Abstract
RNA can be modified in over 100 distinct ways, and these modifications are critical for function. Pseudouridine synthases catalyse pseudouridylation, one of the most prevalent RNA modifications. Pseudouridine synthase 7 modifies a variety of substrates in Saccharomyces cerevisiae including tRNA, rRNA, snRNA, and mRNA, but the substrates for other budding yeast Pus7 homologues are not known. We used CRISPR‐mediated genome editing to disrupt Candida albicans PUS7 and find absence leads to defects in rRNA processing and a decrease in cell surface hydrophobicity. Furthermore, C. albicans Pus7 absence causes temperature sensitivity, defects in filamentation, altered sensitivity to antifungal drugs, and decreased virulence in a wax moth model. In addition, we find C. albicans Pus7 modifies tRNA residues, but does not modify a number of other S. cerevisiae Pus7 substrates. Our data suggests C. albicans Pus7 is important for fungal vigour and may play distinct biological roles than those ascribed to S. cerevisiae Pus7.
Keywords: C. albicans, pseudouridine, pseudouridine synthase 7, pseudouridylation, Pus7, RNA modification, rRNA processing
Abbreviations used
- Pus7
pseudouridine synthase 7
- CaPus7
Candida albicans pseudouridine synthase 7
- ScPus7
Saccharomyces cerevisiae pseudouridine synthase 7
- Ψ
pseudouridine
- ITS1
ribosomal RNA internal‐transcribed spacer 1
- ITS2
ribosomal RNA internal‐transcribed spacer 2
- KTI11
putative Candida albicans Zn‐ribbon protein required for synthesis of diphthamide on translation factor eEF2 and involved in modification of wobble nucleosides in tRNAs
- tE(CUC)
tRNA‐Glu with CUC anticodon
- tE(UUC)
tRNA‐Glu with UUC anticodon
1. INTRODUCTION
RNA bases can be modified in over 100 distinct ways (Machnicka et al., 2013), and these modifications play critical roles in translation, transcription, and splicing (Hoernes & Erlacher, 2017; Roundtree, Evans, Pan, & He, 2017). Pseudouridine (Ψ) is one of the most prevalent RNA modifications and is found in all kingdoms of life (Ge & Yu, 2013). Pseudouridylation requires a pseudouridine synthase cleave the C–N glycosidic bond of a uridine in an RNA chain. The pseudouridine synthase then reattaches the uracil moiety at the C5 position, forming pseudouridine (Cerrudo, Ghiringhelli, & Gomez, 2014; Hammal & Ferre‐D'Amare, 2006). The exposure of N1 following modification permits pseudouridine to participate in additional hydrogen bonds, which have been proposed to confer RNA structural stability (Ofengand, 2002). Pseudouridine is highly abundant in tRNA and the rRNA catalytic centre where it is proposed to play important roles in peptide bond formation (Bakin, Lane, & Ofengand, 1994). The amount of pseudouridylation varies among organisms. For instance, bacteria rRNA contain between five and 15 pseudouridines, whereas mammalian rRNA contains over 70 (Spenkuch, Motorin, & Helm, 2014). In addition, pseudouridine is found in small nuclear RNAs (snRNA), small nucleolar RNAs (snoRNA), and mRNA, (Carlile et al., 2014; Lovejoy, Riordan, & Brown, 2014; Schwartz et al., 2014; Yu, Ge, & Yu, 2011); however, the roles of pseudouridine in these RNA species are less clear.
Pseudouridylation is catalysed by pseudouridine synthases. The number of pseudouridine synthases varies by organism. Some bacteria encode a single pseudouridine synthase, whereas Saccharomyces cerevisiae encodes 10 pseudouridine synthases (Ofengand, 2002; Schwartz et al., 2014) that modify a variety of substrates. The mechanism by which pseudouridine synthases identify their targets varies. Cbf5, for instance, uses snoRNAs to recognize its targets (Lafontaine, Bousquet‐Antonelli, Henry, Caizergues‐Ferrer, & Tollervey, 1998; Watkins et al., 1998). All other S. cerevisiae pseudouridine synthases use a combination of RNA primary and secondary structure to identify their target residues. Some of these residues lie in consensus motifs, but the presence of a consensus motif does not ensure modification will occur (Schwartz et al., 2014). Some pseudouridine synthases target only a single residue or class of residues. For instance, pseudouridine synthase 5 (Pus5) only targets a single residue in the mitochondrial rRNA (Ansmant, Massenet, Grosjean, Motorin, & Branlant, 2000). S. cerevisiae pseudouridine synthase 7 (ScPus7) on the other hand modifies a wide variety of RNA substrates, including snRNA, tRNA, rRNA, and mRNA (Behm‐Ansmant et al., 2003; Decatur & Schnare, 2008; Ma, Zhao, & Yu, 2003; Schwartz et al., 2014). Bacterial Pus7 homologues however modify only tRNAs (Kaya & Ofengand, 2003). Although PUS7 homologues are found in many fungal taxa, it is not known if such substrate promiscuity is conserved.
We characterized the lone Pus7 homologue encoded by the human fungal pathogen Candida albicans (CaPUS7). C. albicans is a member of the CTG clade of fungi that diverged from Saccharomyces roughly 170 million years ago (Massey et al., 2003). Little is known about the process of pseudouridylation in the CTG fungal clade, but genome‐wide expression studies indicate CaPUS7 is induced during biofilm formation, a key fungal virulence factor (Murillo et al., 2005).
To examine the roles of CaPus7, we deleted CaPUS7 from a clinical fungal isolate. Our data suggest CaPus7 is important for rRNA biogenesis and loss of CaPus7 results in changes to C. albicans virulence and fitness. Furthermore, we find CaPus7 and ScPus7 modify distinct substrates. Our data suggest although pseudouridine synthases are conserved between budding yeast taxa, their substrates and roles in vivo are not.
2. METHODS
2.1. Media
YPD: 1% yeast extract, 2% bacto‐peptone, 2% dextrose, 0.15% L‐tryptophan, 0.27‐mM uridine. synthetic complete media (SC), yeast nitrogen broth‐bovine serum albumin (YNB‐BSA) (“Guide to yeast genetics and molecular biology,” 1991), and Spider media, (Liu, Kohler, & Fink, 1994).
2.2. CRISPR‐Cas9‐mediated C. albicans genome editing
pus7/pus7 C. albicans were generated using the method described in Vyas, Barrasa, and Fink (2015). Twenty‐base pair guide sequences (Table S2) were cloned into vector pv1093 (Vyas et al., 2015), which encodes the Cas9 endonuclease as well as nourseothricin yeast and ampicillin bacterial drug resistance markers. One hundred‐base pair repair templates (Table S2) introduced two consecutive stop codons and an EcoRI restriction site to PUS7 upon cleavage repair. Yeast cotransformed with guide and repair template were plated on YPD + Uri supplemented with 200 μg/ml of nourseothricin. Colonies were screened by PCR and EcoRI restriction digestion (Figure 1b). Strain names are listed in Table S3.
Figure 1.
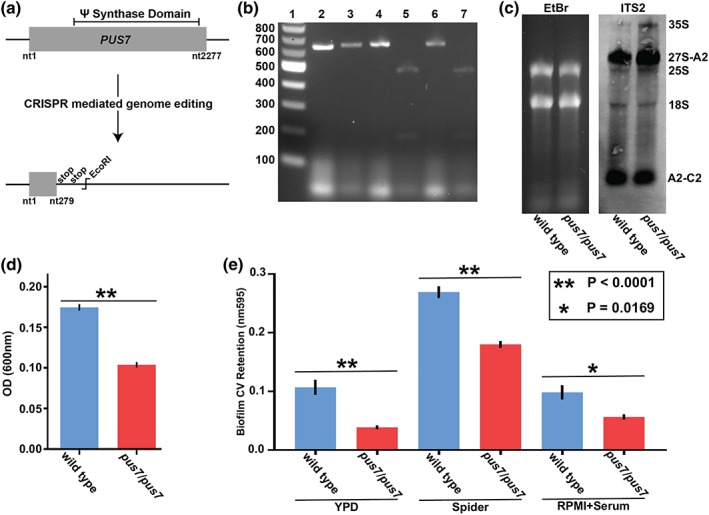
CaPus7 impact on rRNA processing intermediate abundance and cell hydrophobicity. (a) Cartoon depiction of PUS7 disruption. (b) Restriction digestion of colony PCR from potential pus7/pus7 transformants. Lane 1. Ladder 2. PCR product 3. Same PCR product from lane 2 EcoRI‐digested 4. PCR product 5. Same PCR product from lane 4 EcoRI‐digested 6. PCR product 7. Same PCR product from lane 6 EcoRI‐digested (c) Northern blot with ITS2 probe comparing wild type and pus7/pus7 total RNA. Ethidium bromide (EtBr) staining confirms equal loading. Small amounts of 25S and 18S rRNA background staining are labelled; 35S unprocessed rRNA, and 27S‐A2, and A2‐C2 rRNA processing intermediates containing ITS2 are labelled. Four biological replicates were performed, and the image shown is representative. (d) Hydrophobicity was assessed via a xylene hydrocarbon partitioning. Values are the change in aqueous phase absorbance (600 nm) after addition of xylene. (e) Assessment of pus7/pus7 and wild type biofilm formation. Values are absorbance (595 nm) from crystal violet destaining solution retained by biofilms
2.3. RNA sequencing and data analysis
RNA was extracted and purified using acid phenol. Five micrograms of total RNA was polyA‐purified, cloned into libraries, and sequenced by Genewiz on a HighSeq 2500, run in high output mode, for single read of 50 bases. Reference genome SC5314 version A22‐s07‐m01‐r20 was used for analysis. FASTA and gff files were separated by allele. The reference genome was indexed using Bowtie2, and reads were aligned to the indexed genome using Tophat (Trapnell et al., 2012). Alignments were organized into a matrix of read counts using the count function as part of the HTSeq package. Alignments that were ambiguous or not unique were not considered. Using the EdgeR package, genes were filtered from a matrix of counts on the basis of their counts per million reads (CPM). For each gene, a minimum of four libraries were required to have approximately 1.39 CPM to be included in the analysis. Libraries were normalized using EdgeR, and a negative binomial model was fitted for each gene to assess differences in expression level. Each condition had a corresponding coefficient in each model. A likelihood ratio test was used to assess if two given coefficients were significantly different. The cut‐off to consider a gene for differential expression was a p value lower than .001 and an absolute log2FC of greater than 1.3.
2.4. Growth and filamentation
Assays were performed as previously described in (Evans, Smith, et al., 2018). Fourfold serial dilutions were plated using a pin replicator.
2.5. Galleria mellonella infection
Assays were performed as in (Evans, Smith, et al., 2018). Briefly, 235 mg ± 45 mg larvae were randomly assigned to receive 10‐μl sterile PBS, wild type, or pus7/pus7 injections; 107 cfu of wild type or pus7/pus7 log phase cultures grown at 37°C in YPD were washed twice and suspended in 1‐ml sterile PBS. Larvae were incubated at 37°C, and survival was assessed daily by visual inspection. Experiments were discontinued after 7 days. Fifty larvae were subjected to each treatment, and data were assessed via Kaplan–Meier survival analysis and by the log rank test for multiple comparisons in R studio.
2.6. Drug resistance
Wild type or pus7/pus7 yeast were grown overnight in SC (“Guide to yeast genetics and molecular biology,” 1991). One million colony‐forming units of overnight cultures were suspended in 200‐μl SC and plated on SC 2% agar. Fifteen microlitres of 1‐mM fluconazole or 20 μl of 1‐mM caspofungin was added to a sterile paper disc at the centre of each plate. Assays were repeated in triplicate, and disk image R was used to analyse the data as described in Gerstein, Rosenberg, Hecht, and Berman (2016).
2.7. Cell surface hydrophobicity
Cell surface hydrophobicity assays were performed as described previously (Hazen, Plotkin, & Klimas, 1986). Briefly, 10 wild type and pus7/pus7 YPD + Uri log phase cultures grown at 30°C or 37°C were washed and suspended in sterile PBS to yield 5 × 106 cfu. OD600 values were recorded for each replicate. Three millilitres of each suspension was mixed with 1‐ml xylenes. Aqueous phase OD600 values were recorded for each replicate, whereas the aqueous phase of a 3:1 sterile PBS to xylenes solution was used to blank the spectrophotometer. Percent change in OD600 value was calculated for each replicate, and data were assessed via boxplot in R studio. Percent change data were then analysed using analysis of variance Tukey's honestly significant difference test.
2.8. Polystyrene biofilm formation
Biofilm assays were performed as described previously with the following changes (Pierce et al., 2008). YPD + Uri log phase cultures were diluted to 5 × 106 cfu per millilitre in RPMI with 5% serum, Spider, or YPD and added to a 96‐well plate. Each treatment was repeated 16 times, and data were plotted in GraphPad Prism 7. Percent change data were then analysed using analysis of variance Tukey's honestly significant difference test.
2.9. Northern blot
ITS2‐specific probe was generated by PCR incorporating biotin‐16‐dUTP. ITS2 probe was purified using Zymo research gel purification kit. RNA isolation was performed as described above, and 5‐μg RNA was loaded on a denaturing gel. Gel electrophoresis, transfer, and crosslinking were performed according to NorthernMax manual suggestions. Prehybridization, hybridization with ITS2 probe, washes, blocking, IRDye 800CW streptavidin incubation (1:10,000), and imaging were performed as described and recommended by the Odyssey Infrared Imaging System.
2.10. Primer extension
RNA was purified as described above and N‐cyclohexyl‐N′‐(2‐morpholinoethyl)carbodiimide metho‐p‐toluenesulfonate (CMC) treatment was performed as described (Bakin & Ofengand, 1993) with the following modifications. One hundred millimolars of CMC was prepared in BEU buffer. Fifty micrograms of aliquots of RNA was mixed with 30 μl of 100‐mM CMC in BEU or in 30 μl of BEU and incubated at 37°C for 20 min, followed by ethanol precipitation. RNA pellets were suspended in 40 μl of 50‐mM Na2CO3 (pH 10.4) and incubated for 3 hr at 37°C. After incubation, RNA samples were ethanol precipitated and suspended in 10‐μl H2O. Primer extensions were performed as described for avian myeloblastosis virus (AMV) reverse transcriptase (NEB M0277S) with the following modifications. One‐half microgram of CMC/Na2CO3‐treated RNA, 0.5‐mM dATP, 0.5‐mM dCTP, 0.5‐mM dGTP, 0.6‐mM dTTP, 0.2‐mM Biotin‐16‐dUTP, and 20‐μM primer (Table S4) were combined in 10 μl, incubated at 65°C for 5 min, and cooled to 4°C. Two microlitres of AMV buffer, 2‐μl AMV reverse transcriptase, and 6‐μl H2O were added and incubated at 42°C for 1 hr and 80°C for 5 min. Unincorporated biotin‐16‐dUTPs were removed, and separation of extension products was performed on a 12% PAGE Urea gel as previously described (by National Diagnostics). Samples were run at 2 W for 20 min followed by 20 W for 2 hr. RNA was transferred to membrane using idEA Scientific GENIE Transfer Apparatus, and membrane was cross‐linked at 120,000 μJ. Blocking, IRDye 800CW streptavidin incubation (1:10,000), and imaging were performed as recommended by Odyssey Infrared Imaging System manual.
3. RESULTS
To investigate the roles of CaPus7, C. albicans Cas9‐mediated genome editing was used to introduce two stop codons at nucleotide 279 in C. albicans clinical isolate SC5314 (wild type); (Evans, Pickerill, Vyas, & Bernstein, 2018; Vyas et al., 2015). Introduction of stop codons prematurely halts translation at amino acid 93 truncating CaPUS7 to eliminate the pseudouridine synthase domain (Figure 1a). PCR and restriction digestion (Figure 1b) confirmed five homozygous isolates (pus7/pus7).
RNA sequencing was performed to compare gene expression between wild type and pus7/pus7. A number of cell surface genes are down regulated in pus7/pus7 when compared with wild type (Tables 1 and S1). Gene Ontology analysis confirmed oligopeptide transporter genes are significantly down regulated in pus7/pus7 (Table 1, p value 4.79E‐07). The gene that increases in abundance most in pus7/pus7 is KTI11, which has been proposed to play roles in tRNA wobble base modification (Huang, Johansson, & Bystrom, 2005). The transcripts exhibiting the next greatest increase in abundance in pus7/pus7 are Internal Transcribed Spacer 1 (ITS1) and Internal Transcribed Spacer 2 (ITS2). ITS1 and ITS2 must be cleaved from the 35S rRNA transcript before functional 5.8, 18S, and 25S RNA can form (Venema & Tollervey, 1999). Our data suggests rRNA processing intermediates are more abundant in pus7/pus7 and elimination of CaPus7 causes a defect in rRNA processing. The presence of increased unprocessed rRNA and rRNA processing intermediates in pus7/pus7, including 35S rRNA, was confirmed via northern blot probing for ITS2 (Figure 1c). Our results are consistent with previously described C. albicans rRNA processing intermediates (Pendrak & Roberts, 2011). Moreover, a number of cell surface protein transcripts were differentially regulated in pus7/pus7, and we tested if this led to a change in cell surface hydrophobicity and biofilm formation. Using a xylene hydrocarbon‐partitioning assay, pus7/pus7 was found to be less hydrophobic than wild type (Figure 1d). In addition, pus7/pus7 less robustly formed biofilms compared to wild type (Figure 1e).
Table 1.
Transcripts in which abundance changed in pus7/pus7
| Transcripts with increased abundance in pus7/pus7 | IFO3, ITS1, ITS2, KTI11 |
| Transcripts with decreased abundance in pus7/pus7 | ALS2, CAN1, OPT2, OPT3, OPT4, OPT7, OPT9 |
Next, reverse transcriptase primer extension was used to identify CaPus7 substrates (Figure 2a). In S. cerevisiae, Pus7 modifies a variety of RNA substrates, including snRNA, tRNA, rRNA, and mRNA (Behm‐Ansmant et al., 2003; Decatur & Schnare, 2008; Ma et al., 2003; Schwartz et al., 2014). We first tested if CaPus7 is required for modification of tRNA E (CUC), a modification that is conserved among fungi and metazoan (Behm‐Ansmant et al., 2003; Guzzi et al., 2018). CaPus7 is required for modification of residue U13 of tRNA E (CUC) as primer extension terminates at the expected 38 base product (Figure 2b). Additional Pus7‐dependent primer extension termination occurred at extension products corresponding to residues U11 and U8. Next, we tested if CaPus7 is required for modification of U2 snRNA residues 36 or 57. These residues are analogous to sites modified by ScPus7 (Yu et al., 2011). We did not detect modification of either of these residues using primer extension (Figure S6). This suggests CaPus7 modifies some, but not all, of the same substrates as ScPus7.
Figure 2.
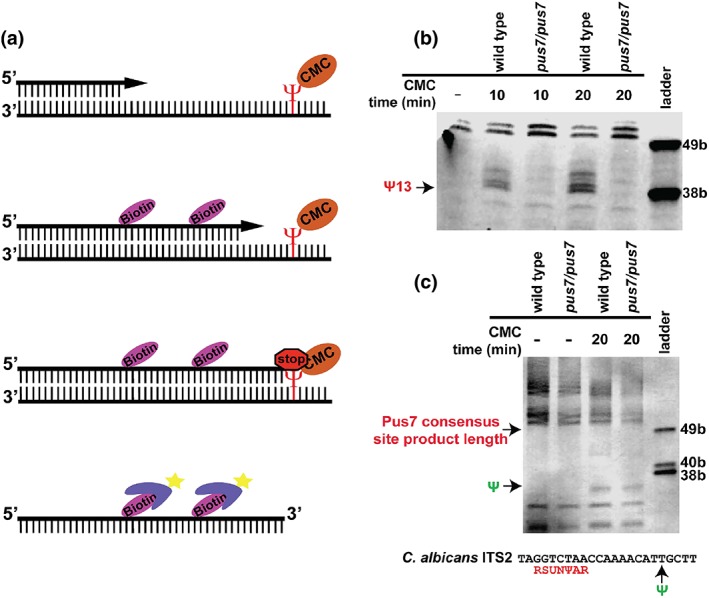
Primer extension mapping of CaPus7 dependent pseudouridylation via a primer extension. (a) Schematic of pseudouridine mapping. (b) Primer extension of tRNA E (CUC) Ψ13. Primer extensions were performed four times. Images shown are representative. (c) Primer extension of ITS2. ScPus7 consensus sequence found in ITS2 is labelled in red. Green Ψ indicates where we see CMC primer extension stoppage that was not dependent upon CaPus7. Primer extensions were performed three times. Images shown are representative
To determine if loss of Pus7 leads to changes in phenotype, serial dilutions of pus7/pus7 and wild type were spotted on YPD or synthetic complete media. (Figures 3a and S1). Qualitative assessment of growth suggests pus7/pus7 grew more slowly than wild type cells at all temperatures and exhibited the most severe growth defect at 40°C. Many genes important for nitrogen import, including peptide transporters (Tables 1 and S1) were down regulated in pus7/pus7. We tested if pus7/pus7 cells were more susceptible to nitrogen limitation by growing them on synthetic complete media that contained BSA as a nitrogen source (YNB‐BSA), but this did not exacerbate pus7/pus7 growth defects (Figure S2).
Figure 3.
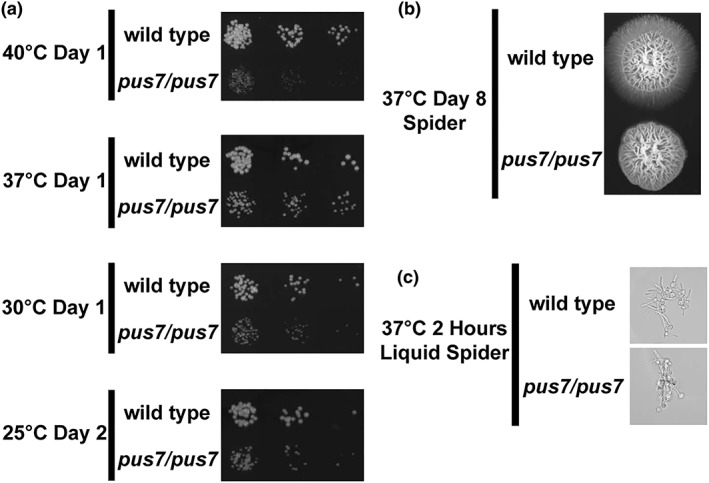
Pus7 impact on Candida albicans growth and filamentation. (a) Comparison of wild type and pus7/pus7 growth on YPD 2% agar at 25°C, 30°C, 37°C, and 40°C. All growth assays were repeated four times and images are representative. (b) Comparison of wild type and pus7/pus7 filamentation on Spider 2% agar. Filamentation assays were repeated four times and images are representative. (c) pus7/pus7 filamentation in liquid Spider media. Liquid filamentation assays were performed four times and multiple fields were assessed for each replicate. Images are representative
We next tested if the absence of Pus7 affects filamentation, a phenotype important for C. albicans virulence. Growth at 37°C on Spider agar robustly induced wild type filamentation but fails to induce pus7/pus7 filamentation after 8 days (Figure 3b). This filamentation defect was also observed at 25°C and 30°C (Figure S3). In liquid Spider media, we find pus7/pus7 filaments as robustly as wild type (Figure 3c). Because filamentation is critical for virulence, we tested if pus7/pus7 has reduced virulence in comparison with wild type in a Galleria mellonella infection model; 175 hours after injection, 75% of G. mellonella larvae treated with pus7/pus7 were still alive whereas only 20% of the worms treated with wild type were alive (p value <.001; Figure 4a). Nearly 100% of worms treated with PBS were alive after 175 hours. These data indicate that pus7/pus7 is significantly less virulent in the G. mellonella infection model.
Figure 4.
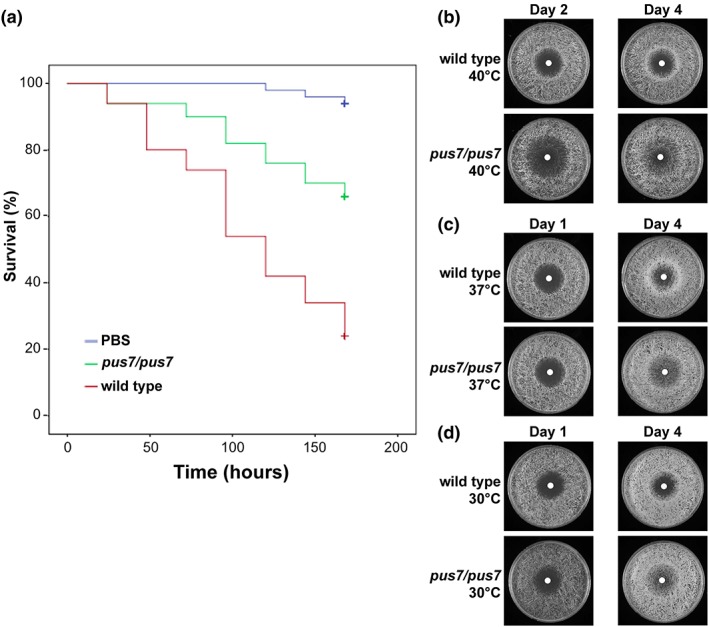
Pus7 impact on Candida albicans virulence and drug susceptibility. (a) Survival of Galleria mellonella larvae following infection by wild type and pus7/pus7. (b) pus7/pus7 and wild type susceptibility to fluconazole at 40°C. (c) pus7/pus7 and wild type susceptibility to fluconazole at 37°C. (d) pus7/pus7 and wild type susceptibility to fluconazole at 30°C
Finally, we tested if the absence of Pus7 had an effect on antifungal drug resistance. A disk diffusion assay was performed with fluconazole, and the pus7/pus7 zone of inhibition was larger than wild type's on day two of incubation at 37°C and 40°C (Figures 4b,c and S4). At 30°C, the zones of inhibition were very similar on day two (Figure 4d). This suggested that after plating, pus7/pus7 cells are initially more susceptible to fluconazole. However, after four days, pus7/pus7 exhibited pronounced trailing growth into the zone of inhibition, suggesting pus7/pus7 cells are better able to overcome the initial fluconazole susceptibility (Figures 4b and S4). Wild type did not display as robust trailing growth into the fluconazole zone of inhibition. Wild type and pus7/pus7 showed similar susceptibility to caspofungin, and neither strain exhibited trailing growth into the zone of inhibition (Figure S5). Pretreatment of wild type or pus7/pus7 with fluconazole led to lower virulence in wax moth larvae (Figure S7), and pus7/pus7 was not found to be more virulent than wild type after fluconazole pretreatment.
4. DISCUSSION
Absence of Pus7 is highly detrimental to C. albicans vigour, leading to slow growth, heat sensitivity, defects in filamentation, and decreased virulence. This is in contrast to ScPUS7 knockout, which leads to moderate slow growth and heat sensitivity (Sinha et al., 2008). We find CaPus7 is essential for some tRNA pseudouridylation. tRNA pseudouridylation is important for translation (Klassen et al., 2016; Klassen & Schaffrath, 2017), and defects in these processes could be responsible for the observed phenotypes. tRNA E (CUC) Ψ13 is catalysed by CaPus7, and other Pus7‐dependent stops at U11 and U8 were also observed. Pus7 may catalyse these additional sites of pseudouridylation, or U13 pseudouridylation may be required for U11 and U8 pseudouridylation via Pus7‐independent mechanisms. ScPus7 often targets a consensus sequence, but this consensus site is not required. U8 lies within sequence RNUNΨAR, similar to the ScPus7 RSUNΨAR consensus sequence. Pus7‐mediated modification sequence specificity has not been extensively studied outside of S. cerevisiae, and as such, requirements for site selection remain unclear in C. albicans.
A second molecular phenotype we observed was a disruption in rRNA processing. Multiple potential models explain how Pus7 absence could lead to defects in rRNA biogenesis. One potential model is direct Pus7‐mediated modification of ITS1/ITS2 is important for rRNA processing and absence of such modification leads to increased ribosome precursors. The lone 8mer conserved between C. albicans ITS1 and ITS2 contains a ScPus7 consensus site (Schwartz et al., 2014). If Pus7 mediated‐pseudouridylation at the consensus site is required for ITS1/ITS2 processing in C. albicans, pseudouridylated ITS1/ITS2 could be efficiently processed from the newly transcribed rRNA and degraded in wild type. In S. cerevisiae, ITS1 and ITS2 are processed and degraded cotranscriptionally or soon after transcription (Kos & Tollervey, 2010). This is consistent with our observation of fewer rRNA ITS2 containing precursors in wild type. However, in the absence of Pus7‐dependent modification, ITS1 and ITS2 will not be modified. This will result in an increased abundance of rRNA precursors in pus7/pus7 that contain nonpseudouridylated ITS2. We tested if CaPus7 modifies the ITS1 or ITS2 Pus7 consensus sites. No CMC‐dependent transcription termination was observed in the ITS1 primer extensions (Figure S8). CMC dependent transcription termination does occur in ITS2, but this termination is nine or 10 bases upstream of the Pus7 consensus site and was not Pus7 dependent (Figure 2c). Termination upstream of the consensus site residue could mask CMC‐dependent stoppage at the predicted Pus7 target, as reverse transcription would terminate before reaching the Pus7 consensus site. Our results are consistent with, but cannot confirm, this model.
Another potential model is Pus7 has an indirect effect on rRNA biogenesis. ScPus7 is promiscuous, and although we do not observe modification by CaPus7 at all analogous sites, it is possible that CaPus7 modifies a broad range of substrates at lower frequency or that modification of a small set of substrates has complex downstream effects. KTI11 an essential gene known to play roles in tRNA wobble base modification was the most up‐regulated gene in pus7/pus7 (Segal et al., 2018). Significant increase in abundance of a key translational regulator such as Kti11 could have dramatic effects on proteins required for rRNA processing (Huang et al., 2005).
Pus7 homologues are found in a variety of fungi, but they have been only minimally characterized outside of S. cerevisiae. Deletion of Schizosaccharomyces pombe Pus7 (SpPus7) leads to a slight decrease in fermentative growth (Malecki & Bahler, 2016), but SpPus7 targets have not been investigated. Humans express two Pus7 homologues and tRNA‐mediated modification by human Pus7 is thought to play important roles in stem cell differentiation (Guzzi et al., 2018). Furthermore, mutation of human PUS7 leads to developmental defects including short stature, microcephaly, and aggressive behaviour (de Brouwer et al., 2018). Our data indicate that the absence of C. albicans Pus7 leads to severe defects in morphological plasticity and slow growth. In addition, our data suggest CaPus7 does not act on U2 snRNA, but does act on tRNA the primary target of mammalian and bacterial Pus7s (Guzzi et al., 2018; Kaya & Ofengand, 2003). Further biochemical characterization of Pus7 homologues is necessary to elucidate Pus7's contributions to health and disease.
FUNDING INFORMATION
Funding was provided by the National Institutes of Health grant 1R15AI130950–01 to Douglas A. Bernstein. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supporting information
Figure S1. Comparison of wild type and pus7/pus7 C. albicans growth on synthetic complete 2% agar plates at 25, 30, 37, and 40°C.
Figure S2. Comparison of wild type and pus7/pus7 C. albicans growth on synthetic complete medium supplemented with BSA 2% agar plates at 25, 30, 37, and 40°C.
Figure S3. Comparison of wild type and pus7/pus7 C. albicans filamentation after 8 days on Spider 2% agar plates at 25 and 30oC.
Figure S4. Quantification of pus7/pus7 impact on C. albicans fluconazole susceptibility.
Figure S5. pus7/pus7 does not impact C. albicans caspofungin susceptibility. pus7/pus7 and wild type exhibited approximately equal day 1 and day 3 susceptibility to caspofungin. Caspofungin resistance was not quantified due to similarity.
Figure S6. Pus7 dependent U2 snRNA pseudouridylation was not detected. (A) Primer extensions using U2 snRNA primer 1 did not yield the predicted 28b or 49b products. (B) Primer extensions using U2 snRNA primer 2 did not yield the predicted 48b or 69b products. Faint Pus7 independent CMC dependent transcriptional termination was observed (A, B). (C) Schematic representation of C. albicans U2 snRNA sequence as well as predicted Pus7 dependent pseudouridine residues and extension primer location.
Figure S7. Pretreatment of C. albicans with fluconazole effect on virulence. 0.5 mg/kg larvae fluconazole was applied. 40 worms were used per treatment.
Figure S8. ITS1 pseudouridylation.
Table S1. Features presented with their respective counts for each sample, along with the log2‐fold change of expression and P‐values for difference in expression between the two groups being compared.
Table S2. Oligonucleotides used for insertion of stop codons to the open reading frame of C. albicans PUS7. Nucleotides added for cloning into the CRISPR‐Cas9 expression vector are in bold and italicized. Nucleotides that encode changes to the C. albicans gDNA are in upper case.
Table S3. Strains used in this study.
Table S4. Oligonucleotides used for primer extension analysis of CMC‐base treated total RNA from wild type and pus7/pus7.
Pickerill ES, Kurtz RP, Tharp A, Guerrero Sanz P, Begum M, Bernstein DA. Pseudouridine synthase 7 impacts Candida albicans rRNA processing and morphological plasticity. Yeast. 2019;36:669–677. 10.1002/yea.3436
Present addressRebecca P. Kurtz, Department of Statistics, University of California Riverside, Riverside, CA 92521.
REFERENCES
- Ansmant, I. , Massenet, S. , Grosjean, H. , Motorin, Y. , & Branlant, C. (2000). Identification of the Saccharomyces cerevisiae RNA:pseudouridine synthase responsible for formation of Ψ2819 in 21S mitochondrial ribosomal RNA. Nucleic Acids Research, 28(9), 1941–1946. 10.1093/nar/28.9.1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin, A. , Lane, B. G. , & Ofengand, J. (1994). Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry, 33(45), 13475–13483. 10.1021/bi00249a036 [DOI] [PubMed] [Google Scholar]
- Bakin, A. , & Ofengand, J. (1993). Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry, 32(37), 9754–9762. 10.1021/bi00088a030 [DOI] [PubMed] [Google Scholar]
- Behm‐Ansmant, I. , Urban, A. , Ma, X. , Yu, Y. T. , Motorin, Y. , & Branlant, C. (2003). The Saccharomyces cerevisiae U2 snRNA:pseudouridine‐synthase Pus7p is a novel multisite‐multisubstrate RNA:Ψ‐synthase also acting on tRNAs. RNA, 9(11), 1371–1382. 10.1261/rna.5520403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile, T. M. , Rojas‐Duran, M. F. , Zinshteyn, B. , Shin, H. , Bartoli, K. M. , & Gilbert, W. V. (2014). Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature, 515(7525), 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo, C. S. , Ghiringhelli, P. D. , & Gomez, D. E. (2014). Protein universe containing a PUA RNA‐binding domain. FEBS Journal, 281(1), 74–87. 10.1111/febs.12602 [DOI] [PubMed] [Google Scholar]
- de Brouwer, A. P. M. , Abou Jamra, R. , Kortel, N. , Soyris, C. , Polla, D. L. , Safra, M. , … Schwartz, S. (2018). Variants in pus7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. American Journal of Human Genetics, 103(6), 1045–1052. 10.1016/j.ajhg.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur, W. A. , & Schnare, M. N. (2008). Different mechanisms for pseudouridine formation in yeast 5S and 5.8S rRNAs. Molecular and Cellular Biology, 28(10), 3089–3100. 10.1128/MCB.01574-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, B. A. , Pickerill, E. S. , Vyas, V. K. , & Bernstein, D. A. (2018). CRISPR‐mediated genome editing of the human fungal pathogen Candida albicans . Journal of Visualized Experiments, 141 10.3791/58764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, B. A. , Smith, O. L. , Pickerill, E. S. , York, M. K. , Buenconsejo, K. J. P. , Chambers, A. E. , & Bernstein, D. A. (2018). Restriction digest screening facilitates efficient detection of site‐directed mutations introduced by CRISPR in C. albicans UME6. PeerJ, 6, e4920 10.7717/peerj.4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, J. , & Yu, Y. T. (2013). RNA pseudouridylation: New insights into an old modification. Trends in Biochemical Sciences, 38(4), 210–218. 10.1016/j.tibs.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein, A. C. , Rosenberg, A. , Hecht, I. , & Berman, J. (2016). diskImageR: Quantification of resistance and tolerance to antimicrobial drugs using disk diffusion assays. Microbiology, 162(7), 1059–1068. 10.1099/mic.0.000295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi, N. , Ciesla, M. , Ngoc, P. C. T. , Lang, S. , Arora, S. , Dimitriou, M. , … Bellodi, C. (2018). Pseudouridylation of tRNA‐derived fragments steers translational control in stem cells. Cell, 173(5), 1204–1216 e1226. 10.1016/j.cell.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Hammal, T. , & Ferre‐D'Amare, A. R. (2006). Pseudouridine synthases. Chemistry & Biology, 13(11), 1125–1135. 10.1016/j.chembiol.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Hazen, K. C. , Plotkin, B. J. , & Klimas, D. M. (1986). Influence of growth conditions on cell surface hydrophobicity of Candida albicans and Candida glabrata . Infection and Immunity, 54(1), 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoernes, T. P. , & Erlacher, M. D. (2017). Translating the epitranscriptome. Wiley Interdiscip Rev RNA, 8(1), e1375 10.1002/wrna.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B. , Johansson, M. J. , & Bystrom, A. S. (2005). An early step in wobble uridine tRNA modification requires the Elongator complex. RNA, 11(4), 424–436. 10.1261/rna.7247705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya, Y. , & Ofengand, J. (2003). A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA, 9(6), 711–721. 10.1261/rna.5230603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen, R. , Ciftci, A. , Funk, J. , Bruch, A. , Butter, F. , & Schaffrath, R. (2016). tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Research, 44(22), 10946–10959. 10.1093/nar/gkw705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen, R. , & Schaffrath, R. (2017). Role of pseudouridine formation by Deg1 for functionality of two glutamine isoacceptor tRNAs. Biomolecules, 7(4). 10.3390/biom7010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos, M. , & Tollervey, D. (2010). Yeast pre‐rRNA processing and modification occur cotranscriptionally. Molecular Cell, 37(6), 809–820. 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D. L. , Bousquet‐Antonelli, C. , Henry, Y. , Caizergues‐Ferrer, M. , & Tollervey, D. (1998). The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes & Development, 12(4), 527–537. 10.1101/gad.12.4.527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Kohler, J. , & Fink, G. (1994). Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science, 266(5191), 1723–1726. 10.1126/science.7992058 [DOI] [PubMed] [Google Scholar]
- Lovejoy, A. F. , Riordan, D. P. , & Brown, P. O. (2014). Transcriptome‐wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae . PLoS ONE, 9(10), e110799 10.1371/journal.pone.0110799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Zhao, X. , & Yu, Y. T. (2003). Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA‐independent mechanism. The EMBO Journal, 22(8), 1889–1897. 10.1093/emboj/cdg191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka, M. A. , Milanowska, K. , Osman Oglou, O. , Purta, E. , Kurkowska, M. , Olchowik, A. , … Grosjean, H. (2013). MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Research, 41(Database issue), D262–D267. 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki, M. , & Bahler, J. (2016). Identifying genes required for respiratory growth of fission yeast. Wellcome Open Res, 1, 12 10.12688/wellcomeopenres.9992.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey, S. E. , Moura, G. , Beltrao, P. , Almeida, R. , Garey, J. R. , Tuite, M. F. , & Santos, M. A. (2003). Comparative evolutionary genomics unveils the molecular mechanism of reassignment of the CTG codon in Candida spp. Genome Research, 13(4), 544–557. 10.1101/gr.811003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- (1991). Guide to yeast genetics and molecular biology. Methods in Enzymology, 194, 1–863. [PubMed] [Google Scholar]
- Murillo, L. A. , Newport, G. , Lan, C. Y. , Habelitz, S. , Dungan, J. , & Agabian, N. M. (2005). Genome‐wide transcription profiling of the early phase of biofilm formation by Candida albicans . Eukaryotic Cell, 4(9), 1562–1573. 10.1128/EC.4.9.1562-1573.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand, J. (2002). Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Letters, 514(1), 17–25. 10.1016/S0014-5793(02)02305-0 [DOI] [PubMed] [Google Scholar]
- Pendrak, M. L. , & Roberts, D. D. (2011). Ribosomal RNA processing in Candida albicans . RNA, 17(12), 2235–2248. 10.1261/rna.028050.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce, C. G. , Uppuluri, P. , Tristan, A. R. , Wormley, F. L. Jr. , Mowat, E. , Ramage, G. , & Lopez‐Ribot, J. L. (2008). A simple and reproducible 96‐well plate‐based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nature Protocols, 3(9), 1494–1500. 10.1038/nport.2008.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree, I. A. , Evans, M. E. , Pan, T. , & He, C. (2017). Dynamic RNA modifications in gene expression regulation. Cell, 169(7), 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S. , Bernstein, D. A. , Mumbach, M. R. , Jovanovic, M. , Herbst, R. H. , Leon‐Ricardo, B. X. , … Regev, A. (2014). Transcriptome‐wide mapping reveals widespread dynamic‐regulated pseudouridylation of ncRNA and mRNA. Cell, 159(1), 148–162. 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, E. S. , Gritsenko, V. , Levitan, A. , Yadav, B. , Dror, N. , Steenwyk, J. L. , … Berman, J. (2018). Gene essentiality analyzed by in vivo transposon mutagenesis and machine learning in a stable haploid isolate of Candida albicans . MBio, 9(5). 10.1128/mBio.02048-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, H. , David, L. , Pascon, R. C. , Clauder‐Munster, S. , Krishnakumar, S. , Nguyen, M. , … Steinmetz, L. M. (2008). Sequential elimination of major‐effect contributors identifies additional quantitative trait loci conditioning high‐temperature growth in yeast. Genetics, 180(3), 1661–1670. 10.1534/genetics.108.092932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spenkuch, F. , Motorin, Y. , & Helm, M. (2014). Pseudouridine: Still mysterious, but never a fake (uridine)! RNA Biology, 11(12), 1540–1554. 10.4161/15476286.2014.992278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Roberts, A. , Goff, L. , Pertea, G. , Kim, D. , Kelley, D. R. , … Pachter, L. (2012). Differential gene and transcript expression analysis of RNA‐seq experiments with TopHat and Cufflinks. Nature Protocols, 7(3), 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J. , & Tollervey, D. (1999). Ribosome synthesis in Saccharomyces cerevisiae . Annual Review of Genetics, 33, 261–311. 10.1146/annurev.genet.33.1.261 [DOI] [PubMed] [Google Scholar]
- Vyas, V. K. , Barrasa, M. I. , & Fink, G. R. (2015). A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Science Advances, 1(3), e1500248 10.1126/sciadv.1500248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, N. J. , Gottschalk, A. , Neubauer, G. , Kastner, B. , Fabrizio, P. , Mann, M. , & Luhrmann, R. (1998). Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA‐binding protein, are present together with Gar1p in all H BOX/ACA‐motif snoRNPs and constitute a common bipartite structure. RNA, 4(12), 1549–1568. 10.1017/s1355838298980761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, A. T. , Ge, J. , & Yu, Y. T. (2011). Pseudouridines in spliceosomal snRNAs. Protein & Cell, 2(9), 712–725. 10.1007/s13238-011-1087-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of wild type and pus7/pus7 C. albicans growth on synthetic complete 2% agar plates at 25, 30, 37, and 40°C.
Figure S2. Comparison of wild type and pus7/pus7 C. albicans growth on synthetic complete medium supplemented with BSA 2% agar plates at 25, 30, 37, and 40°C.
Figure S3. Comparison of wild type and pus7/pus7 C. albicans filamentation after 8 days on Spider 2% agar plates at 25 and 30oC.
Figure S4. Quantification of pus7/pus7 impact on C. albicans fluconazole susceptibility.
Figure S5. pus7/pus7 does not impact C. albicans caspofungin susceptibility. pus7/pus7 and wild type exhibited approximately equal day 1 and day 3 susceptibility to caspofungin. Caspofungin resistance was not quantified due to similarity.
Figure S6. Pus7 dependent U2 snRNA pseudouridylation was not detected. (A) Primer extensions using U2 snRNA primer 1 did not yield the predicted 28b or 49b products. (B) Primer extensions using U2 snRNA primer 2 did not yield the predicted 48b or 69b products. Faint Pus7 independent CMC dependent transcriptional termination was observed (A, B). (C) Schematic representation of C. albicans U2 snRNA sequence as well as predicted Pus7 dependent pseudouridine residues and extension primer location.
Figure S7. Pretreatment of C. albicans with fluconazole effect on virulence. 0.5 mg/kg larvae fluconazole was applied. 40 worms were used per treatment.
Figure S8. ITS1 pseudouridylation.
Table S1. Features presented with their respective counts for each sample, along with the log2‐fold change of expression and P‐values for difference in expression between the two groups being compared.
Table S2. Oligonucleotides used for insertion of stop codons to the open reading frame of C. albicans PUS7. Nucleotides added for cloning into the CRISPR‐Cas9 expression vector are in bold and italicized. Nucleotides that encode changes to the C. albicans gDNA are in upper case.
Table S3. Strains used in this study.
Table S4. Oligonucleotides used for primer extension analysis of CMC‐base treated total RNA from wild type and pus7/pus7.


