Abstract
Background
Previous quantitative synthetic MRI of the brain has been solely performed in 2D.
Purpose
To evaluate the feasibility of the recently developed sequence 3D‐QALAS for brain cortical thickness and volumetric analysis.
Study Type
Reproducibility/repeatability study.
Subjects
Twenty‐one healthy volunteers (35.6 ± 13.8 years).
Field Strength/Sequence
3D T1‐weighted fast spoiled gradient recalled echo (FSPGR) sequence was performed once, and 3D‐QALAS sequence was performed twice with a 3T scanner.
Assessment
FreeSurfer and FIRST were used to measure cortical thickness and volume of subcortical structures, respectively. Agreement with FSPGR and scan–rescan repeatability were evaluated for 3D‐QALAS.
Statistical Tests
Percent relative difference and intraclass correlation coefficient (ICC) were used to assess reproducibility and scan–rescan repeatability of the 3D‐QALAS sequence‐derived measurements.
Results
Percent relative difference compared with FSPGR in cortical thickness of the whole cortex was 3.1%, and 89% of the regional areas showed less than 10% relative difference in cortical thickness. The mean ICC across all regions was 0.65, and 74% of the structures showed substantial to almost perfect agreement. For volumes of subcortical structures, the median percent relative differences were lower than 10% across all subcortical structures, except for the accumbens area, and all structures showed ICCs of substantial to almost perfect agreement. For the scan–rescan test, percent relative difference in cortical thickness of the whole cortex was 2.3%, and 97% of the regional areas showed less than 10% relative difference in cortical thickness. The mean ICC across all regions was 0.73, and 80% showed substantial to almost perfect agreement. For volumes of subcortical structures, relative differences were less than 10% across all subcortical structures except for the accumbens area, and all structures showed ICCs of substantial to almost perfect agreement.
Data Conclusion
3D‐QALAS could be reliably used for measuring cortical thickness and subcortical volumes in most brain regions.
Level of Evidence: 3
Technical Efficacy: Stage 1
J. Magn. Reson. Imaging 2019;50:1834–1842.
TISSUE RELAXOMETRY can provide quantitative values for the evaluation of diseases,1 development,2 and aging,3 as opposed to arbitrary signal intensities of conventional magnetic resonance imaging (MRI) such as T1‐weighted, T2‐weighted, and FLAIR images. However, its use in human brain imaging has mostly been limited to research applications because of additional lengthy scan times. Recently, quantitative synthetic MRI, enabling simultaneous quantification of T1 and T2 relaxation times and proton density (PD) with high reliability,4 has been proposed for whole brain coverage.5, 6 The technique also allows for the creation of any contrast‐weighted image that is used routinely in clinical settings,7 rendering its clinical application highly feasible. Quantitative synthetic MRI has been applied to a variety of diseases, such as multiple sclerosis,8, 9 meningitis,10 and brain infarctions,11 with promising results. The original sequence used for quantitative synthetic MRI was based on a multislice 2D acquisition, providing a relatively low resolution in the slice direction in comparison to conventional 3D T1‐weighted acquisitions. Recently, however, 3D‐QALAS (3D‐quantification using an interleaved Look‐Locker acquisition sequence with T2 preparation pulse) has been developed for simultaneous quantification of T1 and T2 in cardiac imaging, showing high accuracy and precision in the heart and phantoms with various tissue properties.12, 13
As opposed to relaxometry, volumetric analysis of the brain has already been widely performed in clinical settings, such as for the evaluation of patients with neurodegenerative14 and demyelinating disorders.15 Additionally, regional volumetric analysis has been extensively performed in research settings. Changes in cortical thickness and subcortical volumes are related to aging16, 17, 18 and in a wide variety of neurological disorders.14, 16, 19 Taken together, differences in regional cortical thickness and subcortical volume may indicate the state of neurological health, and their accurate measurements may lead to a better understanding of patients' conditions.
Here, we propose application of the 3D‐QALAS sequence for simultaneous acquisition of relaxometry parameters as well as for obtaining volumetric information in high‐resolution 3D. Therefore, the purpose of this study was to show the validity of volumetric information acquired with 3D‐QALAS by 1) evaluating the reproducibility of 3D‐QALAS sequence‐derived volumetric brain measurements using conventional T1‐weighted imaging‐derived measurements as reference standards, and 2) evaluating the repeatability of 3D‐QALAS sequence‐derived measurements by scan–rescan tests, on healthy subjects.
Materials and Methods
Subjects
This study was approved by our Institutional Review Board and written informed consent was acquired from all participants. Twenty‐one healthy volunteers were included in this study (14 women and 7 men; mean age, 35.6 ± 13.8 years). None of the participants had a history of a major medical condition including neurological or psychiatric disorders. Two radiologists (S.F. and A.H.) performed a blind examination on all volunteer exams and confirmed that all had normal structural MRI results.
Image Acquisition
All participants were scanned with a 3T scanner (Discovery 750w; GE Healthcare, Milwaukee, WI) with a 12‐channel head coil. A 3D T1‐weighted fast spoiled gradient recalled echo (FSPGR) sequence was performed once, and the 3D‐QALAS sequence was performed twice (to test scan–rescan) in the same session on all the participants. Between scan–rescan of the 3D‐QALAS sequence, the subjects were taken out of the MRI room and repositioned on the scanner. The scan parameters of FSPGR were as follows: sagittal acquisition; repetition time / echo time / inversion time (TR/TE/TI), 7.7/3.1/400 msec; field of view (FOV), 256 × 256 mm; matrix size, 256 × 256; section thickness, 1.0 mm; flip angle, 11°; receiver bandwidth, 244.1 Hz/pixel; averages, 1; acquisition time, 5 min 45 sec. 3D‐QALAS is based on a multiacquisition 3D gradient echo, with five acquisitions equally spaced in time, interleaved with a T2 preparation pulse and an inversion pulse. Briefly, T1 fitting was performed on four acquisitions after the inversion pulse, and T2 fitting was performed on extrapolation of the signal intensity straddling the T2 prep pulse. Instead of a cardiac trigger, an internal trigger started each of the five acquisitions every 900 msec, making the total cycle time 4.5 sec. Further details of the 3D‐QALAS sequence and its postprocessing are available in a previous study.12 The scan parameters of 3D‐QALAS were as follows: axial acquisition; TR/TE/TI, 8.6/3.5/100 msec; FOV, 256 × 256; matrix size, 256 × 256; section thickness, 1.0 mm; flip angle, 5°; receiver bandwidth, 97.7 Hz/pixel; averages, 1; acquisition time, 11 min 41 sec. We set the spatial resolution of the FSPGR imaging, standard reference in this study, as 1.0 mm isotropic, since the Alzheimer's Disease Neuroimaging Initiative (ADNI)20 study recommended the usage of 1.0 mm isotropic data at 3T. All 3D‐QALAS and FSPGR images were visually examined for artifacts such as ringing, blurring, and ghosting on site upon image acquisition. Images exhibiting these common artifacts were excluded from this study and subjects with such artifacts were rescanned.
Image Postprocessing
Images obtained from the 3D‐QALAS sequence were processed on a prototype version 0.45.5 of the SyMRI software (SyntheticMR, Linkoping, Sweden) to synthesize 3D synthetic T1‐weighted images. TR and TE were virtually set to the default values of 500 msec and 10 msec, respectively. These 3D synthetic T1‐weighted images and FSPGR images were used for subsequent analyses. Noncommercial automatic brain parcellation programs, described below, were used to measure cortical thickness and the volume of subcortical structures on the basis of 3D T1‐weighted images for each subject.
MEASUREMENT OF CORTICAL THICKNESS AND VOLUME
The pipeline of FreeSurfer (v. 5.3.0, http://surfer.nmr.mgh.harvard.edu) was used to obtain cortical thickness and volume for each sequence. FreeSurfer utilizes affine transformations and combines information about voxel intensity relative to a probability distribution for tissue classes with information about the spatial relationship of the voxel to the location of neighboring structures obtained from a manually labeled atlas.21, 22 The Desikan‐Killiany Atlas, consisting of 34 regions per hemisphere, was used to measure average cortical thickness and volume in each area.18 Further details of FreeSurfer are available in previous articles,21, 22 and in the documentation provided by the developers (http://surfer.nmr.mgh.harvard.edu/). The default analysis settings were used in running the “recon‐all” command. Bilateral regional values were averaged for further analysis. Previous research has shown that brain mask cleaning was the only type of manual intervention that improved FreeSurfer‐derived results.23 Therefore, manual brain mask assessment was performed in this study. For each subject, the brain mask was visually assessed on axial, sagittal, and coronal images. Brain masks excluding brain tissue (overcropping) were manually corrected using the Freeview application. A brain mask including extracerebral tissue, such as orbit (undercropping), was not corrected because it still allowed accurate surface demarcation.
VOLUMETRY OF SUBCORTICAL STRUCTURES
Due to high variability in the spatial location and extent of subcortical gray matter segmentations produced by FreeSurfer,21 the volumes of subcortical gray matter structures were obtained using the pipeline of FMRIB Integrated Registration and Segmentation Tool (FIRST, http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/FIRST) implemented in the FMRIB Software Library v. 5.0.9.24 The volumes of subcortical white matter structures were obtained based on FreeSurfer using the Desikan‐Killiany Atlas. Volumes of subcortical structures were measured for each sequence. All segmentation results performed on FreeSurfer and FIRST were visually screened for gross errors.
Statistical Analysis
All statistical analyses were performed with R program v. 3.3.0 (R Core Team [2016]. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.r-project.org/). Agreement to measurements obtained from FSPGR and scan–rescan repeatability were evaluated for 3D‐QALAS. Percent relative difference and intraclass correlation coefficient (ICC) were used to assess reproducibility and repeatability of the 3D‐QALAS sequence‐derived measurements. Within‐subject coefficient of variation (wCV) was also used in assessing repeatability. ICC is a measure of within‐subject relative to between‐subject variability. The ICC estimates of agreement were categorized as the following: slight (0.01–0.20), fair (0.21–0.40), moderate (0.41–0.60), substantial (0.61–0.80), and almost perfect agreement (0.81–1.0).25 Percent relative difference was calculated by dividing the absolute difference by the mean of two measurements, defined as follows:
where X and Y are the measured values. The wCV was defined as follows:
where σw is the within‐subject standard deviation and μ is the overall mean of the measured values.
Results
Representative FreeSurfer and FIRST outputs from 3D‐QALAS sequence‐derived T1‐weighted images are shown in Fig. 1.
Figure 1.
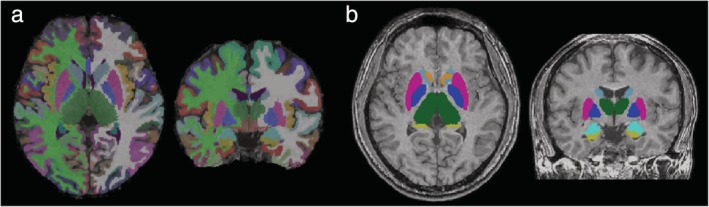
Representative labels created from automated parcellation of brain regions using (a) FreeSurfer and (b) FIRST. Results of segmentation are overlaid on synthetic T1‐weighted images.
Measurement of Cortical Thickness
REPRODUCIBILITY OF 3D‐QALAS SEQUENCE‐DERIVED CORTICAL THICKNESS AND VOLUME: COMPARISON WITH CONVENTIONAL FSPGR
Figure 2 shows a histogram of 3D‐QALAS and FSPGR sequence‐derived cortical thickness estimated using FreeSurfer across all regions in the Desikan‐Killiany Atlas in all subjects. The range of the cortical thicknesses in this study was consistent with previous studies, reporting cortical thickness ranging from 1–4.5 mm (both of postmortem and FreeSurfer‐based findings).18, 23, 26 In Fig. 3a, regional percent relative differences between 3D‐QALAS and FSPGR‐derived cortical thicknesses are overlaid on an inflated brain. Figure 3b shows the boxplots for percent relative differences. Percent relative difference of the whole cortex was 3.1%, and 89% of the regional areas showed less than 10% relative difference in cortical thickness. Cortical thickness of the temporal pole, inferior temporal, pericalcarine, fusiform, and entorhinal cortex showed relatively low agreement. Table 1 shows the ICCs for 3D‐QALAS and FSPGR‐derived cortical thickness. The mean ICC across all regions was 0.65, and 74% of the structures showed substantial to almost perfect agreement. Cortical thickness of the temporal pole, entorhinal, lateral orbitofrontal, pars orbitalis, inferior temporal, pericalcarine, and the fusiform cortex showed particularly low ICC. Supplementary Table 1 shows the ICCs for 3D‐QALAS and FSPGR‐derived cortical volume. The mean ICC across all regions was 86%, and 97% of the structures showed substantial to almost perfect agreement. Cortical thickness of the temporal pole showed particularly low ICC.
Figure 2.
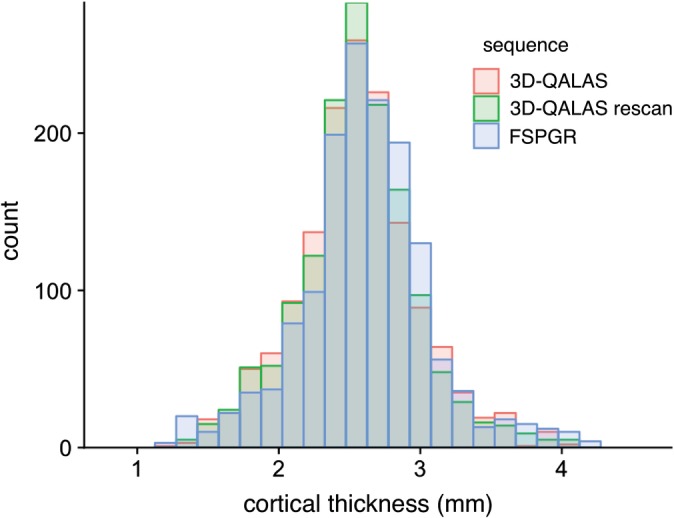
Histograms of cortical thicknesses derived from 3D‐QALAS scan–rescan and FSPGR in all subjects measured using FreeSurfer.
Figure 3.
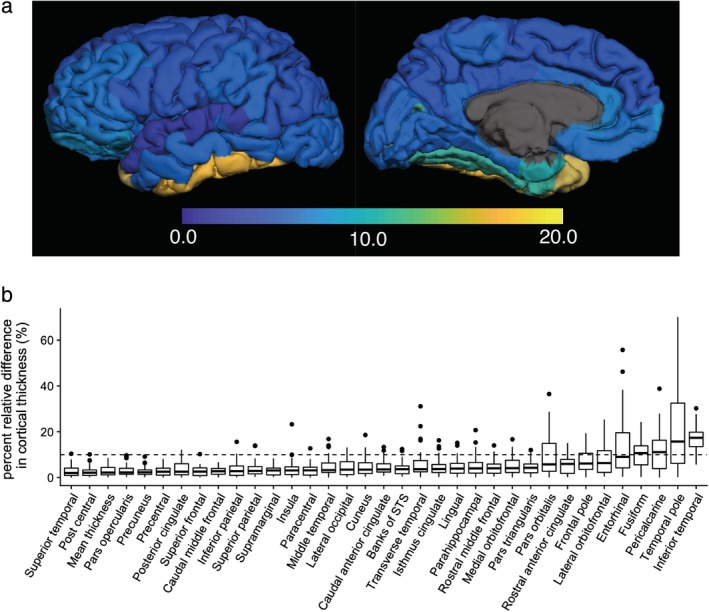
Percent relative difference in cortical thickness for 3D‐QALAS and FSPGR measured using FreeSurfer. Regional percent relative difference is overlaid on an inflated brain surface (a). Median values and interquartile ranges are shown in boxplots (b). Whiskers are set at minimum and maximum, and the horizontal line marks the median. Boxes indicate the interquartile range (25–75%). Dots at the end of the boxplot represent outliers.
Table 1.
Intraclass Correlation Coefficients and Within‐Subject Coefficients of Variation Between 3D‐QALAS and FSPGR, and Scan–Rescan of 3D‐QALAS for Cortical Thicknesses Measured Using FreeSurfer
| FSPGR | Rescan | ||
|---|---|---|---|
| Measurement | ICC | wCV (%) | ICC |
| Thickness | |||
| Mean thickness | 0.79 | 2.1 | 0.81 |
| Caudal anterior cingulate | 0.75 | 3.9 | 0.73 |
| Caudal middle frontal | 0.84 | 2.3 | 0.86 |
| Cuneus | 0.63 | 3.5 | 0.75 |
| Entorhinal | 0.28 | 9.4 | 0.50 |
| Frontal ploe | 0.65 | 8.0 | 0.59 |
| Fusiform | 0.44 | 3.2 | 0.68 |
| Inferior parietal | 0.73 | 3.0 | 0.79 |
| Inferior temporal | 0.47 | 5.0 | 0.36 |
| Insula | 0.66 | 3.9 | 0.65 |
| Isthmus cingulate | 0.79 | 3.9 | 0.83 |
| Lateral occipital | 0.61 | 2.8 | 0.78 |
| Lateral orbitofrontal | 0.38 | 6.1 | 0.52 |
| Lingual | 0.62 | 3.0 | 0.72 |
| Medial orbitofrontal | 0.51 | 4.9 | 0.58 |
| Middle temporal | 0.75 | 4.1 | 0.66 |
| Parahippocampal | 0.86 | 3.4 | 0.90 |
| Paracentral | 0.77 | 3.8 | 0.75 |
| Pars opercularis | 0.79 | 2.2 | 0.87 |
| Pars orbitalis | 0.38 | 9.7 | 0.43 |
| Pars triangularis | 0.76 | 3.5 | 0.80 |
| Pericalcarine | 0.49 | 5.5 | 0.68 |
| Postcentral | 0.87 | 2.5 | 0.86 |
| Posterior cingulate | 0.71 | 3.1 | 0.81 |
| Precentral | 0.70 | 2.3 | 0.77 |
| Precuneus | 0.78 | 1.7 | 0.91 |
| Rosterior anterior cingulate | 0.63 | 4.8 | 0.69 |
| Rostral middle frontal | 0.55 | 3.1 | 0.75 |
| Superior frontal | 0.80 | 1.9 | 0.87 |
| Superior parietal | 0.78 | 1.9 | 0.90 |
| Superior temporal | 0.80 | 2.9 | 0.78 |
| Supramarginal | 0.84 | 2.8 | 0.81 |
| Temporal pole | 0.00 | 12.3 | 0.48 |
| Transverse temporal | 0.62 | 5.0 | 0.70 |
SCAN–RESCAN REPEATABILITY OF 3D‐QALAS SEQUENCE‐DERIVED CORTICAL THICKNESS AND VOLUME
In Fig. 4a, regional percent relative differences between scan and rescan of 3D‐QALAS‐derived cortical thicknesses are overlaid on an inflated brain. Figure 4b shows the boxplots for percent relative differences. Relative percent difference in thickness of the whole cortex was 2.3%, and 97% of the regional cortical thickness showed less than 10% relative difference. Cortical thickness of the temporal pole showed relatively low agreement. Table 1 shows ICCs and wCV for scan–rescan cortical thickness. The mean ICC across all regions was 0.73, and 80% of the structures showed substantial to almost perfect agreement. Temporal pole, entorhinal, pars orbitalis, inferior temporal, and the orbitofrontal cortical thickness showed particularly low ICC and/or wCV. Supplementary Table 1 shows ICCs and wCV for scan–rescan cortical volume. The mean ICC across all regions was 87%, and 94% of the structures showed substantial to almost perfect agreement. Temporal pole and entorhinal cortical volumes showed particularly low ICC and wCV. The 3D‐QALAS sequence‐derived cortical volume of each region is listed in Supplementary Table 2.
Figure 4.
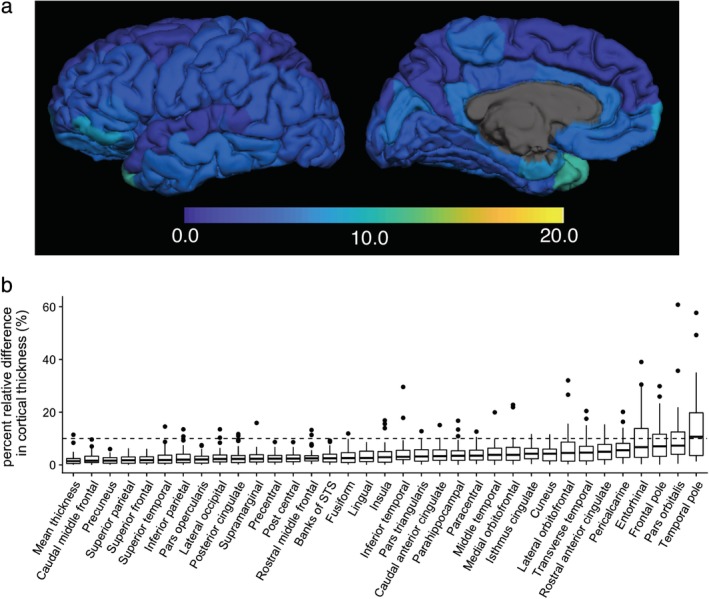
Percent relative difference in cortical thickness for 3D‐QALAS scan–rescan, measured using FreeSurfer. Regional percent relative difference is overlaid on an inflated brain surface (a). Median values and interquartile ranges are shown in boxplots (b). Whiskers are set at minimum and maximum, and the horizontal line marks the median. Boxes indicate the interquartile range (25–75%). Dots at the end of the boxplot represent outliers.
Volumetry of Subcortical Structures
REPRODUCIBILITY OF 3D‐QALAS SEQUENCE‐DERIVED SUBCORTICAL STRUCTURAL VOLUMES: COMPARISON WITH CONVENTIONAL FSPGR
Figure 5a shows the percent relative differences between 3D‐QALAS and FSPGR‐derived measurements in subcortical gray matter structural volumes. The median percent relative differences were lower than 10% across all subcortical structures, except for the accumbens area. ICC for 3D‐QALAS and FSPGR‐derived measurements are shown in Table 2. All structures showed substantial or almost perfect agreement. Supplementary Table 3 shows the ICCs for 3D‐QALAS and FSPGR‐derived white matter volume. All structures except subcortical white matter of the temporal pole showed substantial or almost perfect agreement.
Figure 5.
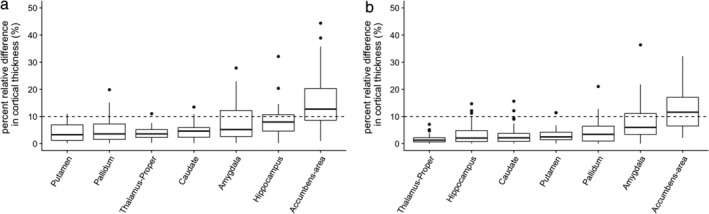
Percent relative difference of subcortical volumes measured using FIRST. (a) and (b) show comparisons between 3D‐QALAS and FSPGR, and scan–rescan of 3D‐QALAS, respectively. Whiskers are set at minimum and maximum, and the horizontal line marks the median. Boxes indicate the interquartile range (25–75%). Dots at the end of the boxplot represent outliers.
Table 2.
Intraclass Correlation Coefficients and Within‐Subject Coefficients of Variation Between 3D‐QALAS and FSPGR, and Scan–Rescan of 3D‐QALAS for Subcortical Volumes Measured Using FIRST
| FSPGR | Rescan | ||
|---|---|---|---|
| Measurement | ICC | wCV (%) | ICC |
| Volume | |||
| Putamen | 0.86 | 2.9 | 0.91 |
| Caudate nucleus | 0.89 | 3.3 | 0.89 |
| Nucleus accumbens | 0.82 | 13.3 | 0.80 |
| Globus pallidus | 0.86 | 4.5 | 0.87 |
| Hippocampus | 0.84 | 3.5 | 0.91 |
| Amygdala | 0.66 | 7.6 | 0.72 |
| Thalamus | 0.91 | 1.5 | 0.96 |
SCAN–RESCAN REPEATABILITY OF 3D‐QALAS SEQUENCE‐DERIVED SUBCORTICAL STRUCTURAL VOLUMES
Figure 5b shows the percent relative difference between scan and rescan of 3D‐QALAS‐derived measurements in subcortical gray matter structural volumes. Percent relative differences were less than 10% across all subcortical structures, except for the accumbens area. ICC and wCV for 3D‐QALAS and FSPGR‐derived measurements are shown in Table 2. All structures showed substantial to almost perfect agreement. The wCVs were lower than 10% across all subcortical structures, except for the accumbens area. Supplementary Table 3 shows the ICCs and wCV for scan–rescan white matter volume. All structures except subcortical white matter of entorhinal and frontal pole showed substantial or almost perfect agreement. The 3D‐QALAS sequence‐derived volume of each subcortical structure is listed in Supplementary Table 4.
Discussion
In this study, 3D synthetic T1‐weighted images showed good agreement with the FSPGR 1.0 mm isotropic images in measuring regional cortical thickness and subcortical volumes in most of the brain regions. High repeatability of the 3D synthetic MRI‐derived brain measurements was demonstrated in the scan–rescan test.
The 3D isotropic acquisition of 3D‐QALAS allows high resolution multiplanar reconstruction, without additional scans from different directions. This capability not only provides the advantage in visual assessment and delineation of lesions, but also enables to accurately segment regional structures. With the quantification of T1, T2, and PD in these regional structures, 3D‐QALAS may enable detecting and describing changes within regional structures, which could be obscured when averaging values over gross anatomic regions. Hence, 3D‐QALAS has a potential to provide thorough and comprehensive characterization of brain lesions as well as the entire brain.
Cortical thickness derived from 3D synthetic T1‐weighted and FSPGR images showed a percent relative difference of 3.1% in the whole cortex, and 89% of the regional areas showed less than 10% relative difference in cortical thickness. Although high agreements were shown in the majority of the brain regions, low agreements were found in cortical thickness of temporal pole, inferior temporal, pericalcarine, and fusiform, as shown by their median percent relative differences of more than 10%. This observation is consistent with previous studies using FreeSurfer that reported a negative relationship between cortical volume/surface area and relative error of measured cortical thickness,23, 27 which can explain low agreement in the small structures in the current study. It has also been reported that the boundaries of the temporal pole and occipital lobe were not defined precisely using FreeSurfer,28 which may explain the low agreement in thickness of the temporal pole and pericalcarine.
Another possibility of the low agreements in lower parts of the brain structure is the effects of B0 inhomogeneities due to susceptibility differences between bone and air, which could cause fitting errors upon quantifying T1, T2, and PD, and affect the subsequent synthetic T1‐weighted images.29
The cortical thickness derived from scan–rescan of 3D synthetic T1‐weighted images showed less than a 10% relative percent difference across all regions except the thickness of the temporal pole. A previous study that included a large collection of cortical thickness data based on scan–rescan of conventional 3D magnetization prepared rapid acquisition with gradient echo (MPRAGE) images showed overall percent relative differences of 2.5–2.8% using FreeSurfer.27 The median percent relative differences of 3D synthetic T1‐weighted image‐derived cortical thickness in our study was 2.9%, which is comparable to the repeatability achieved when using conventional 3D T1‐weighted images.
Subcortical volumes derived from 3D synthetic T1‐weighted and FSPGR images showed a percent relative difference lower than 10% across all structures except the nucleus accumbens, and the scan–rescan test of 3D synthetic T1‐weighted images showed a percent relative difference lower than 10% across all subcortical structures. Segmentation of the accumbens and amygdala showed relatively low agreement both in scan–rescan and comparison with FSPGR in this study, which is consistent with previous studies reporting that segmentation of these area was generally unreliable compared with other subcortical regions.30, 31 One factor that may have contributed to this lower reliability in measurements is that they are the smallest subcortical structures. Morey et al31 reported that the percent relative difference of the accumbens and amygdala volumes, calculated from scan–rescan of 3D T1‐weighted images with 1.0 mm isotropic voxel based on FIRST analysis, were both higher than 10%. Taken together, 3D synthetic T1‐weighted imaging‐based subcortical volume measurement can be assumed to be as reliable as conventional 3D T1‐weighted imaging‐based measurement.
Although the in‐plane resolution of 1.0 mm used in this study is low compared with that of commonly used 2D sequences, high spatial resolution in the slice‐select direction enables reliable detection and reproducible measurements among various slice positions and alignments. In fact, previous studies have shown that 3D imaging could be superior in detecting multiple sclerosis lesions and brain metastases than 2D imaging, even with lower in‐plane resolutions.32, 33
We used T1‐weighted images only with fixed TR and TE for segmentation in this study. Using multichannel inputs (eg, T1, T2, and PD maps), obtained from a single 3D‐QALAS sequence scan, could improve the accuracy of current segmentation algorithms that rely heavily on T1‐weighted image contrasts, without elongating scanning times. Furthermore, even with only T1‐weighting, combining T1‐weighted images with different parameters might improve the overall segmentation, since T1‐weighted images with fixed TR and TE may not be optimal for all brain structures. An additional advantage of synthetic MRI based on relaxation parameters is that the effects of B1 inhomogeneities and coil sensitivity profiles on the T1‐weighted images are removed.34 This is expected to provide a more stable result in volumetric analysis.
Absolute quantification of tissue properties using relaxometry has been previously reported to be useful for the characterization of disease, assessment of disease activity, and monitoring of treatment.35, 36 With accurate volumetric segmentation, 3D synthetic MRI could provide quantitative values of each brain substructure based on a single scan, which could allow for a more quantitative understanding of the brain.
The current study has several limitations. First, we only used a single 3T scanner, hence our results cannot be generalized to scanners with different field strengths. Previous studies have revealed certain biases between 1.5T and 3T for cortical thickness analysis and brain volumetry performed on FreeSurfer using 3D T1‐weighted images.37, 38 Further research is needed to determine whether cortical thickness analysis and subcortical brain volumetry based on 3D‐QALAS differ between different field strengths. Second, we used FSPGR as a standard reference, not with ground truth postmortem values. Although the ground truth for cortical thickness and subcortical volumes were not available, our results showed agreement with previous studies that compared FreeSurfer‐derived measurements with postmortem values. Third, the scanning time in this study was relatively long for a routine clinical scan, making it difficult to use in clinical settings. Combining techniques such as compressed sensing39 and multiband imaging40 may further reduce scan times to a clinically applicable level. Fourth, only healthy volunteers were enrolled in this study. Although our goal in this study was not to compare patients and volunteers, future studies focusing on patients using 3D‐QALAS are desired.
In conclusion, the current study may support the use of 3D quantitative synthetic MRI for reliably measuring cortical thickness and subcortical volumes in human brain, with the exceptions of the nucleus accumbens, and thickness of temporal pole, entorhinal, inferior temporal, pericalcarine, and the fusiform.
Conflict of Interest
Marcel Warntjes is currently employed part‐time at SyntheticMR and has stock in SyntheticMR.
Supporting information
Supplementary Figure S1 Representative series of 3D‐QALAS sequence‐derived T1‐weighted images.
Supplementary Table S1 Intraclass correlation coefficients and within subject coefficients of variation between 3D‐QALAS and FSPGR, and scan‐rescan of 3D‐QALAS for cortical volumes measured using FreeSurfer.
Supplementary Table S2 Regional cortical volumes measured based on 3D‐QALAS. Measurements were obtained from 21 healthy volunteers using FreeSurfer.
Supplementary Table S3 Intraclass correlation coefficients and within subject coefficients of variation between 3D‐QALAS and FSPGR, and scan‐rescan of 3D‐QALAS for white matter volumes measured using FreeSurfer.
Supplementary Table S4 Regional subcortical structure volumes measured based on 3D‐QALAS. Gray matter volumes and white matter volumes were obtained from 21 healthy volunteers using FIRST and FreeSurfer, respectively.
Acknowledgment
Contract grant sponsor: Program for Brain Mapping by Integrated Neurotechnologies for Disease Studies (Brain/MINDS) from Japan Agency for Medical Research and Development, AMED; Contract grant sponsor: ImPACT Program of Council for Science, Technology and Innovation (Cabinet Office, Government of Japan); Contract grant sponsor: JSPS KAKENHI; Contract grant numbers: 16K10327, 19K17150, 19K17177, JP16H06280; Contract grant sponsor: Grant‐in‐Aid for Scientific Research on Innovative Areas, Resource and technical support platforms for promoting research “Advanced Bioimaging Support.”
References
- 1. Bottomley PA, Hardy CJ, Argersinger RE, Allen‐Moore G. A review of 1H nuclear magnetic resonance relaxation in pathology: Are T1 and T2 diagnostic? Med Phys 1987;14:1–37. [DOI] [PubMed] [Google Scholar]
- 2. Lee SM, Choi YH, You SK, et al. Age‐related changes in tissue value properties in children: simultaneous quantification of relaxation times and proton density using synthetic magnetic resonance imaging. Invest Radiol 2018;53:236–245. [DOI] [PubMed] [Google Scholar]
- 3. Kumar R, Delshad S, Woo MA, Macey PM, Harper RM. Age‐related regional brain T2‐relaxation changes in healthy adults. J Magn Reson Imaging 2012;35:300–308. [DOI] [PubMed] [Google Scholar]
- 4. Hagiwara A, Hori M, Cohen‐Adad J, et al. Linearity, bias, intra‐scanner repeatability, and inter‐scanner reproducibility of quantitative multi‐dynamic multi‐echo sequence for rapid simultaneous relaxometry at 3T: A validation study with a standardized phantom and healthy controls. Invest Radiol 2019;54:39–47. [DOI] [PubMed] [Google Scholar]
- 5. Warntjes JB, Leinhard OD, West J, Lundberg P. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magn Reson Med 2008;60:320–329. [DOI] [PubMed] [Google Scholar]
- 6. Hagiwara A, Warntjes M, Hori M, et al. SyMRI of the brain: Rapid quantification of relaxation rates and proton density, with synthetic MRI, automatic brain segmentation, and myelin measurement. Invest Radiol 2017;52:647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanenbaum LN, Tsiouris AJ, Johnson AN, et al. Synthetic MRI for clinical neuroimaging: Results of the Magnetic Resonance Image Compilation (MAGiC) prospective, multicenter, multireader trial. AJNR Am J Neuroradiol 2017;38:1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hagiwara A, Hori M, Yokoyama K, et al. Synthetic MRI in the detection of multiple sclerosis plaques. AJNR Am J Neuroradiol 2017;38:257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Granberg T, Uppman M, Hashim F, et al. Clinical feasibility of synthetic MRI in multiple sclerosis: A diagnostic and volumetric validation study. AJNR Am J Neuroradiol 2016;37:1023–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Andica C, Hagiwara A, Nakazawa M, et al. Synthetic MR imaging in the diagnosis of bacterial meningitis. Magn Reson Med Sci 2017;16:91–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wallaert L, Hagiwara A, Andica C, et al. The advantage of synthetic MRI for the visualization of anterior temporal pole lesions on double inversion recovery (DIR), phase‐sensitive inversion recovery (PSIR), and myelin images in a patient with CADASIL. Magn Reson Med Sci 2018;17:275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kvernby S, Warntjes MJ, Haraldsson H, Carlhall CJ, Engvall J, Ebbers T. Simultaneous three‐dimensional myocardial T1 and T2 mapping in one breath hold with 3D‐QALAS. J Cardiovasc Magn Reson 2014;16:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kvernby S, Warntjes M, Engvall J, Carlhall CJ, Ebbers T. Clinical feasibility of 3D‐QALAS — Single breath‐hold 3D myocardial T1‐ and T2‐mapping. Magn Reson Imaging 2017;38:13–20. [DOI] [PubMed] [Google Scholar]
- 14. Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC. Mapping the evolution of regional atrophy in Alzheimer's disease: Unbiased analysis of fluid‐registered serial MRI. Proc Natl Acad Sci U S A 2002;99:4703–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinnecker T, Granziera C, Wuerfel J, Schlaeger R. Future brain and spinal cord volumetric imaging in the clinic for monitoring treatment response in MS. Curr Treat Options Neurol 2018;20:17. [DOI] [PubMed] [Google Scholar]
- 16. Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL. Normative estimates of cross‐sectional and longitudinal brain volume decline in aging and AD. Neurology 2005;64:1032–1039. [DOI] [PubMed] [Google Scholar]
- 17. Resnick SM, Pham DL, Kraut MA, Zonderman AB, Davatzikos C. Longitudinal magnetic resonance imaging studies of older adults: A shrinking brain. J Neurosci 2003;23:3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–980. [DOI] [PubMed] [Google Scholar]
- 19. Konarski JZ, McIntyre RS, Kennedy SH, Rafi‐Tari S, Soczynska JK, Ketter TA. Volumetric neuroimaging investigations in mood disorders: Bipolar disorder versus major depressive disorder. Bipolar Disord 2008;10:1–37. [DOI] [PubMed] [Google Scholar]
- 20. Jack CR Jr, Bernstein MA, Fox NC, et al. The Alzheimer's Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008;27:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dale AM, Fischl B, Sereno MI. Cortical surface‐based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999;9:179–194. [DOI] [PubMed] [Google Scholar]
- 22. Fischl B. FreeSurfer. Neuroimage 2012;62:774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iscan Z, Jin TB, Kendrick A, et al. Test‐retest reliability of Freesurfer measurements within and between sites: Effects of visual approval process. Hum Brain Mapp 2015;36:3472–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 2011;56:907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159–174. [PubMed] [Google Scholar]
- 26. Salat DH, Lee SY, van der Kouwe AJ, Greve DN, Fischl B, Rosas HD. Age‐associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. Neuroimage 2009;48:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tustison NJ, Cook PA, Klein A, et al. Large‐scale evaluation of ANTs and FreeSurfer cortical thickness measurements. Neuroimage 2014;99:166–179. [DOI] [PubMed] [Google Scholar]
- 28. Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 2010;53:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang H, Ceritoglu C, Li X, et al. Correction of B0 susceptibility induced distortion in diffusion‐weighted images using large‐deformation diffeomorphic metric mapping. Magn Reson Imaging 2008;26:1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nugent AC, Luckenbaugh DA, Wood SE, Bogers W, Zarate CA Jr, Drevets WC. Automated subcortical segmentation using FIRST: Test‐retest reliability, interscanner reliability, and comparison to manual segmentation. Hum Brain Mapp 2013;34:2313–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morey RA, Selgrade ES, Wagner HR 2nd, Huettel SA, Wang L, McCarthy G. Scan‐rescan reliability of subcortical brain volumes derived from automated segmentation. Hum Brain Mapp 2010;31:1751–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tan IL, van Schijndel RA, Pouwels PJ, Ader HJ, Barkhof F. Serial isotropic three‐dimensional fast FLAIR imaging: Using image registration and subtraction to reveal active multiple sclerosis lesions. AJR Am J Roentgenol 2002;179:777–782. [DOI] [PubMed] [Google Scholar]
- 33. Takeda T, Takeda A, Nagaoka T, et al. Gadolinium‐enhanced three‐dimensional magnetization‐prepared rapid gradient‐echo (3D MP‐RAGE) imaging is superior to spin‐echo imaging in delineating brain metastases. Acta Radiol 2008;49:1167–1173. [DOI] [PubMed] [Google Scholar]
- 34. Warntjes JB, Dahlqvist O, Lundberg P. Novel method for rapid, simultaneous T1, T2*, and proton density quantification. Magn Reson Med 2007;57:528–537. [DOI] [PubMed] [Google Scholar]
- 35. West J, Aalto A, Tisell A, et al. Normal appearing and diffusely abnormal white matter in patients with multiple sclerosis assessed with quantitative MR. PLoS One 2014;9:e95161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hagiwara A, Hori M, Yokoyama K, et al. Utility of a multiparametric quantitative MRI model that assesses myelin and edema for evaluating plaques, periplaque white matter, and normal‐appearing white matter in patients with multiple sclerosis: A feasibility study. AJNR Am J Neuroradiol 2017;38:237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pfefferbaum A, Rohlfing T, Rosenbloom MJ, Sullivan EV. Combining atlas‐based parcellation of regional brain data acquired across scanners at 1.5 T and 3.0 T field strengths. Neuroimage 2012;60:940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Han X, Jovicich J, Salat D, et al. Reliability of MRI‐derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. Neuroimage 2006;32:180–194. [DOI] [PubMed] [Google Scholar]
- 39. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58:1182–1195. [DOI] [PubMed] [Google Scholar]
- 40. Feinberg DA, Moeller S, Smith SM, et al. Multiplexed echo planar imaging for sub‐second whole brain FMRI and fast diffusion imaging. PLoS One 2010;5:e15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Representative series of 3D‐QALAS sequence‐derived T1‐weighted images.
Supplementary Table S1 Intraclass correlation coefficients and within subject coefficients of variation between 3D‐QALAS and FSPGR, and scan‐rescan of 3D‐QALAS for cortical volumes measured using FreeSurfer.
Supplementary Table S2 Regional cortical volumes measured based on 3D‐QALAS. Measurements were obtained from 21 healthy volunteers using FreeSurfer.
Supplementary Table S3 Intraclass correlation coefficients and within subject coefficients of variation between 3D‐QALAS and FSPGR, and scan‐rescan of 3D‐QALAS for white matter volumes measured using FreeSurfer.
Supplementary Table S4 Regional subcortical structure volumes measured based on 3D‐QALAS. Gray matter volumes and white matter volumes were obtained from 21 healthy volunteers using FIRST and FreeSurfer, respectively.


