Abstract
Background
Accurate assessment of hepatic steatosis is a key to grade disease severity in non‐alcoholic fatty liver disease (NAFLD).
Methods
We developed a digital automated quantification of steatosis on whole‐slide images (WSIs) of liver tissue and performed a validation study. Hematoxylin–eosin stained liver tissue slides were digitally scanned, and steatotic areas were manually annotated. We identified thresholds for size and roundness parameters by logistic regression to discriminate steatosis from surrounding liver tissue. The resulting algorithm produces a steatosis proportionate area (SPA; ratio of steatotic area to total tissue area described as percentage). The software can be implemented as a Java plug‐in in FIJI, in which digital WSI can be processed automatically using the Pathomation extension.
Results
We obtained liver tissue specimens from 61 NAFLD patients and 18 controls. The area under the curve of correctly classified steatosis by the algorithm was 0.970 (95% CI 0.968–0.973), P < 0.001. Accuracy of the algorithm was 91.9%, with a classification error of 8.1%. SPA correlated significantly with steatosis grade (Rs = 0.845, CI: 0.749–0.902, P < 0.001) and increased significantly with each individual steatosis grade, except between Grade 2 and 3.
Conclusions
We have developed a novel digital analysis algorithm that accurately quantifies steatosis on WSIs of liver tissue. This algorithm can be incorporated when quantification of steatosis is warranted, such as in clinical trials studying efficacy of new therapeutic interventions in NAFLD. © 2019 The Authors. Cytometry Part B: Clinical Cytometry published by Wiley Periodicals, Inc. on behalf of International Clinical Cytometry Society.
Key terms: non‐alcoholic fatty liver disease (NAFLD), steatosis, digital image analysis, pathology
Abbreviations
- AUC
area under the curve
- DIA
digital image analysis
- HSB
hue, saturation, brightness color model
- IQR
interquartile range
- NAFLD
non‐alcoholic fatty liver disease
- NAS
NAFLD activity score
- RGB
red, green, blue color model
- SPA
steatotic proportionate area
- WSIs
whole slide images
Non‐alcoholic fatty liver disease (NAFLD), defined as the abnormal accumulation of fat in the liver, has a spectrum that ranges from steatosis to progressive inflammation (steatohepatitis) and eventually fibrosis and cirrhosis 1. NAFLD is seen as the hepatic manifestation of the metabolic syndrome and is associated with an increased mortality risk, mainly from cardiovascular complications 2. Approximately a quarter of the general population is affected by NAFLD, which constitutes a global health problem 3. Severity of steatosis is associated with progression to steatohepatitis and fibrosis 4, 5. Early recognition of severe steatosis is thus imperative to prevent progression of disease. Accurate assessment of steatosis severity bears relevance to other disease states such as in viral hepatitis and liver transplantation, because progressive disease or graft failure is associated with the degree of steatosis 6, 7, 8, 9, 10. Histological grading of steatosis, together with inflammation, ballooning, and fibrosis, is a frequently used endpoint in clinical trials investigating novel therapies 11. The NAFLD activity score (NAS) is the most commonly used semi‐quantitative grading system, and it grades steatosis as an interval‐percentage upon visual examination of histological liver tissue slides 12. This score, as many other grading systems, is subject to interobserver and intra‐observer variability and prone to inaccuracy 13, 14. More precise measurement of steatosis (reduction) will increase power to test efficacy of new drugs. Current research focuses on development of non‐invasive tools to quantify steatosis such as MR‐spectroscopy or elastography, but validation is still hampered by a subjective and semi‐quantitative gold standard 15.
Digital image analysis (DIA) is a promising method for accurate histological steatosis assessment, as it does not suffer from observer variability, possesses high reproducibility, and produces a continuous outcome measure which may be compared to established standards 13. There are a number of digital algorithms and tools constructed over the last years, aiming to objectively quantify steatosis. Different approaches have been used to automatically discriminate steatosis from non‐steatotic tissue, ranging from tools only using RGB color cut‐offs to algorithms incorporating several features, with predetermined cut‐offs, and recently more advanced approaches such as stereological analysis or supervised machine learning 14, 16, 17, 18, 19, 20. Nonetheless, these algorithms these still include bias introducing features, such as manual correction (e.g., post‐analytical visual examination to exclude any missed large non‐steatotic areas, such as vessels or tissue tears) or analysis of sections of the liver biopsy specimen (e.g., random field selection from several high magnification images).
In the present study, we aimed to develop and evaluate an automated steatosis quantification algorithm for whole‐slide image (WSI) analysis, excluding any interobserver and intra‐observer variability.
MATERIAL AND METHODS
Tissue Samples
Tissue slides from liver biopsies were selected from the Radboudumc histopathology archives from the period 2012–2016. All specimens with a minimum of 11 portal fields were considered for inclusion. To represent the full range of steatosis severity, slides from NAFLD patients, representing each NAS steatosis grade (1: 5–33% steatosis / 2: 34–66% steatosis / 3: more than 66% steatosis) were selected on basis of original pathology report, as well as slides of patients without any abnormalities or steatosis that served as controls (Grade 0: <5% steatosis). All slides were stained with a Hematoxylin–Eosin stain according to standard practice. Steatosis and fibrosis grade were revised according to the NAS and Brunt fibrosis score 12 by an experienced liver pathologist, blinded for the outcome of the initial evaluation. Requirement for ethical approval was waived by the institutional review board (no. 2016‐2763). All tissue sections were fully digitized, producing WSIs, using a P250 Flash digital slide scanner (3DHISTECH, Hungary). WSIs were produced using a 20x objective lens (specimen level pixel size 0.24 × 0.24μm2) and JPEG compressed using quality factor 80.
Handling of WSIs
The steatosis quantification algorithm was implemented as a Java plug‐in for the Fiji image analysis platform 21. To handle complete WSI, the plug‐in uses the PMA.start WSI‐viewer (Pathomation, Belgium) 22. A WSI containing the entire slide was loaded in Fiji at reduced resolution, and a square region containing all tissue was selected manually (see Fig. 1A).This greatly reduces computation time as a large part of the WSI generally consists of background.
Figure 1.
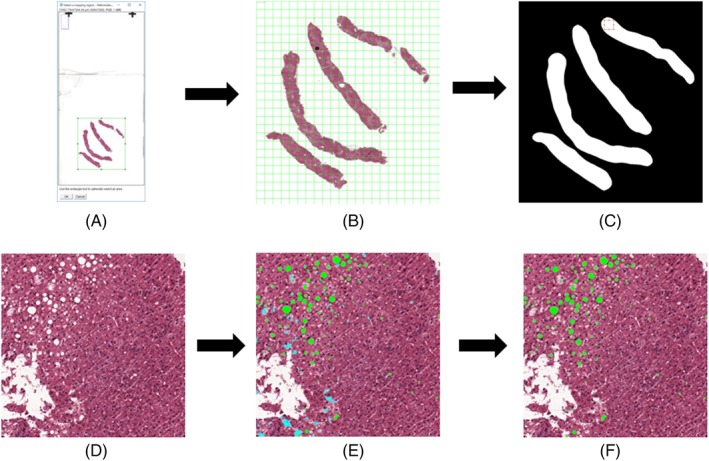
Process flowchart of the steatosis quantification plugin. The whole slide image is presented to the user at a reduced resolution and a smaller region of interest, incorporating all tissue is selected for purpose of saving time (A). The selected region is automatically subdivided by the program into manageable patches (B). The region of interest is split into foreground and background and any patch with foreground in it is handled in turn (C). The patch is loaded at full resolution (D). All white areas are identified and areas that are too small or too large according to the given parameters are immediately discounted. All other white areas are judged further according to the logistic regression analysis. In (E), the green areas pass the logistic regression threshold (i.e., these are steatosis), whereas the cyan areas do not (no steatosis). Finally, only the steatosis areas are kept and measured, after which the tool calculates the steatosis proportionate area of the whole tissue (F). [Color figure can be viewed at http://wileyonlinelibrary.com]
To allow execution of the software on an office grade computer, the selected area is automatically subdivided into consecutive non‐overlapping patches of 3000 by 3000 pixels (729.28 by 729.28 μm at full zoom) (see Fig. 1B). The automated steatosis detection and area measurements are consecutively performed on each individual patch. A foreground detection algorithm is run on every patch to detect if there is any tissue present at all 23. To reduce the analysis time, the patch is not processed if no tissue is detected. The plugin is publically available via this website: https://github.com/Mverp/Steatosis-Measurer.
Steatosis Measurements
Steatotic hepatocytes are typically characterized by 1 a white color, 2 a specific size range, and 3 a round shape 20. The steatosis quantification algorithm detects potential steatotic hepatocytes and distinguishes these from similar objects (e.g., blood vessels, bile ducts, and tissue tearing) on the basis of color, size, and roundness features. The algorithm first establishes a set of potential steatotic hepatocytes in the image by applying a threshold to the saturation channel of the HSB color space. We found that a threshold that optimally distinguishes between white steatotic hepatocytes and more pinkish areas (e.g., cytoplasm within the hepatocyte) is relatively high and will underestimate the surface area of the hepatocytes (see Fig. 2). We therefore first applied a less conservative threshold to identify steatotic hepatocytes with the correct surface area after which the stricter, optimal threshold is used to confirm that the area is indeed a good candidate.
Figure 2.
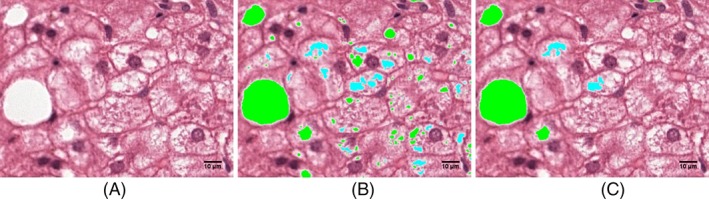
Color saturation thresholding. A part of a whole‐slide image that contains a lot of small lightly colored areas that are not steatotic (A). Without a minimum size, all these areas will be classified as either steatosis or normal tissue and must be corrected manually (B). By introducing a minimum‐size threshold the smaller sized objects are automatically judged to be non‐steatotic tissue and no longer need correction (C). [Color figure can be viewed at http://wileyonlinelibrary.com]
To distinguish between true steatotic hepatocytes and other white areas detected by the procedure described above, we used the size and roundness of detected objects. As a first step, we removed very large (tears in the tissue and non‐tissue background parts of the WSI) and very small (e.g., vacuoles within a hepatocyte, interstitial spaces) objects, while taking care not to remove any true steatotic hepatocytes. To perform a granular separation between steatotic hepatocytes and other objects remaining after the previous steps, we constructed a statistical classifier using logistic regression analysis (SPSS, version 22 Chicago, Illinois). The classifier used measures expressing the size and roundness of objects, as expressed in the following quantitative features 24:
where major axis is the length of the major axis of the ellipse best fitted to the area).
Solidity: (the amount to which an area is convex or concave).
The classifier was constructed using manually classified objects in representative areas from 20 randomly picked biopsies from different patients (five from each NAS grade of steatosis: 0–3). The dataset was split up in a training and test set (50–50%). The classifier resulting from logistic regression analysis was used for the final discrimination, applying a threshold of 0.5 on the classifier output.
After applying the above procedure, the summed surface area of all detected steatotic hepatocytes as well as the total tissue area was calculated. These sums were aggregated over all image patches in a WSI and the steatosis proportionate area (SPA) per WSI (the ratio of the steatotic area to the total tissue area described as a percentage) was calculated.
Correlation to NAS Grading
Biopsies were revised by an experienced pathologist, assessing steatosis grade and fibrosis stage 12. Correlation between SPA, resulting from the algorithm and NAS grade by the pathologist, was analyzed with Spearman Rank correlation coefficient and independent‐samples Kruskall‐Wallis test with post hoc analyses to test significance of differences between independent grades. Distribution of SPA (interquartile range), fat droplet size (in μm2), and number of droplets (per patch and per um2) were analyzed to assess heterogeneity of steatosis within slides and per steatosis grade.
RESULTS
Identification and Classification Parameters
Candidate steatotic hepatocytes were identified by application of two thresholds in the saturation channel after transforming the image from an RGB to HSB color representation. The threshold values were established based on visual effect estimation of applying up to 10 different threshold cut‐offs in a random selection of WSI samples. In a comparable manner, we established feature value thresholds to filter out obvious non‐steatotic areas on the basis of their size (see Table 1).
Table 1.
Feature Thresholds
| Feature | Threshold |
|---|---|
| Saturation for correct surface area | 29 (of 256) |
| Saturation for optimal separation of steatosis | 15 (of 256) |
| Minimal size for optimal separation of steatosis | 12 μm2 |
| Minimum area size | 25 μm2 |
| Maximum area size | 6000 μm2 |
Results of the Logistic Regression Analysis
Feature coefficients for roundness parameters and size were calculated using logistic regression analysis, resulting in the following classifier:
The area under the curve (AUC) of correctly classified steatosis by the algorithm on WSIs in the test set was 0.970 (95% CI 0.968–0.973), P < 0.001 (see Fig. 3). For the present study, an operating point of 0.5 showed the best overall performance of the classifier (see Supplementary Information Table 1).The classifier yields an accuracy of 91.9%, with a classification error of 8.1%. This accuracy shows the performance of the classifier on numbers of objects within the set of pre‐selected candidates. As we are interested in the SPA rather than the numbers of objects, we also calculated the error in summed steatotic cells areas as a result of false positives and false negatives. From the areas calculated on the test set (see Table 2), we can calculate the relative area of false positives as 167,779/3,561,981 × 100% = 4.71% and that of false negatives as 94,011/3,561,981 × 100% = 2.64%.
Figure 3.
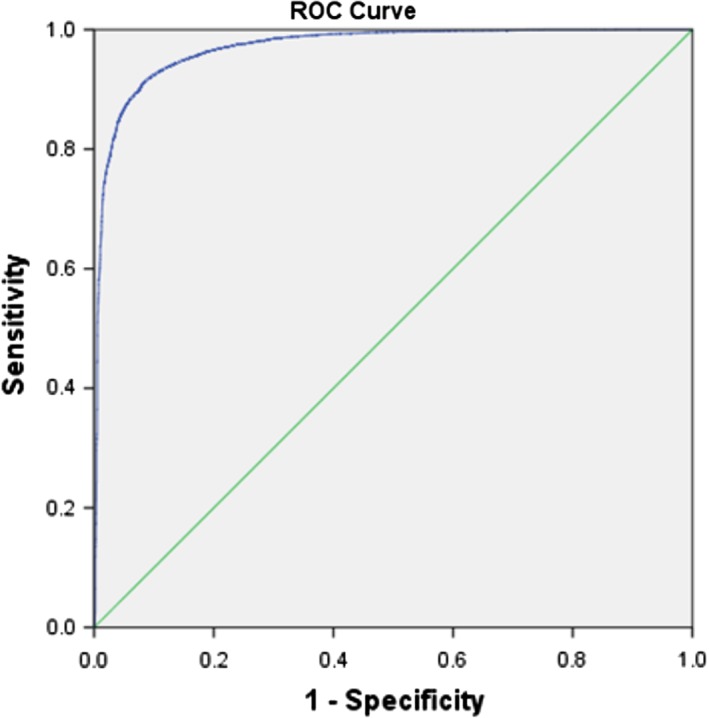
AUC for classification of steatosis. Cut offs of included size and roundness parameters were determined by logistic regression analysis on the test set. [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 2.
Classification Results in Both the Number of Areas and in Total Surface Area
| Classification | Number of areas | Total measured area (in μm2) |
|---|---|---|
| True positive | 16,612 | 3,561,981 |
| False negative | 585 | 94,011 |
| False positive | 850 | 167,779 |
Correlation Between SPA and Steatosis Grading
A total of 79 biopsies were included in the correlation between SPA and steatosis grade (61 patients with NAFLD and 18 controls without hepatic steatosis). The median SPA for Grade 0 steatosis was 1.41% (IQR 1.03–1.80%), in Grade 1: 4.99% (IQR 2.97–9.31%), in Grade 2: 13.65% (IQR 10.90–16.10%), and in Grade 3: 16.34% (IQR 14.48–20.54%) (see Table 3). A strong and significant positive correlation was observed between SPA and steatosis grading (Rs = 0.845, CI: 0.749–0.902, P < 0.001). The SPA also differed significantly between steatosis grades in overall analysis (P < 0.001) and in post hoc analysis between individual grades, except between Grade 2 and 3 (see Fig. 4). In a subgroup analysis (n = 26) of patients with severe fibrosis (BRUNT fibrosis stage ≥2), the SPA also correlated significantly with steatosis grading (Rs = 0.821, P < 0.001).
Table 3.
Results of Automated Image Analysis for Each Grade of Steatosis
| Steatosis grade | N | SPA (%) | IQR of SPA | No. of droplets per mm2 | Droplet size (μm2) | IQR of droplet size |
|---|---|---|---|---|---|---|
| 0 | 18 | 1.41 (1.03–1.80) | 1.22 (0.82–1.53) | 179.5 (151.9–243.6) | 108 (94–124) | 40 (35–55) |
| 1 | 25 | 4.99 (2.97–9.31) | 4.04 (2.62–7.42) | 169.5 (131.9–278.6) | 170 (158–233) | 105 (90–226) |
| 2 | 25 | 13.65 (10.90–16.10) | 8.58 (7.51–11.12) | 169.4 (119.7–288.9) | 249 (218–309) | 279 (239–336) |
| 3 | 11 | 16.34 (14.48–20.54) | 9.26 (8.84–13.09) | 169.8 (136.1–304.2) | 226 (207–321) | 265 (206–406) |
N = number of biopsies analyzed. Median values + IQR (25–75th percentile) are shown.
Figure 4.
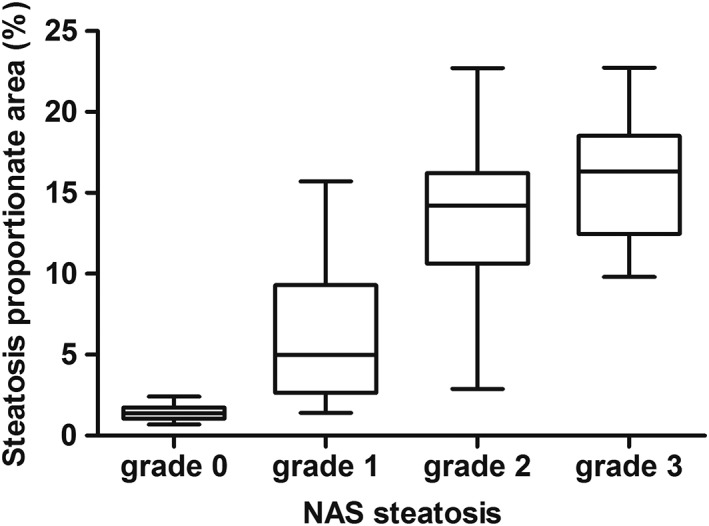
SPA per NAS grade. SPA differs significantly in all post hoc analyses between grades (Grade 0 vs. 1; 0 vs. 2; 0 vs. 3; 1 vs. 2; 1 vs. 3) with P < 0.01, except between Grade 2 and 3 (P = 1.000). Corresponding SPA cut offs were calculated (weighing the mean + SD between grades): Grade 0–1: SPA 3.21%, Grade 1–2: SPA 9.73%, Grade 2–3: SPA 15.63%.
Patch Distribution of Steatosis
We found a significant correlation between IQR of the SPA and steatosis grade (Rs = 0.822, P < 0.001). Similarly, droplet size (in μm2) and total number of droplets per slide correlated significantly with steatosis grade (Rs = 0.723, P < 0.001 and Rs = 0.689, P < 0.001, respectively). Number of droplets per mm2 remained equal in all steatosis grades (see Table 3). This results in an increasing heterogeneity in distribution of SPA with progressing steatosis grades, despite the equal number of droplets per mm2 in all grades. Because every tissue slide is divided in hundreds of smaller patches for measurement purposes the measured SPA per patch differs greatly, as is shown by the heterogeneity of individual patch SPA percentages in a randomly chosen tissue slide for every grade of steatosis (see Fig. 5).
Figure 5.
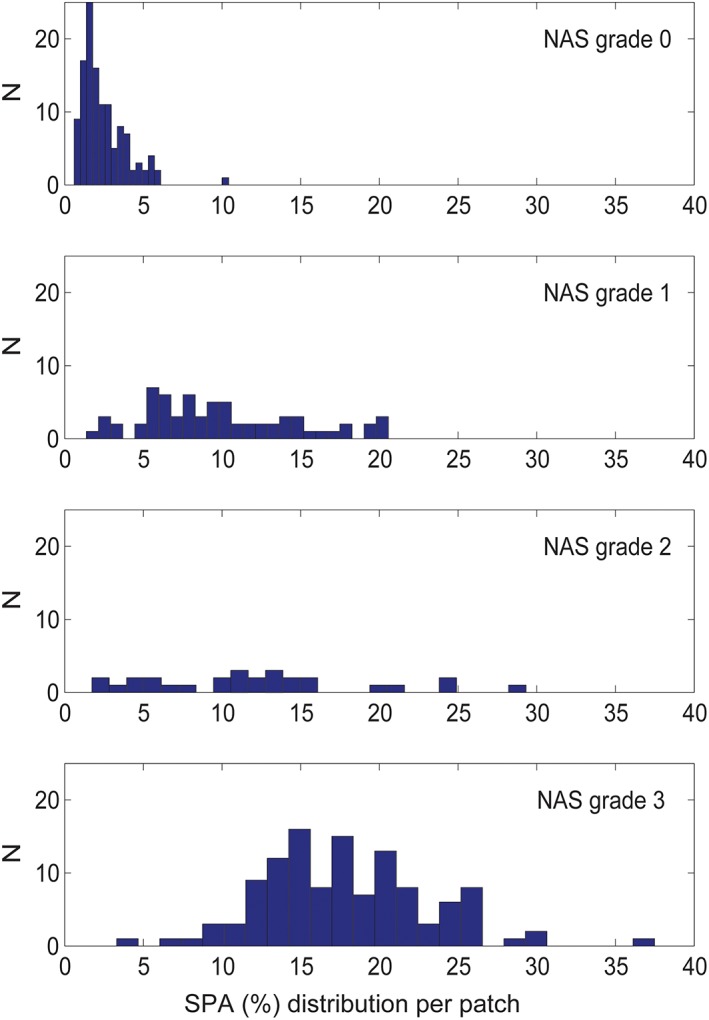
Distribution of total fat percentage per patch. This figure shown for a representative WSI of each NAS steatosis grade (0–1–2–3). On the X‐axis, the SPA % per patch is shown, and on the Y‐axis the number of patches in the representative WSI. [Color figure can be viewed at http://wileyonlinelibrary.com]
DISCUSSION
We describe the development of a novel automated digital analysis algorithm that quantifies liver steatosis on WSIs as a SPA. The algorithm shows good accuracy for discrimination between steatosis and non‐steatotic tissue.
There is an unmet need for an objective quantification of hepatic steatosis. Therapeutical drug trials include reduction of steatosis as study outcome. In the transplantation setting, donor livers with moderate to severe steatosis (approximately exceeding 30%) are considered low quality grafts 25, 26. Quantitative analysis can increase reliability of steatosis assessment and truly determine the clinico‐pathological correlations between severity of steatosis and graft failure. Furthermore, development of non‐invasive alternatives to assess steatosis severity will benefit from a reference standard that has a continuous scale.
Several attempts have been made to establish DIA for this goal but thresholds showed to be insufficient to facilitate automated discrimination, as these required an additional manual correction or special lipid (Oil Red O) staining procedures 14, 18, 27, 28, 29, 30, 31.
By identifying and combining optimal thresholds for several roundness and size parameters with logistic regression, we were able to accurately and automatically quantify steatosis. From the feature coefficients of the logit function, it appears that the size feature has a lesser impact on the classification than the shape features. This is due to the large difference in scale. The shape features are all ranged between 0 and 1, whereas size can reach into the thousands (μm2). For the shape features, it is interesting to note that the solidity has the most influence on the outcome of the classification. This indicates that for steatotic areas it is relatively rare to have protrusions and cavities while flatness and elongation are less disqualifying factors.
An additional benefit of this algorithm is its ability to process WSIs. Tools that rely on (random) selection of fragments of the whole slide are at risk for over‐ or underestimation of steatosis 14, 16, 27, 30, 31, 32. This is particularly relevant as steatosis is heterogeneously distributed, which further increases as the amount of steatosis rises 30.
It takes the algorithm approximately 10 min to analyze the whole slide and produce the SPA. In addition, multiple slides, with no limitation in number, can be imported into the image analysis program to be analyzed automatically in subsequent order. The program can be managed by a technician, leaving the pathologist free handed for other tasks.
The digitally quantified SPA showed to correlate well with NAS steatosis grading; however, SPA was in all cases lower than the percentage range resembled by the pathologists grade. This observation, as well as the absolute percentages measured with our algorithm, corroborates findings from the other DIA methods quantifying steatosis 14, 16, 18, 20, 30, 33. Although scoring by the pathologist is currently the gold standard, it suffers from interobserver and intra‐observer variability 13. Steatotic hepatocytes are identified as white droplets upon histological examination of hematoxylin–eosin stained liver specimens 34. Nonetheless, accurately estimating total percentage of steatosis has been shown to be difficult for pathologists 35. Ideally, the pathologist visually estimates the relative percentage of steatotic hepatocytes compared to all hepatocytes present. However, individual hepatocytes are too small, numerous, and indistinctive to be counted at low magnification. Furthermore, visual overestimation of steatosis tends to increase with progressing severity 14. A possible explanation for such overestimation is proposed by Rawlins et al. and comes from psychological studies that investigated how people judge quantity. When there is a high number of identical items arranged in an area, as is the case with many steatotic hepatocytes in liver parenchyma, people tend to overestimate quantity 18, 36, 37. Currently, thresholds for no, mild, moderate and severe steatosis are based upon the NAS score. As a result of the much lower steatosis percentages measured with DIA, such thresholds need to be re‐established before used in the clinical setting.
There are some limitations to our algorithm. First, visual examination by pathologists allows detection and grading of more abnormalities than only steatosis. We therefore suggest this algorithm to be supplemental to a pathologist's assessment.
A second limitation of the algorithm comes from the automated division of the slide image in smaller patches. Steatotic hepatocytes aligned at the edge of the patch will be cleaved, affecting area size and roundness features, possibly causing misclassification. Nevertheless, in our cohort misclassification (false positives and negatives) led to only a small fractional error on total SPA.
Lastly, with establishing thresholds, we only incorporated clear‐cut features such as color, size, and roundness in a logistic regression analysis. We cannot exclude that in the future other features will turn to be clinically relevant. More advanced techniques such as deep learning, may contribute to future algorithms investigating multiple histopathological features at once.
To conclude, we present a novel automated digital analysis algorithm that accurately and objectively quantifies liver steatosis on WSIs, and has the potential to be incorporated as an addition to visual examination by pathologists.
Conflict of Interests
All authors state they have no conflicts of interest.
AUTHORS' CONTRIBUTIONS
E. Tjwa, I. Munsterman, M. Van Erp, J. van der Laak, and J. Drenth conceived and designed the study, and wrote, edited, and reviewed the manuscript. I. Munsterman, M. Van Erp, G. Weijers, C. Bronkhorst, and C. de Korte researched and analyzed and interpreted data and wrote, edited, and reviewed the manuscript. All authors gave final approval for publication. IM takes full responsibility for the work as a whole, including the study design, access to data and the decision to submit and publish the manuscript.
COMPLIANCE WITH ETHICAL STANDARDS
Requirement for ethical approval was waived by the institutional review board (no. 2016‐2763). Patient material was used according the “Code of Conduct for responsible use of Human tissue and Medical Research” 38.
FUNDING
No funding for conduct of this study was received.
Supporting information
Supplementary table 1 Analysis of threshold cut‐offs on performance of the classifier
ACKNOWLEDGMENTS
The authors thank Irene Otte and Rob van de Loo from the Department of Pathology for assistance in digitizing slides.
How to cite this article: Munsterman ID, van Erp M, Weijers G, Bronkhorst C, de Korte CL, Drenth JPH, van der Laak JAWM, and Tjwa ETTL. A Novel Automatic Digital Algorithm that Accurately Quantifies Steatosis in NAFLD on Histopathological Whole‐Slide Images. Cytometry Part B 2019;96B:521–528.
LITERATURE CITED
- 1. Wree A, Broderick L, Canbay A, Hoffman HM, Feldstein AE. From NAFLD to NASH to cirrhosis‐new insights into disease mechanisms. Nat Rev Gastroenterol Hepatol. 2013;10:627–636. 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 2. Tapper EB, Loomba R (2017) NAFLD, Metabolic Syndrome, and the Fight That Will Define Clinical Practice for a Generation of Hepatologists Hepatology. doi: 10.1002/hep.29722 [DOI] [PMC free article] [PubMed]
- 3. Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease‐meta‐analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 4. Bugianesi E, Marchesini G, Gentilcore E, Cua IH, Vanni E, Rizzetto M, George J. Fibrosis in genotype 3 chronic hepatitis C and nonalcoholic fatty liver disease: Role of insulin resistance and hepatic steatosis. Hepatology. 2006;44:1648–1655. 10.1002/hep.21429. [DOI] [PubMed] [Google Scholar]
- 5. Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: A systematic review and meta‐analysis of paired‐biopsy studies. Clin Gastroenterol Hepatol. 2015;13:643–654 e641‐649; quiz e639‐640. 10.1016/j.cgh.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bedossa P, Moucari R, Chelbi E, Asselah T, Paradis V, Vidaud M, Cazals‐Hatem D, Boyer N, Valla D, Marcellin P. Evidence for a role of nonalcoholic steatohepatitis in hepatitis C: A prospective study. Hepatology. 2007;46:380–387. 10.1002/hep.21711. [DOI] [PubMed] [Google Scholar]
- 7. Blonsky JJ, Harrison SA. Review article: Nonalcoholic fatty liver disease and hepatitis C virus‐‐partners in crime. Aliment Pharmacol Ther. 2008;27:855–865. 10.1111/j.1365-2036.2008.03672.x. [DOI] [PubMed] [Google Scholar]
- 8. de Graaf EL, Kench J, Dilworth P, Shackel NA, Strasser SI, Joseph D, Pleass H, Crawford M, McCaughan GW, Verran DJ. Grade of deceased donor liver macrovesicular steatosis impacts graft and recipient outcomes more than the donor risk index. J Gastroenterol Hepatol. 2012;27:540–546. 10.1111/j.1440-1746.2011.06844.x. [DOI] [PubMed] [Google Scholar]
- 9. Spitzer AL, Lao OB, Dick AA, Bakthavatsalam R, Halldorson JB, Yeh MM, Upton MP, Reyes JD, Perkins JD. The biopsied donor liver: Incorporating macrosteatosis into high‐risk donor assessment. Liver Transpl. 2010;16:874–884. 10.1002/lt.22085. [DOI] [PubMed] [Google Scholar]
- 10. Stern C, Castera L. Non‐invasive diagnosis of hepatic steatosis. Hepatol Int. 2017;11:70–78. 10.1007/s12072-016-9772-z. [DOI] [PubMed] [Google Scholar]
- 11. Sumida Y, Yoneda M. Current and future pharmacological therapies for NAFLD/NASH. J Gastroenterol. 2017;53:362–376. 10.1007/s00535-017-1415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp‐Arida A, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 13. El‐Badry AM, Breitenstein S, Jochum W, Washington K, Paradis V, Rubbia‐Brandt L, Puhan MA, Slankamenac K, Graf R, Clavien PA. Assessment of hepatic steatosis by expert pathologists: The end of a gold standard. Ann Surg. 2009;250:691–697. 10.1097/SLA.0b013e3181bcd6dd. [DOI] [PubMed] [Google Scholar]
- 14. Hall AR, Dhillon AP, Green AC, Ferrell L, Crawford JM, Alves V, Balabaud C, Bhathal P, Bioulac‐Sage P, Guido M, et al. Hepatic steatosis estimated microscopically versus digital image analysis. Liver Int. 2013;33:926–935. 10.1111/liv.12162. [DOI] [PubMed] [Google Scholar]
- 15. EASL‐ALEH . Clinical practice guidelines: Non‐invasive tests for evaluation of liver disease severity and prognosis. J Hepatol. 2015;63:237–264. 10.1016/j.jhep.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 16. Li M, Song J, Mirkov S, Xiao SY, Hart J, Liu W. Comparing morphometric, biochemical, and visual measurements of macrovesicular steatosis of liver. Hum Pathol. 2011;42:356–360. 10.1016/j.humpath.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nativ NI, Chen AI, Yarmush G, Henry SD, Lefkowitch JH, Klein KM, Maguire TJ, Schloss R, Guarrera JV, Berthiaume F, et al. Automated image analysis method for detecting and quantifying macrovesicular steatosis in hematoxylin and eosin‐stained histology images of human livers. Liver Transpl. 2014;20:228–236. 10.1002/lt.23782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rawlins SR, El‐Zammar O, Zinkievich JM, Newman N, Levine RA. Digital quantification is more precise than traditional semiquantitation of hepatic steatosis: Correlation with fibrosis in 220 treatment‐Naïve patients with chronic hepatitis C. Dig Dis Sci. 2010;55:2049–2057. 10.1007/s10620-010-1254-x. [DOI] [PubMed] [Google Scholar]
- 19. St. Pierre TG, House MJ, Bangma SJ, Pang W, Bathgate A, Gan EK, Ayonrinde OT, Bhathal PS, Clouston A, Olynyk JK, et al. Stereological analysis of liver biopsy histology sections as a reference standard for validating non‐invasive liver fat fraction measurements by MRI. PLoS One. 2016;11:e0160789 10.1371/journal.pone.0160789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vanderbeck S, Bockhorst J, Komorowski R, Kleiner DE, Gawrieh S. Automatic classification of white regions in liver biopsies by supervised machine learning. Hum Pathol. 2014;45:785–792. 10.1016/j.humpath.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: An open‐source platform for biological‐image analysis. Nat Methods. 2012;9:676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suceat Y, Angelos P, Waelput W (2017) Free Whole Slide Image Viewer – PMA.Start. Universal Digital Microscopy Software. Free Pathomation, pp.
- 23. Bug D, Feuerhake F, Merhof D. Foreground extraction for histopathological whole slide imaging In: Handels H, Deserno TM, Meinzer H‐P, Tolxdorff T, editors. Bildverarbeitung für die Medizin 2015: Algorithmen ‐ Systeme ‐ Anwendungen. Proceedings des Workshops vom 15. bis 17. März 2015 in Lübeck. Berlin, Heidelberg: Springer Berlin Heidelberg, 2015; p. 419–424. [Google Scholar]
- 24.Image J User Guide (2012). https://imagej.nih.gov/ij/docs/guide/146-30.html
- 25. D'Alessandro AM, Kalayoglu M, Sollinger HW, Hoffmann RM, Reed A, Knechtle SJ, Pirsch JD, Hafez GR, Lorentzen D, Belzer FO. The predictive value of donor liver biopsies on the development of primary nonfunction after orthotopic liver transplantation. Transplant Proc. 1991;23:1536–1537. [PubMed] [Google Scholar]
- 26. Garcia Urena MA, Colina Ruiz‐Delgado F, Moreno Gonzalez E, Jimenez Romero C, Garcia Garcia I, Loinzaz Segurola C, Gonzalez P, Gomez Sanz R. Hepatic steatosis in liver transplant donors: Common feature of donor population? World J Surg. 1998;22:837–844. [DOI] [PubMed] [Google Scholar]
- 27. Fiorini RN, Kirtz J, Periyasamy B, Evans Z, Haines JK, Cheng G, Polito C, Rodwell D, Shafizadeh SF, Zhou X, et al. Development of an unbiased method for the estimation of liver steatosis. Clin Transplant. 2004;18:700–706. 10.1111/j.1399-0012.2004.00282.x. [DOI] [PubMed] [Google Scholar]
- 28. Levene AP, Kudo H, Thursz MR, Anstee QM, Goldin RD. Is oil red‐O staining and digital image analysis the gold standard for quantifying steatosis in the liver? Hepatology. 2010;51:1859–1859. 10.1002/hep.23551 1859; author reply 1860. [DOI] [PubMed] [Google Scholar]
- 29. Riva G, Villanova M, Cima L, Ghimenton C, Bronzoni C, Colombari R, Crestani M, Sina S, Brunelli M, D'Errico A, et al. Oil red O is a useful tool to assess donor liver steatosis on frozen sections during transplantation. Transplant Proc. 2018;50:3539–3543. 10.1016/j.transproceed.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 30. Turlin B, Ramm GA, Purdie DM, Laine F, Perrin M, Deugnier Y, Macdonald GA. Assessment of hepatic steatosis: Comparison of quantitative and semiquantitative methods in 108 liver biopsies. Liver Int. 2009;29:530–535. 10.1111/j.1478-3231.2008.01874.x. [DOI] [PubMed] [Google Scholar]
- 31. Zaitoun AM, Al Mardini H, Awad S, Ukabam S, Makadisi S, Record CO. Quantitative assessment of fibrosis and steatosis in liver biopsies from patients with chronic hepatitis C. J Clin Pathol. 2001;54:461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Levene AP, Kudo H, Armstrong MJ, Thursz MR, Gedroyc WM, Anstee QM, Goldin RD. Quantifying hepatic steatosis ‐ more than meets the eye. Histopathology. 2012;60:971–981. 10.1111/j.1365-2559.2012.04193.x. [DOI] [PubMed] [Google Scholar]
- 33. Franzen LE, Ekstedt M, Kechagias S, Bodin L. Semiquantitative evaluation overestimates the degree of steatosis in liver biopsies: A comparison to stereological point counting. Mod Pathol. 2005;18:912–916. 10.1038/modpathol.3800370. [DOI] [PubMed] [Google Scholar]
- 34. Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander‐Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: A proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 35. Hall AR, Green AC, Luong T‐V, Burroughs AK, Wyatt J, Dhillon AP. The use of guideline images to improve histological estimation of hepatic steatosis. Liver Int. 2014;34:1414–1427. 10.1111/liv.12614. [DOI] [PubMed] [Google Scholar]
- 36. Redden JP, Hoch SJ. The presence of variety reduces perceived quantity. J Consum Res. 2009;36:406–417. 10.1086/598971. [DOI] [Google Scholar]
- 37. Vos PG, van Oeffelen MP, Tibosch HJ, Allik J. Interactions between area and numerosity. Psychol Res. 1988;50:148–154. 10.1007/bf00310175. [DOI] [PubMed] [Google Scholar]
- 38. Organisation F (2011) Code of Conduct for responsible use of Human Tissue and Medical Research
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1 Analysis of threshold cut‐offs on performance of the classifier


