Summary
Mayotte is an island located in the Mozambique Channel, between Mozambique and Madagascar, in the South Western Indian Ocean region. A severe syndrome of unknown aetiology has been observed seasonally since 2009 in cattle (locally named “cattle flu”), associated with anorexia, nasal discharge, hyperthermia and lameness. We sampled blood from a panel of those severely affected animals at the onset of disease signs and analysed these samples by next‐generation sequencing. We first identified the presence of ephemeral bovine fever viruses (BEFV), an arbovirus belonging to the genus Ephemerovirus within the family Rhabdoviridae, thus representing the first published sequences of BEFV viruses of African origin. In addition, we also discovered and genetically characterized a potential new species within the genus Ephemerovirus, called Mavingoni virus (MVGV) from one diseased animal. Finally, both MVGV and BEFV have been identified in cattle from the same herd, evidencing a co‐circulation of different ephemeroviruses on the island. The clinical, epidemiological and virological information strongly suggests that these viruses represent the etiological agents of the observed “cattle flu” within this region. This study highlights the importance of the strengthening and harmonizing arboviral surveillance in Mayotte and its neighbouring areas, including Africa mainland, given the importance of the diffusion of infectious diseases (such as BEFV) mediated by animal and human movements in the South Western Indian Ocean area.
Keywords: Africa, arbovirus, ephemerovirus, Mayotte, next‐generation sequencing, rhabdovirus
1. INTRODUCTION
Bovine ephemeral fever (BEF), also called three‐day sickness, is an arthropod‐borne disease caused by the infection with bovine ephemeral fever virus (BEFV), a rhabdovirus belonging to the genus Ephemerovirus. BEFV can infect a large range of domestic and wild ungulates, though cattle and water buffalo are considered the most clinically susceptible animals (Walker & Klement, 2015). The duration of symptoms is usually short, characterized by bi‐phasic acute hyperthermia, lameness, ocular and nasal discharge before recovery, with a fleeting viraemia of about 1–3 days. Further prolonged and more severe symptoms may be observed, such as recumbency, muscle stiffness, anorexia, ataxia, paralysis and death. The morbidity rate is usually high (80%–100%), whilst the mortality rate remains low (<1%) (Walker & Klement, 2015), although higher rates have been sometimes reported in China (Zheng & Qiu, 2012) and in Turkey (Tonbak et al., 2013). Despite its limited mortality, BEF may have a major economic impact, especially on dairy herds with reduced milk production, on beef cattle with loss of production, temporary infertility in males and on draught animals with their temporary disablement (Walker, 2005).
To date, BEFV infection has not been reported in Europe, in the Americas, nor in the islands of the Pacific, but it is present in many tropical and sub‐tropical regions worldwide, including Australia, Asia (South, South‐East and China), Middle East and Africa. For the latter, the disease is enzootic and seasonally epizootic (Walker, 2005). The first documented epizootic was described in Zimbabwe in 1906, and the disease has been commonly reported in many other sub‐Saharan African countries including South Africa, Sudan, Kenya, Uganda and Tanzania (Bevan, 1907; Walker, 2005). However, historical reports suggest that the regional distribution of BEF in Africa is probably larger, including Madagascar and the Indian Ocean region.
Mayotte is a small island belonging to the French overseas territories (374 km2), located in the Mozambique Channel, between Madagascar and Mozambique. Cattle production is extensive, with a population size of about 17,000 heads. Several vector‐borne cattle diseases have been reported on the island over the past decade, including Rift Valley fever, bluetongue and epizootic haemorrhagic disease (Dommergues et al., 2019; Métras et al., 2016). In addition, due to its proximity and close connection to its neighbouring areas through active trade and human immigration, Mayotte has experienced several introductions of other arboviruses, such as dengue or chikungunya viruses (Tortosa et al., 2012). Since 2009, a clinical syndrome locally named “cattle flu” of an unknown aetiology, and causing symptoms such as anorexia, nasal discharge, hyperthermia and lameness, has also been affecting Mayotte cattle (Girard, Favre, Madi, & Cardinale, 2009). The “cattle flu” was reported all year‐round, with a marked seasonal peak appearing at the end of the rainfall period (55% of cases occur between March and May), suggesting the occurrence of another arthropod‐borne disease. The observed symptoms, the seasonal dynamics, and its proximity to Africa mainland suggested that BEFV may also be present in Mayotte, contributing to the overall cattle disease burden.
In this study, we investigate for the presence of ephemeroviruses, and most especially BEFV in the cattle population of Mayotte.
2. METHODS
As part as the animal diseases surveillance system (SESAM, Système d'Epidémiosurveillance Animale à Mayotte), veterinarians collected blood samples with minimal distress on cattle affected with “cattle flu” as soon as possible after the onset of symptoms, and they ensured the maintenance of the welfare of these animals throughout the study. In this context, Ethical Statement is not applicable. The samples were stored at −80°C. Ten of these samples, collected during the acute symptomatic phase from animals with severe symptoms, were tested for the presence of ephemeroviruses (Table S1). For laboratory testing, we used first a reverse transcription PCR (RT‐PCR) presenting a large spectrum of detection among these viruses (Blasdell et al., 2013). Total RNA was extracted from 200 μl of whole blood using TRI Reagent BD Sigma‐Aldrich, then 8 μl of RNA was used for the complementary DNA (cDNA) step (Dacheux et al., 2008) and PCR was performed with 2 μl of cDNA using Ex Taq TaKaRa. None of the samples tested were found positive, probably due to a lack of sensitivity of this technique which is not nested‐based. The samples were then submitted to next‐generation sequencing (NGS) analysis. Briefly, total RNA was depleted from DNA and purified using RNeasy Mini Kit Qiagen, followed by a ribosomal RNA depletion step with Terminator 5′‐Phosphate‐Dependent Exonuclease Epicentre. Double‐stranded DNA was synthetized according to Matranga et al., 2016, and both NGS libraries and sequencing were done as previously described (Troupin et al., 2016). Pre‐processing of reads was done in the Galaxy platform (Mareuil, Doppelt‐Azeroual, & Ménager, 2017) according to Troupin et al., 2016;. Contigs were then generated by de novo assembly using CLC Assembly Cell in Galaxy and submitted to BLASTx in GenBank. In parallel, contigs were also generated by a dedicated workflow for NGS data analysis locally implemented, which includes the following steps: (a) depletion of host genome reads, (b) de novo generation of contigs using Spades, (c) blast analysis using BLASTn and BLASTx on GenBank and Uniprot databases, respectively, and (d) taxonomic classification and visualization using Krona, adapted from Dacheux et al., 2014;. Contigs related to ephemerovirus were selected and used for genome editing, completed with Sanger sequencing and a final read mapping step using cleaned pre‐processing reads (Troupin et al., 2016).
3. RESULTS AND DISCUSSION
The presence of ephemerovirus was detected in five out of the 10 samples tested, with the identification of BEFV in four of these samples, and a potential new ephemerovirus species in the last one, provisionally named Mavingoni virus (MVGV) (Figure 1a, Table S1) based on its sampling location. Apart from the 5′UTR and 3′UTR regions, the nearly complete genome sequences were obtained for all BEFV and exhibited the classical genome organization for this virus species, expect for the sequence of the putative alpha 3 protein which was not observed (GenBank accession numbers MN148800–MN148803) (Table S2). We obtained MVGV genome from the nucleoprotein gene to the partial sequence for the gamma protein gene (GenBank accession number MN148799). Phylogenetic analyses demonstrated that the four African BEFV had nearly identical genome sequences, with a high percentage of identity (>99%). They were clustered apart from the BEFV originating from the Middle East, Asia and Australia, thus probably representing the first genome representative of African BEFV clade (Figure 1a). Among the other ephemerovirus species, MVGV was distinct from the most closely representatives were Obodhiang virus (OBOV) and Adelaide river virus (ARV), isolated from mosquitoes in Sudan and from cattle in Australia, respectively (Figure 1a).
Figure 1.
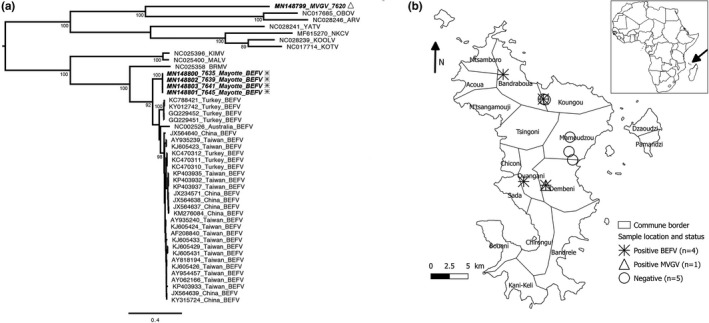
Phylogenetic analysis (a) and geographical location (b) and of the ephemeroviruses from Mayotte. (a) The tree was based on the full‐length glycoprotein nucleotide sequence and was constructed using the maximum‐likelihood approach based on the generalized time‐reversible model proportion of invariable sites plus gamma‐distributed rate heterogeneity (GTR+I+Γ4) utilizing SPR branch‐swapping, as estimated with PhyML 3.0 (Guindon et al., 2010). The robustness of individual nodes was estimated using 100 bootstrap replicates using a Bayesian‐like transformation of aLRT (aBayes). Only bootstrap values ≥ 90 are indicated. Scale bar indicates nucleotide substitutions per site. Ephemeroviruses from Mayotte are indicated in bold and in italic, with the same legend as (b). (b) The map of Mayotte shows the sample locations and their infected ephemerovirus status (the farm in Koungou reported two positive samples)
Specific PCR designed for the detection of BEFV and MVGV was used to test the panel of samples and confirmed the results obtained with NGS (Table S1). Attempts to isolate the virus for three positive samples on newborn suckling mice or repeated Vero cell passages were unsuccessful, probably due to the type of the materials used. Indeed, we used frozen (haemolytic) whole blood which presented cytotoxicity and is not the best suitable sample for BEFV virus isolation compared with frozen buffy coat (Table S2).
Based on this study, we obtained the first African sequences of BEFV and a potential new species among the genus Ephemerovirus, Mavingoni virus (MVGV). All of them were detected in symptomatic cattle, strongly suggesting that they represent the aetiological agent for the “cattle flu” observed in these animals. Indeed, the clinical symptoms observed, associated with their seasonal dynamics, are compatible with this hypothesis. In addition, the short viremia (1–3 days) usually observed with BEF can explain why only half of the samples collected during the acute stage was positive. The geographical distribution of BEFV appears to be large in Mayotte Island, with at least four different communes affected (Figure 1b). Interestingly, BEFV and MVGV viruses were found in animals from the same herd in the commune of Dembeni within two months, evidencing co‐circulation (Figure 1b).
Cattle vector‐borne diseases surveillance should be enhanced in Mayotte and its neighbouring territories. In addition, further epidemiological and entomological studies shall be conducted to understand the extent and the impact of BEF and other cattle vector‐borne infections and to identify the vectors of these viruses on the island.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Supporting information
ACKNOWLEDGEMENTS
The authors wish to acknowledge the staff of the Plate‐forme de microbiologie mutalisée (P2M) of Institut Pasteur, Paris for technical assistance with NGS sequencing and ali Ben M'sa from CoopADEM for his support in the logistics on the field. This study was partly supported by the DP One health Indian Ocean (http://www.onehealth-oi.org). This study was also supported by the European Virus Archive goes Global (EVAg) project that has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No. 653316. Raphaëlle Métras initiated this work whilst funded by a Sir Henry Wellcome Fellowship (Grant number 101581/Z/13/Z).
Dacheux L, Dommergues L, Chouanibou Y, et al. Co‐circulation and characterization of novel African arboviruses (genus Ephemerovirus) in cattle, Mayotte island, Indian Ocean, 2017. Transbound Emerg Dis. 2019;66:2601–2604. 10.1111/tbed.13323
The copyright line for this article was changed on 27 September after original online publication
REFERENCES
- Bevan, L. E. W. (1907). Preliminary report on the so‐called stiff‐sickness or 3‐day‐sickness of cattle. Journal of Comparative Pathology and Therapeutics, 20, 104–113. 10.1016/s0368-1742(07)80022-1 [DOI] [Google Scholar]
- Blasdell, K. R. , Adams, M. M. , Davis, S. S. , Walsh, S. J. , Aziz‐Boaron, O. , Klement, E. , … Walker, P. J. (2013). A reverse‐transcription PCR method for detecting all known ephemeroviruses in clinical samples. Journal of Virological Methods, 191, 128–135. 10.1016/j.jviromet.2013.04.011 [DOI] [PubMed] [Google Scholar]
- Dacheux, L. , Cervantes‐Gonzalez, M. , Guigon, G. , Thiberge, J. M. , Vandenbogaert, M. , Maufrais, C. , … Bourhy, H. (2014). A preliminary study of viral metagenomics of French bat species in contact with humans: Identification of new mammalian viruses. PLoS ONE, 9(1), e87194 10.1371/journal.pone.0087194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacheux, L. , Reynes, J. M. , Buchy, P. , Sivuth, O. , Diop, B. M. , Rousset, D. , … Bourhy, H. (2008). A reliable diagnosis of human rabies based on analysis of skin biopsy specimens. Clinical Infectious Diseases, 47, 1410–1417. 10.1086/592969 [DOI] [PubMed] [Google Scholar]
- Dommergues, L. , Viarouge, C. , Métras, R. , Youssouffi, C. , Sailleau, C. , Zientara, S. , … Cêtre‐Sossah, C. (2019). Evidence of bluetongue and Epizootic Haemorrhagic disease circulation on the island of Mayotte. Acta Tropica, 191, 24–28. 10.1016/j.actatropica.2018.12.037 [DOI] [PubMed] [Google Scholar]
- Girard, S. , Favre, J. , Madi, T. , & Cardinale, E. (2009). Premiers résultats. Bulletin épidémiologique du SESAM Mai 2009 (Système d’épidémiosurveillance animale à Mayotte)[First results. Epidemiological Bulletin update of SESAM (Animal Epidemiosurveillance System in Mayotte)].
- Guindon, S. , Dufayard, J. F. , Lefort, V. , Anisimova, M. , Hordijk, W. , & Gascuel, O. (2010). New algorithms and methods to estimate maximum‐likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biology, 59, 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Mareuil, F. , Doppelt‐Azeroual, O. , & Ménager, H. (2017). A public Galaxy platform at Pasteur used as an execution engine for web services [version 1; not peer reviewed]. F1000Research, 6, 1030 10.7490/f1000research.1114334.1 [DOI] [Google Scholar]
- Matranga, C. B. , Gladden‐Young, A. , Qu, J. , Winnicki, S. , Nosamiefan, D. , Levin, J. Z. , & Sabeti, P. C. (2016). Unbiased deep sequencing of RNA viruses from clinical samples. Journal of Vizualised Experiments, 113, 54117 10.3791/54117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métras, R. , Cavalerie, L. , Dommergues, L. , Mérot, P. , Edmunds, W. J. , Cardinale, E. (2016). The epidemiology of rift valley fever in Mayotte: Insights and perspectives from 11 years of data. PLoS Neglected Tropical Diseases, 10(6), e0004783 10.1371/journal.pntd.0004783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonbak, S. , Berber, E. , Yoruk, M. D. , Azkur, A. K. , Pestil, Z. , & Bulut, H. (2013). A large‐scale outbreak of bovine ephemeral fever in Turkey. Journal of Veterinary Medicine and Sciences, 75, 1511–1514. 10.1292/jvms.13-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortosa, P. , Pascalis, H. , Guernier, V. , Cardinale, E. , Le Corre, M. , Goodman, S. M. , & Dellagi, K. (2012). Deciphering arboviral emergence within insular ecosystems. Infection, Genetics and Evolution, 12, 1333–1339. 10.1016/j.meegid.2012.03.024 [DOI] [PubMed] [Google Scholar]
- Troupin, C. , Dacheux, L. , Tanguy, M. , Sabeta, C. , Blanc, H. , Bouchier, C. , … Bourhy, H. (2016). Large‐scale phylogenomic analysis reveals the complex evolutionary history of rabies virus in multiple carnivore hosts. PLoS Pathogens, 12, e1006041 10.1371/journal.ppat.1006041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, P. J. (2005). Bovine ephemeral fever in Australia and the world. Current Topics in Microbiology and Immunology, 292, 57–80. [DOI] [PubMed] [Google Scholar]
- Walker, P. J. , & Klement, E. (2015). Epidemiology and control of bovine ephemeral fever. Veterinary Research, 46, 124 10.1186/s13567-015-0262-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, F. , & Qiu, C. (2012). Phylogenetic relationships of the glycoprotein gene of bovine ephemeral fever virus isolated from mainland China, Taiwan, Japan, Turkey, Israel and Australia. Virology Journal, 9, 268 10.1186/1743-422X-9-268 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


