Summary
RNA sequencing analyses are often limited to identifying lowest p value transcripts, which does not address polygenic phenomena. To overcome this limitation, we developed an integrative approach that combines large-scale transcriptomic meta-analysis of patient brain tissues with single-cell sequencing data of CNS neurons, short RNA sequencing of human male- and female-originating cell lines, and connectomics of transcription factor and microRNA interactions with perturbed transcripts. We used this pipeline to analyze cortical transcripts of schizophrenia and bipolar disorder patients. Although these pathologies show massive transcriptional parallels, their clinically well-known sexual dimorphisms remain unexplained. Our method reveals the differences between afflicted men and women and identifies disease-affected pathways of cholinergic transmission and gp130-family neurokine controllers of immune function interlinked by microRNAs. This approach may open additional perspectives for seeking biomarkers and therapeutic targets in other transmitter systems and diseases.
Keywords: bipolar disorder, cholinergic systems, circadian regulation, connectomics, microRNA, miRNet, network analysis, RNA-sequencing, schizophrenia, sexual dimorphism
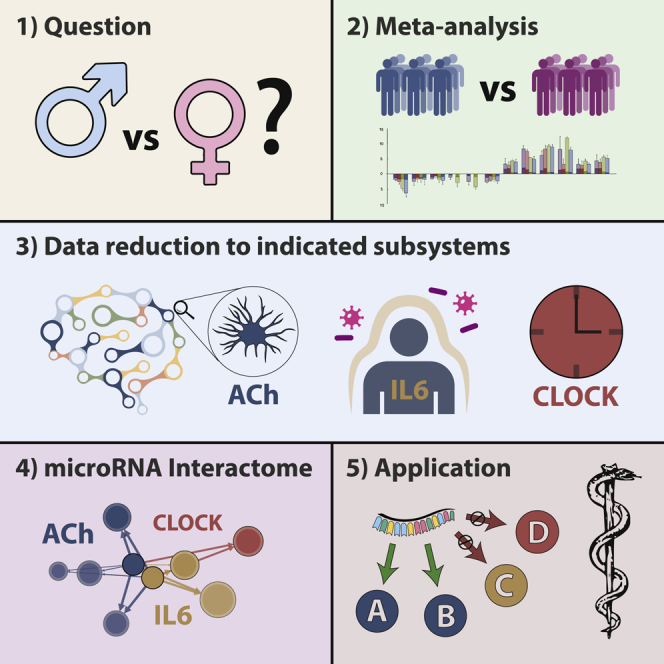
Graphical Abstract
Highlights
-
•
Single-cell transcriptomes reveal a unique profile of cortical cholinergic neurons
-
•
Female- and male-derived cells show distinct neurokine-induced miRNA responses
-
•
Differentially enriched microRNA families constitute a self-organizing network
-
•
Integrative analysis identifies mir-10/mir-199 regulators of cholinergic function
Lobentanzer et al. show how bioinformatically supported high-throughput techniques such as short RNA sequencing can bridge the gap between traditional molecular interaction studies and purely bioinformatic prediction paradigms in an example focused on disentangling the sexual dimorphism in microRNA regulation of the cholinergic/neurokine interface in mental disorders.
Introduction
Recent large-scale genomic (Anttila et al., 2018) and transcriptomic (Gandal et al., 2018) analyses uncovered previously unknown magnitudes of transcriptional correlation (71%) in cortical tissues of schizophrenia (SCZ) and bipolar disorder (BD) patients. This suggests a shared SCZ/BD spectrum, but leaves the causes for the widely divergent sex-specific manifestations of these clinical pathologies largely unclear. Compared with women, men present a higher prevalence of SCZ (odds ratio [OR] = 1.4), a 10 years earlier mean age of highest disease risk (15–25 versus 25–35 years of age) and a worse prognosis (Leger and Neill, 2016). In comparison, BD incidence is not dissimilar in men and women, but 80%–90% of “rapid cyclers” with a particularly bad prognosis are women, and major depressive disorder (MDD), which is a prerequisite for BD diagnosis, affects women more often (OR = 2) (Berger, 2014). Both diseases involve premorbid cognitive impairments and reduced intelligence, but those are more frequent and severe in SCZ than in BD (Bortolato et al., 2015), compatible with its original description as “dementia praecox” (premature dementia) early in the 20th century (Kraepelin, 1913).
The genomic origin of SCZ and BD is tremendously complex. Genome-wide association studies (GWASs) have identified several genotype markers distinguishing the (sub)phenotypes of BD and SCZ (Ruderfer et al., 2018) in genes encoding receptors (e.g., for dopamine, glutamate, or acetylcholine [ACh]), scaffolding proteins (e.g., DISC1, “disrupted in schizophrenia”), transcription factors (TFs), microRNAs (miRNAs), or non-coding genomic regions without known function (Harrison, 2015, Henriksen et al., 2017, Kanazawa et al., 2017). However, the effect of non-coding genomic regions on disease phenotypes is low, possibly indicating that they exert minor effects or do not change expression levels but, rather, modify the expression patterns of secondary gene products (Gulyas-Kovacs et al., 2018).
Notably, central cholinergic processes are closely associated with disease characteristics and their sexual dimorphism: male SCZ patients self-administer more nicotine by smoking than females (7.2 versus 3.3 weighted average OR with 90% lifetime prevalence; de Leon and Diaz, 2005), and pharmacological intervention targeting cholinergic processes is subject to substantial sexual dimorphism (Giacobini and Pepeu, 2018). More specifically, cognitive deficits in SCZ and BD have both been associated with cholinergic dysfunction (van Enkhuizen et al., 2015, Smucny and Tregellas, 2017) and with the sum of anticholinergic medications (Gray et al., 2015, Eum et al., 2017), and a polymorphism in the α5 nicotinic ACh receptor associates with both SCZ and smoking in humans, with parallel manifestations in engineered male mice (Koukouli et al., 2017) and rats (Forget et al., 2018). Correspondingly, cholinergic activation may improve cognition (Sacco et al., 2004, Rowe et al., 2015, Lewis et al., 2017) and mood (Higley and Picciotto, 2014) but provokes schizotypic behavior in Alzheimer’s disease patients (Değirmenci and Keçeci, 2016).
Pinpointing the specific cholinergic perturbations in SCZ and BD is challenging. Although it is well established that cholinergic projection neurons in the basal forebrain (nuclei Ch1–Ch4) (Li et al., 2018) depend on trophic support from their target neurons by retrograde supply of nerve growth factor (NGF) (Levi-Montalcini et al., 1996, Mufson et al., 2009), not much is known about their cortical counterparts. Other neurotrophic factors repeatedly associated with cholinergic function are the neurokines (McManaman and Crawford, 1991), including interleukin-6 (IL-6), ciliary neurotrophic factor (CNTF), and leukemia-inhibiting factor (LIF). This subgroup of cytokines shares receptors and second messenger pathways. The receptors for IL-6 and CNTF are soluble, secreted proteins whose signaling depends on binding the other, dimeric transmembrane receptors (Erta et al., 2012): the gp130 co-receptor (also known as IL6ST [IL-6 signal transducer]) and the LIF receptor. Gp130-family neurokines unite properties of neurotrophic (e.g., on dopaminergic neurons) and immunogenic nature, both prominent facets in the molecular etiology of SCZ and BD (Harrison, 2015) and in cholinergic signaling (Stanke et al., 2006). Neurokines can activate JAK1/2, TYK2, and STAT1/3/5A/5B (Rawlings et al., 2004), affecting neurotransmission as well as immunity. However, to the best of our knowledge, their roles in SCZ/BD have not yet been studied.
SCZ/BD hallmarks also include circadian perturbations. The cholinergic-catecholaminergic imbalance in BD (van Enkhuizen et al., 2015) follows variable transcriptionally regulated rhythms (e.g., CLOCK, ARNTL, and RORA), and affected individuals exhibit decreased rapid eye movement (REM) latency (the duration from onset of sleep to the first REM phase) and increased vulnerability to disease that can be modulated by muscarinic agonists/antagonists (Ising et al., 2005). Correspondingly, the muscarinic M1/M3 receptor genes are essential for REM sleep (Niwa et al., 2018), and sleep deprivation exerts short-term antidepressant effects (Wu and Bunney, 1990), reduced cortical ACh levels (Boonstra et al., 2007), and vast transcriptional changes in basal forebrain cholinergic neurons (Nikonova et al., 2017).
Quantitative measurement of disease relevance of individual perturbed genes is a common methodological problem in transcriptomics analyses. To reduce complexity, transcripts are often filtered by their p values, leading to bias toward few highly expressed and differentially regulated genes. This does not agree with a polygenic model where each component contributes a small effect. To alleviate this bias while still attempting the necessary complexity reduction, we developed an integrative approach of multiple perspectives.
Results
We first explored the neuronal transcriptomic properties of SCZ and BD in a meta-analysis of deposited cortical male and female patient samples; Gene Ontology (GO) enrichment analysis of diverging transcripts helped define an objective set of ontological categories to guide further studies. Next, we ascertained co-expression of the corresponding subsystems in cortical tissues by single-cell sequencing analysis, investigating the putative trophic role of neurokine signaling in cortical cholinergic systems and their transcriptional regulation by TFs and miRNAs. Using pro-cholinergic intervention (stimulation by neurokines) in two closely related cellular models of male and female neuronal origin, we validated the predicted controllers of this molecular interface and identified sexual dimorphisms and affected pathways.
SCZ/BD Transcriptome Meta-analysis
Sex-Independent Pathways Discriminating between SCZ and BD Associate with Immunity
Replicating recent reports of sex-independent overlap in patient brain transcriptomes (Gandal et al., 2018), we validated the high correlation of SCZ/BD expression beta values (139 SCZ and 82 BD brains with matched controls; Spearman’s rho = 0.7100, p < 0.001). To identify transcript subgroups distinguishing between diseased men and women, we then segregated data from males and females. In both cases, this yielded lower correlations between SCZ and BD than sex-independent data (female [F:] 0.6150, p < 0.001; male [M]: 0.5783, p < 0.001). We sought the most discriminating molecular pathways (i.e., those exhibiting the largest difference in Spearman’s ranks between SCZ and BD) and examined their function. Sex-independently, GO enrichment of the top 100 diverging genes yielded numerous terms connected to inflammation and immunity (“acute inflammatory response,” p = 0.003; “cellular response to cytokine stimulus,” p = 0.01).
Transcriptional Sexual Dimorphism Differs between SCZ and BD
Given the possible male or female biases, we studied all of the 2 × 2 combinations of the four possible groups (SCZ males, SCZ females, BD males, and BD females) by calculating expression beta values inside of each group (against matched controls). We then subjected the most diverging beta values (i.e., the most biased genes) in any meaningful combination to GO enrichment analysis (Figure 1; Data S1). The results indicated a larger divergence between sexes in SCZ than in BD; SCZ-biased genes of males and females showed no overlapping GO terms (Figure 1A), but the top 100 BD-biased genes of males and females showed large GO term overlap, particularly in inflammatory components (Figure 1B). Notably, specific components of neurokine signaling (Rawlings et al., 2004) were elevated in both males (IL-6, p = 0.007) and females (JAK/STAT, p = 0.01) with BD.
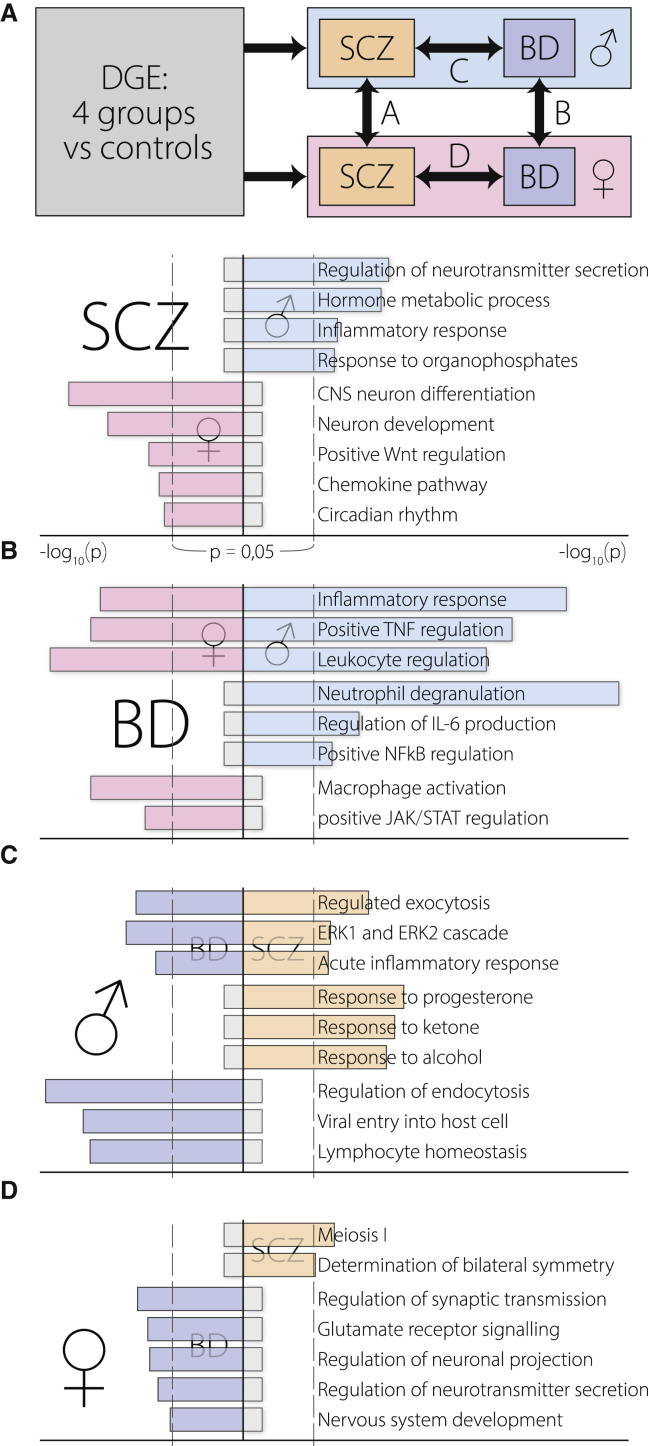
Figure 1.
Diverging Brain Transcriptomes in Males and Females with SCZ/BD
Differential gene expression results of meta-analysis were dually compared: SCZ versus BD or male versus female. GO enrichment of the top 100 distinguishing genes in one dimension was compared with the other for each pair of combinations.
(A) SCZ-biased genes diverge between males and females.
(B) BD-biased genes share immunological ontology in both males and females.
(C) Male-biased genes share immunological ontology in BD and SCZ.
(D) Female-biased genes diverge between SCZ and BD.
Male-Biased Genes Overlap between SCZ and BD
GO terms shared by SCZ and BD patients emerged for male-biased but not female-biased genes. Males with either disease showed elevated inflammation- and immunity-related genes (Figure 1C). In contrast, female-biased BD genes were enriched in terms associated with CNS function or development (Figure 1D), but enrichment analysis of female-biased SCZ genes yielded no CNS-relevant terms. GO terms pertaining to immune processes were very specific, with many referring to single mechanistic components (e.g., IL-6), whereas terms focused on neuronal processes failed to implicate specific neurotransmitters. Because the cholinergic systems present significant overlaps with facets of both SCZ and BD as well as involvement with neurokine signaling (McManaman and Crawford, 1991) and inflammation (Chavan et al., 2017), we chose to focus on the diverging cholinergic transcriptomes in males and females with SCZ/BD.
Cholinergic Systems in the CNS
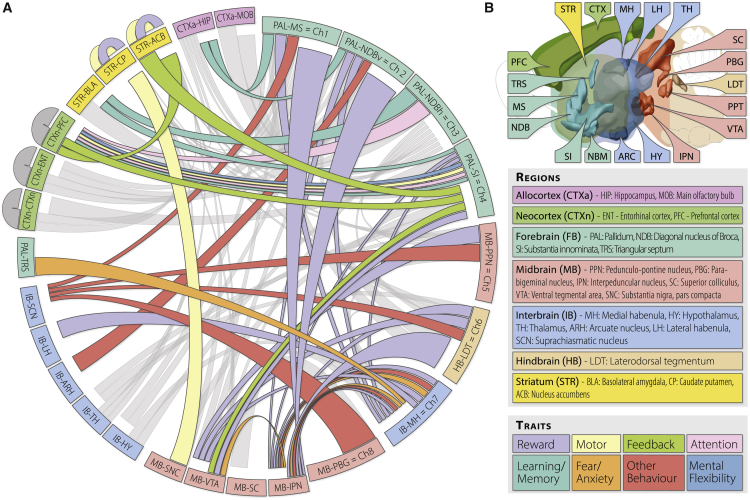
Circos plot analysis of curated functional and anatomical research (Figure 2) and 3D tracing experiments (Oh et al., 2014; Video S1) demonstrate a wide-spread influence of cholinergic systems on both rudimentary and higher cognitive processes throughout the entire mammalian brain (Woolf, 1991, Bina et al., 1993, Sarter et al., 2009, Mesulam, 2013, Luchicchi et al., 2014, Eskow Jaunarajs et al., 2015, Gonzales and Smith, 2015, Lin et al., 2015, Ballinger et al., 2016, Herman et al., 2016, Prado et al., 2017, Haam and Yakel, 2017, McLaughlin et al., 2017). However, trophic factor dependency has only been proven for basal forebrain projection neurons Ch1–Ch4, leaving many open questions about the nature of cholinergic interneurons in the striatum and cortex (Mufson et al., 2009) and their transcriptomic features.
Figure 2.
The Complexity of Cholinergic Networks in the Mammalian Brain
(A) Circos overview demonstrating the widespread interactions of the mammalian brain’s cholinergic systems with physiologic and cognitive processes. Projection origin is denoted by the closeness of the connector and ideogram (first half clockwise) and projection termini by spacing between the connector and ideogram (second half clockwise). Cholinergic neurons have been shown in nuclei Ch1–Ch8 (smaller right half of the ideogram) and as interneurons in the striatum and cortex (outside of the ideogram). Functional traits are indicated by the color of the connectors (connector width is determined by geometry and has no implied meaning).
(B) Brain regions and projection trait legend for (A). Grey: anatomical cholinergic structure with unknown function.
Single-Cell Transcriptome Analysis
Cortical Cholinergic Cells Are Mainly Neurons and Possess Neurokine Receptors
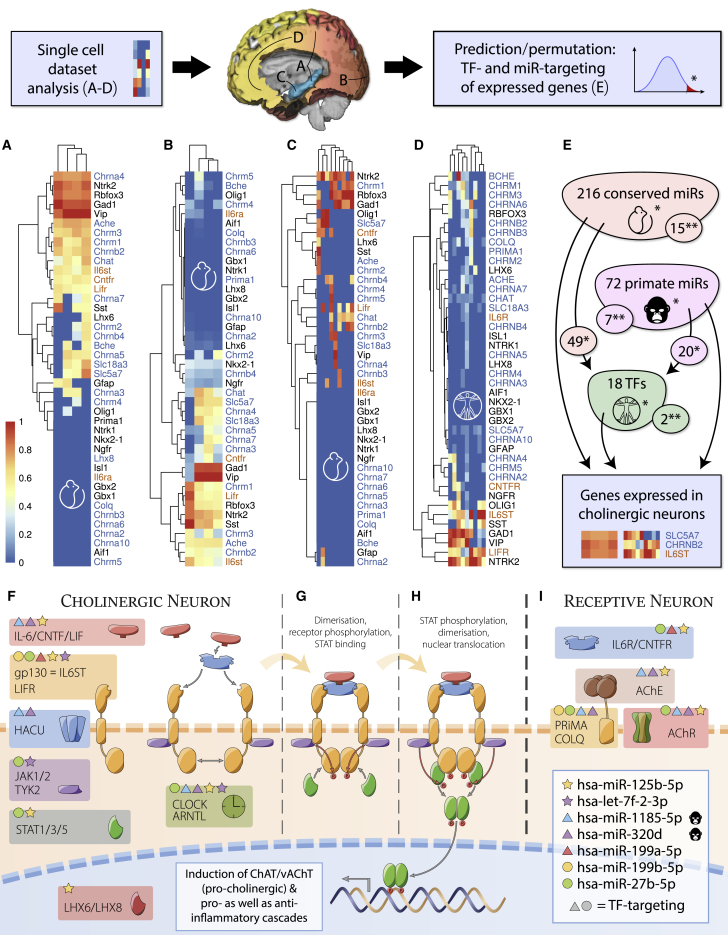
The predicted transcriptomic interaction between cholinergic and trophic factor systems can only be pathologically relevant when the key elements of both pathways coexist in the same cell. However, patient tissues are almost exclusively collected from brain regions (cortex, hippocampus, seldom the striatum) where cholinergic cells are vastly underrepresented (von Engelhardt et al., 2007), which complicates direct retrieval of information on cholinergic processes from total transcriptomes. To examine trophic influences, we turned to web-available single-cell sequencing datasets (Darmanis et al., 2015, Zeisel et al., 2015, Habib et al., 2016, Tasic et al., 2016) and identified putative cholinergic cells in these datasets as those expressing the cholinergic markers choline acetyltransferase (CHAT) and/or the vesicular ACh transporter SLC18A3 (also known as vAChT). Notably, most of these cells or clusters expressed the neuronal marker RBFOX3 (also known as Neu-N) but not the microglial marker AIF1. The oligodendrocyte and astrocyte markers OLIG1 and GFAP were both detected in a minority of samples, indicating sparse cholinergic functions in non-neuronal cells. In both mouse and human brains, the identified cells co-expressed the low-affinity neurotrophin receptor Ngfr (also known as p75) and the two transmembrane neurokine receptor proteins gp130 (also known as IL6ST) and LIFR (Figures 3A–3D) but not the high-affinity receptor for NGF, NTRK1. In summary, the identified cortical neurons distinguished themselves from basal forebrain cholinergic projection neurons by lacking NGF receptive ability but possessing the molecular machinery to process neurokine signaling.
Figure 3.
Single-Cell Sequencing of ChAT/vAChT-Positive Cortical Cells and Analysis of Their Expressed Transcripts
Expression values were normalized (0–1) for each dataset. The order of genes in each heatmap reflects transcript clustering rather than level differences. Columns represent individual samples from original data (see column names in Figure S1); cholinergic genes are denoted in blue and neurokine receptors in orange.
(A) Clustered single-cell sequences of the transgenic mouse somatosensory cortex and hippocampus (Zeisel et al., 2015).
(B) Clustered single-cell sequences from the transgenic mouse visual cortex (Tasic et al., 2016).
(C) Single-nucleus sequencing of adult mouse hippocampus (Habib et al., 2016).
(D) Single-cell sequencing of the human developing neocortex (Darmanis et al., 2015).
(E) Permutation analysis of miRNA and TF targeting data of genes expressed in cholinergic cells (via miRNet) identified putative cholinergic/neurokine co-regulators (∗p < 0.05, ∗∗p < 0.001). Implicated TFs are regulated by a subset of miRNAs targeting cholinergic genes, indicating nested regulation (details in Data S2).
(F–I) Gp130-family neurokine, cholinergic, and circadian signaling pathways are controlled by primate-specific and evolutionarily conserved miRNAs. Components expressed in cholinergic neurons (F), processes following receptor binding (G) and STAT phosphorylation (H), and components of cholinoceptive neurons (I) are shown. miRNA targeting of individual genes (indicated by colored symbols) yields complex transcriptional interactions. Several miRNAs directly targeting the cholinergic pathway also target TFs controlling this pathway (circles and triangles).
Neuronal Transcripts Predict Regulatory Transcription Factor and MicroRNA Circuits
TFs exhibit particularly high complexity in the CNS (Marbach et al., 2016), as do short and long non-coding RNAs (ncRNAs), regulatory elements whose expression is modified in mental diseases (Harrison, 2015). The best-studied ncRNA species are miRNAs, small (18–22 bases) single-stranded RNAs that interfere with translation of transcribed mRNA by guiding an RNA-induced silencing complex (RISC) to the mRNA through sequence complementarity, followed by inhibition and/or degradation of those mRNAs. By targeting multiple genes, miRNAs can exert complex contextual regulation in a temporal and/or spatial fashion (Greenberg and Soreq, 2014), retaining homeostasis, which is paramount for nervous system health (Shaltiel et al., 2013), or leading to nervous system disease when disturbed (Bekenstein et al., 2017, Rajman and Schratt, 2017).
To pursue the coding and non-coding regulatory elements of cholinergic/neurokine processes, we employed targeting analyses of TF-gene and miRNA-gene interactions in a graph database specifically constructed from comprehensive state-of-the-art data (miRNet; STAR Methods) using random permutation to empirically estimate the false discovery rate (FDR). This identified 288 miRNA candidates (of all 2,588 annotated mature miRNAs) and 18 TFs (of 618) with an FDR of less than 5%, and 22 miRNAs and 2 TFs with an FDR of less than 0.1%. The indicated TFs are controlled by a subset of the identified miRNAs (69 predicted, 12 of those with experimental support), indicating nested layers of regulation (Figure 3E; Data S2). This gave rise to the hypothesis outlined in Figures 3F–3I, in which cooperative pathway regulation by miRNAs and TFs connects cholinergic neuronal function with endocrine or paracrine trophic signaling through neurokines. Together, these analyses called for experimental validation of the indicated regulatory pathways linking TFs, cholinergic-neurokine signaling, and miRNAs in male and female cells.
Short RNA Profiling
Neurokine-Induced Cholinergic Differentiation Distinctly Alters Short RNAs in Male and Female Human Neuronal Cells
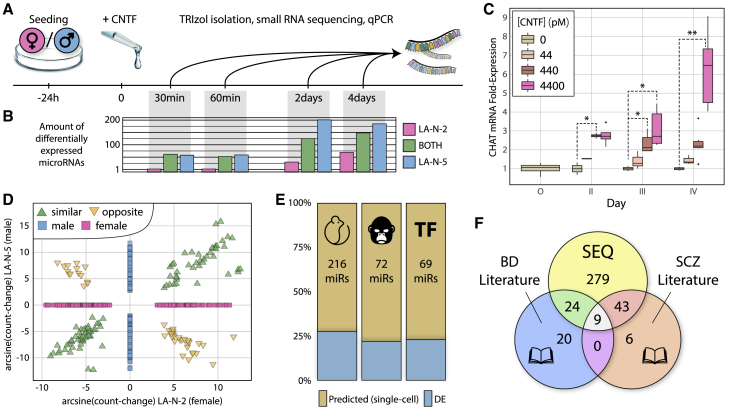
Identifying sexually dimorphic miRNAs of cholinergic relevance in homogenized patient brain samples is challenging because of their tissue-specific and low-level expression (Liu et al., 2016). As an alternative, we used immortalized cell lines of male and female human neuronal origin that undergo cholinergic differentiation when subjected to neurokine stimulation (McManaman and Crawford, 1991). Briefly, we exposed the female cell line LA-N-2 and the male cell line LA-N-5 to CNTF and used short RNA sequencing (GSE132951, 4 biological replicates) to identify the affected miRNAs following exposure to this neurokine (Figure 4A). Both cell lines exhibited an immediate response of miRNA expression as early as 30 min after CNTF onset, with increasing numbers of differentially expressed (DE) miRNAs under prolonged CNTF exposure (Figure 4B; Data S3). In total, we detected 490 DE mature miRNAs: 107 in LA-N-2, 269 in LA-N-5, and 114 in both. However, the frequently used log-fold change metric is not ideally suited for assessing the potential effect of expression changes for individual miRNAs because it does not reflect mean expression levels. To determine the change in expression, we introduced the count-change metric, a combination of base mean expression and log-fold change, to weigh DE miRNAs against one another. Notably, the male-originated LA-N-5 cells responded more strongly to the neurokine stimulus, showing more DE miRNAs with a trend toward higher count-change values (mean of absolute count-change across all DE time points, 20,907 versus 3,066, p = 0.08).
Figure 4.
Cholinergic Response to CNTF Induces Partially Overlapping miRNA Changes in Female- and Male-Originating LA-N-2 and LA-N-5 Cells
(A) Timeline of CNTF differentiation of cells.
(B) Bar graph of miRNAs differentially expressed (DE) during exposure of LA-N-2 cells to 4.4 nM CNTF (pink), LA-N-5 cells to 440 pM CNTF (blue), or in both (green).
(C) Time-dose boxplot of CHAT mRNA expression in LA-N-2 cells during differentiation with CNTF. For LA-N-5 cells, see Figure S2. ∗p < 0.05, ∗∗p < 0.001. Error bars indicate SD.
(D) DE miRNAs overlap between LA-N-2 and LA-N-5 cells. miRNAs DE in same as well as opposite directions show highly correlated count-changes (arcsine transformation was used because negative values prohibit log-fold display).
(E) Neurokine-induced DE miRNAs partly overlap conserved, primate-specific, and TF-targeting miRNAs predicted via single-cell analysis (Figure 3E).
(F) DE miRNA precursors overlap with known BD- and SCZ-relevant miRNA precursors (Beveridge and Cairns, 2012, Fries et al., 2018).
Pico- to nanomolar concentrations of CNTF, well within its physiological range (Sun et al., 2016), elevated the cholinergic marker genes CHAT (p value/day 3 of experiment [pDay3] = 0.005, pDay4 = 4.1E−05) and SLC18A3 (also known as vAChT, pDay2 = 0.002, pDay4 = 0.001). In female-originating LA-N-2 cells, CHAT elevation was visible at low concentrations and significant at medium concentrations (Figure 4C). Also, the effect of 440 pM CNTF appeared to peak at or before 48 h, whereas 4.4 nM elicited a dramatic long-term response. Conversely, the male-originating LA-N-5 cells reacted most strongly to CNTF at 440 pM, showing an “inverted U”-type dose-response curve (Figure S2). As many as 77.8% of the DE miRNAs in LA-N-2 cells reproduced those of a prior sequencing experiment (GSE120520, same layout, only 3 biological replicates).
Apart from a significant male/female overlap, the DE miRNA profiles showed exclusivities, which might translate to sex-specific regulatory processes in neuronal cholinergic differentiation. The count-change values of the 114 miRNAs detected as DE in both cell lines correlated well, whether the change was observed same (76 miRNAs, Spearman’s rho = 0.9066, p < 2.2E−16) or opposite directionally (38 miRNAs, rho = −0.9294, p < 2.2E−16) (Figure 4D). Neurokine-induced differentiation of LA-N-2 and LA-N-5 cells further induced a subset of the conserved, primate-specific, and TF-targeting miRNAs predicted via single-cell analysis of cortical cholinergic neurons (Figure 4E; see also Figure 3E). Literature query of two curations of miRNA precursors in SCZ (Beveridge and Cairns, 2012) and BD (Fries et al., 2018) revealed 76 DE miRNA precursors to be associated with one or both of the diseases (Figure 4F; Data S4). Mature miRNAs in this case could not be assessed because the curated datasets partly lacked strand information.
Transcriptional Interactions
Male and Female Cells Respond to Cholinergic Differentiation by 5 Shared and 12 Diverging miRNA Families
miRBase (http://www.mirbase.org) currently features 151 human miRNA families (designated “mir” with a lowercase “r”) based on homology; we found members of 71 families DE in LA-N-2 and LA-N-5 cells. Gene set enrichment revealed 5 families that were sex-independently enriched in both cell lines, with highest counts of individual members in the families let-7 (Fisher’s exact test, p = 1.6E−08) and mir-30 (p = 0.015); 12 families were only enriched in one of the two lines, with highest counts in the families mir-515 (p = 2.9E−06 in LA-N-2 cells) and mir-154 (p = 3.5E–12 in LA-N-5 cells) (Figure 5A). Of all miRNA families identified in this analysis, five have been associated previously with SCZ (both sexes, let-7 and mir-27; male, mir-181 and mir-199; female, mir-10) and three with BD and SCZ (both sexes, mir-30; male, mir-154; female, mir-17).
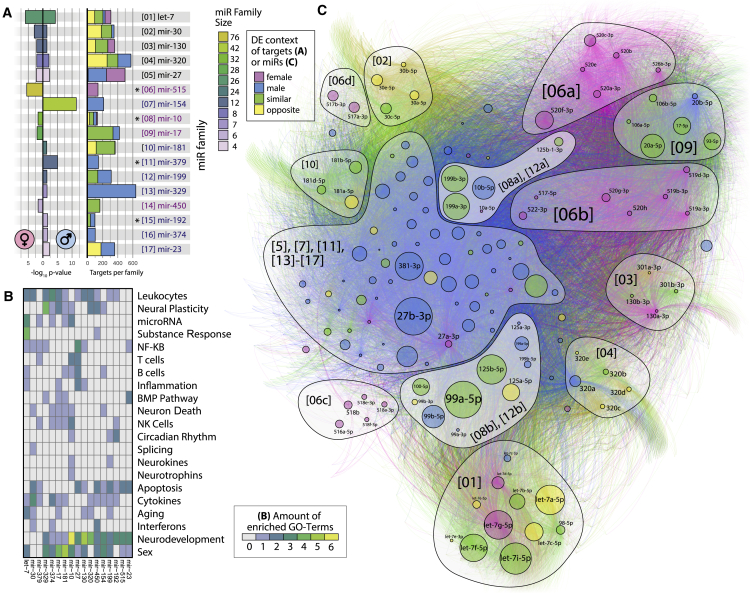
Figure 5.
Differentially Expressed miRNA Families Enriched in LA-N-2 and LA-N-5 Cells Self-Organize in a Comprehensive Targeting Network
(A) The p values of 17 enriched families (left) in LA-N-2 and LA-N-5 cells (female/male symbols) and mean predicted gene target count of each family (right). Color denotes family size (left) or contribution of sex to total target numbers (right), as determined via the DE context of individual family members. Asterisks indicate families with significantly smaller gene target sets (see text).
(B) GO topics curated for enriched families. Frequent terms are immune-, neurodevelopment-, or sex-related. The mir-10 and mir-199 families show rare association with neurokine signaling and circadian rhythm.
(C) The comprehensive network of all DE members of enriched families targeting 12,495 genes self-separates into family-dependent clusters by application of a force-directed algorithm (46,937 unique interactions). miRNA node size denotes absolute count-change, and color denotes DE context. Numbers in brackets correspond to (A). The mir-10 and mir-199 families form two distinct, sexually dimorphic clusters near the center of the network (lighter background color).
Considering the mechanism of action of miRNAs (i.e., their multiple-targeting behavior), it is of interest how broadly these families act on gene targets. Seeking the per-family mean target count (via miRNet), we found that families enriched in only male or female cells had significantly fewer targets than those enriched in both cells (217 versus 378, Welch two-sample t test, p = 0.001). Relative to their size, 4 families showed significantly lower target numbers than all other families: mir-10 (p = 0.016), mir-192 (p = 0.042), mir-379 (p = 0.011), and mir-515 (p < 0.001) (Figure 5A, right). This might indicate a spectrum of functional categories from broadly acting families such as let-7 with sex-independent function to families with a narrow target profile, such as mir-10, whose restricted function can associate with sex-specific effects.
Enriched Families Regulate Genes Involved in Immunity, Neuronal Development, and Sex
To assess their putative functions, we performed GO enrichment analyses of the 300 most-targeted genes in each of the enriched miRNA families. This revealed involvement in 1,124 biological processes whose manual curation enabled us to infer the functional roles of each family (Figure 5B; Data S5). The resulting terms related to neuronal development, cytokine- and leukocyte-mediated immune processes, and sexual dimorphism. Compatible with the hypothesis of “broad to narrow” functioning families, we also found terms with lower incidence and higher specialization in subgroups of families.
Differentially Expressed Families Self-Organize in a Force-Directed Network
Unbiased graphical network analysis of the 212 DE miRNAs from all enriched families and their 12,495 targeted genes (via miRNet) yielded a complex interactome. A force-directed algorithm (Jacomy et al., 2014) yielded apparent clustering and, in some cases, subdivisions of families (Figure 5C). Two main clusters emerged for male and female DE miRNAs from families mir-515 and mir-17 (female) and families mir-154, mir-379, mir-329, mir-129, mir-374, and mir-23 (male). Most other families showed marginal localization, distributed around the network’s edge. Three small families showed high centrality and high count-change: mir-27, targeting particularly many genes, and the neurokine-associated mir-10 and mir-199, with a lower target count.
The mir-10 and mir-199 Families Show Sexual Dimorphism of Expression and Ontological Association with Neurokine and Circadian Mechanisms
Single members of the mir-10 and mir-199 families were detected in each of the DE categories: female only, male only, and same as well as opposite directionally. In the comprehensive network, mir-10 and mir-199 form two distinct clusters (“[08a], [12a]” and “[08b], [12b]”) composed of opposite strands of their respective miRNA precursors. Ontologically, they associate with neurokine and circadian genes. Further, 125a/b-5p (members of mir-10) and 199a/b-5p belong to miRNAs predicted to target genes expressed in single cholinergic neurons (Figures 3E and 4E).
Further analysis requires a closer look into subnetworks (e.g., of individual families), which are astoundingly heterogeneous in size and layout and in their individual sexual dimorphism; a comprehensive assessment is outside of the scope of this study. Fully interactive versions of all individual enriched family networks and further information can be accessed at https://slobentanzer.github.io/cholinergic-neurokine; the entire network in tabular format is provided as a resource (Data S6). Below, we describe the cholinergic/neurokine subnetwork.
The Cholinergic/Neurokine Interface
Creation of a Cholinergic/Neurokine Sub-connectome by Gene and miRNA Filtering
To exemplify the proposed complexity reduction technique, we looked for the common denominator enclosing all aspects of this study; a limited connectome analysis requires a defined set of genes and miRNAs. In the beginning, we performed an unbiased analysis of sexual dimorphism in SCZ and BD, which implicated processes of neuronal, immunological, and circadian origin (Figure 1). Because our experimental data are based on cholinergic processes, we compiled a list of relevant cholinergic genes (Soreq, 2015), adding to it genes from pathways that had emerged in the previous analyses: neurokine signaling and circadian rhythm. Returning to the collection of web-available patient data, we subjected this limited set of 76 genes and their 18 neuronal TFs to differential expression analysis (Data S7).
The miRNAs were gradually filtered by multiple consecutive steps. (1) Permutation analysis of comprehensive miRNA targeting data specific for genes expressed in cholinergic neurons (Figure 3) yielded a list of miRNA candidates that shows overlap with (2) miRNAs DE in our two models of neurokine-induced cholinergic differentiation (Figure 4). (3) We included only families of miRNAs we found to be enriched in differential expression (Figure 5). This filtering process yielded 69 miRNAs from 12 families (Data S8), which were assembled in a force-directed network with the 94 genes of the previously compiled list; as a “spike-in,” we added miR-132-3p (DE in LA-N-5 cells), a well-studied miRNA known to influence cholinergic processes (Shaked et al., 2009, Shaltiel et al., 2013, Hanin et al., 2018). The resulting network (Figure 6A) shows high structural homology to the comprehensive network shown in Figure 5C, with similar groupings and spatial organization of families.
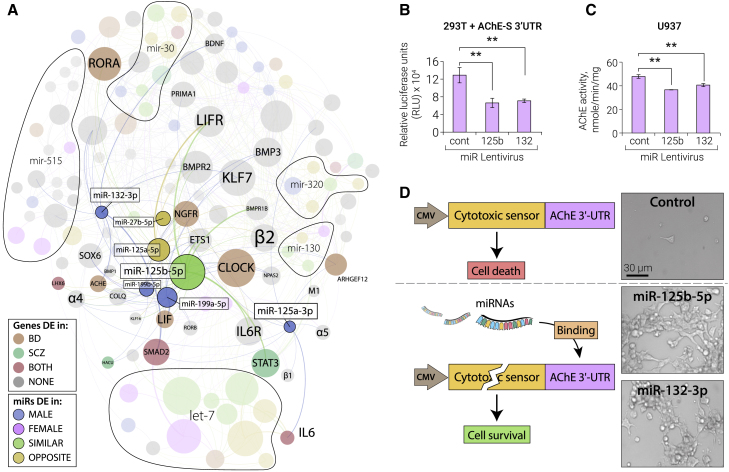
Figure 6.
The Cholinergic/Neurokine Interface and Experimental Validation of AChE Targeting by hsa-miR-125b
(A) The miRNA families mir-10 and mir-199 pose a sexually dimorphic interface of cholinergic, neurokine, and circadian regulation by targeting nicotinic/muscarinic (e.g., α4β2 and M1) and neurokine receptors, transcriptional regulators of cholinergic differentiation (LHX and STAT) and circadian rhythm (CLOCK and RORA), the AChE and the AChE linker proteins PRIMA1/COLQ, and high-affinity choline uptake (HACU). Members of mir-10/199 families, spike-in miR-132-3p, and their targeted genes are shown in color, and other miRNA families that passed the multiple filtering are indicated as areas. miRNA node size corresponds to count-change and gene node size to connectivity; color and thicker edges indicate the DE context and experimentally validated connections.
(B–D) Validation experiments of AChE targeting by miR-125b-5p, with miR-132-3p as a positive control.
(B) Lentiviral expression of miR-132 and miR-125b suppresses luciferase fused to the 3′ UTR of AChE in HEK293T cells. Error bars indicate SE.
(C) Lentiviral expression of miR-132 and miR-125b suppresses the endogenous AChE hydrolytic activity of U937 cells with similar efficacy. Error bars indicate SE.
(D) Life/death assay of stably transfected HEK293T cells carrying the AChE 3′ UTR fused to a cytotoxic sensor and co-transfected with miR-125b-5p, miR-132-3p, or control plasmids. Cells survive in case of binding of miR-132-3p and miR-125-5p to the 3′ UTR.
mir-10/199 Family Members Are Pivotal Factors in the Cholinergic/Neurokine Interface
Of the 23 genes we found to be perturbed in the re-analyzed SCZ/BD patient data, miR-125a-5p and miR-125b-5p targeted 9 genes each, followed by let-7a-3p, let-7f-1-3p, miR-199a-5p, miR-199b-5p, and miR-30a-5p (targeting 7 genes each). In this sub-connectome, the most-targeted DE genes (as indicated by node size) are the circadian regulators CLOCK and RORA and the neurokine pathway genes LIFR and STAT3. Members of mir-10/199 are intricately involved in control of all of these factors, as indicated by their closeness and central location.
The mir-10 Family Member miR-125b-5p Is Highly Differentially Expressed in LA-N-2 and LA-N-5 Cells and Targets AChE
Of all miRNAs in the reduced set, hsa-miR-125b-5p exhibits the highest absolute count-change and displays most experimentally validated targeting relationships with cholinergic/neurokine genes, targeting several inflammation-related neurokine pathway genes (miRTarBase accessions: IL-6, MIRT022105; IL-6R, MIRT006844; JAK2, MIRT734987; LIF, MIRT001037; LIFR, MIRT732494; STAT3, MIRT005006) and other inflammatory pathways (e.g., tumor necrosis factor [TNF], MIRT733472; IRF4, MIRT004534). Additionally, hsa-miR-125b-5p harbors a seed sequence predicting both subunits of the nicotinic α4β2 receptor and AChE as targets.
To experimentally test the capacity of hsa-miR-125b-5p to affect cholinergic signaling, we performed a cell culture luciferase assay and cell death and endogenous enzyme suppression tests on the secreted AChE protein (Figures 6B–6D). This analysis revealed functional suppression of AChE by miR-125b-5p with similar efficacy as that of the positive control miR-132-3p (Shaked et al., 2009, Hanin et al., 2018). Lentiviral infection with miR-132 and miR-125b led to equally efficient suppression of luciferase activity in HEK293T cells stably transfected with a plasmid expressing luciferase fused to the AChE mRNA 3′ UTR (one-way ANOVA, p = 0.004; Figure 6B). Further, lentiviral infection of human monocyte-like U937 cells with miR-125b-5p suppressed endogenous AChE hydrolytic activity (one-way ANOVA, p = 0.005; Figure 6C), and co-transfection of miR-125b-5p with a lentiviral vector expressing a cytotoxic sensor fused to the AChE 3′ UTR resulted in cell survival with a similar efficacy as that of the positive miR-132-3p control in HEK293T cells (n = 3; Figure 6D). In addition to the already experimentally validated interactions, this makes hsa-miR-125b-5p a prime candidate for cholinergic/neurokine mediation.
Discussion
The magnitude of interactions shown by network analyses, further complicated by high between-tissue variability, presents one of today’s largest obstacles in microRNA (miRNA) research. Because miRNAs exert individual effects of low impact, the outcome of their cooperative action can be more adequately represented by a network approach than by traditional molecular interaction studies (Salta and De Strooper, 2017). Although experimental validations are necessary, they often significantly exceed the scope of scientific publications, even for one single miRNA. On the other hand, purely bioinformatical interaction studies lack predictive power, particularly for non-canonical miRNA targeting, with high false positive and false negative rates (Hart et al., 2018). Bioinformatically supported high-throughput techniques such as short RNA sequencing can serve as an integrative middle ground. Our study addresses this purpose by facilitating identification of manageable numbers of interaction partners for deeper analyses of subnetworks involved in specific ontological categories of coding genes. We chose the cholinergic/neurokine interface as an example because of our interest in cholinergic transmission, which is intrinsically linked to many processes relevant in SCZ/BD (Figure 2). Co-expression of cholinergic and neurokine markers in single cortical cells (Figure 3), the pro-cholinergic influence of neurokines on our model cell system (Figure 4), and identification of neurokine signaling and circadian rhythm in GO enrichment analysis of specific miRNA families (Figure 5) highlight the relevance of this choice to our current study.
Limitations
We chose a human cellular model over in vivo experimentation for several reasons. First, implementation of cholinergic differentiation in the brain of a living animal is not straightforward, and individual types of cholinergic neurons are quantitatively inferior to supporting cells in the cortex (von Engelhardt et al., 2007). Second, a recent study (Naqvi et al., 2019) demonstrates that most sex bias in gene expression has arisen since the last common ancestor of boroeutherian mammals (including mouse, rat, dog, macaque, and human), resulting in inadequate representation of sexual dimorphism in any non-human model organism. Third, current animal models of SCZ and BD do not faithfully represent human pathology, showing no predictive power of the clinical efficacy of therapeutic agents (Jones et al., 2011). Additionally, the resolution of differential expression analysis in sequencing is much higher in a homogeneous cell population such as neuronal cell culture because sexual dimorphism can also manifest in distinct tissue composition (Naqvi et al., 2019). However, this choice also entails limitations. Although very similar, LA-N-2 and LA-N-5 cells are immortalized cells derived from two distinct donor individuals, which must be considered when interpreting sexual dimorphisms based on their total transcriptional divergence. Specifically, the stronger response of LA-N-5 cells to CNTF might have influenced the detection limit of miRNAs designated as male by their lack of detection in LA-N-2 cells, partly leading to the dominant male cluster in the comprehensive network. On the other hand, this increases the power of detection of female-designated miRNAs (e.g., mir-515) because they were not detected in LA-N-5 cells in spite of higher sensitivity.
Notwithstanding these limitations, we consider our approach a viable alternative in situations that require a sophisticated perspective on sexual dimorphisms, particularly in diseases where representative animal models are not available. In the present study, we defined ontological categories implicated in SCZ/BD dimorphisms via meta-analysis of deposited patient data, ascertained co-expression of our principal systems of interest in the context of single cortical cells, assessed differential expression via suitable cellular models, and comprehensively analyzed the transcriptional interactions of identified miRNAs in their genome-scale regulatory network. We used and created comprehensive web-available resources that can aid focused studies by generating hypotheses and candidate lists or by putting experimental results into context. We advocate application of this integrative methodology to make use of the many excellent data collections created by modern science.
The Cholinergic/Neurokine Interface
Our approach identified a trophic role for neurokine signaling in cortical cholinergic systems, particularly for IL-6, distinguishing these cortical neurons from the NGF-dependent basal forebrain population. The consistent identification of inflammatory processes across all aspects of our integrative study protocol lends further support to the neuroinflammatory aspect of SCZ/BD; for example, via IL-6-mediated temporary overstimulation (Lurie, 2018). Basal neurokine levels in the brain are comparatively low, and current methods of measurement do not allow analysis of short-term fluctuations in living human subjects. However, it has been shown that CNTF can influence behavior through its effect on cholinergic neurons in the arcuate nucleus (Couvreur et al., 2012). Paracrine control of cholinergic neurons by neurokine-affected supporting cells such as glia or astrocytes is also physiologically feasible, pending further analyses.
Compatible with our previous findings (Cohen et al., 2002), we found a paramount role of CLOCK in the mir-10/mir-199 regulatory network, indicating circadian regulation of neurokine control over cholinergic signaling. In RNA sequencing of 600 prefrontal cortices of SCZ patients and controls, IL6ST (also known as gp130) and the host gene for the IL6ST-targeting hsa-miR-335 were among the top 50 imprinted genes (Gulyas-Kovacs et al., 2018). Although the data used in that study yielded surprisingly few DE genes, they revealed significant depression of the CNTFR and cholinergic receptors, accompanied by an elevation of CLOCK and RORA. Conversely, knockdown of CLOCK in in vitro-differentiated human neurons (Fontenot et al., 2017) caused parallel neurokine receptor and cholinergic transcript perturbations, suggesting an intrinsic relationship of these systems.
miRNA Families
Our tests also implicated several previously uncharacterized factors pivotal for control of cholinergic function in BD and SCZ pathology. Recently, SCZ and BD have come to be recognized as instances of a spectrum of transcriptional perturbations with increasing “transcriptomic severity” from BD to SCZ (Gandal et al., 2018). Our approach identified 3 miRNA families associated with both diseases and 5 associated with SCZ, but none were associated with BD alone, retracing the aspect of increasing severity from a non-coding perspective. The closely related mir-10 and mir-199 families are intrinsically sexually dimorphic, have been associated previously with SCZ (Beveridge and Cairns, 2012, Szatkiewicz et al., 2014), and show a comparatively small number of gene targets, predicting a focused regulatory role. Although individual members of large families such as let-7 or mir-515 often show distinct individual target profiles, mir-10 family members demonstrate overlapping functional features of their targets, even between their −5p and −3p variants. The evolutionarily conserved miR-132-3p, through its suppression of AChE, affects the cholinergic modulation of acute stress (Shaltiel et al., 2013), the immune response (Shaked et al., 2009), and metabolic activities (Hanin et al., 2018) and leads to a transient increase in IL-6, which can be mitigated by nicotine (Shaked et al., 2009). However, miR-125b-5p, but not miR-132-3p, may also target the α4β2 nicotinic receptor subunits so that its modulation could have an additional effect. Additionally, miR-125b-5p can influence inflammatory processes directly by targeting 5-lipoxygenase (Busch et al., 2015) and indirectly via regulation of epigenetic controllers (Zhang et al., 2017). Moreover, miR-125b-5p is directly induced by the vitamin D receptor (Giangreco et al., 2013), and decreased vitamin D levels are thought to be a risk factor for SCZ/BD development (Cieslak et al., 2014). Hence, miR-125b-5p possesses several cholinergic and non-cholinergic features potentially relevant to SCZ/BD pathology.
Our experiments also indicate other miRNAs as potential mediators of sexually dimorphic cholinergic, neurokine, and circadian functions of human neurons. For example, miR-124 is DE between LA-N-2 and LA-N-5 cells after neurokine induction. A recent analysis of complete miR-124 knockout in human induced pluripotent stem cells (iPSCs) (Kutsche et al., 2018) found extensive subsequent transcriptional perturbations, in STAT5B among others, and a shift from glutamate to ACh in the resulting functional neurons. Although the authors did not address this cholinergic aspect, their data and ours co-indicate an influence of miR-124 on the neuronal cholinergic phenotype.
Therapy
At present, treating cognitive malfunctioning through stimulation of the cholinergic system is limited to AChE inhibitors and nicotinic/muscarinic agonists (Rowe et al., 2015). Stimulation of nicotinic (e.g., α4β2, the α5-subunit, and α7; Koukouli et al., 2017) and muscarinic (e.g., M1) receptors (Vijayraghavan et al., 2018) displays a therapy-limiting lack of specificity for individual receptors/subunits. On the other hand, neurokine-based interventions have repeatedly failed because intolerable side effects, even when applied directly to the brain (Mufson et al., 2009). In comparison, antisense oligonucleotide therapeutic agents can simultaneously target multiple disease-relevant genes, be it as mimetic/antagonist of an existing miRNA or as a synthetic oligonucleotide aptamer engineered for a specific target profile. The success of oligonucleotide therapy will depend on calibrating its effect on target genes and achieving a positive balance between target and off-target effects (Greenberg and Soreq, 2014). Therapy with miRNA-like molecules can also potentially ameliorate tachyphylactic effects by targeting a gene and its regulatory elements at the same time (essentially via feedforward loops; Guzzi et al., 2015), as implicated by the nested regulatory circuits we observed. Identification of lead molecules for development of therapeutics with defined profiles largely depends on an integrative approach combining gene and TF targeting.
Recent clinical advances in personalized medicine involve evaluating the benefit of sex-specific therapeutic adjustments in cholinergic medication, at least for Alzheimer’s disease (reviewed in Giacobini and Pepeu, 2018), but molecular examination of cholinergic systems (e.g., with respect to co-expression of sex hormone receptors) has so far been limited to the basal forebrain Ch1–Ch4 nuclei. Although some of the datasets covered in the present study allow sex-specific analyses, many questions concerning sexual dimorphisms remain unaddressed and warrant extensive future studies. The advent of single-cell sequencing will hopefully enable application of this methodology to actual patient tissues. More immediately, our study paves the way for investigation of other processes by application of our method to other ontological categories; e.g., dopaminergic signaling or innate immunity. High-quality datasets large enough to allow statistical analyses can further enable extension of this approach to different brain regions and other psychiatric and non-psychiatric polygenic disorders, such as sporadic Alzheimer’s and Parkinson’s diseases.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Lenti-miR-132-3p | in-house | N/A |
| Lenti-scr | in-house | N/A |
| Lenti-miR-125b-5p | in-house | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| tetraisopropyl pyrophosphoramide | Sigma-Aldrich | T1505 |
| Acetylthiocholine iodide | Sigma-Aldrich | A5751 |
| ciliary neurotrophic factor | Sigma-Aldrich (Merck) | Cat#C3710 |
| TRIzol | ThermoFisher Scientific | Cat#15596018 |
| Critical Commercial Assays | ||
| Dual Luciferase Assay kit | Promega, WI USA | E1910 |
| NEBNext Multiplex Small RNA Library Prep Set for Illumina | New England BioLabs | Cat#E7330 |
| Deposited Data | ||
| Short RNA sequencing of LA-N-2 and LA-N-5 | NCBI GEO | GSE132951 |
| Short RNA sequencing of LA-N-2 | NCBI GEO | GSE120520 |
| Experimental Models: Cell Lines | ||
| U937 | ATCC | CRL-1593.2; RRID:CVCL_0007 |
| HEK293T | ATCC | CRL-3216; RRID:CVCL_0063 |
| LA-N-2 | DSMZ | Cat# ACC-671; RRID:CVCL_1829 |
| LA-N-5 | DSMZ | Cat# ACC-673; RRID:CVCL_0389 |
| Oligonucleotides | ||
| vAChT primer FW+RV | Biorad | PrimePCR “qHsaCED0047922” |
| RPLP0 primer FW+RV | Biorad | PrimePCR “qHsaCED0038653” |
| Custom ChAT primer FW | ThermoFisher Scientific | N/A |
| Custom ChAT primer RV | ThermoFisher Scientific | N/A |
| Custom ACTB primer FW | ThermoFisher Scientific | N/A |
| Custom ACTB primer RV | ThermoFisher Scientific | N/A |
| Software and Algorithms | ||
| R version 3.5.3 | https://www.r-project.org | N/A |
| miRExpress 2.1.4 | http://mirexpress.mbc.nctu.edu.tw/index.php | N/A |
| DESeq2 | http://www.bioconductor.org | N/A |
| Neo4j v3 | https://.neo4j.com/ | N/A |
| limma | http://www.bioconductor.org | N/A |
| circos | http://www.circos.ca | N/A |
| topGO | http://www.bioconductor.org | N/A |
| Other | ||
| Hsa-miR-132-3p plasmid | GeneCopoeia | MmiR3274-MR03 |
| Negative control plasmid (scrambled control for pEZX-MR03) | GeneCopoeia | CmiR0001-MR03 |
| microRNA Target Selection System plasmid | System Biosciences, CA, USA | #MS040-4xxA-1 |
| EcoRI restriction enzyme | New England Biolabs | R0101 |
| NotI restriction enzyme | New England Biolabs | R3189 |
| plate reader | Tecan | GENios Pro Microplate Reader |
| CFX96 real-time PCR cycler | Biorad | Cat#1855196 |
| NextSeq 550 | Illumina | Cat#SY-415-1002 |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hermona Soreq (hermona.soreq@mail.huji.ac.il).
This study did not generate new unique reagents.
Experimental Model and Subject Details
Cell Lines
We employed LA-N-2 (female) (DSMZ Cat# ACC-671, RRID:CVCL_1829) and LA-N-5 (male) (DSMZ Cat# ACC-673, RRID:CVCL_0389) cells as our main experimental model system (McManaman and Crawford, 1991). The cells were purchased at DSMZ (Braunschweig, Germany). These cells respond to differentiation by several neurokines (CNTF, LIF, IL-6) by cholinergic differentiation corresponding with elevation of choline acetyltransferase (ChAT), the central cholinergic marker (mRNA, protein, and activity) as well as its intronic vesicular ACh-transporter gene, vAChT (aka SLC18A3). Cells were maintained at 37°C in 8% CO2 atmosphere in medium consisting of 1:1 DMEM and RPMI 1640, with 20% FCS, with weekly splits. Experiments were performed between splits 2-6 after thawing.
HEK293T and U937 cells were purchased at ATCC and maintained according to ATCC guidelines. Experiments were performed between splits 2-10.
Human Patient Data
We used previously published datasets for several analyses. These comprise several cortical datasets in raw format (Affymetrix) from NCBI GEO: GSE35978 (Chen et al., 2013) (SCZ & BD), GSE53987 (Iwamoto et al., 2005) (SCZ & BD), GSE12649 (Lanz et al., 2015) (SCZ & BD), GSE17612 (Maycox et al., 2009) (SCZ), GSE21138 (Narayan et al., 2008) (SCZ), GSE5392 (Ryan et al., 2006) (BD); next-generation sequencing from NCBI GEO: GSE80655 (Ramaker et al., 2017), GSE106589 (Hoffman et al., 2017), GSE68559 (Webb et al., 2015), GSE96659 (Fontenot et al., 2017), GSE45642 (Li et al., 2013); data of DLPFC sequencing of 600 SCZ patients and controls was obtained from the Common Mind Consortium (http://www.synapse.org/CMC).
Method Details
Methodological Outline
Focusing on a well-defined set of genes aims to avoid some of the data loss that occurs when patient tissues comprising multiple cell types are homogenized for analysis. This is particularly relevant for cortical cholinergic interneurons, where the cell type of interest is numerically inferior, and transcriptomic data can be “diluted” by the other, more prominent cell types, such as glia or astrocytes. The consequent increase in detection threshold can be re-lowered by reducing the number of genes tested. Analysis of single cell datasets can ascertain that the genes analyzed are representative and fairly unique to the cell type in question, as was the case for cholinergic markers in our study, and utmost care should be taken for any gene set of interest and any kind of tissue subjected to this kind of analysis.
Our approach is based on the availability of suitable amounts of patient data in web-available form, combined with a standardized pipeline of statistical pre-processing to equilibrate individual statistical influences. Regardless of how patient data and the subset of genes of interest are selected, the reduction in number of analyzed genes has to be performed after application of the linear model to the batch- and covariate-corrected data to avoid interference with correct linear regression. Once the expression data (be it array- or sequencing-derived) has been corrected for batch effects and covariates, and outliers have been removed, it can be analyzed with a suitable differential gene expression algorithm. The resulting, individual datasets should ideally converge on similar logFC values, but may also show controversy between individual experiments, which can result from a multitude of factors from biological variety to sampling procedures or exact tissue composition, all of which have to be interpreted at the discretion of the scientist.
Neuronal Differentiation/Short RNA Sequencing
Model
LA-N-2 and LA-N-5 cells respond to ciliary neurotrophic factor (CNTF) by cholinergic differentiation corresponding with elevation of choline acetyltransferase (ChAT), the central cholinergic marker (mRNA, protein, and activity) as well as its intronic vesicular ACh-transporter gene, vAChT (aka SLC18A3). We measured the changes in short RNA levels following this intervention at several time points.
Differentiation
To determine effective concentrations of the differentiation agent, we performed a dose-response experiment with both cell lines. Cells were seeded at approximately 200 000 cells per well in 12-well plates, and after 24h incubated with 1, 10, or 100 ng/ml CNTF (Sigma-Aldrich). Dose-response was measured by qPCR of CHAT mRNA, at time points 30 minutes, 60 minutes, 2 days, and 4 days. For each sample, a corresponding control culture was generated.
RNA Extraction for qPCR and Sequencing
RNA was extracted in biological quadruplicates using TRIzol according to the manufacturer′s instructions, as described in Hanin et al. (2018). RNA was precipitated using ethanol, washed, and air-dried before resuspension in RNase-free water. Concentration was measured by Nanodrop 2000 (ThermoFisher Scientific), RNA quality was measured via Bioanalyzer 2100 (Agilent). RNA quality for all samples was near optimal (RIN > 9).
Quantitative Real-time PCR
RNA was analyzed on a BioRad CFX96 real-time PCR cycler using PowerUp SYBR Green Master Mix (Applied Biosystems) in technical duplicates. Primers were designed using primer3 and are as follows (5′ to 3′): ChAT [FW: CAC TTG GTG TCT GAG CA, RV: AGT TTC TGC TGC AGG GTC TC], ACTB (housekeeping) [FW: GCT GTA TTC CCC TCC ATC GT, RV: CTT CTC CAT GTC GTC CCA GT]; additional primers were ordered from BioRad, Germany: vAChT [PrimePCR “qHsaCED0047922”], RPLP0 (housekeeping) [PrimePCR “qHsaCED0038653”]. Data were analyzed with BioRad CFX manager and expression values (normalized to housekeeping genes) exported for statistical testing in R.
Short RNA Sequencing
Short RNA sequencing was performed using Illumina NextSeq 550 according to the manufacturer’s instructions, after cDNA library preparation using the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England BioLabs) as described (Bekenstein et al., 2017). Sequenced reads were aligned to miRBase v21 sequences via miRExpress (Wang et al., 2009) (version 2.1.4). Differential expression was determined via R/DESeq2 (Love et al., 2014).
The Count-Change Metric
We calculated the count-change for individual miRs by combining base mean expression with the de-logarithmized fold-change (from DESeq2 output).
CC: countChange, BM: baseMean, LFC: log2-fold change
It is important to note that the count-change metric, by deriving from the base mean expression across samples, is dependent on sequencing depth, and thus is not instantly generalizable, for instance when comparing different experiments. However, it could be normalized to a degree by considering the total amount of raw reads generated from each sample.
Whole-Transcriptome Meta-analysis
Data Preparation
We processed web-available patient transcriptome datasets by state-of-the-art procedures, analogous to Gandal et al. (2018), to generate transcriptional disease profiles. The following R packages (Bioconductor) were used according to the developer’s instructions: Raw data read, RMA normalization, RNA degradation: affy (Gautier et al., 2004). Batch correction (as per chip scan date) involved: sva (Combat) (Johnson et al., 2007). Outliers were removed as described in Gandal et al. (2018). The array probes were annotated via ENSEMBL gene ID, database version v75, to ensure congruency with prior analyses, using biomaRt (Durinck et al., 2009), and were collapsed via the collapseRows function of WGCNA (Langfelder and Horvath, 2008). Prior to application of the generalized linear mixed model (nlme; Lindstrom and Bates, 1990), datasets were rebalanced and regressed to correct for technical and biological covariate influences.
Regression Analysis
Meta-analysis on all genes present in all datasets (12,391 genes in total) employed a generalized linear mixed model to account for variation per dataset and individual, yielding log2-fold change (logFC) values for each gene between control and disease, which were correlated between SCZ and BD using Spearman’s method (Spearman, 1904). To determine significance, the meta-analysis process was repeated 10,000 times with randomized case/control status, forming a permutation null distribution of the individual correlation coefficients (Figure S3). Of note, several cholinergic and neurokine genes were missing from the whole-genome meta-analysis because of annotation deficits (CHRNA7, CHRM1, LHX8, CHKB, PRIMA1, CNTF), and, at this stage, could not be easily re-introduced.
Gene Ontology (GO) Enrichment Analyses
To focus on the differences, as opposed to the similarities, between BD and SCZ patient brain transcriptomes, we performed GO enrichment analysis on the genes showing the highest rank differences between datasets (Figure S4), using the R package topGO (Alexa et al., 2006). Briefly, we evaluated (smaller) groups of genes for enrichment against a (bigger) background of genes for presence in individual GO terms exceeding statistical estimates. The background comprised of the first 2000 genes according to the applied rank system, computed as a function of Spearman’s rank differences of regression beta values (logFC) between the two compared groups, either as absolute values or “as is” (elevation in one group as opposed to the other). The target genes of the analysis were defined as the 100 top ranked genes (top 5% of background), unless otherwise stated. Statistically significant GO results were compiled and curated for CNS-relevant terms.
Sex Influence on Transcriptomic Differences
Studying web-available datasets of non-degenerative mental disease patients revealed that SCZ and BD datasets possessed sufficient numbers to allow sex-discriminative meta-analysis of statistical significance. Hence, we repeated the above steps for the individual subsets of male and female patients, with the sole change of eliminating the covariate regression for sex, as this would preclude further analysis of this variable, in 4 distinct GO enrichment group comparisons between SCZ-biased, BD-biased, male-biased, and female-biased genes.
miR-gene-TF-targeting: miRNet
To address miR-mRNA targeting relations, we developed an integrated miR-targeting graph database (‘miRNet’) out of publicly available validated and predicted data, implementing a scoring system derived from 10 leading prediction algorithms (Dweep and Gretz, 2015) based on their statistical performance at the whole-genome level. To facilitate the selection process, targeting data were cumulated by summing the amount of positive “hits” of prediction algorithms (1 point) and positive experimental validations (only “strong” evidence, miRTarBase, 10,5 points) of the targeting relationship, yielding a targeting score between 0 and 20,5. We found a minimal score of 6 to suffice for a good balance between type I and II errors. In case of whole-genome interactions, this threshold was raised to 7 to enable computational accessibility in all steps (in this case, graphical analysis was the bottleneck). The database was supplemented by comprehensive transcription factor targeting data via bioinformatically processed “cap analysis of gene expression” (CAGE; Hon et al., 2017), focusing on brain tissues (Marbach et al., 2016), including the tissue-specific transcriptional activities (Figure S5). The code used to create and test this database is available in the accompanying repository. A public release is planned, but not completed at the moment; requests can be directed at the corresponding author.
Analyses performed using this database were implemented in R using the ‘RNeo4j’ package.
Limitations
Through recent methodical and bioinformatical advances, targeting data of transcription factors and miRs has become comprehensive. However, these “complete” datasets are subject to limitations derived from the methods used to accumulate raw data. For TF-targeting, CAGE 5′ peaks were analyzed toward their correlation with gene expression in all available tissues. A cholinergic example of limitations derived from this data involves the CHAT gene, which does not provide a measurable CAGE peak, leading to non-representation in the targeting dataset. In those cases, targeting data acquired through conventional methods have to be substituted, but this cannot be easily done for all affected genes, and is also not comprehensive. In the case of miR-targeting, validated interactions are based on experimental work mainly performed on rodents, leading to a research bias toward evolutionarily conserved miRs. Primate-specific miRs therefore are under-represented, at least in validated data.
Permutation Targeting Analyses
In the current state of comprehensive data on miR-gene (and TF-gene) targeting, no statements can be made with absolute certainty. Thus, an approach which considers relative measures is preferable. For this reason, whenever whole-genome/whole-miRnome targeting was concerned, we employed random permutation of the prediction dataset or each single predicted miR against a randomized background of the same size as the original set (also considering family- and precursor-relationships). The resulting null distribution yields a basis for determination of a false discovery rate.
Neurokine-Induced miRs and the Cholinergic/Neurokine Pathway
Gene targets of the 490 differentially regulated miRs following CNTF exposure were determined by miRNet query (targeting score minimum of 6). The full network, originally comprising ∼160,000 unique relationships, was re-filtered by raising the threshold to score minimum of 7 to be computationally accessible. The resulting network and individual miR-family-subnetworks were plotted using a force-directed layout (Force Atlas 2) in gephi.
Single-Cell Sequencing Dataset Analysis and Permutation
We analyzed 4 web-available datasets of brain single cell gene expression (Darmanis et al., 2015, Zeisel et al., 2015, Habib et al., 2016, Tasic et al., 2016) for neurokine signaling transcripts in cholinergic neurons, identified by their expression of the ACh-synthesizing enzyme choline acetyltransferase (ChAT), and its embedded gene encoding the vesicular acetylcholine transporter SLC18A3, also known as vAChT. Raw gene expression data were normalized and then clustered and plotted via R/pheatmap (Kolde, 2019). The genes expressed in more than one sample per dataset were enriched for targeting of TFs and miRs by random permutation analysis (via miRNet), with 10 000 permutations for conserved and primate miR-target relationships, and human CNS TF interactions.
Subset Analyses
Cholinergic Genes, Transcription Factor, and Neurokine Analyses
To follow our cholinergic interest, we used a recent review (Soreq, 2015) to define a core set of genes, adding to it the neurokine and circadian pathway genes indicated in the previous analyses (to a total of 76 genes). Via miRNet permutation (Data S2), we identified 18 brain-expressed “cholinergic” TFs (p < 0.05) and their CNS transcriptional activity toward each targeted cholinergic gene. These 94 genes (Data S7) were then subjected to differential expression analysis in the deposited patient datasets.
Execution of Subset Analyses
To separately analyze male and female data in the original datasets, we repeated the sex-independent analyses of the identified 94 cholinergic genes and TFs using the limma (Ritchie et al., 2015) pipeline. Pre-processing was identical to the whole-transcriptome approach, and dataset reduction involved restricting the output table (topTable() function) to the studied genes. This further allowed controlling of missing genes of interest by manually solving problems of annotation, which in a whole-genome analysis would have led to loss of information on, e.g., the nicotinic α7 and stress-responding M1 cholinergic receptors.
miR-125b-5p Validation
To test binding of hsa-miR-125b-5p to the acetylcholinesterase (AChE) 3′-UTR, we performed vector-based assays via suppression of a cytotoxic sensor as well as of Renilla luciferase, both fused to the AChE-3′-UTR. Briefly, the 3′ untranslated region (3′-UTR) of human AChE mRNA (Soreq et al., 1990) was cloned into the microRNA Target Selection System plasmid (System Biosciences, CA, USA) multiple cloning site, using EcoRI and NotI restriction enzymes (New England Biolabs). All plasmids were verified by DNA sequencing. For luciferase assays, HEK293T cells were transfected with miRNA Target Selection-AChE-3′UTR, and selected in the presence of Puromycin for 3 weeks. Stably transfected HEK293T (293T-AChE 3′UTR) cells were grown on 12-well plates and infected with lentiviruses expressing miR-125b-5p, miR-132-3p or a negative control sequence. After 48 hours incubation, cells were analyzed using the Dual Luciferase Assay kit (Promega, WI USA) and Luciferase activity was measured using an Envision luminescent plate reader (Perkin-Elmer, Waltham, MA), essentially as previously described (Hanin et al., 2014). For each reporter construct, renilla luciferase activity was normalized according to that of the firefly. Normalized activity after infection of miR-132-3p or miR-125b-5p was expressed as relative to that obtained after infection with the same plasmid with miR negative control. For life/death assay, a similar protocol was used. Stably transfected HEK293T (293T-AChE 3′UTR) cells were infected with lentiviruses expressing miR-125b-5p, miR-132-3p or a negative control sequence. 72 hours post-infection a cytotoxic reporter fused to AChE 3′-UTR was added to the media and cells were kept for an additional 5 days to assess their viability. Statistical significance was determined using ANOVA with correction for multiple testing. To show effects of changes in this miR’s levels on real-life protein activities, we performed an AChE hydrolytic activity assay following infection of human monocyte-like U937 cells with hsa-miR-125b-5p, miR-132-3p or a negative control lentiviral vector. AChE hydrolytic activity levels were assessed by kinetic measurements of the hydrolysis rates of 1 mM acetylthiocholine (ATCh, Sigma) at room temperature, following 20 min incubation with and without 5x10−5 M tetraisopropyl pyrophosphoramide (iso-OMPA, Sigma), a specific inhibitor of butyrylcholinesterase, to selectively assay for AChE-specific or total cholinesterase activity. Each sample was assayed in at least 3 biological replicates. In all cases, hsa-miR-132-3p served as a positive control.
Quantification and Statistical Analysis
Statistical analyses were performed in R. Small sets of continuous variables, such as qPCR and miR-125b-5p validation experiments, were tested using Welch’s Two Sample t test (because in most cases, equal variance could not be assumed; R/t.test) and ANOVA; statistical significance was assumed at p < 0.05. For sequencing count data, the negative binomial generalized linear model was tested using the R/DESeq2 package (“Wald” test including log-fold change shrinkage) to detect differentially expressed miRs, using the supplied independent filtering and correction for multiple testing at an alpha level of 0.1. In cases where additional power was desirable or p values could not be obtained by other means (transcriptome meta-analysis, whole genome miR-targeting), permutation analysis was performed. This comprised random assignment of test variables in the same size as the original test set and repeating the analysis for a large number of times, such that a null distribution of values could be generated, which can be used to determine a false discovery ratio for the original result. Statistical significance was assumed at p < 0.05.
qPCR of ChAT/vAChT mRNA against housekeeping in LA-N-2, p values are found in Results (Welch two-sample t test): Chat, Day 2, 10 ng/ml, t = −3.2436, df = 4.9872; 100 ng/ml, t = −2.349, df = 4.1296; Day 3, 10 ng/ml, t = −2.8481, df = 6.9658; 100 ng/ml, t = −6.3786, df = 3.3998; Day 4, 100 ng/ml, t = −9.0836, df = 6.9835. vAChT, Day 2, t = 5.9222, df = 5.3619; Day 4, t = 7.1016, df = 4.8784. Number of biological replicates: 4.
qPCR of ChAT mRNA against housekeeping in LA-N-5, p values are found in Figure S2 legend (Welch two-sample t test): Day 2, 10 ng/ml, t = −4.5204, df = 3.059; Day 4, 10 ng/ml, t = −4.7639, df = 5.0369; 100 ng/ml, t = −4.9161, df = 2.0262. Number of biological replicates: 4.
Mean absolute count-change LA-N-5 versus LA-N-2, text of Figure 3 (Welch two-sample t test): t = 2.6183, df = 1108. Number of compared miRs: 490.
Target count of families, Figure 5A: enriched in both versus enriched in one cell, t = 3.1831, df = 73; enriched in mir-10 versus enriched in other sex-independent families, t = −3.28, df = 7.1879. Number of compared families: 17 (5 versus 12).
Validation of miR-125b-5p targeting of AChE, p values are found in Results (one-way ANOVA): Figure 6B, n = 4, f-ratio value 9.19882, df between treatments 2, within treatments 9; Figure 6C, control n = 5, 125b n = 4, 132 n = 3, f-ratio value 11.59814, df between treatments 2, within treatments 8. N refers to biological replicates.
Data and Code Availability
The code and additional supplementary materials generated during this study are available at https://github.com/slobentanzer/integrative-transcriptomics.
The sequencing datasets generated during this study are available at NCBI GEO: GSE120520 and GSE132951.
Accompanying website: https://slobentanzer.github.io/cholinergic-neurokine
Acknowledgments
The authors are grateful for in-depth comments by Drs. Naomi Habib (Jerusalem) and Andreas Meisel (Berlin) and grateful to Dr. Ester Bennett (Jerusalem) for assistance with RNA sequencing. The authors acknowledge support of this study by European Research Council advanced award 321501, Israel Science Foundation grant 1016/18, and the Austrian Research Promotion Agency (FFG Bridge 1 Project 378/11) (to H.S.). S.L. received Ph.D. fellowship support from Frankfurt University and travel support to Jerusalem from the Hebrew University Edmond and Lili Safra Center ELSC. G.H. received Ph.D. fellowship support from the Hebrew University.
Author Contributions
Conceptualization, H.S. and S.L.; Methodology, S.L.; Software, S.L.; Investigation, S.L. and G.H.; Resources, J.K. and H.S.; Data Curation, S.L.; Writing – Original Draft, S.L.; Writing – Review and Editing, S.L., J.K., and H.S.; Visualization, S.L.; Supervision, J.K. and H.S.; Project Administration, J.K. and H.S.; Funding Acquisition, J.K. and H.S.
Declaration of Interests
The authors declare no competing interests.
Published: October 15, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.09.017.
Supplemental Information
References
- Alexa A., Rahnenführer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- Anttila V., Bulik-Sullivan B., Finucane H.K., Walters R.K., Bras J., Duncan L., Escott-Price V., Falcone G.J., Gormley P., Malik R. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger E.C., Ananth M., Talmage D.A., Role L.W. Basal Forebrain Cholinergic Circuits and Signaling in Cognition and Cognitive Decline. Neuron. 2016;91:1199–1218. doi: 10.1016/j.neuron.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekenstein U., Mishra N., Milikovsky D.Z., Hanin G., Zelig D., Sheintuch L., Berson A., Greenberg D.S., Friedman A., Soreq H. Dynamic changes in murine forebrain miR-211 expression associate with cholinergic imbalances and epileptiform activity. Proc. Natl. Acad. Sci. USA. 2017;114:E4996–E5005. doi: 10.1073/pnas.1701201114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M. Sixth Edition. Elsevier; 2014. Psychische Erkrankungen: Klinik und Therapie. [Google Scholar]
- Beveridge N.J., Cairns M.J. MicroRNA dysregulation in schizophrenia. Neurobiol. Dis. 2012;46:263–271. doi: 10.1016/j.nbd.2011.12.029. [DOI] [PubMed] [Google Scholar]
- Bina K.G., Rusak B., Semba K. Localization of cholinergic neurons in the forebrain and brainstem that project to the suprachiasmatic nucleus of the hypothalamus in rat. J. Comp. Neurol. 1993;335:295–307. doi: 10.1002/cne.903350212. [DOI] [PubMed] [Google Scholar]
- Boonstra T.W., Stins J.F., Daffertshofer A., Beek P.J. Effects of sleep deprivation on neural functioning: an integrative review. Cell. Mol. Life Sci. 2007;64:934–946. doi: 10.1007/s00018-007-6457-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortolato B., Miskowiak K.W., Köhler C.A., Vieta E., Carvalho A.F. Cognitive dysfunction in bipolar disorder and schizophrenia: a systematic review of meta-analyses. Neuropsychiatr. Dis. Treat. 2015;11:3111–3125. doi: 10.2147/NDT.S76700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S., Auth E., Scholl F., Huenecke S., Koehl U., Suess B., Steinhilber D. 5-lipoxygenase is a direct target of miR-19a-3p and miR-125b-5p. J. Immunol. 2015;194:1646–1653. doi: 10.4049/jimmunol.1402163. [DOI] [PubMed] [Google Scholar]
- Chavan S.S., Pavlov V.A., Tracey K.J. Mechanisms and Therapeutic Relevance of Neuro-immune Communication. Immunity. 2017;46:927–942. doi: 10.1016/j.immuni.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Cheng L., Grennan K., Pibiri F., Zhang C., Badner J.A., Gershon E.S., Liu C., Members of the Bipolar Disorder Genome Study (BiGS) Consortium Two gene co-expression modules differentiate psychotics and controls. Mol. Psychiatry. 2013;18:1308–1314. doi: 10.1038/mp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak K., Feingold J., Antonius D., Walsh-Messinger J., Dracxler R., Rosedale M., Aujero N., Keefe D., Goetz D., Goetz R., Malaspina D. Low vitamin D levels predict clinical features of schizophrenia. Schizophr. Res. 2014;159:543–545. doi: 10.1016/j.schres.2014.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O., Erb C., Ginzberg D., Pollak Y., Seidman S., Shoham S., Yirmiya R., Soreq H. Neuronal overexpression of “readthrough” acetylcholinesterase is associated with antisense-suppressible behavioral impairments. Mol. Psychiatry. 2002;7:874–885. doi: 10.1038/sj.mp.4001103. [DOI] [PubMed] [Google Scholar]
- Couvreur O., Aubourg A., Crépin D., Degrouard J., Gertler A., Taouis M., Vacher C.-M. The anorexigenic cytokine ciliary neurotrophic factor stimulates POMC gene expression via receptors localized in the nucleus of arcuate neurons. Am. J. Physiol. Endocrinol. Metab. 2012;302:E458–E467. doi: 10.1152/ajpendo.00388.2011. [DOI] [PubMed] [Google Scholar]
- Darmanis S., Sloan S.A., Zhang Y., Enge M., Caneda C., Shuer L.M., Hayden Gephart M.G., Barres B.A., Quake S.R. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. USA. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Değirmenci Y., Keçeci H. Visual Hallucinations Due to Rivastigmine Transdermal Patch Application in Alzheimer’s Disease; The First Case Report. Int. J. Gerontol. 2016;10:240–241. [Google Scholar]
- de Leon J., Diaz F.J. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophr. Res. 2005;76:135–157. doi: 10.1016/j.schres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- Erta M., Quintana A., Hidalgo J. Interleukin-6, a major cytokine in the central nervous system. Int. J. Biol. Sci. 2012;8:1254–1266. doi: 10.7150/ijbs.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskow Jaunarajs K.L., Bonsi P., Chesselet M.F., Standaert D.G., Pisani A. Striatal cholinergic dysfunction as a unifying theme in the pathophysiology of dystonia. Prog. Neurobiol. 2015;127–128:91–107. doi: 10.1016/j.pneurobio.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eum S., Hill S.K., Rubin L.H., Carnahan R.M., Reilly J.L., Ivleva E.I., Keedy S.K., Tamminga C.A., Pearlson G.D., Clementz B.A. Cognitive burden of anticholinergic medications in psychotic disorders. Schizophr. Res. 2017;190:129–135. doi: 10.1016/j.schres.2017.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot M.R., Berto S., Liu Y., Werthmann G., Douglas C., Usui N., Gleason K., Tamminga C.A., Takahashi J.S., Konopka G. Novel transcriptional networks regulated by CLOCK in human neurons. Genes Dev. 2017;31:2121–2135. doi: 10.1101/gad.305813.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget B., Scholze P., Langa F., Mourot A., Faure P., Maskos U. A Human Polymorphism in CHRNA5 Is Linked to Relapse to Nicotine Seeking in Transgenic Rats. Curr. Biol. 2018;28:3244–3253.e7. doi: 10.1016/j.cub.2018.08.044. [DOI] [PubMed] [Google Scholar]
- Fries G.R., Carvalho A.F., Quevedo J. The miRNome of bipolar disorder. J. Affect. Disord. 2018;233:110–116. doi: 10.1016/j.jad.2017.09.025. [DOI] [PubMed] [Google Scholar]
- Gandal M.J., Haney J.R., Parikshak N.N., Leppa V., Ramaswami G., Hartl C., Schork A.J., Appadurai V., Buil A., Werge T.M., CommonMind Consortium. PsychENCODE Consortium. iPSYCH-BROAD Working Group Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–697. [Google Scholar]
- Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Giacobini E., Pepeu G. Sex and Gender Differences in the Brain Cholinergic System and in the Response to Therapy of Alzheimer Disease with Cholinesterase Inhibitors. Curr. Alzheimer Res. 2018;15:1077–1084. doi: 10.2174/1567205015666180613111504. [DOI] [PubMed] [Google Scholar]
- Giangreco A.A., Vaishnav A., Wagner D., Finelli A., Fleshner N., Van der Kwast T., Vieth R., Nonn L. Tumor suppressor microRNAs, miR-100 and -125b, are regulated by 1,25-dihydroxyvitamin D in primary prostate cells and in patient tissue. Cancer Prev. Res. (Phila.) 2013;6:483–494. doi: 10.1158/1940-6207.CAPR-12-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales K.K., Smith Y. Cholinergic interneurons in the dorsal and ventral striatum: anatomical and functional considerations in normal and diseased conditions. Ann. NY Acad. Sci. 2015;1349:1–45. doi: 10.1111/nyas.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.L., Anderson M.L., Dublin S., Hanlon J.T., Hubbard R., Walker R., Yu O., Crane P.K., Larson E.B. Cumulative use of strong anticholinergics and incident dementia: a prospective cohort study. JAMA Intern. Med. 2015;175:401–407. doi: 10.1001/jamainternmed.2014.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg D.S., Soreq H. MicroRNA therapeutics in neurological disease. Curr. Pharm. Des. 2014;20:6022–6027. doi: 10.2174/1381612820666140314151924. [DOI] [PubMed] [Google Scholar]
- Gulyas-Kovacs A., Keydar I., Xia E., Fromer M., Hoffman G., Ruderfer D., Consortium C., Sachidanandam R., Chess A. Unperturbed Expression Bias of Imprinted Genes in Schizophrenia. Nat. Commun. 2018;9:2914. doi: 10.1038/s41467-018-04960-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi P.H., Di Martino M.T., Tagliaferri P., Tassone P., Cannataro M. Analysis of miRNA, mRNA, and TF interactions through network-based methods. EURASIP J. Bioinform. Syst. Biol. 2015;2015:4. doi: 10.1186/s13637-015-0023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haam J., Yakel J.L. Cholinergic modulation of the hippocampal region and memory function. J. Neurochem. 2017;142(Suppl 2):111–121. doi: 10.1111/jnc.14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib N., Li Y., Heidenreich M., Swiech L., Avraham-Davidi I., Trombetta J.J., Hession C., Zhang F., Regev A. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science. 2016;353:925–928. doi: 10.1126/science.aad7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin G., Shenhar-Tsarfaty S., Yayon N., Yau Y.H., Bennett E.R., Sklan E.H., Rao D.C., Rankinen T., Bouchard C., Geifman-Shochat S. Competing targets of microRNA-608 affect anxiety and hypertension. Hum. Mol. Genet. 2014;23:4569–4580. doi: 10.1093/hmg/ddu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin G., Yayon N., Tzur Y., Haviv R., Bennett E.R., Udi S., Krishnamoorthy Y.R., Kotsiliti E., Zangen R., Efron B. miRNA-132 induces hepatic steatosis and hyperlipidaemia by synergistic multitarget suppression. Gut. 2018;67:1124–1134. doi: 10.1136/gutjnl-2016-312869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P.J. Recent genetic findings in schizophrenia and their therapeutic relevance. J. Psychopharmacol. 2015;29:85–96. doi: 10.1177/0269881114553647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart M., Kern F., Backes C., Rheinheimer S., Fehlmann T., Keller A., Meese E. The deterministic role of 5-mers in microRNA-gene targeting. RNA Biol. 2018;15:819–825. doi: 10.1080/15476286.2018.1462652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen M.G., Nordgaard J., Jansson L.B. Genetics of Schizophrenia: Overview of Methods, Findings and Limitations. Front. Hum. Neurosci. 2017;11:322. doi: 10.3389/fnhum.2017.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A.M., Ortiz-Guzman J., Kochukov M., Herman I., Quast K.B., Patel J.M., Tepe B., Carlson J.C., Ung K., Selever J. A cholinergic basal forebrain feeding circuit modulates appetite suppression. Nature. 2016;538:253–256. doi: 10.1038/nature19789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley M.J., Picciotto M.R. Neuromodulation by acetylcholine: Examples from schizophrenia and depression. Curr. Opin. Neurobiol. 2014;29:88–95. doi: 10.1016/j.conb.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G.E., Hartley B.J., Flaherty E., Ladran I., Gochman P., Ruderfer D.M., Stahl E.A., Rapoport J., Sklar P., Brennand K.J. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 2017;8:2225. doi: 10.1038/s41467-017-02330-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ising M., Lauer C.J., Holsboer F., Modell S. The Munich vulnerability study on affective disorders: premorbid neuroendocrine profile of affected high-risk probands. J. Psychiatr. Res. 2005;39:21–28. doi: 10.1016/j.jpsychires.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Iwamoto K., Bundo M., Kato T. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum. Mol. Genet. 2005;14:241–253. doi: 10.1093/hmg/ddi022. [DOI] [PubMed] [Google Scholar]
- Jacomy M., Venturini T., Heymann S., Bastian M. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS ONE. 2014;9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jones C.A., Watson D.J., Fone K.C. Animal models of schizophrenia. Br. J. Pharmacol. 2011;164:1162–1194. doi: 10.1111/j.1476-5381.2011.01386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T., Bousman C.A., Liu C., Everall I.P. Schizophrenia genetics in the genome-wide era: a review of Japanese studies. NPJ Schizophr. 2017;3:27. doi: 10.1038/s41537-017-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. 2019. pheatmap: Pretty Heatmaps. R package version 1.0.12.https://CRAN.R-project.org/package=pheatmap [Google Scholar]
- Koukouli F., Rooy M., Tziotis D., Sailor K.A., O’Neill H.C., Levenga J., Witte M., Nilges M., Changeux J.P., Hoeffer C.A. Nicotine reverses hypofrontality in animal models of addiction and schizophrenia. Nat. Med. 2017;23:347–354. doi: 10.1038/nm.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepelin E. Eighth Edition. Barth; 1913. Psychiatrie. Ein Lehrbuch für Studirende und Aerzte. [Google Scholar]
- Kutsche L.K., Gysi D.M., Fallmann J., Lenk K., Petri R., Swiersy A., Klapper S.D., Pircs K., Khattak S., Stadler P.F. Combined Experimental and System-Level Analyses Reveal the Complex Regulatory Network of miR-124 during Human Neurogenesis. Cell Syst. 2018;7:438–452.e8. doi: 10.1016/j.cels.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz T.A., Joshi J.J., Reinhart V., Johnson K., Grantham L.E., 2nd, Volfson D. STEP levels are unchanged in pre-frontal cortex and associative striatum in post-mortem human brain samples from subjects with schizophrenia, bipolar disorder and major depressive disorder. PLoS ONE. 2015;10:e0121744. doi: 10.1371/journal.pone.0121744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger M., Neill J.C. A systematic review comparing sex differences in cognitive function in schizophrenia and in rodent models for schizophrenia, implications for improved therapeutic strategies. Neurosci. Biobehav. Rev. 2016;68:979–1000. doi: 10.1016/j.neubiorev.2016.06.029. [DOI] [PubMed] [Google Scholar]
- Levi-Montalcini R., Skaper S.D., Dal Toso R., Petrelli L., Leon A. Nerve growth factor: from neurotrophin to neurokine. Trends Neurosci. 1996;19:514–520. doi: 10.1016/S0166-2236(96)10058-8. [DOI] [PubMed] [Google Scholar]
- Lewis A.S., van Schalkwyk G.I., Bloch M.H. Alpha-7 nicotinic agonists for cognitive deficits in neuropsychiatric disorders: A translational meta-analysis of rodent and human studies. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2017;75:45–53. doi: 10.1016/j.pnpbp.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Z., Bunney B.G., Meng F., Hagenauer M.H., Walsh D.M., Vawter M.P., Evans S.J., Choudary P.V., Cartagena P., Barchas J.D. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. USA. 2013;110:9950–9955. doi: 10.1073/pnas.1305814110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Yu B., Sun Q., Zhang Y., Ren M., Zhang X., Li A., Yuan J., Madisen L., Luo Q. Generation of a whole-brain atlas for the cholinergic system and mesoscopic projectome analysis of basal forebrain cholinergic neurons. Proc. Natl. Acad. Sci. USA. 2018;115:415–420. doi: 10.1073/pnas.1703601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Brown R.E., Hussain Shuler M.G., Petersen C.C., Kepecs A. Optogenetic Dissection of the Basal Forebrain Neuromodulatory Control of Cortical Activation, Plasticity, and Cognition. J. Neurosci. 2015;35:13896–13903. doi: 10.1523/JNEUROSCI.2590-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom M.L., Bates D.M. Nonlinear mixed effects models for repeated measures data. Biometrics. 1990;46:673–687. [PubMed] [Google Scholar]
- Liu S.J., Nowakowski T.J., Pollen A.A., Lui J.H., Horlbeck M.A., Attenello F.J., He D., Weissman J.S., Kriegstein A.R., Diaz A.A., Lim D.A. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016;17:67. doi: 10.1186/s13059-016-0932-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchicchi A., Mansvelder H.D., Role L.W. Illuminating the role of cholinergic signaling in circuits of attention and emotionally salient behaviors. Front. Synaptic Neurosci. 2014;6:24. doi: 10.3389/fnsyn.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie D.I. An Integrative Approach to Neuroinflammation in Psychiatric disorders and Neuropathic Pain. J. Exp. Neurosci. 2018;12 doi: 10.1177/1179069518793639. 1179069518793639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D., Lamparter D., Quon G., Kellis M., Kutalik Z., Bergmann S. Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat. Methods. 2016;13:366–370. doi: 10.1038/nmeth.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maycox P.R., Kelly F., Taylor A., Bates S., Reid J., Logendra R., Barnes M.R., Larminie C., Jones N., Lennon M. Analysis of gene expression in two large schizophrenia cohorts identifies multiple changes associated with nerve terminal function. Mol. Psychiatry. 2009;14:1083–1094. doi: 10.1038/mp.2009.18. [DOI] [PubMed] [Google Scholar]
- McLaughlin I., Dani J.A., De Biasi M. The medial habenula and interpeduncular nucleus circuitry is critical in addiction, anxiety, and mood regulation. J. Neurochem. 2017;142(Suppl 2):130–143. doi: 10.1111/jnc.14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManaman J.L., Crawford F.G. Skeletal muscle proteins stimulate cholinergic differentiation of human neuroblastoma cells. J. Neurochem. 1991;57:258–266. doi: 10.1111/j.1471-4159.1991.tb02123.x. [DOI] [PubMed] [Google Scholar]
- Mesulam M.M. Cholinergic circuitry of the human nucleus basalis and its fate in Alzheimer’s disease. J. Comp. Neurol. 2013;521:4124–4144. doi: 10.1002/cne.23415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mufson E., Counts S., Perez S., Ginsberg S. Cholinergic system during the progression of AD: therapeutic implications. Expert Rev. Neurother. 2009;8:1703–1718. doi: 10.1586/14737175.8.11.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S., Godfrey A.K., Hughes J.F., Goodheart M.L., Mitchell R.N., Page D.C. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science. 2019;365:eaaw7317. doi: 10.1126/science.aaw7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S., Tang B., Head S.R., Gilmartin T.J., Sutcliffe J.G., Dean B., Thomas E.A. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonova E.V., Gilliland J.D.A., Tanis K.Q., Podtelezhnikov A.A., Rigby A.M., Galante R.J., Finney E.M., Stone D.J., Renger J.J., Pack A.I., Winrow C.J. Transcriptional Profiling of Cholinergic Neurons From Basal Forebrain Identifies Changes in Expression of Genes Between Sleep and Wake. Sleep (Basel) 2017;40:16–20. doi: 10.1093/sleep/zsx059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y., Kanda G.N., Yamada R.G., Shi S., Sunagawa G.A., Ukai-Tadenuma M., Fujishima H., Matsumoto N., Masumoto K.H., Nagano M. Muscarinic Acetylcholine Receptors Chrm1 and Chrm3 Are Essential for REM Sleep. Cell Rep. 2018;24:2231–2247.e7. doi: 10.1016/j.celrep.2018.07.082. [DOI] [PubMed] [Google Scholar]
- Oh S.W., Harris J.A., Ng L., Winslow B., Cain N., Mihalas S., Wang Q., Lau C., Kuan L., Henry A.M. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado V.F., Janickova H., Al-Onaizi M.A., Prado M.A.M. Cholinergic circuits in cognitive flexibility. Neuroscience. 2017;345:130–141. doi: 10.1016/j.neuroscience.2016.09.013. [DOI] [PubMed] [Google Scholar]
- Rajman M., Schratt G. MicroRNAs in neural development: from master regulators to fine-tuners. Development. 2017;144:2310–2322. doi: 10.1242/dev.144337. [DOI] [PubMed] [Google Scholar]
- Ramaker R.C., Bowling K.M., Lasseigne B.N., Hagenauer M.H., Hardigan A.A., Davis N.S., Gertz J., Cartagena P.M., Walsh D.M., Vawter M.P. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017;9:72. doi: 10.1186/s13073-017-0458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings J.S., Rosler K.M., Harrison D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004;117:1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe A.R., Mercer L., Casetti V., Sendt K.-V., Giaroli G., Shergill S.S., Tracy D.K. Dementia praecox redux: a systematic review of the nicotinic receptor as a target for cognitive symptoms of schizophrenia. J. Psychopharmacol. (Oxford) 2015;29:197–211. doi: 10.1177/0269881114564096. [DOI] [PubMed] [Google Scholar]
- Ruderfer D.M., Ripke S., McQuillin A., Boocock J., Stahl E.A., Pavlides J.M.W., Mullins N., Charney A.W., Ori A.P.S., Loohuis L.M.O., Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium. Electronic address: douglas.ruderfer@vanderbilt.edu. Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium Genomic Dissection of Bipolar Disorder and Schizophrenia, Including 28 Subphenotypes. Cell. 2018;173:1705–1715.e16. doi: 10.1016/j.cell.2018.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M.M., Lockstone H.E., Huffaker S.J., Wayland M.T., Webster M.J., Bahn S. Gene expression analysis of bipolar disorder reveals downregulation of the ubiquitin cycle and alterations in synaptic genes. Mol. Psychiatry. 2006;11:965–978. doi: 10.1038/sj.mp.4001875. [DOI] [PubMed] [Google Scholar]
- Sacco K.A., Bannon K.L., George T.P. Nicotinic receptor mechanisms and cognition in normal states and neuropsychiatric disorders. J. Psychopharmacol. (Oxford) 2004;18:457–474. doi: 10.1177/0269881104047273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salta E., De Strooper B. Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci. 2017;18:627–640. doi: 10.1038/nrn.2017.90. [DOI] [PubMed] [Google Scholar]
- Sarter M., Parikh V., Howe W.M. Phasic acetylcholine release and the volume transmission hypothesis: time to move on. Nat. Rev. Neurosci. 2009;10:383–390. doi: 10.1038/nm2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked I., Meerson A., Wolf Y., Avni R., Greenberg D., Gilboa-Geffen A., Soreq H. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity. 2009;31:965–973. doi: 10.1016/j.immuni.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Shaltiel G., Hanan M., Wolf Y., Barbash S., Kovalev E., Shoham S., Soreq H. Hippocampal microRNA-132 mediates stress-inducible cognitive deficits through its acetylcholinesterase target. Brain Struct. Funct. 2013;218:59–72. doi: 10.1007/s00429-011-0376-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smucny J., Tregellas J.R. Targeting neuronal dysfunction in schizophrenia with nicotine: Evidence from neurophysiology to neuroimaging. J. Psychopharmacol. (Oxford) 2017;31:801–811. doi: 10.1177/0269881117705071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq H. Checks and balances on cholinergic signaling in brain and body function. Trends Neurosci. 2015;38:448–458. doi: 10.1016/j.tins.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Soreq H., Ben-Aziz R., Prody C.A., Seidman S., Gnatt A., Neville L., Lieman-Hurwitz J., Lev-Lehman E., Ginzberg D., Lipidot-Lifson Y. Molecular cloning and construction of the coding region for human acetylcholinesterase reveals a G + C-rich attenuating structure. Proc. Natl. Acad. Sci. USA. 1990;87:9688–9692. doi: 10.1073/pnas.87.24.9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spearman C. The Proof and Measurement of Association between Two Things. Am. J. Psychol. 1904;15:72. [PubMed] [Google Scholar]
- Stanke M., Duong C.V., Pape M., Geissen M., Burbach G., Deller T., Gascan H., Otto C., Parlato R., Schütz G., Rohrer H. Target-dependent specification of the neurotransmitter phenotype: cholinergic differentiation of sympathetic neurons is mediated in vivo by gp 130 signaling. Development. 2006;133:141–150. doi: 10.1242/dev.02189. [DOI] [PubMed] [Google Scholar]
- Sun M., Liu H., Min S., Wang H., Wang X. Ciliary neurotrophic factor-treated astrocyte-conditioned medium increases the intracellular free calcium concentration in rat cortical neurons. Biomed. Rep. 2016;4:417–420. doi: 10.3892/br.2016.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatkiewicz J.P., O’Dushlaine C., Chen G., Chambert K., Moran J.L., Neale B.M., Fromer M., Ruderfer D., Akterin S., Bergen S.E. Copy number variation in schizophrenia in Sweden. Mol. Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B., Menon V., Nguyen T.N.T., Kim T.T.K., Jarsky T., Yao Z., Levi B.B., Gray L.T., Sorensen S.A., Dolbeare T. ). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat. Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Enkhuizen J., Janowsky D.S., Olivier B., Minassian A., Perry W., Young J.W., Geyer M.A. The catecholaminergic-cholinergic balance hypothesis of bipolar disorder revisited. Eur. J. Pharmacol. 2015;753:114–126. doi: 10.1016/j.ejphar.2014.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan S., Major A.J., Everling S. Muscarinic M1 Receptor Overstimulation Disrupts Working Memory Activity for Rules in Primate Prefrontal Cortex. Neuron. 2018;98:1256–1268.e4. doi: 10.1016/j.neuron.2018.05.027. [DOI] [PubMed] [Google Scholar]
- von Engelhardt J., Eliava M., Meyer A.H., Rozov A., Monyer H. Functional characterization of intrinsic cholinergic interneurons in the cortex. J. Neurosci. 2007;27:5633–5642. doi: 10.1523/JNEUROSCI.4647-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.C., Lin F.M., Chang W.C., Lin K.Y., Huang H.D., Lin N.S. miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics. 2009;10:328. doi: 10.1186/1471-2105-10-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A., Papp A.C., Curtis A., Newman L.C., Pietrzak M., Seweryn M., Handelman S.K., Rempala G.A., Wang D., Graziosa E. RNA sequencing of transcriptomes in human brain regions: protein-coding and non-coding RNAs, isoforms and alleles. BMC Genomics. 2015;16:990. doi: 10.1186/s12864-015-2207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf N.J. Cholinergic systems in mammalian brain and spinal cord. Prog. Neurobiol. 1991;37:475–524. doi: 10.1016/0301-0082(91)90006-m. [DOI] [PubMed] [Google Scholar]
- Wu J.C., Bunney W.E. The biological basis of an antidepressant response to sleep deprivation and relapse: review and hypothesis. Am. J. Psychiatry. 1990;147:14–21. doi: 10.1176/ajp.147.1.14. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zhang J., Qu P., Zhou C., Liu X., Ma X., Wang M., Wang Y., Su J., Liu J., Zhang Y. MicroRNA-125b is a key epigenetic regulatory factor that promotes nuclear transfer reprogramming. J. Biol. Chem. 2017;292:15916–15926. doi: 10.1074/jbc.M117.796771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code and additional supplementary materials generated during this study are available at https://github.com/slobentanzer/integrative-transcriptomics.
The sequencing datasets generated during this study are available at NCBI GEO: GSE120520 and GSE132951.
Accompanying website: https://slobentanzer.github.io/cholinergic-neurokine