Abstract
Background
The FCER2 gene, via encoding of the CD23 receptor, plays an important role in the regulation of IgE responses. A genetic variant of the FCER2 gene (T2206C) was previously shown to be associated with IgE levels in asthmatic children. IgE sensitization has also been linked to increased levels of fractional exhaled nitric oxide (FENO).
Objective
To investigate whether the FCER2 T2206C variant influences FENO levels in asthmatic children with a reported use of inhaled corticosteroids (ICS).
Methods
This cross‐sectional study involved 593 asthmatic children with a reported use of ICS, availability of FENO measurements and genotyping data on the FCER2 T2206C variant (rs28364072). An additive genetic model was assumed, and the association between the FCER2 T2206C variant and the log‐transformed (ln) FENO levels was evaluated using linear regression analysis, adjusted for age, sex, adapted British Thoracic Society (BTS) treatment steps and atopy.
Results
The mean age of the population was 9.1 ± 2.2 years, and the median of FENO levels was 13.0 ppb with an interquartile range (IQR) of (8.0‐27.5 ppb). The minor allele (G) frequency of rs28364072 was 29.6%, and each extra copy of the G allele was significantly associated with a lower level of the geometric mean of FENO (log scale, β = −0.12, 95% CI: −0.23, −0.02).
Conclusion and Clinical Relevance
Our results showed that the FCER2 T2206C variant was significantly associated with lower FENO levels in carriers of the G allele. Nevertheless, this SNP contributed little to the variability in FENO levels in this patient population. Our findings contribute to the present knowledge on FENO in asthmatic children; however, future replication studies are required to establish the role of this gene in relation to FENO.
Keywords: asthma, FCER2 T2206C variant, fractional exhaled nitric oxide, inhaled corticosteroid
1. INTRODUCTION
Asthma in children is often atopic, IgE‐mediated1, 2, 3 and characterized by chronic airway inflammation.4 The key regulator of IgE in asthma is the low‐affinity IgE receptor, also known as CD23. CD23, a C‐type lectin, is expressed on B cells, macrophages, eosinophils, follicular dendritic cells and platelets.5 Activation of this receptor suppresses the production of IgE6 that results in downregulation of the IgE‐mediated immune response.7 The CD23 receptor is encoded by the Fc fragment of the IgE receptor II (FCER2) gene which is an 11‐exon gene located at chromosome 19p13.2. Variants of FCER2 could modify the IgE synthesis.7 Furthermore, in asthmatic patients, the rs28364072 polymorphism (T2206C) in FCER2 has previously been shown to be associated with elevated IgE,8, 9 exacerbations,8, 9, 10 increased daily need of inhaled corticosteroids (ICS),8, 9 and poor lung function.9, 10
Nitric oxide (NO) is a biomarker used in respiratory disease and results from oxidation of l‐arginine in the presence of nitric oxide synthase (NOS). Three different isoforms of NOS exist: two constitutive NOS (cNOS) isoforms and one inducible NOS (iNOS) isoform. The expression of iNOS is boosted by inflammatory stimulation.11 NO, at higher concentrations, appears to function as an inflammatory agent.12 Previous studies showed that the fractional exhaled nitric oxide (FENO) was raised in the airways of patients with asthma11, 13 and was positively associated with higher IgE levels in asthmatic patients.14, 15 Many factors such as age, height, race, use and adherence to asthma medication, diet, exercise, and environmental factors might affect FENO levels.16, 17 Dugas et al suggested that CD23 has a major regulatory effect on iNOS activation in human monocytes and leads to NO production.18 Furthermore, Kolb et al19 found that the NOS pathway is involved in IgE‐mediated activation of monocytes. They also proposed that the increase of CD23‐driven cAMP (cyclic adenosine monophosphate) in monocytes is partly linked to the NOS pathway.19
A study in Vietnamese children with uncontrolled and untreated asthma (n = 32)20 showed that the levels of FENO were highest in the homozygous carriers (CC) of rs28364072 variation in the FCER2 gene. Their results imply that polymorphisms in the FCER2 gene are associated with FENO levels. However, no studies have evaluated this association in asthmatic children being treated with asthma medication.
In this study, we aimed to investigate the association between the FCER2 T2206C variant and FENO levels in a large cohort of asthmatic children treated with inhaled corticosteroids (ICS).
2. METHODS
2.1. Study setting and population
The study was undertaken in the PACMAN (Pharmacogenetics of Asthma medication in Children: Medication with Anti‐inflammatory effects) cohort study. Details of the study protocol have been described elsewhere.21 In short, the PACMAN study includes children (aged 4‐12 years) who were selected through pharmacies that belonged to the Utrecht Pharmacy Practice Network for Education and Research (UPPER).22 Children who regularly used asthma medication (Anatomical Therapeutic Chemical code R03), namely ≥3 prescriptions within the last 2 years and ≥1 prescription(s) in the last 6 months, were recruited through community pharmacies in the Netherlands.21 Selected children and their parents visited their own pharmacy, and during the visits, parents filled in a questionnaire that provided information about their children: respiratory symptoms, allergy, asthma diagnosis, medication use and adherence, socio‐demographic factors, and environmental factors.21 The Asthma Control Questionnaire (ACQ)23 was also included in the questionnaire. In addition, FENO levels were measured (NIOX Mino; Aerocrine), and saliva samples were collected for DNA extraction (Oragene DNA Self‐Collection kit; DNA Genotek, Inc., Kanata, ON, Canada). In the PACMAN study, daily ICS dosages (defined daily dosages of budesonide equivalent) were based on the last recorded refill prescription in the pharmacy prior to the study visit. The parents provided written informed consent, and the PACMAN cohort study was approved by the Medical Ethics Committee of the University Medical Centre Utrecht. The study population for our analysis consisted of all asthmatic children treated with inhaled corticosteroids (ICS) and who also had FENO measurements, genotyping and questionnaire data.
2.2. Definition of fraction of nitric oxide in exhaled breath
FENO was measured during the baseline visit at the pharmacy by using a single‐breath on‐line (SBOL) technique which was performed with a hand‐held electrochemical analyser (NIOX Mino; Aerocrine, Solna, Sweden) during an exhalation time of 6 seconds.24
2.3. Genotyping
Genotyping was performed using Human Core‐24 BeadChip Marker information, and the quality control (QC) procedures were applied to the genotype data. Data were imputed using the second release of the Haplotype Reference Consortium25 (HRC) (realises 1.1 2016) by mean Michigan Imputation Server.26 We extracted genotype dosage for rs28364072 in the FCER2 gene with a high imputation quality (Rsq score = 0.98). The Rsq score is defined as the ratio of the sample variance of the allele dosage during imputation to the expected variance under Hardy‐Weinberg equilibrium (HWE).27
2.4. Potential confounders/covariables
Potential confounders/covariates consisted of age, sex, (adapted) British Thoracic Society guidelines28 treatment steps (BTS) and atopy. We defined treatment step as follows: step 0, no use of inhaled short‐acting β 2‐agonist; step 1, short‐acting β 2‐agonist as needed; step 2, step 1 plus regular ICS; step 3, step 2 plus long‐acting β 2‐agonist; step 4, step 3 plus oral leukotriene receptor antagonist. Since all patients were required to be on ICS, the study population was on BTS treatment step 2 or above.
Atopy was specified as a reported history of allergic rhinitis, eczema, or food allergy. Asthma control was measured with the help of the 6‐item version of the Asthma Control Questionnaire (ACQ) (investigates respiratory symptoms and need of short‐acting β 2‐agonists).29 A mean score of ACQ ≥ 0.75 indicated ‘not well‐controlled asthma’ and an ACQ score of <0.75 indicated ‘well‐controlled asthma’.23, 30
2.5. Functional annotation of SNPs in strong LD with rs28364072
We retrieved all proxy SNPs in high linkage disequilibrium (LD) (R 2 > 0.8, limit distance 100 kb, and population panel CEU using 1000 Genomes project) with rs28364072 in FCER2 and checked their predicted functions, effects on protein structure, gene regulation, and splicing, using the HaploReg v4.1 (http://www.broadinstitute.org/mammals/haploreg/haploreg.php; in the public domain).
2.6. Expression quantitative trait loci analysis
The correlation of the SNP rs28364072 and its proxies in high LD with the expression level of FCER2 gene in whole blood was checked (expression quantitative trait loci [eQTL] analysis) using publicly available data from (GTEx) portal (GTEx portal:http://www.gtexportal.org/home/) and GeneNetwork.31 Moreover, we checked the effect of the SNP on FCER2 expression across different tissues using the GTEx portal.
2.7. Statistical analyses
Descriptive statistics were used to calculate means and standard deviations for continuous variables and percentages for categorical variables. For continuous variables that were not normally distributed, the median and interquartile range (IQR) were calculated. The chi‐square test and Mann‐Whitney U test were used to determine whether there was a significant difference in characteristics between children with well‐controlled asthma and not well‐controlled asthma. FENO values equal to zero parts per billion (ppb) were set at 2.5 ppb (the detection limit of the NIOX Mino = 5 ppb). Kruskal‐Wallis test was used to assess whether there was a statistically significant difference in the concentration of FENO between three genotype categories of FCER2 rs28364072 namely homozygous (GG), heterozygote (GA) and homozygous (AA). In pairwise comparison, Dunn's pairwise tests (also known as Dunn's post hoc tests) were used on each pair of rs28364072 genotypes (GG vs AG, GG vs AA, and AG vs AA) to investigate the differences in FENO levels by genotype pairs. To control for multiple testing, we divided 0.05 by the number of tests (n = 3) being performed (Bonferroni correction); therefore, we considered a Dunn's P‐value <0.016 (0.05/3) statistically significant. Mann‐Whitney U test was used to evaluate whether there was a significant difference in FENO levels between the two categories: carrier of the mutant allele (GG and AG genotypes) and non‐carrier of the G allele (AA genotype). To assess whether there was any effect of ICS dose on the levels of FENO in our study population, we tested the correlation between FENO levels and ICS dosage using the Spearman's rank test. We also tested this correlation in subgroups of carriers and non‐carriers of the FCER2 T2206C variant. Based on the GINA guideline4 we categorized daily ICS dosage into three categories: low, medium and high (Table S1). Kruskal‐Wallis test was used to assess whether there was a statistically significant difference in the median FENO level between patients treated with low, medium and high ICS dosages.
An additive genetic model was assumed, and the association between the FCER2 T2206C variant and FENO concentrations was assessed using linear regression. As FENO was not normally distributed, FENO levels were used as log‐transformed, natural logarithm (ln), for the regression analysis. The regression analysis was adjusted for age and sex in Model 1, and Model 2 was further adjusted for atopy as a dichotomous variable with two categories (0 = no and 1 = yes) and adapted BTS treatment steps. To investigate whether the association would differ between well‐controlled asthma (ACQ < 0.75) and not well‐controlled asthma (ACQ ≥ 0.75), we estimated the regression models in both. Next, we performed a sensitivity analysis and the FENO values equal to 0 ppb were set at 0 ppb and 5 ppb, respectively and the analyses were repeated. The HardyWeinberg package32 (version 1.6.2) for R was applied to assess HWE using exact test with DOST (Double One‐Sided Tail) P‐value.33
Statistical significance was considered at the P‐value of <0.05 (two‐sided). SPSS 24.0 software (IBM Corporation) was used for the analysis.
3. RESULTS
A data set of 593 children with reported use of ICS, FENO measurements and FCER2 genotype information was available within the PACMAN cohort. The general characteristics of the study population are shown in Table 1. The mean age of the population was 9.1 ± 2.2 years, and the majority of children were boys (62.1%). Of these children; 82% reported a history of atopy and 59% had well‐controlled asthma. The median value of FENO was 13.0 ppb with an interquartile range (IOR) of 8.0‐27.5 ppb. The median value of FENO was significantly lower in well‐controlled asthma 13.0 ppb (IQR = 8.0‐25.0 ppb) compared to not well‐controlled asthma 16.0 ppb (IQR = 9.0‐31.8 ppb) with P = 0.044. FENO values equal to zero ppb were found in 27 subjects (19 boys) with a mean age of 7.9 ± 2.1. The minor allele (G) frequency of rs28364072 was 29.6%, and 10.5% of the participants were homozygous (GG) carriers of the FCER2 T2206C variant. The SNP was in HWE in the total population (P = 0.051), in the category of well‐controlled asthma (P = 0.121) and in the category of not well‐controlled asthma (P = 0.219).
Table 1.
Characteristics of study population
| Baseline characteristics | Total population |
Well‐controlled (ACQ < 0.75) |
Not well‐controlled (ACQ ≥ 0.75) |
P a |
|---|---|---|---|---|
| n | 593 | 341/581b | 240/581b | |
| Age, mean (± SD) | 9.1 ± 2.2 | 9.0 ± 2.1 | 9.2 ± 2.2 | |
| Male, % (n) | 62.1 (368) | 62.8 (127) | 61.7 (148) | 0.795 |
| History of atopyc % (n) | 82 (484/590) | 82.4 (280/340) | 82.5 (198/240) | 0.963 |
| Eczemac, % (n) | 66.9 (394/589) | 65.9 (224/340) | 69.9 (167/239) | 0.313 |
| Food allergyc, % (n) | 52.1 (306/587) | 48.4 (164/339) | 58.0 (138/238) | 0.023 |
| Hayfeverc, % (n) | 46.4 (269/580) | 45.4 (152/335) | 48.5 (114/235) | 0.460 |
| Family history of atopy | ||||
| Paternal asthmac % (n) | 29.9 (163/546) | 27.8 (88/317) | 32.9 (72/219) | 0.203 |
| Paternal eczemac % (n) | 27.6 (147/533) | 28.0 (87/311) | 27.7 (59/213) | 0.945 |
| Paternal hayfeverc % (n) | 38.2 (206/539 | 38.1 (119/312) | 39.2 (85/217) | 0.811 |
| Maternal asthmac % (n) | 26.6 (147/553) | 21.1 (66/313) | 33.2 (76/229) | 0.002 |
| Maternal eczemac % (n) | 39.9 (222/556) | 36.5 (116/318) | 44.1 (100/227) | 0.075 |
| Maternal hayfeverc % (n) | 44.3 (247/557) | 43.2 (137/317) | 46.3 (106/229) | 0.476 |
| Adapted BTS treatment step d % (n) | ||||
| 2 | 69.1 (410) | 74.7 (248) | 66.8 (155) | 0.012 |
| 3 | 20.9 (124) | 20.8 (69) | 22.4 (52) | |
| 4 | 6.9 (41) | 4.5 (15) | 10.8 (25) | |
| Minor allele frequency FCER2 variant % | 29.6 | 29.5 | 29.9 | 0.982 |
| Genotype distribution T2206C variant | ||||
| Homozygous wild‐type (AA) % (n) | 51.4 (305) | 51.6 (176) | 50.8 (122) | |
| Heterozygous (AG) % (n) | 38.1 (226) | 37.8 (129) | 38.3 (92) | |
| Homozygous variant (GG) % (n) | 10.5 (62) | 10.6 (36) | 10.8 (26) | |
| FENO ppb, median (IQR) | 13.0 (8.0‐27.5) | 13.0 (8.0‐25.0) | 16.0 (9.0‐31.8) | 0.044 |
| ACQ b score, median (IQR) | 0.5 (0.17‐1.17) | 0.17 (0.0‐0.5) | 1.3 (1.0‐1.8) | 0.000 |
ACQ, Asthma Control Questionnaire; FENO, fractional exhaled nitric oxide; IQR, Interquartile range
P‐value, To evaluate the significant difference between the two categories, well‐controlled asthma vs not well‐controlled asthma, using chi‐square test or Mann‐Whitney U test.
In the total population, data on the Asthma Control Questionnaire (ACQ) were missing in 12 subjects.
Data were not available for the total population.
18 children were on ICS plus leukotriene receptor antagonist treatment.
3.1. Levels of FENO in different FCER2 variant genotypes
The levels of FENO were compared among the three genotypes of rs28364072 (Table 2 and Figure 1). The FENO levels were lowest in children with GG genotype (10.0 ppb, IQR: 7.0‐22.2 ppb) and highest in AA genotype (16.0 ppb, IQR: 9.0‐28.5 ppb). In pairwise comparison, there was significant evidence (unadjusted P‐value) that the concentration of FENO was statistically significantly different between GG‐AA genotypes and AG‐AA genotypes (Table 2). After adjustment for multiple testing, the FENO levels remained significantly different between genotypes AA and AG (Table 2 and Figure 1). In the category of well‐controlled asthma (n = 341), the FENO levels were significantly different between GG‐AA genotypes and AG‐AA genotypes; however, both were no longer significant after Bonferroni correction (Table 2). In the category of not well‐controlled asthma (n = 240), there were no significant differences in FENO levels at all (P‐value from Kruskal‐Wallis test was equal 0.339; Table 2). We also found a significant difference in the FENO concentrations between the carriers of the mutant allele (GG and AG genotypes) and the non‐carriers of the G allele (AA genotype) in the total population and well‐controlled asthma category (Table 3).
Table 2.
Concentration of FENO among genotypes of rs28364072
|
rs28364072 Genotypes |
FENOappb (IQR) | Pairwise comparison of FENO levels | P b |
|---|---|---|---|
| Total population (n = 593) | |||
| Homozygous variant (GG) | 10.0 (7.0‐22.2) | GG‐AG | 0.541 |
| Heterozygous (AG) | 12.0 (7.0‐27.2) | GG‐AA | 0.029 |
| Homozygous wild‐type (AA) | 16.0 (9.0‐28.5) | AG‐AA | 0.014 |
| Well‐controlled (n = 341) a | |||
| Homozygous variant (GG) | 10.0 (7.0‐20.8) | GG‐AG | 0.526 |
| Heterozygous (AG) | 12.0 (7.0‐23.0) | GG‐AA | 0.041 |
| Homozygous wild‐type (AA) | 14.5 (8.0‐28.8) | AG‐AA | 0.029 |
| Not well‐controlled (n = 240) a | |||
| Homozygous variant (GG) | 14.0 (7.0‐35.8) | GG‐AG | 0.339* |
| Heterozygous (AG) | 13.0 (7.0‐36.0) | GG‐AA | |
| Homozygous wild‐type (AA) | 18.0 (10.0‐28.5) | AG‐AA | |
Abbreviation: FENO, fractional exhaled nitric oxide.
In the total population, data on the Asthma Control Questionnaire (ACQ) were missing in 12 subjects.
Tested with Dunn's P‐value for pairwise comparison and a Bonferroni‐corrected P‐value lower than 0.016 (0.05/3) was considered statistically significant.
P‐value from Kruskal‐Wallis test.
Figure 1.
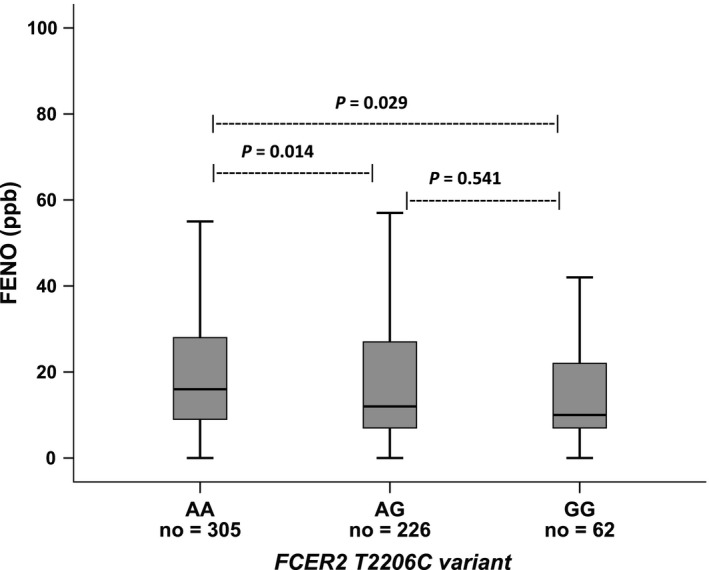
Concentration of FENO among genotypes of FCER2 T2206C variant (rs28364072). FENO, fractional exhaled nitric oxide. The Kruskal–Wallis test was used and in pairwise comparison, a Dunn's P‐value < 0.016 (0.05/3) was considered statistically significant.
Table 3.
Concentration of FENO between carries and non‐carriers of the G allele of rs28364072
| rs28364072 variation | n | FENO ppb median (IQR) |
P a |
|---|---|---|---|
| Total population (n = 593) | |||
| Carriers of the variant (GG/AG) | 288 | 12.0 (7.0‐25.7) | 0.004 |
| Non‐carriers of the variant (AA) | 305 | 16.0 (9.0‐28.5) | |
| Well‐controlled category (n = 341) b | |||
| Carriers of the variant (GG/AG) | 165 | 11.0 (7.0‐23.0) | 0.010 |
| Non‐carriers of the variant (AA) | 176 | 14.5 (8.0‐28.8) | |
| Not well‐controlled category (n = 240) b | |||
| Carriers of the variant (GG/AG) | 118 | 13.0 (7.0‐36.0) | 0.146 |
| Non‐carriers of the variant (AA) | 122 | 18.0 (10.0‐28.5) | |
Abbreviations: FENO, fractional exhaled nitric oxide; IQR, Interquartile range.
P‐value, Tested with Mann‐Whitney U test.
In the total population, data on the Asthma Control Questionnaire (ACQ) were missing in 12 subjects.
3.2. Association of FCER2 variant with FENO levels
The associations of the FCER2 variant with FENO levels are presented in Table 4. In the crude model, each extra copy of the G allele of rs28364072 was significantly associated with a lower level of the geometric mean of FENO (log scale, β = −0.15, 95% CI: −0.26, −0.05). When we adjusted for age and sex (Model 1) and also after further adjustment for BTS treatment steps and a reported history of atopy (Model 2), the estimated effects remained statistically significant.
Table 4.
Regression coefficients and 95% confidence intervals describing the association between rs28364072 variation of the FCER2 gene (per copy of the G allele) and the concentration of FENO
| Crude | Model 1 | Model 2 | |||||
|---|---|---|---|---|---|---|---|
| FENOa | Effect allele | β (95% CI) | P‐value | β (95% CI) | P‐value | β (95% CI) | P‐value |
| Total population (n = 593) | G | −0.15 (−0.26, −0.05) | 0.005 | −0.12 (−0.22, −0.02) | 0.021 | −0.12 (−0.23, −0.02) | 0.018 |
| Well‐controlleda (n = 341) | G | −0.18 (−0.32, −0.05) | 0.007 | −0.16 (−0.29, −0.03) | 0.019 | −0.17 (−0.30, −0.04) | 0.011 |
| Not well‐controlleda (n = 240) | G | −0.12 (−0.29, 0.06) | 0.190 | −0.09 (−0.25, 0.08) | 0.306 | −0.08 (−0.25, 0.09) | 0.331 |
In the total population, data on the Asthma Control Questionnaire (ACQ) were missing in 12 subjects. Crude, only SNP; Model 1, adjusted for age and sex; Model 2, Model 1 further adjusted for (adapted) British Thoracic Society (BTS) treatment steps and atopy.
Abbreviations: ACQ, Asthma Control Questionnaire; CI, Confidence Interval; FENO, Fractional exhaled Nitric Oxide; Not well‐controlled, ACQ ≥ 0.75; Well‐controlled, ACQ < 0.75.
FENO levels were used as log‐transformed (ln).
3.3. Association of FCER2 variant with FENO levels stratified by ACQ scores
We tested the association between the variant and FENO levels when the total population was stratified in well‐controlled asthma (ACQ < 0.75) and not well‐controlled asthma (ACQ ≥ 0.75). In Model 2, adjusted for age, sex, BTS treatment steps and atopy, each extra copy of the G allele of rs28364072 was significantly associated with a lower level of the geometric mean of FENO (log scale, β = −0.17, 95% CI: −0.30, −0.04) among children with well‐controlled asthma. We did not observe any significant association (log scale, β = −0.08, 95% CI: −0.25, 0.09) between the FCER2 variant and FENO levels in the not well‐controlled asthma group (Table 4). However, the point estimate was in the same direction as well‐controlled asthma category.
We performed sensitivity analyses whereby the FENO values equal to 0 ppb were set at 0 and 5 ppb, respectively. The results of these analyses were similar to the results of our original analyses. (Table S2).
3.4. Correlation of FENO levels and ICS daily dosage
Data on ICS dosage were available for 475 out of 593 subjects. There was no correlation between FENO levels and ICS dosage (Spearman's rank correlation coefficient [RS] = −0.016, P = 0.736). When subjects were grouped according to defined daily ICS dosage (low, medium, high), higher FENO levels could be observed in the patients treated with low dosages of ICS category (median FENO: 17.0ppb [IQR: 8.0‐30.0]) compared to patients treated with medium ICS dosages (median FENO: 14.00 ppb [IQR: 8.0‐23.0]) and high ICS dosages (median FENO: 11.5 ppb [IQR: 7.0‐25.7]); however, the differences were not statistically significant. In addition, there was no significant correlation between FENO levels and ICS dosage in carriers or non‐carries of the FCER2 T2206C variant (RS = 0.025, P = 0.703 and RS = −0.038, P = 0.554, respectively).
3.5. Functional annotation and eQTL analysis of rs28364072
Functional annotation, using Haploreg v4.1 data, shows that rs28364072 has several proxy variants (D′ = 1 and R 2 > 0.8), but they are all intronic and synonymous, without any predicted functions, (Table S3). Moreover, the cis‐eQTL data from GTEx portal and GeneNetwork31 showed that the minor allele G of rs28364072 is significantly associated with reduced expression levels of FCER2 in whole blood (Table S4 and Figure S1 and Figure S2). Together, these data may support the notion that rs28364072 has an effect on FCER2 gene expression and function.
4. DISCUSSION
In our population‐based study, we assessed the association between the FCER2 T2206C variant and FENO levels in children with asthma and reported use of ICS. Our results showed that the variation in the FCER2 gene was significantly associated with lower levels of FENO in the total population and in well‐controlled asthmatic patients. There was no statistically significant association between the FCER2 polymorphism and levels of FENO in not well‐controlled asthma group which might be explained by the small sample size of this group.
To the best of our knowledge, the current study is the first to evaluate the association between the FCER2 T2206C variant and FENO levels among asthmatic children using ICS. So far only one study has investigated the same association but in mild‐to‐moderate uncontrolled and untreated asthmatic patients among Vietnamese children (n = 32), and they found that FENO levels were significantly higher in subjects with the FCER2 gene mutation.20 However, due to a small sample size they were not able to define a specific FENO level that was associated with the FCER2 gene variation.20 In the study by Nguyen‐Thi‐Bich et al,20 all subjects with uncontrolled asthma were not on medication for at least one month before inclusion, while in the PACMAN study all subjects were on ICS treatment and ICS therapy is known to decrease FENO levels.34 In addition, the frequency of the rs28364072 homozygous variant of the FCER2 gene was slightly higher among Vietnamese children (15.6%) than (10.5%) in the mainly Caucasian children (91.4%) in the PACMAN study. Moreover, the sample size (n = 593) of the current study was roughly 19 times larger than the Vietnamese study (n = 32).
It has previously been shown that IgE is associated with FENO levels in children with asthma and allergy.14, 15 The low‐affinity IgE receptor, CD23, by nature inhibits IgE production.6 The FCER2 gene affects the inflammatory mechanisms and results in the variability in the response to ICS in asthma.35 The T2206C variant is located in splicing region of intron 9 of FCER2 and might lead to alternative splicing and changes of the gene transcript length. Tantisira et al showed that the T2206C variant was associated with lower FCER2 gene expression.9 This was suggested to be a possible mechanism for the higher IgE levels among carriers of the mutant (C) allele in their findings.9 In animal models, it was confirmed that absence of CD23 (CD23‐/‐) in the knockout mice resulted in elevated IgE‐mediated response compared with wild‐type CD23+/+.36, 37 In addition, use of corticosteroids might be associated with a decrease in CD23 expression,38 as Tantisira et al9 noticed that the highest IgE levels were detected in individuals who were homozygote (CC) for the FCER2 T2206C variant and on ICS treatment. However, in our study population we did not observe a statistically significant effect of ICS dosage on FENO levels. There is some evidence from the literature that during an inflammatory response the CD23 receptor induces iNOS activation that in turn leads to NO production (Figure S3).18 As shown previously, genetic expression of CD23 decreased in individuals homozygous for T2206C variant compared with the two other genotypes,9 which is in line with eQTL data demonstrating decreased expression levels of FCER2 in the carriers of T2206C variant (Table S3). This might lead to less NO production and consequently lower levels of FENO among carriers of the variant; hence, the observed lower FENO levels in carriers of the FCER2 T2206C variant is plausible.
It should be noted that children in our study with not well‐controlled asthma had a high percentage of food allergy and maternal asthma (Table 1). It has been shown that food allergy is associated with an increased risk of hospitalization and use of systematic corticosteroids due to asthma.39, 40 In addition, it has been reported that maternal asthma is a crucial risk factor for developing asthma in childhood.41, 42
In our study, compared to non‐carriers of FCER2 T2206C variant, FENO levels in the carriers of the variant were significantly lower with a median difference of 4ppb (Table 3). Further research is needed to monitor FENO over time in FCER2 T2206C carriers to better understand the clinical relevance of this finding.
A major strength of this study was the availability of information on asthma symptoms and FENO measurements in a large real‐life population of paediatric patients. Still, some limitations need to be addressed. First, atopy was defined based on parental reporting in the questionnaire and was not defined based on a skin prick test. The lack of an objective criterion such as the skin prick test thus might lead to both an underestimation (in case of asymptomatic patients) and an overestimation of atopy. Similarly, asthma symptoms were parent‐reported which might be prone to over‐ or underestimation. Lastly, as gene expression is tissue specific, it would be better to also check the association between T2206C variant and FCER2 gene expression in lung tissue; however, so far, eQTL data from lung tissue is either not available or only in very small sample size.31
To conclude, we observed that the FCER2 T2206C variant was significantly associated with lower levels of FENO in children with well‐controlled asthma treated with ICS. However, this SNP contributed little to the variability in FENO levels in this patient population. We acknowledge that the exact mechanism by which the FCER2 T2206C variant could modify FENO levels needs to be further explored and that future replication studies are required to establish the role of this gene in relation to FENO.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
The PACMAN study was supported by an unrestricted research grant from GSK. Anke H. Maitland‐van der Zee (AHMZ) received unrestricted funding from GSK and Boehringer Ingelheim. AHMZ received honoraria for participating in advisory boards from Astra Zeneca, GSK, and Boehringer Ingelheim. Katia Verhamme works for a research group, who in the past, received unconditional research grants from Pfizer, Boehringer Ingelheim, Yamanouchi and GSK; none of which are related to the content of this paper.
Karimi L, Vijverberg SJH, Farzan N, Ghanbari M, Verhamme KMC, Maitland‐van der Zee AH. FCER2 T2206C variant associated with FENO levels in asthmatic children using inhaled corticosteroids: The PACMAN study. Clin Exp Allergy. 2019;49:1429–1436. 10.1111/cea.13460
REFERENCES
- 1. Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ. Asthma and wheezing in the first six years of life. The Group Health Medical Associates. N Engl J Med. 1995;332:133‐138. [DOI] [PubMed] [Google Scholar]
- 2. Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin‐test reactivity to allergens. N Engl J Med. 1989;320:271‐277. [DOI] [PubMed] [Google Scholar]
- 3. Froidure A, Mouthuy J, Durham SR, Chanez P, Sibille Y, Pilette C. Asthma phenotypes and IgE responses. Eur Respir J. 2016;47:304‐319. [DOI] [PubMed] [Google Scholar]
- 4. Global Initiative for Asthma (GINA) for asthma management and prevention, 2018. Available from: http://www.ginasthma.org/. 2018.
- 5. Kijimoto‐Ochiai S. CD23 (the low‐affinity IgE receptor) as a C‐type lectin: a multidomain and multifunctional molecule. Cell Mol Life Sci. 2002;59:648‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Acharya M, Borland G, Edkins AL, et al. CD23/FcepsilonRII: molecular multi‐tasking. Clin Exp Immunol. 2010;162:12‐23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Laitinen T, Ollikainen V, Lázaro C, et al. Association study of the chromosomal region containing the FCER2 gene suggests it has a regulatory role in atopic disorders. Am J Respir Crit Care Med. 2000;161:700‐706. [DOI] [PubMed] [Google Scholar]
- 8. Koster ES, Maitland‐van der Zee AH, Tavendale R, et al. FCER2 T2206C variant associated with chronic symptoms and exacerbations in steroid‐treated asthmatic children. Allergy. 2011;66(12):1546‐1552. [DOI] [PubMed] [Google Scholar]
- 9. Tantisira KG, Silverman ES, Mariani TJ, et al. FCER2: a pharmacogenetic basis for severe exacerbations in children with asthma. J Allergy Clin Immunol. 2007;120:1285‐1291. [DOI] [PubMed] [Google Scholar]
- 10. Rogers AJ, Tantisira KG, Fuhlbrigge AL, et al. Predictors of poor response during asthma therapy differ with definition of outcome. Pharmacogenomics. 2009;10:1231‐1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pijnenburg MW, De Jongste JC. Exhaled nitric oxide in childhood asthma: a review. Clin Exp Allergy. 2008;38:246‐259. [DOI] [PubMed] [Google Scholar]
- 12. Stewart L, Katial RK. Exhaled nitric oxide. Immunol Allergy Clin North Am. 2012;32:347‐362. [DOI] [PubMed] [Google Scholar]
- 13. Rodway GW, Choi J, Hoffman LA, Sethi JM. Exhaled nitric oxide in the diagnosis and management of asthma: clinical implications. Chron Respir Dis. 2009;6:19‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malinovschi A, Janson C, Holmkvist T, Norback D, Merilainen P, Hogman M. IgE sensitisation in relation to flow‐independent nitric oxide exchange parameters. Respir Res. 2006;7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cardinale F, de Benedictis FM, Muggeo V, et al. Exhaled nitric oxide, total serum IgE and allergic sensitization in childhood asthma and allergic rhinitis. Pediatr Allergy Immunol. 2005;16:236‐242. [DOI] [PubMed] [Google Scholar]
- 16. Turner S. Exhaled nitric oxide and the management of childhood asthma–yet another promising biomarker "has been" or a misunderstood gem. Paediatr Respir Rev. 2015;16:88‐96. [DOI] [PubMed] [Google Scholar]
- 17. Vijverberg SJH, Koster ES, Koenderman L, et al. Exhaled NO is a poor marker of asthma control in children with a reported use of asthma medication: a pharmacy‐based study. Pediatr Allergy Immunol. 2012;23:529‐536. [DOI] [PubMed] [Google Scholar]
- 18. Dugas B, Mossalayi MD, Damais C, Kolb JP. Nitric oxide production by human monocytes: evidence for a role of CD23. Immunol Today. 1995;16:574‐580. [DOI] [PubMed] [Google Scholar]
- 19. Kolb JP, Paul‐Eugene Dugas N, Yamaoka K, Mossalayi MD, Dugas B. Role of CD23 in NO production by human monocytic cells. Res Immunol. 1995;146:684‐689. [DOI] [PubMed] [Google Scholar]
- 20. Nguyen‐Thi‐Bich H, Duong‐Thi‐Ly H, Thom VT, et al. Study of the correlations between fractional exhaled nitric oxide in exhaled breath and atopic status, blood eosinophils, FCER2 mutation, and asthma control in Vietnamese children. J Asthma Allergy. 2016;9:163‐170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koster ES, Raaijmakers JAM, Koppelman GH, et al. Pharmacogenetics of anti‐inflammatory treatment in children with asthma: rationale and design of the PACMAN cohort. Pharmacogenomics. 2009;10:1351‐1361. [DOI] [PubMed] [Google Scholar]
- 22. Koster ES, Blom L, Philbert D, Rump W, Bouvy ML. The Utrecht pharmacy practice network for education and research: a network of community and hospital pharmacies in the Netherlands. Int J Clin Pharm. 2014;36:669‐674. [DOI] [PubMed] [Google Scholar]
- 23. Juniper EF, O'Byrne PM, Guyatt GH, Ferrie PJ, King DR. Development and validation of a questionnaire to measure asthma control. Eur Respir J. 1999;14:902‐907. [DOI] [PubMed] [Google Scholar]
- 24. Koopman M, Arets HG, Uiterwaal CS, van der Ent CK. Comparing 6 and 10 sec exhalation time in exhaled nitric oxide measurements in children. Pediatr Pulmonol. 2009;44:340‐344. [DOI] [PubMed] [Google Scholar]
- 25. McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279‐1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Das S, Forer L, Schönherr S, et al. Next‐generation genotype imputation service and methods. Nat Genet. 2016;48:1284‐1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation‐driven meta‐analysis of genome‐wide association studies. Hum Mol Genet. 2008;17:R122‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levy ML, Thomas M, Small I, Pearce L, Pinnock H, Stephenson P. Summary of the 2008 BTS/SIGN British guideline on the management of asthma. Prim Care Respir J. 2009;18(suppl 1):S1‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Juniper EF, Svensson K, Mork AC, Stahl E. Measurement properties and interpretation of three shortened versions of the asthma control questionnaire. Respir Med. 2005;99:553‐558. [DOI] [PubMed] [Google Scholar]
- 30. Juniper EF, Bousquet J, Abetz L, Bateman ED. Identifying 'well‐controlled' and 'not well‐controlled' asthma using the Asthma Control Questionnaire. Respir Med. 2006;100:616‐621. [DOI] [PubMed] [Google Scholar]
- 31. Westra H‐J, Peters MJ, Esko T, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238‐1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Graffelman J. Exploring diallelic genetic markers: the HardyWeinberg package. Universitat Polit`ecnica de Catalunya. 2019; version 1.6.2.
- 33. Graffelman J. The number of markers in the HapMap project: some notes on chi‐square and exact tests for Hardy‐Weinberg equilibrium. Am J Hum Genet. 2010;86:813‐818; author reply 818–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McNicholl DM, Stevenson M, McGarvey LP, Heaney LG. The utility of fractional exhaled nitric oxide suppression in the identification of nonadherence in difficult asthma. Am J Respir Crit Care Med. 2012;186:1102‐1108. [DOI] [PubMed] [Google Scholar]
- 35. Duong‐Thi‐Ly H, Nguyen‐Thi‐Thu H, Nguyen‐Hoang L, Nguyen‐Thi‐Bich H, Craig TJ, Duong‐Quy S. Effects of genetic factors to inhaled corticosteroid response in children with asthma: a literature review. J Int Med Res. 2017;45:1818‐1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Haczku A, Takeda K, Hamelmann E, et al. CD23 exhibits negative regulatory effects on allergic sensitization and airway hyperresponsiveness. Am J Respir Crit Care Med. 2000;161:952‐960. [DOI] [PubMed] [Google Scholar]
- 37. Riffo‐Vasquez Y, Spina D, Thomas M, Gilbey T, Kemeny DM, Page CP. The role of CD23 on allergen‐induced IgE levels, pulmonary eosinophilia and bronchial hyperresponsiveness in mice. Clin Exp Allergy. 2000;30:728‐738. [DOI] [PubMed] [Google Scholar]
- 38. Wu CY, Sarfati M, Heusser C, et al. Glucocorticoids increase the synthesis of immunoglobulin E by interleukin 4‐stimulated human lymphocytes. J Clin Invest. 1991;87:870‐877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu AH, Jaramillo R, Sicherer SH, et al. National prevalence and risk factors for food allergy and relationship to asthma: results from the National Health and Nutrition Examination Survey 2005–2006. J Allergy Clin Immunol. 2010;126(798‐806):e713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Simpson AB, Glutting J, Yousef E. Food allergy and asthma morbidity in children. Pediatr Pulmonol. 2007;42:489‐495. [DOI] [PubMed] [Google Scholar]
- 41. Lim RH, Kobzik L, Dahl M. Risk for asthma in offspring of asthmatic mothers versus fathers: a meta‐analysis. PLoS ONE. 2010;5:e10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spiegel E, Shoham‐Vardi I, Goldbart A, Sergienko R, Sheiner E. Maternal asthma is an independent risk factor for long‐term respiratory morbidity of the offspring. Am J Perinatol. 2018;35:1065‐1070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


