Abstract
Mycobacterium tuberculosis (Mtb) manipulates multiple host defence pathways to survive and persist in host cells. Understanding Mtb–host cell interaction is crucial to develop an efficient means to control the disease. Here, we applied the Mtb proteome chip, through separately interacting with H37Ra and H37Rv stimulated macrophage lysates, screened 283 Mtb differential proteins. Through primary screening, we focused on fatty acylCoA synthetase FadD13. Mtb FadD13 is a potential drug target, but its role in infection remains unclear. Deletion of FadD13 in Mtb reduced the production of proinflammatory cytokines IL‐1β, IL‐18, and IL‐6. Bimolecular fluorescence complementation and colocalization showed that the binding partner of FadD13 in macrophage was eEF1A1 (a translation elongation factor). Knockdown eEF1A1 expression in macrophage abrogated the promotion of proinflammatory cytokines induced by FadD13. In addition, ΔfadD13 mutant decreased the expression of the NF‐κB signalling pathway related proteins p50 and p65, so did the eEF1A1 knockdown macrophage infected with H37Rv. Meanwhile, we found that deletion of FadD13 reduced Mtb survival in macrophages during Mtb infection, and purified FadD13 proteins induced broken of macrophage membrane. Taken together, FadD13 is crucial for Mtb proliferation in macrophages, and it plays a key role in the production of proinflammatory cytokines during Mtb infection.
Keywords: cytokines, eEF1A1, fatty acylCoA synthetase, Mycobacterium tuberculosis, NF‐kappa B signalling pathway
1. INTRODUCTION
Tuberculosis remains one of the leading causes of morbidity and mortality worldwide. Understanding Mycobacterium tuberculosis (Mtb)–host cell interaction is crucial to develop an efficient means to control the disease. A number of Mtb proteins have been identified their toxicity by interacting with the human macrophage proteins (Griffin et al., 2011; Nair et al., 2009; Parveen et al., 2013; Pathak et al., 2007; Sreejit et al., 2014). As tuberculosis is a complex disease, many proteins may involve in disease development. Building a protein–protein interaction network relies on high throughput tools. To fulfil this demand, we had previously developed an Mtb proteome bank and a proteome microarray (protein chip), which covers about 95% open reading frames of the Mtb genome (Deng et al., 2014). With this unique platform, PPI (Wu et al., 2017), small molecule/drug‐protein binding (Zhang et al., 2017), and the binding partner of Mtb protein BfrB in macrophages (He et al., 2017) were investigated.
In this study, we also used this powerful platform to screen the Mtb virulence‐associated proteins that can interact with macrophage proteins. Over 500 Mtb proteins showed positive signal in the interaction between the macrophage proteome and Mtb proteome chip. Among them, 283 Mtb proteins were differentiated between the avirulent strain H37Ra stimulated and virulent strain H37Rv stimulated macrophage experiments, which maybe virulence related. Gene ontology (GO) categorisation showed that these differential Mtb proteins mainly gathered in cell wall‐associated. So we select several proteins, which are secretion and cell wall related, for further research. In this study, through primary screening, we focused on FadD13 protein.
FadD13 (Rv3089) is a fatty acylcoA synthetase that catalyses the activation of various fatty acids by converting them into fatty acyl‐CoA thioesters and is related with the synthesis of Mtb lipid rich cell envelope (Cleland, 1963). Also, fadD13 is known to be the last gene of the MymA operon of Mtb (Rv3083‐Rv3089), which is required to maintain the appropriate mycolic acid composition and permeability of the Mtb envelope upon exposure to an environment with an acidic pH (Singh et al., 2005). The cell envelope of Mtb is essential to the virulence and resistance of Mtb to hostile environments. The functional loss of the MymA operon of Mtb results in (a) alterations in colony morphology, (b) change of the cell wall structure and thus increase in drug sensitivity, (c) killing of the pathogen by activated macrophages, and (d) a reduction in the ability to persist specifically in the spleen of infected guinea pigs (Singh et al., 2005; Singh, Jain, Gupta, Das, & Tyagi, 2003). The regulatory roles of the FadD13 single protein in host innate immune signalling pathways and cellular functions during mycobacterial infection remain unknown.
Interaction partner of FadD13 in macrophage we identified in this study is eukaryotic translation elongation factor‐1α1 (eEF1A1), which is the second‐most abundant eukaryotic protein, responsible for ribosomal protein synthesis (Burglova et al., 2018). It belongs to the GTP‐binding protein family and promotes the GTP‐dependent binding of aminoacyl‐tRNA (aa‐tRNA) to the A site of the ribosome during the elongation cycle of protein biosynthesis (Kaziro, Itoh, Kozasa, Nakafuku, & Satoh, 1991). In addition, eEF1A1 was found to relate to chaperone‐mediated autophagy (Bandyopadhyay, Sridhar, Kaushik, Kiffin, & Cuervo, 2010) and apoptosis (Duttaroy, Bourbeau, Wang, & Wang, 1998). Importantly, it forms a complex with PARP1 and TXK to act as a T helper 1 (Th1) cell‐specific transcription factor, which binds the promoter of IFN‐gamma to directly regulate IFN‐gamma transcription, and is thus critically involved in Th1 cytokine production (Maruyama, Nara, Yoshikawa, & Suzuki, 2007).
Based on the knowledge, we deduced that eEF1A1 may play a role in the regulation of proinflammatory cytokine production in macrophages infected with Mtb, and interaction of FadD13 with eEF1A1 may have significant effect on it. After a systematic experimental investigation, we found that knockout and overexpression FadD13 significantly altered the amount of eEF1A1 in macrophages and caused downregulation and upregulation of the NF‐κB signalling pathway‐related proteins respectively. The results are presented and discussed in this report.
2. RESULTS
2.1. Screening of Mtb virulence proteins that interact with total proteins from macrophages
To globally identify the Mtb proteins that interact with human macrophage proteins and maybe virulence related, different interaction experiments were designed and performed using Mtb protein chip. Briefly, differentiated THP‐1 macrophages were infected separately with the virulent strain H37Rv and avirulent strain H37Ra, and the cell lysates of the macrophage were incubated with the Mtb proteome microarray (Figure 1a). The Mtb proteins that bind to the macrophage proteins were found by the fluorescence signal readout. Then, the differential binding of Mtb proteins between these two groups were analysed. As a result, a total of 283 differential proteins were identified. Gene ontology (GO) categorisation showed that they are mainly gathered in cell wall‐associated proteins (Figure S1a), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway categorisation revealed that the proteins mainly belong to biosynthesis of secondary metabolites, biosynthesis of amino acids, and purine metabolism (Figure S1b). To further understand the interactions between the 283 differential proteins, a protein–protein network map was established using STRING 10.5 software. The subnetworks were obtained from sorting interaction networks according to the GO and KEGG terms by BiNGO v3.0.3 software. Obviously, there is a good relationship between each subnetwork proteins. GO analysis shows that 58 differential proteins were involved in the cell wall, among which, 33 could link together (Figure 1b). Among the cell wall subnetwork, a number of proteins, such as Rv0120c, GuaA and FadD13, located at the node, which were obvious. The other clusters relating to biosynthesis of secondary metabolites, biosynthesis of amino acids and purine metabolism are shown in Figure S1c,d,e. From these subnetworks, we selected several proteins to evaluate if they are related to virulence or infectivity, we used the overexpression M. smegmatis strains to stimulate macrophages respectively. In the preliminary study, we found FadD13 overexpression M. smegmatis strain promoted the aggregation of M. smegmatis and increased the production of proinflammatory cytokines in macrophages. This led us to decide to conduct a systematic investigation of the function of FadD13.
Figure 1.
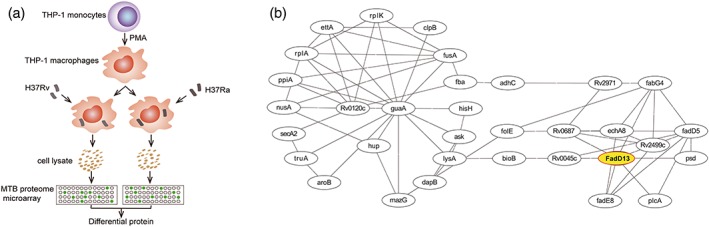
Screening of Mtb virulence proteins that interact with proteins from macrophages. (a) Scheme of protein screening approach. Briefly, differentiated THP‐1 macrophages were infected at a multiplicity of infection (MOI) of 10 with the virulent strain H37Rv and avirulent strain H37Ra for 12 hr, respectively, and the cell lysates of the macrophages were incubated with the Mtb proteome microarray. Then, the differential Mtb proteins that bound to the macrophage proteins were analysed. This presented as the one replicate from two independent experiments. (b) The subnetwork of the differential proteins related to cell wall. The interaction between differential proteins was analysed by STRING (p < .05) and the subnetworks were obtained from sorting interaction networks according to the GO and KEGG terms using BiNGO v3.0.3 software
2.2. Mtb FadD13 promotes proinflammatory cytokines production in macrophages
To investigate the function of FadD13, we failed in construction of the overexpression Mtb strains. To speed up the research process, we constructed overexpression M. smegmatis, which belongs to the same genus of Mtb and is widely used as the model strain in Mtb research, for function study. The verification result of the FadD13 overexpression in M. smegmatis is shown in Figure S2. At the same time, we constructed fadD13 gene knockout mutant in H37Rv, the verification result is shown in Figure S3.
Inflammatory cytokines are essential for the recruitment and activation of cells of the immune system in response to bacterial infection. In the preliminary study, we found FadD13 overexpression M. smegmatis strain promoted the production of proinflammatory cytokines IL‐1β, IL‐18, and IL‐6 in macrophages (Figure 2a), but it could not induce the production of anti‐inflammatory cytokine IL‐10 (data not shown). In line with this, FadD13 knockout mutant decreased the production of proinflammatory cytokines in the macrophage (Figure 2b), but it had no influence on the production of anti‐inflammatory cytokine IL‐10 (Figure 2c). Therefore, the experimental data highly suggest that Mtb FadD13 plays an important role in production of proinflammatory cytokines in macrophages during Mtb infection.
Figure 2.
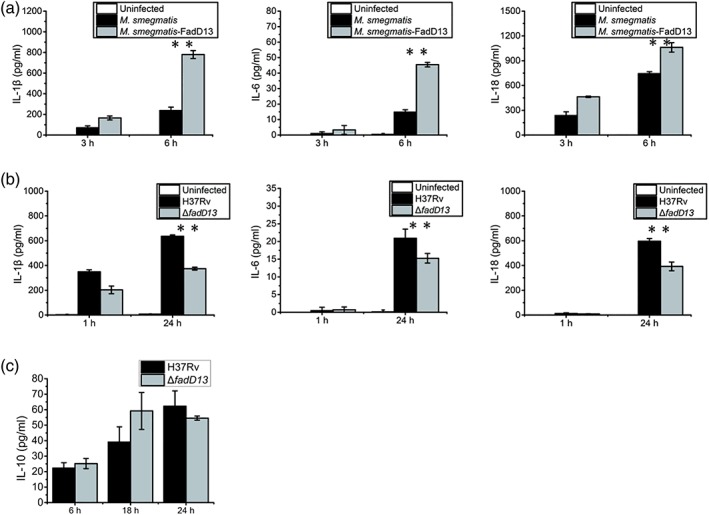
Mtb FadD13 promotes production of proinflammatory cytokines in macrophages. (a) The differentiated THP‐1 macrophages were infected at a multiplicity of infection (MOI) of 10 with M. smegmatis containing the empty vector (pMV261 without H37Rv FadD13 DNA) or pMV261‐FadD13 respectively. (b) The differentiated THP‐1 macrophages were infected at a MOI of 10 with ΔfadD13 mutant and H37Rv respectively. Enzyme‐linked immunosorbent assay (ELISA) was used to detect the production of IL‐1β, IL‐18, and IL‐6 in the supernatant in both (a) and (b). (c) The differentiated THP‐1 macrophages were infected at a MOI of 10 with ΔfadD13 mutant and H37Rv respectively. ELISA was used to detect the production of IL‐10 in the supernatant. The results presented as the one replicate from three independent experiments. * p < .05 and ** p < .01 (two‐tailed unpaired t test)
2.3. eEF1A1 is the binding partner of FadD13 in macrophages
To search for the targets of FadD13 in host cells, we performed coimmunoprecipitation (Co‐IP) experiments to screen for FadD13‐interacting factors in macrophages. We purified His‐tagged FadD13 proteins and incubated them with the cell lysates of the differentiated THP‐1 and RAW264.7 macrophages, respectively, when performing a Co‐IP. A specific band between 55 and 43 kDa was found on the silver stained Co‐IP gels (Figure 3a). Mass spectrometry revealed that a 50 KDa of protein, named eEF1A1, has the highest score in ranking (Tables S1 and S2).
Figure 3.
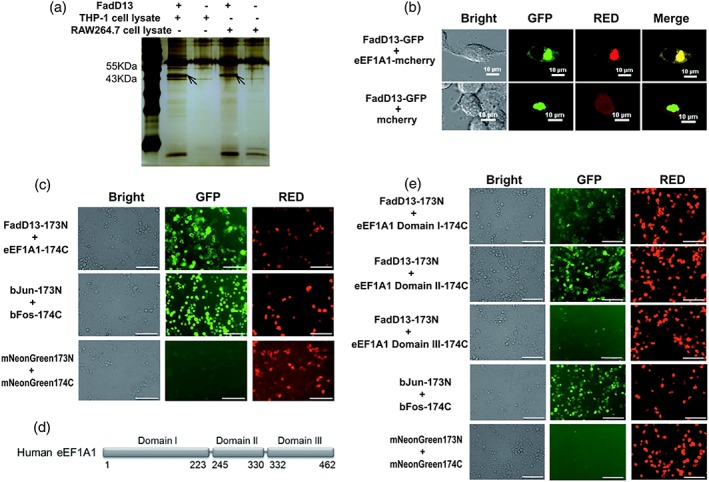
eEF1A1 is the binding partner of FadD13 in macrophages. (a) Coimmunoprecipitation (Co‐IP) was used to screen the binding partner of FadD13 in macrophage. The resin without treatment with FadD13 proteins but separately incubated with the same amount of THP‐1 or RAW264.7 cell lysate was used as the negative control. The band between 55KDa and 43KDa was distinguished from the controls and subject to mass spectrometry. (b) HEK 293T cells were transfected with the vectors encoding GFP tagged FadD13 and mCherry tagged eEF1A1 or GFP tagged FadD13 and mCherry alone for 36 hr. Confocal microscope images showing colocalization of FadD13‐GFP and eEF1A1‐mCherry in HEK 293T cells. (Scale bar: 10 μm). (c) HEK 293 T cells were transfected with the vectors encoding FadD13‐173N and eEF1A1‐174C, respectively, or the vectors encoding bJun‐173N and bFos‐174C, respectively, or the vectors encoding mNeonGreen173N and mNeonGreen174C, respectively, for 48 hr. BiFC results showing interaction between Mtb FadD13 and eEF1A1 in HEK 293T cells. Green fluorescence indicates the interaction and red fluorescence indicates the expression of both target proteins. The groups of bJun‐173N/bFos‐174C and mNeonGreen173N/mNeonGreen174C are the positive control and the negative control respectively. (Scale bar: 100 μm). (d) The structure diagram of the three domains of EF1α (Maruyama et al., 2007). (e) HEK 293T cells were transfected with the vectors encoding FadD13‐173N and eEF1A1 Domain I‐174C respectively or the vectors encoding FadD13‐173N and eEF1A1 Domain II‐174C, respectively, or the vectors encoding FadD13‐173N and eEF1A1 Domain III ‐174C, respectively, or the vectors encoding bJun‐173N and bFos‐174C, respectively, or the vectors encoding mNeonGreen173N and mNeonGreen174C, respectively, for 48 hr. The BiFC results showing interaction between FadD13 and domain II in HEK293T cells. Domains I of eEF1A1 could weakly interact with FadD13. Domains III of eEF1A1 could not interact with FadD13 (Scale bar: 100 μm). The results presented as the one replicate from three independent experiments
Bimolecular fluorescence complementation (BiFC) is a technique for validation of interaction of two protein molecules (a and b) in living cells (Kerppola, 2008). BiFC (Figure 3b) and colocalization (Figure 3c) results illustrate that FadD13 can interact with human eEF1A1 in HEK293T cells. Figure 3b shows the 2‐D images of the green channel (FadD13‐GFP) and the red channel (eEF1A1‐mCherry) as well as their merge are exactly the same, which means a full colocalization of two proteins, implying the interaction of FadD13 and eEF1A1. There might be some self‐aggregation but unlikely occurred to all target proteins; otherwise, the cell cannot produce these images. The results of BiFC further verified the specific interaction of FadD13 and eEF1A1 in cells. eEF1A1 is the second‐most abundant eukaryotic protein (Burglova et al., 2018), which involves in cytoskeletal organisation and other physiological activities (Condeelis, 1995). It consists of three domains (Figure 3d). Through BiFC, we found that Mtb FadD13 could interact with eEF1A1 mainly by binding to its domain II and weakly binding to domain I (Figure 3e). The Mtb FadD13 protein consists of two domains; both could interact with eEF1A1, but the binding of each domain was relatively weak compared with that of the full‐length protein (data not shown). Together, these data confirm that Mtb FadD13 can interact with eEF1A1, mainly by binding to domain II of the eEF1A1 protein.
2.4. Mtb FadD13 promotes the production of proinflammatory cytokines in macrophages through eEF1A1
To test the relationship between eEF1A1 and Mtb virulence, we examined the eEF1A1 expression levels of differentiated THP‐1 macrophages infected with the H37Ra and H37Rv, respectively, at a multiplicity of infection (MOI) of 10. After infection of 24 hr, H37Rv upregulated the expression level of eEF1A1, but the H37Ra did not (Figure 4a). After that, we investigated whether the upregulation of eEF1A1 is related to FadD13. Western blot analysis revealed that deletion of FadD13 in H37Rv resulted in the downregulation of eEF1A1 expression in the macrophages after 24 hr of infection (Figure 4b). Consistent with this, overexpression of FadD13 in HEK293T cells upregulated the expression of eEF1A1 after 24 hr of transfection (Figure 4c). But the transcription level of eEF1A1 in macrophages had no difference between H37Rv‐ or ΔfadD13‐stimulated macrophages and control (Figure S4). So the binding between FadD13 and eEF1A1 may enhance the tolerance of eEF1A1 to degradation.
Figure 4.
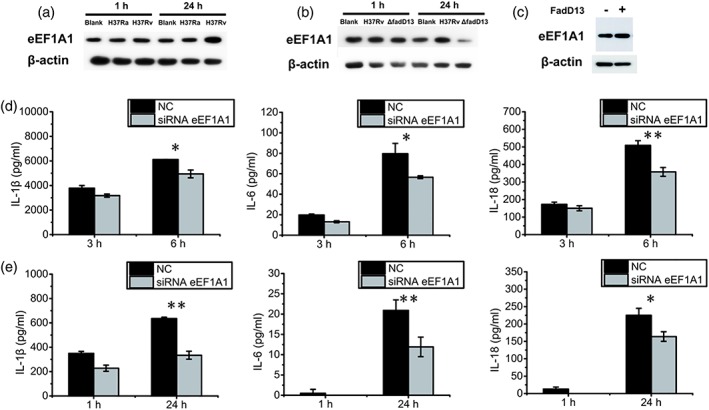
Mtb FadD13 promotes the production of proinflammatory cytokines in macrophages through eEF1A1. (a) Western blot results showing H37Rv upregulated the expression of eEF1A1 in macrophage. The differentiated THP‐1 macrophages were infected at a MOI of 10 with H37Ra and H37Rv, respectively, followed by protein extraction and western blot. Blank: uninfected THP‐1 macrophages; H37Ra: H37Ra infected THP‐1 macrophages; H37Rv: H37Rv infected THP‐1 macrophages. The results presented as the one replicate from two independent experiments. (b) Western blot showing ΔfadD13 mutant decreased the expression of eEF1A1 in the macrophages. The differentiated THP‐1 macrophages were infected at a MOI of 10 with ΔfadD13 mutant and H37Rv respectively, followed by protein extraction and western blot. Blank: uninfected THP‐1 macrophages; ΔfadD13:ΔfadD13 infected THP‐1 macrophages; H37Rv: H37Rv infected THP‐1 macrophages. The results presented as the one replicate from two independent experiments. (c) Western blot result showing FadD13 upregulated the expression of eEF1A1 in overexpressed HEK293T cells. HEK 293T cells were transfected with the vectors encoding FadD13‐GFP or GFP alone respectively for 24 hr, followed by protein extraction and western blot. The results presented as the one replicate from two independent experiments. (d, e) ELISA results showing the production of proinflammatory cytokines (IL‐1β, IL‐18, and IL‐6) decreased in the eEF1A1 knockdown macrophages. NC: THP‐1 macrophages transfected with control siRNA. siRNA eEF1A1: THP‐1 macrophages transfected with eEF1A1 specific siRNA. The differentiated THP‐1 macrophages were infected at a MOI of 10 with FadD13 overexpressed M. smegmatis (d) or H37Rv (e) respectively. The results presented as the one replicate from three independent experiments. * p < .05 and ** p < .01 (two‐tailed unpaired t test)
To investigate whether eEF1A1 is required for production of proinflammatory cytokines in macrophages, we constructed the eEF1A1 expression knockdown macrophages. The verification results of the eEF1A1‐specific siRNA knockdown efficiency are shown in Figure S5, which revealed that this siRNA can efficiently reduce the eEF1A1 expression in macrophages. ELISA analysis showed that the level of the proinflammatory cytokines was significantly reduced in eEF1A1 knockdown macrophages infected with the M. smegmatis‐FadD13‐overexpressing strain and H37Rv respectively (Figure 4d,e). Thus, human eEF1A1 is critical to proinflammatory cytokine production in macrophages during Mtb infection.
The results above indicate that existence of Mtb FadD13 can upregulate expression of eEF1A1 but has no influence on the transcription of eEF1A1 and that the expression level of eEF1A1 is positively correlated with the production of proinflammatory cytokines. We thus conclude that Mtb FadD13 helps to stabilise the eEF1A1 protein in macrophage, which in turn stimulate the production of proinflammatory cytokines.
2.5. Mtb FadD13 and eEF1A1 regulate proinflammatory cytokines production by stimulating NF‐κB signalling pathway in macrophages
To determine which signalling pathway is involved in the increase of proinflammatory cytokines production by FadD13 in macrophages, specific signalling inhibitors were used in our experiment. The links between signalling pathways and FadD13‐mediated induction of cytokine expression were examined by treating the cells with ERK1/2 inhibitor (PD98059), p38 MAPK inhibitor (SB203580), JNK inhibitor (SP600125), Janus kinase inhibitor (AG490), NF‐κB inhibitor (celastrol), and DMSO (blank control) respectively. The NF‐κB inhibitor treatment significantly decreased the production of IL‐1β, IL‐6, and IL‐18 in macrophages infected with FadD13 overexpressed M. smegmatis (Figure 5a). Then, we investigated whether Mtb FadD13 could have influence on the activation of NF‐κB signalling pathway. It is known that TNF and IL‐1β can activate NF‐κB signalling pathway (Imbert et al., 1996; Sethi, Sung, & Aggarwal, 2008). Our experiment show that transient expression of Mtb FadD13 in human HEK293T cells further promoted the activation of NF‐κB (Figure 5b) and increased the phosphorylation of the inhibitory cytoplasmic NF‐κB inhibitor IκBα (Figure 5c). At the same time, western blot results revealed that the ΔfadD13 mutant reduced the expression of the proteins p65, p50, and RelB (Figure 5d), the known NF‐κB transcription factors. The results for p65 and p50 expression were consistent with that of the eEF1A1 knockdown macrophages which infected with H37Rv, but the result for RelB was not (Figure 5e). In Figure 5e, the macrophages containing nontargeting siRNA (NC) or targeting siRNA (siRNA) were all infected with H37Rv; the results indicate that Mtb had induced RelB expression notably. Because the siRNA was designed to silence the expression of eEF1A1, it did not influence the expression of RelB that induced by H37Rv. This result indicates that the induced expression of RelB by FadD13 is not related to the FadD13‐eEF1A1 interaction. Together, these data demonstrates that Mtb FadD13 increases the amount of eEF1A1, through which to increase the activation of NF‐κB signal pathway that to promote the production of proinflammatory cytokines in macrophage during Mtb infection.
Figure 5.
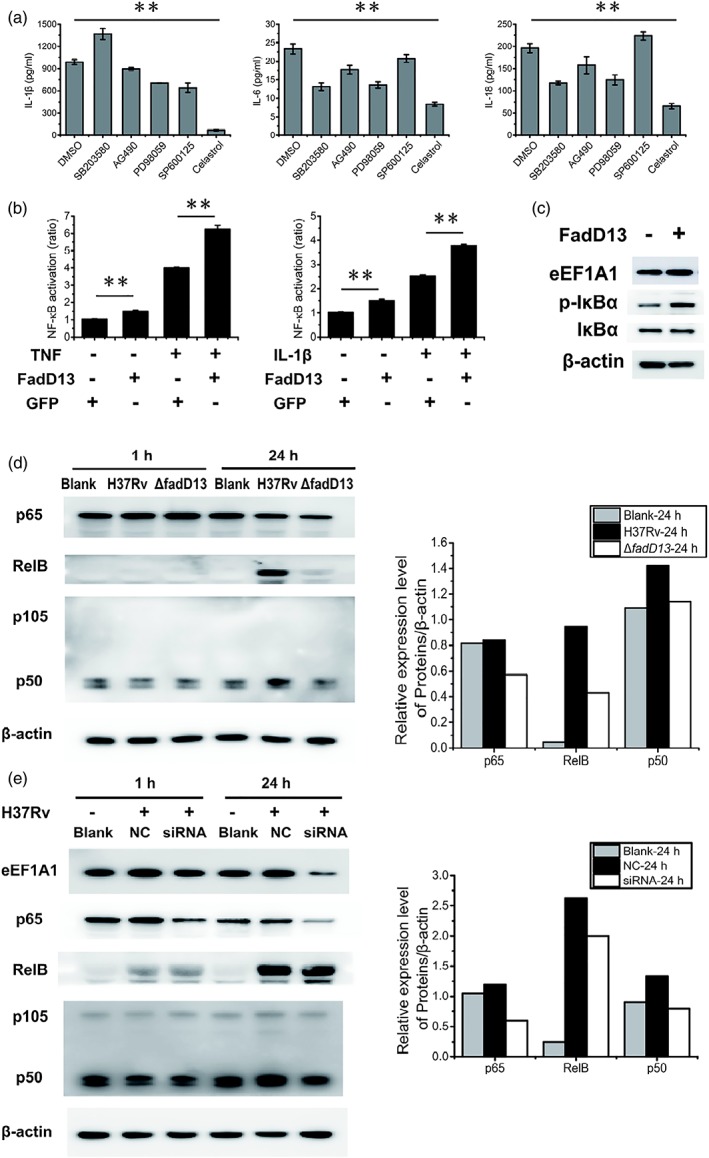
Mtb FadD13 and eEF1A1 regulate proinflammatory cytokines production by stimulating NF‐κB signalling pathway in macrophages. (a) ELISA results showing NF‐κB inhibitor (celastrol) inhibited the production of proinflammatory cytokines (IL‐1β, IL‐18 and IL‐6). Differentiated THP‐1 macrophages were infected at a MOI of 10 with M. smegmatis‐FadD13 for 6 hr. DMSO: DMSO treated macrophages, as the negative control; SB203580: p38 MAPK inhibitor; AG490: Janus kinase inhibitor; PD98059: ERK1/2 inhibitor; SP600125: JNK inhibitor; celastrol: NF‐κB inhibitor. The results presented as the one replicate from three independent experiments. * p < .05 and ** p < .01 (two‐tailed unpaired t test). (b) The results of dual luciferase reporter gene assay showing FadD13 activated NF‐κB signalling pathway in HEK293T cells. HEK 293T cells were transfected with pEGFP‐N1‐FadD13 or pEGFP‐N1 for 24 hr, respectively, then treated with TNF‐α or IL‐1β for 6 hr for activation the NF‐κB pathway. The results presented as the one replicate from two independent experiments. * p < .05 and ** p < .01 (two‐tailed unpaired t test). (c) Western blot results showing FadD13 promoted the phosphorylation of IκBα, the inhibitor of NF‐κB, in HEK293T cells. HEK 293T cells were transfected with pEGFP‐N1‐FadD13 or pEGFP‐N1 for 24 hr, respectively, followed by protein extraction and western blot. The results presented as the one replicate from two independent experiments. (d) Western blot results showing ΔfadD13 mutant decreased the expression of p65, p50 and RelB in the macrophages. Blank: uninfected THP‐1macrophages; H37Rv: H37Rv infected THP‐1 macrophages; ΔfadD13: ΔfadD13 infected macrophages. Macrophages were infected at a MOI of 10 with ΔfadD13 or H37Rv, respectively, for 1 and 24 hr. The right columns stand for the 24 hr western blot results. The results presented as the one replicate from two independent experiments. (e) Western blot results showing eEF1A1 decreased the expression of p65 and p50 (except RelB) in macrophages. Blank: normal THP‐1 macrophages; NC: THP‐1 macrophages transfected with control siRNA; siRNA: THP‐1 macrophages transfected with eEF1A1 specific siRNA. Macrophages were infected at a MOI of 10 with H37Rv for 1 and 24 hr. The right columns stand for the 24 hr western blot results. The results presented as the one replicate from two independent experiments
2.6. Mtb FadD13 proteins can induce the macrophage membrane broken
THP‐1 macrophages were infected by ΔfadD13 mutant and H37Rv wild type, respectively, and the amount of survived Mtb in macrophages is shown in Figure 6a. The result indicates that deletion of FadD13 decreased the Mtb survival in macrophages during infection.
Figure 6.
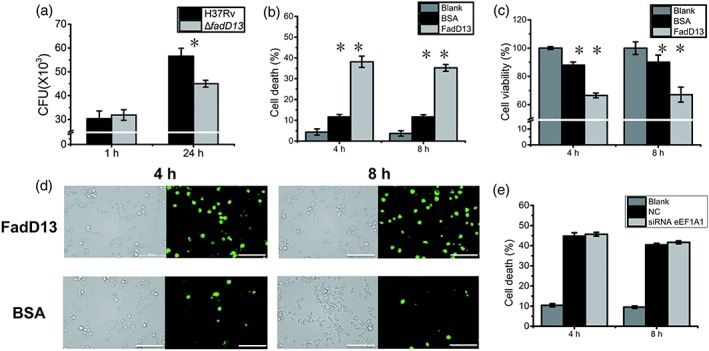
Mtb FadD13 proteins induce the macrophage membrane broken. (a) Plate counting showing FadD13 promoted the survival of Mtb in macrophages. Differentiated THP‐1 macrophages were infected at a MOI of 10 with ΔfadD13 mutant and H37Rv for 1 and 24 hr, respectively. (b) FadD13 induced cell death of THP‐1 macrophage, indicating by release of LDH. Differentiated THP‐1 macrophages were stimulated with 10 μg/ml FadD13 purified proteins for 4 and 8 hr. The release of LDH (b) and cell viability (c) were detected. Blank: untreated THP‐1 macrophage; BSA: BSA treated THP‐1 macrophages; FadD13: FadD13 treated THP‐1 macrophages. (c) Decreased cell viability of THP‐1 macrophages treated with FadD13 proteins. (d) Membrane broken of macrophages treated with FadD13 proteins, staining with FM1‐43 fluorescent dye. The THP‐1 cells were treated as (b), then the cells were staining with 4 μM FM1‐43 and observed in 5 min using confocal microscope (Scale bar: 100 μm). (e) eEF1A1 had no influence on cell death of THP‐1 macrophage induced by FadD13, indicated by LDH release. THP‐1 macrophages transfected with eEF1A1 specific siRNA and control siRNA respectively, then stimulated with 10 μg/ml FadD13 purified proteins for 4 and 8 hr. The release of LDH in the culture medium was detected. Blank: normal THP‐1 macrophages; NC: THP‐1 macrophages transfected with control siRNA; siRNA eEF1A1: THP‐1 macrophages transfected with eEF1A1 specific siRNA. * p < .05 and ** p < .01 (two‐tailed unpaired t test)
FadD13 is an exported protein (de Souza, Leversen, Malen, & Wiker, 2011), and the overexpression of FadD13 in M. smegmatis could result in the secretion of this protein into the 7H9 culture medium (Figure S6). To stimulate the macrophage with FadD13 proteins resulted in release of lactate dehydrogenase (LDH; Figure 6b), indicating the cell membrane was broken. At the same time, the macrophage cell viability also reduced (Figure 6c). The broken of cell membrane can be indicated by a water‐soluble, membrane‐impermeant fluorescence dye FM1‐43, which becomes brighter when it partitions into membranes (McNeil & Kirchhausen, 2005). The fluorescent dye staining results showed that FadD13 protein induced the macrophage membrane broken compared with the BSA treated macrophages (Figure 6d). However, the LDH release induced by FadD13 had no relationship with eEF1A1 (Figure 6e). Taken together, Mtb FadD13 can promote Mtb survival in macrophages and induce the macrophage membrane broken that leads to macrophage cell death.
3. DISCUSSION
Mycobacterial cell wall biosynthesis has been a vulnerable target for antitubercular drugs, which targeting the synthesis of the mycolic acid or arabinan components of the cell envelope (Barry, 2011; Barry, Crick, & McNeil, 2007; Lange, Locher, Wyss, & Then, 2007). Using the Mtb proteome microarray, we screened 283 differential proteins that could interact with proteins from macrophage. GO categorisation showed that they are mainly gathered in cell wall‐associated proteins. The screened cell wall‐related proteins may be potential drug targets for anti‐tuberculosis.
The cytokines play a key role in host immune response to resistance pathogens. Our results suggest that deletion of FadD13 in Mtb resulted in decreasing of proinflammatory cytokines IL‐1β, IL‐18, and IL‐6 secretion, but not anti‐inflammatory cytokine IL‐10. Cheruvu, Plikaytis, and Shinnick (2007) also found that knocking out the MymA operon significantly decreased the production of IL‐1β, IL‐6, IL‐8, RANTES, and MCP‐1 by THP‐1 cells infected with the mutant bacteria. So we consider that FadD13 is crucial in the function of MymA operon. In some cases, the innate limitations of bacterial replication are completely independent of IL‐1β and IL‐18, such as L. pneumophila (Akhter et al., 2009). IL‐1β and IL‐18 are known to be pyroptosis related cytokines. Studies of casp11‐deficient mice showed that pyroptosis alone is insufficient to limit bacterial replication; it requires additional IL‐1β/IL‐18‐independent function of CASP1 to recruit neutrophils to eliminate extracellular bacteria ejected from the niche by pyroptosis (Broz et al., 2012). Whether FadD13 is related to pyroptosis needs further investigation.
As recently reported, Mtb protein Mce2E could bind with eEF1A1 and block the K48‐linked polyubiquitination of eEF1A1 to inhibit eEF1A1 degradation and promote epithelial cell proliferation (Qiang et al., 2018). Our data reveal that Mtb FadD13 could interact with macrophage eEF1A1 and increase the amount of eEF1A1 in macrophage during infection. But FadD13 has no influence on the transcription of eEF1A1, so we speculate that FadD13 may inhibit eEF1A1 degradation. In Figure 4b, the mycobacterial mutant lacking FadD13 resulted in far less eEF1A1 proteins compared with noninfected cells. The reason may be that infection of macrophages by Mtb is a complex process, which may regulate expression of many proteins, some up‐regulated whereas some down‐regulated. After wild‐type H37Rv entering THP‐1, it expresses FadD13, which tends to bind eEF1A1 to prevent the latter from degradation. The FadD13 mutant lost the function of FadD13 production, eEF1A1 proteins are easy to degrade. eEF1A1 is a multifunction protein involving in various of cellular processes, such as cancers (Amiri et al., 2007; Anand et al., 2002; Lamberti et al., 2004; Lee, 2003; Neckers & Neckers, 2005; Tomlinson et al., 2005), apoptosis (Duttaroy et al., 1998), autophagy (Bandyopadhyay et al., 2010), and the other cell death (Borradaile et al., 2006; Chen, Proestou, Bourbeau, & Wang, 2000), as well as Th1 cytokine production (Maruyama et al., 2007). In this experiment, knockdown of eEF1A1 expression reduced proinflammatory cytokine production, which demonstrating that eEF1A1 is critical for proinflammatory cytokines production in macrophage during Mtb infection.
Transcription factor NF‐κB signalling pathway regulates the immune responses and inflammation by controlling the expression of various immune‐regulatory molecules and thus modulates the intracellular survival of pathogens (Behar et al., 2011; Xu, Wang, Gao, & Liu, 2014). It can be activated, directly or indirectly, by antigens, TNF‐α, LPS, IL‐1, IL‐2, IL‐6, IL‐8, and so forth (Imbert et al., 1996; Sethi et al., 2008). By monitoring the secretion amount of IL‐1β, IL‐18, and IL‐6 in the signalling pathway inhibition experiment, NF‐κB signalling pathway was identified relating to the production of proinflammatory cytokines modulated by Mtb FadD13. It is known that NF‐κB activation has two pathways, the canonical and alternative pathways, and P65 and p50 are related with canonical pathway, whereas RelB is related with alternative pathway (Dolcet, Llobet, Pallares, & Matias‐Guiu, 2005). To study which pathway through which FadD13 activates the NF‐ κB signalling pathway, the expression level of p65, p50, and RelB was detected. Further experiments revealed that overexpression of FadD13 led to an increased level of phosphorylation of the inhibitory cytoplasmic NF‐κB regulator IκBα, and deletion of FadD13 lowed expression level of p50, p65, and RelB, a group of family members of NF‐κB signalling pathway. Considering the eEF1A1 knockdown macrophages infected with H37Rv also reduced the expression of p50 and p65 (with RelB as exception), Mtb FadD13 and human eEF1A1 have a synergistic effect in regulating the production of proinflammatory cytokines by acting on NF‐κB signalling pathway.
In our study, deletion of FadD13 decreased the Mtb survival in macrophages during infection. The finding is in line with previous reports that MymA operon is required for Mtb survival in macrophages and spleens of guinea pigs (Cheruvu et al., 2007; Singh et al., 2005, 2005). It could be explained by the fact that FadD13 mutant can promote macrophage phagolysosomal fusion and apoptosis better than the wild type (Olsen et al., 2016).
Previous study showed that Mtb can translocate from the phagolysosome to the cytosol, which is dependent on vacuole membrane pore formation that induced by ESX‐1 system, which is closely related to the virulence, and causing significant cell death (Beffa, Zellweger, Janssens, Wrighton‐Smith, & Zellweger, 2008; Hsu et al., 2003; Simeone et al., 2012; Stamm et al., 2003; van der Wel et al., 2007). ESAT‐6, secreted by ESX‐1, is able to cause dose‐dependent pore formation in host cell membranes, which play a direct role in producing pores in Mycobacterium‐containing vacuole membrane, facilitating M. marinum escape from the vacuole and cell‐to‐cell spread (Beffa et al., 2008; Houben et al., 2012; Simeone et al., 2012; Stamm et al., 2003). A recent paper presented conclusive evidence that ESX‐1 and phthiocerol dimycocerosates (DIM) act in concert to induce phagosomal membrane damage and rupture in infected macrophages (Augenstreich et al., 2017). Similarly, we found that FadD13 protein also could damage the macrophage cell membrane, which led to LDH release and reduced cell viability. Due to the cell membrane and the membrane of Mycobacterium‐containing vacuole have similar structure, we deduced that when Mtb enter the vacuole in macrophages, FadD13 is secreted, together with ESX‐1 and DIM, causing Mycobacterium‐containing vacuole membrane damage. However, this speculation needs to prove experimentally. This may help further to explain that FadD13 promotes Mtb to survive in macrophages. FadD13 induced cell death and can bind eEF1A1 to facilitate secretion of cytokines. We have not found the linkage between these two biological processes, but it is interesting question and worth to investigate.
Based on our findings and previous reports, we build a model (Figure 7) of the interaction mechanism between Mtb FadD13 and macrophages. We speculate that when Mtb cells enter the phagosome in macrophages through endocytosis, the acidic condition induces Mtb FadD13 upregulation. FadD13 protein may induce the Mycobacterium‐containing vacuole pore formation and release to the cytoplasm, which promotes the Mtb proliferation in macrophages. Meanwhile, the macrophage eEF1A1 binds with FadD13 in cytoplasm to prevent itself from degradation, allowing it promotes the activation of NF‐κB signalling pathway, and thus to promote the production of proinflammatory cytokines. This model may provide new insight into the function of Mtb FadD13 immunology.
Figure 7.
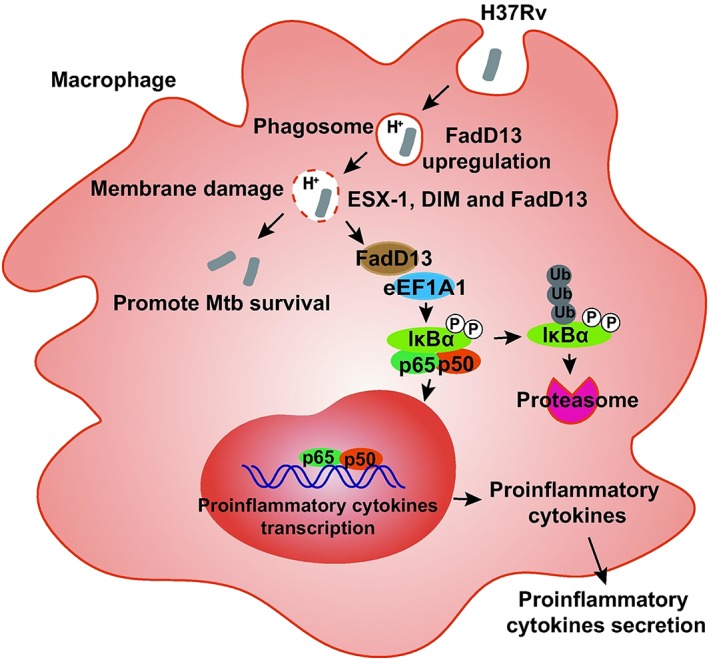
Model of the interaction mechanism between Mtb FadD13 and macrophage. When Mtb cells enter the phagosome in macrophage through endocytosis, the acidic condition induces FadD13 upregulation. Due to the cell membrane and the membrane of Mycobacterium‐containing vacuole have similar structure; we deduced that when Mtb enter the vacuole in macrophages, FadD13 is secreted, together with ESX‐1 and DIM, causing Mycobacterium‐containing vacuole membrane damage. However, this speculation needs to prove experimentally. This may help further to explain that FadD13 promotes Mtb to survive in macrophages. Simultaneously, the macrophage eEF1A1 binds with FadD13 in cytoplasm to prevent itself from degradation, allowing it to promote the activation of NF‐κB signalling pathway, so as to increase the production of proinflammatory cytokines
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains, plasmids, and culture conditions
M. tuberculosis H37Rv (ATCC: American Type Tissue Collection 27294) and H37Ra (ATCC 25177) were grown in Middlebrook 7H9 broth supplemented with 10% albumin dextrose catalase (ADC) enrichment (Becton Dickinson, San Jose, USA), 0.5% glycerol and 0.5% Tween‐80 at 37°C. E. coli DH5α and E. coli HB101 were cultured in flasks using LB medium. E. coli BL21 (DE3) was used for protein purification. E. coli HB101 was used for the gene knockout. The M. smegmatis mc2 155 overexpression strains were cultured in Middlebrook 7H9 broth containing 50 μg/ml kanamycin or on Middlebrook 7H10 agar supplemented with 50 μg/ml kanamycin. The mycobacterium shuttle vector pMV261 (provided by Jiaoyu Deng, WuHan Institute of Virology, CAS) was used to overexpress Mtb FadD13 in M. smegmatis. The Mtb FadD13 full‐length DNA was amplified from Mtb H37Rv genomic DNA. The plasmids p0004s, phAE159, pJH532, and E. coli HB101 were provided by Jiaoyu Deng, WuHan Institute of Virology, CAS.
4.2. Cell culture and differentiation
The human myelomonocytic cell line THP‐1 (ATCC TIB‐202) was cultured in RPMI 1640 medium (LONZA) supplemented with 10% FBS (GIBCO) at 37°C in a 5% CO2 humidified incubator. The mouse macrophage cell line RAW264.7 (Laboratory storage) was maintained in DMEM (LONZA) supplemented with 10% FBS (GIBCO) at 37°C in a 5% CO2 humidified incubator. THP‐1 cells were differentiated using 25 ng/ml phorbol myristate acetate (PMA; Sigma‐Aldrich) for 48 hr. Then, the medium was replaced, and the cells were cultured in fresh RPMI 1640 medium supplemented with 10% FBS and incubated for an additional 12 hr for cell differentiation. HEK293T (Laboratory storage) cells were cultured in DMEM medium (LONZA) supplemented with 10% FBS (GenStar).
4.3. Macrophage protein extraction used for Mtb chip interaction
THP‐1 cells were cultured at 1 × 107 cells/plate in a 10‐cm plate and differentiated with 25 ng/ml of PMA. After 48 hr, the cells were cultured in fresh RPMI 1640 medium with 10% FBS for an additional 12 hr before infection. For the bacteria, each mycobacterial strain was centrifuged, and the supernatant was removed. The pellet was resuspended in PBS and resuspended five times with a 30 G of needle to disperse the pellet. The THP‐1 cells were infected with the H37Rv or H37Ra at an MOI of 10 respectively. Four hours after infection, the medium was discarded, and the cells were washed three times with 1 × PBS to remove noninternalized bacteria. The cells were incubated again with fresh RPMI 1640 medium supplemented with 50 μg/ml gentamicin to inhibit the growth of extracellular bacteria that not removed by washing for another 12 hr, and then lysis buffer (Schroder et al., 2011) was used to extract the whole cell proteins. Every stimulation experiment was performed twice for biological repetition. The blank control group named THP‐1 was treated at the same time as the other groups.
4.4. Screening of Mtb proteins that can interact with macrophage proteins
There were 4,262 proteins spotted in the Mtb proteome microarrays, accounting for >95% of the coding genes (microarray‐tuberculosis‐12, BC Bio). The extracted total cell proteins were biotinylated using an EZ‐Link Sulfo‐NHS‐LC‐Biotinylation kit (Thermo Scientific, Bremen, Germany) according to the manufacturer's instructions. Mtb proteome microarrays were blocked at room temperature for 1 hr with shaking in blocking buffer as described previously (Schroder et al., 2011). To screen for the proteins that can interact with the macrophage proteins, the Mtb proteome microarrays were incubated with the biotinylated THP‐1 whole cell lysates for 12 hr at 4°C with shaking. After that, the Mtb proteome microarrays were washed with PBST three times and then probed with Cy3‐streptavidin for 1 hr at room temperature with shaking. After washing three times, the arrays were dried and scanned with a GenePix 4200A microarray scanner (Molecular Devices, California, USA). The data were analysed with GenePix Pro 6.1 software (Molecular Devices, California, USA).
4.5. Bioinformatics analysis
Differential proteins were analysed at DAVID (https://david.ncifcrf.gov/gene2gene.jsp) for GO category and KEGG pathway category analysis. To further understand the interactions between the differential proteins, a protein–protein network map was established using STRING 10.5 software (https://string-db.org/). The subnetworks were obtained from sorting interaction networks according to the GO and KEGG terms by BiNGO v3.0.3 software.
4.6. Expression and purification of recombinant FadD13 proteins
The open reading frame encoding the Mtb FadD13 protein was amplified by PCR from H37Rv genomic DNA. The PCR product was cloned into the expression vector pET28a and then transduced into E. coli BL21 (DE3). The E. coli BL21 (DE3) cells carrying the recombinant plasmids were grown at 37°C in LB broth to an OD600 of ~0.6, then induced with 0.2 mM of IPTG (isopropyl β‐D‐thiogalactopyranoside) and then incubated for an additional 6 hr at 16°C. After that, the cells were harvested by centrifugation (4,000 g for 20 min). His fusion proteins were purified with Ni‐nitrilotriacetic acid (Ni‐NTA) columns (BioRad). The fractions containing the complexes were further purified by size exclusion chromatography using a Superdex 75 pg gel filtration column (GE Healthcare) equilibrated with 50 mM of Tris (pH 7.0) containing 0.05 M of NaCl. After being concentrated, these fractions were then applied to a Hi‐Trap Heparin HP column (GE Healthcare), and the bound complexes were eluted using a linear NaCl gradient (0–1 M). The protein concentrations were determined by the BCA protein assay (Beyotime), and the protein purity was examined by SDS‐PAGE.
4.7. Gene knockout and overexpression
The unmarked ΔfadD13 mutation in Mtb was constructed by specialised transduction as described earlier (Bardarov et al., 2002). The successful deletion of the gene was demonstrated by PCR and the verification principle as described previously (Figure S3). To express FadD13 in mammalian cells, the FadD13 full‐length DNA fragments were inserted into the vector pEGFP‐N1 and transfected into HEK 293T cells. The bacterial expression plasmids were constructed by cloning FadD13 into pMV261, which allowed the expression of the gene in M. smegmatis mc2 155. The plasmids pMV261‐FadD13 and pMV261 were electroporated into M. smegmatis mc2 155 cultures respectively. 7H10 agar medium supplemented with 50 μg/ml of kanamycin was used for screening the positive clones. There was a His‐tag at the C‐terminal of the Mtb FadD13 protein. Western blotting for His‐tag was used to verify the protein expression. His‐tag antibodies we used in our study are anti‐His‐Tag antibody (MF082‐T; MEI5), HRP Goat anti‐Mouse antibody (AS071; abclonal).
4.8. Infection experiments, CFU counting, western blot, and cytokine detection
THP‐1 cells were cultured at 1 × 106 cells/well in a 12‐well plate and differentiated with 25 ng/ml of PMA for 48 hr. Then, the supernatant was discarded, and fresh RPMI 1640 medium with 10% FBS was added for an additional 12 hr before infection. Infection process was the same as above. At different time points after infection, the cell culture medium was harvested for cytokine detection. Cells were harvested for CFU counting, quantitative PCR or western blot. The cells used for bacterial counting were lysed in 0.1% Triton X‐100 for 15 min, after tenfold dilution of the lysates in 0.05% Tween‐80, plated on 7H10 agar plates. For cytokine detection, the ELISA kits we used were as follows: Human IL‐1β ELISA kit: SEK10139, Sino Biological; Human IL‐6 ELISA kit: SEKB10395, Sino Biological; Human IL‐18 ELISA kit: E‐EL‐H0253C, Elabscience; Human IL‐10 ELISA kit: SEK10947, Sino Biological. The following antibodies we used in western blot are anti‐eEF1A1 antibody (ab202484; abcam), anti‐IκBα antibody (ab32518; abcam), anti‐p‐IκBα antibody (AP0614; abclonal), anti‐p65 antibody (8242R; cell signalling), anti‐p50 antibody (13586S, cell signalling), anti‐RelB antibody (4954R, cell signalling), HRP Goat anti‐Rabbit antibody (AS014; abclonal), and anti‐β‐actin antibody (AC026, abclonal).
4.9. Pull down
Pull down assays were performed according to the manufacturer's instructions using a Pierce Pull Down Biotinylated Protein:Protein Interaction kit. Purified FadD13 proteins were biotinylated using an EZ‐Link Sulfo‐NHS‐LC‐Biotinylation kit. Then, 50 μg/100 μl biotin‐labelled FadD13 was added to the appropriate spin columns, through biotin‐streptavidin binding to the resin. THP‐1 cells were seeded at 5 × 106 cells/plate in a 6‐cm plate and differentiated with 25 ng/ml of PMA for 48 hr. RAW264.7 cells were cultured in a 6‐cm plate for 1 day. Then, the cells were lysed in strong RIPA lysis buffer supplemented with a 1% protease inhibitor “cocktail” and 1 mM of PMSF. Then, the cell lysates incubated with the resin that had been exposed to the FadD13 protein at 4°C for 2 hr. The negative control is the cell lysates incubated with the resin that not exposed to the FadD13 proteins. The resin was extensively washed, and the bound proteins were subjected to SDS‐PAGE and stained with a protein silver staining kit (Pierce). The specific bands were identified by mass spectrometry.
4.10. In vitro coimmunoprecipitation assay
The coimmunoprecipitation assay was performed according to the manufacturer's instructions for the Pierce Co‐Immunoprecipitation Kit. For the coimmunoprecipitation assay, 20 μg of anti‐His antibody (ab18184; abcam) was immobilised on 25 μl of coupling resin. A total of 50 μg of purified FadD13 proteins was added to the antibody immobilised coupling resin. The THP‐1 and RAW264.7 cells were lysed with IP lysis buffer respectively. The cell lysates were separately incubated with the resin for 2 hr with mixing at 4°C. After using 50 μl of elution buffer to wash the bound proteins, the bound proteins were subjected to SDS‐PAGE and stained with a protein silver staining kit (Pierce). The specific bands were identified by mass spectrometry.
4.11. Bimolecular fluorescence complementation
The BiFC plasmid system was constructed in Zhang' lab. The plasmids pLVX‐bJun‐mNeonGreen173N‐T2A‐mCherry‐puro and pLVX‐bFos‐mNeonGreen174C‐T2A‐mCherry‐puro were used as templates. The genes fadD13 and eef1a1 were amplified by PCR from H37Rv and THP‐1 cell cDNA, respectively, and then fadD13 subcloned into the plasmids pLVX‐bJun‐mNeonGreen173N‐T2A‐mCherry‐puro to replace the gene encoding bJun, and eef1a1, eef1a1 domain I/II/III subcloned into the plasmid pLVX‐bFos‐mNeonGreen174C‐T2A‐mCherry‐puro, respectively, to replace the gene encoding bFos. HEK293T cells were seeded in 12‐well plates in growth medium and cultured overnight. To verify protein interaction, HEK293T cells were cotransfected with 0.5 μg of pLVX‐FadD13‐mNeonGreen173N‐T2A‐mCherry‐puro and 0.5 μg of pLVX‐eEF1A1‐mNeonGreen174C‐T2A‐mCherry‐puro using Lipofectamine 3000 (Invitrogen). pLVX‐bJun‐mNeonGreen173N‐T2A‐mCherry‐puro and pLVX‐bFos‐mNeonGreen174C‐T2A‐mCherry‐puro were also cotransfected into the HEK293T cells as the positive control. And pLVX‐mNeonGreen173N‐T2A‐mCherry‐puro and pLVX‐mNeonGreen174C‐T2A‐mCherry‐puro were cotransfected into the HEK293T cells as the negative control. After transfection 48 hr, the cells were analysed by BioTek cytation 3 cell imaging multifunctional detection system.
4.12. Quantitative PCR analysis of gene expression levels
The total RNA of macrophages was extracted using the TRizol reagent method (Life technology). For reverse transcription PCR (RT‐PCR), 2 μg of total RNA was mixed with primers (0.8 pmol each reverse primer), and the RT reaction was performed at 42°C for 30 min using a transcript one‐step gDNA removal and cDNA synthesis supermix kit (Transgen). After the heat inactivation of the RT mixture, transstart Tip green qPCR SuperMix (Transgen) was used to amplify the genes eef1a1 and gapdh with a CFX96 qPCR instrument. The data were analysed using BioRad CFXManager software. The primers used in this study are eef1a1 fwd: TGATCGCCGTTCTGGTAAA, rev: CAGCAAAGCGACCCAAAG; gapdh fwd: GAAGGTCGGAGTCAACGGAT, rev: CCTGGAAGATGGTGATGGG.
4.13. RNA interference
THP‐1 cells were cultured at 1 × 106 cells/well in a 12‐well plate and differentiated with 25 ng/ml of PMA. After 48 hours, the cells were transfected with 0.4 μg eEF1A1 small interfering RNA and 2 μl Lipofectamine™ 3000 (Invitrogen) according to the manufacturer's instructions. At different time, extract the THP‐1 macrophages total RNA and total proteins, using qPCR and western blot to analysis the interference efficiency. The small interfering RNA sequences were as follows: eEF1A1 siRNA, sense‐CCAAGUGCUAACAUGCCUUTT, antisense‐AAGGCAUGUUAGCACUUGGTT; the negative control NC siRNA, sense‐UUCUCCGAACGUGUCACGUTT, antisense‐ACGUGACACGUUCGGAGAATT.
4.14. Luciferase reporter assay
A luciferase reporter assay was performed using a Promega luciferase reporter system. The plasmid pNF‐κBLuc was purchased from Beyotime. HEK293T cells were plated in a 12‐well plate in growth medium and cultured overnight. To evaluate whether FadD13 activates or inhibits the NF‐κB pathway, HEK293T cells were cotransfected with 0.5 μg of pNF‐κB‐Luc, 0.5 μg of pEGFP‐FadD13, and 50 ng of pRL‐TK. The control HEK293T cells were cotransfected with 0.5 μg of pNF‐κB‐Luc, 0.5 μg of pEGFP‐N1, and 50 ng of pRL‐TK. After culture 12 hr, the cells were treated with 20 ng/ml TNF‐α (Invitrogen) or 20 ng/ml IL‐1β (Invitrogen) for 6 hr. After that, the cells were processed according to the promega luciferase reporter system instructions.
4.15. Confocal microscopy
HEK293T cells were plated in a glass bottom cell culture dish at 1 × 106 cells per dish. When necessary, the cells were cotransfected with 0.5 μg of pEGFP‐FadD13 and 0.5 μg of pLVX‐eEF1A1‐mCherry for 36 hr. For the negative control, the cells were transfected with 0.5 μg of pEGFP‐FadD13 and 0.5 μg of pLVX‐T2A‐mCherry. The images were acquired and analysed by confocal microscopy.
4.16. Inhibition of signalling
To identify which signalling pathway was used by FadD13 to promote cytokine secretion, we used signalling inhibitors. All pharmacological inhibitors were obtained from DAKEWE, reconstituted in sterile DMSO (Sigma‐Aldrich), and used according to their manual. The pharmacological inhibitors were as follows: p38 MAP kinase signalling inhibitor (SB203580; InvivoGen), MEK1 and MEK2 signalling inhibitor (PD98059; InvivoGen), JNK signalling inhibitor (SP600125; InvivoGen), NF‐κB signalling inhibitor (celastrol; InvivoGen), and Janus kinase 2 inhibitor (AG490; InvivoGen). An equal amount of DMSO was used as the blank control. The M. smegmatis‐FadD13 overexpressing strain infected the differentiated THP‐1 cells for 4 hr, and then the cells were washed three times with 1 × PBS and incubated again with fresh RPMI 1640 medium supplemented with different signalling inhibitors for 1 hr. After that, the supernatant was discarded, and the cells were incubated again with fresh RPMI 1640 medium supplemented with 50 μg/ml of gentamicin for 6 hr. Then, the supernatants were collected, and cytokine secretion was detected.
4.17. LDH release assay and cell variability assay
Differentiated THP‐1 cells were cultured at 1 × 106 cells/well in a 12‐well plate with RPMI1640 medium with 5% FBS and 10 μg/ml of FadD13 purified proteins, the same concentration of BSA treated cells as the negative control. After treated 4 and 8 hr, the supernatant medium was used to detect the LDH secretion according to LDH cytotoxicity assay kit (Thermo Scientific, USA). The remaining cells were used to detect the cell variability according to celltiter‐blue cell variability assay (Promega, USA).
4.18. FM1‐43 staining assay
Differentiated THP‐1 cells were cultured at 1 × 106 cells in a glass bottom cell culture dish and treated with RPMI1640 medium supplemented with 5% of FBS and 10 μg/ml of FadD13 proteins, the same concentration of BSA treated cells as the negative control. After treated 4 and 8 hr, the cells were stained with 4 μM of FM1‐43 and observed in 5 min using confocal microscope.
4.19. Statistical analysis
All data are presented as the one replicate from three or two independent experiments. Statistical analysis between groups was performed by unpaired two‐tailed Student's t test. Data are presented as mean ± SD. A p value of <.05 was considered to be statistically significant.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
Supporting information
Figure S1. Bioinformatics analysis results of 283 differential proteins screened by Mtb protein chip. (A) The GO category of the 283 differential proteins, which analysed at https://david.ncifcrf.gov/gene2gene.jsp. (Blue: molecular function; Yellow: cell component; Green: biological process.) (P < 0.05). (B) KEGG pathway category of differential proteins, which analyzed at https://david.ncifcrf.gov/gene2gene.jsp. (P < 0.05). The subnetworks of proteins involved in the biosynthesis of amino acids (C), biosynthesis of secondary metabolites (D) and purine metabolism (E). The differential proteins were analyzed by STRING (P < 0.05). The subnetworks were obtained from sorting interaction networks according to the GO and KEGG terms using BiNGO v3.0.3 software.
Figure S2. Western blot result showing FadD13 overexpressed in M. smegmatis. FadD13: M. smegmatis containing pMV261‐FadD13; Blank: M. smegmatis containing pMV261.
Figure S3. Verification results showing fadD13 had been knocked out. Left is the verification schematic, if the target gene has been deleted, the verification sequence between primer A and primer B was 640 bp, if not, the wild type was 1870 bp.
Figure S4. QPCR result showing FadD13 had no influence on the transcription of eEF1A1. The differentiated THP‐1 macrophages were infected at a multiplicity of infection (MOI) of 10 with H37Ra, ΔfadD13 mutant and H37Rv, after infection 1 and 24 hours, extract the RNA of macrophages. Blank: uninfected THP‐1 macrophages; H37Ra: H37Ra infected macrophages; H37Rv: H37Rv infected macrophages; ΔfadD13: ΔfadD13 infected macrophages. The results presented as the one replicate from three independent experiments.
Figure S5. Western blot (left) and qPCR (right) results showing the specific eEF1A1 siRNA reduced the expression of eEF1A1 in THP‐1 macrophage. NC: differentiated THP‐1 macrophages transfected with negative control RNA; siRNA: differentiated THP‐1 macrophages transfected with eEF1A1 specific siRNA. The results presented as the one replicate from three independent experiments.
Figure S6. Western blot result showing FadD13 protein secreted outside the M. smegmatis. The M. smegmatis containing either empty vector (pMV261 without H37Rv FadD13 DNA) or pMV261‐FadD13 were cultured in 7H9 medium for 2 days, when the OD600 was about 1.0, the bacterial suspension was filtered through a 0.22 μm filter to remove the bacteria and then concentrated the culture supernatant. The amount of FadD13 in the culture supernatant was detected with western blot. The results presented as the one replicate from two independent experiments.
Data S7: Supplementary Information
Table 1. Mass spectrometry result of THP‐1.
Table 2. Mass spectrometry result of RAW264.7.
ACKNOWLEDGEMENTS
This work was supported by the National Science and Technology Major Project (2018ZX10731301‐003‐008), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29050100) and a key grant of the Chinese Academy of Sciences (KJZD‐EW‐TZ‐L04). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Wei S, Wang D, Li H, et al. Fatty acylCoA synthetase FadD13 regulates proinflammatory cytokine secretion dependent on the NF‐κB signalling pathway by binding to eEF1A1. Cellular Microbiology. 2019;21:e13090 10.1111/cmi.13090
REFERENCES
- Akhter, A. , Gavrilin, M. A. , Frantz, L. , Washington, S. , Ditty, C. , Limoli, D. , … Amer, A. O. (2009). Caspase‐7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathogens, 5(4), e1000361 10.1371/journal.ppat.1000361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri, A. , Noei, F. , Jeganathan, S. , Kulkarni, G. , Pinke, D. E. , & Lee, J. M. (2007). eEF1A2 activates Akt and stimulates Akt‐dependent actin remodeling, invasion and migration. Oncogene, 26(21), 3027–3040. 10.1038/sj.onc.1210101 [DOI] [PubMed] [Google Scholar]
- Anand, N. , Murthy, S. , Amann, G. , Wernick, M. , Porter, L. A. , Cukier, I. H. , … Lee, J. M. (2002). Gene encoding protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nature Genetics, 31(3), 301–305. 10.1038/ng904 [DOI] [PubMed] [Google Scholar]
- Augenstreich, J. , Arbues, A. , Simeone, R. , Haanappel, E. , Wegener, A. , Sayes, F. , … Astarie‐Dequeker, C. (2017). ESX‐1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cellular Microbiology, 19(7). 10.1111/cmi.12726 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, U. , Sridhar, S. , Kaushik, S. , Kiffin, R. , & Cuervo, A. M. (2010). Identification of regulators of chaperone‐mediated autophagy. Molecular Cell, 39(4), 535–547. 10.1016/j.molcel.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardarov, S. , Bardarov, S. Jr. , Pavelka, M. S. Jr. , Sambandamurthy, V. , Larsen, M. , Tufariello, J. , … Jacobs, W. R. Jr. (2002). Specialized transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis . Microbiology, 148(Pt 10), 3007–3017. 10.1099/00221287-148-10-3007 [DOI] [PubMed] [Google Scholar]
- Barry, C. E. (2011). Lessons from seven decades of antituberculosis drug discovery. Current Topics in Medicinal Chemistry, 11(10), 1216–1225. 10.2174/156802611795429158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry, C. E. , Crick, D. C. , & McNeil, M. R. (2007). Targeting the formation of the cell wall core of M. tuberculosis . Infectious Disorders Drug Targets, 7(2), 182–202. 10.2174/187152607781001808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beffa, P. , Zellweger, A. , Janssens, J. P. , Wrighton‐Smith, P. , & Zellweger, J. P. (2008). Indeterminate test results of T‐SPOT.TB performed under routine field conditions. The European Respiratory Journal, 31(4), 842–846. 10.1183/09031936.00117207 [DOI] [PubMed] [Google Scholar]
- Behar, S. M. , Martin, C. J. , Booty, M. G. , Nishimura, T. , Zhao, X. , Gan, H. , … Remold, H. G. (2011). Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis . Mucosal Immunology, 4(3), 279–287. 10.1038/mi.2011.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borradaile, N. M. , Buhman, K. K. , Listenberger, L. L. , Magee, C. J. , Morimoto, E. T. , Ory, D. S. , & Schaffer, J. E. (2006). A critical role for eukaryotic elongation factor 1A‐1 in lipotoxic cell death. Molecular Biology of the Cell, 17(2), 770–778. 10.1091/mbc.e05-08-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, P. , Ruby, T. , Belhocine, K. , Bouley, D. M. , Kayagaki, N. , Dixit, V. M. , & Monack, D. M. (2012). Caspase‐11 increases susceptibility to Salmonella infection in the absence of caspase‐1. Nature, 490(7419), 288–291. 10.1038/nature11419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglova, K. , Rylova, G. , Markos, A. , Prichystalova, H. , Soural, M. , Petracek, M. , … Hlavac, J. (2018). Identification of eukaryotic translation elongation factor 1‐α 1 gamendazole‐binding site for binding of 3‐hydroxy‐4(1&ITH&IT)‐quinolinones as novel ligands with anticancer activity. Journal of Medicinal Chemistry, 61(7), 3027–3036. 10.1021/acs.jmedchem.8b00078 [DOI] [PubMed] [Google Scholar]
- Chen, E. , Proestou, G. , Bourbeau, D. , & Wang, E. (2000). Rapid up‐regulation of peptide elongation factor EF‐1α protein levels is an immediate early event during oxidative stress‐induced apoptosis. Experimental Cell Research, 259(1), 140–148. 10.1006/excr.2000.4952 [DOI] [PubMed] [Google Scholar]
- Cheruvu, M. , Plikaytis, B. B. , & Shinnick, T. M. (2007). The acid‐induced operon Rv3083‐R0089 is required for growth of Mycobacterium tuberculosis in macrophages. Tuberculosis, 87(1), 12–20. 10.1016/j.tube.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Cleland, W. W. (1963). The kinetics of enzyme‐catalyzed reactions with two or more substrates or products. I. Nomenclature and rate equations. Biochimica et Biophysica Acta, 67, 104–137. 10.1016/0926-6569(63)90211-6 [DOI] [PubMed] [Google Scholar]
- Condeelis, J. (1995). Elongation factor 1α, translation and the cytoskeleton. Trends in Biochemical Sciences, 20(5), 169–170. 10.1016/S0968-0004(00)88998-7 [DOI] [PubMed] [Google Scholar]
- de Souza, G. A. , Leversen, N. A. , Malen, H. , & Wiker, H. G. (2011). Bacterial proteins with cleaved or uncleaved signal peptides of the general secretory pathway. Journal of Proteomics, 75(2), 502–510. 10.1016/j.jprot.2011.08.016 [DOI] [PubMed] [Google Scholar]
- Deng, J. , Bi, L. , Zhou, L. , Guo, S. J. , Fleming, J. , Jiang, H. W. , … Zhang, X. E. (2014). Mycobacterium tuberculosis proteome microarray for global studies of protein function and immunogenicity. Cell Reports, 9(6), 2317–2329. 10.1016/j.celrep.2014.11.023 [DOI] [PubMed] [Google Scholar]
- Dolcet, X. , Llobet, D. , Pallares, J. , & Matias‐Guiu, X. (2005). NF‐kB in development and progression of human cancer. Virchows Archiv, 446(5), 475–482. 10.1007/s00428-005-1264-9 [DOI] [PubMed] [Google Scholar]
- Duttaroy, A. , Bourbeau, D. , Wang, X. L. , & Wang, E. (1998). Apoptosis rate can be accelerated or decelerated by overexpression or reduction of the level of elongation factor‐1α. Experimental Cell Research, 238(1), 168–176. 10.1006/excr.1997.3819 [DOI] [PubMed] [Google Scholar]
- Griffin, J. E. , Gawronski, J. D. , Dejesus, M. A. , Ioerger, T. R. , Akerley, B. J. , & Sassetti, C. M. (2011). High‐resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathogens, 7(9), e1002251 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X. , Jiang, H. W. , Chen, H. , Zhang, H. N. , Liu, Y. , Xu, Z. W. , … Tao, S. C. (2017). Systematic identification of Mycobacterium tuberculosis effectors reveals that BfrB suppresses innate immunity. Molecular & Cellular Proteomics, 16(12), 2243–2253. 10.1074/mcp.RA117.000296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, D. , Demangel, C. , van Ingen, J. , Perez, J. , Baldeon, L. , Abdallah, A. M. , … Peters, P. J. (2012). ESX‐1‐mediated translocation to the cytosol controls virulence of mycobacteria. Cellular Microbiology, 14(8), 1287–1298. 10.1111/j.1462-5822.2012.01799.x [DOI] [PubMed] [Google Scholar]
- Hsu, T. , Hingley‐Wilson, S. M. , Chen, B. , Chen, M. , Dai, A. Z. , Morin, P. M. , … Jacobs, W. R. (2003). The primary mechanism of attenuation of bacillus Calmette–Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proceedings of the National Academy of Sciences of the United States of America, 100(21), 12420–12425. 10.1073/pnas.1635213100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert, V. , Rupec, R. A. , Livolsi, A. , Pahl, H. L. , Traenckner, E. B. M. , MuellerDieckmann, C. , … Peyron, J. F. (1996). Tyrosine phosphorylation of I kappa B‐α activates NF‐kappa B without proteolytic degradation of I kappa B‐α. Cell, 86(5), 787–798. 10.1016/S0092-8674(00)80153-1 [DOI] [PubMed] [Google Scholar]
- Kaziro, Y. , Itoh, H. , Kozasa, T. , Nakafuku, M. , & Satoh, T. (1991). Structure and function of signal‐transducing GTP‐binding proteins. Annual Review of Biochemistry, 60, 349–400. 10.1146/annurev.bi.60.070191.002025 [DOI] [PubMed] [Google Scholar]
- Kerppola, T. K. (2008). Bimolecular fluorescence complementation: Visualization of molecular interactions in living cells. Methods in Cell Biology, 85, 431–470. 10.1016/S0091-679X(08)85019-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti, A. , Caraglia, M. , Longo, O. , Marra, M. , Abbruzzese, A. , & Arcari, P. (2004). The translation elongation factor 1A in tumorigenesis, signal transduction and apoptosis: Review article. Amino Acids, 26(4), 443–448. 10.1007/s00726-004-0088-2 [DOI] [PubMed] [Google Scholar]
- Lange, R. P. , Locher, H. H. , Wyss, P. C. , & Then, R. L. (2007). The targets of currently used antibacterial agents: Lessons for drug discovery. Current Pharmaceutical Design, 13(30), 3140–3154. 10.2174/138161207782110408 [DOI] [PubMed] [Google Scholar]
- Lee, J. M. (2003). The role of protein elongation factor eEF1A2 in ovarian cancer. Reproductive Biology and Endocrinology, 1, 69 10.1186/1477-7827-1-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, T. , Nara, K. , Yoshikawa, H. , & Suzuki, N. (2007). Txk, a member of the non‐receptor tyrosine kinase of the Tec family, forms a complex with poly (ADP‐ribose) polymerase 1 and elongation factor 1α and regulates interferon‐gamma gene transcription in Th1 cells. Clinical and Experimental Immunology, 147(1), 164–175. 10.1111/j.1365-2249.2006.03249.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, P. L. , & Kirchhausen, T. (2005). An emergency response team for membrane repair. Nature Reviews. Molecular Cell Biology, 6(6), 499–505. 10.1038/nrm1665 [DOI] [PubMed] [Google Scholar]
- Nair, S. , Ramaswamy, P. A. , Ghosh, S. , Joshi, D. C. , Pathak, N. , Siddiqui, I. , … Mukhopadhyay, S. (2009). The PPE18 of Mycobacterium tuberculosis interacts with TLR2 and activates IL‐10 induction in macrophage. Journal of Immunology, 183(10), 6269–6281. 10.4049/jimmunol.0901367 [DOI] [PubMed] [Google Scholar]
- Neckers, L. , & Neckers, K. (2005). Heat‐shock protein 90 inhibitors as novel cancer chemotherapeutics ‐ an update. Expert Opinion on Emerging Drugs, 10(1), 137–149. 10.1517/14728214.10.1.137 [DOI] [PubMed] [Google Scholar]
- Olsen, A. , Chen, Y. , Ji, Q. , Zhu, G. , De Silva, A. D. , Vilcheze, C. , … Chan, J. (2016). Targeting Mycobacterium tuberculosis Tumor Necrosis Factor Alpha‐Downregulating Genes for the Development of Antituberculous Vaccines. MBio, 7(3). 10.1128/mBio.01023-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parveen, N. , Varman, R. , Nair, S. , Das, G. , Ghosh, S. , & Mukhopadhyay, S. (2013). Endocytosis of Mycobacterium tuberculosis heat shock protein 60 is required to induce interleukin‐10 production in macrophages. Journal of Biological Chemistry, 288(34), 24956–24971. 10.1074/jbc.M113.461004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, S. K. , Basu, S. , Basu, K. K. , Banerjee, A. , Pathak, S. , Bhattacharyya, A. , … Basu, J. (2007). Direct extracellular interaction between the early secreted antigen ESAT‐6 of Mycobacterium tuberculosis and TLR2 inhibits TLR signaling in macrophages. Nature Immunology, 8(6), 610–618. 10.1038/ni1468 [DOI] [PubMed] [Google Scholar]
- Qiang, L. , Wang, J. , Zhang, Y. , Ge, P. , Chai, Q. , Li, B. , … Liu, C. H. (2018). Mycobacterium tuberculosis Mce2E suppresses the macrophage innate immune response and promotes epithelial cell proliferation. Cellular & Molecular Immunology, 16, 380–391. 10.1038/s41423-018-0016-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, C. , Alhamdani, M. S. , Fellenberg, K. , Bauer, A. , Jacob, A. , & Hoheisel, J. D. (2011). Robust protein profiling with complex antibody microarrays in a dual‐colour mode. Methods in Molecular Biology, 785, 203–221. 10.1007/978-1-61779-286-1_14 [DOI] [PubMed] [Google Scholar]
- Sethi, G. , Sung, B. , & Aggarwal, B. B. (2008). Nuclear factor‐kB activation: From bench to bedside. Experimental Biology and Medicine, 233(1), 21–31. 10.3181/0707-MR-196 [DOI] [PubMed] [Google Scholar]
- Simeone, R. , Bobard, A. , Lippmann, J. , Bitter, W. , Majlessi, L. , Brosch, R. , & Enninga, J. (2012). Phagosomal rupture by mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathogens, 8(2). ARTN), e1002507 10.1371/journal.ppat.1002507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. , Gupta, R. , Vishwakarma, R. A. , Narayanan, P. R. , Paramasivan, C. N. , Ramanathan, V. D. , & Tyagi, A. K. (2005). Requirement of the mymA operon for appropriate cell wall ultrastructure and persistence of Mycobacterium tuberculosis in the spleens of guinea pigs. Journal of Bacteriology, 187(12), 4173–4186. 10.1128/Jb.187.12.4173-4186.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. , Jain, S. , Gupta, S. , Das, T. , & Tyagi, A. K. (2003). mymA operon of Mycobacterium tuberculosis: Its regulation and importance in the cell envelope. FEMS Microbiology Letters, 227(1), 53–63. 10.1016/S0378-1097(03)00648-7 [DOI] [PubMed] [Google Scholar]
- Sreejit, G. , Ahmed, A. , Parveen, N. , Jha, V. , Valluri, V. L. , Ghosh, S. , & Mukhopadhyay, S. (2014). The ESAT‐6 protein of Mycobacterium tuberculosis interacts with beta‐2‐microglobulin (beta2M) affecting antigen presentation function of macrophage. PLoS Pathogens, 10(10), e1004446 10.1371/journal.ppat.1004446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm, L. M. , Morisaki, J. H. , Gao, L. Y. , Jeng, R. L. , McDonald, K. L. , Roth, R. , … Brown, E. J. (2003). Mycobacterium marinum escapes from phagosomes and is propelled by actin‐based motility. Journal of Experimental Medicine, 198(9), 1361–1368. 10.1084/jem.20031072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson, V. A. L. , Newbery, H. J. , Wray, N. R. , Jackson, J. , Larionov, A. , Miller, W. R. , … Abbott, C. M. (2005). Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two‐thirds of breast tumours. BMC Cancer, 5 Artn, 113 10.1186/1471-2407-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel, N. , Hava, D. , Houben, D. , Fluitsma, D. , van Zon, M. , Pierson, J. , … Peters, P. J. (2007). M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell, 129(7), 1287–1298. 10.1016/j.cell.2007.05.059 [DOI] [PubMed] [Google Scholar]
- Wu, F. L. , Liu, Y. , Jiang, H. W. , Luan, Y. Z. , Zhang, H. N. , He, X. , … Tao, S. C. (2017). The Ser/Thr protein kinase protein‐protein interaction map of M. tuberculosis. Molecular & Cellular Proteomics, 16(8), 1491–1506. 10.1074/mcp.M116.065771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. H. , Wang, J. , Gao, G. F. , & Liu, C. H. (2014). Insights into battles between Mycobacterium tuberculosis and macrophages. Protein & Cell, 5(10), 728–736. 10.1007/s13238-014-0077-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. N. , Xu, Z. W. , Jiang, H. W. , Wu, F. L. , He, X. , Liu, Y. , … Tao, S. C. (2017). Cyclic di‐GMP regulates Mycobacterium tuberculosis resistance to ethionamide. Scientific Reports, 7(1), 5860 10.1038/s41598-017-06289-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Bioinformatics analysis results of 283 differential proteins screened by Mtb protein chip. (A) The GO category of the 283 differential proteins, which analysed at https://david.ncifcrf.gov/gene2gene.jsp. (Blue: molecular function; Yellow: cell component; Green: biological process.) (P < 0.05). (B) KEGG pathway category of differential proteins, which analyzed at https://david.ncifcrf.gov/gene2gene.jsp. (P < 0.05). The subnetworks of proteins involved in the biosynthesis of amino acids (C), biosynthesis of secondary metabolites (D) and purine metabolism (E). The differential proteins were analyzed by STRING (P < 0.05). The subnetworks were obtained from sorting interaction networks according to the GO and KEGG terms using BiNGO v3.0.3 software.
Figure S2. Western blot result showing FadD13 overexpressed in M. smegmatis. FadD13: M. smegmatis containing pMV261‐FadD13; Blank: M. smegmatis containing pMV261.
Figure S3. Verification results showing fadD13 had been knocked out. Left is the verification schematic, if the target gene has been deleted, the verification sequence between primer A and primer B was 640 bp, if not, the wild type was 1870 bp.
Figure S4. QPCR result showing FadD13 had no influence on the transcription of eEF1A1. The differentiated THP‐1 macrophages were infected at a multiplicity of infection (MOI) of 10 with H37Ra, ΔfadD13 mutant and H37Rv, after infection 1 and 24 hours, extract the RNA of macrophages. Blank: uninfected THP‐1 macrophages; H37Ra: H37Ra infected macrophages; H37Rv: H37Rv infected macrophages; ΔfadD13: ΔfadD13 infected macrophages. The results presented as the one replicate from three independent experiments.
Figure S5. Western blot (left) and qPCR (right) results showing the specific eEF1A1 siRNA reduced the expression of eEF1A1 in THP‐1 macrophage. NC: differentiated THP‐1 macrophages transfected with negative control RNA; siRNA: differentiated THP‐1 macrophages transfected with eEF1A1 specific siRNA. The results presented as the one replicate from three independent experiments.
Figure S6. Western blot result showing FadD13 protein secreted outside the M. smegmatis. The M. smegmatis containing either empty vector (pMV261 without H37Rv FadD13 DNA) or pMV261‐FadD13 were cultured in 7H9 medium for 2 days, when the OD600 was about 1.0, the bacterial suspension was filtered through a 0.22 μm filter to remove the bacteria and then concentrated the culture supernatant. The amount of FadD13 in the culture supernatant was detected with western blot. The results presented as the one replicate from two independent experiments.
Data S7: Supplementary Information
Table 1. Mass spectrometry result of THP‐1.
Table 2. Mass spectrometry result of RAW264.7.


