Abstract
Purpose
The purpose of the paper was to use a virtual phantom to identify a set of radiomic features from T1‐weighted and T2‐weighted magnetic resonance imaging (MRI) of the brain which is stable to variations in image acquisition parameters and to evaluate the effect of image preprocessing on radiomic features stability.
Methods
Stability to different sources of variability (time of repetition and echo, voxel size, random noise and intensity non‐uniformity) was evaluated for both T1‐weighted and T2‐weighted MRI images. A set of 107 radiomic features, accounting for shape and size, first order statistics, and textural features was used. Feature stability was quantified using intraclass correlation coefficient (ICC). For each source of variability, stability was evaluated before and after preprocessing (Z‐score normalization, resampling, gaussian filtering and bias field correction). Features that have ICC > 0.75 in all the analysis of variability are selected as stable features. Last, the robust feature sets were tested on images acquired with random simulation parameters to assess their generalizability to unseen conditions.
Results
Preprocessing significantly increased the robustness of radiomic features to the different sources of variability. When preprocessing is applied, a set of 67 and 61 features resulted as stable for T1‐weighted and T2‐wieghted images respectively, over 80% of which were confirmed by the analysis on the images acquired with random simulation parameters.
Conclusion
A set of MRI‐radiomic features, robust to changes in TR/TE/PS/ST, was identified. This set of features may be used in radiomic analyses based on T1‐weighted and T2‐weighted MRI images.
Keywords: features stability, MRI, phantoms, radiomics
1. Introduction
Radiomics is one of the most recent fields in medical image analysis and it consists in the extraction of a large number of features from radiological images such as computed tomography (CT), magnetic resonance imaging (MRI) and positron emission tomography (PET) with the final aim of providing “imaging biomarkers” that can be acquired in an inexpensive and non‐invasive way.1 Research on radiomics has already been performed in oncology for different purposes like tumor prognosis,2 staging,3 and prediction of response to treatment,4 often with success.
One of the main limitations of radiomics is that the feature value may depend on several factors (type of scanner, resolution, etc…), making the comparison of results from different studies very difficult. This problem becomes particularly relevant for studies that analyze radiological images collected with different imaging acquisition protocols (retrospective or multi‐center studies). Although it is well known that differences in protocols may reduce the reliability of radiomic features,5, 6, 7, 8, 9, 10, 11, 12, 13 research on association between differences in imaging conditions and radiomic features variability is still ongoing. This is also due to the high number of possible confounding factors (which may also be dependent on the specific imaging technique that is considered) whose exhaustive analysis is not trivial.
Phantoms are useful tools to assess variability to imaging conditions, allowing to evaluate the impact of the different acquisition parameters. In particular, online available phantom datasets (such as the one of Kalendralis et al.14 for CT) and virtual phantoms (such as BrainWeb15 or SimuBloch11 for MRI) can further provide a common ground truth for reliability analyses, increasing the reproducibility of the results. Many phantom‐based variability analyses have been reported in literature,5, 8, 10, 11, 12, 16 most of which focused on CT and PET, with only a few studies related to MRI instead.11, 16 Moreover, the effect of image preprocessing (like resampling and intensity standardization) on feature stability was never properly quantified for MRI images.
In this study different variability analyses were performed on a virtual phantom (BrainWeb) to identify a set of stable radiomic features from T1‐weighted and T2‐weighted MRI. In particular, the following imaging acquisition parameters and sources of variability were considered: time of repetition (TR), and time of echo (TE); pixel spacing (PS) and slice thickness (ST); intensity non‐uniformity (INU); image noise. The analysis was focused on T1‐weighted and T2‐weighted because they are the most used in the clinical practice and are therefore often considered in radiomics studies.4, 17 Effect of different preprocessing on features stability was also evaluated, in order to quantify the improvements on features stability.
1.A. Virtual phantom and simulated datasets
BrainWeb,15 an online available three‐dimensional (3D) virtual phantom was used to simulate the MRI‐scans used for the different variability analyses. The digital phantom was obtained by averaging 27 co‐registered MRI images of real patients, acquired with a 1.5 T MRI scanner.15, 18 Starting from a digital phantom, BrainWeb allows to perform custom MRI acquisitions, by controlling different parameters such as TR, TE, ST, INU and noise level. T1‐weighted and T2‐weighted MRI were simulated in this work.
1.B. Segmentation of the regions of interest
The regions of interest (ROIs) used were obtained directly from the BrainWeb website. In particular, 10 3D ROIs of different sizes representing different tissues (cerebrospinal fluid, gray matter, white matter, fat, muscle, skin, skull, glial matter and connective tissue) were considered. Since the ROIs are very large, to reduce the computational complexity of the radiomic features extraction we used only the 11 central slices of each ROI. Since all the images simulated with BrainWeb shared the same physical space, it was possible to use the same three ROIs for all the MRI acquisitions.
1.C. Radiomic features extraction
For each MRI acquisition, radiomic features were extracted from each of the 10 ROIs using Pyradiomics 2.1.0.19 A total of 107 3D features were computed for each ROI. Among those, 14 were related to shape and size (SS), 18 referred to first order statistics (FOS), 75 were related to texture. Textural features were computed using five different textural matrices: gray level co‐occurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level size zone matrix (GLSZM), neighborhood gray tone difference matrix (NGTDM) and gray level dependence matrix (GLDM). The default extraction parameters of Pyradiomics were used, except for gray‐values discretization. Instead of the fixed bin size discretization (the default of Pyradiomics), we used an intensity discretization with a fixed bin number, because the fixed bin size discretization may not be the best choice in case of images with arbitrary intensity units such as MRI.20 In particular, a 32 bins histogram discretization was used. For a more detailed description of the features used, refer to Pyradiomics documentation.21
1.D. Stability analyses of radiomic features
To test the stability of the features, four different analyses were performed, covering the possible sources of signal variability in MRI images: (a) Analysis of stability to variations in TR/TE; (b) Analysis of stability to variation of voxel size; (c) Analysis of stability to image noise; (d) Analysis of stability to intensity nonuniformity. The different simulation parameters for the different analysis are reported in Table 1. The ranges of interest for TR/TE, PS and ST were chosen to be similar to the ones that can be encountered in the clinical practice. Noise and inhomogeneities were controlled by setting BrainWeb parameters noise percentage and INU percentage, that were chosen to be equal or higher to the ones of analogous studies of literature using BrainWeb dataset.22, 23 The resampling of the images to different resolution was done in MatLab 2018a, since BrainWeb does not allow changes in PS. All the other acquisition parameters (TR, TE, noise level and intensity non‐uniformity) were set directly from BrainWeb.
Table 1.
Simulation parameters for the datasets used for the stability analyses of the study: stability to changes in repetition and echo time; stability to changes in voxel size; stability to image noise; stability to intensity non‐uniformities; stability to random variation of the simulation parameters.
| Stability analyses | |||||
|---|---|---|---|---|---|
| Analysis #1: TR/TE | Analysis #2: voxel size | Analysis #3: noise | Analysis #4: intensity non‐uniformity | Analysis #5: random variations | |
| Number of images | T1w: 42 | T1w: 28 | T1w: 10 | T1w: 4 | T1w: 50 |
| T2w: 48 | T2w: 28 | T2w: 10 | T2w: 4 | T2w: 50 | |
| Pulse sequence | Spin echo | Spin echo | Spin echo | Spin echo | Spin echo |
| Time of repetition (TR) | T1w: 350–650 ms, (50 ms step) | T1w: 500 ms | T1w: 500 ms | T1w: 500 ms | T1w: 350–650 ms, (random) |
| T2w: 2000–9000 ms (1000 ms step) | T2w: 6000 ms | T2w: 6000 ms | T2w: 6000 ms | T2w: 2000–9000 ms, (random) | |
| Time of echo (TE) | T1w: 5–15 ms, (2 ms step) | T1w: 9 ms | T1w: 9 ms | T1w: 9 ms | T1w: 5–15 ms, (random) |
| T2w: 80–130 ms (10 ms step) | T2w: 100 ms | T2w: 100 ms | T2w: 100 ms | T2w: 80–130 ms, (random) | |
| Slice thickness | 1 mm | 1–7 mm, (1 mm step) | 1 mm | 1 mm | 1–7 mm, (random) |
| Pixel spacing | 1 mm | 1–4 mm, (1 mm step) | 1 mm | 1 mm | 1–4 mm, (random) |
| Noise percentage | 0% | 0% | 9% | 0% | 0–9% (random) |
| Intensity non‐uniformity | None | None | None |
3 inhomogeneity field + 1 reference INU%: 40% |
Random inhomogeneity field (3 available) INU%: 0–40% (random) |
For each feature, stability was assessed using the intraclass correlation coefficient (ICC).24 In particular, the type of ICC used was the one to measure agreement in mixed‐effect models, equivalent to the (A,1) model described in McGraw et al.24 If a feature has an ICC of 1, it means that the changes in the parameter of interest caused no changes in the features, otherwise the lower the value of ICC the lower the stability of the feature.
Feature stability was evaluated for both the features extracted from the original images and for the features extracted from images that underwent a specific preprocessing which was aimed at reducing the effect of a particular source of variability, namely: (a) To correct for changes in the signal due to different TR/TE, Z‐score normalization25 was chosen as intensity standard standardization method. (b) Resampling to common isotropic resolution of 1 mm was adopted to correct variability due to voxel size. Isotropic resampling is preferable to anisotropic resampling because it is a better choice for the calculation of 3D textural features.20 (c) Image noise was corrected by applying a gaussian filter (3 × 3 × 3 voxel kernel, σ = 0.5). (d) Bias field correction was obtained through the commonly used N4ITK bias field correction algorithm.26 In each analysis, Wilcoxon signed rank tests were used to assessed significant variations in the values of ICC between the original and preprocessed images.
1.E. Stable features set identification
The results of the previous analyses were used to select a set of stable features. First, a value of 0.75 for the ICC was used as a lower threshold for stability. The value of 0.75 was chosen in agreement to Koo et al.27 Second, the features that, considering the preprocessed images, are stable in all the four analyses are included in the final set of stable features.
To compare the stable feature sets obtained from T1‐weighted and T2‐weighted images the Jaccard index (Jacc) was used:
| (1) |
where the numerator of Eq. (1) is the number of features in the intersection of the two stable features sets and the denominator is the number of features in the union of the two sets.
1.F. Robustness of stable features set on random simulations
The stability of the selected set features was tested on two datasets (one for each image type) composed by 50 images each. The images in those datasets were simulated using random variations of the analyzed parameters (TR, TE, INU, etc…). Details of the T1‐weighted and T2‐weighted datasets are presented in Table 1. This analysis was used to assess what percentage of the selected stable features are stable even when multiple sources of variability are considered, which is typically the case of the images of the clinical practice.
2. Results
2.A. Stability analyses of radiomic features
2.A.1. Stability to changes in TR and TE
Figure 1 shows boxplots representing the values of ICC of the features when the same phantom is acquired with different TR/TE. Values are shown for both T1‐weighted and T2‐weighted images, and for both original and preprocessed images. Only FOS and textural features are considered, since TR/TE variations do not affect the shape and size features.
Figure 1.
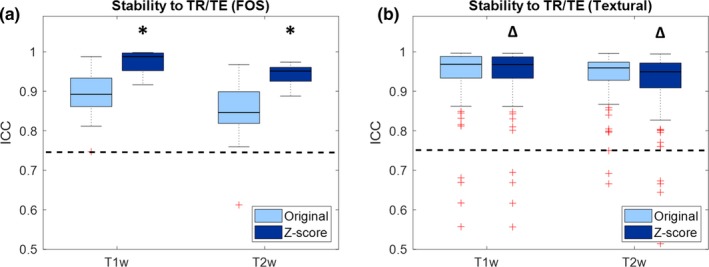
Boxplot representing the intraclass correlation coefficient (ICC) of the radiomic features for stability to variations of repetition and echo time (TR and TE): (a) First order statistics; (b) textural features. Significant differences due to preprocessing are reported with asterisks (ICC increase) or triangles (ICC decrease). The dashed line represents the threshold of stability (ICC = 0.75). [Color figure can be viewed at http://wileyonlinelibrary.com]
From Fig. 1 it can be seen that the majority of features are above the threshold of stability. In both T1‐weigthed and T2‐weighted intensity standardization increases ICC values of FOS features (T1w: median increase 0.09 [0.06–0.11], P = 7.37*10−4; T2w: median increase 0.11 [0.05–0.14], P = 8.44*10−3) and causes a small but significant decrease in ICC of textural features (T1w: median decrease 5.00*10−4 [2.30*10−5–1.50*10−3], P = 3.67*10‐7; T2w: median decrease 1.40*10−4 [3.21*10−4–1.66*10−3], P = 1.93*10−11).
2.A.2. Stability to changes in voxel size
Figure 2 shows boxplots representing the values of ICC of the features when the same phantom is acquired with different voxel sizes. Values are shown for both T1‐weighted and T2‐weighted images, and for both original and isotropically resampled images.
Figure 2.
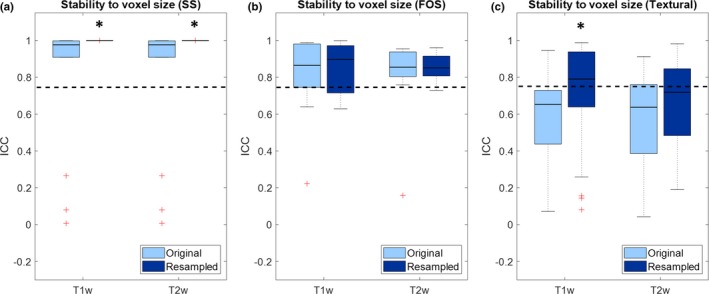
Boxplot representing the intraclass correlation coefficient (ICC) of the radiomic features for stability to variations of voxel size: (a) shape and size; (b) first order statistics; (c) textural features. Significant increase in ICC due to preprocessing are reported with asterisks. The dashed line represents the threshold of stability (ICC = 0.75). [Color figure can be viewed at http://wileyonlinelibrary.com]
From Fig. 2 it can be seen that voxel size has an impact on feature stability, particularly for textural features, whose majority is below the threshold of stability for the original images. In both T1‐weighted and T2‐weighted, resampling to common isotropic resolution significantly increases the stability of shape and size features (T1w/T2w: median increase 0.02 [2.20*10−3–0.09], P = 1.22*10−4). Resampling also significantly increases the stability of and textural features in T1‐weighted images (median increase 0.14 [0.01–0.28], P = 1.87*10−7) but not in T2‐weighted images (median increase 0.01 [−0.11 to 0.18], P = 0.17).
2.A.3. Stability to random noise
Figure 3 shows boxplots representing the values of ICC of the features when the same phantom is acquired with different random noise. Values are shown for both T1‐weighted and T2‐weighted images, and for both original and gaussian filtered images. Only FOS and textural features are considered, since noise does not affect the shape and size features.
Figure 3.
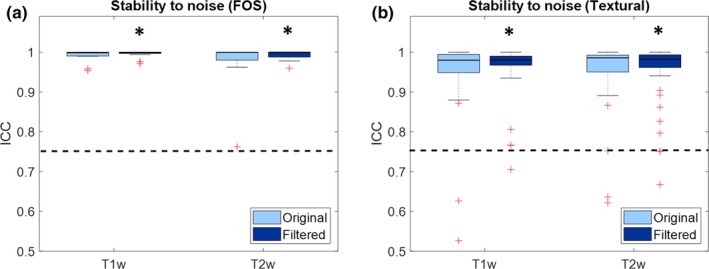
Boxplot representing the intraclass correlation coefficient (ICC) of the radiomic features for stability to image noise: (a) First order statistics; (b) textural features. Significant increase in ICC due to preprocessing are reported with asterisks. The dashed line represents the threshold of stability (ICC = 0.75). [Color figure can be viewed at http://wileyonlinelibrary.com]
From Fig. 3 it can be seen that noise does not have much effect on the stability of radiomic features. In fact, most of the features are above the threshold of stability even before denoising. Gaussian filtering causes a small but significant increase in ICC for both FOS (T1w: median increase 7.76*10−4 [4.32*10−6–4.50*10−3], P = 1.96*10−4; T2w: median increase 1.84*10−4 [5.80*10−6–7.90*10−3], P = 3.26*10−4) and textural features (T1w: median increase 7.70*10−3 [5.17*10−5–0.03], P = 8.53*10−6; T2w: median increase 2.10*10−3 [−1.00*10−3–9.30*10−3], P = 0.03).
2.A.4. Stability to intensity non‐uniformity
Figure 4 shows boxplots representing the values of ICC of the features when the same phantom is acquired with different intensity non‐uniformity fields. Values are shown for both T1‐weighted and T2‐weighted images, and for both original and bias field corrected images. Only FOS and textural features are considered, since intensity inhomogeneity does not affect the shape and size features.
Figure 4.
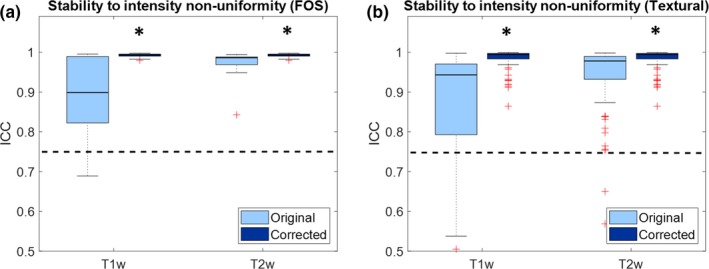
Boxplot representing the intraclass correlation coefficient (ICC) of the radiomic features for stability to intensity non‐uniformity: (a) First order statistics; (b) textural features. Significant increase in ICC due to preprocessing are reported with asterisk. The dashed line represents the threshold of stability (ICC = 0.75). [Color figure can be viewed at http://wileyonlinelibrary.com]
From Fig. 4 it can be seen that the majority of features are above the threshold of stability. In both T1‐weigthed and T2‐weighted intensity standardization significantly increases ICC values of FOS features (T1w: median increase 0.09 [6.40*10−3–0.18], P = 5.35*10−4; T2w: median increase 7.40*10−3 [5.50*10−3−0.02], P = 1.96*10−4) and causes significant increase in ICC of textural features (T1w: median increase 0.05 [0.02–0.15], P = 3.38*10−11; T2w: median increase 0.01 [4.90*10−3–0.04], P = 2.48*10−11).
2.B. Stable features set identification
The features with ICC > 0.75 in all the previous analyses were included in the stable feature set. Table 2 shows the list of selected features, grouped by features class. In total, 67 stable features were identified for T1‐weighted images and 61 were identified for T2‐weighted images. As it can be seen from the Table 2, most of the stable features are stable in both the image types. The Jaccard index between the two features sets is 0.80.
Table 2.
List of selected stable radiomic features grouped by features class and image type: shape and size, first order statistics (FOS), and textural. The red bold names refer to features that are not stable in the random parameter variations test.
| Stable features | ||||
|---|---|---|---|---|
| Image type | Shape and size | FOS | Textural | |
| T1‐weighted and T2‐weighted |
|
|
|
|
| T1‐weighted only |
|
|
||
| T2‐weighted only |
|
|||
2.C. Robustness of stable features set on random simulations
After the stability analysis to random variations of the simulation parameters, most of the stable radiomic features were confirmed in both T1‐weighted and T2‐weighted images.
In the T1‐weighted images, 60 out of the 67 stable features (89.55%) were confirmed. Excluded features were the following: maximum (FOS); maximum probability and sum squares (GLCM); gray level non‐uniformity normalized and long run low gray level emphasis (GLRLM); large dependence low gray level emphasis (GLDM).
In T2‐weighted images, 51 out of the 61 stable features (83.61%) were confirmed. Excluded features were the following: kurtosis, minimum and skewness (FOS); inverse difference, inverse difference moment and joint energy (GLCM); gray level non‐uniformity normalized (GLRLM); large dependence low gray level emphasis and small dependence emphasis (GLDM).
3. Discussion
This work confirms that a non‐negligible percentage of radiomic features may vary considerably when image acquisition parameters are changed. Even when all preprocessing methods are applied), 46 out of 107 features (around 43%) had ICC < 0.75. It is fundamental to reduce the variability due to imaging settings and to define a set of stable features.
Variations in voxel size (PS and ST) represent the major source of variability especially for the textural features. This result is in line with previous studies performed on CT.8, 10 Variations in TR/TE reduce the reliability of the features, especially for FOS. However, for most of them the threshold of ICC remains above the threshold of stability. The same can be stated for variability caused by field strength inhomogeneity and image noise.
The optimal way to reduce image variability would be the standardization of the protocols for image acquisition. However, this is rather difficult when dealing with multi‐center studies and it is not an option when dealing with retrospective cases. Performing image preprocessing before any radiomic features extraction and defining a set of stable features seems to be the best way to standardize results in absence of a unique acquisition protocol. Resampling to a common resolution increases the reliability of the shape and size and textural features to variations of voxel size, even if some of them remain unreliable (ICC < 0.75). This is in line with previous phantom studies on CT.8, 10 Reliability of FOS features to variations of TR/TE can be significantly improved if intensity standardization is applied. Textural feature reliability is reduced by Z‐score normalization, but the effect, although significant, is very small. Preprocessing also helps increasing the stability to intensity non‐uniformities caused by field strength inhomogeneity. In particular, when the N4ITK algorithm is applied, stability is improved in both FOS and textural features. Last, image denoising through gaussian filtering causes small but significant improvements in stability of features to image noise.
Based on the results of the first four stability analysis two stable sets of features, one for T1‐weighted images and the other for T2‐weighted images, were identified. Over 80% of the stable features confirmed their stability when tested on MRI images simulated varying all the four parameters of interest, which of course is a further proof of the robustness of our feature sets. Moreover, many of the stable features were selected for both the image types (Jaccard index 0.80). This may suggest that there is a subset of MRI features whose stability is mainly independent on the image type considered. Further investigations may be performed to understand how the stable feature set that was identified in this study can be suitable for other types of MRI, such as diffusion‐weighted MRI or apparent diffusion coefficient maps. Based on our results, we may assume that those features are suitable for radiomic analysis of the brain using T1‐weighted and T2‐weighted MRI only.
The use of a virtual phantom helped in providing an easy and reproducible way to perform a large number of MRI simulations that were necessary to fully analyze the variability of radiomic features. This is not the first study in which MRI virtual phantoms are used.11, 16 In their experiment Ford et al.11 investigated variability of 29 radiomic features from T1‐weighted images due to changes in TR and TE with two univariate analysis. Although the metrics used are not the same (coefficient of variation instead of ICC) it is possible to notice that only a few radiomics features results as unstable (coefficient of variation > 30% as in defined in Shafiq‐ul‐Hassan et al.10), which is coherent with our findings. To the knowledge of the authors, our study is the most exhaustive in terms of variability analysis, because four different types of stability analyses were performed accounting for common sources of variability of the clinical practice.
This study is not exempt from limitations. One limitation is the fact that the custom MRI simulation was performed only on one phantom, in one district (the brain) and without considering pathological tissue. Additionally, other factors such as the type of scanner used, that are known to influence the radiomic features,5, 8 were not considered. These limitations can be partially addressed. Since many different tissues were considered for the stability analyses that were performed, we can assume that the results that were found in this study can be translated also to other district of the body and to pathological tissues, even though this assumption has to be verified in future studies. As far as the scanner variability and the inter‐phantom variability are concerned, those required necessarily studies on multiple real phantoms representing multiple districts. Such an extensive analysis is non‐trivial and requires much effort in the design of both the proper phantom and experiments to be done. To the extent of our knowledge, such an extensive analysis has not been done yet in any radiomic‐related studies. In fact, most of the phantom studies in literature involve only one phantom.5, 8, 10 Such a complex analysis is beyond the scope of this work but is definitely a goal for future ones. Both virtual and real phantoms may be useful for the assessment of reliability of radiomic features, since the former allows to better isolate the effects of the single sources of variability, while the latter allows more challenging and complete analysis that will be impossible otherwise. Therefore, the two types of analyses should be used complementarily rather than alternatively. However, experiment on virtual phantoms represent an easy way (sometimes the only feasible way) to obtain data that, although preliminary, may guide the selection of features for radiomic analysis.
4. Conclusions
In this study, stability of MRI‐radiomic features to image acquisition parameters was investigated for both T1‐weighted and T2‐weighted images. The effect of image preprocessing was evaluated and a set of stable features (ICC > 0.75) was evaluated for both the image types. The results of this study could provide knowledge to guide future studies on brain radiomics based on T1‐weighted and T2‐weighted MRI and to perform radiomic analyses even in those situations in which a protocol for image acquisition parameters does not exist.
Conflict of interest
The authors have no conflict of interest.
Acknowledgments
The methods used in this study have been developed for the BD2Decide Project of the Horizon 2020 Research and Innovation Program (grant agreement No. 689715).
References
- 1. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corino VDA, Montin E, Messina A, et al. Radiomic analysis of soft tissues sarcomas can distinguish intermediate from high‐grade lesions. J Magn Reson Imaging. 2018;47:829–840. [DOI] [PubMed] [Google Scholar]
- 4. Wang G, He L, Yuan C, Huang Y, Liu Z, Liang C. Pretreatment MR imaging radiomics signatures for response prediction to induction chemotherapy in patients with nasopharyngeal carcinoma. Eur J Radiol. 2018;98:100–106. [DOI] [PubMed] [Google Scholar]
- 5. Mackin D, Fave X, Zhang L, et al. Measuring CT scanner variability of radiomics features. Invest Radiol. 2015;50:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fave X, Zhang L, Yang J, et al. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non‐small cell lung cancer. Transl Cancer Res. 2016;5:349–363. [Google Scholar]
- 7. He L, Huang Y, Ma Z, Liang C, Liang C, Liu Z. Effects of contrast‐enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep. 2016;6:34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Larue RTHM, van Timmeren JE, de Jong EEC, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017;56:1544–1553. [DOI] [PubMed] [Google Scholar]
- 9. Mackin D, Fave X, Zhang L, et al. Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS ONE. 2017;12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shafiq‐ul‐hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44:1050–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ford J, Dogan N, Young L, Yang F. Quantitative radiomics: impact of pulse sequence parameter selection on mri‐based textural features of the brain. Contrast Media Mol Imaging. 2018;2018:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mackin D, Ger R, Dodge C, et al. Effect of tube current on computed tomography radiomic features. Sci Rep. 2018;8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fave X, MacKin D, Yang J, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non‐small cell lung cancer? Med Phys. 2015;42:6784–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kalendralis P, Traverso A, Shi Z, et al. Multicenter CT phantoms public dataset for radiomics reproducibility tests. Med Phys. 2019;46:1512–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collins DL, Zijdenbos AP, Kollokian V, et al. Design and construction of a realistic digital brain phantom. IEEE Trans Med Imaging. 1998;17:463–468. [DOI] [PubMed] [Google Scholar]
- 16. Yang F, Dogan N, Stoyanova R, Ford JC. Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: a simulation study utilizing ground truth. Phys Medica. 2018;50:26–36. [DOI] [PubMed] [Google Scholar]
- 17. Wang Q, Li Q, Mi R, et al. Radiomics nomogram building from multiparametric MRI to predict grade in patients with glioma: A Cohort Study. J Magn Reson Imaging. 2019;49:825–833. [DOI] [PubMed] [Google Scholar]
- 18. Kwan RK, Evans AC, Pike GB. An Extensible MRI Simulator for Post‐Processing Evaluation. In Int. Conf. Vis. Biomed. Comput.1996.
- 19. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zwanenburg S, Leger MV, Löck S. Image biomarker standardization initiative. Reference manual, 2016. https://arxiv.org/abs/1612.07003
- 21. Pyradiomics features description; 2018. https://pyradiomics.readthedocs.io/en/2.1.0/features.html
- 22. Modat M, Cash DM, Winston GP, Duncan JS. Global image registration using a symmetric block‐matching approach. J Med Imaging. 2014;1:024003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alia OM, Mandava R, Aziz ME. A hybrid harmony search algorithm for MRI brain segmentation. Evol Intell. 2011;4:31–49. [Google Scholar]
- 24. Mcgraw KO. Forming inferences about some intraclass correlation coefficients. Psychol Methods. 1996;1:30–46. [Google Scholar]
- 25. Ellingson BM, Zaw T, Cloughesy TF, et al. Comparison between intensity normalization techniques for dynamic susceptibility contrast (DSC)‐MRI estimates of cerebral blood volume (CBV) in human gliomas. J Magn Reson Imaging. 2012;35:1472–1477. [DOI] [PubMed] [Google Scholar]
- 26. Tustison NJ, Cook PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]


