Abstract
Objective
Epstein‐Barr virus‐positive diffuse large B‐cell lymphoma (EBV‐pos DLBCL) is a recently identified entity. Data regarding outcome to frontline immuno‐chemotherapy are conflicting. Although the prognostic impact of the tumour microenvironment (TME) in EBV‐neg DLBCL is well‐established, it remains untested whether the TME influences survival in EBV‐pos DLBCL. There are no data with new digital gene expression technologies that simultaneously interrogate the virus, B cells and the tumour microenvironment (TME).
Methods
We used the NanoString™ platform in a population‐based cohort of 433 patients to establish if the technology could detect EBV in the tumour biopsies and to investigate the influence that EBV has on the complex tumour microenvironment of DLBCL.
Results
Incidence of EBV‐pos DLBCL was 6.9% with 5‐year survival of 65% vs 82% in EBV‐neg DLBCL (P = 0.018). EBV‐pos tissues had similar expression of T‐cell genes compared to EBV‐neg DLBCL but higher levels of the antigen‐presenting molecule B2M. This was countered by elevated PD‐L1, PD‐L2, LAG3 and TIM3 immune checkpoints and a higher CD163/CD68 “M2” macrophage score.
Conclusion
In EBV‐pos DLBCL, the TME is immuno‐tolerogenic and may explain the poor outcomes seen in this subtype of DLBCL.
Keywords: diffuse large B‐cell lymphoma, Epstein‐Barr virus, immune checkpoints, tumour microenvironment, tumour‐associated macrophages
1. INTRODUCTION
Diffuse large B‐cell lymphoma (DLBCL) not otherwise specified (NOS) without evidence of underlying immunodeficiency and that is positive for Epstein‐Barr virus (EBV‐pos DLBCL) was recognised as a distinct entity in the early to mid‐2000s.1, 2, 3 In some studies, but not in others, it has been associated with poor outcome with standard “R‐CHOP” (rituximab‐cyclophosphamide/doxorubicin/vincristine/prednisolone) immuno‐chemotherapy.4, 5, 6, 7, 8, 9 Recent studies indicate EBV‐pos DLBCL occurs in both younger and older patients and that age does not influence pathological characteristics.4, 10
Epstein‐Barr virus represents a foreign antigen against which healthy EBV‐specific cytotoxic T‐cell (CTL) immunity has been extensively characterised.11 This response plays a pivotal role in controlling outgrowth of virus‐infected cells. CTLs scan the surface of virus‐infected cells to detect EBV peptides bound to MHC molecules and eliminate these cells by direct lysis. Conversely, the presence of EBV within the lymphoma cell acts as a potential target, strongly implicating mechanisms of immune evasion. Indeed, EBV‐pos DLBCL cases are associated with alterations in the tumour microenvironment (TME), which in turn may be modulated by the virus. Biopsies are frequently enriched in histiocytes that show high levels of programmed death ligand 1 (PD‐L1) and indoleamine 2,3‐dioxygenase.12, 13 It is postulated that all these factors contribute to a tolerogenic environment that promotes tumour immune escape,14 much as iatrogenic immunosuppression does in EBV‐pos post‐transplant lymphoproliferative disorders.15, 16, 17, 18 Interestingly, we have previously shown that the T‐cell receptor repertoire is a key determinant of the TME and is associated with outcome.19 Furthermore, substantially higher clonal T‐cell responses were observed in EBV‐pos vs EBV‐neg DLBCL.19 Although the prognostic impact of the TME in de novo DLBCL is well‐established,20, 21, 22, 23, 24 it remains untested as to whether the TME is associated with differential survival in EBV‐pos DLBCL.
Conventionally, EBV‐tissue status is determined by EBV‐encoded small RNA in situ hybridisation (“EBER‐ISH”) testing. Digital multiplex gene expression technologies such as NanoString™ are applicable to formalin‐fixed paraffin‐embedded (FFPE) tissues, but this approach is yet to be applied to EBV‐pos DLBCL. Not only can individual EBER molecules be digitally quantified (“EBER‐digital”), but these technologies offer the advantage of simultaneous interrogation of key viral and tumour microenvironment (TME) parameters, all of which are relevant to this unique pathobiological entity.25 In this study, we firstly demonstrate that EBER‐digital is suitable for detecting EBV‐pos DLBCL. Next, the platform was used to concurrently quantify other tumour‐related and TME factors with prognostic and/or biological importance, in a large population‐based multi‐centre series of DLBCL. This indicated that the TME is a principal determinant of survival in EBV‐pos DLBCL.
2. MATERIALS AND METHODS
2.1. Patients
There were 120 patients with a histological diagnosis of DLBCL (excluding follicular lymphoma IIIB, transformed lymphoma or immunosuppression‐related lymphoma) presenting between 2003 and 2014 at the Princess Alexandra Hospital (PAH) that had sufficient tissue for EBER‐digital testing. Of these, 81 had sufficient tissue for EBER‐ISH, and these patients were included in the initial test cohort.
The extension cohort combined the initial test patients with an additional 352 patients with DLBCL, making 433 in total. They were drawn from Canberra Hospital (120) and Royal North Shore Hospital, Sydney (97), patients from the Australasian Leukaemia and Lymphoma Group (ALLG) Discovery Centre (96), and the remaining 39 patients from the PAH. Inclusion criteria differed from the initial test cohort only in that patients required either an EBER‐ISH test result or sufficient tissue for EBER‐digital testing. The additional patients were 309 with EBER‐digital and 43 with only EBER‐ISH. Of the 433 extension cohort patients, 383 had survival data available; 362 of these were treated with R‐CHOP; and 21 patients were treated with alternate regimens. The study was approved by Metro South (Brisbane) Ethics Committee.
2.2. RNA quantification
RNA was extracted from FFPE tumour biopsies using RecoverAll Total Nucleic Acid Extraction Kit for FFPE (Ambion, Life Technologies) as per manufacturer’s instructions and stored at −80°C. Genes were quantified using the nCounter platform (NanoString™) as previously outlined.22, 26 A custom code set was used consisting of selected immune effectors, immune checkpoints, macrophage and antigen‐presenting molecules (CD4, CD8, CD137, PD‐1, PD‐L1, PD‐L2, LAG3, TIM3, CD68, CD163, B2M), the EBV‐related genes (EBER‐1, EBER‐2, latent membrane protein‐1 [LMP1]), the full Lymph2Cx gene set for cell‐of‐origin (COO) categorisation and finally BCL6 and CD30. Additionally, in order to quantify sensitivity of nCounter platform for detection of EBV, RNA was extracted from the EBV‐positive Burkitt’s lymphoma cell line, Namalwa, and serially diluted and mixed (dilution range 1:1‐10−9) with extracted PBMC RNA from an EBV‐negative donor. 150 ng of total RNA from both sources was run on a NanoString® PanCancer Immune Panel spiked in with the following EBV‐specific genes: EBER‐1, EBER‐2, EBNA2 and LMP1.
Hybridisations were carried out according to the NanoString™ Gene Expression Assay Manual. From each RNA sample (100‐300 ng), 5 μL was mixed with 20 μL of nCounter Reporter probes in hybridisation buffer and 5 μL of nCounter Capture probes for a total reaction volume of 30 μL. The hybridisations incubated at 65°C for approximately 16‐20 hours. Raw data were imported and analysed in the NanoString™ data analysis tool nSolver. For normalisation, gene expression data were internally controlled to the mean of the positive control probes to account for inter‐assay variability. Gene normalisation was then performed using the geometric mean of four‐housekeeper genes to account for factors that affect RNA quality and quantity (PGK1, GAPDH, PGAM1 and OAZ1) as previously published22, 24, 26 with the exception of COO categorisation, where Lymph2Cx‐specific normalisers were used in accordance with guidelines. EBER‐ISH was performed as previously described.2, 4, 8, 16, 27
2.3. Statistical analysis
Values between groups of data were tested for statistical significance using the 2‐tailed Mann‐Whitney tests. Categorical data were compared using Fisher’s exact test or chi‐squared test as appropriate. Overall survival (OS) was measured from diagnosis to date of last follow‐up or death. Progression‐free survival (PFS) was measured from the date of diagnosis to the date of last follow‐up, disease progression or death. Survival analysis used Kaplan‐Meier curves and the log‐rank test. Multivariate analysis was performed using Cox regression. All tests were 2‐sided at the threshold of P = 0.05. Multiple hypothesis testing for gene expression associations of immune effectors and checkpoints was corrected using the Bonferroni method. All analyses were prepared using GraphPad Prism platform (version 7, GraphPad Software) and Statistical Package for the Social Sciences SPSS version 24 (International Business Machines Corporation).
3. RESULTS
3.1. Defining EBV‐pos DLBCL by digital multiplex gene expression
The initial aim was to establish a threshold for EBER‐digital positivity to be used for further analysis. Firstly, EBER expression was quantified by NanoString™ in the 81 DLBCL biopsies tested with both EBER‐digital and EBER‐ISH. The median EBER gene count was 34 295 digital counts (range 1900‐459 327) for the 10 cases that were classified EBER‐ISH positive. The remaining 71 cases had a median digital count EBER of 3 (range 1‐1681). A threshold of 500 normalised digital counts was selected for further testing. A receiver operating curve at this level indicated a sensitivity of 100% and specificity of 94%. This level was almost 15 times above background expected levels, and no EBER‐ISH‐positive cases had any EBER‐1 digital counts below this level. In a confirmatory experiment, an EBV + Burkitt’s cell line was diluted at various concentrations (10‐fold dilutions) with PBMC from an EBV‐negative donor. The undiluted cell line sample and 1:10 dilution samples had high EBER‐1 counts of 28 637 and 3854, respectively. The 1:100 sample demonstrated a detectable level of EBER‐1 at 122 digital counts, but all other dilutions below this (6 further dilution levels) recorded EBER‐1 levels below 30 gene counts with similar gene counts for all these dilution levels, that is confirming that counts <30 should be considered as background/undetectable level.
Eighty‐six per cent of cases had an EBER‐1 gene count <30, which is considered below background levels for the assay as described above. Duplicate EBER‐1 assays showed r = 0.98 (P < 0.0001) when EBER‐1 was >30 counts. EBER‐1 correlated with EBER‐2 (r = 0.92, P < 0.001) and LMP1 (r = 0.74, P < 0.001).
3.2. Incidence of EBV‐pos DLBCL in an Australian population
In the extension cohort, an additional 309 cases had sufficient tissue available for NanoString™ (ie, 390 cases total), with 26 cases classified as EBV‐pos DLBCL (6.6%). Similarly, in the 43 cases tested with EBER‐ISH, there were four EBV‐pos cases (9.3%). In the combined 433 cases, 30 EBV‐pos cases were identified giving an overall rate of 6.9%. EBV‐pos cases did not differ by the international prognostic index (IPI) or any of the individual IPI parameters (chi‐squared test) including age category >60 years of age (Table 1). However, patients with EBV‐pos DLBCL were older than EBV‐neg cases (median 66.7 years, range 38.5‐90, vs 61 years, range 18‐90, P = 0.018) despite no statistical difference for the IPI‐based age cut‐off of 60. Of note, 28% of EBV‐pos tumours occurred in patients ≤60 years, and 2 cases were below the age of 50. Thus, whilst patients with EBV‐pos DLBCL were significantly older, it does not preclude the possibility of EBV‐pos DLBCL occurring in younger patients.
Table 1.
Patient characteristics of the cohort
| Characteristics | EBV‐neg (n = 403) | EBV‐pos (n = 30) | P Value |
|---|---|---|---|
| Sex(M) | 177 (54%) | 15 (65%) | NS |
| Age | 61 (18‐89.95) | 66.7 (38.5‐90) | 0.018 |
| Age > 60 | 215 (58%) | 21 (72%) | NS |
| Stage > 2 | 211 (58%) | 18 (75%) | NS |
| ECOG > 1 | 69 (24%) | 9 (38%) | NS |
| LDH > N | 179 (59%) | 12 (48%) | NS |
| EN > 1 | 90 (31%) | 8 (35%) | NS |
| IPI (n = 371) | |||
| 0 | 38 (11%) | 1 (4%) | NS |
| 1,2 | 156 (45%) | 11 (41%) | |
| 3,4,5 | 150 (44%) | 15 (55%) | |
| COO (L2Cx) (n = 307) | |||
| GCB | 174 (61%) | 16 (69%) | NS |
| ABC | 75 (26%) | 2 (9%) | |
| UC | 35 (13%) | 5 (22%) | |
| Therapy | |||
| R‐CHOP | 335 (83%) | 27 (90%) | |
| Other therapy (with outcome) | 19 (5%) | 3 (10%) | |
| Other (no outcome/trial therapy etc) | 49 (12%) | ||
3.3. Relationship between EBV‐pos DLBCL and COO
Tissue was available to establish COO by NanoString™ Lymph2Cx in 307 cases. (284 EBV‐neg tumours and 23 EBV‐pos tumours). In EBV‐neg cases, COO was 61% germinal‐centre B cell (GCB), 26% activated B cell (ABC) and 13% unclassified (UC). The COO distribution for EBV‐pos DLBCL was 69% GCB, 9% ABC and 22% UC, that is similar to EBV‐neg cases (p = NS, chi‐squared test). As expected, EBV‐pos DLBCL (n = 23) had lower levels of the GCB‐associated gene BCL6 than EBV‐neg (n = 284) cases (median gene count 281 vs 507, respectively, P = 0.0009, Mann‐Whitney test). As expected, when subdivided by COO, GCB DLBCL had higher BCL6 than ABC/UC, but interestingly EBV‐pos GCB tumours had a lower expression of BCL6 vs EBV‐neg GCB cases (median gene count 647 (n = 16) vs 982 (n = 174), respectively, P = 0.013, Mann‐Whitney test), whereas no difference was observed for EBV‐pos ABC/UC tumours vs EBV‐neg ABC/UC cases (median gene count 310 (n = 7) vs 237 (n = 110), respectively, P = 0.09).
Seventy‐five cases had both Lymph2Cx and Hans performed: 50 of 75 (67%) samples were concordant between the methods. Thirty‐three of 36 classified as GCB by Hans were GCB by Lymph2Cx, with 1 UC and 2 ABC. Hans classification was less accurate with non‐GCB classification with only 17/39 concordant. The remaining 22 cases were assigned as GCB by Lymph2Cx. These results are consistent with the previously described performance of the Hans classifier in comparison with gene expression analysis.25, 29
3.4. EBV‐pos DLBCL has an adverse impact upon survival independent of IPI and COO
With a median follow‐up of 45 months, 362 patients in the extension cohort received R‐CHOP immuno‐chemotherapy. Of these, 27 (7.4%) had an EBER‐digital gene count >500 or were EBER‐ISH positive. EBV positivity was associated with poorer outcome, with OS significantly inferior in EBV‐pos cases compared to EBV‐neg cases with 5‐year OS 65% vs 82%, P = 0.018 (Figure 1), respectively, and a trend towards inferior 5‐year PFS of 55% vs 72%, P = 0.09. When including all 383 patients with follow‐up data irrespective of treatment, EBV‐pos DLBCL remained very similar with inferior 5‐year overall survival of 65% vs 80% for EBV‐neg cases (P = 0.026) and a trend towards poorer 5‐year PFS of 56.5% vs 71.8%, P = 0.13.
Figure 1.

Kaplan‐Meier estimates of overall survival (OS) in DLBCL stratified by EBV‐tissue status. EBV‐pos DLBCL has significantly inferior (A) OS when treated with R‐CHOP and (B) inferior OS when all EBV‐pos DLBCL cases are included irrespective of therapy received
There were 307 cases with COO assigned by Lymph2Cx. Patients with tumours classified as GCB had an improved outcome compared to those classified as ABC (5‐year OS 84.6% vs 69.7%, P = 0.033, Figure S1). By multivariate analysis including COO, IPI and EBER‐digital, only EBV status (P = 0.045) and IPI (P = 0.001) but interestingly not COO (P = 0.25) were independent predictors of OS. Similarly, in a multivariate model including the 342 patients with EBV status (EBER‐digital or EBER‐ISH) and IPI data available therapy, both were independent predictors of overall survival (P = 0.031 and P < 0.001, respectively). EBV status was not an independent predictor of PFS in any model.
3.5. Epstein‐Barr virus associations with host and viral genes
In the 390 cases with EBER‐digital, host gene expression results were compared, stratified by EBER‐digital status. EBV‐pos tumours showed strong associations with immune genes. Whilst effector markers such as CD4, CD8 and CD137 did not differ, there were significant differences with regard to immune checkpoint molecules. The expression of immune checkpoints PD‐L1 (P < 0.0001), PD‐L2 (P = 0.08), LAG3 (P = 0.01) and TIM3 (P = 0.05) were all higher in EBV‐pos cases (Figure 2—gene expression and Figure S2—immunohistochemistry), but PD‐1 expression did not differ. CD163 gene counts were higher in EBV‐pos DLBCL than EBV‐neg DLBCL, median 700 (range 67‐3281) vs 280 (range 1‐6192) gene counts, respectively, P = 0.008. The ratio of CD163/CD68 (a more specific M2 signature) was also significantly associated with EBV‐pos disease (P = 0.005).
Figure 2.
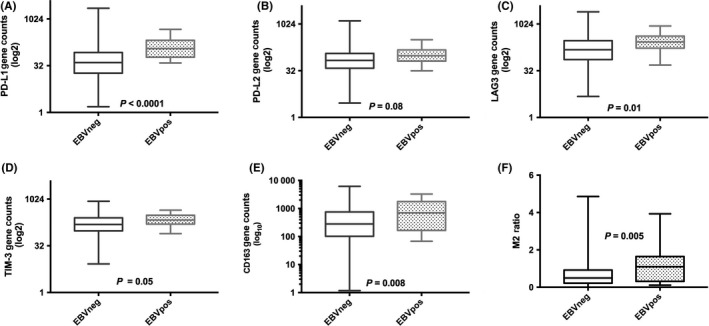
Host immune gene expression in DLBCL stratified by EBV‐tissue status. EBV‐pos DLBCL has significantly higher mRNA levels of immune checkpoint/macrophage markers (A) PD‐L1, (B) PD‐L2, (C) LAG‐3, (D) TIM‐3, (E) CD163 and (F) M2 macrophage ratio (CD163/CD68) (all error bars, SEM). (For all graphs, EBV‐neg DLBCL, n = 364 and EBV‐pos DLBCL, n = 26)
Effective antigen presentation by the malignant B cell is required for effective T cell–mediated elimination. The B2M molecule is vital for the recognition of antigen by cytotoxic T cells as part of the MHC class I structure. Interestingly, we found that B2M gene expression by NanoString™ was significantly higher in EBV‐pos cases (P = 0.015). Consistent with previous IHC‐based CD30 observations,4, 6 CD30 was strongly associated with EBV‐pos tumours with a ~4‐fold increase (P < 0.0001). LMP1, an EBV‐related oncogene with immunomodulatory properties, showed significant correlations with CD163 (r = 0.61, P = 0.003) and the M2 signature (r = 0.6, P = 0.003) as well as PD‐L1 (r = 0.49, P = 0.014).
3.6. Impact of EBV‐pos DLBCL of prognosis could potentially be influenced M2 macrophages
We then tested whether host gene expression might in part explain the adverse outcomes observed in our small cohort of 23 EBV‐pos DLBCL EBER‐digital cases treated with R‐CHOP immuno‐chemotherapy. The CD163/CD68 M2 ratio (cut‐off ratio CD163/CD68 = 0.75) divided patients into two distinct prognostic groups for 5‐year PFS (P = 0.004) and 5‐year OS (P = 0.01) with high levels of M2 associated with inferior outcome (Figure 3). Results should be interpreted with caution given small numbers in our cohort. This M2 ratio was also predictive of poor outcome in EBV‐neg cases, but a higher level of M2 infiltration was found in significantly more EBV‐pos DLBCL cases (56% vs. 34%, P = 0.038, Fisher’s test).
Figure 3.
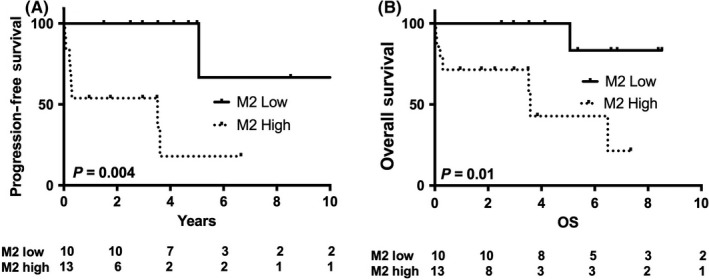
Kaplan‐Meier estimates of progression‐free survival (PFS) and overall survival (OS) in EBV‐pos DLBCL stratified by M2 ratio. Twenty‐three cases of EBV‐pos DLBCL had EBER‐digital and were treated with R‐CHOP. High levels of the M2 macrophage ratio were associated with (A) inferior PFS and (B) OS in EBV‐pos DLBCL compared to EBV‐pos cases with a low M2 ratio
4. DISCUSSION
In a large population‐based Australian cohort, the incidence of EBV‐pos DLBCL was 6.9%. Outcome was inferior to EBV‐neg DLBCL after treatment with first‐line immuno‐chemotherapy. By digital multiplex gene expression, EBV‐pos DLBCL had a distinct TME, with elevated immune checkpoint expression. The CD163/CD68 “M2” ratio segregated EBV‐pos DLBCL into groups with highly contrasting survival outcomes to R‐CHOP, indicating that the TME is a principal determinant of survival. The differences in the TME between patients have not been accounted for in previous studies and may explain why the inferior survival in patients with EBV‐pos DLBCL treated by R‐CHOP has been demonstrated in some studies but not confirmed in all.4, 5, 6, 7, 8, 9
Application of NanoString™ enabled genes reflective of the TME to be simultaneously interrogated along with EBV‐tissue status. Intratumoral T‐cell infiltration has previously been shown to be prognostic in DLBCL treated with R‐CHOP.21 However, we observed that both CD4 and CD8 were equivalent between EBV‐pos and EBV‐neg tumours. Against this, PD‐L1, PD‐L2, LAG3 and TIM3 immune checkpoint expression levels were elevated in EBV‐pos relative to EBV‐neg biopsies. This is in keeping with our previous observations that EBV is also associated with up‐regulated LAG3 in classical Hodgkin lymphoma (cHL).30 LMP1 is a key viral oncogene with established immunomodulatory abilities, and its levels correlated with PD‐L1—consistent with its known ability to induce PD‐L1 in other EBV‐pos lymphomas.31 Importantly, the antigen‐presenting molecule B2M was present at higher levels in EBV‐pos disease. These data are consistent with antigen presentation typically being intact in EBV‐pos disease 32, 33 and that viral‐induced immune evasion occurs at least in part via an immuno‐tolerogenic TME.
We demonstrated significantly higher levels of M2‐type macrophages compared to M1‐type macrophages in the tumour in EBV‐pos DLBCL vs EBV‐neg DLBC using the ratio of CD163:CD68 to indicate the intratumoral level of this macrophage subset. Results should be interpreted with caution given small numbers in our cohort. M2 immunosuppressive macrophages are associated with inferior outcome in many cancers including DLBCL.22 In cHL, we have previously demonstrated that EBV levels correlate with CD163.34 Similarly, in EBV‐pos DLBCL LMP1 correlated with M2. Next, we tested to see whether M2 could stratify outcome within EBV‐pos DLBCL. Patients with low levels of M2 macrophages had improved outcome compared to those with high levels of M2 macrophages. However, numbers are small and must be interpreted with caution. These results require validation in larger patient groups before definitive conclusions can be drawn.
Rates of EBV‐pos DLBCL appear to vary geographically. However, many centres apply EBER‐ISH in a targeted fashion, such as elderly patients or those with specific histological features. Results of large population‐based series that have formally examined the impact of EBV‐tissue status in patients with de novo DLBCL treated with frontline R‐CHOP immuno‐chemotherapy have been conflicting.5, 6, 7, 35 It remains unclear to what extent these differences reflect ethnic variations between studies conducted in either predominantly Asian or Caucasian population, and further large‐scale studies in new geographic localities are required. However, addition of rituximab to CHOP (“R‐CHOP”) improves response and survival.36 The incidence of EBV‐pos disease in Australia (6.9%) lies between rates observed in Europe and North American of 2%‐5%7, 37 and that seen in South‐East Asia and South America where 4%‐15% of newly diagnosed DLBCL can be EBV‐pos.35, 38 One explanation is that this reflects the country’s evolving demographics, including increasing immigration from South‐East Asia. The ethnicity of patients in this study was not obtained (to our knowledge, ethnicity has not been specifically been included in prior studies of EBV‐pos DLBCL), and these data should be collected in future studies of EBV‐pos DLBCL. Our findings also confirm recent descriptions of the disease in younger age groups with approximately a quarter of our cohort under the age of 60 and 2 cases occurring in patients <50 years of age. However, it remains a disease of predominantly older patients.4, 10, 12
Although the Hans classifier demonstrates that EBV‐pos DLBCL occurs in both GCB and non‐GCB cases, many studies show enrichment in non‐GCB tumours.6, 38 In contrast, in one of the largest studies to date there was no significant difference in the frequency of non‐GCB‐ to GCB‐typed patients.7 In another study in which the majority of COO was performed by gene expression,39 with the remaining performed by Hans, the proportions of GCB vs ABC were almost identical.4 Due to its inherent subjectivity and variability in scoring, the concordance between IHC and gene expression is modest and the prognostic value of the Hans method has been challenged.25, 29, 40 To our knowledge, the present study is the first to use the Lymph2Cx COO classifier in EBV‐pos DLBCL. We observe that the rate of GCB in EBV‐pos DLBCL biopsies was higher than previously described using the Hans classifier and that there was no enrichment of one subtype of COO over another. Consistent with reported BCL6 IHC findings,3 EBV‐pos DLBCL had lower levels of the Hans/GCB‐related gene BCL6 than EBV‐neg cases. However, this difference was confined to the EBV‐pos GCB DLBCL subset only. These findings appear to be consistent with evidence suggesting EBV induces an atypical GCB reaction associated with persistent infection in a latent form,41 and to the known ability of EBV‐miR‐BART9 and BART17‐5p to down‐regulate BCL6 expression.42 It is unclear what impact (if any) this might cause upon discrepancies in COO classification by IHC and gene expression in EBV‐pos DLBCL. It is also possible that the Lymph2Cx assay may not be as accurate in subsets of DLBCL cases with unique tumour microenvironments as occurs in EBV‐pos DLBCL given the assay is not performed specifically on tumour cells only. Additional studies are required before firm conclusions can be made.
In summary, we demonstrate that whilst the effector immune response in EBV DLBCL appears intact, it is counterbalanced by high levels of immune suppression in the TME with both elevated immune checkpoints and very high levels of tumour‐associated macrophages that appear to impact survival. Future clinical trials that focus on targeting these pathways may lead to improved outcomes for this poor prognosis subgroup of DLBCL.
Supporting information
ACKNOWLEDGEMENTS
This study was supported by the Kasey‐Anne Lymphoma Giving Fund—a giving account of the Lord Mayor’s Charitable Foundation—and the Pathology Queensland Study, Education and Research Committee. CK is supported by a Leukaemia Foundation Bridgestone Award, an NHMRC Early Career Fellowship, a Princess Alexandra Hospital Award, a Haematology Society of Australia and New Zealand new investigator grant, and Cancer Australia and Cancer Cure Australia; MKG is supported by the Leukaemia Foundation. Use of samples from the ACT Haematology Research Tissue Bank is acknowledged.
Keane C, Tobin J, Gunawardana J, et al. The tumour microenvironment is immuno‐tolerogenic and a principal determinant of patient outcome in EBV‐positive diffuse large B‐cell lymphoma. Eur J Haematol. 2019;103:200–207. 10.1111/ejh.13274
REFERENCES
- 1. Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375‐2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuze T, Nakamura N, Hashimoto Y, Sasaki Y, Abe M. The characteristics of Epstein‐Barr virus (EBV)‐positive diffuse large B‐cell lymphoma: comparison between EBV(+) and EBV(‐) cases in Japanese population. Jpn J Cancer Res. 2000;91(12):1233‐1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ok CY, Papathomas TG, Medeiros LJ, Young KH. EBV‐positive diffuse large B‐cell lymphoma of the elderly. Blood. 2013;122(3):328‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ok CY, Ye Q, Li L, et al. Age cutoff for Epstein‐Barr virus‐positive diffuse large B‐cell lymphoma–is it necessary? Oncotarget. 2015;6(16):13933‐13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hong JY, Yoon DH, Suh C, et al. EBV‐positive diffuse large B‐cell lymphoma in young adults: is this a distinct disease entity? Ann Oncol. 2015;26(3):548‐555. [DOI] [PubMed] [Google Scholar]
- 6. Tracy SI, Habermann TM, Feldman AL, et al. Outcomes among North American patients with diffuse large B‐cell lymphoma are independent of tumor Epstein‐Barr virus positivity or immunosuppression. Haematologica. 2018;103(2):297‐303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ok CY, Li L, Xu‐Monette ZY, et al. Prevalence and clinical implications of epstein‐barr virus infection in de novo diffuse large B‐cell lymphoma in Western countries. Clin Cancer Res. 2014;20(9):2338‐2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahn J‐S, Yang D‐H, Duk Choi Y, et al. Clinical outcome of elderly patients with Epstein‐Barr virus positive diffuse large B‐cell lymphoma treated with a combination of rituximab and CHOP chemotherapy. Am J Hematol. 2013;88(9):774‐779. [DOI] [PubMed] [Google Scholar]
- 9. Sato AI, Nakamura N, Kojima M, et al. Clinical outcome of Epstein‐Barr virus‐positive diffuse large B‐cell lymphoma of the elderly in the rituximab era. Cancer Sci. 2014;105(9):1170‐1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu T‐X, Liang J‐H, Miao YI, et al. Epstein‐Barr virus positive diffuse large B‐cell lymphoma predict poor outcome, regardless of the age. Sci Rep. 2015;5:12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein‐Barr virus. Annu Rev Immunol. 2007;25:587‐617. [DOI] [PubMed] [Google Scholar]
- 12. Nicolae A, Pittaluga S, Abdullah S, et al. EBV‐positive large B‐cell lymphomas in young patients: a nodal lymphoma with evidence for a tolerogenic immune environment. Blood. 2015;126(7):863‐872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen BJ, Chapuy B, Ouyang J, et al. PD‐L1 expression is characteristic of a subset of aggressive B‐cell lymphomas and virus‐associated malignancies. Clin Cancer Res. 2013;19(13):3462‐3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nicholas NS, Apollonio B, Ramsay AG. Tumor microenvironment (TME)‐driven immune suppression in B cell malignancy. Biochim Biophys Acta. 2016;1863(3):471‐482. [DOI] [PubMed] [Google Scholar]
- 15. Gandhi MK. Epstein‐Barr virus‐associated lymphomas. Expert Rev Anti Infect Ther. 2006;4(1):77‐89. [DOI] [PubMed] [Google Scholar]
- 16. Jones K, Nourse JP, Morrison L, et al. Expansion of EBNA1‐specific effector T cells in posttransplantation lymphoproliferative disorders. Blood. 2010;116(13):2245‐2252. [DOI] [PubMed] [Google Scholar]
- 17. Nourse JP, Jones K, Gandhi MK. Epstein‐Barr Virus‐related post‐transplant lymphoproliferative disorders: pathogenetic insights for targeted therapy. Am J Transplant. 2011;11(5):888‐895. [DOI] [PubMed] [Google Scholar]
- 18. Shannon‐Lowe C, Rickinson AB, Bell AI. Epstein‐Barr virus‐associated lymphomas. Philos Trans R Soc Lond B Biol Sci. 2017;372(1732):20160271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keane C, Gould C, Jones K, et al. The T‐cell receptor repertoire influences the Tumor microenvironment and is associated with survival in aggressive B‐cell lymphoma. Clin Cancer Res. 2017;23(7):1820‐1828. [DOI] [PubMed] [Google Scholar]
- 20. Fowler NH, Cheah CY, Gascoyne RD, et al. Role of the tumor microenvironment in mature B‐cell lymphoid malignancies. Haematologica. 2016;101(5):531‐540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Keane C, Gill D, Vari F, Cross D, Griffiths L, Gandhi M. CD4(+) tumor infiltrating lymphocytes are prognostic and independent of R‐IPI in patients with DLBCL receiving R‐CHOP chemo‐immunotherapy. Am J Hematol. 2013;88(4):273‐276. [DOI] [PubMed] [Google Scholar]
- 22. Keane C, Vari F, Hertzberg M, et al. Ratios of T‐cell immune effectors and checkpoint molecules as prognostic biomarkers in diffuse large B‐cell lymphoma: a population‐based study. Lancet Haematol. 2015;2(10):e445‐455. [DOI] [PubMed] [Google Scholar]
- 23. Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. 2014;14(8):517‐534. [DOI] [PubMed] [Google Scholar]
- 24. Vari F, Arpon D, Keane C, et al. Immune evasion via PD‐1/PD‐L1 on NK‐cells and monocyte/macrophages is more prominent in Hodgkin lymphoma than DLBCL. Blood. 2018;131(16):1809‐1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scott DW, Wright GW, Williams PM, et al. Determining cell‐of‐origin subtypes of diffuse large B‐cell lymphoma using gene expression in formalin‐fixed paraffin‐embedded tissue. Blood. 2014;123(8):1214‐1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones K, Wockner L, Brennan RM, et al. The impact of HLA class I and EBV latency‐II antigen‐specific CD8(+) T cells on the pathogenesis of EBV(+) Hodgkin lymphoma. Clin Exp Immunol. 2016;183(2):206‐220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoeller S, Tzankov A, Pileri SA, Went P, Dirnhofer S. Epstein‐Barr virus‐positive diffuse large B‐cell lymphoma in elderly patients is rare in Western populations. Hum Pathol. 2010;41(3):352‐357. [DOI] [PubMed] [Google Scholar]
- 28. Nourse JP, Crooks P, Keane C, et al. Expression profiling of Epstein‐Barr virus‐encoded microRNAs from paraffin‐embedded formalin‐fixed primary Epstein‐Barr virus‐positive B‐cell lymphoma samples. J Virol Methods. 2012;184(1–2):46‐54. [DOI] [PubMed] [Google Scholar]
- 29. Yoon N, Ahn S, Yong Yoo H, Jin Kim S, Seog Kim W, Hyeh KY. Cell‐of‐origin of diffuse large B‐cell lymphomas determined by the Lymph2Cx assay: better prognostic indicator than Hans algorithm. Oncotarget. 2017;8(13):22014‐22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gandhi MK, Lambley E, Duraiswamy J, et al. Expression of LAG‐3 by tumor‐infiltrating lymphocytes is coincident with the suppression of latent membrane antigen‐specific CD8+ T‐cell function in Hodgkin lymphoma patients. Blood. 2006;108(7):2280‐2289. [DOI] [PubMed] [Google Scholar]
- 31. Green MR, Rodig S, Juszczynski P, et al. Constitutive AP‐1 activity and EBV infection induce PD‐L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: implications for targeted therapy. Clin Cancer Res. 2012;18(6):1611‐1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee SP, Constandinou CM, Thomas WA, et al. Antigen presenting phenotype of Hodgkin Reed‐Sternberg cells: analysis of the HLA class I processing pathway and the effects of interleukin‐10 on epstein‐barr virus‐specific cytotoxic T‐cell recognition. Blood. 1998;92(3):1020‐1030. [PubMed] [Google Scholar]
- 33. Murray PG, Constandinou CM, Crocker J, Young LS, Ambinder RF. Analysis of major histocompatibility complex class I, TAP expression, and LMP2 epitope sequence in Epstein‐Barr virus‐positive Hodgkin’s disease. Blood. 1998;92(7):2477‐2483. [PubMed] [Google Scholar]
- 34. Jones K, Vari F, Keane C, et al. Serum CD163 and TARC as disease response biomarkers in classical Hodgkin lymphoma. Clin Cancer Res. 2013;19(3):731‐742. [DOI] [PubMed] [Google Scholar]
- 35. Oyama T, Yamamoto K, Asano N, et al. Age‐related EBV‐associated B‐cell lymphoproliferative disorders constitute a distinct clinicopathologic group: a study of 96 patients. Clin Cancer Res. 2007;13(17):5124‐5132. [DOI] [PubMed] [Google Scholar]
- 36. Beltran BE, Quiñones P, Morales D, et al. Response and survival benefit with chemoimmunotherapy in Epstein‐Barr virus‐positive diffuse large B‐cell lymphoma. Hematol Oncol. 2017;36(1):93-97. [DOI] [PubMed] [Google Scholar]
- 37. Hofscheier A, Ponciano A, Bonzheim I, et al. Geographic variation in the prevalence of Epstein‐Barr virus‐positive diffuse large B‐cell lymphoma of the elderly: a comparative analysis of a Mexican and a German population. Mod Pathol. 2011;24(8):1046‐1054. [DOI] [PubMed] [Google Scholar]
- 38. Park S, Lee J, Ko YH, et al. The impact of Epstein‐Barr virus status on clinical outcome in diffuse large B‐cell lymphoma. Blood. 2007;110(3):972‐978. [DOI] [PubMed] [Google Scholar]
- 39. Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression‐based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100(17):9991‐9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castillo JJ, Beltran BE, Song M‐K, et al. The Hans algorithm is not prognostic in patients with diffuse large B‐cell lymphoma treated with R‐CHOP. Leuk Res. 2012;36(4):413‐417. [DOI] [PubMed] [Google Scholar]
- 41. Thorley‐Lawson DA, Gross A. Persistence of the Epstein‐Barr virus and the origins of associated lymphomas. N Engl J Med. 2004;350(13):1328‐1337. [DOI] [PubMed] [Google Scholar]
- 42. Martín‐Pérez D, Vargiu P, Montes‐Moreno S, et al. Epstein‐Barr virus microRNAs repress BCL6 expression in diffuse large B‐cell lymphoma. Leukemia. 2012;26(1):180‐183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


