Abstract
Background
Over 2,000 people a year in the United Kingdom need a bone marrow or blood stem cell transplant. It is important to accurately quantify the hematopoietic stem cells to predict whether the transplant will be successful in replenishing the immune system. However, they are present at low frequency, which complicates accurate quantification. The current gold standard method is single‐platform flow cytometry using internal reference counting beads to determine the concentration of CD34 cells. However, volumetric flow cytometers have the ability to measure the acquisition volume, which removes the need for reference beads for calculation of cell concentrations.
Method
In this study, we compared both methods for calculating CD34 cell concentrations in volumetric cytometers, using either the volume reading or the number of reference beads for calculation. In addition, the uncertainty of measurement for each method was estimated.
Results
The results show that both methods have similar uncertainties of measurement. Regression analysis showed low to no statistical difference in CD34 cell concentrations obtained with each method.
Conclusions
Overall, this study suggests that the volumetric method is a valid approach but that the adoption of this technology may be hindered without some form of external calibration of volume readings to increase confidence in the measurement. © 2019 The Authors. Cytometry Part B: Clinical Cytometry published by Wiley Periodicals, Inc. on behalf of International Clinical Cytometry Society.
Keywords: flow cytometry, immunotherapy, rare event detection, single platform, standardization, stem cells, transplantation
INTRODUCTION
Hematopoietic stem cell cells are used clinically to reconstitute the hematopoietic system after radiation or chemotherapy and express the cell‐surface antigen CD34. The number of viable CD34+ cells in transplants is routinely quantified by flow cytometry.
Standardization efforts for the quantification of CD34 cells over time converged on a set of guidelines commonly referred to as the CD34 ISHAGE method 1 which is still the method of choice today. In addition, the current “gold standard” for quantifying CD34 concentrations in hematopoietic stem cell products is single‐platform CD34 enumeration using internal reference counting beads 2. However, some flow cytometers are equipped with the ability to provide the absolute volume of cell suspension analyzed 3. In principle, the known volume would be sufficient to calculate a CD34 concentration, without the need for internal reference counting beads. In this study, we compared the concentrations obtained with both methods using the same instruments, at various laboratories worldwide, using a CD34 reference material as a comparator sample.
MATERIALS AND METHODS
This technology comparison was part of a metrological study organized through the Bureau International des Poids et Mesures, an intergovernmental organization where Member States cooperate on matters related to measurement science. Twelve participants from seven different countries took part in the study looking at measurement uncertainty in CD34 cell enumeration by flow cytometry.
Each participant was sent six vials of CD34 reference material (CD34+ Cell Enumeration System Suitability RS, Catalog # 1084292, United States Pharmacopeia, MD) and six Trucount™ tubes from the same lot (Lot: 14304, BD Biosciences, Germany). Participants reconstituted three vials of reference material in 0.5 mL of distilled water (CD34 High concentration) and three vials in 1.5 mL of distilled water (CD34 Low concentration). The reference material was thoroughly mixed by inversion, and 100 μl was sampled and transferred to Trucount™ tubes by reverse‐pipetting using a calibrated pipette. Each participant was asked to record the weight of the sample at each stage of sample processing in a calibrated balance to measure pipetting variability. Participants recorded the weight of the reconstitution volume added to reference material vials, the sample volume added to the Trucount™ tubes, and the weights of antibody reagents and dilution buffer added to each Trucount™ tube.
The cells were stained by adding CD45‐FITC and CD34‐PE antibodies to each Trucount™ tube. Participants were given free choice of which antibodies to use, provided they met the criteria of being a FITC‐conjugated antibody that recognizes all isoforms of CD45 and a PE‐conjugated antibody clone II or clone III, as these stain all glycosylation forms of the CD34 antigen. Various antibodies were used in the study: CD45‐FITC/CD34‐PE clones 2D1/8G12 (Catalog # 341071, BD Biosciences, Germany), Stem Cell Enumeration kit (Catalog # 344563, BD Biosciences, Germany), CD34‐PE clone 581 (Catalog # 555822, BD Biosciences, Germany), CD34‐PE clone AC136 (Catalog # 130‐113‐179, Miltenyi Biotec, Germany), CD45‐FITC clone HI30 (Catalog # 555482, BD Biosciences, Germany; Catalog # 304006, Biolegend, San Diego, CA; and also Catalog # MHCD4501‐4, eBioscience, San Diego, CA), CD45‐FITC clone REA747 (Catalog # 130‐110‐631, Miltenyi Biotec, Germany).
After antibody staining, the sample was diluted in 2 mL of PBS 0.5–1% (w/v) bovine or human serum albumin and analyzed in volumetric cytometers. This “lyse no wash” method has been shown to be more accurate for CD34 cell enumeration than washing and resuspending the sample 1, except here PBS is used instead of red blood cell lysis buffer as the CD34 reference material contains no red blood cells. The volumetric flow cytometers used were: two BD FACSVerse™ (BD Biosciences, Germany), one CyFlow Cube 8 (Sysmex Partec, Germany), two CyFlow ML (Sysmex Partec, Germany), one optical reference flow cytometer (Physikalisch‐Technische Bundesanstalt, Germany), two Attune (Thermo Fisher Scientific, Waltham, MA), and one BD Accuri™ (BD Biosciences, Germany).
Each flow cytometer was set up using the instrument manufacturer's microspheres. Participants then set up a compensation matrix with single stained controls. The default threshold on Forward Scatter (FSC) excludes small objects such as counting beads and so was adjusted to be set on FITC as all beads and white blood cells (CD45‐FITC+) were fluorescent in FITC. A stopping gate of 75,000 CD45+ events was set with a maximum acquisition time of 15 min per tube to avoid the possibility of loss of homogenous mixing of cells and beads during long acquisition times.
The data were analyzed by each laboratory using the CD34 ISHAGE gating strategy provided (Supporting Information Fig. S1). The CD34 cell concentrations were calculated for the same sample using either the number of bead events or the acquisition volume (Equations 1–3 in Supporting Information). Central data analysis was then performed at NIBSC (statistical analysis) and PTB (uncertainty of measurement estimates).
This study aimed to estimate measurement uncertainty. Measurement uncertainty is a parameter associated with the result of a measurement that characterizes the dispersion of the values that could reasonably be attributed to the measurand. Participants used calibrated pipettes and recorded the weight of the sample at different stages of sample processing on a calibrated balance, which was incorporated in the uncertainty of measurement calculations (Equations 7 and 9 in Supporting Information). In addition, the number of beads in 50 BD Trucount™ bead tubes from the same Lot was assessed by impedance‐based reference particle counters and an optical reference flow cytometer (Physikalisch‐Technische Bundesanstalt, Germany) to incorporate the contribution of tube‐to‐tube variability in the bead standard in the uncertainty of measurement calculation. Equations can be found in Supporting Information.
Statistical analysis
The data were tested for outliers using the Grubb's test 4 in GraphPad Prism (v5 for Windows, GraphPad Software, La Jolla, CA, http://www.graphpad.com). The CD34 cell concentrations obtained using both methods were compared: a Pearson's correlation coefficient was calculated, and regression analysis was performed with R 5 using RStudio (v3.4). A Deming regression analysis was chosen because it considers the uncertainties of measurement 6. The matched data sets for each individual laboratory/instrument were then compared with a paired t test (Graph Pad Prism 5).
RESULTS
The mean value and uncertainty of measurement estimated with the Trucount method was 26.2 ± 0.9 CD34 cells per microliter and 8.6 ± 0.4 CD34 cells per microliter for the high and low concentration samples, respectively. For the volumetric method, they were 26.0 ± 1.5 for the high concentration sample and 8.1 ± 0.5 for the low concentration sample (Table 1).
Table 1.
Comparison of Measurement Uncertainty Using Trucount and Volumetric Methods
| Data set | Participant range (CD34 cells/μL) | Weighed mean value (CD34 cells/μL) | Uncertainty of mean value (CD34 cells/μL) |
|---|---|---|---|
| CD34 High, Trucount | 19.2–34.5 | 26.2 | 0.9 |
| CD34 Low, Trucount | 6.2–10.8 | 8.6 | 0.4 |
| CD34 High, volumetric | 16.9–34.5 | 26.0 | 1.5 |
| CD34 Low, volumetric | 6.2–10.6 | 8.1 | 0.5 |
The table depicts the uncertainty of measurement estimate for CD34 cell concentrations obtained using either the Trucount method or the volumetric method. The components taken into account in the estimate were the variability within a batch of Trucount beads and the variability in sample preparation volumes recorded gravimetically at each stage of sample processing.
Three outliers were found in the matched Trucount/volumetric cell concentrations when analyzing the lower concentration samples (displayed in gray, Fig. 1). However, two of the three outliers came from the same participant and instrument. The data files were reviewed and show significant fluorescence intensity drop during the acquisition of one of the outliers (Supporting Information, Fig. 2) which could have been caused by laser drift and likely meant disproportional loss of beads and cells from the beads and hematopoietic stem cell gates and therefore inaccurate cell concentrations. After exclusion of the three outliers, there was a good correlation with a Pearson's coefficient of r = 0.98.
Figure 1.
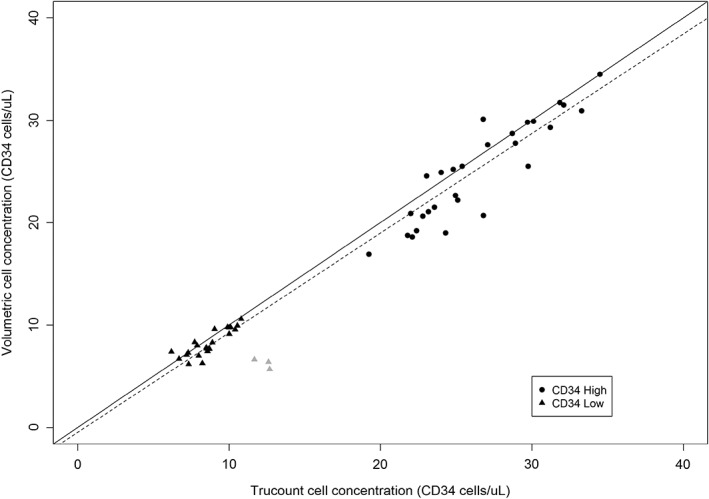
Comparison of volumetric and bead‐based counting of CD34 cells by single‐platform flow cytometry. CD34 cells were enumerated using BD Trucount™ tubes in volumetric cytometers. A CD34 reference material was assessed at two concentrations: CD34 High shown in circles and CD34 Low shown in triangles. The cell concentrations obtained with the number of bead events are shown in the x‐axis and with the recorded acquisition volume in the y‐axis. Outliers are shown in gray. The solid line indicates agreement and the dashed line indicates fitted Deming regression which accounts for error in both variables. The slope is 0.972 with a confidence interval of 0.934–1.02. The intercept is −0.58 with a confidence interval of −1.17 to −0.008.
The data were next analyzed using regression analysis to compare both methods. The solid line in Figure 1 represents the hypothetical scenario of perfect agreement between the matched values. A Deming regression was performed which takes into account error in both variables, as calculated in the uncertainty of measurement (dashed line in Fig. 1). The slope of the Deming regression line (dashed line in Fig. 1) is 0.972 with a confidence interval of 0.934–1.02. The fact that the slope of the line includes 1, which corresponds to perfect agreement, means no statistical difference was found between the two methods by regression analysis. The Deming regression line intercepts the y‐axis at −0.58 CD34 cells per microliter with a confidence interval of −1.17 to −0.008. This suggests a worst‐case scenario of lower volumetric counts by less than two cells per microliter, which is not likely clinically significant.
The matched Trucount and volumetric values for each instrument in the study were compared with a paired t test. Volumetric results were significantly lower (P < 0.05) for four of the nine instruments used in the study but not statistically different for the other five instruments. However, due to the low number of laboratories using the same instrument (n = 1 or 2 per instrument), it is not possible to extrapolate instrument‐specific performance from these data.
DISCUSSION
The study set out to compare volumetric and bead‐based counting of CD34 cells. It showed a good correlation between the two methods with a Pearson's correlation similar to other references in the field which report r values between 0.93 7 and 0.99 8, 9. In the laboratory, the validity of volumetric approaches has been demonstrated for other instruments in the past such as hematology analyzers. However, bead‐based flow cytometry still seems more popular than volumetric flow cytometry for CD34 cell enumeration, despite its higher cost. We discuss sources of uncertainty shared and specific to each of the two methods, Trucount and volumetric, and suggest ways to minimize them.
Taking our study as an example, we found three outliers in the data, in the lower cell concentration samples. In comparison to the pooled data in Figure 1, the outliers in gray seem to have an unexpectedly low volumetric reading and/or an unexpectedly high Trucount reading. One possibility would be an inaccurately high volume reading from the cytometer. Poor reproducibility for concentrations lower than nine CD34 cells per microliter in volumetric cytometry has been reported elsewhere 10 and one publication also reports consistently higher than accurate volume readings in a volumetric cytometer 11. However, there are other possibilities that can first be ruled out. Bergeron et al. described how inspecting the listmode files on time plots can identify laser drift during sample acquisition, which is not apparent when analyzing the data in the standardized gating strategy 12. Reviewing the participant's listmode files in time plots shows significant fluorescence intensity drop, probably caused by laser instability, during acquisition of one of the outliers (Supporting Information, Fig. 2), which likely caused disproportionate loss of cells and beads from their respective gates, and inaccurate cell concentrations. However, there was no evidence of laser instability during acquisition of the other two outliers.
The bead‐based method relies on an accurate bead count. There are various possible causes of inaccurate bead quantification. One possibility is poor mixing prior to acquisition or changes in the local concentration of beads during the longer acquisition time required for the lower concentration samples (about 15 min). Again, a way to investigate the possibility of nonhomogenous mixing of cells and beads is to inspect the ungated data in time plots. As Trucount™ beads have higher fluorescence than cells in all the channels, two separate streams can be seen when plotting the data in a time versus fluorescent channel plot (Supporting Information, Fig. 2). The proportion of bead and cell events detected over time should remain stable during sample acquisition if homogenous mixing is maintained. No nonhomogenous mixing was evident in the analysis of the remaining two outliers in time plots. Another cause of occasional bead loss can be adherence to the sample tube but this is preventable by the inclusion of protein in the buffer 13. For this reason, we included 0.5–1% albumin in the dilution buffer. In addition, a common new operator mistake that will result in partial loss of beads is to acquire the samples using the default threshold in the cytometer. The default threshold is set to exclude debris but also excludes some beads due to their small size. For this reason, participants were instructed to set the threshold to FITC so as to include all white blood cells (which are stained with a pan‐CD45 FITC antibody) and beads (which are fluorescent in all channels including FITC). There could also be a contribution from tube‐to‐tube variation in Trucount™ beads. In our study, the number of beads per tube was lower than that reported by the manufacturer, and there was a small tube‐to‐tube variation. However, the results did not change significantly after recalculating with the lower number of beads per tube we found in our assessment which implies that other sources of uncertainty have a more significant impact. In conclusion, the ability to obtain an accurate CD34 concentration with the established bead‐based method relies on an accurate bead count. To minimize uncertainty, users should ensure homogenous mixing, include serum proteins in the buffer, and ensure that beads are not excluded by the default threshold during acquisition.
In summary, both methods have their limitations. Shared factors that contribute to uncertainty of measurement include cellular reference material reconstitution, sample processing, data analysis, and flow cytometry platforms. The cell reconstitution can vary due to slight differences in pipetting and vial‐to‐vial variation within a batch. Sampling by reverse pipetting with adequately calibrated pipettes will minimize pipetting error. The choice of antibodies is particularly important in CD34 enumeration. CD34 antibodies should be of class II and class III to ensure detection of all glycosylation variants of the molecule, and a pan‐CD45 antibody clone should be used. CD34 should additionally be conjugated to a bright fluorochrome, typically phycoerythrin (PE) 1, 14. A whole blood lysis technique should be employed to prevent loss of cells during additional processing. Differences relating to data analysis can result from population gating, number of events collected, and inappropriate compensation. Users should collect a minimum number of 75,000 total white blood cells and 100 CD34 events which has been shown to ensure statistical significance of the rare CD34 population 1. Although these shared sources of uncertainty cannot be avoided, they can be minimized through the various measures described above.
The uncertainty of measurement obtained in the metrological study was similar for both methods and we feel both proved equally valid as tools to assess absolute CD34 cell concentrations. Discrepancies from the expected value should be investigated through examination of the data in time plots to rule out laser instability and then followed by repeat testing. If they remain, then sources of uncertainty particular to each method can be evaluated. With the reference bead method, acquisition time and loss of beads to the tube can be the issues. With the volumetric method, the accuracy of the volume readings should be investigated. Volume is a unit traceable to a higher order standard (mass) using the gravimetric method and therefore it seems plausible that the uncertainty of volume measurement could be calculated in different cytometer platforms in a similar way to pipette calibrations. Such a method would be traceable to the International System of Units (SI) and would potentially allow for better agreement of absolute measurements of cell concentrations in different volumetric flow cytometers and different laboratories.
Supporting information
Appendix S1: Supporting Information
ACKNOWLEDGMENT
The authors are thankful to Dr Jayne Morrow as committee chair for driving this project to completion and to Dr Sandra Diebold for critical review of the manuscript.
How to cite this article: Saraiva L, Wang L, Kammel M, Kummrow A, Atkinson E, Lee JY, Yalcinkaya B, Akgöz M, Höckner J, Ruf A, Engel A, Zhang Y‐Z, O'Shea O, Sassi MP, Divieto C, Lekishvili T, Campbell J, Liu Y, Wang J, Stebbings R, Gaigalas AK, Rigsby P, Neukammer J, Vessillier S. Comparison of Volumetric and Bead‐Based Counting of CD34 Cells by Single‐Platform Flow Cytometry. Cytometry Part B 2019;96B:508–513.
LITERATURE CITED
- 1. Sutherland DR, Anderson L, Keeney M, Nayar R, Chin‐Yee I. The ISHAGE guidelines for CD34+ cell determination by flow cytometry. International Society of Hematotherapy and Graft Engineering. J Hematother. 1996;5:213–226. [DOI] [PubMed] [Google Scholar]
- 2. Keeney M, Barnett D, Gratama JW. Impact of standardization on clinical cell analysis by flow cytometry. J Biol Regul Homeost Agents. 2004;18:305–312. [PubMed] [Google Scholar]
- 3. Fukuda J, Kaneko T, Egashira M, Oshimi K. Direct measurement of CD34+ blood stem cell absolute counts by flow cytometry. Stem Cells. 1998;16(4):294–300. [DOI] [PubMed] [Google Scholar]
- 4. Grubbs FE. Procedures for detecting outlying observations in samples. Technometrics. February, 1969;11(4):1–21. [Google Scholar]
- 5. R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2018. [Google Scholar]
- 6. Cornbleet PJ, Gochman N. Incorrect least‐squares regression coefficients in method‐comparison analysis. Clin Chem. 1979;25:432–438. [PubMed] [Google Scholar]
- 7. Gutensohn K, Anikolitsis A, Gramatzki M, Spitzer D, Buwitt‐Beckmann U, Humpe A. Direct volumetric flow cytometric quantitation of CD34+ stem and progenitor cells. Transfus Med. 2012;22:205–210. [DOI] [PubMed] [Google Scholar]
- 8. Mariani M, Colombo F, Assennati SM, Frugoni C, Cattaneo A, Trombetta E, Rebulla P, Porretti L, Porretti L. Evaluation of an easy and affordable flow cytometer for volumetric haematopoietic stem cell counting. Blood Transfus. 2014;12:416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mortazavi S, Ardalan FA, Nodehi SR, Karder FF, Miraliakbari N. True volumetric method for flow cytometric enumeration of CD34 + stem cells and its agreement with a standard bead‐based single‐platform protocol. Cytotherapy. 2012;14:621–629. [DOI] [PubMed] [Google Scholar]
- 10. Gisselo CG, Roer O, Hoffmann MH, Hansen MB, Taaning E, Johnsen HE. Assessing agreement between CD34 enumeration by flow cytometry and volumetric analysis. Bone Marrow Transplant. 2002;29:699–703. [DOI] [PubMed] [Google Scholar]
- 11. Wang L, Zhang YZ, Choquette S, Gaigalas AK. Measurement of microsphere concentration using a flow cytometer with volumetric sample delivery. J Res Natl Inst Stand Technol. 2014;119:629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bergeron M, Lustyik G, Phanuef S, Ding T, Nicholson JKA, Janossy G, Shapiro H, Barnett D, Mandy F. Stability of currently used cytometers facilitates the identification of pipetting errors and their volumetric operation: Time can tell all. Cytometry B. 2003;52B:37–40. [DOI] [PubMed] [Google Scholar]
- 13. Brando B, Gohde W Jr, Scarpati B, D'avanzo G. The “vanishing counting bead” phenomenon: Effect on absolute CD34+ cell counting in phosphate‐buffered saline‐diluted leukapheresis samples. Cytometry. 2001;43:154–160. [DOI] [PubMed] [Google Scholar]
- 14. Gratama JW, Orfao A, Barnett D, Brando B, Huber A, Janossy G, Johnsen HE, Keeney M, Marti GE, Preijers F, et al. Flow cytometric enumeration of CD34+ hematopoietic stem and progenitor cells. European Working Group on Clinical Cell Analysis. Cytometry. 1998;34(3):128–142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information


