Summary
Plants endure challenging environments in which they are constantly threatened by diverse pathogens. The soil‐borne fungus Verticillium dahliae is a devastating pathogen affecting many plant species including cotton, in which it significantly reduces crop yield and fiber quality. Melatonin involvement in plant immunity to pathogens has been reported, but the mechanisms of melatonin‐induced plant resistance are unclear. In this study, the role of melatonin in enhancing cotton resistance to V. dahliae was investigated. At the transcriptome level, exogenous melatonin increased the expression of genes in phenylpropanoid, mevalonate (MVA), and gossypol pathways after V. dahliae inoculation. As a result, lignin and gossypol, the products of these metabolic pathways, significantly increased. Silencing the serotonin N‐acetyltransferase 1 (GhSNAT1) and caffeic acid O‐methyltransferase (GhCOMT) melatonin biosynthesis genes compromised cotton resistance, with reduced lignin and gossypol levels after V. dahliae inoculation. Exogenous melatonin pre‐treatment prior to V. dahliae inoculation restored the level of cotton resistance reduced by the above gene silencing effects. Melatonin levels were higher in resistant cotton cultivars than in susceptible cultivars after V. dahliae inoculation. The findings indicate that melatonin affects lignin and gossypol synthesis genes in phenylpropanoid, MVA, and gossypol pathways, thereby enhancing cotton resistance to V. dahliae.
Keywords: cotton, gossypol, lignin, melatonin, Verticillium dahliae
Significance Statement
This study unveils that melatonin plays a role in the interaction between cotton and a fungal pathogen. Melatonin affects lignin and gossypol synthesis genes in phenylpropanoid, MVA, and gossypol pathways, thereby enhancing cotton resistance to Verticillium dahliae.
Introduction
The bio‐molecule melatonin (N‐acetyl‐5‐methoxytryptamine) is found in almost all living kingdoms, including animals and plants (Arnao and Hernandez‐Ruiz, 2014; Nawaz et al., 2015). It has been characterized as a neurohormone with wide‐ranging roles in animals, including the regulation of circadian rhythms, immune system, reactive oxygen species (ROS), sleep, food intake, mood, and body temperature (Dollins et al., 1994; Rodriguez et al., 2004; Brainard et al., 2011; Carrillo‐Vico et al., 2013). Melatonin was first identified in plants in 1995 (Dubbels et al., 1995; Hattori et al., 1995) and has drawn increased attention from phytologists due to its wide existence and versatile functions in the plant kingdom. In plant development, for example, it regulates seed germination, root development, photoprotection, flowering, leaf senescence, seed yield, and fruit ripening (Wang et al., 2012; Byeon and Back, 2014; Byeon et al., 2014b; Zhang et al., 2014; Sun et al., 2015). Plant melatonin also alleviates abiotic stresses from cold, heat, drought, salt, to heavy metals (Shi et al., 2015b,d; Wei et al., 2015; Li et al., 2016b,c; Ding et al., 2018). Studies have implicated melatonin in plant immunity (Yin et al., 2013; Lee et al., 2015; Shi et al., 2015c,d; Mandal et al., 2018), but the underlying mechanisms remain unclear.
Regulating plant innate immunity offers a promising and sustainable approach for controlling microbial diseases. Plants have evolved two innate immunity strategies against pathogens (Chisholm et al., 2006; Jones and Dangl, 2006). The first strategy, pattern‐triggered immunity, involves recognition of microbial‐associated molecular patterns by host pattern‐recognizing receptors. The second strategy, known as effector‐triggered immunity (ETI), confers resistance through the recognition of effectors released from pathogens by intracellular resistance (R) proteins (Boller and Felix, 2009; Dodds and Rathjen, 2010; Cook et al., 2015). The downstream responses triggered by these two innate immunity strategies are not independent but instead partially overlap, including expression induction of pathogenesis‐related (PR) genes, phytohormone homeostasis regulation, ROS production, and secondary metabolite accumulation (La Camera et al., 2004; Pieterse et al., 2009; Feng and Shan, 2014).
Phenylpropanoids function as inducible antimicrobial compounds and as signaling molecules in plant–pathogen responses (Dixon et al., 2002; Naoumkina et al., 2010). Moreover, phenylpropanoid metabolism is the most important plant metabolic pathway during plant defense against biotic stress (La Camera et al., 2004; Cass et al., 2015). Lignin biosynthesis is a downstream branch of the phenylpropanoid pathway. Specifically, the pathway synthesizes monolignols, which are substrates of lignin polymerization (Boerjan et al., 2003; Mottiar et al., 2016). Before pathogen inoculation, lignin can enhance plant mechanical strength and thicken cell walls to form a physical barrier and inhibit pathogen invasion and colonization (Hu et al., 2018). At the same time, lignin accumulation in the infected cells not only inhibits the spread of pathogens and pathogen‐produced toxins and enzymes but also prevents pathogens from extracting water and nutrients from host plants (Mottiar et al., 2016).
Most plants produce specialized secondary metabolites that confer pathogen resistance (Dixon, 2001). Gossypol is a phytoalexin exclusively biosynthesized in Gossypium plants and represents a group of cadinene‐type sesquiterpene aldehydes with defense functions. In cotton (Gossypium spp.), the presence of gossypol can contribute to pathogen defense (Sunilkumar et al., 2006; Mao et al., 2007). Gossypol is synthesized via the mevalonate (MVA) and gossypol pathways (Heinstein et al., 1970; Chen et al., 1995, 1996; Liu et al., 1999; Luo et al., 2001; Tian et al., 2018). Although the MVA pathway that converts acetyl‐CoA to farnesyl pyrophosphate (FPP) is found in almost all plants, the conversion of FPP to hemigossypol in the gossypol pathway is exclusive to Gossypium. Gossypol is subsequently biosynthesized by the free‐radical coupling of two molecules of hemigossypol (Benedict et al., 2006).
Cotton is an important industrial and economic crop and is cultivated widely around the world (Sunilkumar et al., 2006). Verticillium wilt caused by the hemibiotrophic fungus Verticillium dahliae is a soil‐borne vascular disease affecting many plant species, including cotton (Wang et al., 2004; Mo et al., 2015; Zhang et al., 2016b,c). Because its hyphae reside in the vascular tissue of plants, V. dahliae is extremely difficult to control using fungicides. The dormant microsclerotia of V. dahliae can survive in soil for many years (Klosterman et al., 2009; Gao et al., 2011b; Zhao et al., 2016). The typical symptoms of Verticillium wilt in cotton plants include leaf chlorosis and wilt, leaf defoliation, vascular tissue browning, and plant death (Xu et al., 2011; Li et al., 2016a; Zhang et al., 2018a,b). Its widespread and destructive effect has led to huge economic losses for the cotton industry in China (Xu et al., 2011; Li et al., 2017b; Gong et al., 2018). Verticillium wilt is therefore a critical issue for the cotton industry and will require new research breakthroughs for the development of effective disease control measures.
In this study, we demonstrate that exogenous melatonin enhanced cotton resistance to V. dahliae through changes in the metabolic flux into phenylpropanoid, MVA, and gossypol pathways, subsequently affecting lignin and gossypol accumulation. Meanwhile, suppressing endogenous melatonin levels led to compromised resistance, with reduced lignin and gossypol biosynthesis after inoculation with V. dahliae. The findings therefore reveal that the metabolic mechanism underlying melatonin‐mediated resistance to V. dahliae in cotton involves accelerated lignin biosynthesis and the antifungal activity of the phytoalexin gossypol.
Results
Exogenous melatonin enhanced cotton resistance to V. dahliae
After pre‐treatments with different melatonin concentrations (0, 10, 25, 50 or 100 μm), 2‐week‐old TM‐1 cotton seedlings were inoculated with a V. dahliae spore suspensions. Different responses to the disease infection were observed in melatonin‐pre‐treated and control seedlings (Figure 1a). The symptoms, including wilt, chlorosis, and dark brown streaks in stems, were much more severe in the control than in pre‐treated seedlings at 22 days post inoculation (dpi) (Figure 1a,b). The disease index increased with prolonged dpi, but disease development was slower in the pre‐treated seedlings. At 22 dpi, the disease index was 56, 50, 15, 21 and 34% for the control, 10, 25, 50 and 100 μm melatonin‐pre‐treated seedlings, respectively (Figure 1d). While almost all control seedlings were dead at 28 dpi, seedlings pre‐treated with 25 or 50 μm melatonin showed less stunting and chlorosis and had a lower disease index (36 and 47%, respectively) (Figure 1d). In addition, fungal recovery assays indicated reduced fungal hyphae in the stems of pre‐treated seedlings (Figure 1c), and real‐time quantitative polymerase chain reaction (PCR) analysis showed a significantly lower quantity of V. dahliae in the leaves of pre‐treated seedlings compared with the control (Figure 1e).
Figure 1.
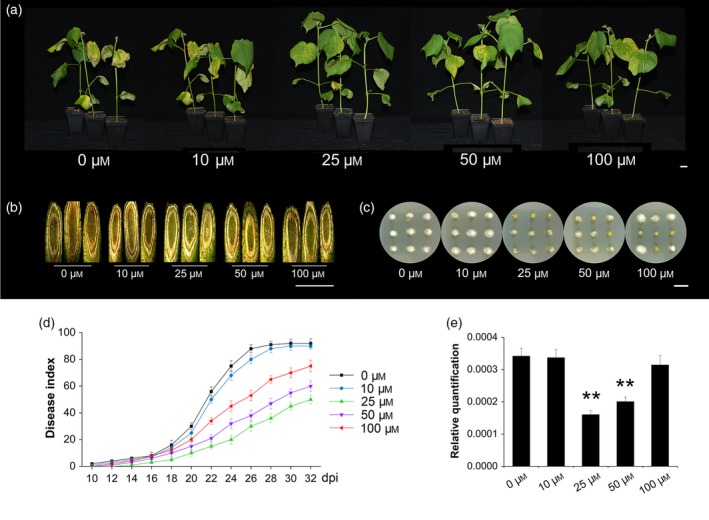
Effect of exogenous melatonin pre‐treatment on cotton resistance to Verticillium dahliae.
(a) Disease symptoms of cotton plants pre‐treated with different melatonin concentrations (0, 10, 25, 50, and 100 μm) following inoculation with V. dahliae strain Vd086 at 22 dpi. Bar = 1 cm.
(b) Fungal accumulation in the stems of cotton plants pre‐treated with different concentrations of melatonin at 22 dpi. Bar = 1 cm.
(c) Growth of hyphae recovered from the V. dahliae‐infected cotton. The stem sections were plated on PDA medium, incubated at 25°C, and photographed at 5 days post plating. Bar = 1 cm.
(d) Disease indexes of the melatonin‐pre‐treated cotton were determined from 10 dpi to 32 dpi. The values are the means ± SD; n = 32.
(e) Detection for the relative quantification of fungal biomass. Total DNA from the leaves of melatonin‐pre‐treated cotton plants inoculated with V. dahliae for 22 days was extracted as the template for fungal biomass detection by qPCR. GhUBQ7 was used as the internal control. The values are the means ± SD; n = 3. Statistical analyses were performed using Student's t test. *, P < 0.05; **, P < 0.01.
To determine whether exogenous melatonin acted as a fungicide and protected the pre‐treated cotton seedlings against the disease, V. dahliae was cultured on potato dextrose agar (PDA) medium with different melatonin concentrations (0, 10, 25, 50 or 100 μm) (Figure S1a). There were no significant differences in the diameters of V. dahliae mycelium and sclerotium between the treatments (Figure S1b,c). Therefore, melatonin itself did not have a fungicidal effect against the disease, suggesting instead that melatonin might act as a regulator to enhance cotton host resistance against Verticillium wilt. Based on the above assay results, the 25 μm melatonin concentration was used for the subsequent analyses.
Transcriptome sequencing
To gain insights into how exogenous melatonin might induce a defensive mechanism leading to enhanced resistance to V. dahliae, a comparative transcriptome analysis was performed. Complex perception, signal transduction, and exchange of chemicals usually occur in the early stages of pathogen inoculation (Kunkel and Brooks, 2002; Jones and Dangl, 2006), so the selected sampling time points were 0, 6, 12, 24, and 48 h post V. dahliae inoculation. Ten root samples were obtained at each time point from the control (V_0, V_6, V_12, V_24, and V_48) and from seedlings pre‐treated with 25 μm melatonin (M_V_0, M_V_6, M_V_12, M_V_24, and M_V_48). All samples were used for transcriptome sequencing with three biological replicates (Table S1). In total, 870.22 million raw reads were obtained for the V libraries (V_0, V_6, V_12, V_24, and V_48), and 833.47 million raw reads were obtained for the M_V libraries (M_V_0, M_V_6, M_V_12, M_V_24, and M_V_48). After removing adapter and low‐quality sequences along with contaminated reads, 124.93 Gb and 119.94 Gb high‐quality clean bases remained from the V and M_V libraries, respectively. Using the G. hirsutum genome, the number of mapped clean reads was 46.88–57.18 million for the V libraries (93.38–94.93% mapped rate) and 41.91–56.47 million for the M_V libraries (93.63–95.00% mapped rate) (Table S2). Comparing the melatonin‐pre‐treated and control seedlings after V. dahliae inoculation, 9114 differentially expressed genes (DEGs) were identified, with 4577 significantly upregulated and 4537 significantly downregulated genes (Table S3).
Exogenous melatonin promoted lignin accumulation after V. dahliae inoculation
Comparative transcriptome changes for genes involved in phenylpropanoid pathway were examined. Figure 2(a) shows an overview of phenylpropanoid pathway and the melatonin‐induced expression pattern changes of nine genes in this pathway during the defense response of cotton to V. dahliae infection. Phenylpropanoids derive from the amino acid phenylalanine, with different steps of the pathway catalyzed by nine types of enzymes: ammonia lyase (PAL), cinnamate 4‐hydroxylase (C4H), 4‐coumarate‐CoA ligase (4CL), cinnamoyl‐CoA reductase (CCR), hydroxycinnamoyl transferase (HCT), caffeoyl‐CoA O‐methyltransferase (CCoAMT), ferulic acid 5‐hydroxylase (F5H), O‐methyltransferase 1 (OMT1), and cinnamyl alcohol dehydrogenase (CAD). The pathway produces guaiacyl (G), syringyl (S), and p‐hydroxyphenyl (H) lignin, and the three types of monolignols polymerize to form lignin. Gene expression levels were illustrated using a heatmap (Figure 2a) and estimated by log2(fold change); fold change indicates the fragments per kilobase of transcript per million (FPKM) reads sequenced ratio of the melatonin‐pre‐treated samples to the control samples. Although there were some cases of downregulation or non‐significant upregulation for some of the genes at different time points, most of the nine types of genes in this pathway were upregulated in melatonin‐pre‐treated seedlings within 48 h post inoculation (hpi) compared with the non‐melatonin‐pre‐treated control. Generally, the upregulation of phenylpropanoid pathway genes began at 6 hpi, except for GhC4H which was upregulated at an earlier time point after V. dahliae infection. The peaks of relative expression of these genes occurred between 12 and 48 hpi.
Figure 2.
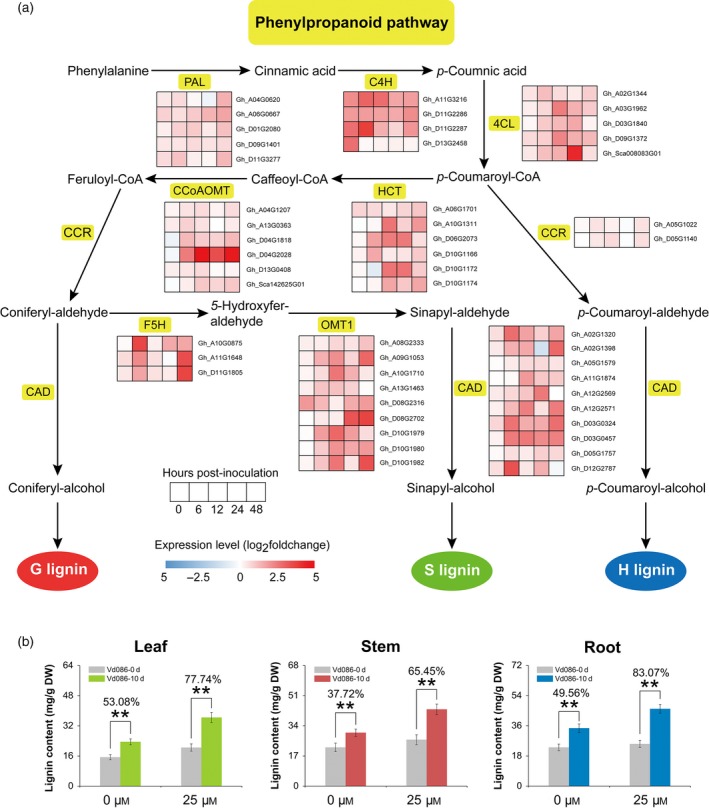
Expression profiles of phenylpropanoid pathway genes and the lignin content in different cotton tissues.
(a) Overview of phenylpropanoid pathway and the melatonin‐induced changes in the expression of phenylpropanoid pathway genes after Verticillium dahliae inoculation. Expression levels are indicated by the heatmap at 0, 6, 12, 24, and 48 h post inoculation, estimated using log2(fold change) for each transcript. Fold change is the ratio of FPKM (fragments per kilobase of transcript per million reads sequenced) for melatonin‐pre‐treated samples to non‐pre‐treated control samples. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4‐hydroxylase; 4CL, 4‐coumarate:CoA ligase; HCT, hydroxycinnamoyl transferase; CCoAOMT, caffeoyl‐CoA O‐methyltransferase; F5H, ferulic acid 5‐hydroxylase; OMT1, O‐methyltransferase 1; CCR, cinnamoyl‐CoA reductase; CAD, cinnamyl alcohol dehydrogenase.
(b) Melatonin‐induced changes in lignin content in different tissues after V. dahliae inoculation. Values are the means ± SD; n = 6. Statistical analyses were performed using Student's t‐test. *, P < 0.05; **, P < 0.01.
To validate the transcriptome expression data, four phenylpropanoid pathway genes, Gh4CL (Gh_Sca008083 g01), GhHCT (Gh_D06G2073), GhCCoAMT (Gh_D04G2028), and GhOMT1 (Gh_D10G1979), were randomly selected for qPCR analysis. The fold changes for the qPCR analysis indicated the relative expression ratio of the melatonin‐pre‐treated samples to the control, and the transcriptome profile fold changes were similar to those from the qPCR analysis (Figure S2a).
Based on the upregulation of genes related to phenylpropanoid pathway, we also investigated changes in lignin in stems by safranin and fast green staining at 10 dpi (Figure S3). The red staining area in stems after V. dahliae inoculation was larger than in the non‐inoculation control, indicating that the pathogen infection induced lignin biosynthesis (Figure S3a,b). The red staining area was much larger in the stems of melatonin‐pre‐treated samples compared the non‐melatonin‐pre‐treated control after V. dahliae inoculation (Figure S3c,d), suggesting a combined effect of melatonin pre‐treatment and V. dahliae inoculation resulting in accelerated lignin biosynthesis. Figure 2(b) shows the lignin contents for different tissues at 0 dpi and 10 dpi. Although lignin levels were induced by pathogen infection in both melatonin‐pre‐treated and control seedlings, the levels were higher in the tissues of the pre‐treated seedlings compared with the control after V. dahliae inoculation. At 10 dpi, lignin content increased by 77.74% in leaves, 65.45% in stems, and 83.07% in roots of the melatonin‐pre‐treated samples, while the increases in the control were 53.08, 37.72, and 49.56%, respectively. Furthermore, lignin content was higher in the melatonin‐pre‐treated samples than in the control at 0 dpi, especially in leaves and stems, indicating that melatonin pre‐treatment increased lignin biosynthesis in cotton seedlings before V. dahliae inoculation (Figure 2b). Therefore, exogenous melatonin led to increased lignin biosynthesis through phenylpropanoid pathway, thereby enhancing the resistance of cotton to V. dahliae.
Exogenous melatonin promoted gossypol accumulation after V. dahliae inoculation
To investigate whether gossypol, a specialized phytoalexin in Gossypium, was involved in the exogenous melatonin‐induced resistance of cotton to V. dahliae, the expression of genes in MVA and gossypol pathways was investigated. Figure 3(a) illustrates these pathways and shows the expression pattern changes of genes in both pathways when cotton seedlings were infected with V. dahliae. The acetyl‐CoA precursor enters MVA pathway, and the production of FPP is catalyzed by acyl CoA‐cholesterol acyltransferase (ACAT), 3‐hydroxy‐3‐methylglutaryl‐CoA (HMG‐CoA) synthase (HMGS), HMG‐CoA reductase (HMGR), mevalonate kinase (MVK), phosphomevalonate kinase (MVP), diphosphomevalonate decarboxylase (PMD), and FPP synthase (FPS). The heatmap shows that only GhHMGR, the key gene for the conversion of HMG‐CoA to MVA in MVA pathway, was significantly upregulated in melatonin‐pre‐treated samples at certain infection time points compared with the control (Figure 3a).
Figure 3.
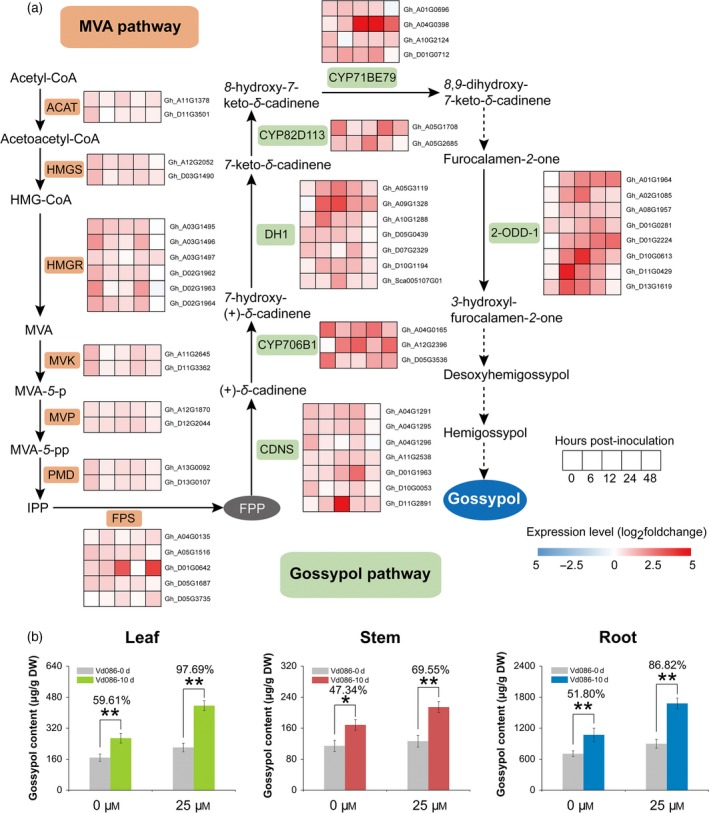
Expression profiles of MVA and gossypol pathway genes and gossypol content in different cotton tissues.
(a) Overview of MVA and gossypol pathways and the melatonin‐induced changes in the expression profiles of MVA and gossypol pathway genes after Verticillium dahliae inoculation. The expression levels are indicated by the heatmap at 0, 6, 12, 24, and 48 h post inoculation, estimated using log2(fold change) for each transcript. Fold change is the ratio of FPKM (fragments per kilobase of transcript per million reads sequenced) for melatonin‐pre‐treated samples to non‐pre‐treated control samples. Dashed arrows indicate unidentified reactions. ACAT, acyl CoA‐cholesterol acyltransferase; HMGS, 3‐hydroxy‐3‐methylglutaryl‐coenzyme‐A (HMG‐CoA) synthase; HMGR, HMG‐CoA reductase; MVK, mevalonate kinase; MVP, phosphomevalonate kinase; PMD, diphosphomevalonate decarboxylase; FPS, farnesyl diphosphate synthase; CDNS, (+)‐δ‐cadinene synthase; CYP706B1, CYP82D113, and CYP71BE79DH1, three cytochrome P450 monooxygenases; DH1, alcohol dehydrogenase; 2‐ODD‐1, 2‐oxoglutarate/Fe(II)‐dependent dioxygenase.
(b) Melatonin‐induced changes in gossypol content in different tissues after V. dahliae inoculation. The values are the means ± SD; n = 6. Statistical analyses were performed using Student's t‐test. *, P < 0.05; **, P < 0.01.
In Gossypium, gossypol pathway is one of the downstream branches of MVA pathway. Although gossypol pathway is not fully characterized, six critical enzymes have been identified, including (+)‐δ‐cadinene synthase (CDNS), CYP706B1, alcohol dehydrogenase (DH1), CYP82D113, CYP71BE79 and 2‐oxoglutarate/Fe(II)‐dependent dioxygenase (2‐ODD‐1). Expression analysis of these six genes revealed that they were upregulated in melatonin‐pre‐treated samples during V. dahliae infection (Figure 3a), and the peaks of relative expression occurred between 6 and 48 hpi. Moreover, we observed more significant upregulation of genes in the downstream positions of gossypol pathway, suggesting that the metabolic flux from MVA pathway was more prone to enter gossypol pathway to biosynthesize gossypol in melatonin‐pre‐treated seedlings after V. dahliae inoculation.
To validate the transcriptome expression data for genes involved in both MVA and gossypol pathways, the following genes were randomly selected for qPCR analysis: GhCYP706B1 (Gh_A12G2396), GhCYP82D113 (Gh_A05G2685), GhCYP17BE79 (Gh_A04G0398), and Gh2‐ODD‐1 (Gh_D10G0613). The transcriptome profile fold changes were consistent with those from the qPCR analysis (Figure S2b).
The gossypol contents in different cotton tissues at 0 dpi and 10 dpi are listed in Figure 3(b). Pathogen inoculation induced gossypol accumulation with or without exogenous melatonin pre‐treatment, but melatonin pre‐treatment led to a much higher gossypol level after V. dahliae inoculation. In the control, gossypol content increased by 59.61% in leaves, 47.34% in stems, and 51.80% in roots at 10 dpi; in contrast, the increases were 97.69, 69.55, and 86.82%, respectively, with melatonin pre‐treatment. Therefore, gossypol biosynthesis was induced and intensified by exogenous melatonin plus V. dahliae inoculation, suggesting that MVA pathway, gossypol pathway, and the synthesized product gossypol are involved in melatonin‐mediated resistance to V. dahliae in cotton.
Identification of melatonin biosynthesis genes in cotton
Melatonin is biosynthesized from tryptophan in plants through the sequential action of several enzymes including tryptophan decarboxylase, tryptamine 5‐hydroxylase (T5H), serotonin N‐acetyltransferase (SNAT), and caffeic acid O‐methyltransferase (COMT) (Figure S4) (Kang et al., 2013; Byeon et al., 2014a; Lee et al., 2014; Arnao and Hernandez‐Ruiz, 2018). Using AtSNAT1 (AT1G32070) and AtCOMT (AT5G54160) as queries, BLAST analyses were performed to identify their homologs in the cotton genome. We obtained a putative SNAT1 gene (Gh_A02G0898 in the A subgenome and Gh_D02G1063 in the D subgenome) and a putative COMT gene (Gh_A12G2227 in the A subgenome and Gh_D12G2714 in the D subgenome). The putative SNAT1 gene encoded a predicted protein of 251 amino acid residues with a conserved N‐acetyltransferase motif (pfam13508) from amino acid 159 to 232; the sequence identity with the AtSNAT1 protein was 60.75% (Figure S5a). The putative COMT gene encoded a predicted protein of 365 amino acid residues with a conserved dimerization motif (pfam08100) and a conserved O‐methyltransferase motif (pfam00891) from amino acid residues 34 to 83 and 127 to 347, respectively; the sequence identity with the AtCOMT protein was 78.90% (Figure S6a). Functional complementation experiments were performed by transforming the putative SNAT1 (Gh_D02G1063) and COMT (Gh_D12G2714) genes into Arabidopsis snat1 and comt mutants, respectively. The melatonin‐deficient phenotypes of the snat1 and comt mutants were both complemented by the introduced sequences (Figure S7), indicating the involvement of the putative SNAT1 (Gh_D02G1063) and COMT (Gh_D12G2714) genes in melatonin biosynthesis in cotton. The genes are hereafter referred to as GhSNAT1 and GhCOMT.
Phylogenetic analysis was performed for the GhSNAT1 and GhCOMT proteins using 20 homologs from 16 and 15 species, respectively, representing algae, fern, moss, gymnosperms, monocotyledons, and dicotyledons (Tables S4 and S5). The analysis indicated that both SNAT1 and COMT proteins are ancient and conserved in plants (Figures S5b and S6b). Expression profiling in different cotton tissues revealed higher expression of the D subgenome homologs than those from the A subgenome (Figures S5c and S6c). We therefore selected the D subgenome homologs of both genes for further analysis.
Suppressing endogenous melatonin led to susceptibility of cotton to V. dahliae
To investigate the effect of endogenous melatonin on cotton resistance to V. dahliae, we reduced endogenous melatonin levels by suppressing GhSNAT1 and GhCOMT expression using TRV‐based virus‐induced gene silencing (VIGS) technology. TRV:CLA1, TRV:PGF, and TRV:CLA1+PGF plants were used as positive controls to validate the effects of the gene silencing and co‐silencing systems. TRV:CLA1 and TRV:PGF plants had bleaching and glandless phenotypes, respectively, from the first true leaves. TRV:CLA1+PGF plants exhibited both the bleaching and glandless phenotypes from the first true leaves (Figure S8). These observed phenotypes confirmed that the gene silencing and co‐silencing systems used in this study were stable and effective. Two weeks after Agrobacterium infiltration, GhSNAT1 and GhCOMT expression levels were significantly suppressed in the leaves, stems, and roots of TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants compared with the TRV:00 negative control, according to qPCR and reverse transcription PCR (RT‐PCR) assays (Figure S9a,b). Similarly, melatonin levels in the three plant tissues were also significantly reduced (Figure S9c). When the VIGS cotton seedlings were inoculated with V. dahliae at the two true‐leaf stage, the disease infection in TRV:SNAT1 and TRV:COMT plants was more severe than in the TRV:00 control at 22 dpi (Figure 4a,b). Most significantly, TRV:SNAT1+COMT plants were almost dead at this time point. The disease indexes increased sharply from 10 to 28 dpi, but the index always remained higher in TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT than in TRV:00 plants (Figure 4c). Accordingly, the amount of fungal biomass was significantly higher in TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT plants (Figure 4d). Therefore, suppression of endogenous melatonin biosynthesis led to an increased susceptibility of cotton to V. dahliae.
Figure 4.
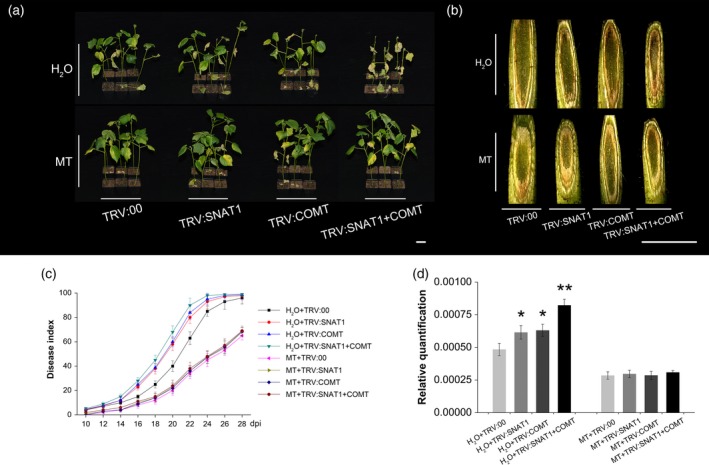
Recovery of the compromised resistance to Verticillium dahliae in VIGS plants by exogenous melatonin pre‐treatment.
(a) Disease symptoms of H2O‐pre‐treated and melatonin‐pre‐treated TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT inoculated with V. dahliae; photographs were taken at 20 dpi. Bar = 1 cm.
(b) Fungal accumulation in the stems of H2O‐pre‐treated and melatonin‐pre‐treated TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT inoculated with V. dahliae at 20 dpi. Bar = 1 cm.
(c) Disease indexes of H2O‐pre‐treated and melatonin‐pre‐treated TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT inoculated with V. dahliae were determined from 10 dpi to 28 dpi. The values are the means ± SD; n = 32.
(d) Relative quantification of fungal biomass. Total DNA from leaves of H2O‐pre‐treated and melatonin‐pre‐treated TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT inoculated with V. dahliae for 20 days was extracted as the template for fungal biomass detection by qPCR. GhUBQ7 was used as the internal control. The values are the means ± SD; n = 3. Statistical analyses were performed using Student's t‐test. *, P < 0.05; **, P < 0.01.
The lignin contents of different tissues in TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants were measured before and after V. dahliae inoculation. At 0 dpi, the lignin contents did not differ among TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants (Figure 5a). In contrast to the 55.29% increase in lignin accumulation in TRV:00 roots at 10 dpi, the increases were only 16.97, 14.63, and 9.69% in TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT roots, respectively (Figure 5a). The lignin accumulation in other tissues of TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT plants followed a trend similar to that observed in 10 dpi roots (Figure 5a). These results may be attributable to the inhibited expression of lignin biosynthesis genes in phenylpropanoid pathway, including GhPAL, Gh4CL, GhHCT, GhCCoAMT, GhOMT1, and GhCAD, as these genes had reduced expression levels in TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants at 12 hpi (Figure 5b).
Figure 5.
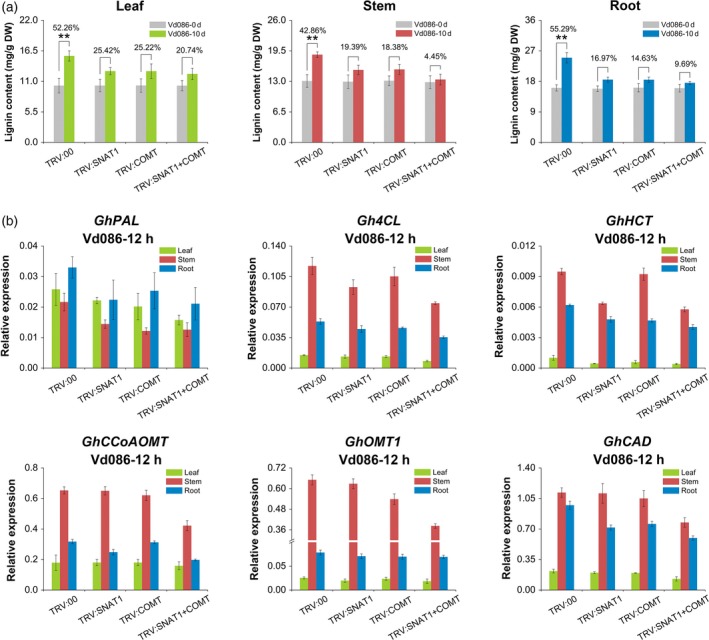
Lignin content and expression levels of phenylpropanoid pathway genes in VIGS plants. (a) Melatonin‐induced changes in lignin content in different tissues of TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT inoculated with V. dahliae for 10 days. The values are the means ± SD; n = 3. (b) Melatonin‐induced changes in the expression levels of phenylpropanoid pathway genes in TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT at 12 dpi. The values are the means ± SD; n = 3. Statistical analyses were performed using Student's t‐test. *, P < 0.05; **, P < 0.01.
Gossypol contents were not significantly different in TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants at 0 dpi (Figure 6a). At 10 dpi, gossypol accumulation in response to V. dahliae infection was suppressed in the leaves, stems, and roots of TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT plants compared with TRV:00 plants (Figure 6a). Specifically, the gossypol content only increased by 15.39, 14.86, and 8.51% in TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT roots at 10 dpi, respectively, in contrast to the 55.92% increase in TRV:00 roots. The expression levels of six known gossypol pathway genes (GhHMGR, GhFPS, GhCDNS, GhCYP706B1, GhCYP82D113, and Gh2‐ODD‐1) were also decreased in TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants compared with TRV:00 plants at 12 hpi (Figure 6b).
Figure 6.
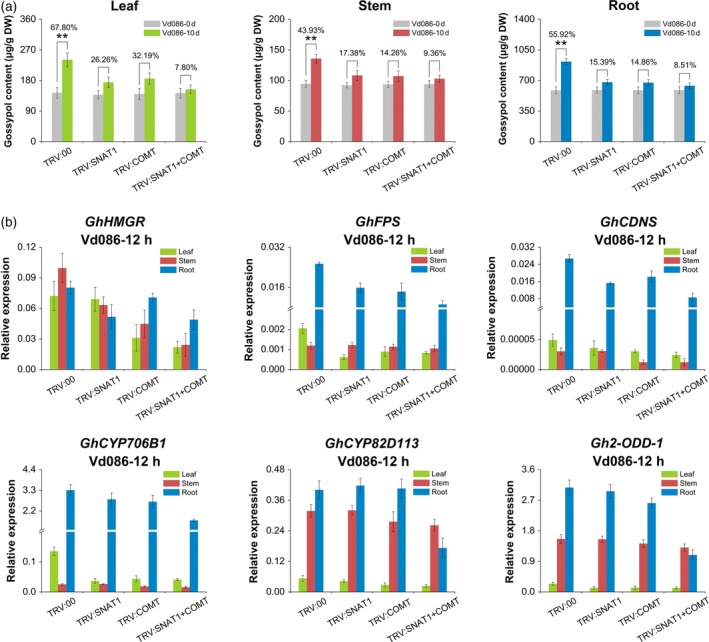
Gossypol content and expression levels of MVA and gossypol pathway genes in VIGS plants.
(a) Melatonin‐induced changes in gossypol content in different tissues of TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT inoculated with V. dahliae for 10 days. The values are the means ± SD; n = 3.
(b) Melatonin‐induced changes in the expression levels of MVA and gossypol pathway genes in TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1 + COMT at 12 days dpi. The values are the means ± SD; n = 3. Statistical analyses were performed using Student's t‐test. *, P < 0.05; **, P < 0.01.
Exogenous melatonin pre‐treatment of the above VIGS plants restored their resistance to the disease. When TRV:00, TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants were pre‐treated with 25 μm melatonin before V. dahliae inoculation, both the exhibited disease symptoms and severity resembled that observed in TRV:00 plants (Figure 4a and b). Notably, their disease indexes followed a similar trend, and there were no significant differences in the amount of fungal biomass in the plant tissues (Figure 4c and d). Lignin and gossypol levels in different tissues of TRV:SNAT1, TRV:COMT, and TRV:SNAT1+COMT plants after V. dahliae inoculation were also restored by exogenous melatonin pre‐treatment (Figure S10a,b). All these findings suggested that suppressing endogenous melatonin in cotton plants increased the susceptibility of cotton to V. dahliae through the downregulation of lignin and gossypol biosynthesis.
Melatonin levels in different cotton cultivars after V. dahliae inoculation
To investigate whether endogenous melatonin content is related to V. dahliae resistance in cotton, we compared the melatonin contents of two V. dahliae‐resistant (JM‐958 and ZZM‐2) and two V. dahliae‐susceptible (Ejing‐1 and JM‐1) cultivars. Figure 7(a) shows their disease symptoms at 22 dpi. At this time point, the susceptible plants were almost all dead, with disease indexes of 78 and 80% for Ejing‐1 and JM‐1, respectively. In contrast, the resistant cultivars exhibited minimal wilt and chlorosis, with disease indexes of 21 and 18% for JM‐958 and ZZM‐2, respectively (Figure 7b). The difference in disease resistance was confirmed by an assay for fungal biomass (Figure 7c).
Figure 7.
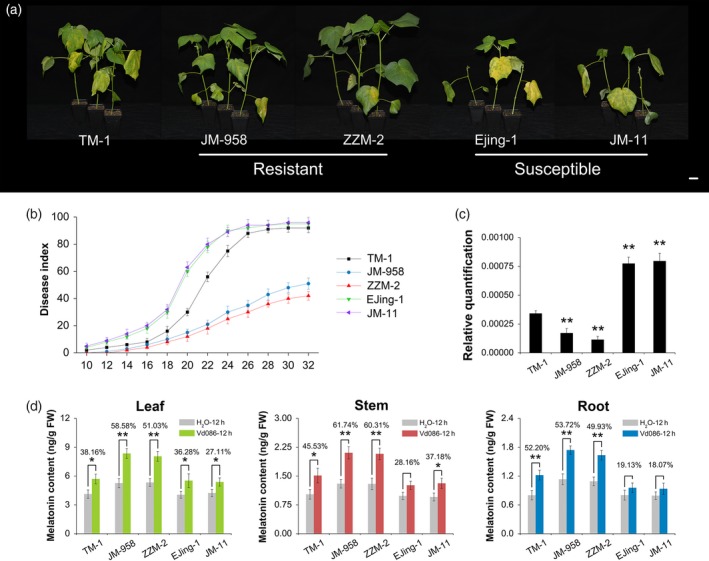
Correlation between melatonin level and resistance to Verticillium dahliae.
(a) Disease symptoms of V. dahliae‐resistant (JM‐958 and ZZM‐2) and V. dahliae‐susceptible (Ejing‐1 and JM‐11) cotton cultivars inoculated with V. dahliae; photographs were taken at 22 days post inoculation (dpi). Bar = 1 cm.
(b) Disease indexes of the resistant and susceptible cotton cultivars were determined from 10 to 32 dpi. The values are the means ± SD; n = 32.
(c) Relative quantification of fungal biomass. Total DNA from leaves of the resistant and susceptible cotton cultivars inoculated with V. dahliae for 22 days was extracted as template for fungal biomass detection by qPCR. GhUBQ7 was used as the internal control. The values are the means ± SD; n = 3.
(d) Pathogen‐induced melatonin levels of the resistant and susceptible cotton cultivars at 12 dpi. The values are the means ± SD; n = 3. Statistical analyses were performed using Student's t‐test. *, P < 0.05; **, P < 0.01.
The melatonin levels in different tissues were higher in the resistant cultivars than in the susceptible cultivars (Figure 7d). Although melatonin was induced in all of the cultivars after V. dahliae inoculation, the induction was more pronounced in the resistant cultivars. At 12 hpi, the melatonin content in the leaves of JM‐958 and ZZM‐2 increased by 58.58 and 51.03%, respectively; in contrast, the increases were only 36.28 and 27.11% in Ejing‐1 and JM‐11 leaves, respectively (Figure 7d). These results indicated that endogenous melatonin is involved in the resistance of these cotton cultivars to V. dahliae.
Discussion
Verticillium wilt is one of the most destructive fungal diseases affecting plant species, and in cotton, it causes great reductions in yield and fiber quality (Xu et al., 2011; Hu et al., 2018). Understanding the mechanisms underlying cotton resistance to V. dahliae is critical for minimizing production loss in the cotton industry, and the related research questions have therefore drawn much interest.
Melatonin helps enhance resistance to V. dahliae in cotton
Melatonin is a biomolecule with a wide range of roles in plants under stress conditions, including plant immunity (Arnao and Hernandez‐Ruiz, 2014). In this study, we pre‐treated cotton seedlings with different concentrations of melatonin (0, 10, 25, 50, and 100 μm) before V. dahliae infection. While the disease symptoms became less severe as the pre‐treatment concentration increased from 0 to 25 μm, the symptoms worsened as the concentration increased from 25 to 100 μm, indicating that the 25 μm concentration was optimal for enhancing cotton resistance to V. dahliae (Figure 1). Suppressing endogenous melatonin by VIGS technology led to susceptibility of cotton to V. dahliae (Figure 4). Co‐silenced TRV:SNAT1+COMT plants with extremely low endogenous melatonin content were more susceptible than TRV:SNAT1 and TRV:COMT plants during V. dahliae infection, indicating a dosage effect of endogenous melatonin on cotton resistance to V. dahliae. In other words, cotton plants became more susceptible to V. dahliae as the endogenous melatonin levels decreased. The compromised resistance to V. dahliae was restored when these VIGS plants were pre‐treated with melatonin before V. dahliae inoculation (Figure 4). We also observed that endogenous melatonin was induced by V. dahliae infection in different cotton cultivars (Figure 7). Furthermore, melatonin itself has no inhibitory effect on the growth of V. dahliae colonies in vitro (Figure S1). These results suggest that melatonin is a regulator involved in the cotton response to V. dahliae infection. This study uncovers that melatonin plays an important role in the interaction between cotton and a fungal pathogen.
Diverse mechanisms of melatonin‐mediated plant resistance to pathogens
Previous studies have shown that melatonin contributes to enhanced plant immunity to different pathogens. Exogenous melatonin enhanced the resistance of Malus to Marssonina apple blotch (Diplocarpon mali) by maintaining steady‐state hydrogen peroxide (H2O2) levels and increasing the activity of plant defense‐related enzymes (Yin et al., 2013). Melatonin pre‐treatment increased nitric oxide (NO) levels and the expression of SA‐related genes to enhance the innate immunity of Arabidopsis to the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 (Shi et al., 2015a,c). Melatonin alters the expression of genes involved in both PAMP‐mediated and ETI‐mediated defense in watermelon leaves during Phytophthora capsici infection (Mandal et al., 2018). Therefore, we propose that melatonin mediates broad‐spectrum pathogen resistance in plants via diverse mechanisms, possibly depending on the disease mechanism of the pathogen. For example, the infection of cotton roots by V. dahliae hyphae in soil leads to the colonization of vascular tissues; from the parasitic to the saprophytic phases, the hyphae and dormant microsclerotia propagate in the infected cotton, resulting in vessel blockage and cotton wilt disease (Gerik and Huisman, 1988).
Secondary metabolites play an important role in plant–pathogen interactions (Piasecka et al., 2015; Kumari et al., 2016). Phenylpropanoid pathway and its product lignin exist in all plant species and enhance resistance to pathogens (Lange et al., 1995; Menden et al., 2007; Bhuiyan et al., 2009). Our transcriptome and metabolic studies show that melatonin pre‐treatment upregulated the expression of phenylpropanoid pathway genes and increased lignin accumulation in cotton seedlings after V. dahliae inoculation (Figures 2 and S3). Consistently, overexpression of GhLac1 and GhLac15, genes related to phenylpropanoid pathway and lignin biosynthesis, was previously found to enhance V. dahliae resistance in cotton (Hu et al., 2018; Zhang et al., 2018a,b). Transcriptional and histochemical analysis of resistant cotton (G. barbadense cv. 7124) revealed that genes involved in phenylpropanoid pathway and lignin accumulation were significantly induced after V. dahliae infection (Xu et al., 2011). Gossypol is a major secondary metabolite specific to Gossypium plants, and it plays a crucial role in the defense against pathogen invasion (Luo et al., 2001). In cotton, V. dahliae infection induces the expression of the gene encoding (+)‐δ‐cadinene synthase (CDNS), a key gossypol pathway enzyme (Townsend et al., 2005). Silencing the GbCAD1 gene, which encodes another key gossypol pathway enzyme, compromises cotton resistance to V. dahliae (Gao et al., 2013). In the present study, exogenous melatonin pre‐treatment upregulated the expression of genes involved in MVA and gossypol pathways and increased gossypol accumulation in cotton seedlings after V. dahliae inoculation (Figure 3). VIGS assays confirmed the involvement of lignin and gossypol in melatonin‐mediated cotton resistance, as lignin induction and gossypol induction were inhibited in melatonin‐suppressed cotton seedlings during pathogen infection (Figure 5). As expected, exogenous melatonin pre‐treatment rescued the inhibition of lignin and gossypol biosynthesis after V. dahliae inoculation (Figure S10). We therefore concluded that melatonin regulates phenylpropanoid, MVA, and gossypol pathways to promote lignin and gossypol biosynthesis, therefore enhancing cotton resistance to V. dahliae.
It is worth noting that plant small‐molecule hormones, including jasmonic acid (JA) and salicylic acid (SA), play key roles in the plant response to V. dahliae infection (Glazebrook, 2005; Campos et al., 2014; Zhang et al., 2016a, 2017). However, we did not observe any significant differences in JA and SA levels (Figure S11) or changes in the expression of genes involved in these hormone signaling pathways between melatonin‐pre‐treated and control samples during V. dahliae infection (Figure S12). We therefore presume that melatonin‐mediated cotton resistance to V. dahliae is likely to be independent of these hormone signaling pathways.
The present findings shed light on the metabolic mechanisms of melatonin‐mediated resistance of cotton to V. dahliae. However, further molecular and genetic evidence is needed to support the identified resistance mechanisms. Wei et al. (2018) recently reported the first melatonin receptor (CAND2/PMTR1) in plants. In our transcriptome data, 86 differentially expressed transcription factor (TF) genes were identified at 12 hpi. Among these TF genes, 59 were significantly upregulated in the melatonin‐pre‐treated samples and included bHLH, bZIP, NAC, and MYB family members (Table S6). Based on these lines of evidence, future studies should focus on the melatonin signaling pathway to uncover melatonin‐mediated mechanisms important for the plant–pathogen response.
Plant–pathogen interactions have led to the evolution of sophisticated defense systems in plants to recognize pathogens and limit their invasion, colonization, and infection. In the context of crop production, inducing or manipulating certain plant systems may enhance the ability of plants to overcome the destructive impact of diseases. In this study, we have demonstrated that melatonin is an important factor regulating plant immunity and that external melatonin application can promote lignin and gossypol synthesis, therefore enhancing cotton resistance to Verticillium wilt. Our findings may help establish available avenues for plant breeding strategies aimed at fungal pathogen resistance via melatonin‐mediated enhanced plant immunity.
Experimental procedures
Plant materials and growth conditions
Gossypium hirsutum cv. TM‐1 is a standard genetic line of upland cotton. Gossypium hirsutum cv. JM‐958 and ZZM‐2 were used as the positive controls and are resistant to V. dahliae. G. hirsutum cv. EJ‐1 and JM‐11 were used as the negative controls and are susceptible to V. dahliae. All cotton accessions were cultivated in a growth chamber at 25°C under a 16 h light/8 h dark cycle. Arabidopsis thaliana Columbia ecotype (Col‐0) and the T‐DNA insertion mutants snat1 (SALK_032239) and comt (SALK_135290) were obtained from the Arabidopsis Biological Resource Center (ABRC, Columbus, OH, USA). Arabidopsis plants were grown in a growth chamber at 22°C under a 16 h light/8 h dark cycle. Nicotiana benthamiana was used for subcellular localization and was grown in a growth chamber at 28°C under a 16 h light/8 h dark cycle. All biological samples used for further analysis, including RNA and DNA extraction and content determination of melatonin, gossypol, and lignin, were frozen in liquid nitrogen and stored at −80°C.
Melatonin pre‐treatment
To study the effects of melatonin on cotton seedlings, 2‐week‐old cotton seedlings with two true leaves visible were treated with different melatonin concentrations (0, 10, 25, 50, or 100 μm) by spraying the aerial parts once a day for 3 days. Control seedlings were treated with 0 μm melatonin concentration or sterile distilled water. After melatonin pre‐treatment, cotton seedlings were inoculated with V. dahliae.
Effect of melatonin on the growth of V. dahliae
To investigate the effect of melatonin on the growth of V. dahliae, the PDA medium containing different concentrations of melatonin, including 0, 10, 25, 50, or 100 μm, was prepared. Next, 0.5‐cm diameter stained blocks cultured for 5 days were plated onto the prepared PDA medium and grown at 25°C in the dark. Each treatment was repeated at least nine times and observed 2 weeks later.
Fungal culture conditions and infection of plants
Vd086, a virulent defoliating V. dahliae strain, was isolated from cotton in Hangzhou, China and grown on PDA medium at 25°C for 5 days. Mycelia were collected and cultured in Czapek's medium for 5 days at 25°C with shaking (150 rpm).
Cotton seedlings at the two true‐leaf stage were inoculated with Vd086 using a sterile needle to inject 20 ml spore suspensions (107 conidia/ml) into the soil. Control plants in each treatment were inoculated with an equal volume of sterile distilled water. The plants inoculated with V. dahliae were cultured in a growth chamber at 25°C under a 16 h light/8 h dark cycle.
Disease assessment after V. dahliae inoculation
The disease index was calculated by assessing at least 32 individual plants per treatment and repeated three times with the following formula: disease index = ((∑disease grade × the number of infected plants)/(total assessed plants × 4)) × 100. The disease severity was assessed according to the following point scale: 0, no visible wilting or yellowing symptoms; 1, one or two cotyledons wilted or dropped off; 2, one true leaf wilted or dropped off; 3, two true leaves wilted or dropped off; and 4, all true leaves dropped off or the whole plant has died (Xu et al., 2012). The assessment was performed every 3 days for at least 1 month over the experiment period.
For the fungal recovery assay, the stems 1 cm above the cotyledons of cotton seedlings were collected at 22 dpi, surface‐sterilized for 20 min using 5% NaClO, then rinsed with sterile distilled water five times. The stem sections were plated on PDA medium and cultured for 5 days at 25°C.
To assess the fungal accumulation in the cotton stems, the stems 1 cm above the cotyledons were collected at 22 dpi, hand sectioned, then observed using a Leica MZ95 microscope (Leica, Wetzlar, Germany).
To detect and quantify the biomass of V. dahliae in cotton seedlings, the first true leaf of cotton plants at 22 dpi was collected for DNA extraction. qPCR was used to quantify the relative amount of fungal DNA (Chai et al., 2017).
Transcriptome sequencing
The root samples of cotton seedlings pre‐treated with 0 or 25 μm melatonin were collected for transcriptome sequencing at 0, 6, 12, 24, and 48 h post inoculation (hpi). The cotton seedlings pre‐treated with 0 μm melatonin served as the control. Three biological replicates were performed for each treatment. Total RNA was extracted using the RNAprep Pure Plant Kit (Tiangen, Beijing, China) according to the manufacturer's instructions. RNA degradation, purity, concentration, and integrity were detected using electrophoresis, a NanoPhotometer spectrophotometer (IMPLEN, Los Angeles, CA, USA), the Qubit RNA Assay Kit in Qubit Fluorometer 2.0 (Life Technology, Carlsbad, CA, USA), and the RNA Nano 6000 Assay Kit for the Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA). Sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, Beverly, CA, USA) following the manufacturer's recommendations. Sequencing was performed on the Illumina HiSeq4000 platform with 150 bp paired‐end reads. The transcriptome sequencing raw reads were processed to remove adaptors and low‐quality reads using Trimmomatic (Bolger et al., 2014). All further analysis was performed using only the high‐quality paired‐end clean reads. The clean reads were aligned to the G. hirsutum cv. TM‐1 genome (Zhang et al., 2015) using hisat 2.0.4 software (Kim et al., 2015). HTSeq v0.6.1 (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) was used to count the read numbers mapped to each gene. The expected number of fragments per kilobase of genes per million mapped reads (FPKM) of each gene was calculated based on the length of the gene and the read count mapped to the gene (Trapnell et al., 2010). Differentially expressed genes were defined using the DESeq R package (Anders and Huber, 2010) with an adjusted P‐value (q‐value) <0.05. Pathway enrichment analysis was based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/) (Kanehisa et al., 2008) using KOBAS (Mao et al., 2005) to test the statistical enrichment of DEGs in the KEGG pathways.
DNA and RNA extraction, RT‐PCR, and qPCR
Total DNA was extracted from plant leaves using the Plant DNA Mini Kit (Aidlab, Beijing, China) according to the manufacturer's instructions. Total RNA was extracted from different tissues using the RNAprep Pure Plant Kit (Tiangen). Total RNA (500 ng) was reverse transcribed using the ReverTra Ace qPCR RT Kit (with gDNA remover) (Toyobo, Osaka, Japan). qPCR analysis was performed using a LightCycler FastStart DNA Master SYBR Green I kit (Roche, Basel, Switzerland) on a LightCycler96 Real‐Time PCR detection system (Roche). Three independent biological replicates were used for each analysis with at least three technical replicates each. The relative expression levels were evaluated using the comparative cycle threshold method according to Livak and Schmittgen (2001). GhUBQ7 (Tan et al., 2013) was used as the internal control, which was stably expressed in cotton plants and not affected by treatments and genotypes. The gene‐specific primers were designed using Primer Premier 5 software. All primers are listed in Table S6.
Phylogenetic analysis and amino acid sequence alignment of SNAT1 and COMT orthologs
For constructing the phylogenetic trees, the SNAT1 and COMT were extracted from 16 and 15 species, respectively, including fern, moss, gymnosperm, algae, monocotyledons, and dicotyledons. The two gene genomic data of G. arboreum, G. raimondii, and G. hirsutum was obtained from http://cgp.genomics.org.cn, http://www.phytozome.net, and http://mascotton.njau.edu.cn, respectively. The genomic data of other species were obtained from http://www.phytozome.net. mega 5.1 software (Tamura et al., 2011) was used for constructing the phylogenetic trees using Poisson correction model in the neighbor‐joining (NJ) method with 1000 bootstrap resamplings.
clustalX 2.0 software (Larkin et al., 2007) was used for amino acid sequence alignment of SNAT1 and COMT proteins, including G. hirsutum, G. arboreum, G. raimondii, Theobroma cacao, A. thaliana, and Oryza sativa. genedoc software (http://www.nrbsc.org/gfx/genedoc/ebinet.htm) was used for visualizing the alignment results. All the amino acid sequences were checked against Pfam (http://pfam.sanger.ac.uk/) (Finn et al., 2014) and the NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/cdd) (Marchler‐Bauer et al., 2015) to confirm the conserved domains.
Construction of overexpression vectors and transformation of Arabidopsis
Full‐length coding sequences of GhSNAT1 and GhCOMT were amplified by RT‐PCR from cDNA of TM‐1 using gene‐specific primers (Table S6). Both PCR fragments were inserted into a modified pCAMBIA1301–GFP binary vector to generate pCAMBIA1301‐SNAT1–GFP and pCAMBIA1301‐COMT–GFP, respectively.
Arabidopsis mutants, snat1 and comt, were sown in pots containing peat and vermiculite (1:1) in a growth chamber at 22°C under a 16 h light/8 h dark cycle. The transgenic Arabidopsis plants were created via the floral dip method (Clough and Bent, 1998). The transgenic plants were selected for kanamycin resistance. After selection for four generations, the homozygous GhSNAT1 and GhCOMT overexpression lines were obtained.
Virus‐induced gene silencing system construction
Tobacco rattle virus (TRV)‐based vectors (pTRV1 and pTRV2) were used for VIGS in the study. The GhSNAT1 and GhCOMT inserts were amplified from G. hirsutum cv. TM‐1 cDNA by PCR. The primers are listed in Table S7. The GhSNAT1 PCR fragment was double digested with EcoRI and BamHI enzymes then ligated into pTRV2. Similarly, the GhCOMT PCR fragment was ligated into BamHI−KpnI‐digested pTRV2. The pTRV1, pTRV2:GhSNAT1, and pTRV2:GhCOMT vectors were introduced into Agrobacterium strain GV3101 by electroporation. Agrobacterium colonies carrying the desired vectors were cultured overnight at 28°C in an antibiotic selection medium containing 50 mg L−1 rifampicin and 50 mg L−1 kanamycin. The transformed Agrobacterium cells were collected and resuspended in infiltration medium (10 mm MgCl2, 10 mm MES, and 200 mm acetosyringone) then adjusted to an OD600 value of 1. Agrobacterium strains containing pTRV1 and pTRV2 vectors were mixed at a ratio of 1:1. For virus‐induced co‐silencing, Agrobacterium strains containing pTRV1, pTRV2:GhSNAT1, and pTRV2:GhCOMT vectors were mixed at a ratio of 2:1:1. The Agrobacterium suspension was injected into the cotyledons of 7‐day‐old G. hirsutum cv. TM‐1 seedlings. After incubation for 24 h in darkness, the cotton seedlings were transferred to a growth chamber at 25°C under a 16 h light/8 h dark cycle. The empty pTRV2 vector was used as the negative control. CLOROPLASTOS ALTERADOS 1 (CLA1) (Gao et al., 2011a) and PIGMENT GLAND FORMATION (PGF) (Ma et al., 2016) were used as positive controls to validate the silencing effect of our VIGS system. At least 32 cotton seedlings were used for each analysis, and all VIGS experiments were performed three times.
Determination of melatonin content using HPLC‐MS
The melatonin contents in various plant tissues, including leaves, stems, and roots, were determined according to a previously described method (Arnao and Hernandez‐Ruiz, 2009) with some modifications. Fresh samples (0.5 g) were ground in liquid nitrogen and homogenized in 3 ml methanol containing 1 μl 50 ng ml−1 [2H6]‐melatonin (M215002, Toronto Research Chemicals Ltd, Toronto, ON, Canada) as an internal standard. After shaking for 12 h at 4°C in darkness and centrifugation at 1500 g and 4°C for 20 min, the supernatant was transferred to another tube. The remaining residue was washed with 2 ml methanol and combined with the supernatant fraction. The supernatant fraction was then concentrated by drying with nitrogen. The residue was dissolved in 0.5 ml 70% methanol and filtered with a 0.22 μm syringe filter (Agela, Newark, NJ, USA). HPLC electrospray ionization/MS‐MS analysis was performed on an Agilent 6460 triple quad liquid chromatography/mass spectrometry (LC/MS) (Agilent) equipped with an Agilent‐XDB C18 column at 35°C.
Measurement of gossypol content by HPLC
To measure the gossypol content in plant tissues, leaf, stem, and root samples were dried in a lyophilizer and ground to a fine powder. The powder (50 mg) was suspended in 3 ml acetone, incubated in an ultrasonic bath for 30 min, and centrifuged at 1500 g for 20 min. The supernatant was transferred to another tube. The residue was washed with 3 ml methanol twice and combined with the supernatant fraction. The extract was adjusted to 25 ml with acetone, filtered through a 0.4‐ mm syringe filter (Agela), then analyzed using an Agilent 1100 HPLC (Agilent) according to previously described methods (Li et al., 2017a).
Histochemical analysis and lignin content determination
Cotton stems at the same position above the cotyledons were collected from cotton seedlings at 10 dpi. Staining with safranin and fast green was used to visualize lignin deposition. The stems were dehydrated with different concentrations of ethanol, embedded in paraffin, then hand‐cut into cross sections. After dewaxing with xylene and ethanol, the sections were stained with 1% safranin for 1–2 h and 0.5% fast green for 60 s, oven‐dried at 60°C, then photographed under a Leica MZ95 microscope (Leica).
Plant tissues, including leaves, stems, and roots, were collected from cotton seedlings at 0 dpi and 10 dpi. The lignin content was determined by the lignin‐thioglycolic acid (LTGA) reaction (dos Santos et al., 2004). After the different compounds in the plant parts were removed with phosphate buffer, Triton® X‐100, NaCl, water, and acetone, the protein‐free cell wall fraction was retained. The LTGA reaction was used to measure lignin content in this fraction, and the lignin content was expressed as the weight/dry weight.
Determination of phytohormones using HPLC‐MS
To determine the endogenous contents of JA, JA‐Ile, and SA, approximately 100 mg of cotton root samples were homogenized with methanol and shaken at 4°C overnight in the dark. The dissolution, filtration, storage, and quantification of the combined extract were performed as described in Hu et al. (2016). To each sample were added 7.5 ng (±) of 9‐,10‐dihydro‐JA, and 75 ng of 1‐naphthaleneacetic acid as internal standards for the JA, JA‐Ile, and SA content assays.
Data statement
The data that support the findings of this study are openly available within this manuscript and its supporting materials.
Author contributions
SZ, JC and CL conceived and designed the project. CL, QH, FZ, JY, CL, YZ, QX and BS performed the experiments. CL, TZ and LM developed libraries and performed sequencing. CL and QH analyzed the data. CL wrote the manuscript draft, which was revised by JC and SZ.
Conflicts of interest
The authors declare no conflicts of interest.
Supporting information
Figure S1. Effects of melatonin on the growth of V. dahliae.
Figure S2. Comparison of transcriptome sequencing data and qPCR results.
Figure S3. Histochemical analysis of lignin deposition in stem cross‐sections of cotton seedlings with or without V. dahliae inoculation.
Figure S4. Biosynthetic pathway of melatonin.
Figure S5. Alignment and phylogenetic analysis of the SNAT1 family and the GhSNAT1 expression profile in different cotton tissues.
Figure S6. Alignment and phylogenetic analysis of the COMT family and the GhCOMT expression profile in different cotton tissues.
Figure S7. Determination of melatonin content in transgenic Arabidopsis.
Figure S8. Positive controls for the virus‐induced gene silencing (VIGS) system.
Figure S9. GhSNAT1 and GhCOMT expression levels in VIGS lines.
Figure S10. Recovery of lignin and gossypol levels in different tissues of melatonin‐pre‐treated VIGS plants after V. dahliae inoculation.
Figure S11. Changes in JA, JA‐Ile, and SA levels in cotton roots during V. dahliae inoculation.
Figure S12. The expression of representative transcription factors in the JA and SA signaling pathways.
Table S1. Summary of transcriptome sequencing data.
Table S2. Summary of mapped reads in transcriptome analysis.
Table S3. Summary of differentially expressed genes in transcriptome analysis.
Table S4. Information for SNAT used in phylogenetic analysis.
Table S5. Information for COMT used in phylogenetic analysis.
Table S6. Differentially expressed transcription factor genes between melatonin‐pre‐treated and control samples at 12 h post inoculation.
Table S7. Primers used in this study.
Acknowledgments
We are grateful to Dr. Xiaodan Wu for kindly helping us with the determination of melatonin and phytohormone contents in plant tissues. We also acknowledge Professor Shiming Liu of CSIRO Agriculture and Food for proofreading the manuscript. This work was supported by the National Key Technology R&D program of China (2016YFD0101404) and China Agriculture Research System (CARS‐18‐25).
Contributor Information
Shuijin Zhu, Email: shjzhu@zju.edu.cn.
Jinhong Chen, Email: jinhongchen@zju.edu.cn.
References
- Anders, S. and Huber, W. (2010) Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnao, M.B. and Hernandez‐Ruiz, J. (2009) Assessment of different sample processing procedures applied to the determination of melatonin in plants. Phytochem. Anal. 20, 14–18. [DOI] [PubMed] [Google Scholar]
- Arnao, M.B. and Hernandez‐Ruiz, J. (2014) Melatonin: plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 19, 789–797. [DOI] [PubMed] [Google Scholar]
- Arnao, M.B. and Hernandez‐Ruiz, J. (2018) Melatonin and its relationship to plant hormones. Annu. Bot. 121, 195–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict, C.R. , Liu, J. and Stipanovic, R.D. (2006) The peroxidative coupling of hemigossypol to (+)‐ and (‐)‐gossypol in cottonseed extracts. Phytochemistry, 67, 356–361. [DOI] [PubMed] [Google Scholar]
- Bhuiyan, N. , Selvaraj, G. , Wei, Y. and King, J. (2009) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W. , Ralph, J. and Baucher, M. (2003) Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. and Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Brainard, G.C. , Hanifin, J.P. , Greeson, J.M. , Byrne, B. , Glickman, G. , Gerner, E. and Rollag, M.D. (2011) Action spectrum for melatonin regulation in humans: evidence for a novel circadian photoreceptor. J. Neurosci. 21, 6405–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon, Y. and Back, K. (2014) An increase in melatonin in transgenic rice causes pleiotropic phenotypes, including enhanced seedling growth, delayed flowering, and low grain yield. J. Pineal Res. 56, 408–414. [DOI] [PubMed] [Google Scholar]
- Byeon, Y. , Lee, H. , Lee, K. and Back, K. (2014a) Caffeic acid O‐methyltransferase is involved in the synthesis of melatonin by methylating N‐acetylserotonin in Arabidopsis . J. Pineal Res. 57, 219–227. [DOI] [PubMed] [Google Scholar]
- Byeon, Y. , Park, S. , Lee, H. , Kim, Y. and Back, K. (2014b) Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J. Pineal Res. 56, 275–282. [DOI] [PubMed] [Google Scholar]
- Campos, M.L. , Kang, J.H. and Howe, G.A. (2014) Jasmonate‐triggered plant immunity. J. Chem. Ecol. 40, 657–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo‐Vico, A. , Lardone, P.J. , Alvarez‐Sanchez, N. , Rodriguez‐Rodriguez, A. and Guerrero, J.M. (2013) Melatonin: buffering the immune system. Int. J. Mol. Sci. 14, 8638–8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass, C.L. , Peraldi, A. , Dowd, P.F. et al. (2015) Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium . J. Exp. Bot. 66, 4317–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Q. , Shang, X. , Wu, S. , Zhu, G. , Cheng, C. , Cai, C. , Wang, X. and Guo, W. (2017) 5‐Aminolevulinic acid dehydratase gene dosage affects programmed cell death and immunity. Plant Physiol. 175, 511–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Chen, Y. , Heinstein, P. and Davisson, V. (1995) Cloning, expression, and characterization of (+)‐δ‐cadinene synthase a catalyst for cotton phytoalexin biosynthesis. Arch. Biochem. Biophys. 324, 255–266. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Wang, M. , Davisson, V. and Heinstein, P. (1996) Cloning and heterologous expression of a second (+)‐δ‐cadinene synthase from Gossypium arboreum . J. Nat. Prod. 59, 944–951. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T. , Coaker, G. , Day, B. and Staskawicz, B.J. (2006) Host‐microbe interactions: shaping the evolution of the plant immune response. Cell, 124, 803–814. [DOI] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cook, D.E. , Mesarich, C.H. and Thomma, B.P. (2015) Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 53, 541–563. [DOI] [PubMed] [Google Scholar]
- Ding, F. , Wang, G. , Wang, M. and Zhang, S. (2018) Exogenous melatonin improves tolerance to water deficit by promoting cuticle formation in tomato plants. Molecules, 23, 1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R. (2001) Natural products and plant disease resistance. Nature, 411, 843–847. [DOI] [PubMed] [Google Scholar]
- Dixon, R. , Achnine, L. , Kota, P. , Liu, C. , Reddy, M. and Wang, L. (2002) The phenylpropanoid pathway and plant defence a genomics perspective. Mol. Plant Pathol. 3, 371–390. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dollins, A.B. , Zhdanova, I.V. , Wurtman, R.J. , Lynch, H.J. and Deng, M.H. (1994) Effect of inducing nocturnal serum melatonin concentrations in daytime on sleep, mood, body temperature, and performance. Proc. Natl Acad. Sci. USA, 91, 1824–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbels, R. , Reiter, R.J. , Klenke, E. , Goebel, A. , Schnakenberg, E. , Ehlers, C. , Schiwara, H.W. and Schloot, W. (1995) Melatonin in edible plants identified by radioimmunoassay and by high performance liquid chromatography‐mass spectrometry. J. Pineal Res. 18, 28–31. [DOI] [PubMed] [Google Scholar]
- Feng, B. and Shan, L. (2014) ROS open roads to roundworm infection. Sci. Signal. 7, pe10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn, R.D. , Bateman, A. , Clements, J. et al. (2014) Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Britt, R.C. Jr , Shan, L. and He, P. (2011a) Agrobacterium‐mediated virus‐induced gene silencing assay in cotton. J. Vis. Exp. 54, e2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Wheeler, T. , Li, Z. , Kenerley, C.M. , He, P. and Shan, L. (2011b) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, W. , Long, L. , Zhu, L.F. , Xu, L. , Gao, W.H. , Sun, L.Q. , Liu, L.L. and Zhang, X.L. (2013) Proteomic and virus‐induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae . Mol. Cell Proteomics, 12, 3690–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik, J. and Huisman, O. (1988) Study of field‐grown cotton roots infected with Verticillium dahliae using an immunoenzymatic staining technique. Phytopathology, 78, 1174–1178. [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Gong, Q. , Yang, Z. , Chen, E. et al. (2018) A phi‐class glutathione S‐transferase gene for Verticillium wilt resistance in Gossypium arboreum identified in a genome‐wide association study. Plant Cell Physiol. 59, 275–289. [DOI] [PubMed] [Google Scholar]
- Hattori, A. , Migitaka, H. , Iigo, M. , Itoh, M. , Yamamoto, K. , Ohtani‐Kaneko, R. , Hara, M. , Suzuki, T. and Reiter, R.J. (1995) Identification of melatonin in plants and its effects on plasma melatonin levels and binding to melatonin receptors in vertebrates. Int. J. Biochem. Mol. Biol. 35, 627–634. [PubMed] [Google Scholar]
- Heinstein, P. , Herman, D. , Tove, S. and Smith, F. (1970) Heinstein Biosynthesis of gossypol. Incorporation of mevalonate‐2‐14C and isoprenyl pyrophosphates. J. Biol. Chem. 145, 4658–4665. [PubMed] [Google Scholar]
- Hu, H. , He, X. , Tu, L. , Zhu, L. , Zhu, S. , Ge, Z. and Zhang, X. (2016) GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3‐MYB transcription factor GhMYB25‐like. Plant J. 88, 921–935. [DOI] [PubMed] [Google Scholar]
- Hu, Q. , Min, L. , Yang, X. et al. (2018) Laccase GhLac1 modulates broad‐spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and jasmonic acid synthesis. Plant Physiol. 176, 1808–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M. , Araki, M. , Goto, S. et al. (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, K. , Lee, K. , Park, S. , Byeon, Y. and Back, K. (2013) Molecular cloning of rice serotonin N‐acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J. Pineal Res. 55, 7–13. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Langmead, B. and Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods, 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman, S.J. , Atallah, Z.K. , Vallad, G.E. and Subbarao, K.V. (2009) Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. [DOI] [PubMed] [Google Scholar]
- Kumari, I. , Ahmed, M. and Akhter, Y. (2016) Multifaceted impact of trichothecene metabolites on plant‐microbe interactions and human health. Appl. Microbiol. Biotechnol. 100, 5759–5771. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. and Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- La Camera, S. , Gouzerh, G. , Dhondt, S. , Hoffmann, L. , Fritig, B. , Legrand, M. and Heitz, T. (2004) Metabolic reprogramming in plant innate immunity the contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 198, 267–284. [DOI] [PubMed] [Google Scholar]
- Lange, B. , Lapierre, C. and Sandermann, H. (1995) Elicitor‐induced spruce stress lignin (structural similarity to early developmental lignins). Plant Physiol. 108, 1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, M.A. , Blackshields, G. , Brown, N.P. et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics, 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lee, H. , Byeon, Y. , Lee, K. , Lee, H. and Back, K. (2014) Cloning of Arabidopsis serotonin N‐acetyltransferase and its role with caffeic acid O‐methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localizations. J. Pineal Res. 57, 418–426. [DOI] [PubMed] [Google Scholar]
- Lee, H. , Byeon, Y. , Tan, D.X. , Reiter, R.J. and Back, K. (2015) Arabidopsis serotonin N‐acetyltransferase knockout mutant plants exhibit decreased melatonin and salicylic acid levels resulting in susceptibility to an avirulent pathogen. J. Pineal Res. 58, 291–299. [DOI] [PubMed] [Google Scholar]
- Li, F. , Shen, H. , Wang, M. , Fan, K. , Bibi, N. , Ni, M. , Yuan, S. and Wang, X. (2016a) A synthetic antimicrobial peptide BTD‐S expressed in Arabidopsis thaliana confers enhanced resistance to Verticillium dahliae . Mol. Genet. Genomics, 291, 1647–1661. [DOI] [PubMed] [Google Scholar]
- Li, M.Q. , Hasan, M.K. , Li, C.X. et al. (2016b) Melatonin mediates selenium‐induced tolerance to cadmium stress in tomato plants. J. Pineal Res. 61, 291–302. [DOI] [PubMed] [Google Scholar]
- Li, X. , Tan, D.X. , Jiang, D. and Liu, F. (2016c) Melatonin enhances cold tolerance in drought‐primed wild‐type and abscisic acid‐deficient mutant barley. J. Pineal Res. 61, 328–339. [DOI] [PubMed] [Google Scholar]
- Li, C. , Zhao, T. , Li, C. , Mei, L. , Yu, E. , Dong, Y. , Chen, J. and Zhu, S. (2017a) Determination of gossypol content in cottonseeds by near infrared spectroscopy based on Monte Carlo uninformative variable elimination and nonlinear calibration methods. Food Chem. 221, 990–996. [DOI] [PubMed] [Google Scholar]
- Li, T. , Ma, X. , Li, N. , Zhou, L. , Liu, Z. , Han, H. , Gui, Y. , Bao, Y. , Chen, J. and Dai, X. (2017b) Genome‐wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Benedict, C.R. , Stipanovic, R.D. and Bell, A. (1999) Purification and characterization of s‐adenosyl‐l‐methionine desoxyhemigossypol‐6‐O‐methyltransferase from plants. an enzyme capable of methylating the defense cotton. Plant Physiol. 121, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luo, P. , Wang, Y. , Wang, G. , Essenberg, M. and Chen, X. (2001) Molecular cloning and functional identification of (+)‐δ‐cadinene‐8‐hydroxylase, a cytochrome P450 mono‐oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 28, 95–104. [DOI] [PubMed] [Google Scholar]
- Ma, D. , Hu, Y. , Yang, C. et al. (2016) Genetic basis for glandular trichome formation in cotton. Nat. Commun. 7, 10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M.K. , Suren, H. , Ward, B. , Boroujerdi, A. and Kousik, C. (2018) Differential roles of melatonin in plant‐host resistance and pathogen suppression in cucurbits. J. Pineal Res. 65, e12505. [DOI] [PubMed] [Google Scholar]
- Mao, X. , Cai, T. , Olyarchuk, J.G. and Wei, L. (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics, 21, 3787–3793. [DOI] [PubMed] [Google Scholar]
- Mao, Y.B. , Cai, W.J. , Wang, J.W. , Hong, G.J. , Tao, X.Y. , Wang, L.J. , Huang, Y.P. and Chen, X.Y. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant‐mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Marchler‐Bauer, A. , Derbyshire, M.K. , Gonzales, N.R. et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menden, B. , Kohlhoff, M. and Moerschbacher, B. (2007) Wheat cells accumulate a syringyl‐rich lignin during the hypersensitive resistance response. Phytochemistry, 68, 513–520. [DOI] [PubMed] [Google Scholar]
- Mo, H. , Wang, X. , Zhang, Y. , Zhang, G. , Zhang, J. and Ma, Z. (2015) Cotton polyamine oxidase is required for spermine and camalexin signalling in the defence response to Verticillium dahliae . Plant J. 83, 962–975. [DOI] [PubMed] [Google Scholar]
- Mottiar, Y. , Vanholme, R. , Boerjan, W. , Ralph, J. and Mansfield, S.D. (2016) Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37, 190–200. [DOI] [PubMed] [Google Scholar]
- Naoumkina, M.A. , Zhao, Q. , Gallego‐Giraldo, L. , Dai, X. , Zhao, P.X. and Dixon, R.A. (2010) Genome‐wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 11, 829–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz, M.A. , Huang, Y. , Bie, Z. , Ahmed, W. , Reiter, R.J. , Niu, M. and Hameed, S. (2015) Melatonin: Current status and future perspectives in plant science. Front. Plant Sci. 6, 1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka, A. , Jedrzejczak‐Rey, N. and Bednarek, P. (2015) Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytol. 206, 948–964. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M. , Leon‐Reyes, A. , Van der Ent, S. and Van Wees, S.C. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Rodriguez, C. , Mayo, J.C. , Sainz, R.M. , Antolin, I. , Herrera, F. , Martin, V. and Reiter, R.J. (2004) Regulation of antioxidant enzymes: a significant role for melatonin. J. Pineal Res. 36, 1–9. [DOI] [PubMed] [Google Scholar]
- dos Santos, W.D. , Ferrarese, M.L.L. , Finger, A. , Teixeira, A.C.N. and Ferrarese‐Filho, O. (2004) Lignification and related enzymes in Glycine max root growth‐inhibition by ferulic acid. J. Chem. Ecol. 30, 1203–1212. [DOI] [PubMed] [Google Scholar]
- Shi, H. , Chen, Y. , Tan, D.X. , Reiter, R.J. , Chan, Z. and He, C. (2015a) Melatonin induces nitric oxide and the potential mechanisms relate to innate immunity against bacterial pathogen infection in Arabidopsis. J. Pineal Res. 59, 102–108. [DOI] [PubMed] [Google Scholar]
- Shi, H. , Jiang, C. , Ye, T. , Tan, D.X. , Reiter, R.J. , Zhang, H. , Liu, R. and Chan, Z. (2015b) Comparative physiological, metabolomic, and transcriptomic analyses reveal mechanisms of improved abiotic stress resistance in bermudagrass [Cynodon dactylon (L). Pers.] by exogenous melatonin. J. Exp. Bot. 66, 681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Qian, Y. , Tan, D.X. , Reiter, R.J. and He, C. (2015c) Melatonin induces the transcripts of CBFDREB1s and their involvement in both abiotic and biotic stresses in Arabidopsis. J. Pineal Res. 59, 334–342. [DOI] [PubMed] [Google Scholar]
- Shi, H. , Tan, D.X. , Reiter, R.J. , Ye, T. , Yang, F. and Chan, Z. (2015d) Melatonin induces class A1 heat‐shock factors (HSFA1s) and their possible involvement of thermotolerance in Arabidopsis . J. Pineal Res. 58, 335–342. [DOI] [PubMed] [Google Scholar]
- Sun, Q. , Zhang, N. , Wang, J. et al. (2015) Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. J. Exp. Bot. 66, 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar, G. , Campbell, L.M. , Puckhaber, L. , Stipanovic, R.D. and Rathore, K.S. (2006) Engineering cottonseed for use in human nutrition by tissue‐specific reduction of toxic gossypol. Proc. Natl Acad. Sci. USA, 103, 18054–18059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Peterson, D. , Peterson, N. , Stecher, G. , Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J. , Tu, L. , Deng, F. , Hu, H. , Nie, Y. and Zhang, X. (2013) A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol. 162, 86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X. , Ruan, J. , Huang, C. et al. (2018) Characterization of gossypol biosynthetic pathway. Proc. Natl Acad. Sci. USA, 115, 5410–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend, B.J. , Poole, A. , Blake, C.J. and Llewellyn, D.J. (2005) Antisense suppression of a (+)‐delta‐cadinene synthase gene in cotton prevents the induction of this defense response gene during bacterial blight infection but not its constitutive expression. Plant Physiol. 138, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M.J. , Salzberg, S.L. , Wold, B.J. and Pachter, L. (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.Y. , Cai, Y. , Gou, J.Y. , Mao, Y.B. , Xu, Y.H. , Jiang, W.H. and Chen, X.Y. (2004) VdNEP, an elicitor from Verticillium dahliae, induces cotton plant wilting. Appl. Environ. Microb. 70, 4989–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P. , Yin, L. , Liang, D. , Li, C. , Ma, F. and Yue, Z. (2012) Delayed senescence of apple leaves by exogenous melatonintreatment: toward regulating the ascorbate–glutathione cycle. J. Pineal Res. 53, 11–20. [DOI] [PubMed] [Google Scholar]
- Wei, W. , Li, Q.T. , Chu, Y.N. et al. (2015) Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J. , Li, D.X. , Zhang, J.R. , Shan, C. , Rengel, Z. , Song, Z.B. and Chen, Q. (2018) Phytomelatonin receptor PMTR1‐mediated signaling regulates stomatal closure in Arabidopsis thaliana . J. Pineal Res. 65, e12500. [DOI] [PubMed] [Google Scholar]
- Xu, L. , Zhu, L. , Tu, L. , Liu, L. , Yuan, D. , Jin, L. , Long, L. and Zhang, X. (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA‐Seq‐dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Yang, L. , Zhang, J. , Guo, X. , Zhang, X. and Li, G. (2012) Prevalence of the defoliating pathotype of Verticillium dahliae on cotton in central China and virulence on selected cotton cultivars. J. Phytopathol. 160, 369–376. [Google Scholar]
- Yin, L. , Wang, P. , Li, M. et al. (2013) Exogenous melatonin improves Malus resistance to Marssonina apple blotch. J. Pineal Res. 54, 426–434. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Zhang, H. , Zhao, B. et al. (2014) The RNA‐seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 56, 39–50. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Hu, Y. , Jiang, W. et al. (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM‐1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Ding, Z. , Wu, K. et al. (2016a) Suppression of jasmonic acid‐mediated defense by viral‐inducible microRNA319 facilitates virus infection in rice. Mol. Plant, 9, 1302–1314. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Jin, Y. , Zhao, J.H. , Gao, F. , Zhou, B.J. , Fang, Y.Y. and Guo, H.S. (2016b) Host‐Induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae . Mol. Plant, 9, 939–942. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Zhao, Y.L. , Zhao, J.H. , Wang, S. , Jin, Y. , Chen, Z.Q. , Fang, Y.Y. , Hua, C.L. , Ding, S.W. and Guo, H.S. (2016c) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants, 2, 16153. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Corwin, J.A. , Copeland, D. , Feusier, J. , Eshbaugh, R. , Chen, F. , Atwell, S. and Kliebenstein, D.J. (2017) Plastic transcriptomes stabilize immunity to pathogen diversity: the jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell, 29, 2727–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Wang, M. , Li, N. et al. (2018a) Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 16, 1172–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Wu, L. , Wang, X. et al. (2018b) The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence‐induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 20, 309–322. 10.1111/mpp.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.L. , Zhou, T.T. and Guo, H.S. (2016) Hyphopodium‐specific VdNoxB/VdPls1‐dependent ROS‐Ca2+ signaling is required for plant infection by Verticillium dahliae . PLoS Pathog. 12, e1005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Effects of melatonin on the growth of V. dahliae.
Figure S2. Comparison of transcriptome sequencing data and qPCR results.
Figure S3. Histochemical analysis of lignin deposition in stem cross‐sections of cotton seedlings with or without V. dahliae inoculation.
Figure S4. Biosynthetic pathway of melatonin.
Figure S5. Alignment and phylogenetic analysis of the SNAT1 family and the GhSNAT1 expression profile in different cotton tissues.
Figure S6. Alignment and phylogenetic analysis of the COMT family and the GhCOMT expression profile in different cotton tissues.
Figure S7. Determination of melatonin content in transgenic Arabidopsis.
Figure S8. Positive controls for the virus‐induced gene silencing (VIGS) system.
Figure S9. GhSNAT1 and GhCOMT expression levels in VIGS lines.
Figure S10. Recovery of lignin and gossypol levels in different tissues of melatonin‐pre‐treated VIGS plants after V. dahliae inoculation.
Figure S11. Changes in JA, JA‐Ile, and SA levels in cotton roots during V. dahliae inoculation.
Figure S12. The expression of representative transcription factors in the JA and SA signaling pathways.
Table S1. Summary of transcriptome sequencing data.
Table S2. Summary of mapped reads in transcriptome analysis.
Table S3. Summary of differentially expressed genes in transcriptome analysis.
Table S4. Information for SNAT used in phylogenetic analysis.
Table S5. Information for COMT used in phylogenetic analysis.
Table S6. Differentially expressed transcription factor genes between melatonin‐pre‐treated and control samples at 12 h post inoculation.
Table S7. Primers used in this study.


