Abstract
Mitochondria are as highly specialized organelles and masters of the cellular energy metabolism in a constant and dynamic interplay with their cellular environment, providing adenosine triphosphate, buffering Ca2+ and fundamentally contributing to various signaling pathways. Hence, such broad field of action within eukaryotic cells requires a high level of structural and functional adaptation. Therefore, mitochondria are constantly moving and undergoing fusion and fission processes, changing their shape and their interaction with other organelles. Moreover, mitochondrial activity gets fine‐tuned by intra‐ and interorganelle H+, K+, Na+, and Ca2+ signaling. In this review, we provide an up‐to‐date overview on mitochondrial strategies to adapt and respond to, as well as affect, their cellular environment. We also present cutting‐edge technologies used to track and investigate subcellular signaling, essential to the understanding of various physiological and pathophysiological processes.
Keywords: calcium, intracellular signaling, mitochondria, mitochondria‐associated ER membranes, mitochondrial membrane potential, mitochondrial structure, potassium
This review provides an up‐to‐date overview of the intra‐ and interorganelle signaling within mitochondria, highlighting their role as highly specialized working units and signaling hubs. Moreover, this review presents cutting‐edge technologies to track the kinetics and dynamics of these molecular processes, which are essential to the understanding of various physiological and pathophysiological processes. For instance, super‐resolution microscopy enables us to follow morphological changes within mitochondria under assorted conditions.

Abbreviations
- CJ
cristae junction
- CM
cristae membrane
- DRP1
dynamin‐related protein 1
- dSTORM
direct stochastic optical reconstruction microscopy
- EMRE
essential MCU regulator
- ETC
electron transport chain
- FADH2
flavin adenine dinucleotide
- FRET
Förster resonance energy transfer
- GE
genetically encoded
- GPS2
G protein pathway suppressor 2
- IMM
inner mitochondrial membrane
- IP3R
inositol triphosphate receptor
- LETM1
leucine zipper‐EF‐hand‐containing transmembrane protein 1
- LIT
Light‐inducible tethering
- MAMs
mitochondria‐associated ER membranes
- MCU
mitochondrial calcium uniporter
- MFN1
mitofusin 1
- MFN2
mitofusin 2
- MICOS complex
mitochondrial contact site and cristae organizing complex
- MICU1
mitochondrial calcium uptake 1
- MICU2
mitochondrial calcium uptake 2
- mPTP
mitochondrial permeability transition pore
- mtUPR
mitochondrial unfolded protein response
- NADH
nicotinamide adenine dinucleotide
- OMA1
metalloprotease‐related protein 1
- OMM
outer mitochondrial membrane
- OPA1
optic atrophy 1
- OXPHOS
oxidative phosphorylation
- PALM
photo‐activated localization microscopy
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- ROS
reactive oxygen species
- SIM
structured illumination microscopy
- STED
stimulated emission depletion
- TMRM
tetramethylrhodamine methyl ester
- TOM
translocase of the outer membrane
- UCP2
uncoupling protein 2
- UCP3
uncoupling protein 3
- VDAC
voltage‐dependent anion channel
- YME1L
ATP‐dependent metalloprotease
- Δψm
Mitochondrial membrane potential
Introduction
Mitochondria have undergone a drastic transition from free‐living bacteria to fundamental compartments within eukaryotic cells 1. Therefore, the mitochondria's broad field of contributions, ranging from production of ATP by oxidative phosphorylation (OXPHOS), buffering of Ca2+, and contribution to various cellular signaling pathways, was supplemented with highly dynamic structural and functional adaptations 2, 3. Stretching throughout the cell as a highly dynamic network along the cytoskeleton, mitochondria are constantly undergoing fusion and fission to meet the requirements of cellular subdomains under various conditions 2, 3, as discussed in our ‘Mitochondria as highly specialized working unit’ section. Besides by structure and shape, mitochondria's activity is largely controlled by the homeostasis of ions (Fig. 1), including H+ 4, 5, 6, K+ 7 and Ca2+ 8. These ions are not just powerful tools to fine‐tune mitochondrial activity, but also operate as messengers for intra‐organelle and intercellular communication, as explained in the chapter ‘Fine‐tuning of mitochondrial activity’. While fulfilling their multiple tasks, mitochondria strongly rely on the support of their cellular environment. This results in a busy interplay between mitochondria and various organelles 9, as summarized in ‘Mitochondria as signaling hubs: Give&Get’. Having the majority of proteins encoded by nuclear DNA makes a constant communication between mitochondria and the nucleus indispensable 10, for instance. Moreover, mitochondria form highly specialized signaling hubs with the endoplasmic reticulum (ER) to ensure and control lipid and Ca2+ transfer in restricted subdomains 11. Therefore, it is obvious that interplay between mitochondria and different cellular compartments takes place at specific contact sites or through the exchange of second messengers. To track these subcellular processes, cutting‐edge techniques are required to make investigation possible, including high‐resolution microscopy as well as highly sensitive organelle‐targeted biosensors. We provide an overview about techniques that enable us to study all of these processes at each chapter as well as in the table (Table 1), highlighting the importance of technological progress to reveal further mysteries about our cellular powerplants.
Figure 1.
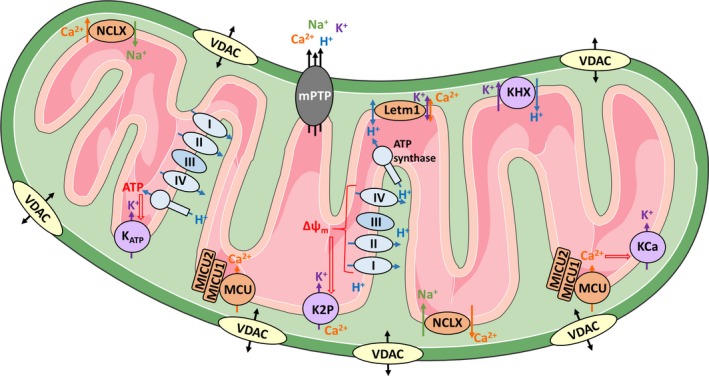
An overview of various types of mitochondrial ion channels. The OMM is largely permeable due to the VDAC, which allows the transport of small metabolites and ions across the OMM. In contrast, ion transport across the IMM has to be highly constricted. Proton pumping from the mitochondrial matrix into the intermembrane space generates the Δψm, boosting ATP generation and regulating the transfer of ions across the IMM. K+ influx is modulated by the Δψm triggering K+ influx via two‐pore domain K+ (K2P) channels and by mitochondrial ATP production affecting ATP‐sensitive K+ channels (KATP). Moreover, mitochondrial Ca2+ levels boost mitochondrial K+ uptake by affecting the activity of Ca2+‐activated K+ (KCa) channels. Extrusion of K+ is ensured by the K+/H+ transporter (KHX). The homeostasis of mitochondrial Ca2+ levels is also highly regulated. The MCU ensures MICU and gets tightly controlled by various proteins, including the gatekeepers MICU1 and MICU2. Furthermore, mitochondrial Ca2+ level is kept in check through an exchange with other ions by, for instance, the NCLX. Moreover, the LETM1 was proposed to act as Ca2+/H+ and/or K+/H+ antiporter, in either cases influencing Ca2+ influx and extrusion. Tight control of Ca2+ homeostasis is essential, since overwhelming accumulation of Ca2+ induces death‐bringing opening of the mPTP.
Table 1.
Technical approaches to track mitochondrial structure, activity, and interorganelle interplay.
| Technical approach | Advantages/disadvantages | References |
|---|---|---|
| Mitochondrial structure and shape | ||
| Fluorescence microscopy | ||
| Confocal microscopy |
○ Conventional resolution of ~ 280 nm + Imaging of living cells and fixed cells possible − Resolution of maximal 150 nm (4Pi) |
230 |
| SIM |
+ Live cell and time lapse imaging + Superior spatial resolution over confocal microscopy + Analysis of submitochondrial structures |
66 |
| STED | ||
| PALM/dSTORM |
+ Highest spatial resolution possible with fluorescence microscopy + Easy specific targeting with high labeling density using fluorescent marker − Fixation of the sample is often necessary − Temporal resolution of PALM displays a problem for moving structures in living cells |
65 |
| Electron microscopy |
+ Precise analysis of submitochondrial structures possible + Very high spatial resolution + Immunogold preparation allows protein localization but lacks in labeling density − Fixation and embedding of the sample necessary |
20 |
| Mitochondrial energy production | ||
| Oroboros O2k |
○ Oxygen consumption is measured by a polarographic oxygen electrode + Analysis of cells, tissues, and isolated mitochondria possible + Sequential injection/titration of compounds possible − Not suitable for high‐throughput screening |
74 |
| Seahorse technology |
+Oxygen consumption and extracellular acidification are measured in parallel by fluorescent sensors + Analysis of adherent cells, suspensions cells, permeabilized cells, isolated mitochondria, and—using specific tissue plates—tissues possible − Only four injections possible − 24‐ and 96‐well‐based assay platform |
75 |
| GE ATP probes |
+ Organelle‐targeting makes analysis of ATP levels in various cellular compartments possible in real time − Proper transfection/infection efficiency is required |
76, 77 |
| ATeams | ||
| pHmito | ||
| GE pH probes | ||
| mtAlpHi |
○ Organelle‐targeted pH sensor that allows pH measurements in the lumen of mitochondria + Excitation of the probe at 498 nm reduces phototoxicity − Sensor provides intensiometric read‐out, hampering pH quantification − Proper transfection/infection efficiency is required |
99 |
| SypHer |
○ Organelle‐targeted pH sensor that allows pH measurements in the lumen of mitochondria + Excitation at 430 nm and 480 nm with the detection of a constant emission wavelength at 530 nm allows a ratiometric read‐out and an easy pH quantification − Excitation of the probe at 430 nm might cause phototoxicity − Proper transfection/infection efficiency is required |
92 |
| Fluorescent dyes | ||
| BCECF |
○ Chemical pH sensor allowing global intracellular pH measurements + Cell loading with BCECF‐AM yields high fluorescent cell number + pKa of ~ 6.98 is close to the cytosolic pH + Excitation at 440 and 490 nm with the detection of a constant emission wavelength at 530 nm allows a ratiometric read‐out and an easy pH quantification − Excitation of the probe at 430 nm might cause phototoxicity − Presence of cellular esterases required for BCECF‐AM to BCECF cleavage |
101 |
| SNARF |
○ Chemical pH sensor allowing global intracellular pH measurements + Cell loading with SNARF‐AM yields high fluorescent cell number + pKa of ~ 7.5 is close to the cytosolic intracellular pH + Excitation at ~ 500 nm with the detection of two emission wavelengths at 580 and 640 nm allows a ratiometric read‐out and an easy pH quantification + Long excitation wavelength of the probe reduces phototoxicity − Presence of cellular esterases required for SNARF‐1‐AM to SANRF‐1 cleavage − Although global intracellular staining, probe was used to measure pHmito using high‐resolution microscopy − Measurements require a sophisticated microscope setup due to separation of two emission wavelengths |
102 |
| Mito‐pH |
○ Chemical pH sensor allowing specific pHmito measurements + Cell loading with Mito‐pH yields high fluorescent cell number + pKa of ~ 7.33 is close to the cytosolic pH + Excitation at 490 and 560 nm with the detection of emission wavelengths at 520 and 600 nm allows a ratiometric read‐out and an easy pH quantification − Long excitation wavelengths of the probe reduce phototoxicity |
103 |
| Δψm | ||
| Fluorescent dyes | ||
| TMRM |
○ Monochromatic dye (λex = 555 nm, λem = 570 nm) for semiquantitative analysis of Δψm + Cell loading with TMRM yields high fluorescent cell number − Mitochondria with low Δψm are possibly not stained and cannot be measured − Alteration of cellular respiration and binding to mitochondrial membrane might affect results |
107 |
| TMRE |
○ Monochromatic dye (λex = 549 nm, λem = 574 nm) for semiquantitative analysis of Δψm + Cell loading with TMRE yields high fluorescent cell number − Mitochondria with low Δψm are possibly not stained and cannot be measured − Alteration of cellular respiration and binding to mitochondrial membrane might affect results |
105 |
| JC‐1 |
○ Chemical, ratiometric dye (λex = 488 nm, λem = 530 and 595 nm) for semiquantitative analysis of Δψm + All mitochondria are stained, independent of their Δψm + Cell loading with JC‐1 yields high fluorescent cell number − Photosensitive − Fluorescence may be changed independently of Δψm by, for instance, H2O2 or disturbed equilibrium between monomers and aggregates |
106 |
| Mitochondrial K+ homeostasis | ||
| Patch‐clamp |
+ Gold standard method for K+ fluctuation measurements + Very sensitive method − Isolation of mitochondria and preparation of mitoblasts required to measure K+ fluctuations across the IMM − Usage of isolated mitochondria might be far from the physiologic intracellular situation |
118, 126, 128, 137 |
| GE K+ probes | ||
| GEPII |
○ Organelle‐targeted K+ sensor that allows K+ measurements in the lumen of mitochondria + EC50 of 60.95 mm is suitable for measurements of [K+] within the mitochondrial matrix + Excitation at 430 nm with the detection of two emission wavelengths at 475 and 525 nm allows a ratiometric read‐out and an easy K+ quantification − Excitation of the probe at 430 nm might cause phototoxicity − Proper transfection/infection efficiency is required − Measurements require a rather sophisticated microscope setup as two emission wavelengths have to be properly separated |
111 |
| Mitochondrial Ca2+ homeostasis | ||
| Fluorescent dyes | ||
| Fura‐2 |
○ Indirect measurement of mitochondrial Ca2+ movement + Simple experimental preparation − Experimental preparation limited to simple cellular incubation − Not suitable for direct measurement of intra‐organelle Ca2+ |
174 |
| Fluo‐3/Fluo‐4 |
○ Indirect measurement of mitochondrial Ca2+ movement + Simple experimental preparation − Significant leak in certain cell types resulting in lowered Ca2+ signals |
179 |
| Rhod‐2 |
+ AM ester dye that allows for mitochondria‐specific Ca2+ measurement − Only suited to short experimental protocols |
180 |
| GE Ca2+ probes | ||
| 4mtD3cpv |
○ Organelle‐targeted, FRET‐based (λex = 430 nm, λem = 480 and 535 nm) mitochondrial Ca2+ sensor ○ K D for Ca2+ around 600 nm + Highly sensitive cameleon with a wide monitoring range − Proper transfection/infection efficiency is required |
185 |
| 4mtD1GO‐CAM |
○ Organelle‐targeted, FRET‐based (λex = 477 nm, λem = 510 and 560 nm) mitochondrial Ca2+ sensor ○ K D for Ca2+ around 1.53 μm + Red‐shifted cameleon with a wide monitoring range + Very well suitable for combination with other organelle‐targeted Ca2+ indicators and to be used simultaneously with Fura‐2 to correlate cytosolic and mitochondrial Ca2+ signals − Proper transfection/infection efficiency is required |
186 |
|
mtGEM ‐GECO1 |
○ Organelle‐targeted, FRET‐based (λex = 394 nm, λem = 455 and 511 nm) mitochondrial Ca2+ sensor ○ K D for calcium around 340 nm + Very well suitable for combination with other organelle‐targeted Ca2+ indicators − Proper transfection/infection efficiency is required |
187 |
| mtCAR‐GECO1 |
○ Organelle‐targeted, intensiometric‐based (λex = 565 nm, λem = 620 nm) mitochondrial Ca2+ sensor + Very well suitable for combination with other organelle‐targeted Ca2+ indicators − Proper transfection/infection efficiency is required |
188 |
Mitochondria as highly specialized working units
Dynamic changes in structure and shape of mitochondria
The mitochondrial network is a highly specialized working unit capable of undergoing dynamic adaptation in order to meet metabolic needs and to allow internal and external signaling 2, 3. Thereby, constant fission and fusion of the inner (IMM) and outer mitochondrial membrane (OMM) play a crucial role in maintaining mitochondrial integrity. The fusion and fission homeostasis affects the opening probability of the mPTP, facilitating uncontrolled efflux of ions 12, as well as oxidative capacity 13, 14, production of reactive oxygen species (ROS) 15, mitophagy 16, apoptosis 17, 18, Ca2+ signaling 18, 19, and mitochondrial interactions with other cell compartments 20, 21, 22.
Mitochondrial fission is mainly mediated by the cytosolic dynamin‐related protein 1 (DRP1). It binds to the OMM and constricts the mitochondrion, an action facilitated by its GTPase activity 23. Furthermore, dynamin is necessary to achieve complete fission, as DRP1 activity leads to constriction, but not cleavage, during the fission process 24, 25. Nevertheless, it is still unknown whether a specific process or piece of protein machinery is necessary for the cleavage of the IMM.
Mitochondrial fusion is mainly driven by the proteins optic atrophy 1 (OPA1) and dynamin‐like protein mitofusin 1 and 2 (MFN1 and MFN2) 26, 27. MFN1 and MFN2 are located at the OMM and serve to facilitate fusion of two organelles. The necessary energy to fuse membranes is provided predominantly by the GTPase activity of MFN1 28. Extracellular‐signal‐regulated kinase is able to reduce MFN1 activity by phosphorylation, resulting in the fragmentation of mitochondria 29. Analogous to the OMM‐located MFN1 and MFN2, OPA1 is one of the main regulators responsible for the fusion of the IMM 30. Two forms of OPA1, L‐OPA1 and S‐OPA, are kept in balance by the metalloprotease‐related protein 1 (OMA1) and ATP‐dependent metalloprotease YME1L via proteolytic cleavage of the IMM bound L‐OPA1 to soluble S‐OPA1. A predominance of the L‐form yields mitochondrial fusion, while metabolic stimuli or cell stress signals activate L‐OPA1 proteolysis by OMA1 and YME1L, respectively, resulting in fission 31, 32.
Recently, intermitochondrial signaling by either nanotubes or direct interaction via intermitochondrial junctions has been intensively studied. These intermitochondrial contact sites have a high electron density and are coordinated pairs of cristae oriented orthogonally to the OMM in two adjoined mitochondria 33. Nano tunnels are either a result of stalled and incomplete fission events 34 or are de novo generated mitochondrial protrusions formed by members of the kinesin family, like the protein kinesin‐1 heavy chain along the cellular tubulin cytoskeleton 35. These intermitochondrial contact sites are thought to transmit Ca2+ or apoptotic signals across mitochondria 36 or even a coupling of the mitochondrial membrane potential (Δψm) of neighboring mitochondria 37. Thereby, kissing and nanotunneling of mitochondria represent an alternative form of intermitochondrial communication 38, 39. Notably, exchange of, for instance, matrix proteins through nanotunnels follows a slower kinetic compared to conventional fusion most likely due to their small diameter of < 100 nm 38.
The IMM is morphologically separated into two compartments, divided by the cristae junction (CJ): (a) the inner boundary membrane directly facing the inner leaflet of the OMM; and (b) the cristae membrane (CM), forming the protrusions and invaginations of the IMM into the mitochondrial matrix 40, 41. Both compartments differ in protein composition and functional activity 42, 43. OPA1 is involved in the stabilization of the CJ and is interconnected with the mitochondrial contact site and cristae organizing complex (MICOS complex) 44. Loss of OPA1 by knockdown or knockout leads to widened cristae lumen and CJ 17, 45, reduction of IMM potential 46, increase in basal mitochondrial Ca2+ levels 47, and apoptosis induction 48. Similar to OPA1, the inner boundary membrane localized mitochondrial Ca2+ uptake 1 (MICU1) is involved in CJ stabilization, maintenance of IMM potential, and cytochrome c restriction to the cristae lumen. The Ca2+ sensing ability of MICU1 and its thereby affected quaternary structure might influence the CJ stability and permeability 49. The MICOS complex is composed of several subunits with mitofilin representing the biggest 50. MICOS proteins are especially enriched in the CJ and form IMM‐IMM and IMM‐OMM contact sites with proteins like OPA1 44, 51, translocase of the outer membrane (TOM) 52, 53 and S‐adenosylmethionine synthetase 54 to ensure CJ biogenesis and stabilization. The structure of the CJ also restricts the FOF1‐ATP synthase and the respiratory chain complexes to the CM, leading to a closed compartment of the cristae lumen 55, 56. Since the OMM is generally permeable due to voltage‐dependent anion channel (VDAC) 57, 58, the CJ seems to form a diffusion barrier for protons, creating an isolated space for the activity of FOF1‐ATP synthase and respiratory chain complexes with lateral pH gradients 59. Computational models and in vitro experiments have shown that low pH causes cristae invaginations of the IMM based on the electrostatic induced negative curvature of the outer CM leaflet 60, 61. Besides the direct involvement of the FOF1‐ATP synthase in cristae formation and morphology 42, 56, 62, an indirect effect of CM localized respiratory supercomplexes mediated by the decreased pH in the CJ might be an important factor in cristae biogenesis and shape definition 63.
Techniques to analyze structure and shape
While the current knowledge about the ultrastructure of mitochondria relies mainly on electron microscopy data, recent advancements in fluorescence microscopy, in particular super‐resolution microscopy techniques like structured illumination microscopy (SIM), stimulated emission depletion (STED), or photo‐activated localization microscopy (PALM), enable researchers to visualize and investigate mitochondrial ultrastructure in living cells.
direct stochastic optical reconstruction microscopy (dSTORM) is an alternative to immunogold labeling and electron microscopy for analysis of submitochondrial localization of proteins like the FOF1‐ATP synthase 64 or uncoupling protein 4 (UCP4) with high spatial resolution 65.
However, dSTORM is not suitable for live cell imaging and PALM approaches in live cells do not reach the necessary temporal resolution necessary to analyze dynamics of the IMM. Therefore, STED 66 or SIM 67 microscopy have been used to image, for instance, binding of fluorescent dyes dependent on Δψm, such as Mitotracker GreenFM and tetramethylrhodamine methyl ester (TMRM), to the IMM. By using these techniques, cristae structure can be resolved and dynamic rearrangement of the IMM can be investigated due to high temporal resolution 67, 68. The dynamics of the IMM in regions of close mitochondria–ER contact could be analyzed, for instance. While silencing of OPA1 reduced IMM kinetics globally, ER Ca2+ release decreased IMM kinetics exclusively in mitochondrial areas in close proximity to the ER, pointing to a Ca2+‐regulated mechanism of IMM rearrangement 67. The submitochondrial dynamic and MICU1‐dependent relocalization of mitochondrial Ca2+ uniporter (MCU) and UCP2 upon ER‐Ca2+ release form the entire IMM into the inner boundary membrane, which was visualized recently using SIM 49. Similar observations of MCU inner boundary membrane localization under oxidizing conditions were made with STED and SIM showing the superior performance of super‐resolution over conventional microscopy in live cells 69.
The STED technique has been used to visualize spiral MICOS‐cluster arrays along the inner boundary membrane of yeast and human mitochondria, making an estimation of mitochondrial microarchitecture in combination with the use of immunogold labeling possible 43, 70. However, while STED microscopy achieves better resolution than SIM, it comes with the drawback of generally higher excitation intensity, leading to photobleaching and toxicity and, thus, limited acquisition time. SIM can be used with low excitation intensities, high frame rate, and over extended periods of time 71. Also, nonsaturation fluorescence microscopy along with intensive deconvolution was used to analyze the cristae kinetics in live cells and to distinguish between the IMM and matrix structures 68.
Energy production as principal task
The mitochondria's core working unit is the mitochondrial respiratory chain located in the IMM within the cristae 55, 56. Oxidation of glycolysis derived pyruvate and nicotinamide adenine dinucleotide (NADH) by mitochondria yields about 15 times more ATP than glycolysis itself 72. Thereby, acetyl coenzyme A, derived from oxidative decarboxylation of pyruvate or beta‐oxidation, fuels the citric acid cycle to produce NADH and flavin adenine dinucleotide (FADH2).
These reducing equivalents serve as electron donors for the electron transport chain (ETC), consisting of NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), ubiquinone, cytochrome bc1 complex (complex III), and cytochrome c and cytochrome c oxidase (complex IV). After transfer of electrons, derived from the NADH and FADH2 as hydrogen molecules, to the ETC via complex I and II, the electron transport through complex I to complex IV is coupled to proton pumping from the mitochondrial matrix into the locally restricted cristae lumen. This process causes a negative charge in the matrix and a positive charge in the IMS resulting in an electrochemical gradient, used for the proton transport back from the IMS into the mitochondrial matrix through ATP synthase (complex V). The released energy is, finally, utilized by FOF1‐ATP synthase to gain the cellular energy carrier, ATP, by phosphorylation of adenosine diphosphate 73.
Techniques to measure activity of mitochondrial activity
Besides a broad range of biochemical approaches such as western blot, ROS, and ATP assays, there are specific measurements for mitochondrial bioenergetics available. Based on a polarographic oxygen electrode measuring the concentration and consumption of oxygen before and after injection of various substrates, the Oroboros O2k has been in use since the 1990s. This machine offers the possibility to analyze cells, tissues as well as isolated mitochondria with high resolution. Furthermore, sequential injection and titration of compounds inhibiting different complexes of the mitochondrial respiratory chain or boosting maximal mitochondrial activity can be done during the ongoing measurement. However, the Oroboros O2k is not suitable for high‐throughput screenings as only two samples can be analyzed at the same time 74. Therefore, the Seahorse XF Extracellular Flux Analyzer has been introduced about 10 years ago, offering 24‐ and 96‐well‐based assay platforms. Thereby, the oxygen consumption rate and the extracellular acidification are measured in parallel by fluorescent sensors. However, this brings the limitation that injectable compounds may interfere with the fluorescent sensor. Moreover, in contrast to the Oroboros O2k, only four compounds can be injected during the measurements. Since the sensor‐containing biocartridge has to be loaded with the injectable compounds before starting the actual measurement, it is not possible to change or adapt the compound concentrations during the measurement. Notably, various cell and tissues plates and protocol are meanwhile available, making the analysis of not just adherent cells but also tissues, suspension cells, permeabilized cells, and isolated mitochondria possible 75. Another appealing approach is the usage of genetically encoded (GE) ATP indicators based on a Förster resonance energy transfer (FRET) and equipped with an ATP‐sensing subunit of the bacterial FOF1‐ATP synthase. Usage of these so‐called ATeams sensors enabled tracking of ATP levels ranging from 7.4 µm to 3.3 mm in different cellular compartments in real time 76, 77.
Fine‐tuning of mitochondrial activity
The H+ ion
Mitochondrial activity and function are not solely controlled by mitochondrial structure, but also by various fine‐tuning mechanisms, including homeostasis of ions (Fig. 1). In mitochondria, the H+ has a unique role. It is well known that the concentration of protons (H+) needs to be tightly regulated to preserve essential functions on a single cell level as well as in organisms 78, 79. While the cytosol, the ER, and the nucleus have a neutral pH of 7.0 to 7.4, lysosomes or secretory vesicles maintain an extremely acidic pH for degradation or secretion purposes 80, 81. Strikingly, we can find acidic as well as alkaline areas in mitochondria 82. While the mitochondrial intermembrane space represents a slightly acidic environment with a pH of ~ 6.8, the mitochondrial matrix is the most alkaline compartment within a cell with pH values around 7.6–8.0 82. This difference in the proton concentration is mainly caused by the activity of the respiratory chain, transporting electrons along respiratory complexes. The serial reduction of electrons provides enough energy to shuttle protons via complex I, III, and IV from the matrix into the IMS against their concentration gradient. The accumulation of H+ in the intermembrane space is essential for building up a driving force to activate the ATP generating F1/F0 ATP synthase, while re‐entering the mitochondrial matrix 4, 5, 6. Protons are forced back into the mitochondrial matrix by the pH gradient, a chemical or concentration gradient, and the Δψm, representing a charge or electrical gradient 83, 84.
The Δψm is not only essential for the generation of ATP 6, but also to regulate transfer of ions like K+ 83, 85, 86, Na+ 87, 88, Cl− 89, 90, and Ca2+ 91 across the mitochondrial membrane. Dysregulation of Ca2+ homeostasis and mitochondrial Ca2+ overload results in increased permeability of the IMM to protons, decreasing Δψm as well as the mitochondrial pH gradient 92 and initiating cell death 93.
Since the mitochondrial metabolism is tightly regulated, it is not surprising that dysregulations and sustained changes of the Δψm lead to severe mitochondrial dysfunctions and have been associated with various disease conditions, including cancer 94, 95 and neurodegeneration 96, 97. For instance, some types of tumors have been associated with elevated Δψm linked to increased glycolytic rates and resistance to regulated cell death 98. Consistent with these reports, some lung cancer cell lines (A549, H446, SPC), breast cancer MCF7 cells, and glioblastoma MO59K cells exhibited higher Δψm than the correspondent healthy, noncancerous cell types 94. Moreover, higher Δψm was linked to increased tumorigenicity and malignancy of cancer stem cells, linked to development and (re)occurrence of malignant tumors, while cells with lower Δψm were more prone to differentiation. Since reduction of Δψm by rapamycin decreased tumorigenicity significantly in these cells, targeting Δψm might be a potential strategy to prevent development of malignant tumors 94.
Techniques to measure pHmito and Δψm
Nowadays, a huge variety of indicators and GE sensors is available to determine pHmito and Δψm and the most prominent candidates will be presented below.
mtAlpHi was described as one of the first encoded pHmito indicators, visualizing and characterizing metabolic changes within mitochondria 99. The calmodulin of the Ca2+ indicator camgaroo was substituted by a portion of aequorin comprising only two EF hands, resulting in a Ca2+ ‐insensitive probe with an estimated pKa of 8.5, excitation at 498 nm, and emission at 522 nm.
One of the most commonly used GE pHmito sensors is SypHer 92, based on a circular permutated yellow fluorescent proteins (YFP) derived from mutating the cysteine residues of the H2O2 sensor Hyper 100. SypHer exhibits ratiometric responses at 430 nm and 480 nm upon changing the pH, but is insensitive to H2O2, Ca2+, and PO3−. SypHer proved perfectly suitable for the detection of cytosolic as well as mitochondrial pH values due to a pKa of 8.71, a 20‐fold increase upon switching from pH 7 to 10, and a fourfold increase in the range between pH 7 and 8. Furthermore, SypHer has been also used in simultaneous measurements in combination with other sensors performing two‐ or multi‐color imaging 92.
The development of small chemical fluorescent dyes to specifically monitor pHmito has been challenging, but was finally accomplished a few years ago. While dyes like BCECF 101 or SNARF 102 have been used to measure pH in the cytosol, probes like Mito‐pH specifically stain mitochondria. Mito‐pH consists of a pH‐sensitive FITC fluorophore fused to a pH‐insensitive hemicyanine group. This pH‐insensitive part of the probe not only acts as the reference fluorophore for a ratiometric read‐out, but allows for localization in mitochondria, due to their lipophilic cationic nature. The sensor reacts reversibly to changes in pH, thereby exhibiting a double ratiometric read‐out of dual excitation/dual emission and dual excitation between pH 6.1 and 8.4 103. Another approach to monitor pH changes was made by using a chemical system composed of a piperazine‐linked naphthalimide being responsible for the fluorescent off and on signaling, a cationic triphenylphosphonium group for specific mitochondrial targeting, as well as a reactive benzyl chloride subunit for fixation in mitochondria. This probe accumulates within mitochondria due to the effect of the Δψm on the cationic triphenylphosphonium group. Additionally, the benzyl chloride was thought to undergo nucleophilic substitution with reactive thiols of mitochondrial proteins, ensuring mitochondrial localization even upon acidification or membrane depolarization 104.
While several GE pH sensors have been developed in the last decade, to the best of our knowledge, there is no GE sensor available for measuring Δψm.
Frequently used indicators for semiquantitative analysis of Δψm are small fluorescent dyes based on a lipophilic cation structure like TMRM, tetramethylrhodamine ethyl ester (TMRE) 105, and 5,5′,6,6′‐tetrachloro‐1,1′,3,3′‐tetraethylbenzimi‐ dazolylcarbocyanine iodide (JC‐1) 106. TMRM and TMRE are single wavelength indicators emitting red light upon Δψm‐dependent accumulation within mitochondria. The excitation and emission wavelength of these monochromatic dyes are quite similar, with excitation at 555 nm and emission at 570 nm for TMRM and excitation at 549 nm and emission at 574 nm for TMRE 105. If used in higher concentrations, TMRM and TMRE might alter the cellular respiration and bind to mitochondrial membranes 105, 107. In contrast, JC‐1 (λex: 488 nm) is a ratiometric dye, existing as green fluorescent monomer at depolarized membrane potential (λem: 530 nm) and forming red aggregates (λem: 595 nm) at hyperpolarized membrane potential 106, 108. Notably, JC‐1 is very photosensitive and the fluorescence may be changed independently of Δψm by, for instance, H2O2 or disturbed equilibrium between monomers and aggregates. All these dyes are differently permeant and require specifically adjusted loading protocols dependent on the respective cell type 108. Moreover, complete depolarization of mitochondria, by for instance FCCP, might be necessary to achieve a baseline, which makes comparison of different Δψm measurements possible 109.
The K+ ion
Potassium ions (K+) are essentially involved in various processes and represent the most abundant intracellular cation. The cytosolic K+ concentration is important for the maintenance of the cell's membrane potential, works as cofactor for various enzymes, and regulates cell volume as well as endo‐ and exocytosis. Also, mitochondrial functions rely on an intact mitochondrial K+ () homeostasis. While cytosolic K+ concentration is close to 140 mm in vital cells, the concentration of ranges between 20 and 60 mm 110, 111.
Since mitochondrial volume 112, Δψm 113, mitochondrial metabolism 114, and ROS production 115 are tightly coupled to , the transport of this ion has to be strictly controlled by K+ channels and transporters located in the IMM 116.
Δψm drives K+ influx by diffusion across the membrane, referred to as K+ leak, mostly via two‐pore domain K+ (K2P) channels, and via ATP‐sensitive K+ channels located in the IMM (mitoKATP). Besides, voltage gated K+ (Kv) channels and Ca2+ activated K+ (KCa) channels are located within the IMM 117, 118.
The role of mitoKATP, in particular, has been extensively studied. The generation of high levels of ATP by FOF1‐ATP synthase causes a decrease in Δψm and leads to a decreased flux of K+ across the mitoKATP channels into the matrix, probably preventing devastating mitochondrial depolarization. However, under conditions of oxidative stress, increased levels of ROS activate mitoKATP channels, dissipating Δψm and counteracting further ROS production. The interplay between K+ and H+ becomes evident when considering the presence of K+/H+ transporters (KHX) in the IMM, transporting K+ from and H+ into the mitochondrial matrix. One of these transporters is the leucine zipper‐EF‐hand‐containing transmembrane protein 1 (LETM1), facilitating K+ extrusion from the mitochondrial matrix and, thereby, also modulating Na+ and Ca2+ homeostasis 119.
High levels of matrix K+ assist in modulating the transmembrane H+ gradient, altering ATP production, but may also promote the controlled re‐generation of the H+ gradient toward the IMM via the ETC 120. Interestingly, the administration of nonselective KATP channel blockers such as glibenclamide, widely used for the treatment of type 2 diabetes, was found to ameliorate ischemia reperfusion injury in the brain, kidney, intestine, and lungs. The effect was associated with a modulation of the oxidative stress caused by releasing ischemia, after starting reperfusion 121, 122, 123, thus highlighting the importance of proper homeostasis.
Besides the interplay of with Δψm and H+, is fundamentally affected by mitochondrial Ca2+ homeostasis. The most prominent example of Ca2+ activated K+ channels represents the large conductance KCa (BKCa) channel, found in the plasma membrane of excitable cells such as neurons and skeletal muscle cells. Several types of KCa channels could be also found in the IMM in various cell types, including the mitochondrial localized large conductance KCa (mitoBKCa) in LN299 human glioma cells 124. Activation of these channel types is caused by elevated cytosolic Ca2+ levels as well as by changes in the Δψm, leading to K+ fluctuations across the membrane. While the pore vestibule of mitoBKCa faces the intermembrane space, the Ca2+ sensitive domain is located in the mitochondrial matrix, indicating that activation of mitoBKCa requires elevation of mitochondrial matrix Ca2+ 125, 126, 127. Notably, charybdotoxin, which strongly inhibits BKCa channels at the plasma membrane, failed to affect mitoBKCa, raising the question whether mitoBKCa and BKCa are structurally possibly unrelated to each other 127, 128, 129.
Notably, application of compounds acting as plasma membrane BKCa channel openers like NS1619 126, accelerating mitochondrial K+ uptake twofold, halved the size of a myocardial infarct in guinea pig hearts after reperfusion of ischemic regions. These results suggest the participation of mitoBKCa channels against necrotic and apoptotic cell injury after ischemic tissue damage, possibly by modulation of the mitochondrial respiratory chain 7, prevention of [Ca2+]mito overload, maintenance of Δψm, and/or normalization of ROS levels 126, 130, 131. Moreover, NS1619 as well as CGS7184, another BKCa channel opener, also exhibited protective effects on neuronal cells 132, 133. Application of these compounds resulted in K+ flux from the extracellular space into the mitochondrial matrix in isolated rat brain mitochondria, causing Δψm depolarization and reduced ROS production 134. In addition, application of the BKCa channel activator NS11021 was reported to inhibit glutamate‐induced oxidative stress by attenuating ER stress and mitochondrial dysfunction 135. Notably, CGS7184 was shown to directly activate the mitoBKCa by single‐channel recordings. While this compound boosted oxygen consumption rate in isolated rat brain mitochondria, it exhibited cytotoxic effects by increase of cytoplasmic Ca2+ concentration and consequent activation of calpains in intact neuronal HT22 cells 136. These results highlight the therapeutic potential but also the risk for side effects of BKCa channel modulating compounds.
Techniques to measure mitochondrial K+
Considering the importance of intact cellular K+ homeostasis, scientists have been eager to find ways to investigate cellular and mitochondrial K+ levels. dynamics are frequently measured via patch‐clamp approaches using isolated mitochondria or mitoblasts 118, 126, 128, 137. A valuable alternative to investigate subcellular and especially dynamics within single living cells is provided by fluorescent indicators 138, 139, 140. These indicators represent small chemical dyes, either unspecifically labeling the whole cell or specifically localizing within mitochondria. K+ binding to the sensors results in an alteration of their fluorescence depending on the K+ concentration 138, 139, 140. Recently, the first GE indicators suitable for intracellular K+ measurements have been developed 111, 141. These probes, referred to as GE potassium ion indicators (GEPIIs), in combination with available chemical fluorescent indicators sensitive for K+ will in future deepen our understanding of subcellular and particularly mitochondrial K+ homeostasis 142.
The Ca2+ ion
While mitochondrial function and activity is heavily dependent on Ca2+ homeostasis, mitochondria, in turn, also affect the ion's intricate role as a widespread signaling molecule within the cell. Previously thought to function primarily as a regulator of cytosolic Ca2+, it was later determined that influx and efflux machinery are geared more toward control of the organelle´s own Ca2+ levels 143. Changes to concentration are known to have an effect on cellular ATP production 144, 145, respiration 146, ER–mitochondria crosstalk ability (as reviewed by 147), the onset of cellular apoptosis 148, 149, autophagy 150, and many other processes related to cellular health. The mitochondria's function also depends heavily on its ability to send messages to other organelles and receive them from the rest of the cell through Ca2+ movement. For example, the mitochondrion is known to preferentially take up Ca2+ released by the ER 151, a process that usually involves close contact sites between the two organelles 150. Importantly, such association of the mitochondria with other cellular components is not limited to the ER; rather, other interaction has been documented between mitochondria and the nucleus 152, cytoskeleton 142, and plasma membrane 153, among others. Of the signals that pass to and from these parts of the cell, uptake, specifically, has multiple significant consequences pertaining to proper overall function; Ca2+ being sequestered into the matrix ultimately has an effect on local and more widespread cellular Ca2+ signals. Homeostasis between this organelle´s Ca2+ stores and the rest of the cell is crucial, as an overload of intraluminal Ca2+ can lead to the initiation of the apoptotic pathway 8.
Mitochondria are able to activate the intrinsic apoptotic pathway following Ca2+ overload through the release of multiple proteins from their lumen into the cytosol 154. For example, cytochrome c, an important player in the ETC, is under normal circumstances found attached to the IMM. Following Ca2+ overload, mitochondria release this protein into the cytosol, where it stimulates the formation of a caspase‐activating complex otherwise known as the apoptosome. The apoptosome's overall function of activating killer caspases ultimately results in the death of the cell 155. Concerning another important aspect of metabolism and health, cellular respiration, Ca2+ is again vital. It is known that the close positioning of mitochondria to large sources of Ca2+ (namely, the ER or the plasma membrane) allows for significant accumulation of the ion inside the mitochondrial matrix upon physiological Ca2+ release. This increase in matrix Ca2+ levels, in turn, affects mitochondrial metabolism through stimulation of mitochondrial effector molecules such as particular dehydrogenases of the Krebs cycle which are functionally dependent on Ca2+, ultimately leading to ATP production 156. The ability of mitochondria to accumulate Ca2+, and the existence of a uniporter, has been reported on since the 1960s 157, 158, but the molecular identity of the MCU was only determined in 2011 159, 160. MCU is a 40 kilodalton (kDa; 35 kDa in its cleaved form) IMM protein with two transmembrane domains, which forms the core of the uniporter complex 159, 160, 161 uptake occurs primarily through MCU activity, and this uptake is highly selective but has a low apparent K D for Ca2+.
These effects are achieved through MCU regulators. Essential MCU regulator (EMRE) was shown to be a core component of the uniporter complex and is essential for MCU mediated‐ MICU 162. In addition, EMRE was shown to regulate MCU activity based on matrix Ca2+ levels 163. MICU1 and MICU2 are considered to be MCU gatekeepers, setting a uptake threshold, which is higher than that of the MCU 164, 165. Cells with MICU1 knockdown were shown to have elevated basal levels 166. MICU2 is considered to be a negative regulator of MCU 165. Thereby, the stoichiometry of MICU1 to MICU2 was shown be an important factor in Ca2+ uptake and was also reported to vary across different tissues and organs 167. Besides, also UCP2 and UCP3 were shown to influence MCU‐dependent Ca2+ uptake at higher Ca2+ concentrations, whereas LETM1 was shown to influence a potentially different uptake mechanism, likely more pronounced when Ca2+ levels are lower 168. It was shown that UCP2 normalizes MICU in case of protein methyl transferase 1‐driven methylation of MICU1, resulting in sensitivity loss of MICU1 to Ca2+ 169. An important, but as of yet unanswered, aspect of uptake is whether there are different uptake pathways operating in relation to low versus high cellular Ca2+ concentration sources, as well as what the physiological implications of these potential different uptake pathways may be. Another interesting phenomenon that awaits clarification is the spatial resolution of uptake, namely whether it occurs along the full length of the IMM, or whether this activity is restricted to certain membrane regions. Important to consider when discussing uptake of Ca2+ into the mitochondrial matrix is the organelle's ability to balance overall charge through extrusion mechanisms. In the case of the mitochondrion, Ca2+ influx is mainly kept in check through an exchange with other ions. The major protein playing a role in this process was identified to be Na+/Ca2+ exchanger (NCLX) 170. Its presence thus necessitates another mechanism to remove the excess Na+ that accumulates in the matrix following the exchanger's activity. This is proposed to be achieved by members of the Na+/H+ exchanger family, consisting of multiple plasma membrane and organellar transporters 171. LETM1 has also been considered as having a role in Ca2+ movement, though its exact function in this capacity is as yet unclear. It has been proposed to act as a Ca2+/H+ antiporter, which implies a function in Ca2+ extrusion 172. Other publications indicate that it is instead a K+/H+ antiporter, thereby influencing Ca2+ uptake and/or extrusion via secondary means 119, 173.
Techniques to measure mitochondrial Ca2+
Mitochondrial Ca2+ homeostasis is clearly a complex and wide‐ranging process; therefore, the techniques required are also spread across multiple disciplines. In particular, the use of various sensors to study intracellular Ca2+, as well as more specific indicators able to determine the ion's movement to, within and from the mitochondrion itself, provide the quickest insight into the intricate web of Ca2+ signals constantly present throughout the cell.
Probes for overall intracellular Ca2+ measurement can generally be clustered into chemically engineered fluorophores and GE FPs.
In the recent past, the number of Ca2+ sensors specific to mitochondria and other organelles has reached new heights. Sensors used to measure general intracellular Ca2+ movement, including the ubiquitously employed cytosolic Ca2+ sensor Fura‐2 174, as well as the Calcium green family of indicators, are frequently being supplemented, improved upon or replaced by novel developments in sensor technology that allow for a more direct view of the processes occurring within mitochondria and other organelles. With the huge variety currently available, many criteria must be considered to optimally measure intracellular or mitochondrial Ca2+, dependent on the desired experimental read‐out. Choosing the best‐suited Ca2+ sensor necessitates consideration of factors such as the probe´s original form and modification required for expression in cells, its affinity for Ca2+, and its spectral properties, among others (as thoroughly reviewed by 175).
Briefly, chemical indicators, compounds which change their fluorescence properties following binding to Ca2+, are perhaps best suited for observation of the ion's cytoplasmic movement. While they are much simpler to employ than the average GE sensor (no cellular transfection required; simple cell‐loading steps are sufficient), a main disadvantage of chemical sensors for organelle‐specific Ca2+ measurement is the lack of controlled localization once loaded into target cells. Use of such probes will therefore not guarantee mitochondria‐exclusive expression and Ca2+ monitoring. Chemical fluorophores are thus mainly suitable for drawing indirect conclusions on signaling as it pertains to the rest of the cell.
Fura‐2 is a classic example of a ratiometric sensor designed for such purposes. Compared to what was available prior to its characterization, Fura‐2 offered ~ 30× increased fluorescence signals and improved ability to bind Ca2+ specifically over other divalent cations, among other advantages 174. This sensor and its variants continue to appear prominently in cellular Ca2+ research related to crosstalk between mitochondria and the rest of the cell; for example, investigation into the mechanism whereby the ER and mitochondria interact through mitochondria‐associated ER membranes (MAMs) commonly employs the cytosolic Ca2+ sensor Fura‐2 to draw conclusions on Ca2+ signaling between the two organelles 176, 177. Fluo‐3 is another fluorescent dye that has been widely used to measure cytoplasmic Ca2+ movement, but has its own purported disadvantages. For example, Lee et al. 178 showed that in certain cell types this indicator leaks significantly, leading to lowered fluorescence measurements for intracellular Ca2+, combined with increased background fluorescence as leaked dye binds to Ca2+ present in surrounding medium. Fluo‐3's close relative, Fluo‐4, proves similar in structure and other properties, but exhibits increased fluorescence and range for Ca2+ 179. There are also cell‐permeant dyes that target the mitochondria specifically, such as the Rhod‐2 dye of the rhodamine‐based family of indicators. These dyes were first introduced in the late 1980s and include Rhod‐2, X‐rhod‐1, and many variants, all of which exhibit increased fluorescence upon binding Ca2+ 180. The AM ester varieties of these indicators are cationic, which causes ion‐potential‐centric uptake of Ca2+ into the mitochondria. Due to these properties, the rhodamine‐based AM esters have been employed in the literature as mitochondrial‐Ca2+ selective indicators 181, 182. The appeal of Rhod‐2, for example, is easy to see, as just like Fura‐2 and other common cytosolic Ca2+ dyes, only cellular incubation prior to experiments is required. However, Rhod‐2 is known to diffuse out of the mitochondria and into the cytosol after a relatively short period of time, making longer experiments unreliable insofar as accurate mitochondrial Ca2+ measurement goes. These are but a few examples, and, importantly, each of the outlined tools come with their own advantages and drawbacks. Nevertheless, together, they exemplify the broad range of sensors currently available for cytoplasmic Ca2+ measurement.
Among those sensors that are continuously evolving are the new generation of GE fluorescent sensors; compounds comprised of a FP fused to some form of sensing polypeptide. In most cases, the FP is bound to a protein that undergoes a conformational change in response to substrate binding. These sensors are generally considered advantageous over their chemical counterparts due to their substrate specificity, and the fact that their GE nature prevents variance in probe uptake across different cells 183. Touted as being perhaps the most useful advantage of such sensors is the ability to target them to highly specific cellular regions 184, as shown at the turn of the century by Arnaudeau et al. (2001) with the use of a cameleon indicator targeted to each of the cytosol, mitochondrial matrix, and ER lumen.
Development of tools for accurate measurement of levels specifically has provided new insights into fluxes and their regulation.
For example, 4mtD3cpV is a FRET‐based ratiometric Ca2+ sensor that is currently widely used. It is excited with blue light (430 nanometers—nm) and exhibits dual emission at 480 and 535 nm. The ratio of the 480 nm emission to the FRET signal (535 nm) provides investigators with insight into and enhanced understanding of basal levels as well as its fluctuations 185.
Another FRET‐based ratiometric Ca2+ sensor, which can be combined with cytosolic Fura‐2 dye, is 4mtD1GO‐CAM 186. It is a red‐shifted sensor, which allows the researcher to measure Ca2+ in the mitochondria while simultaneously employing Fura‐2 to measure cytosolic Ca2+. As Fura‐2 has a wide excitation–emission spectrum, combined measurement with 4mtD1GO‐CAM proves extremely useful for investigation into the spatiotemporal fluxes of Ca2+. In addition, as both the Fura‐2 dye and mtD1GO‐CAM sensor are ratiometric, accurate observation of basal Ca2+ levels is also possible 186.
Ca2+ sensors of the GECO series provide a good opportunity to measure Ca2+ levels simultaneously in different compartments 187, 188. mtGEM‐GECO1 is a ratiometric sensor with a very low K D for Ca2+ (340 nm) and has an excitation wavelength of 394 nm and dual emission of 455 and 511 nm 187. Its relative mtCAR‐GECO1 is an intensiometric sensor with excitation and emission spectra of 565 and 620 nm, respectively 188. Excitation and emission spectra of CAR‐GECO1 and GEM‐GECO1 sensors allow measurement of Ca2+ levels in two different organelles or mitochondrial compartments simultaneously with almost no spectral overlap.
From this simple examination of a handful of the more prominent cytosolic and mitochondria‐specific sensors in use today, it is clear that investigators have no lack of options when it comes to studying the intricate movements of Ca2+ in this organelle and throughout the cell.
Mitochondria as signaling hubs: Give & Get
Interplay between mitochondria and nucleus
Although mitochondria are equipped with their own circular deoxyribonucleic acid containing 37 genes (i.e., 13 genes encoding for proteins, such as subunits of the respiration complexes and the ATP synthase, 24 genes encoding for tRNAs), almost all mitochondrial proteins are encoded by the nuclear genome, making constant communication between mitochondria and the nucleus indispensable 10. Consequently, mitochondrial biogenesis strongly depends on the contribution of the nucleus, and properly controlled signaling cascades are required to fine‐tune mitochondrial protein synthesis, counteract mitochondrial dysfunction, and initiate compensatory mechanisms 189. Constant mitochondrial status control by the nucleus helps to prevent mitochondrial malfunction, to counteract damage, and to restore mitochondrial homeostasis via situation‐induced activation of transcriptional response 190. The most prominent example of mitochondrial–nucleus crosstalk is retrograde signaling pathways. In these pathways, the mitochondrial unfolded protein response (mtUPR) initiates a protective transcriptional program upon proteotoxic perturbations, aiming to re‐achieve homeostasis in mitochondrial protein biosynthesis and recover the defective organelle 191, 192.
Techniques to measure mitochondrial–nucleus interplay
Dually targeted proteins, localizing to the nucleus as well as to mitochondria, are used as communication indicators for mtUPR (retrograde) signaling. As mitochondrial protein import strongly affects cell viability, the transport of these proteins, visualized by tagging them with a FP, can be used as an indicator of mitochondrial fitness 193, 194, 195. For instance, the mammalian activating transcription factor 5 is imported into mitochondria under physiological conditions, but trapped and consequently translocated to the nucleus to activate transcriptional adaption in case of mitochondrial dysfunction 196. In addition, transcriptional cofactor G protein pathway suppressor 2, another modulator of mtUPR, has also been presented as such an indicator protein, translocating between mitochondria and the nucleus depending on the functionality of mitochondria 197.
Interestingly, cytosolic proteins prone to aggregation are imported into mitochondria for degradation. This translocation of misfolded or aggregated proteins from the cytosol to mitochondria, associated with increased mitochondrial stress levels, can be visualized using split FP techniques 198.
Interplay between mitochondria and ER
Contact between mitochondria and the ER occurs at very specialized junctions stabilized by MAMs, forming locally restricted signaling hubs to restrict and protect the transfer of lipids and Ca2+ between mitochondria and the ER 199. First discovered by electron microscopy in the 1950s 200 but their basic function only described in the 1990s 201, 202, investigation of these structures has been further pushed by the development of cutting‐edge technologies like high‐resolution microscopy over the last 20 years 199. Various proteins stabilizing and modulating mitochondrial–ER interplay, including Ras‐related protein RAB32 203, MFN2 204, or phosphofurin acidic cluster sorting protein 2 205, as well as protein tethering complexes like inositol triphosphate receptor (IP3R), inositol‐requiring enzyme 1 α 206, glucose‐related protein 75 and VDAC, have been identified and characterized 207. Disrupted communication between the ER and mitochondria has been associated with pathological conditions and human diseases 208, such as Alzheimer's disease 209, 210, Charcot‐Marie Tooth 211, Parkinson's disease 212, 213, viral infections 214, cancer 215, diabetes mellitus, and age‐related dysfunction 216.
Techniques to measure ER–mitochondrial interplay
Different approaches have been developed to explore the physical and functional sides of ER‐mitochondria tethering. The current state‐of‐the‐art technique to visualize MAMs is electron microscopy, making quantification of the distance between the membranes of the ER and mitochondria, as well as the number of contact sites, possible 217. Coupling electron microscopy with tomography has provided 3D models and information about the structure and plasticity of the contact points between the two organelles 20. While these techniques come with unbeatable resolution, they also bring the disadvantage of fixation, possibly affecting mitochondrial structure. Therefore, large efforts have recently been placed to develop methods that allow the visualization of MAMs in living cells and still provide high spatial resolution. As discussed in our ‘Techniques to analyze structure and shape’ section, advancements in high‐resolution fluorescence microscopy and in the design of organelle‐targeted FP‐tagged proteins or sensors allow to visualize and investigate structural and functional mitochondrial and ER interplay in living cells 216. Moreover, split FP (split‐FP) approaches or so‐called bimolecular fluorescence complementation technology have been used to study mitochondrial–ER interaction 218. Cieri et al. (2017) have designed split‐GFP‐based contact site sensors (SPLICS), fusing the GFP1–10 moiety to an OMM targeting signal and the GFP11 β‐strand to an ER leading peptide and varying the linker length between the targeting signals and the split‐FP to visualize narrow (approximately 8–10 nm) and wide (approximately 40–50 nm) distances between the ER and mitochondria. As soon as the split‐FPs are in close vicinity due to the interactions between proteins fused to each fragment, they from a full fluorescent FP 219. Using split‐FP technology based on GFP, dynamic changes in the structure of MAMs could be visualized and enhanced formation of ER–mitochondrial contact by mitochondrial uncouplers shown 220. Moreover, Harmon et al. have developed a split‐FP approach to study mitochondrial–ER interaction based on a YFP Venus by fusing the n‐terminal fragment of Venus128 to a mouse ER–protein and targeting the c‐terminal fragment of Venus to the OMM via the n‐terminal leading peptide of TOMM20. As a result, alterations in the MAM structure in response to ER stress, starvation, and protein level changes could be detected 221.
Recently, Ding et al. 222 have developed a novel FP approach using a pair of quenched and nonfluorescent FP‐derived monomers that become a fluorescent heterodimer upon FP association. Alford et al. 223 exploited this FP technology and generated probes by fusing one monomer to the C‐terminus of TOMM20 and targeting the other monomer of the dimerization‐dependent FP pair to the ER–membrane via calnexin. Hajnoczky et al. exploited the FKBPFRB heterodimerization system by fusing FKBP to an OMM targeting signal and combining FRB with an ER retention signal to enable rapamycin inducible tethering between the two organelles. By using this approach, tethering of the ER and mitochondria results in an increase of already in close apposition located contact sites, rather than creating new organelle contacts 224.
Furthermore, recently developed light‐inducible tethering systems allow the induction of ER–mitochondrial interaction, which facilitates the functional study of ER–mitochondrial contacts 225.
Besides various techniques based on cutting‐edge microscopy, an IP3R–VDAC proximity ligation assay has been developed for the quantification of ER–mitochondria interplay. Proximity ligations assays, as an in situ tool, enable the detection of endogenous proteins, protein modifications, and protein interactions with high specificity and sensitivity by using antibodies to detect two unique protein targets 226. Furthermore, Percoll density gradient was used to purify MAMs in order to analyze their composition 227. Functional characterization of MAM regions taking lipid exchange into account has been performed using phosphatidylserine (PS) to phosphatidylethanolamine (PE) conversions or as mitochondrial PS content with regard to lipid transfer 228. Since mitochondria are not governed by classical vesicular trafficking mechanisms, required membrane phospholipids for the mitochondrial membrane biogenesis have to be imported into the organelle. Therefore, mitochondria rely on the efficient supply of lipids from the ER, meaning that biosynthesis of some phospholipids depends on the mitochondria–ER crosstalk. PS, coming from the ER and being directly transferred to mitochondria, is converted in the organelle to PE. Therefore, this reaction can be used for the functional characterization of MAM regions by labeling these phospholipids with radioisotopes 228. Moreover, Ca2+ imaging has offered a very nice approach to determine Ca2+ exchange between mitochondria and the ER 229.
Conclusion
As presented in the current review, mitochondria are versatile organelles within eukaryotic cells, which deliver utilizable energy and function as signaling hubs, communicating with various cellular compartments to maintain their own function but also to provide their service. Their structure and shape as well as their activity undergo dynamic changes in order to fulfill their tasks under various conditions in different cellular subdomains. As discussed in this review, investigation into mitochondrial function advanced alongside cutting‐edge technologies, enabling us to gain new insight into subcellular signaling processes. Further development of these methods, as well as completely new technological strategies, will most likely further broaden our understanding in the upcoming years. This might potentially yield the path to unveiling subcellular processes causing still‐incurable diseases and, thereby, help to develop novel treatment strategies. Through that example, it is obvious that constructive teamwork between people with different expertise is increasingly important and is urgently needed to meet the upcoming health‐related challenges in an aging society.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
CTM, JR, GZ, SB, HB, ZK, and BG contributed chapters to the manuscript. CTM together with WFG and RM planned the manuscript's structure and content.
Acknowledgements
The research of the authors is funded by the Austrian Science Fund (FWF): CTM is currently funded by an Erwin Schroedinger Abroad Fellowship (J4205‐B27). Authors are, furthermore, funded by the FWF‐projects P28529‐B27 and I3716‐B27 to RM and the FWF‐funded Doctoral Program Metabolic and Cardiovascular Disease (DK‐W1226 to WFG). Besides, authors are funded by the Ph.D. program Molecular Medicine (MOLMED) of the Medical University of Graz and by Nikon Austria within the Nikon Center of Excellence, Graz. The Nikon Center of Excellence, Graz, is supported by the Austrian infrastructure program 2013/2014, Nikon Austria Inc., and BioTechMed, Graz.
References
- 1. Zachar I, Szilagyi A, Szamado S & Szathmary E (2018) Farming the mitochondrial ancestor as a model of endosymbiotic establishment by natural selection. Proc Natl Acad Sci USA 115, E1504–E1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sukhorukov VM & Meyer‐Hermann M (2015) Structural heterogeneity of mitochondria induced by the microtubule cytoskeleton. Sci Rep 5, 13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schmitt K, Grimm A, Dallmann R, Oettinghaus B, Restelli LM, Witzig M, Ishihara N, Mihara K, Ripperger JA, Albrecht U et al (2018) Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab 27, 657–666.e5. [DOI] [PubMed] [Google Scholar]
- 4. Reid RA, Moyle J & Mitchell P (1966) Synthesis of adenosine triphosphate by a protonmotive force in rat liver mitochondria. Nature 212, 257–258. [DOI] [PubMed] [Google Scholar]
- 5. Guo R, Gu J, Zong S, Wu M & Yang M (2018) Structure and mechanism of mitochondrial electron transport chain. Biomed J 41, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishi T & Forgac M (2002) The vacuolar (H+)‐ATPases–nature's most versatile proton pumps. Nat Rev Mol Cell Biol 3, 94–103. [DOI] [PubMed] [Google Scholar]
- 7. Kicinska A & Szewczyk A (2004) Large‐conductance potassium cation channel opener NS1619 inhibits cardiac mitochondria respiratory chain. Toxicol Mech Methods 14, 59–61. [DOI] [PubMed] [Google Scholar]
- 8. Giorgi C, Romagnoli A, Pinton P & Rizzuto R (2008) Ca2+ signaling, mitochondria and cell death. Curr Mol Med 8, 119–130. [DOI] [PubMed] [Google Scholar]
- 9. Murley A & Nunnari J (2016) The emerging network of mitochondria‐organelle contacts. Mol Cell 61, 648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kotiadis VN, Duchen MR & Osellame LD (2014) Mitochondrial quality control and communications with the nucleus are important in maintaining mitochondrial function and cell health. Biochim Biophys Acta 1840, 1254–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rieusset J (2018) The role of endoplasmic reticulum‐mitochondria contact sites in the control of glucose homeostasis: an update. Cell Death Dis 9, 388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gazaryan IG & Brown AM (2007) Intersection between mitochondrial permeability pores and mitochondrial fusion/fission. Neurochem Res 32, 917–929. [DOI] [PubMed] [Google Scholar]
- 13. Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ & Capaldi RA (2004) Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res 64, 985–993. [DOI] [PubMed] [Google Scholar]
- 14. Cogliati S, Frezza C, Soriano ME, Varanita T, Quintana‐Cabrera R, Corrado M, Cipolat S, Costa V, Casarin A, Gomes LC et al (2013) Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell 155, 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. NavaneethaKrishnan S, Rosales JL & Lee KY (2018) Loss of Cdk5 in breast cancer cells promotes ROS‐mediated cell death through dysregulation of the mitochondrial permeability transition pore. Oncogene 37, 1788–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G et al (2008) Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 27, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B et al(2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189. [DOI] [PubMed] [Google Scholar]
- 18. Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ & Rizzuto R (2004) Drp‐1‐dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+‐mediated apoptosis. Mol Cell 16, 59–68. [DOI] [PubMed] [Google Scholar]
- 19. Szabadkai G, Simoni AM, Bianchi K, De Stefani D, Leo S, Wieckowski MR & Rizzuto R (2006) Mitochondrial dynamics and Ca2+ signaling. Biochim Biophys Acta 1763, 442–449. [DOI] [PubMed] [Google Scholar]
- 20. Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J & Voeltz GK (2011) ER tubules mark sites of mitochondrial division. Science 334, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arasaki K, Shimizu H, Mogari H, Nishida N, Hirota N, Furuno A, Kudo Y, Baba M, Baba N, Cheng J et al (2015) A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev Cell 32, 304–317. [DOI] [PubMed] [Google Scholar]
- 22. Loson OC, Song Z, Chen H & Chan DC (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell 24, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang CR & Blackstone C (2007) Cyclic AMP‐dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem 282, 21583–21587. [DOI] [PubMed] [Google Scholar]
- 24. Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X & Ramachandran R (2015) Cardiolipin's propensity for phase transition and its reorganization by dynamin‐related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell 26, 3104–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Irajizad E, Ramachandran R & Agrawal A (2019) Geometric instability catalyzes mitochondrial fission. Mol Biol Cell 30, 160–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santel A & Fuller MT (2001) Control of mitochondrial morphology by a human mitofusin. J Cell Sci 114(Pt 5), 867–874. [DOI] [PubMed] [Google Scholar]
- 27. Eura Y, Ishihara N, Yokota S & Mihara K (2003) Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J Biochem 134, 333–344. [DOI] [PubMed] [Google Scholar]
- 28. Ishihara N, Eura Y & Mihara K (2004) Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J Cell Sci 117(Pt 26), 6535–6546. [DOI] [PubMed] [Google Scholar]
- 29. Pyakurel A, Savoia C, Hess D & Scorrano L (2015) Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol Cell 58, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song Z, Chen H, Fiket M, Alexander C & Chan DC (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178, 749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI & Langer T (2009) Regulation of OPA1 processing and mitochondrial fusion by m‐AAA protease isoenzymes and OMA1. J Cell Biol 187, 1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E & Langer T (2014) The i‐AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol 204, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Picard M, McManus MJ, Csordas G, Varnai P, Dorn GW 2nd, Williams D, Hajnoczky G & Wallace DC (2015) Trans‐mitochondrial coordination of cristae at regulated membrane junctions. Nat Commun 6, 6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang L, Trushin S, Christensen TA, Bachmeier BV, Gateno B, Schroeder A, Yao J, Itoh K, Sesaki H, Poon WW et al (2016) Altered brain energetics induces mitochondrial fission arrest in Alzheimer's disease. Sci Rep 6, 18725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang C, Du W, Su QP, Zhu M, Feng P, Li Y, Zhou Y, Mi N, Zhu Y, Jiang D et al (2015) Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res 25, 1108–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pacher P & Hajnoczky G (2001) Propagation of the apoptotic signal by mitochondrial waves. EMBO J 20, 4107–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kurz FT, Aon MA, O'Rourke B & Armoundas AA (2010) Spatio‐temporal oscillations of individual mitochondria in cardiac myocytes reveal modulation of synchronized mitochondrial clusters. Proc Natl Acad Sci USA 107, 14315–14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lavorato M, Iyer VR, Dewight W, Cupo RR, Debattisti V, Gomez L, De la Fuente S, Zhao YT, Valdivia HH, Hajnoczky G et al (2017) Increased mitochondrial nanotunneling activity, induced by calcium imbalance, affects intermitochondrial matrix exchanges. Proc Natl Acad Sci USA 114, E849–E858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang X, Sun L, Ji S, Zhao T, Zhang W, Xu J, Zhang J, Wang Y, Wang X, Franzini‐Armstrong C et al (2013) Kissing and nanotunneling mediate intermitochondrial communication in the heart. Proc Natl Acad Sci USA 110, 2846–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F & Neupert W (2011) The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J 30, 4356–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perkins G, Renken C, Martone ME, Young SJ, Ellisman M & Frey T (1997) Electron tomography of neuronal mitochondria: three‐dimensional structure and organization of cristae and membrane contacts. J Struct Biol 119, 260–272. [DOI] [PubMed] [Google Scholar]
- 42. Davies KM, Anselmi C, Wittig I, Faraldo‐Gomez JD & Kuhlbrandt W (2012) Structure of the yeast F1Fo‐ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA 109, 13602–13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jans DC, Wurm CA, Riedel D, Wenzel D, Stagge F, Deckers M, Rehling P & Jakobs S (2013) STED super‐resolution microscopy reveals an array of MINOS clusters along human mitochondria. Proc Natl Acad Sci USA 110, 8936–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Glytsou C, Calvo E, Cogliati S, Mehrotra A, Anastasia I, Rigoni G, Raimondi A, Shintani N, Loureiro M, Vazquez J et al (2016) Optic atrophy 1 is epistatic to the core MICOS component MIC60 in mitochondrial cristae shape control. Cell Rep 17, 3024–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang Z, Wakabayashi N, Wakabayashi J, Tamura Y, Song WJ, Sereda S, Clerc P, Polster BM, Aja SM, Pletnikov MV et al (2011) The dynamin‐related GTPase Opa1 is required for glucose‐stimulated ATP production in pancreatic beta cells. Mol Biol Cell 22, 2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tezze C, Romanello V, Desbats MA, Fadini GP, Albiero M, Favaro G, Ciciliot S, Soriano ME, Morbidoni V, Cerqua C et al (2017) Age‐associated loss of OPA1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metab 25, 1374–1389.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fulop L, Szanda G, Enyedi B, Varnai P & Spat A (2011) The effect of OPA1 on mitochondrial Ca(2)(+) signaling. PLoS ONE 6, e25199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P & Lenaers G (2003) Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem 278, 7743–7746. [DOI] [PubMed] [Google Scholar]
- 49. Gottschalk B, Klec C, Leitinger G, Bernhart E, Rost R, Bischof H, Madreiter‐Sokolowski CT, Radulović S, Eroglu E, Sattler W et al (2019) MICU1 controls cristae junction and spatially anchors mitochondrial Ca2+ uniporter complex. Nat Commun 10, 3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kozjak‐Pavlovic V (2017) The MICOS complex of human mitochondria. Cell Tissue Res 367, 83–93. [DOI] [PubMed] [Google Scholar]
- 51. Barrera M, Koob S, Dikov D, Vogel F & Reichert AS (2016) OPA1 functionally interacts with MIC60 but is dispensable for crista junction formation. FEBS Lett 590, 3309–3322. [DOI] [PubMed] [Google Scholar]
- 52. Bohnert M, Wenz LS, Zerbes RM, Horvath SE, Stroud DA, von der Malsburg K, Muller JM, Oeljeklaus S, Perschil I, Warscheid B et al (2012) Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol Biol Cell 23, 3948–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska‐Lwowska A, Wiese S, Rao S, Milenkovic D et al (2011) Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell 21, 694–707. [DOI] [PubMed] [Google Scholar]
- 54. Ioakeimidis F, Ott C, Kozjak‐Pavlovic V, Violitzi F, Rinotas V, Makrinou E, Eliopoulos E, Fasseas C, Kollias G & Douni E (2014) A splicing mutation in the novel mitochondrial protein DNAJC11 causes motor neuron pathology associated with cristae disorganization, and lymphoid abnormalities in mice. PLoS ONE 9, e104237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vogel F, Bornhovd C, Neupert W & Reichert AS (2006) Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol 175, 237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Harner ME, Unger AK, Geerts WJ, Mari M, Izawa T, Stenger M, Geimer S, Reggiori F, Westermann B & Neupert W (2016) An evidence based hypothesis on the existence of two pathways of mitochondrial crista formation. Elife 5, e18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mannella CA & Bonner WD Jr (1975) X‐ray diffraction from oriented outer mitochondrial membranes. Detection of in‐plane subunit structure. Biochim Biophys Acta 413, 226–233. [DOI] [PubMed] [Google Scholar]
- 58. Colombini M (1979) A candidate for the permeability pathway of the outer mitochondrial membrane. Nature 279, 643–645. [DOI] [PubMed] [Google Scholar]
- 59. Rieger B, Junge W & Busch KB (2014) Lateral pH gradient between OXPHOS complex IV and F(0)F(1) ATP‐synthase in folded mitochondrial membranes. Nat Commun 5, 3103. [DOI] [PubMed] [Google Scholar]
- 60. Song DH, Park J, Philbert MA, Sastry AM & Lu W (2014) Effects of local pH on the formation and regulation of cristae morphologies. Phys Rev E Stat Nonlin Soft Matter Phys 90, 022702. [DOI] [PubMed] [Google Scholar]
- 61. Khalifat N, Puff N, Bonneau S, Fournier JB & Angelova MI (2008) Membrane deformation under local pH gradient: mimicking mitochondrial cristae dynamics. Biophys J 95, 4924–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Muhleip AW, Joos F, Wigge C, Frangakis AS, Kuhlbrandt W & Davies KM (2016) Helical arrays of U‐shaped ATP synthase dimers form tubular cristae in ciliate mitochondria. Proc Natl Acad Sci USA 113, 8442–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wittig I, Carrozzo R, Santorelli FM & Schagger H (2006) Supercomplexes and subcomplexes of mitochondrial oxidative phosphorylation. Biochim Biophys Acta 1757, 1066–1072. [DOI] [PubMed] [Google Scholar]
- 64. van de Linde S, Sauer M & Heilemann M (2008) Subdiffraction‐resolution fluorescence imaging of proteins in the mitochondrial inner membrane with photoswitchable fluorophores. J Struct Biol 164, 250–254. [DOI] [PubMed] [Google Scholar]
- 65. Klotzsch E, Smorodchenko A, Lofler L, Moldzio R, Parkinson E, Schutz GJ & Pohl EE (2015) Superresolution microscopy reveals spatial separation of UCP4 and F0F1‐ATP synthase in neuronal mitochondria. Proc Natl Acad Sci USA 112, 130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ishigaki M, Iketani M, Sugaya M, Takahashi M, Tanaka M, Hattori S & Ohsawa I (2016) STED super‐resolution imaging of mitochondria labeled with TMRM in living cells. Mitochondrion 28, 79–87. [DOI] [PubMed] [Google Scholar]
- 67. Gottschalk B, Klec C, Waldeck‐Weiermair M, Malli R & Graier WF (2018) Intracellular Ca(2+) release decelerates mitochondrial cristae dynamics within the junctions to the endoplasmic reticulum. Pflugers Arch 470, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dikov D & Bereiter‐Hahn J (2013) Inner membrane dynamics in mitochondria. J Struct Biol 183, 455–466. [DOI] [PubMed] [Google Scholar]
- 69. Dong Z, Shanmughapriya S, Tomar D, Siddiqui N, Lynch S, Nemani N, Breves SL, Zhang X, Tripathi A, Palaniappan P et al (2017) Mitochondrial Ca(2+) uniporter is a mitochondrial luminal redox sensor that augments MCU channel activity. Mol Cell 65, 1014–1028.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stoldt S, Stephan T, Jans DC, Bruser C, Lange F, Keller‐Findeisen J, Riedel D, Hell SW & Jakobs S (2019) Mic60 exhibits a coordinated clustered distribution along and across yeast and mammalian mitochondria. Proc Natl Acad Sci USA 116, 9853–9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shao L, Kner P, Rego EH & Gustafsson MG (2011) Super‐resolution 3D microscopy of live whole cells using structured illumination. Nat Methods 8, 1044–1046. [DOI] [PubMed] [Google Scholar]
- 72. Huttemann M, Lee I, Samavati L, Yu H & Doan JW (2007) Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim Biophys Acta 1773, 1701–1720. [DOI] [PubMed] [Google Scholar]
- 73. Mitchell P (1961) Coupling of phosphorylation to electron and hydrogen transfer by a chemi‐osmotic type of mechanism. Nature 191, 144–148. [DOI] [PubMed] [Google Scholar]
- 74. Horan MP, Pichaud N & Ballard JW (2012) Review: quantifying mitochondrial dysfunction in complex diseases of aging. J Gerontol A Biol Sci Med Sci 67, 1022–1035. [DOI] [PubMed] [Google Scholar]
- 75. Klec C, Madreiter‐Sokolowski CT, Stryeck S, Sachdev V, Duta‐Mare M, Gottschalk B, Depaoli MR, Rost R, Hay J, Waldeck‐Weiermair M et al (2019) Glycogen synthase kinase 3 beta controls presenilin‐1‐mediated endoplasmic reticulum Ca(2)(+) leak directed to mitochondria in pancreatic islets and beta‐cells. Cell Physiol Biochem 52, 57–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato‐Yamada Y, Nagai T & Noji H (2009) Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer‐based genetically encoded indicators. Proc Natl Acad Sci USA 106, 15651–15656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vishnu N, Jadoon Khan M, Karsten F, Groschner LN, Waldeck‐Weiermair M, Rost R, Hallstrom S, Imamura H, Graier WF & Malli R (2014) ATP increases within the lumen of the endoplasmic reticulum upon intracellular Ca2+ release. Mol Biol Cell 25, 368–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Casey JR, Grinstein S & Orlowski J (2010) Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol 11, 50–61. [DOI] [PubMed] [Google Scholar]
- 79. Kellum JA (2000) Determinants of blood pH in health and disease. Crit Care 4, 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wileman T (2013) Autophagy as a defence against intracellular pathogens. Essays Biochem 55, 153–163. [DOI] [PubMed] [Google Scholar]
- 81. Maeda Y & Kinoshita T (2010) The acidic environment of the Golgi is critical for glycosylation and transport. Methods Enzymol 480, 495–510. [DOI] [PubMed] [Google Scholar]
- 82. Santo‐Domingo J & Demaurex N (2012) Perspectives on: SGP symposium on mitochondrial physiology and medicine: the renaissance of mitochondrial pH. J Gen Physiol 139, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mitchell P & Moyle J (1969) Estimation of membrane potential and pH difference across the cristae membrane of rat liver mitochondria. Eur J Biochem 7, 471–484. [DOI] [PubMed] [Google Scholar]
- 84. Zorova LD, Popkov VA, Plotnikov EY, Silachev DN, Pevzner IB, Jankauskas SS, Babenko VA, Zorov SD, Balakireva AV, Juhaszova M et al (2018) Mitochondrial membrane potential. Anal Biochem 552, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nicholls DG (1974) The influence of respiration and ATP hydrolysis on the proton‐electrochemical gradient across the inner membrane of rat‐liver mitochondria as determined by ion distribution. Eur J Biochem 50, 305–315. [DOI] [PubMed] [Google Scholar]
- 86. Szewczyk A, Jarmuszkiewicz W & Kunz WS (2009) Mitochondrial potassium channels. IUBMB Life 61, 134–143. [DOI] [PubMed] [Google Scholar]
- 87. Garcia‐Marcos M, Fontanils U, Aguirre A, Pochet S, Dehaye JP & Marino A (2005) Role of sodium in mitochondrial membrane depolarization induced by P2X7 receptor activation in submandibular glands. FEBS Lett 579, 5407–5413. [DOI] [PubMed] [Google Scholar]
- 88. Alvarez BV & Villa‐Abrille MC (2013) Mitochondrial NHE1: a newly identified target to prevent heart disease. Front Physiol 4, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tomaskova Z & Ondrias K (2010) Mitochondrial chloride channels–what are they for? FEBS Lett 584, 2085–2092. [DOI] [PubMed] [Google Scholar]
- 90. Drummond‐Main CD & Rho JM (2018) Electrophysiological characterization of a mitochondrial inner membrane chloride channel in rat brain. FEBS Lett 592, 1545–1553. [DOI] [PubMed] [Google Scholar]
- 91. Chalmers S & McCarron JG (2008) The mitochondrial membrane potential and Ca2+ oscillations in smooth muscle. J Cell Sci 121(Pt 1), 75–85. [DOI] [PubMed] [Google Scholar]
- 92. Poburko D, Santo‐Domingo J & Demaurex N (2011) Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem 286, 11672–11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Eskes R, Antonsson B, Osen‐Sand A, Montessuit S, Richter C, Sadoul R, Mazzei G, Nichols A & Martinou JC (1998) Bax‐induced cytochrome C release from mitochondria is independent of the permeability transition pore but highly dependent on Mg2+ ions. J Cell Biol 143, 217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang BB, Wang DG, Guo FF & Xuan C (2015) Mitochondrial membrane potential and reactive oxygen species in cancer stem cells. Fam Cancer 14, 19–23. [DOI] [PubMed] [Google Scholar]
- 95. Bonnet S, Archer SL, Allalunis‐Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S et al (2007) A mitochondria‐K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell 11, 37–51. [DOI] [PubMed] [Google Scholar]
- 96. Sai Y, Zou Z, Peng K & Dong Z (2012) The Parkinson's disease‐related genes act in mitochondrial homeostasis. Neurosci Biobehav Rev 36, 2034–2043. [DOI] [PubMed] [Google Scholar]
- 97. Kim H, Perentis RJ, Caldwell GA & Caldwell KA (2018) Gene‐by‐environment interactions that disrupt mitochondrial homeostasis cause neurodegeneration in C. elegans Parkinson's models. Cell Death Dis 9, 555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Porporato PE, Filigheddu N, Pedro JMB, Kroemer G & Galluzzi L (2018) Mitochondrial metabolism and cancer. Cell Res 28, 265–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Abad MF, Di Benedetto G, Magalhaes PJ, Filippin L & Pozzan T (2004) Mitochondrial pH monitored by a new engineered green fluorescent protein mutant. J Biol Chem 279, 11521–11529. [DOI] [PubMed] [Google Scholar]
- 100. Mishina NM, Markvicheva KN, Bilan DS, Matlashov ME, Shirmanova MV, Liebl D, Schultz C, Lukyanov S & Belousov VV (2013) Visualization of intracellular hydrogen peroxide with HyPer, a genetically encoded fluorescent probe. Methods Enzymol 526, 45–59. [DOI] [PubMed] [Google Scholar]
- 101. Ozkan P & Mutharasan R (2002) A rapid method for measuring intracellular pH using BCECF‐AM. Biochim Biophys Acta 1572, 143–148. [DOI] [PubMed] [Google Scholar]
- 102. Ramshesh VK & Lemasters JJ (2012) Imaging of mitochondrial pH using SNARF‐1. Methods Mol Biol 810, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chen Y, Zhu C, Cen J, Bai Y, He W & Guo Z (2015) Ratiometric detection of pH fluctuation in mitochondria with a new fluorescein/cyanine hybrid sensor. Chem Sci 6, 3187–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Lee MH, Park N, Yi C, Han JH, Hong JH, Kim KP, Kang DH, Sessler JL, Kang C & Kim JS (2014) Mitochondria‐immobilized pH‐sensitive off‐on fluorescent probe. J Am Chem Soc 136, 14136–14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Scaduto RC Jr & Grotyohann LW (1999) Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J 76, 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Perelman A, Wachtel C, Cohen M, Haupt S, Shapiro H & Tzur A (2012) JC‐1: alternative excitation wavelengths facilitate mitochondrial membrane potential cytometry. Cell Death Dis 3, e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Floryk D & Houstek J (1999) Tetramethyl rhodamine methyl ester (TMRM) is suitable for cytofluorometric measurements of mitochondrial membrane potential in cells treated with digitonin. Biosci Rep 19, 27–34. [DOI] [PubMed] [Google Scholar]
- 108. Perry SW, Norman JP, Barbieri J, Brown EB & Gelbard HA (2011) Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques 50, 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Roy SS & Hajnoczky G (2009) Fluorometric methods for detection of mitochondrial membrane permeabilization in apoptosis. Methods Mol Biol 559, 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zoeteweij JP, van de Water B, de Bont HJ & Nagelkerke JF (1994) Mitochondrial K+ as modulator of Ca(2+)‐dependent cytotoxicity in hepatocytes. Novel application of the K(+)‐sensitive dye PBFI (K(+)‐binding benzofuran isophthalate) to assess free mitochondrial K+ concentrations. Biochem J 299(Pt 2), 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Bischof H, Rehberg M, Stryeck S, Artinger K, Eroglu E, Waldeck‐Weiermair M, Gottschalk B, Rost R, Deak AT, Niedrist T et al (2017) Novel genetically encoded fluorescent probes enable real‐time detection of potassium in vitro and in vivo . Nat Commun 8, 1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kaasik A, Safiulina D, Zharkovsky A & Veksler V (2007) Regulation of mitochondrial matrix volume. Am J Physiol Cell Physiol 292, C157–C163. [DOI] [PubMed] [Google Scholar]
- 113. Schulz R & Di Lisa F (2016) Mitochondrial potassium homeostasis: a central player in cardioprotection. Cardiovasc Res 110, 4–5. [DOI] [PubMed] [Google Scholar]
- 114. Grover GJ & Garlid KD (2000) ATP‐Sensitive potassium channels: a review of their cardioprotective pharmacology. J Mol Cell Cardiol 32, 677–695. [DOI] [PubMed] [Google Scholar]
- 115. Malinska D, Mirandola SR & Kunz WS (2010) Mitochondrial potassium channels and reactive oxygen species. FEBS Lett 584, 2043–2048. [DOI] [PubMed] [Google Scholar]
- 116. Checchetto V, Teardo E, Carraretto L, Leanza L & Szabo I (2016) Physiology of intracellular potassium channels: a unifying role as mediators of counterion fluxes? Biochim Biophys Acta 1857, 1258–1266. [DOI] [PubMed] [Google Scholar]
- 117. Szabo I & Zoratti M (2014) Mitochondrial channels: ion fluxes and more. Physiol Rev 94, 519–608. [DOI] [PubMed] [Google Scholar]
- 118. Laskowski M, Augustynek B, Kulawiak B, Koprowski P, Bednarczyk P, Jarmuszkiewicz W & Szewczyk A (2016) What do we not know about mitochondrial potassium channels? Biochim Biophys Acta 1857, 1247–1257. [DOI] [PubMed] [Google Scholar]
- 119. Austin S, Tavakoli M, Pfeiffer C, Seifert J, Mattarei A, De Stefani D, Zoratti M & Nowikovsky K (2017) LETM1‐Mediated K(+) and Na(+) homeostasis regulates mitochondrial Ca(2+) efflux. Front Physiol 8, 839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Garlid KD & Paucek P (2003) Mitochondrial potassium transport: the K(+) cycle. Biochim Biophys Acta 1606, 23–41. [DOI] [PubMed] [Google Scholar]
- 121. Abdallah DM, Nassar NN & Abd‐El‐Salam RM (2011) Glibenclamide ameliorates ischemia‐reperfusion injury via modulating oxidative stress and inflammatory mediators in the rat hippocampus. Brain Res 1385, 257–262. [DOI] [PubMed] [Google Scholar]
- 122. Pompermayer K, Souza DG, Lara GG, Silveira KD, Cassali GD, Andrade AA, Bonjardim CA, Passaglio KT, Assreuy J, Cunha FQ et al (2005) The ATP‐sensitive potassium channel blocker glibenclamide prevents renal ischemia/reperfusion injury in rats. Kidney Int 67, 1785–1796. [DOI] [PubMed] [Google Scholar]
- 123. Pompermayer K, Amaral FA, Fagundes CT, Vieira AT, Cunha FQ, Teixeira MM & Souza DG (2007) Effects of the treatment with glibenclamide, an ATP‐sensitive potassium channel blocker, on intestinal ischemia and reperfusion injury. Eur J Pharmacol 556, 215–222. [DOI] [PubMed] [Google Scholar]
- 124. Kaczmarek LK, Aldrich RW, Chandy KG, Grissmer S, Wei AD & Wulff H (2017) International union of basic and clinical pharmacology. C. Nomenclature and properties of calcium‐activated and sodium‐activated potassium channels. Pharmacol Rev 69, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gu XQ, Siemen D, Parvez S, Cheng Y, Xue J, Zhou D, Sun X, Jonas EA & Haddad GG (2007) Hypoxia increases BK channel activity in the inner mitochondrial membrane. Biochem Biophys Res Commun 358, 311–316. [DOI] [PubMed] [Google Scholar]
- 126. Xu W, Liu Y, Wang S, McDonald T, Van Eyk JE, Sidor A & O'Rourke B (2002) Cytoprotective role of Ca2+‐ activated K+ channels in the cardiac inner mitochondrial membrane. Science 298, 1029–1033. [DOI] [PubMed] [Google Scholar]
- 127. Balderas E, Zhang J, Stefani E & Toro L (2015) Mitochondrial BKCa channel. Front Physiol 6, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bednarczyk P, Wieckowski MR, Broszkiewicz M, Skowronek K, Siemen D & Szewczyk A (2013) Putative structural and functional coupling of the mitochondrial BKCa channel to the respiratory chain. PLoS ONE 8, e68125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Singh H, Lu R, Bopassa JC, Meredith AL, Stefani E & Toro L (2013) MitoBK(Ca) is encoded by the Kcnma1 gene, and a splicing sequence defines its mitochondrial location. Proc Natl Acad Sci USA 110, 10836–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Sato T, Saito T, Saegusa N & Nakaya H (2005) Mitochondrial Ca2+‐activated K+ channels in cardiac myocytes: a mechanism of the cardioprotective effect and modulation by protein kinase A. Circulation 111, 198–203. [DOI] [PubMed] [Google Scholar]
- 131. Shi Y, Jiang MT, Su J, Hutchins W, Konorev E & Baker JE (2007) Mitochondrial big conductance KCa channel and cardioprotection in infant rabbit heart. J Cardiovasc Pharmacol 50, 497–502. [DOI] [PubMed] [Google Scholar]
- 132. O'Rourke B (2007) Mitochondrial ion channels. Annu Rev Physiol 69, 19–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Szewczyk A & Marban E (1999) Mitochondria: a new target for K channel openers? Trends Pharmacol Sci 20, 157–161. [DOI] [PubMed] [Google Scholar]
- 134. Kulawiak B, Kudin AP, Szewczyk A & Kunz WS (2008) BK channel openers inhibit ROS production of isolated rat brain mitochondria. Exp Neurol 212, 543–547. [DOI] [PubMed] [Google Scholar]
- 135. Yan XH, Guo XY, Jiao FY, Liu X & Liu Y (2015) Activation of large‐conductance Ca(2+)‐activated K(+) channels inhibits glutamate‐induced oxidative stress through attenuating ER stress and mitochondrial dysfunction. Neurochem Int 90, 28–35. [DOI] [PubMed] [Google Scholar]
- 136. Augustynek B, Koprowski P, Rotko D, Kunz WS, Szewczyk A & Kulawiak B (2018) Mitochondrial BK channel openers CGS7181 and CGS7184 exhibit cytotoxic properties. Int J Mol Sci 19, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Cheng Y, Gulbins E & Siemen D (2011) Activation of the permeability transition pore by Bax via inhibition of the mitochondrial BK channel. Cell Physiol Biochem 27, 191–200. [DOI] [PubMed] [Google Scholar]
- 138. Kong X, Su F, Zhang L, Yaron J, Lee F, Shi Z, Tian Y & Meldrum DR (2015) A highly selective mitochondria‐targeting fluorescent K(+) sensor. Angew Chem Int Ed Engl 54, 12053–12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Jezek P, Mahdi F & Garlid KD (1990) Reconstitution of the beef heart and rat liver mitochondrial K+/H+ (Na+/H+) antiporter. Quantitation of K+ transport with the novel fluorescent probe, PBFI. J Biol Chem 265, 10522–10526. [PubMed] [Google Scholar]
- 140. Rimmele TS & Chatton JY (2014) A novel optical intracellular imaging approach for potassium dynamics in astrocytes. PLoS ONE 9, e109243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Shen Y, Wu SY, Rancic V, Aggarwal A, Qian Y, Miyashita SI, Ballanyi K, Campbell RE & Dong M (2019) Genetically encoded fluorescent indicators for imaging intracellular potassium ion concentration. Commun Biol 2, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Malli R, Frieden M, Osibow K & Graier WF (2003) Mitochondria efficiently buffer subplasmalemmal Ca2+ elevation during agonist stimulation. J Biol Chem 278, 10807–10815. [DOI] [PubMed] [Google Scholar]
- 143. Denton RM, Randle PJ & Martin BR (1972) Stimulation by calcium ions of pyruvate dehydrogenase phosphate phosphatase. Biochem J 128, 161–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Hansford RG & Chappell JB (1967) The effect of Ca2+ on the oxidation of glycerol phosphate by blowfly flight‐muscle mitochondria. Biochem Biophys Res Commun 27, 686–692. [DOI] [PubMed] [Google Scholar]
- 145. Denton RM, McCormack JG & Edgell NJ (1980) Role of calcium ions in the regulation of intramitochondrial metabolism. Effects of Na+, Mg2+ and ruthenium red on the Ca2+‐stimulated oxidation of oxoglutarate and on pyruvate dehydrogenase activity in intact rat heart mitochondria. Biochem J 190, 107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fournier N, Ducet G & Crevat A (1987) Action of cyclosporine on mitochondrial calcium fluxes. J Bioenerg Biomembr 19, 297–303. [DOI] [PubMed] [Google Scholar]
- 147. Griffiths EJ & Rutter GA (2009) Mitochondrial calcium as a key regulator of mitochondrial ATP production in mammalian cells. Biochim Biophys Acta 1787, 1324–1333. [DOI] [PubMed] [Google Scholar]
- 148. Wrogemann K & Pena SD (1976) Mitochondrial calcium overload: a general mechanism for cell‐necrosis in muscle diseases. Lancet 1, 672–674. [DOI] [PubMed] [Google Scholar]
- 149. Kruman II & Mattson MP (1999) Pivotal role of mitochondrial calcium uptake in neural cell apoptosis and necrosis. J Neurochem 72, 529–540. [DOI] [PubMed] [Google Scholar]
- 150. La Rovere RM, Roest G, Bultynck G & Parys JB (2016) Intracellular Ca(2+) signaling and Ca(2+) microdomains in the control of cell survival, apoptosis and autophagy. Cell Calcium 60, 74–87. [DOI] [PubMed] [Google Scholar]
- 151. Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA & Pozzan T (1998) Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280, 1763–1766. [DOI] [PubMed] [Google Scholar]
- 152. Liu Z & Butow RA (2006) Mitochondrial retrograde signaling. Annu Rev Genet 40, 159–185. [DOI] [PubMed] [Google Scholar]
- 153. Ball EH & Singer SJ (1982) Mitochondria are associated with microtubules and not with intermediate filaments in cultured fibroblasts. Proc Natl Acad Sci U S A 79, 123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kroemer G, Galluzzi L & Brenner C (2007) Mitochondrial membrane permeabilization in cell death. Physiol Rev 87, 99–163. [DOI] [PubMed] [Google Scholar]
- 155. Hill MM, Adrain C & Martin SJ (2003) Portrait of a killer: the mitochondrial apoptosome emerges from the shadows. Mol Interv 3, 19–26. [DOI] [PubMed] [Google Scholar]
- 156. Jouaville LS, Pinton P, Bastianutto C, Rutter GA & Rizzuto R (1999) Regulation of mitochondrial ATP synthesis by calcium: evidence for a long‐term metabolic priming. Proc Natl Acad Sci USA 96, 13807–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Vasington FD & Murphy JV (1962) Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem 237, 2670–2677. [PubMed] [Google Scholar]
- 158. Deluca HF & Engstrom GW (1961) Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci USA 47, 1744–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher‐Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL et al (2011) Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476, 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. De Stefani D, Raffaello A, Teardo E, Szabo I & Rizzuto R (2011) A forty‐kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Oxenoid K, Dong Y, Cao C, Cui T, Sancak Y, Markhard AL, Grabarek Z, Kong L, Liu Z, Ouyang B et al (2016) Architecture of the mitochondrial calcium uniporter. Nature 533, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Sancak Y, Markhard AL, Kitami T, Kovacs‐Bogdan E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA et al (2013) EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342, 1379–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Vais H, Mallilankaraman K, Mak DD, Hoff H, Payne R, Tanis JE & Foskett JK (2016) EMRE Is a matrix Ca(2+) sensor that governs gatekeeping of the mitochondrial Ca(2+) uniporter. Cell Rep 14, 403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE & Mootha VK (2010) MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature 467, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, Girgis HS, Kuchimanchi S, De Groot J, Speciner L et al (2013) MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE 8, e55785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Fuente Perez S, Bogorad R et al (2013) MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell Metab 17, 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Paillard M, Csordas G, Szanda G, Golenar T, Debattisti V, Bartok A, Wang N, Moffat C, Seifert EL, Spat A et al (2017) Tissue‐specific mitochondrial decoding of cytoplasmic Ca(2+) signals is controlled by the stoichiometry of MICU1/2 and MCU. Cell Rep 18, 2291–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Waldeck‐Weiermair M, Jean‐Quartier C, Rost R, Khan MJ, Vishnu N, Bondarenko AI, Imamura H, Malli R & Graier WF (2011) Leucine zipper EF hand‐containing transmembrane protein 1 (Letm1) and uncoupling proteins 2 and 3 (UCP2/3) contribute to two distinct mitochondrial Ca2+ uptake pathways. J Biol Chem 286, 28444–28455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Madreiter‐Sokolowski CT, Klec C, Parichatikanond W, Stryeck S, Gottschalk B, Pulido S, Rost R, Eroglu E, Hofmann NA, Bondarenko AI et al (2016) PRMT1‐mediated methylation of MICU1 determines the UCP2/3 dependency of mitochondrial Ca(2+) uptake in immortalized cells. Nat Commun 7, 12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Palty R, Silverman WF, Hershfinkel M, Caporale T, Sensi SL, Parnis J, Nolte C, Fishman D, Shoshan‐Barmatz V, Herrmann S et al (2010) NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc Natl Acad Sci USA 107, 436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Numata M, Petrecca K, Lake N & Orlowski J (1998) Identification of a mitochondrial Na+/H+ exchanger. J Biol Chem 273, 6951–6959. [DOI] [PubMed] [Google Scholar]
- 172. Jiang D, Zhao L & Clapham DE (2009) Genome‐wide RNAi screen identifies Letm1 as a mitochondrial Ca2+/H+ antiporter. Science 326, 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. De Marchi U, Santo‐Domingo J, Castelbou C, Sekler I, Wiederkehr A & Demaurex N (2014) NCLX protein, but not LETM1, mediates mitochondrial Ca2+ extrusion, thereby limiting Ca2+‐induced NAD(P)H production and modulating matrix redox state. J Biol Chem 289, 20377–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Grynkiewicz G, Poenie M & Tsien RY (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem 260, 3440–3450. [PubMed] [Google Scholar]
- 175. Paredes RM, Etzler JC, Watts LT, Zheng W & Lechleiter JD (2008) Chemical calcium indicators. Methods 46, 143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. de Brito OM & Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. [DOI] [PubMed] [Google Scholar]
- 177. Wang W, Xie Q, Zhou X, Yao J, Zhu X, Huang P, Zhang L, Wei J, Xie H, Zhou L et al (2015) Mitofusin‐2 triggers mitochondria Ca2+ influx from the endoplasmic reticulum to induce apoptosis in hepatocellular carcinoma cells. Cancer Lett 358, 47–58. [DOI] [PubMed] [Google Scholar]
- 178. Lee J, Ishihara A, Oxford G, Johnson B & Jacobson K (1999) Regulation of cell movement is mediated by stretch-activated calcium channels. Nature 400, 382–386. [DOI] [PubMed] [Google Scholar]
- 179. Gee KR, Brown KA, Chen WN, Bishop‐Stewart J, Gray D & Johnson I (2000) Chemical and physiological characterization of fluo‐4 Ca(2+)‐indicator dyes. Cell Calcium 27, 97–106. [DOI] [PubMed] [Google Scholar]
- 180. Minta A, Kao JP & Tsien RY (1989) Fluorescent indicators for cytosolic calcium based on rhodamine and fluorescein chromophores. J Biol Chem 264, 8171–8178. [PubMed] [Google Scholar]
- 181. Hoth M, Fanger CM & Lewis RS (1997) Mitochondrial regulation of store‐operated calcium signaling in T lymphocytes. J Cell Biol 137, 633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Hajnoczky G, Robb‐Gaspers LD, Seitz MB & Thomas AP (1995) Decoding of cytosolic calcium oscillations in the mitochondria. Cell 82, 415–424. [DOI] [PubMed] [Google Scholar]
- 183. De Michele R, Carimi F & Frommer WB (2014) Mitochondrial biosensors. Int J Biochem Cell Biol 48, 39–44. [DOI] [PubMed] [Google Scholar]
- 184. Demaurex N (2005) Calcium measurements in organelles with Ca2+‐sensitive fluorescent proteins. Cell Calcium 38, 213–222. [DOI] [PubMed] [Google Scholar]
- 185. Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev‐Ram V, Baker D & Tsien RY (2006) Ca2+ indicators based on computationally redesigned calmodulin‐peptide pairs. Chem Biol 13, 521–530. [DOI] [PubMed] [Google Scholar]
- 186. Waldeck‐Weiermair M, Alam MR, Khan MJ, Deak AT, Vishnu N, Karsten F, Imamura H, Graier WF & Malli R (2012) Spatiotemporal correlations between cytosolic and mitochondrial Ca(2+) signals using a novel red‐shifted mitochondrial targeted cameleon. PLoS ONE 7, e45917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T et al (2011) An expanded palette of genetically encoded Ca(2)(+) indicators. Science 333, 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Wu J, Liu L, Matsuda T, Zhao Y, Rebane A, Drobizhev M, Chang YF, Araki S, Arai Y, March K et al (2013) Improved orange and red Ca(2)+/‐ indicators and photophysical considerations for optogenetic applications. ACS Chem Neurosci 4, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Whelan SP & Zuckerbraun BS (2013) Mitochondrial signaling: forwards, backwards, and in between. Oxid Med Cell Longev 2013, 351613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Eisenberg‐Bord M & Schuldiner M (2017) Ground control to major TOM: mitochondria‐nucleus communication. FEBS J 284, 196–210. [DOI] [PubMed] [Google Scholar]
- 191. Haynes CM & Ron D (2010) The mitochondrial UPR ‐ protecting organelle protein homeostasis. J Cell Sci 123(Pt 22), 3849–3855. [DOI] [PubMed] [Google Scholar]
- 192. Dominy JE & Puigserver P (2013) Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harb Perspect Biol 5, pii: a015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Dolezal P, Likic V, Tachezy J & Lithgow T (2006) Evolution of the molecular machines for protein import into mitochondria. Science 313, 314–318. [DOI] [PubMed] [Google Scholar]
- 194. Neupert W & Herrmann JM (2007) Translocation of proteins into mitochondria. Annu Rev Biochem 76, 723–749. [DOI] [PubMed] [Google Scholar]
- 195. Endo T & Yamano K (2010) Transport of proteins across or into the mitochondrial outer membrane. Biochim Biophys Acta 1803, 706–714. [DOI] [PubMed] [Google Scholar]
- 196. Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW & Haynes CM (2016) The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol 26, 2037–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Cardamone MD, Tanasa B, Cederquist CT, Huang J, Mahdaviani K, Li W, Rosenfeld MG, Liesa M & Perissi V (2018) Mitochondrial retrograde signaling in mammals is mediated by the transcriptional cofactor GPS2 via direct mitochondria‐to‐nucleus translocation. Mol Cell 69, 757–772.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Ruan L, Zhou C, Jin E, Kucharavy A, Zhang Y, Wen Z, Florens L & Li R (2017) Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Csordas G, Weaver D & Hajnoczky G (2018) Endoplasmic reticulum‐mitochondrial contactology: structure and signaling functions. Trends Cell Biol 28, 523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Bernhard W, Haguenau F, Gautier A & Oberling C (1952) Submicroscopical structure of cytoplasmic basophils in the liver, pancreas and salivary gland; study of ultrafine slices by electron microscope. Z Zellforsch Mikrosk Anat 37, 281–300. [PubMed] [Google Scholar]
- 201. Voelker DR (1990) Characterization of phosphatidylserine synthesis and translocation in permeabilized animal cells. J Biol Chem 265, 14340–14346. [PubMed] [Google Scholar]
- 202. Vance JE (1991) Newly made phosphatidylserine and phosphatidylethanolamine are preferentially translocated between rat liver mitochondria and endoplasmic reticulum. J Biol Chem 266, 89–97. [PubMed] [Google Scholar]
- 203. Bui M, Gilady SY, Fitzsimmons RE, Benson MD, Lynes EM, Gesson K, Alto NM, Strack S, Scott JD & Simmen T (2010) Rab32 modulates apoptosis onset and mitochondria‐associated membrane (MAM) properties. J Biol Chem 285, 31590–31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. de Brito OM & Scorrano L (2009) Mitofusin‐2 regulates mitochondrial and endoplasmic reticulum morphology and tethering: the role of Ras. Mitochondrion 9, 222–226. [DOI] [PubMed] [Google Scholar]
- 205. Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM & Thomas G (2005) PACS‐2 controls endoplasmic reticulum‐mitochondria communication and Bid‐mediated apoptosis. EMBO J 24, 717–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Carreras‐Sureda A, Jana F, Urra H, Durand S, Mortenson DE, Sagredo A, Bustos G, Hazari Y, Ramos‐Fernandez E, Sassano ML et al (2019) Non‐canonical function of IRE1alpha determines mitochondria‐associated endoplasmic reticulum composition to control calcium transfer and bioenergetics. Nat Cell Biol 21, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T & Rizzuto R (2006) Chaperone‐mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol 175, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Vance JE (2014) MAM (mitochondria‐associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta 1841, 595–609. [DOI] [PubMed] [Google Scholar]
- 209. Area‐Gomez E, Del Carmen Lara M, Castillo MD, Tambini C, de Guardia‐Laguarta AJ, Groof M, Madra J, Ikenouchi M, Umeda TD, Bird SL Sturley et al (2012) Upregulated function of mitochondria‐associated ER membranes in Alzheimer disease. EMBO J 31, 4106–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Area‐Gomez E, de Groof AJ, Boldogh I, Bird TD, Gibson GE, Koehler CM, Yu WH, Duff KE, Yaffe MP, Pon LA et al (2009) Presenilins are enriched in endoplasmic reticulum membranes associated with mitochondria. Am J Pathol 175, 1810–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Zuchner S, Mersiyanova IV, Muglia M, Bissar‐Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J et al (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause charcot‐marie‐tooth neuropathy type 2A. Nat Genet 36, 449–451. [DOI] [PubMed] [Google Scholar]
- 212. Narendra D, Tanaka A, Suen DF & Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183, 795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Wang X, Winter D, Ashrafi G, Schlehe J, Wong YL, Selkoe D, Rice S, Steen J, LaVoie MJ & Schwarz TL (2011) PINK1 and parkin target miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Williamson CD, DeBiasi RL & Colberg‐Poley AM (2012) Viral product trafficking to mitochondria, mechanisms and roles in pathogenesis. Infect Disord Drug Targets 12, 18–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Morciano G, Marchi S, Morganti C, Sbano L, Bittremieux M, Kerkhofs M, Corricelli M, Danese A, Karkucinska‐Wieckowska A, Wieckowski MR et al (2018) Role of mitochondria‐associated ER membranes in calcium regulation in cancer‐specific settings. Neoplasia 20, 510–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Madreiter‐Sokolowski CT, Waldeck‐Weiermair M, Bourguignon MP, Villeneuve N, Gottschalk B, Klec C, Stryeck S, Radulovic S, Parichatikanond W, Frank S et al (2019) Enhanced inter‐compartmental Ca(2+) flux modulates mitochondrial metabolism and apoptotic threshold during aging. Redox Biol 20, 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA & Hajnoczky G (2006) Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol 174, 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD & Regan L (2005) Detecting protein‐protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc 127, 146–157. [DOI] [PubMed] [Google Scholar]
- 219. Cieri D, Vicario M, Giacomello M, Vallese F, Filadi R, Wagner T, Pozzan T, Pizzo P, Scorrano L, Brini M et al (2018) SPLICS: a split green fluorescent protein‐based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ 25, 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Yang Z, Zhao X, Xu J, Shang W & Tong C (2018) A novel fluorescent reporter detects plastic remodeling of mitochondria‐ER contact sites. J Cell Sci 131, pii: jcs208686. [DOI] [PubMed] [Google Scholar]
- 221. Harmon M, Larkman P, Hardingham G, Jackson M & Skehel P (2017) A Bi‐fluorescence complementation system to detect associations between the Endoplasmic reticulum and mitochondria. Sci Rep 7, 17467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Ding Y, Li J, Enterina JR, Shen Y, Zhang I, Tewson PH, Mo GC, Zhang J, Quinn AM, Hughes TE et al (2015) Ratiometric biosensors based on dimerization‐dependent fluorescent protein exchange. Nat Methods 12, 195–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Alford SC, Ding Y, Simmen T & Campbell RE (2012) Dimerization‐dependent green and yellow fluorescent proteins. ACS Synth Biol 1, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Csordas G, Varnai P, Golenar T, Roy S, Purkins G, Schneider TG, Balla T & Hajnoczky G (2010) Imaging interorganelle contacts and local calcium dynamics at the ER‐mitochondrial interface. Mol Cell 39, 121–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Shi F, Kawano F, Park SE, Komazaki S, Hirabayashi Y, Polleux F & Yazawa M (2018) Optogenetic control of endoplasmic reticulum‐mitochondria tethering. ACS Synth Biol 7, 2–9. [DOI] [PubMed] [Google Scholar]
- 226. Tubbs E & Rieusset J (2016) Study of endoplasmic reticulum and mitochondria interactions by in situ proximity ligation assay in fixed cells. J Vis Exp. 10.3791/54899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Wieckowski MR, Giorgi C, Lebiedzinska M, Duszynski J & Pinton P (2009) Isolation of mitochondria‐associated membranes and mitochondria from animal tissues and cells. Nat Protoc 4, 1582–1590. [DOI] [PubMed] [Google Scholar]
- 228. Vance JE (1990) Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem 265, 7248–7256. [PubMed] [Google Scholar]
- 229. Lynes EM, Raturi A, Shenkman M, Ortiz Sandoval C, Yap MC, Wu J, Janowicz A, Myhill N, Benson MD, Campbell RE et al (2013) Palmitoylation is the switch that assigns calnexin to quality control or ER Ca2+ signaling. J Cell Sci 126(Pt 17), 3893–903. [DOI] [PubMed] [Google Scholar]
- 230. Hell SW & Nagorni M (1998) 4Pi confocal microscopy with alternate interference. Opt Lett 23, 1567–1569. [DOI] [PubMed] [Google Scholar]


