Abstract
G protein–coupled receptors (GPCRs) are encoded by over 800 genes in the human genome. Motivated by different scientific rationales, the two classification systems that are mainly in use, the ABC and GRAFS systems, organize GPCRs according to their pharmacological features and phylogenetic relations, respectively. Within those systems, adhesion GPCRs (aGPCRs) constitute a group of over 30 mammalian homologs, most of which are still orphans with undefined activating signals and signal transduction properties. Previous efforts have further subdivided mammalian aGPCRs into nine subfamilies to indicate phylogenetic relationships. However, this subclassification scheme has shortcomings and inconsistencies that require attention. Here, we have reassessed the phylogenetic relationships of aGPCRs from vertebrate and invertebrate species. Our findings confirm that secretin receptor–like GPCRs most probably emerged from ancestral aGPCRs. We show that reassignment of several aGPCRs to families essentially requires input from functional data. Our analyses establish the need for introducing novel aGPCR subfamilies due to aGPCR sequences from invertebrate species that are not readily assignable to any existing subfamily. We conclude that the current classification systems ought to be updated to consider an unambiguous taxonomy of a hierarchically organized classification and pharmacological properties, and to accommodate phylogenetic affiliations between aGPCR genes within mammals and across the animal kingdom.
Keywords: GPCR, adhesion GPCR, phylogeny, classification
The classification of adhesion G protein–coupled receptors (aGPCRs) into nine subfamilies according to existing systems for classifying GPCRs suffers from various shortcomings. This paper reassesses the phylogenetic relationships of aGPCRs from vertebrates and invertebrates and shows that reassignment of several aGPCRs to families essentially requires input from functional data, and that there is a need to introduce novel aGPCR subfamilies due to invertebrate aGPCRs that are not readily assignable to any existing subfamily.
Introduction
The classification of genes into a hierarchical order is essential to reflect their evolutionary history, but also for pragmatic matters, such as their systematic nomination, which is the basis for unambiguous scientific communication. Historically, most gene classifications are based on mutual structural and functional features of their resulting gene products. However, common structural features, such as folds and domains, as well as common functional properties, such as enzymatic activities and binding properties, may be the result of convergent evolution originating from an entirely different genetic starting point. Moreover, different gene products may arise from a single gene due to alternative splice events and/or transcriptional start sites. In contrast, classifications based on phylogenetic sequence relationships of genes are straightforward and allow for both the assignment and comparison of genes, even from distantly related species. The complexity and importance of thorough classification and meaningful naming of individual genes scale with the size of gene families.
G protein–coupled receptors (GPCRs) are encoded by more than 800 genes in the human genome1 and thus comprise one of the largest gene family in vertebrates. They constitute biosensors participating in a plethora of body functions, and because of their excellent pharmacological tractability are prime targets to combat various diseases.
The Nomenclature Committee of the International Union of Basic and Clinical Pharmacology (NC‐IUPHAR) considers six GPCR classes, based on sequence homology.2 Their prototype members are as follows: class A (rhodopsin‐like), class B (secretin receptor–like), class C (metabotropic glutamate receptor–like), class D (fungal mating pheromone receptor–like), class E (cyclic AMP receptor–like), and class F (frizzled/smoothened–like).3 Therefore, the rhodopsin, adhesion/secretin, and glutamate families are referred to as classes A, B, and C, respectively.3, 4
Alternatively, GPCRs have been ordered based on phylogenetic sequence relations, generating the GRAFS classification,5, 6 which comprises the five Glutamate, Rhodopsin, Adhesion, Frizzled, and Secretin receptor families. Obviously, the use of terms as versatile as “family,” “class,” “subclass,” “subtype,” “group,” “clade,” “cluster,” “branch,” and “superfamily” requires a logical definition when a classification is based on phylogenetic properties. This becomes even more evident when receptors within a “class” or “family” are subclassified and terms such as “groups,” “subfamilies,” or “families” are used for the same collection of receptors.2
Currently, NC‐IUPHAR subclassifies GPCRs within a class (family), leading to more than 50 subfamilies that largely derive from common pharmacological properties, for example, receptivity to the same agonist(s). This functional ordering principle only partially matches phylogenetic relationships. For example, the human repertoire of P2Y receptors contains eight members (P2Y1, 2, 4, 6, and 11–14).7 Based on their preferred agonist, they are further classified into adenine nucleotide–activated (P2Y1, P2Y11, P2Y12, and P2Y13), pyrimidine nucleotide–activated (P2Y4 and P2Y6), ATP/UTP‐activated (P2Y2), and UDP‐sugar–activated (P2Y14) receptors. In contrast, similarities in their amino acid sequences subdivide P2Y receptors into two phylogenetically distinct groups (P2Y1‐like: P2Y1, 2, 4, 6, and 11; and P2Y12‐like: P2Y12–14) that emerged independently in evolution.8, 9 Crystallographic studies demonstrated the different orientation of the adenine group within the ligand binding site of P2Y1 and P2Y12 and are in line with this phylogenetic observation.10, 11, 12 Similarly, phylogenetic analysis clusters muscarinic acetylcholine receptors within rhodopsin‐like GPCRs (agonist specificity), and further into two groups (coupling specificity), M2/M4 and M1/M3/M5 (Fig. 1) that couple to Gαi/o and Gαq/11 proteins, respectively.13, 14 Hence, the classification of GPCRs on a phylogenetic basis mainly preserves their functional characteristics, such as agonist specificity and G protein–coupling specificity. However, the predictive value of a phylogeny‐based agonist‐ and/or G protein–coupling assignment mutually depends on the distance of the receptors’ phylogenetic relationship (clustering level). The distance of the receptors’ phylogenetic relationship, which clusters agonist and/or signaling specificity, is not a defined value but rather a range of distances and can vary between different receptor subfamilies. Furthermore, agonist preference is not always superior to the clustering level of G protein–coupling specificity for a given receptor subfamily. For example, the vasopressin receptors, V1AR and V2R, share the same agonist, arginine vasopressin, but couple mainly to Gq/11 and Gs proteins, respectively.15 However, the oxytocin receptor, which is closely related to both vasopressin receptors rendering all of them one receptor subfamily, couples to Gq/11 proteins, like V1AR, and preferentially binds oxytocin, but also arginine vasopressin.16
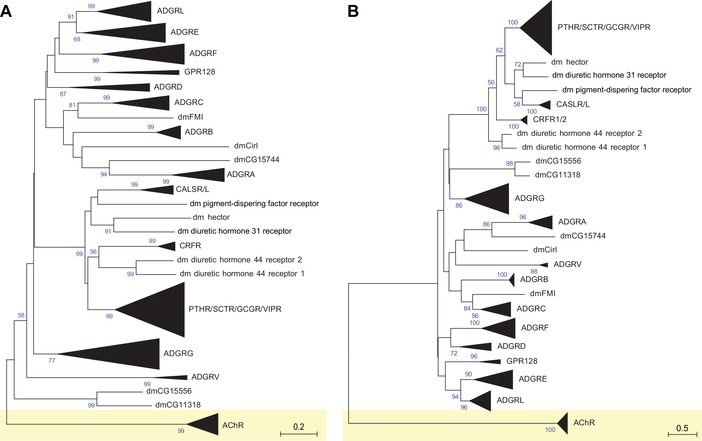
Figure 1.
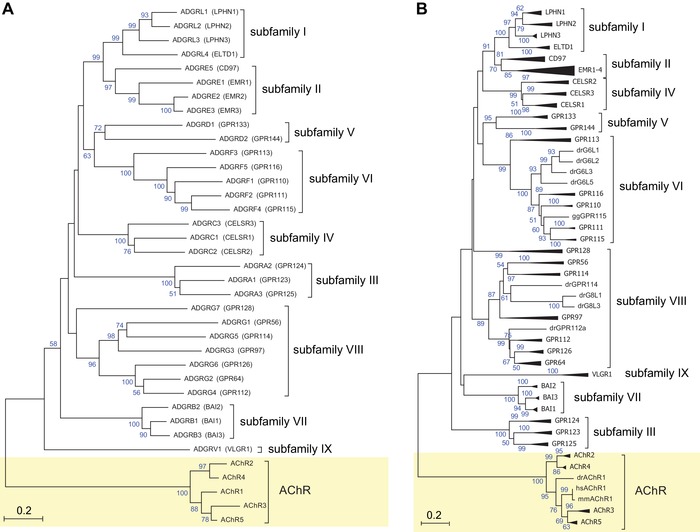
Current nomenclature of aGPCRs based on phylogenetic analyses. The evolutionary relationships of (A) only human aGPCRs and (B) human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors (AChR) served as the outgroup. The evolutionary history was inferred using the neighbor‐joining method.67 The optimal trees with the sum of branch lengths of 14.06117543 (A) and 27.58139556 (B) are shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.68 The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method69 and are represented in the units of the number of amino acid substitutions per site. The analyses involved 37 (A) and 150 (B) amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 216 (A) and 196 (B) positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22
For experimental bioscientists, classification schemes may thus represent a guide or, conversely, become an obstacle. Whether the outcome swings one way or the other merely depends on the available ordering logic, particularly when receptor homologs unavailable for experimental interrogation in humans are investigated in genetic model organisms. For example, assignment of newly identified, functionally elusive receptors to a specific GPCR category with known functions may help tremendously in their deorphanization. In contrast, lack or erroneous placement of individual receptors in a specific category may preclude salient experimental examination.
The family of adhesion GPCRs (aGPCRs) contains 33 mammalian receptor homologs, most of which are orphans with unknown signals and signaling properties.17 aGPCRs possess very large extracellular N‐termini with adhesive structural folds and a G protein–coupled receptor Autoproteolysis‐INducing (GAIN) domain,18 anchored in the plasma membrane via a seven‐transmembrane (7TM) helices domain, which shows some structural resemblance to the 7TM domain of secretin receptor–like GPCRs.19 Recently, a consortium of scientists working on aGPCRs revised the aGPCR nomenclature.17 Based on the phylogeny of the human aGPCR genes, nine subfamilies were defined (Fig. 1A). The phylogenetic relations were determined merely on the basis of the 7TM amino acid sequences of human aGPCRs20 because of the variable length and composition of their extracellular N‐termini.
The current GRAFS classification system and further subclassification efforts that followed in the wake of the GRAFS system were based on vertebrate and human datasets. Emerging problems with the placement of homologs conserved in invertebrate genomes to the aGPCR subclassification reflect the shortcomings and problems of the currently used system. Moreover, the present subclassification of the aGPCR family underestimates the diversity of aGPCRs even at the mammalian level. Similar observations have been made recently for rhodopsin‐like GPCRs,21 further challenging the current subclassification of GPCRs.
These issues could be satisfactorily remedied by employing ordering principles of the different hierarchy levels present in the aGPCR group that are based on strict phylogenetic criteria (bootstraps/cluster and branch lengths). Further guidance can come from the application of phylogenetic thresholds that separate those hierarchy levels. These should be derived from already well‐investigated GPCR groups and applied to aGPCRs.
Using maximum and minimum phylogenetic distances within well‐investigated rhodopsin‐like receptor subfamilies and minimal distance between receptor subfamilies, which do not share agonists, we found that the current subclassification of aGPCRs into “subfamilies” is not phylogenetically comparable to the subclassification of rhodopsin‐like GPCRs. Members of most rhodopsin‐like GPCR “subfamilies” are phylogenetically less divergent than members within individual vertebrate aGPCR “subfamilies.” On the contrary, the evolutionary distance between many orthologous vertebrate aGPCRs defines them as discrete “subfamilies.”
Materials and methods
Alignments and phylogenetic analysis
All amino acid sequences of the 7TM domains of vertebrate and invertebrate aGPCRs were obtained from GenBank by BLASTing with the 7TM amino acid sequence of all human aGPCRs with the default parameters for a protein BLAST (NCBI) search. Multiple sequence alignments were generated using the alignment explorer implemented in MEGA7 with ClustalW and BLOSUM62 as the scoring matrix and using default parameters. The evolutionary history of the 7TM domain of human, mouse, chicken, and zebrafish aGPCRs was inferred using the maximum likelihood method based on the Jones–Taylor–Thornton (JTT) matrix–based model using MEGA7.22 The bootstrap consensus tree was inferred from 1000 replicates. To account for input‐order bias, similar trees were made with at least three different randomized alignments. Of note, we found no major differences in the tree structure by changing the input order. Initial trees for the heuristic search were obtained by applying the neighbor‐joining method to a matrix of pairwise distances estimated using the JTT model.
Results
Current classification systems for GPCRs
There exist a number of classification systems for GPCRs based on their pharmacological, structural, and/or phylogenetic properties. The most commonly used nomenclatures refer to the NC‐IUPHAR and GRAFS ordering system (Table 1). Hierarchically, the GRAFS system6 and the level system23 provide the best resolution; however, they fall short in nonrhodopsin families/classes. For example, in the level system, secretin receptor–like GPCRs and aGPCRs together form a level 2 entity. At level 2 (subfamily), this system lists not only calcitonin receptors, EGF module‐containing, mucin‐like hormone receptor (EMR), and latrophilins, but also individual receptors, such as GPR64, GPR126, and so on. Levels 4 and 5 are not defined for aGPCRs, but only for rhodopsin‐like GPCRs, with level 5 for individual receptors. This may indicate that the discriminators of level 2 define GPR64 and GPR126 as being as diverse as subfamilies of the amine or nucleotide‐like receptors in the rhodopsin family of this classification. Clearly defined discriminators for the hierarchic levels (e.g., which properties assign a grouped receptor set to a family, subfamily, or class) are also missing for all other classifications. In the different classification systems, sometimes the terms “class,” “family,” “subfamily,” and “group” are ambiguously used for the same hierarchical level.
Table 1.
Different GPCR classification systems
| Hierarchy level | Bottom | ↔ | ↔ | ↔ | ↔ | Top | |
|---|---|---|---|---|---|---|---|
| Numerical | 1° | 2° | 3° | 4° | 5° | 6° | |
| Taxonomical | Species | Genus | Family | Order | Class | Phylum | Reference |
| NC‐IUPHAR |
Individual receptor (e.g., HTR2A) |
Subclass/subfamilies (e.g., serotonin receptors) | Class/clan/family (e.g., rhodopsin‐like) | Superfamily (GPCR) | 3 | ||
| Joseffson |
Individual receptor (e.g., HTR2A) |
Subclass (amine receptors) | Class (e.g., rhodopsin‐like) | Clade (e.g., rhodopsin‐like and secretin receptor–like | Superfamily (GPCR) | 21 | |
| GRAFS | Individual receptor (e.g., HTR2A receptor of HTR2 subgroup) | Subgroup (e.g., HTR2 subgroup of amine receptor cluster) | Branch/cluster * (e.g., amine receptor) | Group (e.g., α‐group of rhodopsin receptors) | Family/cluster * (e.g., rhodopsin‐like) | Superfamily (GPCR) | 6 |
| GRAFS/aGPCR | Individual receptor | Group* (aGPCR I–IX) | Subfamily*/family/group* (e.g., aGPCR) | Superfamily (GPCR) | 22 | ||
| Level system |
Individual receptor subtype (level 5) (e.g., HTR2A) |
Subsubfamily (level 3) (e.g., serotonin receptors) |
Subfamily (level 3) (e.g., amine receptors) |
Family (level 2) (e.g., rhodopsin‐like) |
GPCR versus non‐GPCR (level 1) | 20 | |
| GRAFS/aGPCR revised | Individual receptor/subtype | Subfamily (aGPCR I–IX) | Family (e.g., aGPCR) | Superfamily (GPCR) | 15 | ||
| New nomenclature | Individual receptor subtypes (e.g., HTR2A) | Genus of closely related subtypes (e.g., HTR2A, HTR2B, and HTR2C) | Family of different subtypes (e.g., serotonin receptors) | Order of distinct branches (e.g., amine receptors) | Class of GPCR (rhodopsin‐like) | Phylum of GPCR | This study |
Note: Different classification approaches for GPCRs. Bold terms indicate intermediate hierarchy‐level denominators, asterisks denote ambiguous use of hierarchy‐level denominators in the original publications. When we refer to published nomenclature systems, we use the terms employed therein.
Phylogeny does not support the organization of aGPCRs into nine subfamilies
Human aGPCRs have been divided into “groups” (redefined as “subfamilies” in Ref. 17) (2° level) based on the phylogenetic relationships of the 7TM domains and bootstrap analyses of nodes separating branches.24 The bootstrapping value reflects how well a node supports the phylogenetic tree model. This implies that only aGPCRs, which significantly cluster, ought to be considered a distinct group.
In order to provide an unambiguous taxonomic and hierarchically organized description of the set of aGPCRs, we first reanalyzed the human aGPCR dataset using a neighbor‐joining approach (Fig. 1A). We found that most groups, which were previously assigned, cluster by bootstrap values >50% (Fig. 1A) and, as expected, that more closely related to aGPCRs branch from a node with higher bootstrapping values than groups of more distantly related aGPCRs. However, in most cases, the nodes separating groups were not supported by bootstrapping values (>90%), suggesting that their distinct grouping should be dissolved. Consequently, the pairs of groups I and II as well as groups V and VI, which form four individual groups in the current classification, should collapse into one group per pair.
Furthermore, the branch lengths within groups and between groups are not a valuable discriminator. For example, the branch lengths within group VIII are as long as the cumulative branch lengths of groups I and II (Fig. 1A). This may indicate either different sequence constraints or an arbitrary collection of group members. Therefore, we assumed that the strength of support for phylogenetic nodes is simply too low when the analysis is based exclusively on human aGPCR sequences.
Therefore, we extended the phylogenetic testing and included aGPCR sequences from human, mouse, chicken, and zebrafish. However, the outcome (Fig. 1B) was essentially comparable to the results obtained from human‐only dataset (Fig. 1A). Individual vertebrate aGPCRs are well separated in the tree, whereas the groups are poorly supported by significant nodes or group‐specific clustering. For example, groups I and II still form a bootstrapping value–supported cluster. Furthermore, we considered that the phylogenetic method employed to generate and analyze the trees may impact the outcome of receptor clustering. However, when we generated trees using a maximum likelihood approach, we obtained the same conclusions (Fig. S1A and B, online only).
Our analysis therefore confirms results reported in the original and follow‐up publications that introduced the aGPCR group classification, in which many group‐separating nodes are not significantly supported by bootstrapping values (<90%).19, 24, 25 Thus, it remains unclear whether additional criteria other than phylogenetic parameters were employed in the definition of “group” level in this previous work.
An additional criticism pertains to the currently used numerical order of individual members, which does not reflect the relative relation and distance of members within a group/subfamily. For example, ADGRF2 and F4 are more closely related than F4 and F5. A similar inconsistency is found in the current nomenclature for rhodopsin‐like GPCRs. Here, for example, muscarinic acetylcholine receptors M2 and M4 are phylogenetically and functionally more closely related than M1 and M2 (Fig. 1).
In sum, due to the current lack of known endogenous agonists, pharmacology‐based classification criteria cannot be applied to the aGPCR family. Therefore, its subclassification must rest on rules governing the ordering of well‐defined rhodopsin‐like GPCRs.
Branch length comparison shows lower diversity within the aGPCR family than in individual rhodopsin‐like GPCR groups
To maintain agonist and signaling specificity and thus the fitness of a species during evolution, only a certain degree of freedom in structural diversity is tolerated (evolutionary plasticity of protein structure).26 Branch lengths of a phylogenetic tree reflect the number of substitutions per site across sequences and are therefore a measure for the structural conservation of a given protein. Proteins may differ in their structural conservation (constraint) and respective branch lengths even when identical species, and therefore the same evolutionary time, are compared. Faster evolving genes may reflect adaptation processes or a complete loss of constraint (pseudogenization).27 The comparison of evolutionarily well‐characterized GPCRs, indicated by preserved agonist and signaling specificity, offers an average of constraints (branch lengths) in GPCRs. Thus, we performed phylogenetic analyses for selected rhodopsin‐like GPCRs, secretin receptor–like GPCRs, and aGPCRs in parallel. To this end, we retrieved homologous sequences for all human GPCRs and, if available from GenBank, their orthologs from mouse, chicken, and zebrafish.
As shown in Figure 2 and previously reported,19, 28 the secretin receptor family branches off from the aGPCR family. Interestingly, the total branch length of the entire secretin receptor–like GPCR cluster compares well with the cumulative branch lengths of individual aGPCR subfamilies, for example, VI (ADGRF) or VIII (ADGRG), although they contain a lower number of individual sequences (signified by base size of the triangles in Fig. 2).
Figure 2.
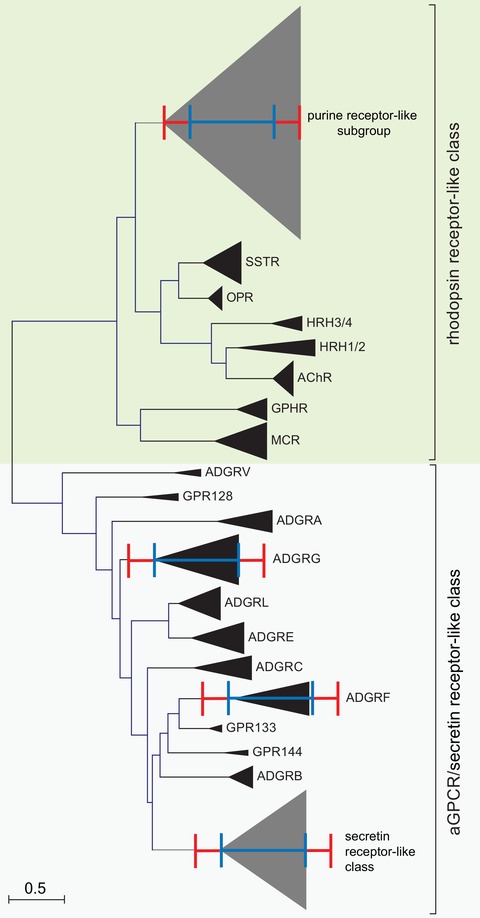
Molecular phylogenetic analysis of selected rhodopsin‐like GPCRs, secretin receptor–like GPCRs, and aGPCRs by the maximum likelihood method. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix–based model.70 The tree with the highest log likelihood (–52401.93) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 425 amino acid sequences, where orthologous sequences of human, mouse, chicken, and zebrafish were included when available. All positions containing gaps and missing data were eliminated. There was a total of 129 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22 The orthologs and paralogs of individual receptors and receptor subgroups, respectively, were condensed and depicted as triangles. The red and blue scale bars represent the branch lengths of the purine cluster within the delta group and secretin class members, respectively.
The rhodopsin‐like receptor classification defines 4 groups (α, β, γ, and δ) and 13 clusters.6 Importantly, the entire aGPCR/secretin receptor–like set shows a total branch length similar to that of the purine receptor‐like subgroup of the δ group (Fig. 2), which includes receptors with diverse agonist specificities (e.g., including receptors for carbonic acids, phospholipids, nucleotides, and peptides6). Of note, the δ group also contains a cluster of glycoprotein hormone receptors (GPHR, see Fig. 2) and the MAS‐related receptor cluster,6 which we excluded from our analysis.
Taken together, our analysis shows that aGPCRs and secretin receptor–like GPCRs show comparable diversity profiles, which are lower than those found in individual rhodopsin‐like receptor groups. This demonstrates that the current “subfamily” designation of aGPCRs is not consistent with the standards and approaches employed to establish clusters (3° level) and groups (4° level) within rhodopsin‐like GPCRs.
Branch length comparison of aGPCRs with pharmacologically different rhodopsin‐like GPCRs
Our data show that the subclassification of the aGPCR family into nine “subfamilies”/“groups” has no equivalent in the NC‐IUPHAR–supported rhodopsin‐like GPCR designation (see also, Table 1). Individual receptor subclasses/subfamilies (3° level) comprise the next lower hierarchical level in rhodopsin‐like GPCRs. Typically, subclass members are activated by the same or structurally related agonists.2 In order to compare the structural diversity of the aGPCR family members at a pharmacologically defined hierarchy level, we built a phylogenetic tree, where GPCRs with similar endogenous agonists were collapsed into a single branch (Fig. 3). We observed that histamine receptor ortho‐/paralogs, which constituted the most distantly related receptors within a pharmacological group in the dataset, resulted in the largest branch length within this tree, as expected. In contrast, the β, γ, and κ opioid receptor ortho‐/paralogs, which are highly conserved among fish, chicken, mouse, and human, amounted to the smallest branch length in the tree (Fig. 3A). All other subclass branch lengths ranged between these minimum and maximum values.
Figure 3.
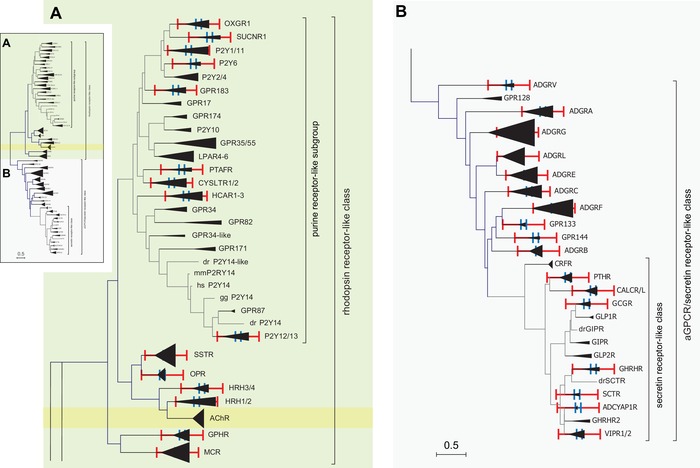
Molecular phylogenetic analysis of selected rhodopsin‐like GPCRs, secretin receptor–like GPCRs, and aGPCRs by the maximum likelihood method. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix–based model.70 The tree with the highest log likelihood (–52401.93) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 425 amino acid sequences, where orthologous sequences of human, mouse, chicken, and zebrafish were included when available. All positions containing gaps and missing data were eliminated. There was a total of 129 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22 The inset depicts the full phylogenetic tree, which was enlarged for rhodopsin‐like GPCRs (A) and the aGPCR/secretin receptor‐like class (B). The orthologs and paralogs of individual receptors and receptor subgroups, respectively, were condensed and depicted as triangles. The red and blue scale bars represent the branch lengths of the opioid receptor family (OPR) and the histamine type 1 and type 2 family (HRH1/2), respectively.
Projection of the so derived rhodopsin‐like receptor diversity range onto secretin receptor–like GPCRs revealed a lower sequence diversity for each secretin receptor–like subfamily or individual receptor (Fig. 3B). In the case of aGPCRs, we found that “subfamilies” ADGRA, ADGRB, ADGRC, ADGRE, and ADGRL, as well as the individual aGPCR VLGR1/ADGRV1, are placed within the range of structural conservation found in rhodopsin‐like GPCRs. Remarkably, however, the diversity of “subfamilies” ADGRD, ADGRF, and ADGRG was exceeding the maximum of those observed in rhodopsin‐like GPCRs (Fig. 3B). This finding may reflect the variability of the number of amino acids required to determine the activity profile of a given GPCR, as is the case for peptide‐ versus amine‐activated rhodopsin‐like receptors.29
Several species contain novel subfamilies not covered in the current adhesion GPCR classification
The wealth of genomic data generated by next‐generation sequencing approaches allowed for the extraction and comparison of the GPCR repertoire that includes aGPCRs from numerous invertebrate and vertebrate species.28, 30, 31, 32, 33 This approach facilitated the reconstruction of a phylogenetic tree encompassing all GPCR classes. Based on this tree, aGPCRs appear to have evolved from the cAMP receptor family before the split of the Unikonts from the common ancestor of eukaryotes about 1275 million years ago.28 Further, several invertebrate species contain aGPCRs and aGPCR subfamilies without close relatives in vertebrate genomes.20, 28, 32 However, since invertebrates (e.g., arthropods, nematodes, and cephalopods) serve as prominent and widely used, genetically amenable animal models enabling in‐depth interrogation of molecular mechanisms, it is of great interest to establish orthology between individual invertebrate and vertebrate aGPCRs.
To evaluate their phylogenetic relationships, aGPCR sequences derived from several model organisms were first placed into an established phylogenetic tree. As an example, the Drosophila genome contains at least 10 potential aGPCR/secretin receptor–like family members. Introduction of these aGPCR candidates into the phylogenetic tree of vertebrate aGPCRs and secretin receptors revealed that five of them are assigned to secretin receptor–like GPCR branches. Of the five remaining receptors that were assigned as aGPCRs, CG15744 and CG11895/Flamingo/Starry night are most likely related to ADGRA and ADGRC members, respectively (Fig. 4). Both CG15556 and CG11318 34 are not clearly assignable to any of the current “subfamilies,” but rather form a novel, additional “subfamily” in the aGPCR/secretin receptor–like family (Fig. 4). Most interestingly, CG8639/Cirl, an aGPCR, that was formally assigned as a latrophilin homolog,35 appears to be unrelated to the ADGRL subfamily based on its 7TM domain sequence but is most closely related to the ADGRA subfamily of aGPCRs. These results clearly highlight the importance and necessity of phylogenetic trees that include other invertebrate aGPCRs.
Figure 4.
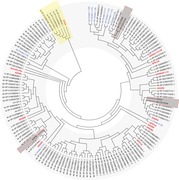
Phylogenetic relationship of Drosophila melanogaster aGPCR‐like sequences with vertebrate aGPCRs. The evolutionary relationships of D. melanogaster, human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors served as the outgroup. (A) The evolutionary history was inferred using the neighbor‐joining method.67 The optimal tree with the sum of branch lengths of 42.71172243 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.68 The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method69 and are in the units of the number of amino acid substitutions per site. (B) The evolutionary history was inferred the maximum likelihood method based on the JTT matrix–based model.70 The tree with the highest log likelihood (–29511.43) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The analyses involved 225 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 193 (A) and 141 (B) positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22 dm, D. melanogaster.
Figure 5 shows a phylogenetic tree generated from more than 500 aGPCR sequences from vertebrates and invertebrates. This tree demonstrates that the current nine subfamilies of aGPCRs cover only a small receptor spectrum of this diverse GPCR class/family. Secretin receptor–like GPCRs are very likely a subgroup within aGPCRs and should be classified as such. Interestingly, in this phylogenetic tree, the large subfamily VIII (ADGRG) splits into three groups (GPR56/97/114, GPR64/112/126, and GPR128), and both members of subfamily ADGRD, GPR133 and GPR144, are only distantly related to each other and do not form a joint cluster. The distant relationships between GPR133 and GPR144, both of which are currently ADGRD members, and between GPR128 and the remaining ADGRG members have been previously discussed.28 In contrast, the individual clustering of subfamilies ADGRL and ADGRE is still weakly supported by the bootstrapping values (>50%), reiterating the need to fuse both subfamilies (Fig. 1).
Figure 5.
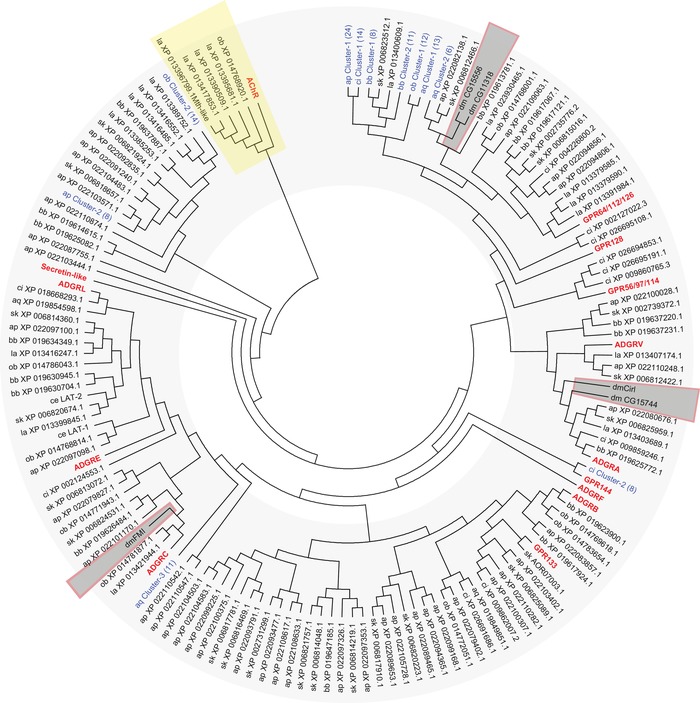
Phylogenetic analysis of selected invertebrate and vertebrate aGPCRs. The evolutionary relationships of selected invertebrate aGPCRs and human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors (AChR) served as the outgroup. The evolutionary history was inferred using the neighbor‐joining method.67 The optimal tree with the sum of branch length = 138.25837393 is shown. The evolutionary distances were computed using the Poisson correction method69 and are in the units of the number of amino acid substitutions per site. The analyses involved 525 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 183 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22 Current subfamilies of aGPCRs are condensed and shown in bold red characters. aGPCR clusters found in invertebrates are shown in blue and the number of receptors included in the cluster is given in parenthesis. Species included in the analysis (aGPCRs from D. melanogaster, including the newly identified genes, are boxed): vertebrate: Homo sapiens (hs), Mus musculus (mm), Gallus gallus (gg), Danio rerio (dr); Cephalochordata: Branchiostoma belcheri (bb); Tunicata: Ciona intestinalis (ci); Hemichordata: Saccoglossus_kowalevskii (sk); Brachiopoda: Lingula anatina (la); Echinodermata: Acanthaster planci (ap); Mollusca Octopus bimaculoides (ob); Neoptera: Drosophila melanogaster (dm); Nematoda: Caenorhabditis elegans (ce); Parazoa: Amphimedon queenslandica (aq).
With regard to aGPCRs from Drosophila melanogaster, the clustering of CG15744 with the ADGRA subfamily is again supported by the bootstrapping value (84%), and so is the relationship of CG8639/Cirl to this cluster (25%) (Fig. 5; Fig. S2, online only). Similarly, CG11895/Flamingo/Starry night is related to ADGRC. However, CG15556 and CG11318 are related to the aq‐Cluster‐2 that contains six aGPCRs found in the genome of the parazoan Amphimedon queenslandica (Fig. 5; Fig. S2, online only).
Discussion
Current classification of GPCRs is mainly based on pharmacological and functional properties collected in GPCRdb.36 However, about 100 of the nonodorant GPCRs are considered orphans,37 including all aGPCRs. A better understanding of the phylogeny may help in deorphanizing GPCRs, whose physiological receptor‐activating signals are still unknown, and therefore may represent new druggable targets for pharmaceutical research.38 Phylogenetic classifications of GPCRs are mainly based on sequence alignments.6, 23 Indeed, there is a significant overlap of the chemical nature of endogenous agonists and the clustering of GPCRs in phylogenetic analyses,29, 39, 40, 41 and alignment‐free classifications, such as 7TMRmine,42 GPCR Tree,43 and a proteochemometric approach,44 endorse this classification logic. The availability of GPCR structures now enables the assessment of the evolutionary history of GPCRs by combining both sequence and structural properties in phylogenetic approaches.41, 45, 46 Therefore, the classification not only improves naming and cataloguing of GPCRs, but can also help to pinpoint experimental approaches for in‐depth functional analyses.
In general, predictions from distance‐based phylogenetic methods (e.g., neighbor‐joining method) appear more consistent with the agonist‐based classification of GPCRdb than those from character‐based methods (e.g., maximum likelihood method).29 However, a distance‐based clustering approach causes hierarchical problems, for example, the distance between Flamingo‐like receptors (adhesion/secretin family) and glycoprotein hormone rhodopsin‐like GPCRs is lower than between odorant receptors and amine receptor (both rhodopsin‐like).29 This indicates that for classification attempts structural and/or sequence hallmarks are required to assign given sequences first into classes (5° level) and then into subgroups (1°–4° levels) reflecting their phylogenetic relationships within the GPCR superfamily (6° level) (Table 1).
The aGPCR family of receptors counts 33 homologs in mammals and constitutes the second largest family within the GPCR superfamily. It has been noted that aGPCRs are found in every organ system in humans; however, thus far general concepts of how this molecule class operates have remained largely unknown. One possibility to attain a better understanding of the functional underpinnings of aGPCRs is to understand the evolutionary history and the genetic relationships of aGPCRs to each other, as well as to other GPCR families.
According to the currently used GRAFS classification, aGPCRs form an individual entity next to the glutamate, rhodopsin, frizzled, secretin receptor “families.”5, 6 In contrast to this classification, our analysis suggests that the secretin receptor–like and aGPCR families cluster, a finding that is consistent with previous studies.19 Consequently, the secretin receptor–like GPCR family should be merged with the aGPCR family and subcategorized accordingly.
The current subclassification of the aGPCR family is based on the phylogenetic profile of 7TM sequences of the 33 human genes20 (Fig. 1A). Several aspects relating to this internal ordering scheme are controversial. First, the scientific rationale of the subfamily assembly is not clear. For example, according to bootstrap values and branch length values, subfamilies ADGRL and ADGRE cluster together and should not be divided into two separate subfamilies. In addition, ADGRG7/GPR128, although not closely related to other subfamily VIII members, is grouped into this subfamily. Second, the current subfamily classification completely neglects aGPCR homologs from other species and renders the assignment of newly discovered genes difficult, if not impossible. By screening the D. melanogaster genome using the combination of a 7TM and GAIN domain‐encoding sequence as a query, we identified three novel putative aGPCRs: CG15556, CG11318, and CG15744. While dmCG15744 clustered to subfamily III (ADGRA), dmCG15556 and dmCG11318 could not be assigned to any of the existing subfamilies but rather constitute a novel aGPCR subfamily. This is particularly noteworthy in light of efforts that use genetic models, such as the nematode Caenorhabditis elegans,47, 48 the vinegar fly D. melanogaster,34, 35, 49 and the zebrafish Danio rerio 50, 51, 52 to unravel the physiological roles and pharmacological underpinnings of aGPCR functions.
As evident from a recent genomic analysis of the zebrafish aGPCR repertoire25 (Fig. 1B), a one‐to‐one ortholog assignment of aGPCRs between human and the zebrafish is problematic. This is due to the fact that several zebrafish aGPCRs are phylogenetically placed in a basal position to more than one human aGPCR (e.g., in subfamily VI and subfamily VIII), and because the zebrafish often possesses multiple orthologs (e.g., LPHN1, CD97, and CELSR1) that are related to a single human aGPCR ortholog. This is not an aGPCR‐specific issue, but also evident in many GPCRs when homologs of distantly related species are compared. Here, an intermediate category between the individual member (1° level) and “subfamilies” (currently 3° level) is required.
Information about the degree of relationship of receptors is important for in vitro analyses of aGPCRs as much as for the investigation of aGPCR function in their native biological environments. Moreover, ligand specificity and putative functional redundancy are highly relevant in in vivo settings and it is therefore important to know whether closely related genes can be considered as “subtypes” because they share the same endogenous agonist(s). For instance, thus far the Drosophila homolog CG8639/Cirl has been associated with vertebrate latrophilins. Several ligands have been described for mammalian latrophilins (teneurins,53 FLRTs,54, 55 and neurexin‐1β54); however, to date, no evidence for an interaction between the orthologous invertebrate ligand candidates and invertebrate latrophilins/dmCirl has been obtained yet.56 While the extracellular region of vertebrate latrophilins and dmCIRL contains rhamnose‐binding lectin and hormone receptor motif domains, the latter lacks the olfactomedin domain,57 which is required for biochemical interactions of vertebrate latrophilins with neurexin‐1β and FLRT. This observation renders neurexin‐1β and FLRT unlikely interactors for dmCIRL (corroborated by the fact that the Drosophila genome does not encode FLRT49). Thus, our phylogenetic assessment, which separates dmCirl from vertebrate latrophilins, may also reflect the differing ligand preferences between the phyla.
Our analysis has further highlighted the dissimilarities in classification systems applied to GPCRs, and their differing, inconsistent, and incongruent terminologies. The parallelism of classifications, such as the NC‐IUPHAR and GRAFS systems, has caused much confusion with respect to their internal hierarchies, and consequently regarding the basis for the relationships between aGPCRs. Advocating a bottom‐up ordering logic in the phylogenetic classification of GPCRs, such a system uses stacked hierarchy levels denominated by taxonomic terms, which distinctly separate species (1°), genus (2°), family (3°), order (4°), class (5°), and phylum (6°). Applied to GPCRs, this established system could be used as in the following example: species (1°): metabotropic serotonin 5‐HT1A; genus (2°): metabotropic serotonin 5‐HT1, comprising 5‐HT1A/5‐HT1B/5‐HT1D/5‐HT1F; family (3°): all metabotropic serotonin receptors, comprising 5‐HT1/5‐HT2/5‐HT4‐7; order (4°): for example, alpha group6 or aminergic receptor; class (5°): rhodopsin‐like GPCRs; and phylum (6°): GPCRs.
Collectively, our data demand a revision of the current GRAFS classification‐based assignment of aGPCRs into nine subfamilies. The current lack of sufficient functional information does not allow for equivalent discriminators as we have for many rhodopsin‐like GPCRs. Currently, we can only rely on the comparison of evolutionary distances found for the classification levels in rhodopsin‐like GPCRs and the evolutionary distances of aGPCRs determined from sequence alignments. Following this constraint, one would consider, for example, the relationship of the three ADGRA members (GPR123/ADGRA1, GPR124/ADGRA2, and GPR125/ADGRA3) as a joint family, a 3° relationship. Currently, a 2° level in this family is missing since there is a strict one‐to‐one orthology between fish and human and no further subdivision into, for example, GPR123a and GPR123b is required (Fig. S2, online only). Similarly, the three members of ADGRC (CELSR1‐3/ADGRC1‐3) should be considered a family too (3°), but there are two zebrafish CELSR1 homologs, CELSR1a and CELSR1b, introducing the genus level (2°) in this family (Fig. S2, online only). Our data also suggest to divide the current subfamily VIII (ADGRG) into at least three families (3°), with GPR56/GPR97/GPR114, GPR64/GPR112/GPR126, and GPR128 (Fig. 5). Similarly, GPR133/ADGRD1 and GPR144/ADGRD2 do not belong to the same “subfamily” but even constitute separate 3° entities (Fig. 5). The former ADGRE “subfamily” is most probably an offspring of the ADGRL “subfamily,” and both subfamilies robustly cluster together with branch lengths comparable to those within ADGRF. Therefore, the ADGRE and ADGRL members should be considered as one family (3°) with several genera (2°), since there is not always a one‐to‐one orthology (e.g., four CD97/ADGRE5 and two LPHN1/ADGRL1 homologs in zebrafish). Alternatively, and maintaining the internal phylogenetic distance‐based logic of our novel classification system, if one considers both ADGRE and ADGRL as two families (3°), then ADGRF and GPR56/GPR97/GPR114 need to be rearranged into multiple families because of comparable or even higher branch length differences between the members. The secretin receptor–like GPCRs and several other separate branches may form separate orders (4°) within the class (5°) of aGPCR‐like and secretin receptor–like GPCRs.
Conclusions
The current subclassification of aGPCRs into several “subfamilies”17 bears a number of ambiguities, which may mislead the prediction of the functional relationships between aGPCRs. Although the hierarchical classification structure we suggest (Table 1) can readily be applied on a 1° level (individual subtype, e.g., zebrafish CELSR1a), 2° level (genus of closely related subtypes, e.g., zebrafish CELSR1a and CELSR1b), 5° level (class of aGPCRs), and 6° level (phylum of GPCRs), the 3° level (family—identical agonist/activation mode) and 4° level (order—chemically related agonists/comparable activation modes) cannot be defined yet by the current state of research based on meaningful phylogenetic and/or pharmacological parameters. The following tasks need to be solved in the future in order to replace the current classification of aGPCRs with an easy adaptable and extendable classification and nomenclature.
First and most important, the current receptor clustering and the subsequent nomenclature are only partially supported by phylogenetic analyses, but not by pharmacological data. Projecting the mainly pharmacology‐driven classification of rhodopsin‐like and secretin receptor–like GPCRs onto a phylogenetic tree (Fig. 3), the average sequence distances within pharmacologically defined receptor families (3° level) are lower than in the current “subfamilies” of aGPCRs. This indicates that structurally diverse aGPCR “subfamilies,” such as ADGRD, ADGRF, and ADGRG, most probably accommodate several families (3° level). Therefore, the deduction of the mode of activation and/or signal transduction of the receptor homologs within these “subfamilies” by apparent relationship will yield mixed results at best. The only way to solve this issue is to provide more functional data with respect to activation and signaling mechanisms of aGPCRs. With analogy to rhodopsin‐ and secretin receptor–like GPCRs, this will allow defining pharmacology‐based families (3° level), which may then be assembled into orders (4° level). It will be interesting to see how pharmacological families project onto the phylogenetic distances of aGPCRs. Depending on the signal/agonists and activation mechanism, we may see very different constraints and, therefore, branch lengths within families (3° level).
Second, since secretin receptor–like GPCRs seem to be an offspring of ancient aGPCRs, the members of this currently termed “class” or “family” should be placed into the hierarchical system. If one considers aGPCRs as a class (5° level), secretin receptor–like GPCRs would appear as one order (4° level) of this class.
Third, a revised aGPCR nomenclature should logically renumber aGPCR family members (3° level) to accurately depict evolutionary context and relationships. For example, BAI1/ADGRB1 is more closely related to BAI3/ADGRB3 than to BAI2/ADGRB2 (Fig. 1). Renumbering should read like BAI1/ADGRB1, BAI3/ADGRB2, and BAI2/ADGRB3.
Fourth, the nomenclature system should account for the diversity of vertebrate and invertebrate aGPCR sequences.
Fifth, a revised aGPCR nomenclature should consider the expected multitude of aGPCR transcript variants. Recent studies have already highlighted that numerous variants are derived from single aGPCR genes by alternative promoters and splicing.53, 58, 59, 60, 61, 62, 63, 64, 65, 66
Author contributions
T.L. and T.S. conceived the study, provided supervision, validated the experimental results and visualized the data together, and administered the project. N.S., T.L., and T.S. performed the investigation, data curation, and analysis; provided resources; and wrote the manuscript.
Competing interests
The authors declare no competing interests.
Supporting information
Figure S1. Current nomenclature of aGPCRs based on phylogenetic analysis by the maximum likelihood method. The evolutionary relationships of (A) only human aGPCRs and (B) human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors served as the outgroup. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix–based model.70 The trees with the highest log likelihood (A: –11604.80; B: –21663.01) are shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The analyses involved 37 (A) and 150 (B) amino acid sequences. All positions containing gaps and missing data were eliminated. There was a total of 195 (A) and 158 (B) positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22
Suppl. File. Scholz et al. fas contains all sequences analyzed in this study (in FASTA format).
Figure S2. Higher resolution of the phylogenetic analysis of selected invertebrate and vertebrate aGPCRs from Figure 5 of the main text. The evolutionary relationships of selected invertebrate aGPCRs and human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors (AChR) served as the outgroup. The evolutionary history was inferred using the neighbor‐joining method.67 The optimal tree with the sum of branch length = 138.25837393 is shown. The evolutionary distances were computed using the Poisson correction method69 and are in the units of the number of amino acid substitutions per site. The analyses involved 525 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 183 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22 Current subfamilies of aGPCRs are condensed and shown in bold red characters. aGPCR clusters found in invertebrates are shown in blue and the number of receptors included in the cluster is given in parenthesis. Species included in the analysis: vertebrate: Homo sapiens (hs), Mus musculus (mm), Gallus gallus (gg), Danio rerio (dr); Cephalochordata: Branchiostoma belcheri (bb); Tunicata: Ciona intestinalis (ci); Hemichordata: Saccoglossus_kowalevskii (sk); Brachiopoda: Lingula anatina (la); Echinodermata: Acanthaster planci (ap); Mollusca Octopus bimaculoides (ob); Neoptera: Drosophila melanogaster (dm); Nematoda: Caenorhabditis elegans (ce); Parazoa: Amphimedon queenslandica (aq).
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft to N.S. (FOR 2149/ P01, SCHO 1791/1‐2), T.L. (FOR 2149/P01, LA2861/4‐2; FOR 2149/P03, LA2861/5‐2; LA2861/7‐1; TRR166/C03), and T.S. (FOR 2149/P04; SFB 1052/B6). N.S. was supported by a Junior Research Grant awarded from the Medical Faculty of Leipzig University.
Contributor Information
Tobias Langenhan, Email: tobias.langenhan@gmail.com.
Torsten Schöneberg, Email: schoberg@medizin.uni-leipzig.de.
References
- 1. Lagerstrom, M.C. & Schioth H.B.. 2008. Structural diversity of G protein‐coupled receptors and significance for drug discovery. Nat. Rev. Drug Discov. 7: 339–357. [DOI] [PubMed] [Google Scholar]
- 2. Alexander, S.P. , et al 2017. The concise guide to pharmacology 2017/18: G protein‐coupled receptors. Br. J. Pharmacol. 174(Suppl. 1): S17–S129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolakowski, L.F., Jr. 1994. GCRDb: a G‐protein‐coupled receptor database. Receptors Channels 2: 1–7. [PubMed] [Google Scholar]
- 4. Attwood, T.K. & Findlay J.B.. 1994. Fingerprinting G‐protein‐coupled receptors. Protein Eng. 7: 195–203. [DOI] [PubMed] [Google Scholar]
- 5. Schioth, H.B. & Fredriksson R.. 2005. The GRAFS classification system of G‐protein coupled receptors in comparative perspective. Gen. Comp. Endocrinol. 142: 94–101. [DOI] [PubMed] [Google Scholar]
- 6. Fredriksson, R. , et al 2003. The G‐protein‐coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63: 1256–1272. [DOI] [PubMed] [Google Scholar]
- 7. Abbracchio, M.P. , et al 2006. International Union of Pharmacology LVIII: update on the P2Y G protein‐coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol. Rev. 58: 281–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Duc, D. , et al 2017. P2Y receptors in immune response and inflammation. Adv. Immunol. 136: 85–121. [DOI] [PubMed] [Google Scholar]
- 9. Schoneberg, T. , et al 2007. Structural and functional evolution of the P2Y(12)‐like receptor group. Purinergic Signal. 3: 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang, D. , et al 2015. Two disparate ligand‐binding sites in the human P2Y1 receptor. Nature 520: 317–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang, J. , et al 2014. Agonist‐bound structure of the human P2Y12 receptor. Nature 509: 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang, K. , et al 2014. Structure of the human P2Y12 receptor in complex with an antithrombotic drug. Nature 509: 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caulfield, M.P. 1993. Muscarinic receptors—characterization, coupling and function. Pharmacol. Ther. 58: 319–379. [DOI] [PubMed] [Google Scholar]
- 14. Wess, J. , et al 1995. Muscarinic acetylcholine receptors: structural basis of ligand binding and G protein coupling. Life Sci. 56: 915–922. [DOI] [PubMed] [Google Scholar]
- 15. Schoneberg, T. , et al 1998. Molecular aspects of vasopressin receptor function. Adv. Exp. Med. Biol. 449: 347–358. [DOI] [PubMed] [Google Scholar]
- 16. Chini, B. , et al 1996. Two aromatic residues regulate the response of the human oxytocin receptor to the partial agonist arginine vasopressin. FEBS Lett. 397: 201–206. [DOI] [PubMed] [Google Scholar]
- 17. Hamann, J. , et al 2015. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein‐coupled receptors. Pharmacol. Rev. 67: 338–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arac, D. , et al 2012. A novel evolutionarily conserved domain of cell‐adhesion GPCRs mediates autoproteolysis. EMBO J. 31: 1364–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nordstrom, K.J. , et al 2009. The secretin GPCRs descended from the family of adhesion GPCRs. Mol. Biol. Evol. 26: 71–84. [DOI] [PubMed] [Google Scholar]
- 20. Krishnan, A. , et al 2016. Classification, nomenclature, and structural aspects of adhesion GPCRs. Handb. Exp. Pharmacol. 234: 15–41. [DOI] [PubMed] [Google Scholar]
- 21. Rinne, M. , et al 2019. Cartography of rhodopsin‐like G protein‐coupled receptors across vertebrate genomes. Sci. Rep. 9: 7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar, S. , Stecher G. & Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peng, Z.L. , Yang J.Y. & Chen X.. 2010. An improved classification of G‐protein‐coupled receptors using sequence‐derived features. BMC Bioinformatics 11: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bjarnadottir, T.K. , et al 2004. The human and mouse repertoire of the adhesion family of G‐protein‐coupled receptors. Genomics 84: 23–33. [DOI] [PubMed] [Google Scholar]
- 25. Harty, B.L. , et al 2015. Defining the gene repertoire and spatiotemporal expression profiles of adhesion G protein‐coupled receptors in zebrafish. BMC Genomics 16: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panchenko, A.R. , et al 2005. Evolutionary plasticity of protein families: coupling between sequence and structure variation. Proteins 61: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coin, L. & Durbin R.. 2004. Improved techniques for the identification of pseudogenes. Bioinformatics 20(Suppl. 1): i94–i100. [DOI] [PubMed] [Google Scholar]
- 28. Krishnan, A. , et al 2014. The GPCR repertoire in the demosponge Amphimedon queenslandica: insights into the GPCR system at the early divergence of animals. BMC Evol. Biol. 14: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu, G.M. , Mai T.L. & Chen C.M.. 2017. Visualizing the GPCR network: classification and evolution. Sci. Rep. 7: 15495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krishnan, A. , et al 2013. Remarkable similarities between the hemichordate (Saccoglossus kowalevskii) and vertebrate GPCR repertoire. Gene 526: 122–133. [DOI] [PubMed] [Google Scholar]
- 31. Haitina, T. , et al 2008. Expression profile of the entire family of Adhesion G protein‐coupled receptors in mouse and rat. BMC Neurosci. 9: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nordstrom, K.J. , Fredriksson R. & Schioth H.B.. 2008. The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein‐coupled receptors. BMC Evol. Biol. 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gloriam, D.E. , Fredriksson R. & Schioth H.B.. 2007. The G protein‐coupled receptor subset of the rat genome. BMC Genomics 8: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blanco‐Redondo, B. & Langenhan T.. 2018. Parallel genomic engineering of two Drosophila genes using Orthogonal attB/attP sites. G3 (Bethesda) 8: 3109–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scholz, N. , et al 2015. The adhesion GPCR latrophilin/CIRL shapes mechanosensation. Cell Rep. 11: 866–874. [DOI] [PubMed] [Google Scholar]
- 36. Pandy‐Szekeres, G. , et al 2018. GPCRdb in 2018: adding GPCR structure models and ligands. Nucleic Acids Res. 46: D440–D446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laschet, C. , Dupuis N. & Hanson J.. 2018. The G protein‐coupled receptors deorphanization landscape. Biochem. Pharmacol. 153: 62–74. [DOI] [PubMed] [Google Scholar]
- 38. Chung, S. , Funakoshi T. & Civelli O.. 2008. Orphan GPCR research. Br. J. Pharmacol. 153(Suppl. 1): S339–S346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vassilatis, D.K. , et al 2003. The G protein‐coupled receptor repertoires of human and mouse. Proc. Natl. Acad. Sci. USA 100: 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Joost, P. & Methner A.. 2002. Phylogenetic analysis of 277 human G‐protein‐coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol. 3: RESEARCH0063. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kakarala, K.K. & Jamil K.. 2014. Sequence‐structure based phylogeny of GPCR Class A Rhodopsin receptors. Mol. Phylogenet. Evol. 74: 66–96. [DOI] [PubMed] [Google Scholar]
- 42. Lu, G. , et al 2009. 7TMRmine: a Web server for hierarchical mining of 7TMR proteins. BMC Genomics 10: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davies, M.N. , et al 2008. GPCRTree: online hierarchical classification of GPCR function. BMC Res. Notes 1: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lapinsh, M. , et al 2005. Improved approach for proteochemometrics modeling: application to organic compound–amine G protein‐coupled receptor interactions. Bioinformatics 21: 4289–4296. [DOI] [PubMed] [Google Scholar]
- 45. Wolf, S. & Grunewald S.. 2015. Sequence, structure and ligand binding evolution of rhodopsin‐like G protein‐coupled receptors: a crystal structure‐based phylogenetic analysis. PLoS One 10: e0123533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cvicek, V. , W.A. Goddard, 3rd & Abrol R.. 2016. Structure‐based sequence alignment of the transmembrane domains of all human GPCRs: phylogenetic, structural and functional implications. PLoS Comput. Biol. 12: e1004805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Promel, S. , et al 2012. The GPS motif is a molecular switch for bimodal activities of adhesion class G protein‐coupled receptors. Cell Rep. 2: 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Muller, A. , et al 2015. Oriented cell division in the C. elegans embryo is coordinated by G‐protein signaling dependent on the adhesion GPCR LAT‐1. PLoS Genet. 11: e1005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scholz, N. , et al 2017. Mechano‐dependent signaling by Latrophilin/CIRL quenches cAMP in proprioceptive neurons. elife 6: e28360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liebscher, I. , et al 2014. A tethered agonist within the ectodomain activates the adhesion G protein‐coupled receptors GPR126 and GPR133. Cell Rep. 9: 2018–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Petersen, S.C. , et al 2015. The adhesion GPCR GPR126 has distinct, domain‐dependent functions in Schwann cell development mediated by interaction with laminin‐211. Neuron 85: 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ackerman, S.D. , et al 2018. GPR56/ADGRG1 regulates development and maintenance of peripheral myelin. J. Exp. Med. 215: 941–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boucard, A.A. , Maxeiner S. & Sudhof T.C.. 2014. Latrophilins function as heterophilic cell‐adhesion molecules by binding to teneurins: regulation by alternative splicing. J. Biol. Chem. 289: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Boucard, A.A. , Ko J. & Sudhof T.C.. 2012. High affinity neurexin binding to cell adhesion G‐protein‐coupled receptor CIRL1/latrophilin‐1 produces an intercellular adhesion complex. J. Biol. Chem. 287: 9399–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jackson, V.A. , et al 2015. Structural basis of latrophilin–FLRT interaction. Structure 23: 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schoneberg, T. & Promel S.. 2019. Latrophilins and teneurins in invertebrates: no love for each other? Front. Neurosci. 13: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Langenhan, T. , et al 2009. Latrophilin signaling links anterior–posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Dev. Cell 17: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bjarnadottir, T.K. , et al 2007. Identification of novel splice variants of Adhesion G protein‐coupled receptors. Gene 387: 38–48. [DOI] [PubMed] [Google Scholar]
- 59. Salzman, G.S. , et al 2016. Structural basis for regulation of GPR56/ADGRG1 by its alternatively spliced extracellular domains. Neuron 91: 1292–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wilde, C. , et al 2016. The constitutive activity of the adhesion GPCR GPR114/ADGRG5 is mediated by its tethered agonist. FASEB J. 30: 666–673. [DOI] [PubMed] [Google Scholar]
- 61. Aust, G. , et al 2003. Detection of alternatively spliced EMR2 mRNAs in colorectal tumor cell lines but rare expression of the molecule in colorectal adenocarcinomas. Virchows Arch. 443: 32–37. [DOI] [PubMed] [Google Scholar]
- 62. Stacey, M. , et al 2001. Human epidermal growth factor (EGF) module‐containing mucin‐like hormone receptor 3 is a new member of the EGF‐TM7 family that recognizes a ligand on human macrophages and activated neutrophils. J. Biol. Chem. 276: 18863–18870. [DOI] [PubMed] [Google Scholar]
- 63. Matsushita, H. , Lelianova V.G. & Ushkaryov Y.A.. 1999. The latrophilin family: multiply spliced G protein‐coupled receptors with differential tissue distribution. FEBS Lett. 443: 348–352. [DOI] [PubMed] [Google Scholar]
- 64. Sugita, S. , et al 1998. Alpha‐Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G‐protein‐linked receptors. G‐protein coupling not required for triggering exocytosis. J. Biol. Chem. 273: 32715–32724. [DOI] [PubMed] [Google Scholar]
- 65. Kwakkenbos, M.J. , et al 2006. An unusual mode of concerted evolution of the EGF‐TM7 receptor chimera EMR2. FASEB J. 20: 2582–2584. [DOI] [PubMed] [Google Scholar]
- 66. Rothe, J. , et al 2019. Involvement of the adhesion GPCRs latrophilins in the regulation of insulin release. Cell Rep. 26: 1573–1584.e5. [DOI] [PubMed] [Google Scholar]
- 67. Saitou, N. & Nei M.. 1987. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- 68. Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 69. Zuckerkandl, E. & Pauling L.. 1965. Evolutionary Divergence and Convergence in Proteins. New York, NY: Academic Press. [Google Scholar]
- 70. Jones, D.T. , Taylor W.R. & Thornton J.M.. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8: 275–282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Current nomenclature of aGPCRs based on phylogenetic analysis by the maximum likelihood method. The evolutionary relationships of (A) only human aGPCRs and (B) human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors served as the outgroup. The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix–based model.70 The trees with the highest log likelihood (A: –11604.80; B: –21663.01) are shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The analyses involved 37 (A) and 150 (B) amino acid sequences. All positions containing gaps and missing data were eliminated. There was a total of 195 (A) and 158 (B) positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22
Suppl. File. Scholz et al. fas contains all sequences analyzed in this study (in FASTA format).
Figure S2. Higher resolution of the phylogenetic analysis of selected invertebrate and vertebrate aGPCRs from Figure 5 of the main text. The evolutionary relationships of selected invertebrate aGPCRs and human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors (AChR) served as the outgroup. The evolutionary history was inferred using the neighbor‐joining method.67 The optimal tree with the sum of branch length = 138.25837393 is shown. The evolutionary distances were computed using the Poisson correction method69 and are in the units of the number of amino acid substitutions per site. The analyses involved 525 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 183 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22 Current subfamilies of aGPCRs are condensed and shown in bold red characters. aGPCR clusters found in invertebrates are shown in blue and the number of receptors included in the cluster is given in parenthesis. Species included in the analysis: vertebrate: Homo sapiens (hs), Mus musculus (mm), Gallus gallus (gg), Danio rerio (dr); Cephalochordata: Branchiostoma belcheri (bb); Tunicata: Ciona intestinalis (ci); Hemichordata: Saccoglossus_kowalevskii (sk); Brachiopoda: Lingula anatina (la); Echinodermata: Acanthaster planci (ap); Mollusca Octopus bimaculoides (ob); Neoptera: Drosophila melanogaster (dm); Nematoda: Caenorhabditis elegans (ce); Parazoa: Amphimedon queenslandica (aq).


