Summary
Mutations in SOCS1 are frequent in primary mediastinal B‐cell lymphoma and classical Hodgkin lymphoma. In the latter, SOCS1 mutations affect the length of the encoded protein (major mutations) and are associated with shorter patient survival. Two independent studies examined the prognostic impact of SOCS1 mutations in diffuse large B‐cell lymphoma (DLBCL) and showed differing results. This may be due to the small number of included patients, the heterogeneity of patients’ demographics and the distinct treatment schemes in these studies. To overcome the size limitations of these previous studies, we assessed SOCS1 mutations in the RICOVER‐60 cohort. The cohort uniformly consists of elderly patients (aged 61–80 years) treated with the CHOP‐14 scheme (cyclophosphamide, hydroxydaunorubicin, vincristine, prednisolone at 14‐day intervals) with or without an additional rituximab treatment. Patient outcomes were analysed with regard to overall SOCS1 mutation frequency, major and minor mutations and a novel impact‐based classifier – against the treatment modalities. Patients harbouring putative pathogenic SOCS1 mutations showed significant reduced overall survival within the CHOP plus rituximab group. Hence, putative pathogenic SOCS1 mutations seem to efface the beneficial effect of the therapeutic CD20 antibody. Comparing published data of whole exome and transcriptome sequencing of a large DLBCL cohort confirmed that predicted deleterious SOCS1 mutations forecast pre‐eminent survival in early onset DLBCL.
Keywords: SOCS1, diffuse large B‐cell lymphoma, CHOP, R‐CHOP
Accounting for 30–40%, the diffuse large B‐cell lymphomas (DLBCLs) are a major group of non‐Hodgkin lymphomas (NHL) (Reiter & Klapper, 2008; Roman & Smith, 2011). In recent years, it has been shown that gene expression patterns of DLBCLs play a critical role for treatment outcome resulting in a sub‐classification of activated B‐cell (ABC) or germinal centre B‐cell (GCB) ‐like DLBCLs (Alizadeh et al, 2000). A third pattern identifies primary mediastinal B‐cell lymphoma (PMBL). Receiving state‐of‐the‐art treatment with cyclophosphamide, hydroxydaunorubicin, vincristine and prednisone (CHOP regimen) in combination with the anti‐CD20 antibody, rituximab (Pfreundschuh et al, 2006; Pfreundschuh et al, 2008), results in differing 3‐year survival rates, with ABC‐like DLBCL having the least favourable and PMBL having the best prognosis (Rosenwald & Staudt, 2003; Lenz et al, 2008; Dunleavy et al, 2013; Roschewski et al, 2014). These differences clearly indicate the importance of the transcriptome in DLBCL. The oncogenic programmes in DLBCL also differ and are largely governed by their genetic background. We were the first to draw attention to mutations in SOCS1 occurring in B‐cell lymphomas (Barth et al, 2005; Weniger et al, 2006; Ritz et al, 2008; Schif et al, 2013), a finding that has since been confirmed by others (Juskevicius et al, 2017). A recent study for driver mutations in 1001 DLBCLs revealed that SOCS1 mutations are amongst the 15 most frequent mutations, affirming the relevance of SOCS1 mutations in DLBCL (Reddy et al, 2017).
SOCS1 (synonyms include JAB, SSI‐1 and TIP3) was described as a negative regulator of the STAT‐mediated expression of proliferation‐ and survival‐associated genes by interacting with Janus kinases (JAKs) (Endo et al, 1997; Naka et al, 1997; Ohya et al, 1997; Starr et al, 1997). Mutations in SOCS1 were shown to be common in PMBL, in both cell line models and primary tumours, and were associated with a delayed degradation of JAK2 (Melzner et al, 2005; Melzner et al, 2006; Ritz et al, 2008). Furthermore, SOCS1 mutations were identified in 3/5 classical Hodgkin lymphoma (cHL) cell lines and 8/19 patients (Weniger et al, 2006). This high frequency was confirmed in 64/105 cases of another cHL cohort. Here, classifying the mutations into minor (those that do not alter the length of the encoded protein) and major mutations (indels and/or truncating mutations) were found to be useful as a prognostic factor for early relapse and overall survival (OS) (Lennerz et al, 2015). cHL patients harbouring tumour cells with major mutations in SOCS1 had a significantly shorter OS and freedom from disease progression.
To further clarify the impact of SOCS1 mutations in NHL, Schif et al (2013) showed an inverted effect of SOCS1 mutations in a historical cohort of 154 majoritarian CHOP‐treated DLBCL patients. In contrast to the other lymphoma entities, mutations resulting in severe changes to the SOCS1 protein led to good outcomes compared to patients harbouring tumours with wildtype SOCS1. Paradoxically, minor mutations in SOCS1 were found in tumour tissue of patients with the shortest OS, which was even shorter than the SOCS1 wildtype group.
In a recent study, the SOCS1 mutation effects were examined in an independent cohort of DLBCL patients (Juskevicius et al, 2017). A group of 21/76 R‐CHOP‐treated DLBCL patients was shown to have mutations in the SOCS1 gene. Only parts of the results of the initial study could be confirmed. Juskevicius et al (2017) also showed that SOCS1 mutations led to longer progression‐free survival (PFS) in this cohort, but no differences were seen between major and minor mutations, indicating that the negative effects of minor mutations are unimportant for the patients’ survival in this cohort.
We tested a homogeneous cohort of elderly DLBCL patients [RICOVER‐60 trial (Pfreundschuh et al, 2008)] treated with the CHOP regimen, either with or without additional rituximab treatment, in order to ascertain whether the impact of SOCS1 mutations in DLBCL in CHOP‐treated patients could be confirmed and whether the addition of rituximab affected the impact of SOCS1 mutations as a prognosticator.
Methods
Patients’ data
The DLBCL patients in this study belonged to the rituximab‐with‐CHOP‐over‐age‐60 years trial (RICOVER‐60; DSHNHL 1999‐1A) (Pfreundschuh et al, 2008). The patients were treated with 6 or 8 cycles of the CHOP‐14 treatment regimen either with or without 8 cycles of rituximab (CHOP vs. R‐CHOP). Follow‐up data included event‐free (EFS), progression‐free (PFS), and OS together with rates of response and frequencies of toxicity. The study was carried out in compliance with the Helsinki declaration and confirmed by the Ethics Committees of the participating centres. The examination of the SOCS1 mutation status and correlation to the patients’ data was confirmed by a separate ethics vote. Patient characteristics, immunohistochemical and fluorescence in situ hybridization data were collected as described previously (Pfreundschuh et al, 2008; Ott et al, 2010; Staiger et al, 2017).
SOCS1 sequencing
To deal with to the high fragmentation of DNA isolated from formalin‐fixed and paraffin‐embedded (FFPE) material, the open reading frame of SOCS1 was examined using primer pairs amplifying overlapping amplicons of 177–360 bp (see Figure S1, Table SI). Sanger sequencing was performed, and the data were analysed using the Seqscape 2.5 software (Applied Biosystems, Foster City, CA, USA). The limit of mutation detection was set to 25%. All sequences were checked manually in order to identify insertions or deletions.
Classification of mutations
We used three separate mutations classification schemes: (i) We distinguished between SOCS1 mutant and wildtype mutations; (ii) We used a distinction of SOCS1 major, minor, and wildtype mutations following prior definitions (minor = non‐length altering) and major (indels and/or truncating mutations) (Lennerz et al, 2015); (iii) The impact of point mutations on the protein function was also estimated using a composite prediction composed of polyphen‐2 (Adzhubei et al, 2010), Fathmm (Shihab et al, 2013), Provean (Choi et al, 2012), SIFT (Sim et al, 2012), Panther (Mi et al, 2013) and SusPect (Yates et al, 2014). Briefly, the results were scored ranging from 0 (=predicted neutral) to predicted pathogenic (1) and a z score was calculated as the arithmetic mean across the six individual prediction scores. A z score of <0·5 was ranked as a predicted neutral, whereas a score of >0·7 was ranked as a predicted pathogenic mutation. In the cases with a score between 0·5 and 0·7, the mutation was tested using the Meta‐SNP prediction tool (Capriotti et al, 2013) for a final classification. Specifically, for the third classification, patients harbouring at least one putative pathogenic SOCS1 mutations were combined with those patients harbouring insertions, deletions or premature stop codons and are referred to collectively as “putative pathogenic”; the remaining patients (putative neutral SOCS1 mutations and wildtype) were grouped and we refer to these as “putative neutral”.
Statistical analysis
Event‐free survival (events: progression, start of salvage treatment, additional (unplanned) treatment, relapse or death from any cause), PFS (events: progression, relapse or death from any cause), and OS (event: death from any cause) were measured from the time of randomization. EFS, PFS and OS were estimated according to the Kaplan–Meier method and log‐rank tests were performed. Multivariate Cox regression models for SOCS1 results were adjusted for the factors of the International Prognostic Index (IPI, factors: age >60 years, lactate dehydrogenase >N, EAStern Cooperative Oncology Group performance status >1, Stage III/IV, extranodal involvement >1). Hazard ratios (HRs) with 95% confidence intervals (CIs) and P‐values are presented. For the correlation of SOCS1 results with qualitative data (morphology, immunohistochemistry, fluorescence in situ hybridisation and Lymph2Cx data) and for differences regarding patient characteristics, we used chi‐square and Fisher’s exact test. The two‐sided significance level was P < 0·05. No P‐value correction for multiple tests were done due to the explorative character of the analyses. Statistical analyses were performed using SPSS Statistics 23 (IBM Corp., Armonk, NY, USA).
Re‐evaluation of published next generation sequencing data
To validate our findings, published exome and transcriptome data and OS of 1001 DLBCL patients (Reddy et al, 2017) were re‐evaluated with respect to SOCS1 mutation status. After the exclusion of patients with missing data (SOCS1 mutation status, OS), SOCS1 mutations were classified as described earlier. Survival curves and statistics (Log‐rank tests) were carried out using the Prism software (GraphPad Software Inc., San Diego, CA, USA).
Results
Evaluable subgroup of the patients represents the whole RICOVER‐60 cohort
About 30% (266 of an initial 949 patients with confirmed reference diagnosis of DLBCL) of the DLBCL patients of the cohort could be examined. This reduced number was due to the availability of remaining tumour tissue and the grade of DNA degradation within the samples and fits to novel data of TP53 mutations within this cohort (Zenz et al, 2017).
We tested for differences in EFS, PFS (PFS) and OS between the cases with or without SOCS1 mutation data. No significant differences were found, either in the total group or after splitting into treatment‐based sub‐groups (Figure S2). This was also true for the gender distribution (with: 143 males (54%): 123 females (46%) and without: 368 males (54%): 315 femails (46%), respectively; P = 0·973) and the median of the patients’ age (with: 68 years [61–80] versus without: 69 [61–80]; P = 0·436). No clinically relevant difference for the distribution of the IPI score between patients with SOCS1 mutation data and the whole RICOVER‐60 cohort were observed (IPI 1, 2, 3, 4–5: 39, 25, 25, 11% and 32, 27, 25, 16%, respectively). Therefore, the subgroup of 266 patients investigated is representative of the total of 949 patients of the cohort.
SOCS1 mutations in the Ricover‐60 cohort
Mutations in the SOCS1 gene were detected by polymerase chain reaction (PCR) amplification of the open reading frame followed by Sanger sequencing of the amplicons. Eighty‐five of the 266 (32%) patient DNAs showed aberrations compared to the reference SOCS1 sequence. Exclusively silent single nucleotide exchanges were detected in 20 of 266 (8%) and single nucleotide polymorphisms resulting in amino acid changes were found in 48/266 (18%) of the patient DNAs examined. Mutations affecting the length of the resulting SOCS1 protein caused by insertions, deletions or premature stop codons were seen in 17/266 (6%) patients. Therefore, the frequency of non‐silent SOCS1 mutations in the RICOVER‐60 cohort is 24%. SOCS1 single nucleotide exchanges found in the RICOVER‐60 cohort were distributed throughout the entire open reading frame in a homogeneous manner (Fig 1A). Therefore, no mutation hotspots could be identified. We point out that the lack of hotspots defers specifically to a lack of recurrent positional accumulation across cases, which we confirmed by comparison with publicly available data across different cancer types (Figure S3). In contrast, we acknowledge that there is an accumulation of SOCS1 variants in nucleotide motifs that follow the somatic hypermutation motifs RGYW/WRCY, DGYW/WRCH and WA/TW (Mottok et al, 2009). The patients were grouped using different mutation classifiers with respect to the preceding studies (1).
Figure 1.
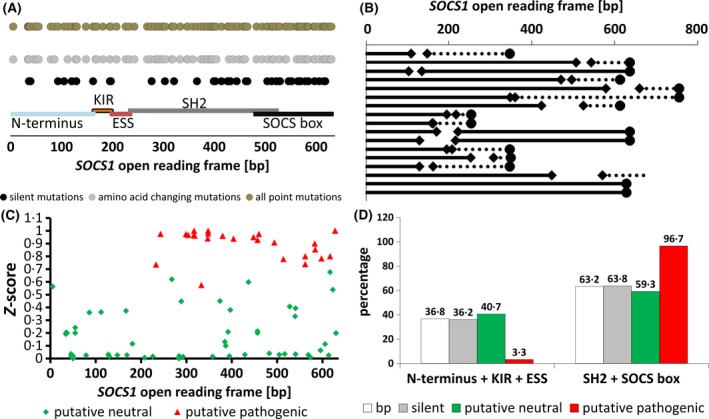
Distribution and location of SOCS1 mutations found in the patient DNA of the RICOVER‐60 cohort. (A) Distribution of point mutations over the open reading frame of SOCS1. Silent mutations are presented in black, mutations resulting in amino acid changes are light grey. The total of all point mutations is shown as dark grey dots. Special domains of the SOCS1 protein are displayed as coloured lines covering the corresponding base pairs (bp) as indicated. (B) Diagram of the SOCS1 mutations (n = 17) affecting the length of the SOCS1 protein. Diamonds indicate deletions. Circles indicate stop codons. Dotted lines mark changed sequences as a result of frameshifts. (C) Diagram of the location of amino acid changing point mutations throughout the SOCS1 open reading frame with respect to the calculated impact on protein function (z score). Mutations predicted to affect the protein function are displayed in red. Mutations calculated to be neutral for the protein function are coloured in green. (D) Presentation of the point mutations with respect to the functional domains of the SOCS1 protein as percentages. White bars show the allocation of the base pairs attributed to the domain groups “N‐terminus, KIR and ESS domain” and “SH2 domain and SOCS box”, to clarify if the mutation distributions are due to the different numbers of base pairs within the groups. Grey bars represent silent mutations. Green bars illustrate mutations calculated not to affect the protein function (putative neutral). There were no differences compared to the number of base pairs within the two groups, suggesting a random occurrence of the mutations. Red bars give the distribution of mutations affecting the function of the SOCS1 protein. These mutations were almost exclusively found in the SH2 domain and the SOCS‐box.
Table 1.
SOCS1 mutation status and grouping of the DLBCL patients of the RICOVER‐60 sub‐cohort. Grouping of the patients with respect to the OCS1 mutation status, three different classifiers and the treatment with or without rituximab.
| SOCS1 status | Classifier (Schif et al, 2013) | Classifier (Juskevicius et al, 2017) | Novel classifier | CHOP‐treated (n = 136) | R‐CHOP‐treated (n = 130) | Total (n = 266) |
|---|---|---|---|---|---|---|
| Wildtype | Wildtype | Wildtype | Putative neutral | 90 (66%) | 91(70%) | 181 (68%) |
| Silent mutations | 9 (7%) | 11 (9%) | 20 (8%) | |||
| Point mutations (tolerated) | minor mutations | mutated | 16 (12%) | 11 (9%) | 27 (10%) | |
| Point mutations (damaging) | putative pathogenic | 13 (10%) | 8 (6%) | 21 (8%) | ||
| Indels (in frame) | major mutations | 3 (2%) | 0 (0%) | 3 (1%) | ||
| Indels (out of frame) or premature stop codons | 5 (4%) | 9 (7%) | 14 (5%) |
For comparison of the results with the first DLBCL study (Schif et al, 2013), the patients were grouped as SOCS1 wildtype (n = 201), SOCS1 minor mutated (n = 48) and SOCS1 major mutated patients (n = 17) and split into CHOP‐ (n = 136) and R‐CHOP (n = 130) subgroups to reduce bias provoked by the treatment. To compare the results with the second DLBCL study (Juskevicius et al, 2017), the patients were grouped into SOCS1 wildtype (n = 201) and SOCS1 mutated patients (n = 65). Additionally, the prediction of the severity of mutational impact on the function of the SOCS1 protein was used as a novel classifier, resulting in patients with putative neutral (n = 228, wildtype SOCS1 included) and putative pathogenic (n = 38) SOCS1 mutations. About 37% of the base pairs of the SOCS1 open reading frame can be assigned to the N‐terminus, the KIR and the ESS domain, whereas about 63% refer to the SH2 domain and the SOCS box. Silent mutations found in the N‐terminus, the KIR and the ESS domain account for 36%, leaving 64% of the silent mutations located in the SH2 domain and SOCS box. The distribution of putative neutral mutations was comparable (41% and 59% respectively). Therefore, silent and putative neutral mutations were found to be distributed randomly throughout the entire open reading frame. Interestingly, the putative pathogenic point mutations were mainly located in the SH2 domain and the SOCS box of the SOCS1 protein (97%; Fig 1C,D). Deletions and point mutations causing premature stop codons affect the SH2 domain or the SOCS box in 14/17 (82%) of cases (Fig 1B), highlighting the importance of these domains for the protein’s function. Comparing patient characteristics, IHC and FISH data with regard to the differing classifiers of SOCS1 mutations, no striking differences or correlations were detected (Table SII).
The paradoxical effect on patients’ survival of minor and major SOCS1 mutations in CHOP‐treated DLBCL cannot be confirmed
The initial SOCS1 study on DLBCL revealed a paradoxical effect of minor and major SOCS1 mutations compared to cHL cases (Schif et al, 2013). To elucidate whether the results of this study of SOCS1 mutation effects in DLBCL could be confirmed, the cases of the novel cohort were grouped into SOCS1 wildtype, SOCS1 minor mutation and SOCS1 major mutation cases. Comparable classification standards were used as reported by Schif et al (2013). No significant differences were detected in EFS, PFS or OS of the groups in the corresponding group of CHOP‐treated DLBCL patients (n = 136; Figure S4B). The same results were observed in the R‐CHOP‐treated patients (n = 130; Figure S4C) and the total number of patients (n = 266; Figure S4A). Therefore, the finding that minor SOCS1 mutations lead to reduced survival times compared to major mutations in DLBCL patients was not confirmed in this DLBCL trial, also after adjusting for IPI factors in multivariate Cox regression models.
The uniform positive effect on patients’ outcome of mutated SOCS1 in R‐CHOP‐treated DLBCL cannot be confirmed
A recent study on mutations in DLBCL indicated a uniform positive effect of SOCS1 mutations regardless of a sub‐classification of the mutations (Juskevicius et al, 2017). To test if these positive effects of mutated SOCS1 in R‐CHOP‐treated DLBCL patients could be confirmed, all mutated cases (except silent mutations) were grouped together and tested for differences in EFS, PFS and OS against the group with wildtype SOCS1. Among the patients treated with R‐CHOP (the corresponding comparison group to the recent SOCS1 mutation study), no significant differences were found (P = 0·573; P = 0·790; P = 0·521; Figure S5C). Comparable results were observed for the CHOP‐treated patients (P = 0·114; P = 0·230; P = 0·595; Figure S5B) and the total number of patients (P = 0·473; P = 0·588; P = 0·937; Figure S5A). Therefore, the results of this recently published cohort were also not confirmed by this trial, also after adjusting for IPI factors in multivariate Cox regression model.
SOCS1 mutations predicted to have negative impact on protein function are associated with shorter overall survival
Regarding several published data stating that SOCS1 mutation effects are dependent on the type of mutation (Schif et al, 2013; Lennerz et al, 2015), we tested the nucleotide changes with respect to the severity of their impact on protein function using different protein prediction tools. Using this refined method, the patients were divided into patients harbouring tumours with putative regular (wildtype SOCS1 and neutral SOCS1 mutations) or putative pathogenic SOCS1 protein. Using this new classifier, we tested for differences in EFS, PFS and OS in a total of 266 patients and for the treatment‐adjusted subgroups. Although no differences could be observed in the total number of patients or in the CHOP‐treated group, we found a significantly shorter OS in patients with putative pathogenic SOCS1 mutations treated with the R‐CHOP regimen (P = 0·037; Fig 2). In a multivariate Cox regression model adjusted for IPI factors, these results were confirmed with a HR of 1·9 (95% CI (0·9–4·2); however, these differences did not reach significance (P = 0·105). The reduced survival in elderly patients with putative pathogenic SOCS1 mutations may be a consequence of the known and superimposed strong prognostic effects of higher IPI stages. Nonetheless, after adjustment for the IPI factors and cell of origin ABC vs. GCB subtype (Lymph2Cx), a significant hazard ratio in R‐CHOP treated patients with putative pathogenic SOCS1 mutations (HR = 2·8 95% CI (1·0–7·4) P = 0·042) was observed for OS.
Figure 2.
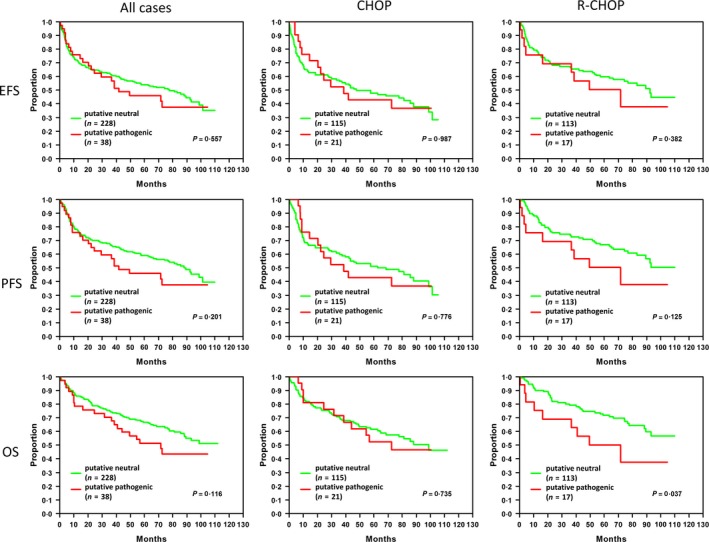
Survival of DLBCL patients related to the predicted effect of SOCS1 mutations. The survival time analyses of the patients of the RICOVER‐60 trial are shown with putative neutral (green) or putative pathogenic (red) SOCS1 with respect to all cases (left panel) or patient treatments [middle panel: CHOP (cyclophosphamide, hydroxydaunorubicin, vincristine, prednisolone)‐treated; right panel: R‐CHOP (CHOP + rituximab)‐treated]. The analyses were calculated for event‐free survival (EFS), progression‐free survival (PFS) and overall survival (OS). Patients harbouring putative pathogenic SOCS1 mutations show a significantly shorter overall survival (P = 0·037). Abbreviations: n, numbers of patients; P, P‐value from log‐rank test.
Younger age at diagnosis and putative pathogenic SOCS1 mutations as prediction of pre‐eminent survival
We conducted a re‐evaluation of a large fourth cohort of published and online available sequencing data (Reddy et al, 2017) of DLBCL patients with respect to the SOCS1 mutation status, to validate the prognostic associations. No differences in OS were seen comparing patients by SOCS1 mutation status (n = 91 vs. SOCS1 wildtype n = 871, Fig 3A). Classifying the SOCS1 mutations into minor and major mutations revealed that patients with major mutations tended to have longer OS compared to patients with minor mutations or wildtype SOCS1. However, these differences were not significant (P = 0·153). The same was shown by classifying into putative neutral or putative pathogenic SOCS1 mutations (Fig 3B,C). As the patients of the RICOVER‐60 cohort were uniformly elderly patients aged over 60 years, we excluded the patients younger than 60 years from the cohort reported by Reddy et al (2017). In these age‐based subsets we did not see differences in OS in the remaining subgroup of elderly patients; neither by SOCS1 mutation status, nor by using the other SOCS1 classifiers (Fig 3D–F). Examination in the subgroup of patients younger than 60 years at diagnosis showed a trend for a longer OS regarding all SOCS1 mutations and for major mutations compared to the corresponding subgroup of patients with SOCS1 wildtype (P = 0·197 and P = 0·072). The novel classifier (putative neutral and putative pathogenic SOCS1) revealed a significant difference in OS in this subgroup of younger patients (under 60 years of age at diagnosis, Fig 3G–I; P = 0·014).
Figure 3.
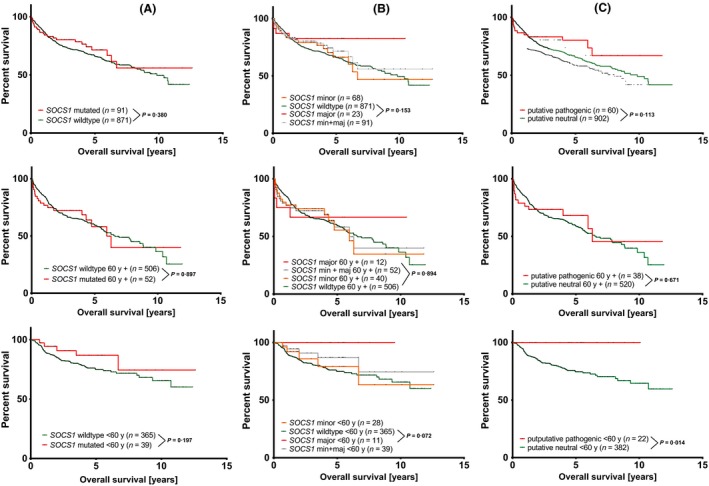
Re‐evaluation of publicly available sequence and survival data of DLBCL patients (Reddy et al, 2017). Overall survival of patients with mutated SOCS1 and SOCS1 wildtype. A, B, C: total evaluable patient data (n = 962); D, E, F: patients aged over 60 years (60y) at diagnosis (n = 558); G, H, I: patients aged under 60 y at diagnosis (n = 404). Comparisons were illustrated either as SOCS1 wildtype versus mutated SOCS1 (A,D,G), SOCS1 wildtype versus SOCS1 major and/or minor mutations (B, E, H), and putative neutral SOCS1 versus putative pathogenic SOCS1 (C, F, I). P‐values represent log rank tests (Mantel‐Cox) comparing 2 survival curves.
Putative pathogenic SOCS1 mutations stall the benefit of Rituximab on overall survival of DLBCL patients
Knowing that harbouring a pathogenic SOCS1 mutation is associated with shorter OS, we compared the EFS, PFS and OS of patients with putative regular SOCS1 protein treated with or without rituximab. The same comparison was made for patients with putative pathogenic SOCS1 proteins. Log rank tests showed that the R‐CHOP‐treated patients with putative regular SOCS1 protein had significant differences in EFS and PFS and showed a trend to longer OS (P = 0·045, P = 0·011, and P = 0·088) compared to the corresponding CHOP‐treated patients. No significant differences were seen when comparing the patients with putative pathogenic SOCS1 with respect to rituximab treatment (P = 0·863, P = 0·884, P = 0·611). Therefore, it seems that SOCS1 pathogenic mutations stall the benefit gained by rituximab treatment (Fig 4) in elderly patients. These results were confirmed in multivariate Cox regression models for rituximab‐containing treatment adjusted for IPI factors (within putative neutral: HREFS = 0·7 (95%CI: 0·4–1·0) P = 0·028, HRPFS = 0·6 (95%CI: 0·4–0·8) P = 0·004, HROS = 0·7 (95%CI: 0·4–1·0) P = 0·053; within putative pathogenic: HREFS = 1·2 (95%CI: 0·4–3·1) P = 0·749, HRPFS = 1·2 (95%CI: 0·4–3·3) P = 0·723, HROS = 2·2 (95%CI: 0·7–6·5) P = 0·156). This supports the hypothesis that putative pathogenic SOCS1 mutations counteract the beneficial effect of a rituximab treatment.
Figure 4.
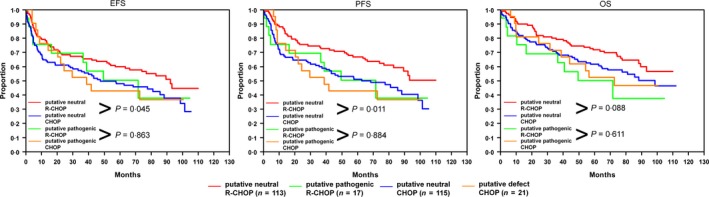
Putative pathogenic SOCS1 mutations distinguish rituximab responder and non‐responder. The survival curves for the DLBCL subgroups of patients with putative regular SOCS1 treated with rituximab (red line), patients with putative neutral SOCS1 treated without rituximab (blue line), patients with putative pathogenic SOCS1 treated with rituximab (green line) and without rituximab (orange line) are shown for event‐free (EFS), progression‐free (PFS) and overall survival (OS). P‐values indicate the results of log ranks tests. CHOP: cyclophosphamide, hydroxydaunorubicin, vincristine, prednisolone; R‐CHOP: CHOP + rituximab.
Discussion
Here, we report results of a large‐scale assessment of SOCS1 genotyping as a prognostic biomarker in a large cohort of clinically well‐annotated DLBCL patients. We show that putative pathogenic SOCS1 mutations seem to efface the beneficial effect of the therapeutic CD20 antibody, rituximab. We have validated these findings in an independent cohort and consider these findings important for several reasons. The functions of the SOCS1 protein have been examined over the last two decades and a crucial role found in the cytokine signalling pathway mediated by JAKs and STAT proteins. As the relevance of mutations in SOCS1 was clearly demonstrated for primary mediastinal B‐cell lymphoma (Barth et al, 2005; Ritz et al, 2008) and cHL (Weniger et al, 2006; Lennerz et al, 2015), findings concerning the role of mutated SOCS1 in DLBCL were controversial (Schif et al, 2013; Juskevicius et al, 2017). Our first study of a DLBCL cohort (Schif et al, 2013) revealed that subgrouping, of major and minor mutated SOCS1, which had previously been seen in cHL patients, can also be found in DLBCL patients. But the impact of the mutations seemed to be inverted compared to the impact found in cHL. While major SOCS1 mutations in cHL were a prognostic factor for shorter survival, in DLBCL major mutations paradoxically predicted an extremely good outcome. DLBCL patients with minor SOCS1 mutations were predicted to have extremely short survival times. The results of the second study (Juskevicius et al, 2017) confirm that SOCS1 mutations in DLBCL may be used to predict a better outcome, at least for PFS in these patients. But here, the devastating effect of minor SOCS1 mutations was not seen at all. To clarify the situation, we embarked to verify the data in a cohort that not only reflects both studies but also overcomes some of the limitations of the previous studies. To overcome the limitation of low patient numbers, 266 samples of the RICOVER‐60 trial were shown to be representative of the 949 confirmed DLBCL patients of the whole study. All patients of this prospective and randomized cohort were elderly, aged between 61 and 80 years, eliminating the issue of extreme age variability of onset of the disease in the other studies. The age of the majority (n = 154) of the CHOP‐treated patients in the study reported by Schif et al (2013), as well as the age of the 76 R‐CHOP‐treated patients of the study reported by Juskevicius et al (2017), was extremely heterogeneous, ranging from 3 to 93 years in the first and 18 to 81 years in the latter, which is a confounder on its own (Klapper et al, 2012). As the patients’ treatment may be a relevant factor for the differing impact of SOCS1 mutations, it is a great benefit of the RICOVER‐60 trial that it includes both CHOP‐ and R‐CHOP‐treated patients. The mutation frequency of SOCS1 in our cohort is 24% (65/266) and is hereby in between the frequencies determined in former studies (16% and 28%, respectively).
In general, we have to state that our results do not confirm those of either Schif et al (2013) or Juskevicius et al (2017). This may be for different reasons: patient age, non‐uniformity of treatment in the cohort reported by Schif et al (2013) and probably a technical reason because two patients of their cohort with no further mutations than silent mutations in SOCS1 were grouped as minor mutations. Nevertheless, we could establish that mutations affecting protein function (putative pathogenic SOCS1) can be used to predict a significantly shorter OS in R‐CHOP‐treated elderly DLBCL patients. This corresponds to mutated SOCS1 effects seen in PMBL and cHL. Additionally, we found that putative pathogenic SOCS1 mutations might predict whether a DLBCL patient benefits from additional rituximab treatment given in elderly patients. Comparing our results of CHOP‐ vs. R‐CHOP treated patients, the hypothesis arises that the positive effect of rituximab on patient outcome is no longer noticeable in R‐CHOP‐treated patients with putative pathogenic mutations in SOCS1. Therapeutic CD20 antibodies act in different ways: (i), induction of non‐classical apoptosis by crosslinking multiple CD20 molecules, (ii), activation of complement‐dependent cytotoxicity, (iii), induction of cell‐mediated cytotoxicity, (iv), antigen signalling enhancement via FcγR crosslinking, or (v), complement‐enhanced antibody dependent cell‐mediated cytotoxicity [reviewed in (Boross & Leusen, 2012)]. The net effect of pathogenic mutations of SOCS1 was shown to prolong the JAK‐STAT signals inducing proliferation in B lymphoma cells (Melzner et al, 2006). A hypothetical bridge between these processes might be that tonic JAK‐STAT activity confers some kind of resistance to at least one of the rituximab effects. This is a topic to be investigated further in in vitro studies.
In order to validate our findings within an independent cohort of age‐mixed DLBCL patients with published whole exome and transcriptome data (Reddy et al, 2017), we found that the inconsistent beneficial effects of SOCS1 defect mutations are due to the broad range in the age at diagnosis of DLBCL patients in the studies published earlier (Schif et al, 2013; Juskevicius et al, 2017). Splitting these patients to subgroups with early and late onset of disease (age of diagnosis, <60 and 60+ years, respectively), we clearly confirmed the beneficial SOCS1 defect mutation effect described in the first two studies (Schif et al, 2013; Juskevicius et al, 2017) within the younger patients. Elderly patients did not show an altered survival related to SOCS1 mutation status. This emerged as a trend when stratifying the mutations into minor and major. Using the novel classifiers (putative neutral and putative pathogenic SOCS1 mutations) significantly enhanced this effect. Therefore, the initial concept of sub‐classifying the SOCS1 mutations is useful in DLBCL as a classifier for the patients’ outcome albeit in a more nuanced manner and is very likely to be especially useful for outcome prediction in early onset DLBCL patients.
In summary, the distinct role of SOCS1 in DLBCL remains uncertain; three independent studies of differing cohorts failed to show a uniform effect of SOCS1 mutations. Possibly, the effects of SOCS1 mutations are partly superimposed by effects of other mutated genes. Nonetheless, the results of this study deliver the first data on elderly patients within a highly controlled prospective and randomized trial of DLBCL. Furthermore, it could be shown that the 266 patients analysed reflect the larger number of 949 DLBCL patients. The issue of patients undergoing differing treatments was overcome by having subgroups of the two main treatment regimens used in recent decades. This study is the first on SOCS1 mutation effects in DLBCL patients revealing a comparable impact of SOCS1 mutations as seen in PMBL or cHL. Nonetheless, it has to be noted that the shorter OS of R‐CHOP treated patients with putative pathogenic SOCS1 mutations retained significance in the multivariate analysis against the stronger prognostic factor of IPI staging only after adjustment for IPI factors and cell of origin ABC vs. GCB subtype (Lymph2Cx). Furthermore, the inferior OS of elderly patients with putative pathogenic SOCS1 mutations could not be confirmed in an independent cohort. It is conceivable that this SOCS1 mutation effect is restricted to elderly patients treated with the R‐CHOP regimen.
Our analysis of the published next generation sequencing data (Reddy et al, 2017), showed that the age of onset is a very important factor for consideration when using SOCS1 mutations as a predictor for survival in DLBCL patients treated with CHOP, and R‐CHOP or other rituximab‐containing treatment schemes. Although further research has yet to be performed to consolidate this, the results of our study imply a difference between elderly DLBCL patients who benefit or not from rituximab treatment in addition to the classical CHOP therapy regime. The survey of four independent DLBCL studies implies that putative pathogenic SOCS1 mutations may be used to forecast an extremely favourable OS of early onset DLBCL patients (aged <60 years).
Competing financial interest statement
All authors declare no competing financial interests.
Authors’ contributions
PM conceived the study. MM and KM performed experiments and analysed the data. ML analysed data. JL collected patients’ DNA. MP, LT, NS and PM took part in initial patient recruitment and ACF, SH, M‐LH, HS, AR and WK confirmed the diagnosis. Statistics were performed by MZ, MK and ML. KM and PM wrote the manuscript. All authors had final approval of the submitted and published versions.
Supporting information
Figure S1. Schematic position and length of amplicons used to cover the SOCS1 open reading frame.
Figure S2. The subgroup examined represents the whole RICOVER‐60 cohort.
Figure S3. Comparison of positional accumulation of variants across SOCS1 using publicly available databases across different cancer types.
Figure S4. Survival curves of DLBCL patients related to SOCS1 mutation subtypes.
Table SI. Primer sequences of PCR primers used to cover the SOCS1 open reading frame.
Table SII. A: Summary of patient characteristics, immunohistochemistry and fluorescence in situ hybridization data.
Acknowledgements
This work is dedicated to Michael Pfreundschuh, the senior scientist of the RICOVER‐ 60 trial, who suddenly left us on 5 March, 2018. The technical assistance of Karola Dorsch, Elena Moser, and Lena Kelsch, Beate Mann and Katja Rillich is gratefully acknowledged.
Clinicaltrials.gov identifier: NCT00052936
References
- Adzhubei, I.A. , Schmidt, S. , Peshkin, L. , Ramensky, V.E. , Gerasimova, A. , Bork, P. , Kondrashov, A.S. & Sunyaev, S.R. (2010) A method and server for predicting damaging missense mutations. Nature Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh, A.A. , Eisen, M.B. , Davis, R.E. , Ma, C. , Lossos, I.S. , Rosenwald, A. , Boldrick, J.C. , Sabet, H. , Tran, T. , Yu, X. , Powell, J.I. , Yang, L. , Marti, G.E. , Moore, T. , Hudson, J. Jr , Lu, L. , Lewis, D.B. , Tibshirani, R. , Sherlock, G. , Chan, W.C. , Greiner, T.C. , Weisenburger, D.D. , Armitage, J.O. , Warnke, R. , Levy, R. , Wilson, W. , Grever, M.R. , Byrd, J.C. , Botstein, D. , Brown, P.O. & Staudt, L.M. (2000) Distinct types of diffuse large B‐cell lymphoma identified by gene expression profiling. Nature, 403, 503–511. [DOI] [PubMed] [Google Scholar]
- Barth, T.F. , Melzner, I. , Wegener, S. , Bucur, A.J. , Bruderlein, S. , Dorsch, K. , Hasel, C. , Leithauser, F. & Moller, P. (2005) Biallelic mutation of SOCS‐1 impairs JAK2 degradation and sustains phospho‐JAK2 action in MedB‐1 mediastinal lymphoma line. Verhandlungen der Deutschen Gesellschaft fur Pathologie, 89, 234–244. [PubMed] [Google Scholar]
- Boross, P. & Leusen, J.H. (2012) Mechanisms of action of CD20 antibodies. American Journal of Cancer Research, 2, 676–690. [PMC free article] [PubMed] [Google Scholar]
- Capriotti, E. , Altman, R.B. & Bromberg, Y. (2013) Collective judgment predicts disease‐associated single nucleotide variants. BMC Genomics, 14(Suppl. 3), S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y. , Sims, G.E. , Murphy, S. , Miller, J.R. & Chan, A.P. (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS One, 7, e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy, K. , Pittaluga, S. , Maeda, L.S. , Advani, R. , Chen, C.C. , Hessler, J. , Steinberg, S.M. , Grant, C. , Wright, G. , Varma, G. , Staudt, L.M. , Jaffe, E.S. & Wilson, W.H. (2013) Dose‐adjusted EPOCH‐rituximab therapy in primary mediastinal B‐cell lymphoma. New England Journal of Medicine, 368, 1408–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, T.A. , Masuhara, M. , Yokouchi, M. , Suzuki, R. , Sakamoto, H. , Mitsui, K. , Matsumoto, A. , Tanimura, S. , Ohtsubo, M. , Misawa, H. , Miyazaki, T. , Leonor, N. , Taniguchi, T. , Fujita, T. , Kanakura, Y. , Komiya, S. & Yoshimura, A. (1997) A new protein containing an SH2 domain that inhibits JAK kinases. Nature, 387, 921–924. [DOI] [PubMed] [Google Scholar]
- Juskevicius, D. , Jucker, D. , Klingbiel, D. , Mamot, C. , Dirnhofer, S. & Tzankov, A. (2017) Mutations of CREBBP and SOCS1 are independent prognostic factors in diffuse large B cell lymphoma: mutational analysis of the SAKK 38/07 prospective clinical trial cohort. Journal of Hematology & Oncology, 10, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapper, W. , Kreuz, M. , Kohler, C.W. , Burkhardt, B. , Szczepanowski, M. , Salaverria, I. , Hummel, M. , Loeffler, M. , Pellissery, S. , Woessmann, W. , Schwanen, C. , Trumper, L. , Wessendorf, S. , Spang, R. , Hasenclever, D. , Siebert, R. & Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe (2012) Patient age at diagnosis is associated with the molecular characteristics of diffuse large B‐cell lymphoma. Blood, 119, 1882–1887. [DOI] [PubMed] [Google Scholar]
- Lennerz, J.K. , Hoffmann, K. , Bubolz, A.M. , Lessel, D. , Welke, C. , Ruther, N. , Viardot, A. & Moller, P. (2015) Suppressor of cytokine signaling 1 gene mutation status as a prognostic biomarker in classical Hodgkin lymphoma. Oncotarget, 6, 29097–29110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz, G. , Wright, G. , Dave, S.S. , Xiao, W. , Powell, J. , Zhao, H. , Xu, W. , Tan, B. , Goldschmidt, N. , Iqbal, J. , Vose, J. , Bast, M. , Fu, K. , Weisenburger, D.D. , Greiner, T.C. , Armitage, J.O. , Kyle, A. , May, L. , Gascoyne, R.D. , Connors, J.M. , Troen, G. , Holte, H. , Kvaloy, S. , Dierickx, D. , Verhoef, G. , Delabie, J. , Smeland, E.B. , Jares, P. , Martinez, A. , Lopez‐Guillermo, A. , Montserrat, E. , Campo, E. , Braziel, R.M. , Miller, T.P. , Rimsza, L.M. , Cook, J.R. , Pohlman, B. , Sweetenham, J. , Tubbs, R.R. , Fisher, R.I. , Hartmann, E. , Rosenwald, A. , Ott, G. , Muller‐Hermelink, H.K. , Wrench, D. , Lister, T.A. , Jaffe, E.S. , Wilson, W.H. , Chan, W.C. , Staudt, L.M. , Lymphoma, Leukemia Molecular & Profiling, P. (2008) Stromal gene signatures in large‐B‐cell lymphomas. New England Journal of Medicine, 359, 2313–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzner, I. , Bucur, A.J. , Bruderlein, S. , Dorsch, K. , Hasel, C. , Barth, T.F. , Leithauser, F. & Moller, P. (2005) Biallelic mutation of SOCS‐1 impairs JAK2 degradation and sustains phospho‐JAK2 action in the MedB‐1 mediastinal lymphoma line. Blood, 105, 2535–2542. [DOI] [PubMed] [Google Scholar]
- Melzner, I. , Weniger, M.A. , Bucur, A.J. , Bruderlein, S. , Dorsch, K. , Hasel, C. , Leithauser, F. , Ritz, O. , Dyer, M.J. , Barth, T.F. & Moller, P. (2006) Biallelic deletion within 16p13.13 including SOCS‐1 in Karpas1106P mediastinal B‐cell lymphoma line is associated with delayed degradation of JAK2 protein. International Journal of Cancer, 118, 1941–1944. [DOI] [PubMed] [Google Scholar]
- Mi, H. , Muruganujan, A. & Thomas, P.D. (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Research, 41, D377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottok, A. , Renne, C. , Seifert, M. , Oppermann, E. , Bechstein, W. , Hansmann, M.L. , Kuppers, R. & Brauninger, A. (2009) Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B‐cell lymphoma entities. Blood, 114, 4503–4506. [DOI] [PubMed] [Google Scholar]
- Naka, T. , Narazaki, M. , Hirata, M. , Matsumoto, T. , Minamoto, S. , Aono, A. , Nishimoto, N. , Kajita, T. , Taga, T. , Yoshizaki, K. , Akira, S. & Kishimoto, T. (1997) Structure and function of a new STAT‐induced STAT inhibitor. Nature, 387, 924–929. [DOI] [PubMed] [Google Scholar]
- Ohya, K. , Kajigaya, S. , Yamashita, Y. , Miyazato, A. , Hatake, K. , Miura, Y. , Ikeda, U. , Shimada, K. , Ozawa, K. & Mano, H. (1997) SOCS‐1/JAB/SSI‐1 can bind to and suppress Tec protein‐tyrosine kinase. Journal of Biological Chemistry, 272, 27178–27182. [DOI] [PubMed] [Google Scholar]
- Ott, G. , Ziepert, M. , Klapper, W. , Horn, H. , Szczepanowski, M. , Bernd, H.W. , Thorns, C. , Feller, A.C. , Lenze, D. , Hummel, M. , Stein, H. , Muller‐Hermelink, H.K. , Frank, M. , Hansmann, M.L. , Barth, T.F. , Moller, P. , Cogliatti, S. , Pfreundschuh, M. , Schmitz, N. , Trumper, L. , Loeffler, M. & Rosenwald, A. (2010) Immunoblastic morphology but not the immunohistochemical GCB/nonGCB classifier predicts outcome in diffuse large B‐cell lymphoma in the RICOVER‐60 trial of the DSHNHL. Blood, 116, 4916–4925. [DOI] [PubMed] [Google Scholar]
- Pfreundschuh, M. , Schubert, J. , Ziepert, M. , Schmits, R. , Mohren, M. , Lengfelder, E. , Reiser, M. , Nickenig, C. , Clemens, M. , Peter, N. , Bokemeyer, C. , Eimermacher, H. , Ho, A. , Hoffmann, M. , Mertelsmann, R. , Trumper, L. , Balleisen, L. , Liersch, R. , Metzner, B. , Hartmann, F. , Glass, B. , Poeschel, V. , Schmitz, N. , Ruebe, C. , Feller, A.C. & Loeffler, M. , German High‐Grade Non‐Hodgkin Lymphoma Study Group (2008) Six versus eight cycles of bi‐weekly CHOP‐14 with or without rituximab in elderly patients with aggressive CD20+ B‐cell lymphomas: a randomised controlled trial (RICOVER‐60). The Lancet Oncology, 9, 105–116. [DOI] [PubMed] [Google Scholar]
- Pfreundschuh, M. , Trumper, L. , Osterborg, A. , Pettengell, R. , Trneny, M. , Imrie, K. , Ma, D. , Gill, D. , Walewski, J. , Zinzani, P.L. , Stahel, R. , Kvaloy, S. , Shpilberg, O. , Jaeger, U. , Hansen, M. , Lehtinen, T. , Lopez‐Guillermo, A. , Corrado, C. , Scheliga, A. , Milpied, N. , Mendila, M. , Rashford, M. , Kuhnt, E. , Loeffler, M. & MabThera International Trial Group (2006) CHOP‐like chemotherapy plus rituximab versus CHOP‐like chemotherapy alone in young patients with good‐prognosis diffuse large‐B‐cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) Group. The Lancet Oncology, 7, 379–391. [DOI] [PubMed] [Google Scholar]
- Reddy, A. , Zhang, J. , Davis, N.S. , Moffitt, A.B. , Love, C.L. , Waldrop, A. , Leppa, S. , Pasanen, A. , Meriranta, L. , Karjalainen‐Lindsberg, M.L. , Norgaard, P. , Pedersen, M. , Gang, A.O. , Hogdall, E. , Heavican, T.B. , Lone, W. , Iqbal, J. , Qin, Q. , Li, G. , Kim, S.Y. , Healy, J. , Richards, K.L. , Fedoriw, Y. , Bernal‐Mizrachi, L. , Koff, J.L. , Staton, A.D. , Flowers, C.R. , Paltiel, O. , Goldschmidt, N. , Calaminici, M. , Clear, A. , Gribben, J. , Nguyen, E. , Czader, M.B. , Ondrejka, S.L. , Collie, A. , Hsi, E.D. , Tse, E. , Au‐Yeung, R.K.H. , Kwong, Y.L. , Srivastava, G. , Choi, W.W.L. , Evens, A.M. , Pilichowska, M. , Sengar, M. , Reddy, N. , Li, S. , Chadburn, A. , Gordon, L.I. , Jaffe, E.S. , Levy, S. , Rempel, R. , Tzeng, T. , Happ, L.E. , Dave, T. , Rajagopalan, D. , Datta, J. , Dunson, D.B. & Dave, S.S. (2017) Genetic and functional drivers of diffuse large B cell lymphoma. Cell, 171, e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter, A. & Klapper, W. (2008) Recent advances in the understanding and management of diffuse large B‐cell lymphoma in children. British Journal of Haematology, 142, 329–347. [DOI] [PubMed] [Google Scholar]
- Ritz, O. , Guiter, C. , Dorsch, K. , Dusanter‐Fourt, I. , Wegener, S. , Jouault, H. , Gaulard, P. , Castellano, F. , Moller, P. & Leroy, K. (2008) STAT6 activity is regulated by SOCS‐1 and modulates BCL‐XL expression in primary mediastinal B‐cell lymphoma. Leukemia, 22, 2106–2110. [DOI] [PubMed] [Google Scholar]
- Roman, E. & Smith, A.G. (2011) Epidemiology of lymphomas. Histopathology, 58, 4–14. [DOI] [PubMed] [Google Scholar]
- Roschewski, M. , Staudt, L.M. & Wilson, W.H. (2014) Diffuse large B‐cell lymphoma‐treatment approaches in the molecular era. Nature Reviews Clinical Oncology, 11, 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald, A. & Staudt, L.M. (2003) Gene expression profiling of diffuse large B‐cell lymphoma. Leukaemia & Lymphoma, 44(Suppl. 3) S41–47. [DOI] [PubMed] [Google Scholar]
- Schif, B. , Lennerz, J.K. , Kohler, C.W. , Bentink, S. , Kreuz, M. , Melzner, I. , Ritz, O. , Trumper, L. , Loeffler, M. , Spang, R. & Moller, P. (2013) SOCS1 mutation subtypes predict divergent outcomes in diffuse large B‐Cell lymphoma (DLBCL) patients. Oncotarget, 4, 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab, H.A. , Gough, J. , Cooper, D.N. , Stenson, P.D. , Barker, G.L. , Edwards, K.J. , Day, I.N. & Gaunt, T.R. (2013) Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human Mutation, 34, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, N.L. , Kumar, P. , Hu, J. , Henikoff, S. , Schneider, G. & Ng, P.C. (2012) SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Research, 40, W452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger, A.M. , Ziepert, M. , Horn, H. , Scott, D.W. , Barth, T.F.E. , Bernd, H.W. , Feller, A.C. , Klapper, W. , Szczepanowski, M. , Hummel, M. , Stein, H. , Lenze, D. , Hansmann, M.L. , Hartmann, S. , Moller, P. , Cogliatti, S. , Lenz, G. , Trumper, L. , Loffler, M. , Schmitz, N. , Pfreundschuh, M. , Rosenwald, A. , Ott, G. & German High‐Grade Lymphoma Study Group (2017) Clinical impact of the cell‐of‐origin classification and the MYC/BCL2 dual expresser status in diffuse large B‐cell lymphoma treated within prospective clinical trials of the german high‐grade non‐Hodgkin's Lymphoma Study Group. Journal of Clinical Oncology, 35, 2515–2526. [DOI] [PubMed] [Google Scholar]
- Starr, R. , Willson, T.A. , Viney, E.M. , Murray, L.J. , Rayner, J.R. , Jenkins, B.J. , Gonda, T.J. , Alexander, W.S. , Metcalf, D. , Nicola, N.A. & Hilton, D.J. (1997) A family of cytokine‐inducible inhibitors of signalling. Nature, 387, 917–921. [DOI] [PubMed] [Google Scholar]
- Weniger, M.A. , Melzner, I. , Menz, C.K. , Wegener, S. , Bucur, A.J. , Dorsch, K. , Mattfeldt, T. , Barth, T.F. & Moller, P. (2006) Mutations of the tumor suppressor gene SOCS‐1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho‐STAT5 accumulation. Oncogene, 25, 2679–2684. [DOI] [PubMed] [Google Scholar]
- Yates, C.M. , Filippis, I. , Kelley, L.A. & Sternberg, M.J. (2014) SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. Journal of Molecular Biology, 426, 2692–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenz, T. , Kreuz, M. , Fuge, M. , Klapper, W. , Horn, H. , Staiger, A.M. , Winter, D. , Helfrich, H. , Huellein, J. , Hansmann, M.L. , Stein, H. , Feller, A. , Moller, P. , Schmitz, N. , Trumper, L. , Loeffler, M. , Siebert, R. , Rosenwald, A. , Ott, G. , Pfreundschuh, M. , Stilgenbauer, S. & German High‐Grade Non‐Hodgkin Lymphoma Study Group (DSHNHL) (2017) TP53 mutation and survival in aggressive B cell lymphoma. International Journal of Cancer, 141, 1381–1388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Schematic position and length of amplicons used to cover the SOCS1 open reading frame.
Figure S2. The subgroup examined represents the whole RICOVER‐60 cohort.
Figure S3. Comparison of positional accumulation of variants across SOCS1 using publicly available databases across different cancer types.
Figure S4. Survival curves of DLBCL patients related to SOCS1 mutation subtypes.
Table SI. Primer sequences of PCR primers used to cover the SOCS1 open reading frame.
Table SII. A: Summary of patient characteristics, immunohistochemistry and fluorescence in situ hybridization data.


