cDNA was diluted to 10 ng/μl and stored -80°C for subsequent use.
Quantitative gene expression analysis was performed using SYBR Green Supermix (Biorad) in CFX connect light cycler (Biorad), according to the manufacturer’s instructions. Gene relative expression was calculated using the ΔΔct method (Livak and Schmittgen, 2001) and normalized to a reference control (GAPDH or ß-Actin). Primers for qRT-PCR were designed with the assistance of online tools (Primer 3Plus) using at least one exon junction binding-site per primer pair where possible. A complete list of primers is available in Key Resources Table. Size and specificity of PCR products were confirmed by gel electrophoresis.
RNA Sequencing Analysis
Detailed methodology and analysis of whole RNA-Seq dataset are described in Lewis et al., 2019). RNA was extracted from synovial tissue homogenised at 4°C in Trizol reagent. Library preparation was performed using TruSeq RNA Sample Preparation Kit v2 (Illumina). Multiplexed libraries were sequenced on Illumina HiSeq2500 to generate 50 million paired-end 75 base pair reads per sample. Synovium transcript abundances were quantified from RNA-Seq FASTQ files over GENCODE v24/GRCh38 transcripts using Kallisto v0.43.0. Estimated read counts generated using tximport 1.6.0 were normalised using DESeq2 1.18.1, accounting for average transcript length correction, incorporating batch, sex and pathotype as model covariates. Transcript abundances were normalised and converted to regularised log expression (RLE). Differential gene expression analysis was performed using DESeq2 with likelihood ratio test between pathotype or ELS group. Q-values were calculated using Benjamini-Hochberg false discovery rate (FDR), with a cut-off of Q ≤ 0.05 to define differentially expressed genes.
Gene sets highly specific to immune cell tissue types were derived based on CAGE sequencing data from the FANTOM5 project (Dimont et al., 2014). Module scores specific for T cell subsets were analysed for correlation with metabolic gene expression in synovial tissue.
Hierarchical clustering within seven groups of metabolic genes differentially expressed between synovial biopsies classified as positive or negative for ELS by histological analysis (FDR ≤ 0.05) was performed using Euclidean distance metric and Ward’s linkage method and plotted using ComplexHeatmap 1.17.1.
Metabolic genes were selected via the use of the KEGG pathway database.
ELISA
Secreted IL17A and IFNγ were measured in cell culture supernatants from 2-4 x 106 CD4+ T cells/well with a human IL17A (homodimer) and IFNγ ELISA Ready-SET-Go Assay (fisherscientific) respectively, according to the manufacturer’s instructions.
Quantification and Statistical Analysis
Statistical details of experiments can be found in the figure legends. All data are expressed as ± SD or ± SEM as indicated in figure legends. Statistical tests were selected based on appropriate assumptions with respect to data distribution and variance characteristics. Statistical significance was determined using unpaired Student's t test or ANOVA (one- or two-way). ll statistical analysis were carried out in GraphPad Prism7. Significant differences are indicated as follows: ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001.
Data and Code Availability
RNA-seq data are deposited at ArrayExpress and are accessible via accession E-MTAB-6141 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6141).
Acknowledgments
The work was performed with funds from Versus Arthritis (fellowship 21386) to V.P.; British Heart Foundation (fellowship FS/12/38/29640), Queen Mary Innovation Ltd (proof of concept fund), and University of Birmingham (start-up grant) to C.M.; Fondazione Cariplo (2015-0552) to M.R. and C.M.; Cancer Research UK (fellowship C50242/A17728) to J.J.K.; V.P. is supported by a Versus Arthritis Fellowship (21386). D.C. was supported by a fellowship from the Institut Pasteur Foundation Cenci-Bolognetti. R.H. was supported by a Medical Research Council UK PhD studentship. S.E.H. was supported by an Oliver Bird PhD Studentship from the Nuffield Foundation. J.J.K. is supported by a Cancer Research UK Career Development fellowship (C50242/A17728). C.M. is supported by a British Heart Foundation Intermediate Basic Science research fellowship (FS/12/38/29640). We thank Frederik Radvan for his contribution toward some experimental aspects of the revision of our paper. We thank the Wellcome Trust Sanger Institute Mouse Genetics Project (Sanger MGP) and its funders for providing the mutant mouse line (Allele: Slc5a12em1(IMPC)Wtsi) and INFRAFRONTIER/EMMA (www.infrafrontier.eu). Funding information may be found at www.sanger.ac.uk/mouseportal and associated primary phenotypic information at www.mousephenotype.org.
Author Contributions
Conceptualization, V.P., M.C., R.H., J.J.K., M.B., C.P., and C.M.; Methodology, V.P., M.C., V.B., K.G., E.P., J.S., and S.E.H.; Investigation, V.P., M.C., V.B., D.C., K.G., E.P., R.H., J.S., and S.E.H.; Analysis, V.P., M.C., V.B., K.G., E.P., R.H., J.S., K.B., M.J.L., J.J.K., M.B., and C.M.; Resources, M.R., F.H., M.J.L., and C.P.; Writing – Original Draft, V.P., M.C., and C.M.; Writing – Review and Editing, all authors; Visualization, V.P., M.C., and C.M.; Supervision, M.J.L., J.J.K., M.B., C.P., and C.M.; Project Administration, C.M.; Funding Acquisition, V.P., M.R., J.J.K., M.B., C.P., and C.M.
Declaration of Interests
J.J.K. is an employee of and shareholder in Rheos Medicines, Inc.
Published: November 7, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cmet.2019.10.004.
Supplemental Information
References
- Abu-Elheiga L., Brinkley W.R., Zhong L., Chirala S.S., Woldegiorgis G., Wakil S.J. The subcellular localization of acetyl-CoA carboxylase 2. Proc. Natl. Acad. Sci. USA. 2000;97:1444–1449. doi: 10.1073/pnas.97.4.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Elheiga L., Matzuk M.M., Abo-Hashema K.A., Wakil S.J. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- Aletaha D., Neogi T., Silman A.J., Funovits J., Felson D.T., Bingham C.O., 3rd, Birnbaum N.S., Burmester G.R., Bykerk V.P., Cohen M.D. 2010 Rheumatoid arthritis classification criteria: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann. Rheum. Dis. 2010;69:1580–1588. doi: 10.1136/ard.2010.138461. [DOI] [PubMed] [Google Scholar]
- Amorini A.M., Nociti V., Petzold A., Gasperini C., Quartuccio E., Lazzarino G., Di Pietro V., Belli A., Signoretti S., Vagnozzi R. Serum lactate as a novel potential biomarker in multiple sclerosis. Biochim. Biophys. Acta. 2014;1842:1137–1143. doi: 10.1016/j.bbadis.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Anastasiou D., Poulogiannis G., Asara J.M., Boxer M.B., Jiang J.K., Shen M., Bellinger G., Sasaki A.T., Locasale J.W., Auld D.S. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M., Marayati R., Moffitt R., Yeh J.J. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget. 2017;8:56081–56094. doi: 10.18632/oncotarget.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anflous-Pharayra K., Cai Z.J., Craigen W.J. VDAC1 serves as a mitochondrial binding site for hexokinase in oxidative muscles. Biochim. Biophys. Acta. 2007;1767:136–142. doi: 10.1016/j.bbabio.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Angelin A., Gil-de-Gómez L., Dahiya S., Jiao J., Guo L., Levine M.H., Wang Z., Quinn W.J., 3rd, Kopinski P.K., Wang L. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017;25:1282–1293. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C.N., Bähre H. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- Biniecka M., Connolly M., Gao W., Ng C.T., Balogh E., Gogarty M., Santos L., Murphy E., Brayden D., Veale D.J. Redox-mediated angiogenesis in the hypoxic joint of inflammatory arthritis. Arthritis Rheumatol. 2014;66:3300–3310. doi: 10.1002/art.38822. [DOI] [PubMed] [Google Scholar]
- Bombardieri M., Lewis M., Pitzalis C. Ectopic lymphoid neogenesis in rheumatic autoimmune diseases. Nat. Rev. Rheumatol. 2017;13:141–154. doi: 10.1038/nrrheum.2016.217. [DOI] [PubMed] [Google Scholar]
- Bradley A., Anastassiadis K., Ayadi A., Battey J.F., Bell C., Birling M.C., Bottomley J., Brown S.D., Bürger A., Bult C.J. The mammalian gene function resource: the international knockout mouse consortium. Mamm. Genome. 2012;23:580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A., Singer K., Koehl G.E., Kolitzus M., Schoenhammer G., Thiel A., Matos C., Bruss C., Klobuch S., Peter K. LDHA-associated lactic acid production blunts tumour immunosurveillance by T and NK cells. Cell Metab. 2016;24:657–671. doi: 10.1016/j.cmet.2016.08.011. [DOI] [PubMed] [Google Scholar]
- Bruns L., Frey O., Morawietz L., Landgraf C., Volkmer R., Kamradt T. Immunization with an immunodominant self-peptide derived from glucose-6-phosphate isomerase induces arthritis in DBA/1 mice. Arthritis Res. Ther. 2009;11:R117. doi: 10.1186/ar2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., O'Sullivan D., Klein Geltink R.I., Curtis J.D., Chang C.H., Sanin D.E., Qiu J., Kretz O., Braas D., van der Windt G.J. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante M.F., Oliveira P.G., Garcia-Carbonell R., Croft A.P., Smith J.M., Serrano R.L., Sanchez-Lopez E., Liu X., Kisseleva T., Hay N. Hexokinase 2 as a novel selective metabolic target for rheumatoid arthritis. Ann. Rheum. Dis. 2018;77:1636–1643. doi: 10.1136/annrheumdis-2018-213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañete J.D., Celis R., Moll C., Izquierdo E., Marsal S., Sanmartí R., Palacín A., Lora D., de la Cruz J., Pablos J.L. Clinical significance of synovial lymphoid neogenesis and its reversal after anti-tumour necrosis factor alpha therapy in rheumatoid arthritis. Ann. Rheum. Dis. 2009;68:751–756. doi: 10.1136/ard.2008.089284. [DOI] [PubMed] [Google Scholar]
- Chen D.Y., Chen Y.M., Chen H.H., Hsieh C.W., Lin C.C., Lan J.L. Increasing levels of circulating Th17 cells and interleukin-17 in rheumatoid arthritis patients with an inadequate response to Anti-TNF-α therapy. Arthritis Res. Ther. 2011;13:R126. doi: 10.1186/ar3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.Y., Shiau A.L., Wu C.L., Wang C.R. Amelioration of experimental arthritis by intra-articular injection of an epidermal growth factor receptor tyrosine kinase inhibitor. Clin. Exp. Rheumatol. 2015;33:839–843. [PubMed] [Google Scholar]
- Cheung E.C., Ludwig R.L., Vousden K.H. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc. Natl. Acad. Sci. USA. 2012;109:20491–20496. doi: 10.1073/pnas.1206530109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala S.S., Wakil S.J. Structure and function of animal fatty acid synthase. Lipids. 2004;39:1045–1053. doi: 10.1007/s11745-004-1329-9. [DOI] [PubMed] [Google Scholar]
- Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., Cyrus N., Brokowski C.E., Eisenbarth S.C., Phillips G.M. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell N.W., Lund P., Hems R., Krebs H.A. Acceleration of gluconeogenesis from lactate by lysine (short communication) Biochem. J. 1973;134:671–672. doi: 10.1042/bj1340671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton J.M., Gillespie J.G., Hawley S.A., Hardie D.G. 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- Dimont E., Hofmann O., Ho Sui S.J., Forrest A.R., Kawaji H., FANTOM Consortium. Hide W. CAGExploreR: an R package for the analysis and visualization of promoter dynamics across multiple experiments. Bioinformatics. 2014;30:1183–1184. doi: 10.1093/bioinformatics/btu125. [DOI] [PubMed] [Google Scholar]
- Eigenbrodt E., Reinacher M., Scheefers-Borchel U., Scheefers H., Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit. Rev. Oncog. 1992;3:91–115. [PubMed] [Google Scholar]
- Endo Y., Asou H.K., Matsugae N., Hirahara K., Shinoda K., Tumes D.J., Tokuyama H., Yokote K., Nakayama T. Obesity drives Th17 cell differentiation by inducing the lipid metabolic kinase, ACC1. Cell Rep. 2015;12:1042–1055. doi: 10.1016/j.celrep.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Ene-Obong A., Clear A.J., Watt J., Wang J., Fatah R., Riches J.C., Marshall J.F., Chin-Aleong J., Chelala C., Gribben J.G. Activated pancreatic stellate cells sequester CD8+ T cells to reduce their infiltration of the juxtatumoral compartment of pancreatic ductal adenocarcinoma. Gastroenterology. 2013;145:1121–1132. doi: 10.1053/j.gastro.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B., Li K.Y., Cai L., Hensley C.T., Kim J., Zacharias L.G., Yang C., Do Q.N., Doucette S., Burguete D. Lactate metabolism in human lung tumours. Cell. 2017;171:358–371. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Yang H., Zhang Y., Wei H., Zhu Z., Zhu B., Yang M., Cao W., Wang L., Wu Z. Tumour cell-derived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene. 2017;36:5829–5839. doi: 10.1038/onc.2017.188. [DOI] [PubMed] [Google Scholar]
- Fiek C., Benz R., Roos N., Brdiczka D. Evidence for identity between the hexokinase-binding protein and the mitochondrial porin in the outer membrane of rat liver mitochondria. Biochim. Biophys. Acta. 1982;688:429–440. doi: 10.1016/0005-2736(82)90354-6. [DOI] [PubMed] [Google Scholar]
- Fransen J., van Riel P.L. The disease activity score and the EULAR response criteria. Rheum. Dis. Clin. North Am. 2009;35:745–757. doi: 10.1016/j.rdc.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Fujii W., Kawahito Y., Nagahara H., Kukida Y., Seno T., Yamamoto A., Kohno M., Oda R., Taniguchi D., Fujiwara H. Monocarboxylate transporter 4, associated with the acidification of synovial fluid, is a novel therapeutic target for inflammatory arthritis. Arthritis Rheumatol. 2015;67:2888–2896. doi: 10.1002/art.39270. [DOI] [PubMed] [Google Scholar]
- Garcia-Carbonell R., Divakaruni A.S., Lodi A., Vicente-Suarez I., Saha A., Cheroutre H., Boss G.R., Tiziani S., Murphy A.N., Guma M. Critical role of glucose metabolism in rheumatoid arthritis fibroblast-like synoviocytes. Arthritis Rheumatol. 2016;68:1614–1626. doi: 10.1002/art.39608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl E.J., Falchuk K.H., Zeiger L.S., Sullivan A.L., Hebert C.L., Adams J.P., Decker J.L. A physiological approach to the assessment of disease activity in rheumatoid arthritis. J. Clin. Invest. 1971;50:1167–1180. doi: 10.1172/JCI106594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal E., Umapathy N.S., Martin P.M., Ananth S., Gnana-Prakasam J.P., Becker H., Wagner C.A., Ganapathy V., Prasad P.D. Cloning and functional characterization of human SMCT2 (SLC5A12) and expression pattern of the transporter in kidney. Biochim. Biophys. Acta. 2007;1768:2690–2697. doi: 10.1016/j.bbamem.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon G., Mackow M.C., Levy H.R. On the mechanism of interaction of steroids with human glucose 6-phosphate dehydrogenase. Arch Biochem Biophys. 1995;318:25–29. doi: 10.1006/abbi.1995.1199. [DOI] [PubMed] [Google Scholar]
- Haas R., Cucchi D., Smith J., Pucino V., Macdougall C.E., Mauro C. Intermediates of metabolism: from bystanders to signalling molecules. Trends Biochem. Sci. 2016;41:460–471. doi: 10.1016/j.tibs.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Haas R., Smith J., Rocher-Ros V., Nadkarni S., Montero-Melendez T., D'Acquisto F., Bland E.J., Bombardieri M., Pitzalis C., Perretti M. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 2015;13:e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap A.P., Wilson M.C. The monocarboxylate transporter family - role and regulation. IUBMB Life. 2012;64:109–119. doi: 10.1002/iub.572. [DOI] [PubMed] [Google Scholar]
- Haworth O., Hardie D.L., Burman A., Rainger G.E., Eksteen B., Adams D.H., Salmon M., Nash G.B., Buckley C.D. A role for the integrin alpha6beta1 in the differential distribution of CD4 and CD8 T-cell subsets within the rheumatoid synovium. Rheumatology. 2008;47:1329–1334. doi: 10.1093/rheumatology/ken263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Tsuboi H., Iizuka-Koga M., Takahashi H., Asashima H., Yokosawa M., Kondo Y., Ohta M., Wakasa Y., Matsumoto I. Suppression of glucose-6-phosphate-isomerase induced arthritis by oral administration of transgenic rice seeds expressing altered peptide ligands of glucose-6-phosphate-isomerase. Mod. Rheumatol. 2017;27:457–465. doi: 10.1080/14397595.2016.1218598. [DOI] [PubMed] [Google Scholar]
- Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., Esparza L.A., Reya T., Zhan Le, Yanxiang Guo J. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humby F., Lewis M., Ramamoorthi N., Hackney J.A., Barnes M.R., Bombardieri M., Setiadi A.F., Kelly S., Bene F., DiCicco M. Synovial cellular and molecular signatures stratify clinical response to csDMARD therapy and predict radiographic progression in early rheumatoid arthritis patients. Ann. Rheum. Dis. 2019;78:761–772. doi: 10.1136/annrheumdis-2018-214539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang C.S., Shemorry A., Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanami K., Matsumoto I., Tanaka Y., Inoue A., Goto D., Ito S., Tsutsumi A., Sumida T. Arthritogenic T cell epitope in glucose-6-phosphate isomerase-induced arthritis. Arthritis Res. Ther. 2008;10:R130. doi: 10.1186/ar2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.W., Bombardieri M., Greenhill C.J., McLeod L., Nerviani A., Rocher-Ros V., Cardus A., Williams A.S., Pitzalis C., Jenkins B.J. Interleukin-27 inhibits ectopic lymphoid-like structure development in early inflammatory arthritis. J. Exp. Med. 2015;212:1793–1802. doi: 10.1084/jem.20132307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G.W., Jones S.A. Ectopic lymphoid follicles: inducible centres for generating antigen-specific immune responses within tissues. Immunology. 2016;147:141–151. doi: 10.1111/imm.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S., Humby F., Filer A., Ng N., Di Cicco M., Hands R.E., Rocher V., Bombardieri M., D'Agostino M.A., McInnes I.B. Ultrasound-guided synovial biopsy: a safe, well-tolerated and reliable technique for obtaining high-quality synovial tissue from both large and small joints in early arthritis patients. Ann. Rheum. Dis. 2015;74:611–617. doi: 10.1136/annrheumdis-2013-204603. [DOI] [PubMed] [Google Scholar]
- Krenn V., Morawietz L., Häupl T., Neidel J., Petersen I., König A. Grading of chronic synovitis–a histopathological grading system for molecular and diagnostic pathology. Pathol. Res. Pract. 2002;198:317–325. doi: 10.1078/0344-0338-5710261. [DOI] [PubMed] [Google Scholar]
- Kugyelka R., Kohl Z., Olasz K., Mikecz K., Rauch T.A., Glant T.T., Boldizsar F. Enigma of IL-17 and Th17 cells in rheumatoid arthritis and in autoimmune animal models of arthritis. Mediators Inflamm. 2016;2016:6145810. doi: 10.1155/2016/6145810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegren U., Andersson J., Wigzell H. Mechanisms of T lymphocyte activation by Okt3 antibodies. A general model for T cell induction. Eur. J. Immunol. 1984;14:325–328. doi: 10.1002/eji.1830140409. [DOI] [PubMed] [Google Scholar]
- Le Goffe C., Vallette G., Charrier L., Candelon T., Bou-Hanna C., Bouhours J.F., Laboisse C.L. Metabolic control of resistance of human epithelial cells to H2O2 and NO stresses. Biochem. J. 2002;364:349–359. doi: 10.1042/BJ20011856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M.J., Barnes M.R., Blighe K., Goldmann K., Rana S., Hackney J.A., Ramamoorthi N., John C.R., Watson D.S., Kummerfeld S.K. Molecular portraits of early rheumatoid arthritis identify clinical and treatment response phenotypes. Cell Rep. 2019;28:2455–2470. doi: 10.1016/j.celrep.2019.07.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu J., Zhu J., Kuei C., Yu J., Shelton J., Sutton S.W., Li X., Yun S.J., Mirzadegan T. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J. Biol. Chem. 2009;284:2811–2822. doi: 10.1074/jbc.M806409200. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mackay G.M., Zheng L., van den Broek N.J., Gottlieb E. Analysis of cell metabolism using LC-MS and isotope tracers. Methods Enzymol. 2015;561:171–196. doi: 10.1016/bs.mie.2015.05.016. [DOI] [PubMed] [Google Scholar]
- Magistretti P.J., Allaman I. Lactate in the brain: from metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 2018;19:235–249. doi: 10.1038/nrn.2018.19. [DOI] [PubMed] [Google Scholar]
- Mathupala S.P., Ko Y.H., Pedersen P.L. Hexokinase-2 bound to mitochondria: cancer's Stygian link to the "Warburg Effect" and a pivotal target for effective therapy. Semin. Cancer Biol. 2009;19:17–24. doi: 10.1016/j.semcancer.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.Y., Yan M., Du Y., Wu T., Khobahy E., Kwon S.R., Taneja V., Bashmakov A., Nukala S., Ye Y. Intra-articular nuclear factor-κB blockade ameliorates collagen-induced arthritis in mice by eliciting regulatory T cells and macrophages. Clin. Exp. Immunol. 2013;172:217–227. doi: 10.1111/cei.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Vaknin N., Saha A., Legendre M., Carmona-Rivera C., Amin M.A., Rabquer B.J., Gonzales-Hernandez M.J., Jorns J., Mohan S., Yalavarthi S. DEK-targeting DNA aptamers as therapeutics for inflammatory arthritis. Nat. Commun. 2017;8:14252. doi: 10.1038/ncomms14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsholme E.A., Sugden P.H., Williams T. Effect of citrate on the activities of 6-phosphofructokinase from nervous and muscle tissues from different animals and its relationships to the regulation of glycolysis. Biochem. J. 1977;166:123–129. doi: 10.1042/bj1660123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olloquequi J., Ferrer J., Montes J.F., Rodríguez E., Montero M.A., García-Valero J. Differential lymphocyte infiltration in small airways and lung parenchyma in COPD patients. Respir. Med. 2010;104:1310–1318. doi: 10.1016/j.rmed.2010.03.002. [DOI] [PubMed] [Google Scholar]
- O'Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Beckett O., Flavell R.A., Li M.O. An essential role of the Forkhead-box transcription factor FoxO1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino J.G., Shulga N., Hoek J.B. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 2002;277:7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- Patel D.D., Kuchroo V.K. Th17 cell pathway in human immunity: lessons from genetics and therapeutic interventions. Immunity. 2015;43:1040–1051. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Pellerin L., Magistretti P.J. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc. Natl. Acad. Sci. USA. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M., Yin N., Chhangawala S., Xu K., Leslie C.S., Li M.O. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354:481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., Fowler K.D., Chalmin F., Merkler D., Kuchroo V.K., Pot C. IL-27 induces Th17 differentiation in the absence of STAT1 signalling. J. Immunol. 2015;195:4144–4153. doi: 10.4049/jimmunol.1302246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., Pitcher L.A., Sullivan J.M., Mitsdoerffer M., Acton S.E., Franz B., Wucherpfennig K., Turley S., Carroll M.C., Sobel R.A. Th17 cells induce ectopic lymphoid follicles in central nervous system tissue inflammation. Immunity. 2011;35:986–996. doi: 10.1016/j.immuni.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettitt S.J., Liang Q., Rairdan X.Y., Moran J.L., Prosser H.M., Beier D.R., Lloyd K.C., Bradley A., Skarnes W.C. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods. 2009;6:493–495. doi: 10.1038/nmeth.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitzalis C., Jones G.W., Bombardieri M., Jones S.A. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat. Rev. Immunol. 2014;14:447–462. doi: 10.1038/nri3700. [DOI] [PubMed] [Google Scholar]
- Pitzalis C., Kelly S., Humby F. New learnings on the pathophysiology of RA from synovial biopsies. Curr. Opin. Rheumatol. 2013;25:334–344. doi: 10.1097/BOR.0b013e32835fd8eb. [DOI] [PubMed] [Google Scholar]
- Pucino V., Bombardieri M., Pitzalis C., Mauro C. Lactate at the crossroads of metabolism, inflammation, and autoimmunity. Eur. J. Immunol. 2017;47:14–21. doi: 10.1002/eji.201646477. [DOI] [PubMed] [Google Scholar]
- Pucino V., Lucherini O.M., Perna F., Obici L., Merlini G., Cattalini M., La Torre F., Maggio M.C., Lepore M.T., Magnotti F. Differential impact of high and low penetrance TNFRSF1A gene mutations on conventional and regulatory CD4+ T cell functions in TNFR1-associated periodic syndrome. J. Leukoc. Biol. 2016;99:761–769. doi: 10.1189/jlb.3A0915-399R. [DOI] [PubMed] [Google Scholar]
- Raineri R., Levy H.R. On the specificity of steroid interaction with mammary gland glucose-6-phosphate dehydrogenase. Biochemistry. 1970;9:2233–2243. doi: 10.1021/bi00813a003. [DOI] [PubMed] [Google Scholar]
- Reina-Campos M., Moscat J., Diaz-Meco M. Metabolism shapes the tumor microenvironment. Curr. Opin. Cell Biol. 2017;48:47–53. doi: 10.1016/j.ceb.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland C.L., Arumugam T., Deng D., Liu S.H., Philip B., Gomez S., Burns W.R., Ramachandran V., Wang H., Cruz-Monserrate Z. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res. 2014;74:5301–5310. doi: 10.1158/0008-5472.CAN-14-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D., Maier B., Morawietz L., Krenn V., Kamradt T. Immunization with glucose-6-phosphate isomerase induces T cell-dependent peripheral polyarthritis in genetically unaltered mice. J. Immunol. 2004;172:4503–4509. doi: 10.4049/jimmunol.172.7.4503. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Shen Y., Wen Z., Li Y., Matteson E.L., Hong J., Goronzy J.J., Weyand C.M. Metabolic control of the scaffold protein TKS5 in tissue-invasive, proinflammatory T cells. Nat. Immunol. 2017;18:1025–1034. doi: 10.1038/ni.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.Z., Wang R., Huang G., Vogel P., Neale G., Green D.R., Chi H. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai T., Nazarewicz R.R., Wallis B.B., Yanes R.E., Watanabe R., Hilhorst M., Tian L., Harrison D.G., Giacomini J.C., Assimes T.L. The glycolytic enzyme PKM2 bridges metabolic and inflammatory dysfunction in coronary artery disease. J. Exp. Med. 2016;213:337–354. doi: 10.1084/jem.20150900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarnes W.C., Rosen B., West A.P., Koutsourakis M., Bushell W., Iyer V., Mujica A.O., Thomas M., Harrow J., Cox T. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S.R., Gopal E., Zhuang L., Itagaki S., Martin P.M., Fei Y.J., Ganapathy V., Prasad P.D. Cloning and functional identification of slc5a12 as a sodium-coupled low-affinity transporter for monocarboxylates (SMCT2) Biochem. J. 2005;392:655–664. doi: 10.1042/BJ20050927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuhaft P.S., MCCarty D.J. Synovial fluid pH, lactate, oxygen and carbon dioxide partial pressure in various joint diseases. Arthritis Rheum. 1971;14:475–484. doi: 10.1002/art.1780140407. [DOI] [PubMed] [Google Scholar]
- Tumanov S., Bulusu V., Kamphorst J.J. Analysis of fatty acid metabolism using stable isotope tracers and mass spectrometry. Methods Enzymol. 2015;561:197–217. doi: 10.1016/bs.mie.2015.05.017. [DOI] [PubMed] [Google Scholar]
- van Baarsen L.G., Lebre M.C., van der Coelen D., Aarrass S., Tang M.W., Ramwadhdoebe T.H., Gerlag D.M., Tak P.P. Heterogeneous expression pattern of interleukin 17A (IL-17A), IL-17F and their receptors in synovium of rheumatoid arthritis, psoriatic arthritis and osteoarthritis: possible explanation for nonresponse to anti-IL-17 therapy? Arthritis Res. Ther. 2014;16:426. doi: 10.1186/s13075-014-0426-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wauwe J.P., De Mey J.R., Goossens J.G. Okt3: a monoclonal anti-humanT lymphocyte antibody with potent mitogenic properties. J. Immunol. 1980;124:2708–2713. [PubMed] [Google Scholar]
- Vyssokikh M.Y., Brdiczka D. The function of complexes between the outer mitochondrial membrane pore (VDAC) and the adenine nucleotide translocase in regulation of energy metabolism and apoptosis. Acta Biochim. Pol. 2003;50:389–404. [PubMed] [Google Scholar]
- Weyand C.M., Zeisbrich M., Goronzy J.J. Metabolic signatures of T-cells and macrophages in rheumatoid arthritis. Curr. Opin. Immunol. 2017;46:112–120. doi: 10.1016/j.coi.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.K., Gerdin A.K., Karp N.A., Ryder E., Buljan M., Bussell J.N., Salisbury J., Clare S., Ingham N.J., Podrini C. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldetsadik A.D., Vogel M.C., Rabeh W.M., Magzoub M. Hexokinase II-derived cell-penetrating peptide targets mitochondria and triggers apoptosis in cancer cells. FASEB J. 2017;31:2168–2184. doi: 10.1096/fj.201601173R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabu M., Shime H., Hara H., Saito T., Matsumoto M., Seya T., Akazawa T., Inoue N. IL-23-dependent and -independent enhancement pathways of IL-17A production by lactic acid. Int. Immunol. 2011;23:29–41. doi: 10.1093/intimm/dxq455. [DOI] [PubMed] [Google Scholar]
- Yang X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- Yang Z., Fujii H., Mohan S.V., Goronzy J.J., Weyand C.M. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J. Exp. Med. 2013;210:2119–2134. doi: 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Shen Y., Oishi H., Matteson E.L., Tian L., Goronzy J.J., Weyand C.M. Restoring oxidant signalling suppresses proarthritogenic T cell effector functions in rheumatoid arthritis. Sci. Transl. Med. 2016;8:331ra38. doi: 10.1126/scitranslmed.aad7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Mikami N., Matsushima Y., Miyawaki M., Endo H., Banno R., Tsuji T., Fujita T., Kohno T. Combination treatment with fingolimod and a pathogenic antigen prevents relapse of glucose-6-phosphate isomerase peptide-induced arthritis. Immun. Inflam. Dis. 2016;4:263–273. doi: 10.1002/iid3.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are deposited at ArrayExpress and are accessible via accession E-MTAB-6141 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6141).