Abstract
Donor‐derived cell‐free DNA (dd‐cfDNA) is a noninvasive biomarker for comprehensive monitoring of allograft injury and rejection in kidney transplantation (KTx). dd‐cfDNA quantification of copies/mL plasma (dd‐cfDNA[cp/mL]) was compared to dd‐cfDNA fraction (dd‐cfDNA[%]) at prespecified visits in 189 patients over 1 year post KTx. In patients (N = 15, n = 22 samples) with biopsy‐proven rejection (BPR), median dd‐cfDNA(cp/mL) was 3.3‐fold and median dd‐cfDNA(%) 2.0‐fold higher (82 cp/mL; 0.57%, respectively) than medians in Stable Phase patients (N = 83, n = 408) without rejection (25 cp/mL; 0.29%). Results for acute tubular necrosis (ATN) were not significantly different from those with biopsy‐proven rejection (BPR). dd‐cfDNA identified unnecessary biopsies triggered by a rise in plasma creatinine. Receiver operating characteristic (ROC) analysis showed superior performance (P = .02) of measuring dd‐cfDNA(cp/mL) (AUC = 0.83) compared to dd‐cfDNA(%) (area under the curve [AUC] = 0.73). Diagnostic odds ratios were 7.31 for dd‐cfDNA(cp/mL), and 6.02 for dd‐cfDNA(%) at thresholds of 52 cp/mL and 0.43%, respectively. Plasma creatinine showed a low correlation (r = 0.37) with dd‐cfDNA(cp/mL). In a patient subset (N = 24) there was a significantly higher rate of patients with elevated dd‐cfDNA(cp/mL) with lower tacrolimus levels (<8 μg/L) compared to the group with higher tacrolimus concentrations (P = .0036) suggesting that dd‐cfDNA may detect inadequate immunosuppression resulting in subclinical graft damage. Absolute dd‐cfDNA(cp/mL) allowed for better discrimination than dd‐cfDNA(%) of KTx patients with BPR and is useful to avoid unnecessary biopsies.
Keywords: biomarker, clinical decision‐making, clinical research/practice, immunosuppressant, immunosuppression/immune modulation, kidney failure/injury, kidney transplantation/nephrology, rejection
Short abstract
Donor‐derived cell‐free DNA concentrations in combination with fractions measured repeatedly after kidney transplantation allow for clinical laboratory monitoring of graft damage, including rejection, to aid personalized patient care.
Abbreviations
- AMR
antibody‐mediated rejection
- ATN
acute tubular necrosis
- BPR
biopsy‐proven rejection
- D
days
- dd‐cfDNA
donor‐derived cell‐free DNA
- ddPCR
droplet digital PCR
- ddPCR‐LI
droplet digital PCR length index
- DSA
donor-specific antibody
- IF/TA
interstitial fibrosis/tubular atrophy
- IQR
interquartile range
- ISD
immunosuppressive drug
- KTx
kidney transplantation
- M
months
- N
number of patients
- n
number of samples
- PCR
polymerase chain reaction
- RCV
reference change value
- ROC
receiver operating characteristic
- TCMR
T cell–mediated rejection
- W
weeks
- Y
year
1. INTRODUCTION
Previous studies have evaluated the clinical validity of donor‐derived cell‐free DNA fraction (dd‐cfDNA[%]) as a noninvasive biomarker for comprehensive monitoring of allograft injury,1, 2, 3, 4, 5, 6 including in kidney transplantation (KTx). Since organ transplants are also genome transplants, this enables the discrimination of donor‐ from host‐derived cfDNA and noninvasive monitoring for allograft injury.2 This new approach could be useful to personalize immunosuppression and thereby improve outcomes.
There are about 19 000 KTx per year with over 200 000 living kidney allograft recipients in the United States.7 The 10‐year US kidney allograft survival rate is on average only 55% (47% for deceased, 63% for living donor transplants).8 This suboptimal graft survival rate illustrates the limitations of current methods used to monitor and treat KTx patients. About 100 000 patients are on waiting lists for KTx and 5000 to 10 000 kidney patients die prematurely each year.9 The median waiting time for a kidney is 3.6 years. The shortage of donor organs makes it especially important to reduce premature graft loss. Long‐term patient outcome after KTx is limited by multiple factors, namely irreversible chronic allograft dysfunction, acute or chronic rejection, and adverse effects of standard immunosuppression such as nephrotoxicity, cardiovascular disease, opportunistic infection, and malignancies.
Current diagnostic measures are unreliable for the early detection of kidney graft injury including acute and chronic rejection as well as subclinical rejection. Changes in plasma creatinine are most commonly used to decide whether a renal biopsy should be done to detect rejection. However, plasma creatinine is a measure of glomerular function rather than kidney tissue damage. An increase of plasma creatinine may also be due to exsiccation, the use of angiotensin converting enzyme (ACE) inhibitors, or immunosuppressive drug (ISD) toxicity. By the time a rejection‐related increase in plasma creatinine is evident, a significant degree of tissue damage has already occurred within the kidney.10 Therefore, interventions based on plasma creatinine may be too late or even inappropriate. ISD monitoring mainly indicates potential toxicity, but is a poor biomarker of graft damage.11 A further limitation of the current standard of care is that rejection episodes can be confirmed only by biopsies. However, serial biopsies to assess graft integrity (eg, to adjust and individualize ISD treatment) are clinically impractical, cost‐prohibitive, and a major burden for patients since biopsies have uncommon, but potentially serious complications. In addition, the reliability of biopsies is also limited, being prone to sampling and interpretation errors, and also have turnaround times that limit their usefulness for making rapid decisions.12
Against this background, rapid dd‐cfDNA determination has great potential as a noninvasive biomarker for early detection of acute or chronic rejection after KTx. Further important areas of application include the detection of asymptomatic graft injury leading to irreversible damage, the detection of under‐immunosuppression associated with the risk of de novo donor‐specific antibody (DSA) formation and subsequent graft loss,13 including the assessment of minimal necessary exposure to guide tapering and prevent immune activation.
At least 47 published studies in solid organ transplantation have investigated dd‐cfDNA(%) using shotgun or targeted next‐generation sequencing, or droplet digital PCR (ddPCR).5 For noninvasive routine monitoring, ddPCR seems to be a very practical approach with a clinically acceptable turnaround time (same day).
dd‐cfDNA(%) has recently been reported to be a sensitive, noninvasive biomarker of kidney transplant rejection superior to serum creatinine.4 Methods determining dd‐cfDNA(%), however, have the potential disadvantage of being affected by changes in the circulating recipient cfDNA.14 This limitation can be overcome by absolute quantification of dd‐cfDNA (eg, as genomic copies of dd‐cfDNA per mL of patient's plasma[cp/mL]).
A blinded, prospective, single center study was conducted in adult KTx patients in whom the time course of both fractional and absolute amount of dd‐cfDNA in plasma were determined.
2. MATERIALS AND METHODS
2.1. Study design
A total of 218 adult KTx patients were assessed for eligibility in a blinded, prospective, single‐center (Klinikum Stuttgart, Stuttgart, Germany), noninterventional study to examine the clinical validity of both dd‐cfDNA(%) and absolute number of dd‐cfDNA copies per mL of plasma (dd‐cfDNA[cp/mL]), compared to plasma creatinine for the detection of graft injury and acute rejection (Figure 1). Patients’ data evaluated for inclusion in the analyses included both types of dd‐cfDNA determination, clinical observations, immunosuppressant drug (ISD) treatment, as well as both biopsy and laboratory results used to detect graft injury.
Figure 1.
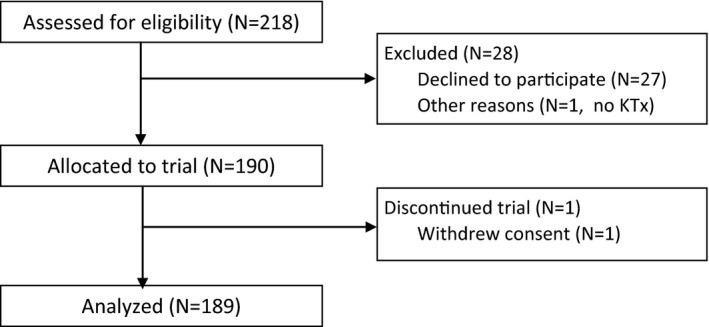
Selection of patients with samples included in statistical analyses based on compliance with predetermined inclusion criteria. KTx, kidney transplant
Of the 218 assessed patients, a total of 29 were excluded (Figure 1). The remaining 189 patients had made predetermined surveillance visits between September 2013 and October 2017. Study data collection was completed in October 2017 resulting in varying follow‐up times for individual patients. Daily samples were drawn on the exact given day (first 2 weeks); thereafter, samples were assigned to the closest scheduled sampling time point (ie, 3 weeks [W], 4W, months [M] 2, 3, 4, 5, 6, 8, 10, and 1 year [Y]) (Figure 2). During the first year post‐KTx, three of the 189 evaluated patients died.
Figure 2.
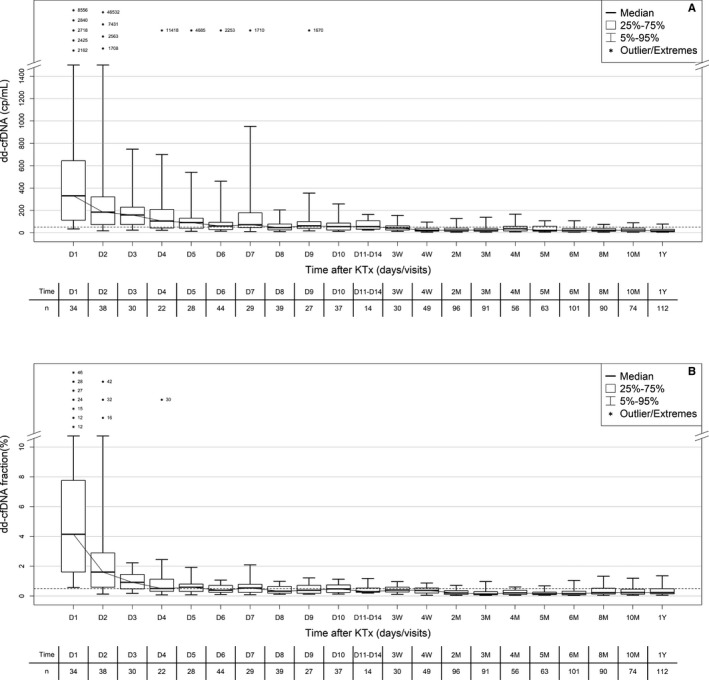
Time course in plasma dd‐cfDNA(cp/mL) (A) and dd‐cfDNA(%) (B) during the first year after KTx in patients' samples of Non‐rejecting Phase. Boxes represent median with interquartile range, with whiskers showing the 5th‐95th percentile. Predetermined visits and number of samples (n) are given below each time point. Values for outliers are shown as numbers (either as cp/mL or %). dd‐cfDNA, donor‐derived cell‐free DNA; KTx, kidney transplant
Demographic data for all included patients treated as part of this transplant center's regular KTx program are shown in Table 1.
Table 1.
Patient demographics, indications for KTx, graft number, compatibility data and induction agents used for the 189 patients who contributed data to the analyses presented
| Characteristic | Overall | Sample subgroups, N or mean ± SD | ||
|---|---|---|---|---|
| N | Percent or mean ± SD | Stable Phase | BPR | |
| Patients evaluated | 189 | 100% | 83 | 15 |
| Age (years) | 189 | 52 ± 14 | 49 ± 13 | 56 ± 10 |
| Gender | ||||
| Female | 69 | 36.5% | 33 | 7 |
| Male | 120 | 63.5% | 50 | 8 |
| Race | ||||
| Caucasian | 184 | 97.4% | 80 | 15 |
| Asian | 5 | 2.6% | 3 | 0 |
| Indication for KTx | ||||
| IgA nephropathy | 25 | 13.2% | 17 | 1 |
| Reflux nephropathy | 14 | 7.4% | 6 | 1 |
| FSGS | 3 | 1.6% | 1 | 0 |
| Polycystic kidney disease | 42 | 22.2% | 18 | 4 |
| Diabetes | 7 | 3.7% | 3 | 0 |
| Hypertension/nephrosclerosis | 11 | 5.8% | 4 | 2 |
| Alport syndrome | 5 | 2.6% | 1 | 0 |
| Interstitial nephropathy | 7 | 3.7% | 3 | 0 |
| Glomerulonephritis | 32 | 16.9% | 15 | 2 |
| Lupus erythematosus | 4 | 2.1% | 2 | 0 |
| Other | 38 | 20.1% | 13 | 5 |
| Prior grafts | ||||
| 0 | 161 | 85.2% | 71 | 12 |
| 1 | 23 | 12.2% | 10 | 3 |
| 2 | 4 | 2.1% | 1 | 0 |
| >2 | 1 | 0.5% | 1 | 0 |
| Donor type | ||||
| Living | 71 | 37.6% | 42 | 5 |
| Deceased | 118 | 61.9% | 41 | 10 |
| AB0 compatible | ||||
| Compatible | 165 | 87.3% | 72 | 12 |
| Incompatible | 24 | 12.7% | 11 | 3 |
| HLA mismatch | ||||
| 0 | 22 | 11.6% | 14 | 1 |
| 1‐2 | 42 | 22.2% | 23 | 1 |
| 3‐4 | 77 | 40.7% | 28 | 7 |
| 5‐6 | 48 | 25.4% | 18 | 6 |
| Induction agent | ||||
| Basiliximab | 139 | 73.5% | 59 | 10 |
| Rituximab | 21 | 11.1% | 9 | 2 |
| Thymoglobulin | 27 | 14.3% | 14 | 2 |
| Rituximab & Thymoglobulin | 2 | 1.1% | 1 | 1 |
| Delayed graft function | ||||
| Yes | 55 | 29.1% | 0 | 11 |
| No | 134 | 70.9% | 83 | 4 |
Abbreviations: BPR, biopsy‐proven rejection; FSGS, focal segmental glomerulosclerosis; HLA, human leukocyte antigen; IgA, immunoglobulin A; KTx, kidney transplant; SD, standard deviation.
The basic immunosuppressive regimens used typically consisted of induction therapy with basiliximab, rituximab or antithymocyte globulin (ATG), corticosteroids (variable withdrawal posttransplant), and maintenance therapy with calcineurin inhibitors tacrolimus (N = 189) or cyclosporine (N = 13) in combination with mycophenolate sodium (N = 189). Administration of everolimus was also documented in 12 patients. ISD dosing was adapted to achieve this center's therapeutic target trough ISD concentrations. Biopsy‐proven or clinically suspected rejection were treated with steroids (250 mg/day to 1 g/day over up to 3 days) and/or increased dosage or switches of standard immunosuppression.
Institutional ethics approval was obtained at the Medical Faculty University Tuebingen, Germany (approval number: 616/2013BO2‐), and all participants provided written, informed consent. Unblinding of dd‐cfDNA results and clinical data was done after the study was completed.
Missing a per‐protocol visit was not an exclusion criterion for a patient. An average of nine of the 15 per‐protocol visits was recorded for the 189 evaluated patients. All laboratory testing, including ISD concentrations, were measured as per this center's routine laboratory analyses, and all biopsies were performed, analyzed, and reported by experienced pathologists as per standard of care at this center. The indication for biopsy was based on increasing creatinine in plasma and clinical evaluation by the physicians. Protocol biopsies are not performed at the participating center because of risk/benefit considerations.
2.2. Blood collection and cfDNA extraction
Blood (~9 mL) was collected in ethylenediaminetetraacetic acid tubes. Plasma was separated within 2 h of collection from blood cells and stored frozen at −20°C until DNA extraction. Median storage time until DNA extraction was 16 weeks (IQR: 8‐44). Furthermore, an ethylenediaminetetraacetic acid–stabilized urine sample was collected once from a subset of patients (N = 158). Before extraction all plasma samples were centrifuged at 4000 g for 20 min at 4°C. cfDNA was extracted using the High Pure Viral Extraction Large Volume Kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's recommendations without the use of carrier RNA. The elution volume was 50 μL. An artificial spike was added to the plasma (1‐2 mL per sample) immediately before adding the protease/binding buffer. The spike consisted of a B.taurus derived 320 bp DNA (B.tau 4.61 BTA3:124.5 MB) that was prepared by PCR on a vector‐cloned fragment. The product was stored frozen and added to the plasma at ~6000 cp/mL freshly thawed immediately prior to extraction.
2.2.1. Measurement of dd‐cfDNA fraction(%)
The fractional abundance of the circulating donor‐derived DNA was determined by the dd‐cfDNA‐ddPCR method version‐1 as previously described15, 16 and detailed data on the analytical validity are given in the Supporting Information. Different from the original method, urine DNA was used for the determination of the informative SNP set for 158 of the 189 patients, since it is enriched with graft DNA.
2.2.2. dd‐cfDNA(cp/mL) quantification
Absolute quantification of haploid GcDNA genomic copies per mL plasma(cp/mL) is calculated by multiplying the total concentration of cfDNA(cp/mL) in a sample by the dd‐cfDNA fraction(%). The concentration of the total cfDNA is determined by a droplet digital PCR and is corrected for extraction efficiency and the reduced PCR efficiency resulting from the fragmentation of the cfDNA (see Supporting Information for further details).
2.3. Criteria for inclusion of samples and subgroup assignments
A total of 189 patients (N) had samples (n) drawn at time points that met preestablished inclusion criteria for use in the statistical analyses. The same patients could have samples assigned to various subcategories, depending on time after transplantation and their clinical condition around the sampling time (Supporting Information, Figure S1).
Criteria for inclusion and numbers in sample subcategories defined as “Non‐rejecting Phase,” “Stable Phase,” “Negative Biopsy,” “Borderline TCMR,” “BPR,” and “IF/TA” were:
“Non‐rejecting Phase” (N = 184; n = 1104): samples from patients with pathological biopsy findings within 15 days before or after dd‐cfDNA determination were excluded. Samples collected starting from day 1 postengraftment up to 1 year.
“Stable Phase” (N = 83; n = 408): samples collected during at least three consecutive visits at all of which the patient had none of the following exclusion criteria: clinical suspicion of rejection, received dialysis, received dialysis at the visit, BK‐virus (BK‐polyomavirus) infection diagnosed after KTx, any active infection, received a steroid bolus, plasmapheresis performed, change in function or medication, biopsy done, sample collected within 4 days after KTx; samples collected until day 10 postengraftment if an abnormal dd‐cfDNA decline was evident (outliers in Figure 2, D1‐D4), no renal surgery within 4 days before blood sampling. Category includes samples from Negative Biopsy group.
“Borderline TCMR” (N = 25; n = 34): samples collected ≤6 days before time of biopsy. Samples collected after a steroid bolus was given to the patient and samples collected within 4 days after KTx were excluded.
“BPR” (N = 15; n = 22): same as for borderline T cell–mediated rejection (TCMR).
“Negative Biopsy” (N = 7; n = 12)—biopsy without pathological result: same as for borderline TCMR, additionally samples from 1 to 6 days after a biopsy were included.
“IF/TA” (N = 24; n = 30)—interstitial fibrosis/tubular atrophy (IFTA): same as for borderline TCMR.
“ATN” (N = 29; n = 31)—acute tubular necrosis: same as for borderline TCMR. Combinations with other organ injuries were not included.
To avoid the influence of reperfusion injury, plasma samples for dd‐cfDNA determinations were included in the comparisons only if they were collected at least 5 days post‐KTx or later. In all patients with biopsies, no samples collected after a steroid bolus or abnormal dd‐cfDNA decline until day 10 were included.
2.4. Histopathologic diagnosis
All percutaneous biopsies were evaluated by pathologists who were unaware of the dd‐cfDNA results. Biopsies (N = 15) classified as biopsy‐proven rejection (BPR) included: Acute TCMR (N = 4), Active antibody‐mediated rejection (AMR) (N = 6), Chronic Active AMR (N = 1), mixed Acute TCMR, and Active AMR (N = 4). In general, BPRs were not severe. Allograft pathology was classified according to the Banff schema.17, 18 Biopsies reported as normal (N = 7, n = 12) were classified as biopsy negative; those showing borderline TCMR (N = 25, n = 38) as Borderline TCMR. Biopsies categorized as IF/TA (N = 24, n = 30), BK‐virus infection (N = 6, n = 10), and acute tubular necrosis (ATN) (N = 29, n = 31) were also included.
2.5. Statistical analysis
All statistical analyses were performed using R 3.5.1 (2018‐07‐02). Continuous data were presented with median and interquartile range (IQR) or mean and standard deviation, with 95% confidence intervals (CI) where feasible, whereas frequencies are reported as proportions. Box plots show the 5th and 95th percentiles as whiskers, 25th and 75th percentiles as a box and a median line. Observations of dd‐cfDNA fraction(%) and dd‐cfDNA(cp/mL) from subgroups such as Negative Biopsy, Stable Phase, Borderline TCMR, and BPR were compared using linear mixed‐effects models with random intercept, to account for repeated subject measurements. dd‐cfDNA values were log‐transformed prior to the linear mixed‐effects regression. Resulting P‐values of pairwise comparisons were reported unadjusted. All P < .05 were considered statistically significant.
Spearman correlation coefficients were calculated for dd‐cfDNA fraction(%), dd‐cfDNA(cp/mL) and plasma creatinine (mg/dL). Additionally, 95% bootstrap CI were reported.19 Association between nominal categories derived from dd‐cfDNA(cp/mL) and tacrolimus(μg/L) was assessed using Fisher's exact test.
Outliers in the Stable Phase group were statistically identified using a generalized extreme studentized deviate (GESD) test for outliers.20 dd‐cfDNA(cp/mL) and dd‐cfDNA fraction(%) were simultaneously considered in this univariate test by dimensionality reduction via principal component analysis (PCA). All analyses on diagnostic values (AUC, optimal thresholds, sensitivity, specificity, positive predictive value, negative predictive value) were performed excluding the identified outliers. Following ICH E9 guideline, analysis results for the uncleaned data set were also provided (Supporting Information, Table S1 and Figure S6).
ROC analyses were performed to assess how well dd‐cfDNA fraction(%) and dd‐cfDNA(cp/mL) discriminated between Stable Phase and BPR subgroups. ROC curves and AUCs were reported with 95% bootstrap CI.21, 22 Optimal thresholds were calculated using simultaneous maximization of sensitivity and specificity.23, 24 Sensitivity and specificity were reported with 95% bootstrap CI. Diagnostic odds ratios at optimal thresholds were reported with 95% CI.
A sensitivity analysis using one randomly chosen value per patient to evaluate the effect of repeated measures on the ROC analysis was conducted. Reference change values were calculated as described under Supporting Information.
3. RESULTS
3.1. Time dependence of dd‐cfDNA
The time dependence of dd‐cfDNA after KTx in the absence of clinically suspected rejection showed the pattern typically seen after solid organ transplantation. All 1104 samples from the “Non‐rejecting Phase” were used for this evaluation. Elevated values seen in the first days post‐KTx (presumably due to ischemia/reperfusion injury), were typically followed by a rapid decline, reaching baseline fractions and copy numbers about 4 to 6 days and 8 to 10 days, respectively, after engraftment (Figure 2). Outliers in Figure 2 represent such patients with abnormal delayed dd‐cfDNA decline after kidney engraftment.
In living donor patients, lower initial (first 5 days post‐KTx) dd‐cfDNA fractions and concentrations were seen compared to recipients of grafts from deceased donors, presumably due to less ischemia/reperfusion damage (Table 2). About 5 days after transplantation, in both types of donors, median dd‐cfDNA fraction(%) reached a value of about 0.6%. Median dd‐cfDNA(cp/mL) at 5 days post‐KTx results were still numerically higher (98 vs. 68 cp/mL, P = .5) in recipients of grafts from deceased vs living donors, respectively (Table 2).
Table 2.
Median dd‐cfDNA fractions and total amounts in patients during Non‐rejection Phase who received organs from deceased vs living donors
| Characteristic | Days (D) after KTx, median (IQR; n) | |||||
|---|---|---|---|---|---|---|
| Measurement | Donor type | D1 | D2 | D3 | D4 | D5 |
| dd‐cfDNA(%) | Deceased | 7.60 (4.54‐16.88) n = 16 | 2.02 (1.25‐3.05) n = 19 | 1.00 (0.71‐1.44) n = 17 | 0.48 (0.32‐1.21) n = 21 | 0.59 (0.37‐0.70) n = 18 |
| Living | 2.08 (1.45‐4.12) n = 18 | 1.41 (0.47‐2.47) n = 19 | 0.65 (0.26‐1.16) n = 13 | 0.68 (0.68‐0.68) n = 1 | 0.60 (0.22‐1.43) n = 10 | |
| dd‐cfDNA (cp/mL) | Deceased | 617 (332‐1291) n = 16 | 214 (166‐318) n = 19 | 180 (93‐361) n = 17 | 103 (43‐219) n = 21 | 98 (41‐180) n = 18 |
| Living | 143 (80‐370) n = 18 | 145 (52‐316) n = 19 | 102 (61‐170) n = 13 | 136 (136‐136) n = 1 | 68 (42‐111) n = 10 | |
Abbreviations: dd‐cfDNA, donor‐derived cell‐free DNA; IQR, interquartile range; KTx, kidney transplant.
Number of samples (n) equal to number of patients (N).
3.2. Diagnostic performance of dd‐cfDNA concentration vs fraction
For diagnostic discrimination analysis, the defined groups of Stable Phase samples were compared to BPR samples. dd‐cfDNA fraction(%) and dd‐cfDNA(cp/mL) values of 0.43% and 52 cp/mL, respectively, were identified as providing optimal discrimination between results obtained from Stable Phase and BPR patient groups by simultaneous optimization of sensitivity and specificity. However, threshold values of 0.50% (the 71st percentile for stable patients) and 50 cp/mL (the 70th percentile for stable patients) are suggested to allow for clinically more practical use.
Beginning at day 5, dd‐cfDNA results for Stable Phase sample group and Negative Biopsy samples were compared to results for samples of biopsy‐proven injuries (Figure 3, Supporting Information, Table S1). Seventy‐two percent of BPR episodes occurred between 5 and 14 days post‐KTx. Median dd‐cfDNA(%) in BPR samples was 0.57% (IQR: 0.36%‐0.89%) and median dd‐cfDNA(cp/mL) was 82 cp/mL (IQR: 53 cp/mL‐147 cp/mL). In Stable Phase patients, median dd‐cfDNA(%) was 0.29% (IQR: 0.17%‐0.56%) and median dd‐cfDNA (cp/mL) was 25 cp/mL (IQR: 11 cp/mL‐60 cp/mL). Median dd‐cfDNA(%) in BPR samples was 2.0‐fold higher than the median in Stable Phase patients (Figure 3). When data for absolute quantification of dd‐cfDNA(cp/mL) were compared between the same sample groups results showed a much better discrimination of BPR patients; median dd‐cfDNA(cp/mL) was 3.3‐fold higher than in the Stable Phase group. Biopsy proven Borderline TCMR patients also showed elevated dd‐cfDNA(%) and dd‐cfDNA(cp/mL) values, respectively. In patients with IF/TA median dd‐cfDNA(%) was 0.46% (IQR: 0.28%‐0.87%) and median dd‐cfDNA(cp/mL) was 35 cp/mL (IQR: 23‐84 cp/mL). In patients with ATN, median dd‐cfDNA(%) was 0.46% (IQR: 0.33%‐0.83%) and median dd‐cfDNA(cp/mL) was 64 cp/mL (IQR: 43 cp/mL‐126 cp/mL). Results for ATN were not significantly different from BPR (Figure 3, Supporting Information, Table S1). Median dd‐cfDNA(%) in BK‐virus positive samples was 0.40% (IQR: 0.13%‐1.56%) and median dd‐cfDNA(cp/mL) was 28 cp/mL (IQR: 13 cp/mL‐53 cp/mL).
Figure 3.
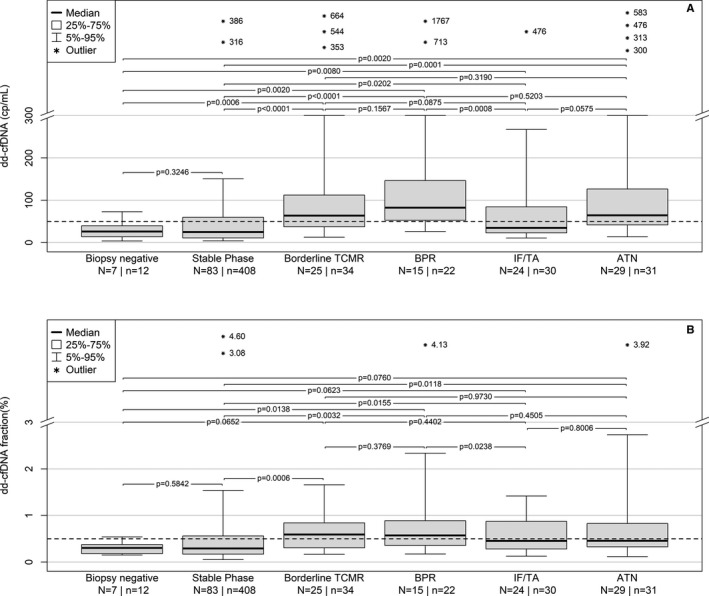
Comparison of post KTx dd‐cfDNA(cp/mL) (A) and dd‐cfDNA(%) (B) data from samples beginning at day 5 post‐KTx. ATN, acute tubular necrosis; BPR, biopsy‐proven rejection; dd‐cfDNA, donor‐derived cell‐free DNA; KTx, kidney transplant; TCMR, T cell‐mediated rejection
ROC analyses for the ability of dd‐cfDNA to discriminate acute rejection from Stable Phase samples excluding outliers (Figure 4) showed significantly (P = .022) better diagnostic accuracy for absolute quantification (AUC = 0.83; 95% CI, 0.74‐0.89) compared to dd‐cfDNA(%) (AUC = 0.73; 95% CI, 0.61‐0.82). ROC curve analysis was also performed including outliers (Supporting Information, Figure S6), where also a superior performance (P = .019) of dd‐cfDNA(cp/mL) compared to dd‐cfDNA(%) was observed. The reference change value (RCV) obtained in 24 patients was 58% for dd‐cfDNA(%) and 67% for dd‐cfDNA(cp/mL).
Figure 4.
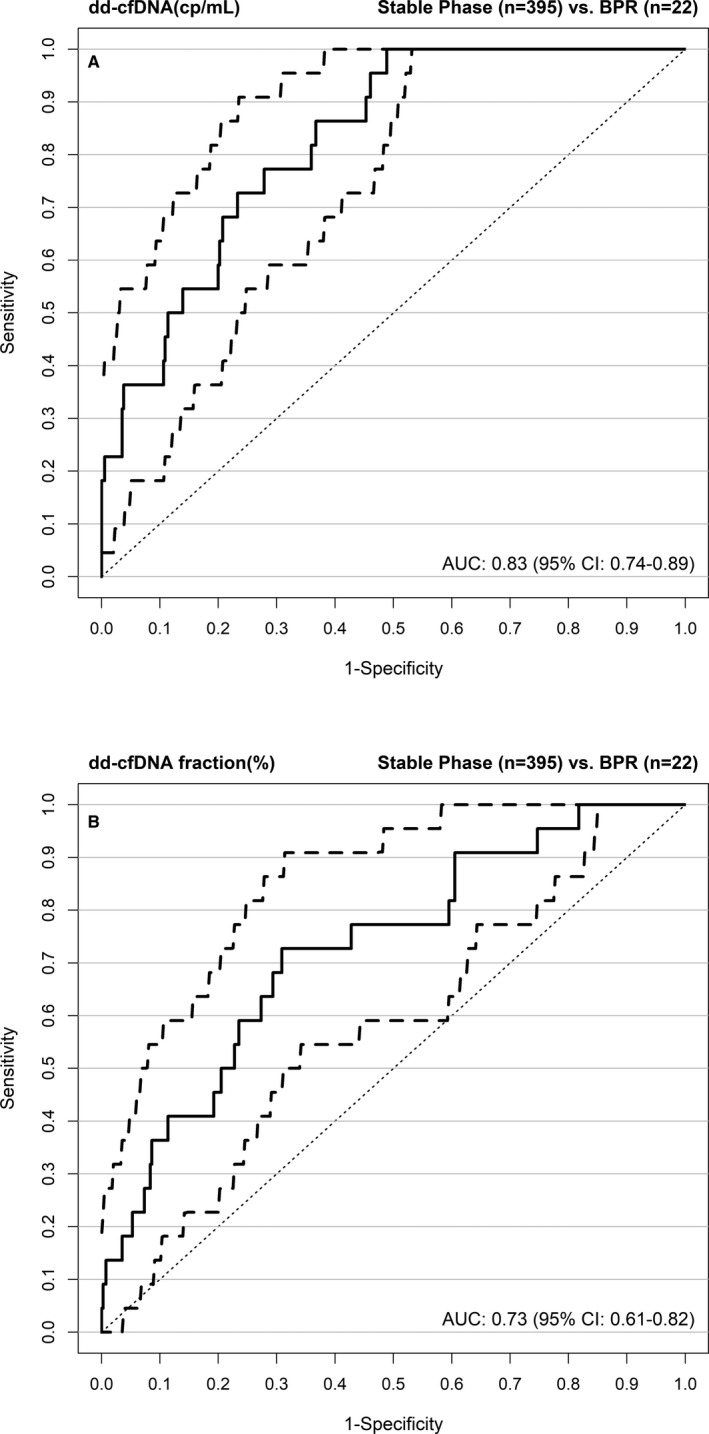
ROC curves for dd‐cfDNA(cp/mL) (A) and dd‐cfDNA(%) (B) showing the superior performance (P = .02) of measuring the absolute amount (cp/mL) of dd‐cfDNA rather than the dd‐cfDNA fraction(%). Outliers in the Stable Phase group were excluded from analysis. AUC, area under the curve; BPR, biopsy‐proven rejection; CI, confidence interval; dd‐cfDNA, donor‐derived cell‐free DNA; ROC, receiver operating characteristic
Results of simultaneous maximization of sensitivity and specificity obtained from ROC curves in BPR vs Stable Phase samples are shown in Table 3 and Supporting Information, Table S2. When outliers in the Stable Phase group were excluded prior to analysis, sensitivity was 73% for dd‐cfDNA(cp/mL) and dd‐cfDNA(%), while specificity was 73% for dd‐cfDNA(cp/mL) vs 69% for dd‐cfDNA(%) at thresholds of 52 cp/mL and 0.43%, respectively. Consequently, a comparison of dd‐cfDNA(cp/mL) vs dd‐cfDNA(%) revealed a higher diagnostic odds ratio of 7.31 (95% CI, 2.8‐19.2) for dd‐cfDNA(cp/mL) vs 6.02 (95% CI, 2.4‐15.1) for dd‐cfDNA(%) but very similar negative predictive values (98% in both cases). A sensitivity analysis with one randomly chosen observation per patient did not reveal any notable differences.
Table 3.
Diagnostic performance of dd‐cfDNA(cp/mL) and dd‐cfDNA(%) at calculated thresholds attained by simultaneous maximization of sensitivity and specificity. When feasible, values are given with 95% confidence interval. Outliers in the Stable Phase patient group were excluded prior to analysis
| dd‐cfDNA(cp/mL) | dd‐cfDNA(%) | |
|---|---|---|
| Threshold | 52 | 0.43 |
| Sensitivity | 0.73 (0.55‐0.91) | 0.73 (0.45‐0.86) |
| Specificity | 0.73 (0.59‐0.88) | 0.69 (0.37‐0.79) |
| PPV | 0.13 | 0.12 |
| NPV | 0.98 | 0.98 |
| DOR | 7.31 (2.8‐19.2) | 6.02 (2.4‐15.1) |
Abbreviations: BPR, biopsy‐proven rejection; DOR, diagnostic odds ratio; NPV, negative predictive value; PPV, positive predictive value; ROC, receiver operating characteristic.
Obtained from ROC curves in BPR (N = 15; n = 22) vs Stable Phase patients samples (N = 82; n = 395).
3.3. Detection of inadequate immunosuppression
The very high negative predictive values for both dd‐cfDNA results suggest that these tests can allow avoidance of unnecessary biopsies triggered by elevated or increasing plasma creatinine. This possibility is also supported by a comparison of results for plasma creatinine, dd‐cfDNA(%) and dd‐cfDNA(cp/mL) in patients (N = 7) who had negative biopsies and elevated plasma creatinine (Figure 5). In six of the seven cases both dd‐cfDNA values were below their respective thresholds. Of note, the correlation coefficient between dd‐cfDNA(cp/mL) and dd‐cfDNA(%) was 0.67 (95% CI, 0.65‐0.70) while creatinine showed only a low correlation coefficient of 0.37 (95% CI, 0.32‐0.41) with dd‐cfDNA(cp/mL).
Figure 5.
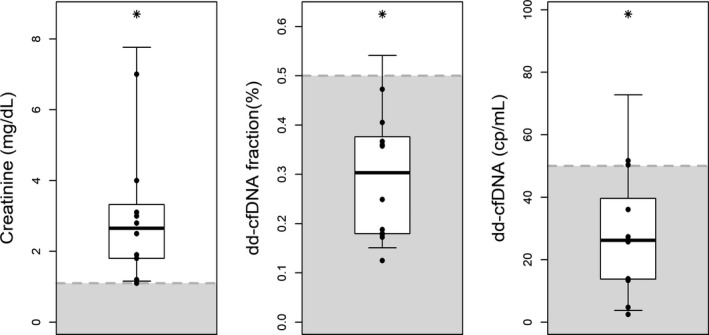
Same‐day plasma creatinine, dd‐cfDNA(%) and dd‐cfDNA(cp/mL) results in samples drawn during the period from 6 days before to 6 days after the biopsy, but excluding the day of the biopsy, in seven patients who had normal biopsy findings. Shaded areas show values below the upper limit of the plasma creatinine reference interval and thresholds for dd‐cfDNA. dd‐cfDNA, donor‐derived cell‐free DNA
The possibility that inadequate immunosuppression was responsible for at least some of the variations in dd‐cfDNA values was explored in a selected patient subgroup (N = 24) without clinically suspected rejection, each of whom had both, a >60% change in tacrolimus concentrations along with a change in the opposite direction for dd‐cfDNA(cp/mL) in samples collected during at least three consecutive visits. There was a significantly higher proportion of samples having elevated dd‐cfDNA(cp/mL) and lower tacrolimus levels (<8 μg/L) compared to samples with higher tacrolimus concentrations (63% vs. 27%, P = .0036), as shown in Figure 6. A similar association (data not shown) was found for tacrolimus concentrations and dd‐cfDNA(%).
Figure 6.
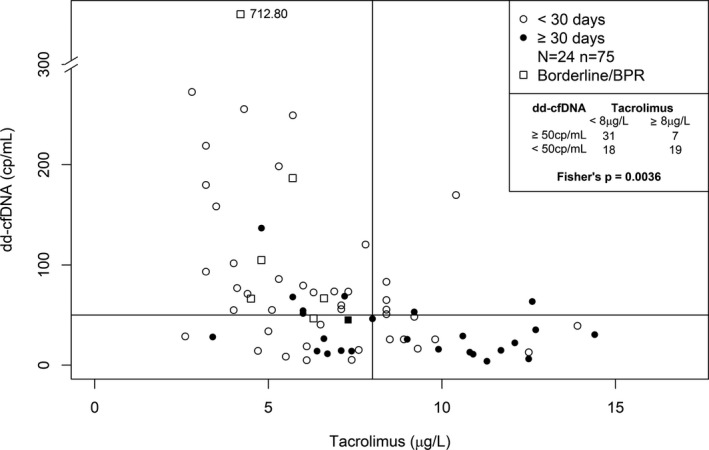
Association between low predose tacrolimus concentrations and increased dd‐cfDNA(cp/mL) in patients (N = 24) with a change of tacrolimus concentration >60% in samples (n = 75) collected at three consecutive visits. Lines represent the cut‐off points for Fisher test (Tacrolimus: 8 μg/L; dd‐cfDNA: 50 cp/mL). BPR, biopsy‐proven rejection; dd‐cfDNA, donor‐derived cell‐free DNA
4. DISCUSSION
There are no existing biomarkers in kidney transplantation that adequately measure the status of active injury to the allograft, or indicate a sufficient immunosuppression. In contrast to plasma creatinine, which is a widely used marker of renal function, dd‐cfDNA is a biomarker of allograft injury. The current standard is to perform invasive graft biopsies which are subject to variability of interpretation and are unsuited for frequent monitoring.12 dd‐cfDNA in combination with ISD monitoring has the potential to reduce premature graft loss and reduce costs. This is important for KTx due to the shortage of donor organs. In addition, there are health economic implications, since acute rejection events are associated with significant increases in the cost of care (17 000 to 28 000 USD per year).11 If the patient has to be retransplanted, the average cost is 111 891 USD. Kidney graft failure with return to dialysis causes an average annual expense of 75 836 USD, a multiple of annual costs for a patient with a functioning graft (19 364 USD).
There is emerging evidence for the clinical validity of dd‐cfDNA determination for early detection of rejection episodes at an actionable stage.4, 5, 25, 26 Elevation of dd‐cfDNA is not rejection specific, but reveals the degree of graft cell injury, complements histologic findings, and most importantly can help to avoid unnecessary biopsies, based on the high negative predictive value shown herein. As described by Whitlam14 and Bloom,4 we also observed elevations of dd‐cfDNA in patients with IF/TA. In general, results can be used to monitor individual responses to rejection treatments, to detect under‐immunosuppression (eg, noncompliance), and can be helpful to achieve personalized immunosuppression. Therefore, dd‐cfDNA monitoring can increase the effectiveness of transplant recipient surveillance and allows a shift in emphasis from reaction to prevention.
The use of relative dd‐cfDNA fraction—usually calculated as dd‐cfDNA(%)—however, has the disadvantage that results can be influenced by the amounts of cfDNA from the recipient. Certain conditions (eg, infections, exercise, non–graft‐associated vascular compromise, medications) can result in recipient cell damage causing an increased release of host DNA into the blood stream and thus lowering dd‐cfDNA(%). Most importantly, the vast majority of cfDNA (~90%) in a plasma sample stems from white blood cells (ie, neutrophils and lymphocytes) undergoing natural apoptosis in the blood.27 Eliminating the influence of variability in total cfDNA concentration (denominator for fraction calculation) seems to be important in a setting where the backbone of therapy is drug interventions targeting the immune system which cause variations in both the number and types of recipient blood cells present. Avoiding this biasing influence can be achieved by using absolute quantification as copies per mL plasma, which is not affected by changes in recipient cfDNA.
In this study, dd‐cfDNA(cp/mL) showed statistically significant superior ability to discriminate between KTx patients with vs without graft damage or rejection (Figures 3 and 4) compared with dd‐cfDNA(%). Results for ATN (reflecting tubular damage) were not significantly different from BPR.
The high negative predictive value of dd‐cfDNA for BPR is the reason why this test can be helpful to avoid unnecessary biopsies. In our study, six of seven negative biopsies triggered by elevated plasma creatinine could have been avoided as dd‐cfDNA fraction and concentration were below the respective thresholds. This is particularly important in those patients with a high biopsy risk.
The association found between increased dd‐cfDNA values and low tacrolimus concentrations post KTx (Figure 6) suggests that dd‐cfDNA can be useful to detect subclinical (ie, clinically unsuspected) graft damage as a result of immune activation triggered by under‐immunosuppression. This is consistent with observations in liver transplantation.28 Persistent under‐immunosuppression can cause de novo DSA formation, which increases the risk of subsequent graft loss.13, 29, 30, 31 Early diagnosis and treatment of subclinical AMR may improve outcomes after KTx.32, 33 However, the detection of IgG DSA has failed to reliably predict ongoing AMR.34 AMR is associated with 20% to 30% allograft loss.35 The results shown in Figure 6 are also consistent with the hypothesis that the dd‐cfDNA elevations seen in some “Stable Phase” samples (Figures 2 and 3) were in fact caused by clinically unsuspected graft damage. Premature or individually too aggressive ISD tapering is one possible explanation for some dd‐cfDNA elevations seen in our patients who were classified as clinically stable. In our study site, tapering of tacrolimus was started after 3 months with a reduction of the target range to 4‐6 μg/L (AB0/HLA incompatible patients: 6‐8 μg/L).
So far, studies directly comparing different dd‐cfDNA determination methods are lacking. It is encouraging, however, that the median values of clinically stable patients were very similar in different studies using different methods for dd‐cfDNA determination.4, 36, 37, 38 In our cohort, the median was 0.29% (IQR: 0.17%‐0.56%). Bromberg36 published a value of 0.21% (IQR: 0.12%‐0.39%), Bloom4 of 0.30% (IQR: 0.14%‐0.77%), Sigdel37 of 0.40%, and Gielis38 a mean value of 0.46% (±0.21%). Bloom and Sigdel proposed a dd‐cfDNA(%) cut‐off value at 1%. The 97.5th percentile of Stable Phase patients reported by Bromberg et al36 was 1.20% and in our study 1.03% excluding ties in a sensitivity analysis. We suggest a cut‐off at 0.5% based on simultaneous maximization of sensitivity and specificity from our study data.
The authors are aware of only one recent report by Whitlam et al, regarding absolute quantification of dd‐cfDNA in KTx, wherein a median for patients with negative biopsy of 7 cp/mL (IQR: 5‐11 cp/mL) was observed.14 This is substantially lower than the median value we observed in clinically stable patients of 25 cp/mL (IQR: 11‐60 cp/mL) in our data. The obvious discrepancy appears to be due to methodological differences. In contrast to the method used herein, Whitlam et al did not account for cfDNA extraction and ddPCR amplification efficiency. Together, both effects account for a 44%‐55% underestimation of the real copy numbers/mL compared to our standardized method (which eliminates both negatively biasing effects). These methodological differences explain the difference between the cut‐off point of 21 cp/mL selected by Whitlam et al and the rounded cut‐off at 50 cp/mL we used.
Limitations of this study include the lack of protocol biopsies and the fact that the vast majority of patients were of Caucasian origin.
The data from our study support the role of dd‐cfDNA as a pivotal addition to the methods currently used to achieve personalized immunosuppression in KTx, such as immunological monitoring, therapeutic drug monitoring, microbial screening, and biopsy. Based on all this information, in agreement with other reports,5, 37 we are confident that dd‐cfDNA can be recommended for clinical translation as a valid tool to aid personalized patient care for the benefit of transplanted patients and healthcare payers. To achieve this, methods providing a fast turnaround time, such as the ddPCR used herein, will allow frequent monitoring and provide actionable results (eg, to aid in decision‐making for biopsy).
In conclusion, the absolute quantification of dd‐cfDNA(cp/mL) was superior to dd‐cfDNA fraction (%) as a biomarker of BPR. This is presumably because dd‐cfDNA(cp/mL), unlike dd‐cfDNA(%), is not affected by changes in recipient DNA present in the circulation. We, however, concur with Whitlam et al14 that considering both absolute quantification of dd‐cfDNA(cp/mL) and dd‐cfDNA fraction together may provide additional diagnostic information. The findings strengthen the evidence that dd‐cfDNA determinations after transplantation are useful to avoid unnecessary biopsies triggered by plasma creatinine elevations and to detect asymptomatic, subclinical graft damage, including from inadequate immunosuppression.
AUTHOR CONTRIBUTIONS
E.W., M.S., V.S., M.K., and G.H. were responsible for recruitment, sample collection, and documentation of clinical data. N.M. and J.B. performed the laboratory work. T.A. and T.F. performed the statistical data analysis. M.O., P.D.W., E.S., and J.B. wrote the manuscript and edited the paper, and provided scientific advice. H.J.G., V.S., E.W., and M.S. provided clinical and scientific advice.
DISCLOSURE
M.O. acts as consultant and advisor to Chronix Biomedical. E.S. and J.B. are employees of and own stock and intellectual property rights at Chronix Biomedical. The other authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting information
ACKNOWLEDGMENTS
The project was supported by the German Federal Ministry of Education and Research, Project Management Jülich to ES, MO/TF, and EW (KMU‐innovativ‐13: FKZ 031A584A+B). The funders had no role in study design, data collection and analyses, decision to publish, or preparation of the manuscript.
Oellerich M, Shipkova M, Asendorf T, et al. Absolute quantification of donor‐derived cell‐free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant. 2019;19:3087‐3099. 10.1111/ajt.15416
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Snyder TM, Khush KK, Valantine HA, et al. Universal noninvasive detection of solid organ transplant rejection. Proc Natl Acad Sci USA. 2011;108:6229‐6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell‐free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schütz E, Fischer A, Beck J, et al. Graft‐derived cell‐free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: a prospective, observational, multicenter cohort study. PLoS Med. 2017;14:e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloom RD, Bromberg JS, Poggio ED, et al. Cell‐free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221‐2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Knight SR, Thorne A, Faro MLL. Donor‐specific cell‐free DNA as a biomarker in solid organ transplantation. A systematic review. Transplantation. 2018;103:273‐283. [DOI] [PubMed] [Google Scholar]
- 6. Agbor‐Enoh S, Jackson AM, Tunc I, et al. Late manifestation of alloantibody‐associated injury and clinical pulmonary antibody‐mediated rejection: evidence from cell‐free DNA analysis. J Heart Lung Transplant. 2018;37:925‐932. [DOI] [PubMed] [Google Scholar]
- 7. OPTN/SRTR Annual Data Report 2016. Am J Transplant. 2018;18(S1):1‐503. [Google Scholar]
- 8. Arnaud CH, C&EN . Uncovering the hidden signs of organ transplant rejection. 2018;96(5): 26‐30. https://cen.acs.org [Google Scholar]
- 9. Held PJ, McCormick F, Ojo A, et al. A cost‐benefit analysis of government compensation of kidney donors. Am J Transplant. 2016;16:877‐885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. American Society of Nephrology . American Society of Nephrology renal research report. J Am Soc Nephrol. 2005;16:1886‐1903. [DOI] [PubMed] [Google Scholar]
- 11. First MR, Lee D, Lewis P, et al. An economic analysis of the cost effectiveness of blood gene expression profiling in kidney transplant recipients. J Health Med Econ. 2017;3:3. [Google Scholar]
- 12. Miller CA, Fildes JE, Ray SG, et al. Non‐invasive approaches for the diagnosis of acute cardiac allograft rejection. Heart. 2013;99:445‐453. [DOI] [PubMed] [Google Scholar]
- 13. Wiebe C, Gibson IW, Blydt‐Hansen TD, et al. Evolution and clinical pathologic correlations of de novo donor‐specific HLA antibody post kidney transplant. Am J Transplant. 2012;12:1157‐1167. [DOI] [PubMed] [Google Scholar]
- 14. Whitlam JB, Ling L, Skene A, et al. Diagnostic application of kidney allograft‐derived absolute cell‐free DNA levels during transplant dysfunction. Am J Transplant. 2019;19:1037‐1049. [DOI] [PubMed] [Google Scholar]
- 15. Beck J, Bierau S, Balzer S, et al. Digital droplet PCR for rapid quantification of donor DNA in the circulation of transplant recipients as a potential universal biomarker of graft injury. Clin Chem. 2013;59:1732‐1741. [DOI] [PubMed] [Google Scholar]
- 16. Beck J, Oellerich M, Schütz E. A universal droplet digital PCR approach for monitoring of graft health after transplantation using a preselected SNP set In: Karlin‐Neumann G, Bizouarn F, eds. Methods in Molecular Biology: Digital PCR. Methods and Protocols, 1st edn New York: Humana Press; 2018:335‐348. [DOI] [PubMed] [Google Scholar]
- 17. Haas M, Loupy A, Lefaucheur C, et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell‐mediated rejection, antibody‐mediated rejection, and prospects for integrative endpoints for next‐generation clinical trials. Am J Transplant. 2018;18:293‐307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roufosse C, Simmonds N, Clahsen‐van Groningen M, et al. A 2018 reference guide to the Banff classification of renal allograft pathology. Transplantation. 2018;102:1795‐1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hervé M. RVAideMemoire: testing and plotting procedures for biostatistics. R package version 0.9‐70. 2018. https://CRAN.R-project.org/package=RVAideMemoire. Accessed January 10, 2019.
- 20. Rosner B. Percentage points for a generalized ESD many‐outlier procedure. Technometrics. 1983;25:165‐172. [Google Scholar]
- 21. DeLong ER, DeLong DM, Clarke‐Pearson DL. Comparing the areas under two or more correlated receiver operating characteristics curves: a nonparametric approach. Biometrics. 1988;44:837‐845. [PubMed] [Google Scholar]
- 22. Konietschke F, Placzek M, Schaarschmidt F, et al. nparcomp: an R software package for nonparametric multiple comparisons and simultaneous confidence intervals. J Stat Softw. 2015;64:1‐17. [Google Scholar]
- 23. Riddle DL, Stratford PW. Interpreting validity indexes for diagnostic tests: an illustration using the Berg balance test. Phys Ther. 1999;79:939‐948. [PubMed] [Google Scholar]
- 24. Gallop RJ, Crits‐Christoph P, Muenz LR, et al. Determination and interpretation of the optimal operating point for ROC curves derived through generalized linear models. Underst Stat. 2003;2:219‐242. [Google Scholar]
- 25. Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical‐grade assay to measure donor‐derived cell‐free DNA in solid organ transplant recipients. J Mol Diagn. 2016;18:890‐902. [DOI] [PubMed] [Google Scholar]
- 26. Gielis EM, Ledeganck KJ, De Winter BY, et al. Cell‐free DNA: an upcoming biomarker in transplantation. Am J Transplant. 2015;15:2541‐2551. [DOI] [PubMed] [Google Scholar]
- 27. Kim S, Jiang P, Chan KCA, et al. Plasma DNA tissue mapping by genome‐wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci USA. 2015;112:E5503‐E5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oellerich M, Schütz E, Kanzow P, et al. Use of graft‐derived cell‐free DNA as an organ integrity biomarker to reexamine effective tacrolimus trough concentrations after liver transplantation. Ther Drug Monit. 2014;36:136‐140. [DOI] [PubMed] [Google Scholar]
- 29. Hoshino J, Kaneku H, Everly MJ, et al. Using donor‐specific antibodies to monitor the need for immunosuppression. Transplantation. 2012;93:1173‐1178. [DOI] [PubMed] [Google Scholar]
- 30. Garg N, Samaniego MD, Clark D, et al. Defining the phenotype of antibody‐mediated rejection in kidney transplantation: advances in diagnosis of antibody injury. Transplant Rev. 2017;31:257‐267. [DOI] [PubMed] [Google Scholar]
- 31. Sellarés J, de Freitas DG, Mengel M, et al. Understanding the causes of kidney transplant failure: the dominant tole of antibody‐mediated rejection and nonadherence. Am J Transplant. 2012;12:388‐399. [DOI] [PubMed] [Google Scholar]
- 32. Loupy A, Vernerey D, Tinel C, et al. Subclinical rejection phenotypes at 1 year post‐transplant and outcome of kidney allografts. J Am Soc Nephrol. 2015;26:1721‐1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parajuli S, Joachim E, Alagusundaramoorthy S, et al. Subclinical antibody mediated rejection after kidney transplantation: treatment outcomes. [published online ahead of print 2019] Transplantation. 2019. 10.1097/TP.00000000000002566. [DOI] [PubMed] [Google Scholar]
- 34. Eskandary F, Bond G, Kozakowski N, et al. Diagnostic contribution of donor‐specific antibody characteristics to uncover late silent antibody‐mediated rejection – results of a cross‐sectional screening study. Transplantation. 2017;101:631‐641. [DOI] [PubMed] [Google Scholar]
- 35. Kim M, Martin ST, Townsend KR, et al. Antibody‐mediated rejection in kidney transplantation: a review of pathophysiology, diagnosis, and treatment options. Pharmacotherapy. 2014;34:733‐744. [DOI] [PubMed] [Google Scholar]
- 36. Bromberg J, Brennan DC, Poggio E, et al. Biological variation of donor‐derived cell‐free DNA in renal transplant recipients: clinical implications. J Appl Lab Med. 2017;02:309‐321. [DOI] [PubMed] [Google Scholar]
- 37. Sigdel TK, Acosta Archila F, Constantin T, et al. Optimizing detection of kidney transplant injury by assessment of donor‐derived cell‐free DNA via massively multiplex PCR. J Clin Med. 2019;8:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gielis EM, Beinaert C, Dendooven A, et al. Plasma donor‐derived cell‐free DNA kinetics after kidney transplantation using a single tube multiplex PCR assay. PLoS ONE. 2018;13(12):e0208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


