Abstract
MicroRNAs (miRNAs) are attracting a growing interest in the scientific community due to their central role in the etiology of major diseases. On the other hand, nanoparticle carriers offer unprecedented opportunities for cell specific controlled delivery of miRNAs for therapeutic purposes. This review critically discusses the use of nanoparticles for the delivery of miRNA-based therapeutics in the treatment of cancer and neurodegenerative disorders and for tissue regeneration. A fresh perspective is presented on the design and characterization of nanocarriers to accelerate translation from basic research to clinical application of miRNA-nanoparticles. Main challenges in the engineering of miRNA-loaded nanoparticles are discussed, and key application examples are highlighted to underline their therapeutic potential for effective and personalized medicine.
Keywords: Nanoparticles, Physicochemical characterization, Biomaterials, MicroRNA delivery and release, Personalized medicine
1. Introduction
MicroRNAs (miRNAs) are non-coding endogenous RNAs composed of short nucleotide sequences, 20–24 nucleotides, that act in the post-transcriptional regulation of gene expression [1]. As soon as miRNA activity was confirmed in humans [2,3], significant attention was generated on their clinical application, because of their impact on critical cellular activities, such as metabolism [4], differentiation/proliferation [5,6] and programmed cell death [6]. Several studies have described the influence of miRNAs on the onset and progression of diseases, including cancer [[7], [8], [9]], neurodegenerative disorders [10,11], cardiovascular diseases [[12], [13], [14]], and other pathologies [[15], [16], [17]]. Such studies have led to research that focused on identifying changes in miRNA expression between normal and diseased states, resulting in miRNAs being proposed as diagnostic markers or therapeutic targets [[18], [19], [20]]. However, despite the excitement generated by miRNAs, their efficacy is limited by poor targeting ability, short circulation time and off-target effects of naked miRNA-based agents. To overcome these barriers, miRNA-loaded nanoparticles (NPs) have been proposed [[21], [22], [23]], by virtue of NPs ability to shield the loaded agent from the external environment, thereby reducing inactivation or degradation, and enhancing circulation time and targeted accumulation [24]. Additionally, the use of miRNAs for therapeutic purpose carries distinctive advantages over traditional gene therapy that involves the delivery of larger molecules, such as mRNAs, or DNA. Firstly, miRNAs have a broader range of therapeutic targets, since a single miRNA can simultaneously regulate a multitude of mRNAs. Furthermore, miRNAs in the form of miRNA inhibitor and miRNA mimic can regulate both the expression and the repression of multiple genes, while the activity of siRNAs and mRNAs is limited to repression or upregulation of one specific gene, respectively. miRNAs also have a small size, which may facilitate their encapsulation into NPs with high efficiency, their delivery into cells and their ability to act at the cytoplasmic level [25,26].
Herein, we discuss the preparation, characterization and application of NPs for delivering miRNA-based agents, which include miRNA mimics (miR mimics) or miRNA inhibitors/anti-miRNAs (miR inhibitors/anti-miRs) and focus on the advantages of NP-mediated methods as compared to naked miRNA delivery. Key examples of miRNA-loaded NPs for specific disease settings are presented, including their applications in tissue engineering (TE), cancer treatment, cancer immunotherapy and neurodegenerative disorders.
2. Physiological relevance of microRNAs (miRNAs)
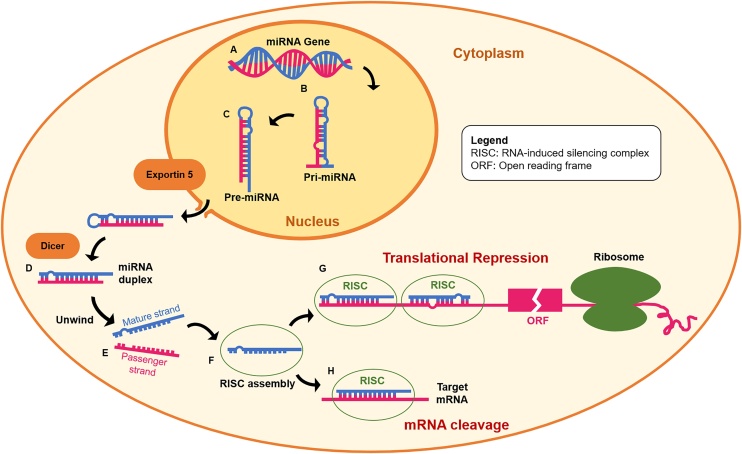
MiRNAs are proposed to act primarily by binding to the 3’ untranslated regions (UTRs) of messenger RNA (mRNA) without requiring perfect base pairing [[27], [28], [29]]. As such, one miRNA can simultaneously regulate several genes, while a single mRNA can be repressed by several miRNAs [25,26]. As shown in Fig. 1, miRNA formation begins in the nucleus with the DNA transcription of primary miRNA precursor (pri-miRNA) [30]. The transcribed pri-miRNAs are further processed to form ∼70 nucleotide hairpin structures (pre-miRNAs), that are exported to the cytoplasm to be condensed into 20–24 nt double-stranded miRNA duplexes [31]. These duplexes then enter the miRNA-induced silencing complex (miRISC) where they unwind to form a mature strand and a passenger strand. The miRISC retains the mature strand and releases the passenger strand which is degraded [32]. miRISC binds target mRNAs, thereby inducing either interference with protein synthesis [33,34], or degradation of the target mRNA [35,36].
Fig. 1.
Endogenous production of miRNA and their mechanism of regulation of gene expression: (A) genes encoding for miRNA are transcribed and (B) miRNA precursor (pri-miRNA) form double-stranded structures. Pri-miRNAs are further transformed into (C) pre-miRNAs that are exported by Exportin into the cytoplasm to be (D) condensed into miRNA duplexes. (E) miRNA duplexes unwind to form the passenger and guide strands. The mature strand stays in the miRNA-induced silencing complex (miRISC), forming a (F) temporary asymmetric RISC assembly. Finally, miRNAs bind target mRNAs, thereby inducing (G) translational repression or (H) degradation of target mRNAs. This figure is adapted from https://www.sigmaaldrich.com/life-science/functional-genomics-and-rnai/mirna/learning-center/mirna-introduction.html.
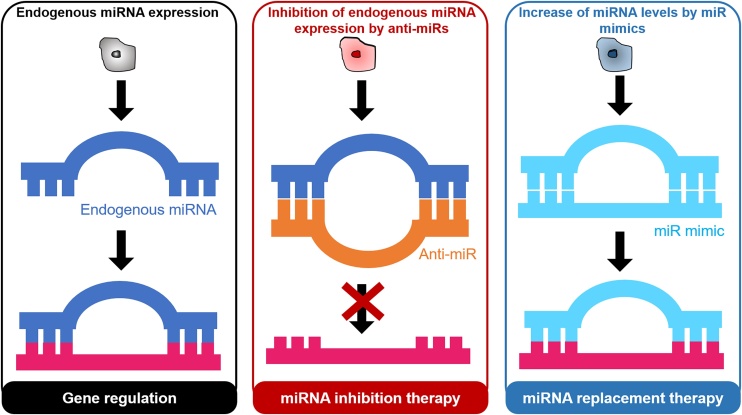
Depending on the target miRNA expression, miRNA therapy can take the form of: miRNA inhibition therapy [37,38], or miRNA replacement or reinforcement therapy (Fig. 2) [39,40]. In the former approach, an anti-miR or miRNA inhibitor is used, consisting of a single-stranded oligonucleotide with a complementary sequence to mature miRNA. In the latter approach, miR mimics that have an identical sequence as the endogenous mature miRNA are introduced into cells to provide an exogenous source of additional miRNAs.
Fig. 2.
(A) In a normal tissue state, expression of endogenous microRNAs (miRNAs) allows for gene regulation. In the treatment of certain diseases, (B) inhibition of specific endogenous miRNAs is achieved through miRNA inhibition therapy with anti-miRNAs (anti-miRs). (C) Alternatively, miRNA replacement therapy utilizes miR mimics to provide an exogenous source of additional miRNAs.
3. Design criteria of nanoparticles (NPs) as efficient delivery vectors for miRNAs
Despite the progressively increasing knowledge of miRNA ability to mediate biological processes both in vitro and in vivo, critical hurdles related to miRNA delivery should be overcome to promote their clinical application [41,42]. Naked miRNAs are poorly taken up by cells due to their negative charge [43], undesired off-target [44] or on-target [45] effects, short half-life in systemic circulation [46,47], and limited stability in blood due to their rapid degradation or inactivation by nucleases that are abundantly present in the blood stream [48,49]. Herein, the main advantages and disadvantages of the different strategies to overcome the challenges of efficient miRNA delivery are discussed.
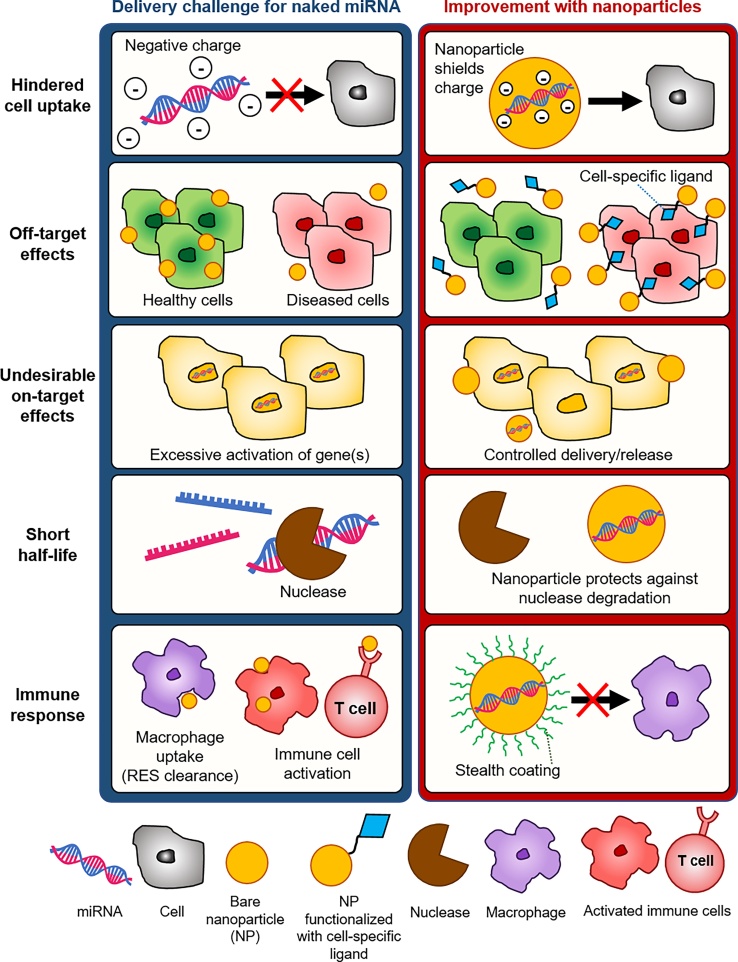
Both miR mimics and anti-miRs have been delivered in vitro using commercially available transfection agents, such as DharmaFECT™ and Lipofectamine™ [[50], [51], [52]], or by electroporation [53,54]. Alternatively, chemical modifications can be introduced to miRNAs to augment stability and allow carrier-free in vivo delivery of modified anti-miRs and miR mimics which are also known as antagomiRs [55] and agomiRs [56], respectively. For example, in the case of anti-miRs, in vivo silencing of endogenous miRNAs has been enhanced by integrating locked nucleic acids (LNA) or peptide nucleic acids (PNA), as reviewed elsewhere [57]. As an alternative to chemical modification, anti-miRs and miR mimics have been encapsulated into NPs. Due to their favorable transport properties, NPs have been reported to improve the in vivo delivery of miRNA agents; NPs protect their payload and enhance target specificity, [58] thus limiting adverse effects and improving therapeutic outcomes, as illustrated in Fig. 3 [59].
Fig. 3.
Key challenges of miRNA delivery in vivo. The challenges of delivering naked miRNAs include hindered miRNA uptake by cells due to negatively charged groups of miRNAs, undesirable off-target or on-target effects, short miRNA half-life under physiological conditions and unfavorable immune response. NPs encapsulate miRNAs, thus shielding charge groups and allowing their uptake by cells. Functionalizing NPs with cell-specific ligands allows NPs to deliver miRNAs to specific cells, thus reducing off-target effects. NPs allow for controlled miRNA release, avoiding excessive activation of multiple gene targets. NPs also increase the half-life of miRNA in vivo, by protecting the payload from degradation. Finally, addition of a stealth coating around NPs prevents their clearance by the reticuloendothelial system (RES) and avoids unfavorable immune cell stimulation.
The main challenges in miRNA delivery with nanocarriers are related to their low encapsulation efficiency [60], and the need for cell targeting, as shown schematically in Table 1 [61,62]. Because of their high affinity with water, miRNAs quickly diffuse into the water phase when nanoprecipitation or emulsion-based preparation methods are used, resulting in low encapsulation efficiency [60]. NPs improve the tissue distribution and site-specific localization of miRNA. However, the degree of enhancement is generally poor. As such, numerous studies have focused on modifying the NP surface with ligands for specific receptors on target cells, thus facilitating NP uptake by receptor-mediated endocytosis and reducing the required dosage and side effects of treatment [21,63,64].
Table 1.
Main general design criteria for miRNA-loaded nanoparticles (NPs).
| NP requirements for in vivo delivery | Delivery challenges | Suggested improvements | Reference(s) |
|---|---|---|---|
| High encapsulation efficiency | miRNA water solubility and negative charge. | miRNA complexation with positively charged molecules prior to nanoprecipitation or emulsification. | [71,75,76,77] |
| Colloidal stability | Protein corona formation and possible aggregation. | Decoration of NP surface with anti-fouling molecules. | [72,73,74] |
| Cell targeting | miRNA off-target effects. | Surface functionalization of NPs with specific ligands for cell targeting. | [41,78,79] |
| Cargo release in cytoplasm | miRNA degradation in low pH of endosome. | Use of materials with proton-accepting groups, which enable miRNA complexation and protection from degradation (i.e. proton sponge effect). | [66,67] |
| Controlled and sustained release, and increased half-life | Fast NP degradation rate and burst-release. | Control degradation and/or trigger miRNA release with stimuli-responsive materials (e.g. containing pH-sensitive histidine-, tertiary amine-, and sulphonamide groups; or nitroimidazole or azobenzene groups for hypoxia-driven disassembly). | [80] |
Moreover, colloidal stability of NPs in complex physiological media is demanded for cell-targeted delivery of miRNAs [65]. After administration, NPs should ideally circulate until they reach the desired site, and should be designed to undergo endosomal escape in order to guarantee the proper interaction between the miRNA and its intra-cellular target (for example by exploiting the proton sponge effect) [66,67]. However, circulation time depends on NP interactions with the biological microenvironment that could lead to their fast clearance. Specifically, once NPs are exposed to body fluids, their surface is covered by plasma proteins [68,69], resulting in masked surface ligands, non-specific uptake and reduced stability. There are different factors affecting NP circulation half-life, sequestration by the mononuclear phagocyte system (MPS) and biodistribution, including surface charge and hydrophobicity, size and shape [24]. Previous studies showed that neutral particles are less subjected to opsonization than highly charged particles especially if positively charged (cationic) [70,71]. Similarly, high hydrophobicity is related to a higher likelihood of clearance, which can be reduced by modifying the surface with polyethylene glycol (PEG), or by surface-camouflaging strategies, resulting in enhanced circulation half-life [[72], [73], [74]].
Importantly, the disease setting crucially determines the physical and biological barriers that the NP must overcome in addition to the basic hurdles that already impede miRNA delivery [41]. Based on these considerations, different strategies can be developed to prepare NPs that can effectively deliver miRNA to the target cells.
4. Methods to prepare miRNA-loaded NPs
Various preparation techniques, such as single or double emulsions, nanoprecipitation, and interfacial polymerization, have been employed for the preparation miRNA-loaded NPs. The selection of the most appropriate method is influenced by the constituent material and the desired surface characteristics [81]. Emulsion-based methods are the most commonly used to prepare miRNA-loaded NPs. These methods utilize high-speed homogenization or ultrasonication [82]. In the single-emulsion version, an oil-in-water (o/w) emulsion is formed by homogenizing or sonicating a polymer solution into an external, surfactant-containing, water phase. The double-emulsion technique, typically used to encapsulate hydrophilic payloads, utilizes two emulsification steps to obtain water-in-oil-in-water (w/o/w) or oil-in-water-in-oil (o/w/o) emulsions [81,83].
Emulsion methods have been used to prepare monomethoxy(polyethylene glycol)-poly(d,l-lactide-co-glycolide)-poly(l-lysine)-lactobionic acid (mPEG-PLGA-PLL-LA) [77], PLGA and poly(glycerol adipate-co-ω-pentadecalactone) (PGA-co-PDL) NPs for miRNA delivery [84]. For instance, miR-99a-loaded mPEG-PLGA-PLL-LA NPs have been obtained via the double emulsion method. For this purpose, miRNA is dissolved in water and subsequently dropped into a PLL-LA solution in dichloromethane, followed by sonication. The w/o/w emulsion was then dropped in water containing Pluronic-F68 and sonicated to obtain a w/o/w double emulsion. A reduction in the surface charge from 25 mV for blank NPs to 3 mV for miRNA-loaded NPs was taken as evidence of successful miRNA loading. The authors also demonstrate 80% of sustained payload release at 132 h, suggesting extended duration for the interactions between miR-99a and target genes.
Polymer NPs can be formed via nanoprecipitation, by dropwise addition to water of a polymer solution in a water-miscible solvent, causing its rapid displacement [81,85,86]. For instance, miRNA-loaded PLGA/chitosan (PLGA/CS) NPs with 150–180 nm size have been prepared via the nanoprecipitation method by dropwise addition of PLGA solution into a water solution of CS and miR-34 s, in the presence of Poloxamer surfactant [75]. This method could achieve miR-34 entrapment efficiency (EE) between 50% and 95%, depending on the amount of miR-34 in the formulation, and controlled release up to 48 h.
Unconventional techniques, such as combination of click chemistry (CC) and controlled radical polymerization (CRP), electrospinning, and particle replication in non-wetting template (PRINT), can potentially be used to prepare miRNA-loaded NPs because of their versatility, ease of implementation and low cost. Although these techniques have not been used to prepare miRNA-loaded NPs, we advocate that they are advantageous methods, since harsh temperatures, extreme pH conditions or potentially-toxic solvents are not required. Combination of CC and CRP can allow the preparation of NPs with controlled size and improved stability, as shown with DNA-conjugated NPs [[87], [88], [89]], as well as in vivo active cell recognition, as demonstrated by Koo et al. with dibenzylcyclooctyne (DBCO)-modified liposomes that were able to bind the azide groups expressed on cancer cells [90]. PRINT is known to generate particles with precise structure through the use of elastomeric molds of predefined shape and size, into which a polymer solution containing the drug is poured. NPs obtained by PRINT have been used in mice studies to investigate specific tumour cell uptake [91], and to test the treatment of brain metastasis when loaded with docetaxel [92].
Finally, recent advances in microfluidics have enabled the synthesis of libraries of NPs by rapid mixing of droplet streams controlled by micro pumps into silicon or glass microchannels [93]. These micro-reactors provide homogeneous conditions for NP nucleation and growth, resulting in higher reproducibility [94]. The microfluidic technology has been used to engineer small interfering RNA-loaded NPs (siRNA-loaded NPs) [95], but to the best of our knowledge, no evidence of their use to prepare miRNA-loaded NPs has been reported.
5. Selection of the biomaterial for miRNA encapsulation
The choice of the biomaterial that constitutes the NP is of paramount importance, as it dictates the choice of the preparation technique as well as the final properties and structure of the NP [96,97]. With the exciting advent of stimuli-responsive biomaterials, that change their properties upon modification of external conditions (including temperature or pH), multi-functional NPs have been developed for an expanding range of therapeutic applications (Table 2) [98,99].
Table 2.
Applications and therapeutic effects of nanoparticles (NPs) for delivering microRNAs (miRNAs) and/or drugs in tissue engineering (TE) and cancer.
| NPs | miRNAs | Application | Therapeutic effect | Stage | Ref |
|---|---|---|---|---|---|
| CS | agomiR-199 | Tibia regeneration | Osteogenic differentiation of MSCs and bone regeneration | Pre-clinical Bone defects on Sprague–Dawley rats | [56] |
| Hyaluronan sulfate | miR-21 | Myocardial infarction | Reduced inflammation and reduced cardiac fibrosis | Pre-clinical Myocardial infarction in C57BL/6 mice | [159] |
| PEI-PLGA | miRNAs targeting COX1 & COX2 | Flexor tendon adhesions | Reduced inflammation and support for tendon healing | Pre-clinical Tendon injury in chicken | [160] |
| PLGA | miR-26a | Calvarian bone regeneration | Increased osteoblastic activity | Pre-clinical Bone defect in C57BL/6 mice | [161] |
| Polyketal | miR-106b, miR-148b and miR-204 | Myocardial infarction | Improved cardiac function and reduced infarct size | Pre-clinical Myocardial infarction in C57BL/6 mice | [162] |
| Fe3O4-PEI | Let-7a | Glioblastoma | PI3K and RAS downregulation | In vivo proof of concept (sub cutaneous breast cancer in nu/nu mice) | [144] |
| Gold | miR-182 | Brain cancer | Reduced tumour burden and increased survival | Pre-clinical Intra-cranial tumours on SCID mice | [163] |
| Gold | miR-145 | Breast cancer, prostate cancer | miR-145 expression recovery | In vitro | [78] |
| HA-CS | miR-34a and doxorubicin (DOX) | Triple negative breast cancer | Increased cell sensitization to DOX | In vitro | [117] |
| Liposomes | miR-101 and DOX | Hepatic carcinoma | Reduced tumour growth | In vivo proof of concept subcutaneous tumour in BALB/c nude mice | [23] |
| Liposomes | Anti-miR-21 | Glioblastoma | Inhibition of miR-21 expression | Pre-clinical Intra-cranial tumours in C57BL/6 mice | [164] |
| PEG-lipids | miR-122 | Hepatic carcinoma | Reduced tumour growth | In vivo proof of concept Flank model of HCC in nude mice | [158] |
| PCL-PEG | miR-200c and docetaxel | Gastric cancer | Tumour growth inhibition | In vivo proof of concept Flank model in Balb/C mice | [102] |
| PEI-PEG | miR-145 | Prostate cancer | Tumour growth inhibition | Pre-clinical Intra-peritoneal tumour in nude mice | [79] |
| PEI | miR-145 and miR-33a | Colon carcinoma | Reduced tumour growth | In vivo proof of concept subcutaneous tumour in nude mice | [101] |
| PEI | miR-145 | Metastatic breast cancer | Reduced cell proliferation in vitro | In vitro | [100] |
| PLGA | miR-99a | Hepatic carcinoma | Tumour growth inhibition | In vivo proof of concept subcutaneous tumour in nude mice | [77] |
| PLGA-PEG | miR-7 and paclitaxel (PTX) | Ovarian cancer | Increased cancer cell apoptosis and sensitization to PTX in vitro | In vivo proof of concept subcutaneous tumour in nude mice | [165] |
| PLGA-PEI-HA | miR-145 | Colon carcinoma | Tumour growth inhibition | In vivo proof of concept subcutaneous tumour in nude mice | [63] |
| PU-PEI | miR-145 | Lung tumour | Reduced tumour growth and prolonged survival | Pre-clinical Intra-bronchial tumours in nude mice | [103,104] |
| PU-PEI | miR-145 | Brain tumour | Reduced tumour growth and prolonged survival | Pre-clinical Intra-cranial tumours in nude mice | [104] |
| Silica | Anti-miR221 Temozolomide | Glioma | Increased cancer cell apoptosis in vitro | In vitro | [146] |
| Silica | miR-34a | Neuroblastoma | Tumour growth delay and apoptosis and reduced vascularization | Pre-clinical Retro-peritoneal tumours in CB-17/SCID mice | [147] |
5.1. Synthetic polymers
The use of synthetic polymer NPs for nucleic acid delivery has been extensively investigated, because of their efficient cargo release within the cell cytoplasm and their ease of manufacturing and functionalization with targeting molecules [41]. Among synthetic polymers, poly(ethylene imine)s (PEIs) [100,101], PLGA [75,76], poly(ε-caprolactone) (PCL) [102], and polyurethanes (PUs) [103,104] have been used for miRNA delivery.
PEI, both in linear and branched form, is a polycationic polymer. The PEI repeating unit consists of a secondary amine group and one ethylene spacer and is commercially available in different molecular weights, having different release efficiency and biocompatibility [105,106]. PEI chains are positively charged, depending on the protonation degree of secondary amines as a function of the pH. The positive charge allows electrostatic complexation with nucleic acids [107]. Due to its proton acceptor behavior, PEI exhibits weak-base buffering properties, reducing nuclease activity [108], and protecting nucleic acids from degradation within endosomal vesicles. As a proton acceptor, PEI also causes the “proton sponge effect”, leading to endosomal lysis and consequent release of nucleic acids into the cytoplasm [66,67,109].
PEI has been shown to deliver miR-145 and miR-33a, and allow the assessment of the in vivo anti-tumour activity of these miRNAs in a mouse model of colon carcinoma [101]. Local or systemic administration of PEI-miRNA complexes could result in reduced tumour growth compared to controls, indicating that such NPs represent a promising strategy for miRNA replacement therapy in cancer treatments [101]. PEI has also successfully delivered miR-145 in vitro to metastatic breast cancer cells [100], and in vivo in a xenograft colon cancer models [63].
However, while PEI NPs show efficient miRNA encapsulation and delivery, their high positive charge density and non-biodegradability can affect cell viability, thus limiting their use [108]. In this context, NPs that are surface-modified or based on blends of PEI and PEG have been proposed to reduce toxicity and improve biodistribution [110]. For instance, Zhang et al. have used branched PEI-PEG NPs to deliver miR-145 to prostate cancer [79]. The PEG chains were surface-modified with poly-arginine showing enhanced uptake in prostate cancer in vivo, reduction of tumour growth and increased survival.
Biocompatible and biodegradable synthetic polyesters, such as PLGA and PCL have also been investigated for nucleic acids delivery, because of their lower toxicity and biodegradability as compared to PEIs [[111], [112], [113]]. PLGA is a biocompatible and non-cytotoxic polymer, known to degrade by hydrolysis, generating lactic and glycolic acid monomers, which are metabolized through the Krebs cycle [111]. By varying the ratio between the constituent monomers, lactic and glycolic acid, the degradation rate of PLGA NPs can be tuned in the range from several months to years [111,114]. Low molecular weight polymers with higher glycolide content are more hydrophilic with a shorter degradation time. In contrast, PLGA NPs with higher lactic acid content have a hydrophobic nature and degrade more gradually.
MiRNA encapsulation is made difficult by the negative surface charge of PLGA NPs [115,116], which reduces complexation efficiency with the phosphate groups of miRNAs. This limitation can be overcome by using cationic compounds combined with PLGA. For instance, CS, a cationic polymer, has been shown to promote the retention of miRNAs inside PLGA NPs [75]. Protamine sulfate (PS) coating of PLGA NPs has also been used to enhance complexation of miRNAs on the NP surface [76]. In another study, Wang et al. have generated PLGA-PEI NPs to co-deliver miR-542-3p and doxorubicin (DOX) in triple negative breast cancer cells [117]. PLGA-PEI NPs could be covalently grafted with hyaluronic acid (HA) to be specifically internalized by cancer cells, thus enhancing their cytotoxicity as assessed in vitro on breast cancer cells.
PCL has attracted high attention for the preparation of biocompatible NPs for nucleic acid delivery. NPs composed of PCL are promising for their high colloidal stability, quick cellular uptake, low toxicity in vitro and in vivo, and controlled release of their drug cargo. The degradation product of PCL is 6-hydroxyhexanoic acid, which is a natural metabolite in the human body [118]. Liu et al. have prepared PEG-peptide-PCL NPs by the double emulsion method to co-deliver miR-200c and docetaxel [102]. A gelatinases-cleavable peptide was selected to facilitate delivery after NPs internalization by tumour cells. In addition, spermidine has been used to complex with miR-200c during NP preparation, achieving about 85% transfection efficiency in vitro.
Among synthetic polymers, PU-based NPs have shown high biocompatibility combined with versatile physical and chemical properties [119]. Cationic PU-short branch PEI nanocomplexes have been developed by Chiou et al. as vehicles for miR-145 delivery to inhibit the stem cell niche in the brain tumour [103]. The authors demonstrate successful miR-145 transfection (nearly 90%) and efficient knockdown of miRNA targets, Sox2 and Oct4, in vitro. Intracranial injection of PU-PEI-miR-145 nanocomplexes could reduce tumour growth by synergizing with chemoradiotherapy. Furthermore, such nanocomplexes show a therapeutic effect on lung adenocarcinoma model [104], with decreased metastases, inhibited epithelial-to-mesenchymal transition (EMT) in vitro, increased survival and delayed tumour growth in mice [104].
5.2. Natural polymers
Among natural polymers, materials that can be obtained from animal or vegetal sources, CS and HA are promising carriers for nucleic acids [[120], [121], [122]]. CS is obtained from chitin and is non-cytotoxic, non-immunogenic, and highly biocompatible [123,124]. At acidic pH, the protonated amino groups of CS rapidly interact with opposite-charged molecules, such as siRNA or miRNA with high loading efficiency [22,125]. In spite of these advantages, CS NPs offer limited control over the delivery of nucleic acids because of their strong interaction with the loaded agent [[126], [127], [128]], which results in inefficient unpacking of the complex in the cytoplasm [127]. To overcome these drawbacks, hydrophobic moieties have been conjugated to CS to weaken the polymer/nucleic acid interaction and enhance cytoplasmic drug delivery. Lipid chains, bile acids or negatively charged polymers, such as PLGA, have also been combined with CS to facilitate miRNA release [[129], [130], [131]].
HA is a highly hydrophilic anionic natural polysaccharide, that can be recognized by HA receptors on cells [132,133]. By virtue of its chemical versatility, HA has been extensively used to prepare NPs for the delivery of cytostatic drugs, proteins, polynucleotides, immunomodulators and imaging agents. HA covalently grafted to cationic polymers or lipid molecules has been proposed to deliver polynucleotides. For example, HA chemically linked to PEI and PEG has been used to release miRNA and DOX [117,134]. HA has also been successfully used to modify the surface of cationic polynucleotide-loaded NPs [135]. However, although cationic polymers efficiently form complexes with nucleic acids due to charge interactions [129], their positive charge contributes to their uptake by the MPS with subsequent rapid elimination and toxicity [136]. For this purpose, several authors have proposed surface-modifications with HA to mask the positive charge of cationic NPs, lipid complexes or liposomes [135]. For instance, HA has been electrostatically attached to the surface of positively charged liposomes [137,138] and calcium phosphate NPs [139], as well as chemically bound to lipids on the NP surface. Liang et al. have used HA-NPs loaded with miR-145 to target colon cancer cells through their overexpressed CD44 receptors. Real-time qPCR and western blot analyses revealed enhanced accumulation and reduced tumour growth [63], suggesting the potential therapeutic success of utilizing such NPs in the clinic.
5.3. Inorganic NPs
Due to their biocompatibility, controllable size and morphology, several inorganic materials have been proposed in nanomedicine, including gold, calcium phosphate, silica and iron oxides [140,141]. Firstly, iron oxide-based NPs have been proposed for imaging purposes, as contrast agents for magnetic resonance imaging (MRI), in thermal therapy, as well as in drug and gene delivery. The combination of magnetic NPs with cationic compounds is needed to achieve high miRNA encapsulation [142]. Streptavidin-coated magnetite (Fe3O4)-based NPs have been modified with biotin-bound miR-335/PEI complexes to deliver the miRNA cargo to human mesenchymal stem cells (hMSCs), achieving enhanced target gene knockdown as compared to a PEI-miR strategy [143]. Yin et al. have used zinc-doped Fe3O4 NPs to deliver let-7a miRNA to glioblastoma cells by magnetofection [144]. Loading of let-7a miRNA into NPs has been achieved via a layer-by-layer approach, using branched PEI as polycation. These complexes showed high cellular uptake (at nearly 98%) and downregulation of the let-7a targets, PI3K and RAS.
Silica NPs and mesoporous silica NPs (MSPs) are silica-based nanostructures which possess high biocompatibility and stability [145]. For these reasons, MSPs have received great attention in clinical applications. MSPs have been used to co-deliver anti-miR-221 and Temozolomide (TMZ) to treat drug-resistant glioma cells [146]. Silica NPs carrying a tumour suppressor miRNA miR-34a have been developed by Tivnan et al. to specifically inhibit neuroblastoma growth [147]. GD2-antibody could further be conjugated on the NP surface to enhance specific targeting, delaying tumour growth, enhancing apoptosis and reducing vascularization in vivo.
Calcium phosphate (CaP) NPs are inexpensive, nontoxic, bioresorbable and easily synthesized nanocarriers. They have been widely used as gene carriers for 40 years, using DNA-CaP co-precipitation as an efficient method to introduce nucleic acids into cells; they remain stable in the endosomal compartment and are rapidly degraded in the mildly acidic lysosome environment, causing the vesicle to burst, releasing their content in the cytoplasm [148]. Banik et al. have shown that large molecular weight gene therapeutics, such as plasmid DNA can successfully interact with CaP-NPs in a spontaneous and cooperative manner, without NP intercalation with DNA base pairs [148]. However, miRNAs might have poor interaction because of low spatial charge density. Accordingly, long-chain miRNAs (lc-miRNAs) have been developed to enhance encapsulation efficiencies into PEI-CaPNPs [149]. Lc-miRNAs have been prepared by adding thiol groups on both the 3’ and the 5’ miRNA end, thus obtaining longer chains with higher encapsulation efficiency.
Gold NPs (Au-NPs) have been used in drug delivery by virtue of their biocompatibility, ease of functionalization, and tunable size and shape [[150], [151], [152]]. The surface of Au-NPs is typically functionalized with thiol or amino groups to enable miRNA entrapment. Ghosh et al. have developed a non-toxic gold-based formulation to obtain excellent miRNA cellular uptake through endocytosis and intracellular delivery [153]. This formulation was based on miRNA/ Au-NPs-S-PEG, with a ratio of 1-10-0.5, respectively. Cystamine functionalization of Au NPs can also be used for miRNA complexation, whereas PEG can prevent NP aggregation and miRNA degradation. Ekin et al. have used Au-NPs to deliver and recover the expression of miR-145 (a tumour suppressor gene) into prostate and breast cancer cells, where miR-145 is usually downregulated [78].
5.4. Lipid-based NPs
Lipids are the main components of the cell membrane, thus making lipid-based NPs (LNPs) capable of interacting with the membrane, promoting cellular uptake of their contents. LNPs are composed of a lipid bilayer that contains the miRNA in the aqueous core or, in the case of multi-lamellar liposomes, between lipid bilayers [154]. Typically, miRNA-loaded LNPs are made with a mixture of cationic lipids, neutral lipids and PEG [155]. Spontaneous electrostatic interaction between cationic lipids and negatively charged miRNA results in an efficient condensation of miRNAs and in their protection from enzymatic degradation. Also, the positive charge facilitates the interaction with the opposite-charged cellular membrane [155]. However, the use of cationic lipids is commonly associated with cell toxicity, as they can disrupt cell membrane integrity, induce cytoplasm vacuolization and reduce cell activity [156]. Furthermore, the presence of cationic lipids can result in the interaction with negatively charged serum proteins, thus inducing aggregate formation, which are subsequently eliminated by the liver and the spleen. For these reasons, several strategies to reduce the cationic charge have been attempted. For instance, combination with neutral lipids, such as cholesterol (Chol), dioleoylphosphatidyl ethanolamine (DOPE) and phosphatidylcholine (PC), known as “helper lipids”, has been proposed to enhance stability and reduce toxicity of LNPs [157]. PEG modifications on the LNP surface have also been implemented to increase their half-life. PEGylated cationic/neutral LNPs have been used for the systemic delivery of liver-specific tumour suppressor miR-122 in hepatocellular carcinoma (HCC), achieving ∼ 50% decreased growth after 30 days [158].
6. Therapeutic application of miRNA-based nanomedicine
6.1. miRNA-based nanomedicine in tissue engineering (TE)
MiRNAs are potent therapeutics for TE applications. In this context, miRNA-activated scaffolds can be prepared for tissue-specific miRNA replacement, or modulation (Fig. 4). Here, we report the recent results with miRNA in TE, focusing mostly on cardiac and bone regeneration.
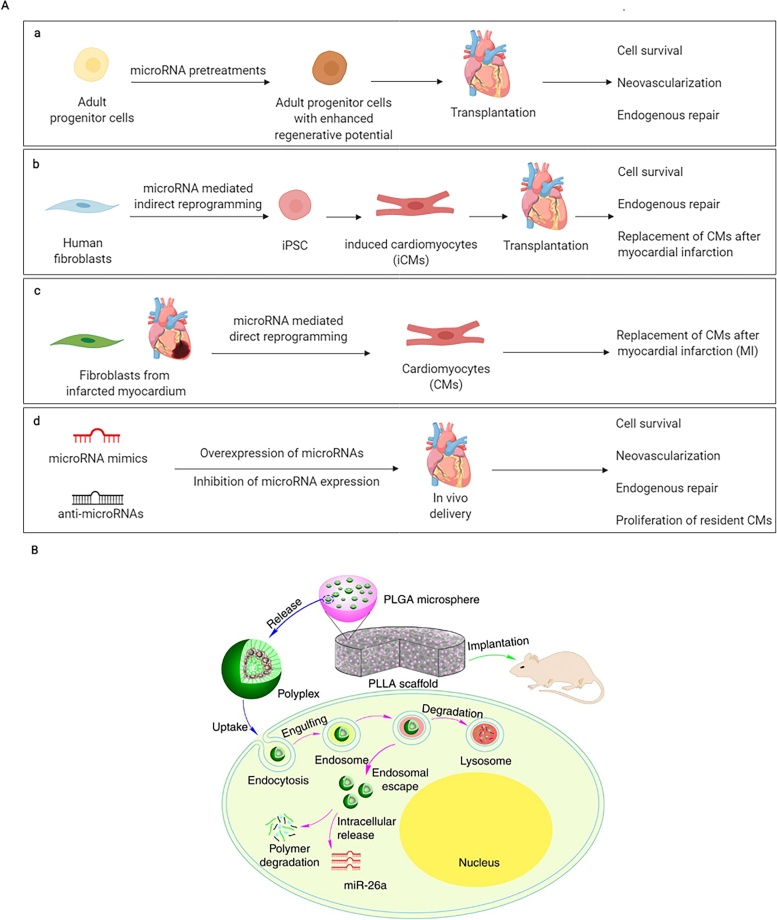
Fig. 4.
microRNA (miRNA) therapeutic approaches in tissue engineering (TE). (A) Different miRNA-based therapeutic strategies for cardiac repair and regeneration after injury. miRNAs can (a) augment the regenerative capacity of adult cardiac progenitor cells, (b) reprogram human fibroblasts into iPSCs (c) directly reprogram human fibroblasts into cardiomyocytes in vivo, and (d) guide gene expression associated with cardiac repair. This figure was created with BioRender. (B) miRNA-based therapy to regenerate bone defects, using a cell-free 3D scaffold loaded with miR-26a-containing PLGA microspheres. Image reproduced with permission from Zhang, X. et al., Nature communications, 7 p.10376 (2016) http://creativecommons.org/licenses/by/4.0/.
Myocardial infarction (MI), a leading cause of death, consists in the irreversible loss of cardiomyocytes, cardiac ECM degradation and the formation of a fibrotic scar. [166] The discovery that miRNA treatment can induce cardiomyocyte proliferation and the direct reprogramming of fibroblasts into cardiomyocytes have raised the interest in using miRNAs as potential therapeutics in MI, as illustrated in Fig. 4A [167,168]. In spite of the excitement raised by these findings, the lack of suitable delivery vehicles for miRNAs have limited their practical application. Recently, Di Mauro et al. have generated CaP-NPs which successfully encapsulated and delivered miR-133 or miR-39-3p into murine cardiac cells without altering their biological function [169]. When administered in vivo, via retro-orbital injection, miRNA-encapsulated CaP-NPs accumulated in the left cardiac ventricle and in the liver, spleen and kidney. This non-specific uptake suggests that surface-modification of NPs for specific tissue targeting can improve the efficacy of miRNA-loaded NPs. Yang et al. have generated polyketal (PK3) NPs loaded with a combination of three miRNAs (miR-106b, miR-148b and miR-204), targeting Nox2, a ROS intermediate, with increased expression after MI [162]. Potentially, the inhibition of Nox2 expression by local treatment with NPs improved cardiac function and reduced infarct size in mouse models. Bejerano et al. also showed tissue remodeling post-MI through the delivery of miR-21 assembled into hyaluronan-sulfate (HAS) NPs [159]. After MI, prolonged macrophage recruitment to the infarct area could result in persistent inflammation and delayed tissue repair, thus inducing further cardiac damage. Also, miR-21 upregulation in peritoneal macrophages has been previously shown to promote inflammation resolution [170]. HAS-mediated delivery of miR-21 in vivo could elicit the natural reparative ability of macrophages, thus reducing cardiac fibrosis and cell apoptosis.
Several studies have elucidated the role of miRNAs in bone remodeling, by controlling osteoblast differentiation and inhibition through the regulation of different signaling pathways [171]. Zhao et al. have used CS-based NPs to deliver anti-miR-138 to bone marrow mesenchymal stem cells (BMSCs) to promote osteogenic differentiation [172]. Anti-miR-138 could regulate the ERK1/2 pathway, inducing osteoblast differentiation. Wang et al. have engineered miR-21-loaded HA/CS NPs (miR-21-HA/CS NPs) that could coat cell culture plates to induce osteogenic differentiation of BMSCs [173]. miR-21 has been selected because it downregulates SOX2 expression, thereby inducing the differentiation of hMSCs towards osteogenic lineages. After BMSC transfection with miR-21-HA/CS NPs, osteogenic-related genes could be upregulated compared to controls, including Collagen type I alpha1 (COL1), osteopontin (OPN), Runt-related transcription factor 2 (RUNX2), and osteocalcin (OCN). Moreover, after 14 days of culture with miR-21-HA/CS NPs, BMSCs that were subsequently cultured in an osteogenic differentiation medium could display significantly higher ability for ECM mineralization, as suggested by a higher calcium accumulation compared to controls.
As TE often requires the presence of three-dimensional (3D) constructs to favour and support new tissue ingrowth, recent approaches combining NPs with injectable hydrogels or scaffolds have also been exploited. For instance, Zhang et al. have proposed a two-stage miRNA delivery strategy to improve controlled release and transfection efficiency of miR-26a for calvarial bone regeneration (Fig. 4B) [161]. This strategy relies on the encapsulation of polyplexes carrying miRNAs into PLGA microspheres and their subsequent immobilization onto poly(l-lactic acid) (PLLA) scaffolds. This two-stage miRNA delivery could regenerate bone defects in mice by activating the osteoblastic activity of endogenous stem cells.
CS hydrochloride NPs loaded with agomiR-199-5p have been used to induce hMSC osteogenic differentiation in vitro through the regulation of the HIF pathway [56]. NPs could allow sustained agomiR-199-5p delivery in vitro for at least 21 days. Here, agomiR-loaded NPs were encapsulated into a fibrin gel and the scaffolds were implanted within a tibia bone defect in rats, allowing successful in situ bone tissue regeneration. In another study, Zhou et al. demonstrate a local sustained gene delivery system by using PEI-modified PLGA-NPs embedded into HA hydrogel, to deliver an engineered miRNA plasmid which targets cyclooxigenase COX-1 and COX-2 to reduce flexor tendon adhesions [160]. Administration of miRNA by PLGA NPs in hydrogel could prevent tendon adhesions by decreasing inflammatory response by directly downregulating COX-1 and COX-2.
6.2. miRNA-based nanomedicine in cancer
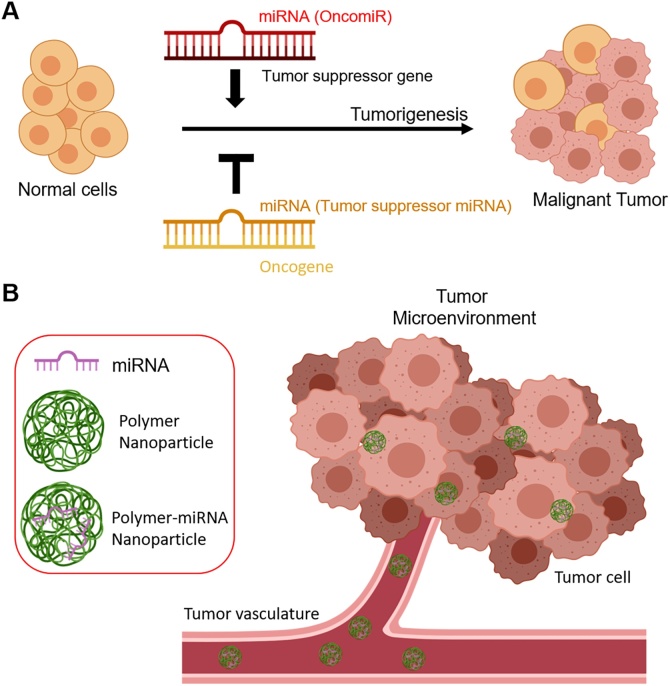
In cancer treatment, miRNA-based therapy can follow two strategies: (i) miRNA inhibition and (ii) miRNA replacement (Fig. 5) [174]. The first approach provides specific anti-miR oligonucleotides (AMOs) that inhibit tumour-promoting miRNAs (oncomiRs), such as miR-519a and miR-32 [175,176]. The latter strategy aims at introducing exogenous miRNAs (miR mimics) known to promote tumour suppression, such as miR-34a and let-7b [177,178]. However, in spite of the evidence of potentiated anti-cancer toxicity of combinational treatments based on anti-cancer drugs and miRNAs, clinical application is limited by practical hurdles. Preclinical trials have highlighted the difficulty of delivering naked miRNAs to tumours and the need for more efficient delivery systems [179,180].
Fig. 5.
(A) Roles of microRNAs (miRNAs) in cancer. Depending on the target gene, miRNAs suppress or stimulate cancer-related genes. Image modified from Oliveto, S. et al., World journal of biological chemistry, 8 (1), p.45 (2017). (B) Therapeutic approach using miRNA-loaded polymer NPs circulating in the tumour microenvironment. Images (A) and (B) were created with BioRender.
Numerous authors have shown that chemotherapeutics and miRNAs can be encapsulated within NPs, representing an effective strategy to achieve potentiated cytotoxicity both in vitro and in vivo. For instance, miRNA-mediated regulation of the p53 network or the extracellular signal-regulated kinase (ERK) pathway has been used in synergy with cytotoxic drugs, such as DOX and Paclitaxel (PTX) [165,181]. Deng et al. have designed CS/HA NPs, exploiting the positive charges of CS to form complexes with miR-34, a regulator of the p53 pathway. DOX was co-encapsulated with miR-34 into CS/HA NPs, showing enhanced efficacy against triple negative breast cancer cells in vitro, as miR-34 restored the normal p53 pathway, sensitizing the cells to DOX chemotherapy [181]. Recently, nano-sized liposomes have also been prepared encapsulating DOX in combination with miR-101, a tumour-suppressive miRNA that is downregulated in HCC [182]. The effect of co-delivery could inhibit the growth of malignant HCC cells, both in vitro and in vivo, due to the synergistic effect of both agents [23].
Cui et al. demonstrate that the co-encapsulation of PTX and miR-7 into mPEG-PLGA-PLL-LA NPs [165]. miRNA-7 suppresses the EGFR/ERK pathway, which is known to drive cell proliferation and resistance to PTX [122,183]. Their developed NPs could enhance PTX release and improve miRNA efficiency, resulting in increased apoptosis of ovarian cancer cells and reduced resistance to PTX [165]. Costa and colleagues demonstrate the preferential accumulation within brain tissue of liposomal NPs that were conjugated with chlorotoxin (CTX), a glioma-specific peptide marker. CTX-NPs could efficiently deliver anti-miR-21 oligonucleotides, with no signs of systemic immunogenicity [164].
Studies on lung adenocarcinoma (LAC) have shown the presence of a small subset of cancer stem cells (CSCs) with altered miRNA expression [184]. miR-145 loaded into PU-PEI NPs showed beneficial effect on LAC metastases through the inhibition of EMT processes and improved sensitization to chemoradiotherapies, prolonging survival of mice. Abnormal expression of miRNAs has also been found in malignant brain tumours. For instance, inhibition of miRNA-21 has been used in GBM treatment to rescue tumour suppressor genes, such as PTEN and PDCD4, while miRNA-182 has been associated with favorable prognosis because of its role as a regulator of apoptosis, growth, and differentiation [[185], [186], [187]]. Anti-miR-21 NPs co-encapsulating DOX have demonstrated enhanced cell apoptosis and reduced tumour growth in brain tumour models [64]. Kouri et al. demonstrate gold NPs covalently functionalized with miR-182 that can selectively deliver miR-182 to brain tumours, reducing tumour size and increasing survival [163]. These studies demonstrate the extensive potential of miRNA-loaded NPs in cancer treatments.
7. Frontiers in miRNA-based therapies
7.1. Cancer immunotherapy
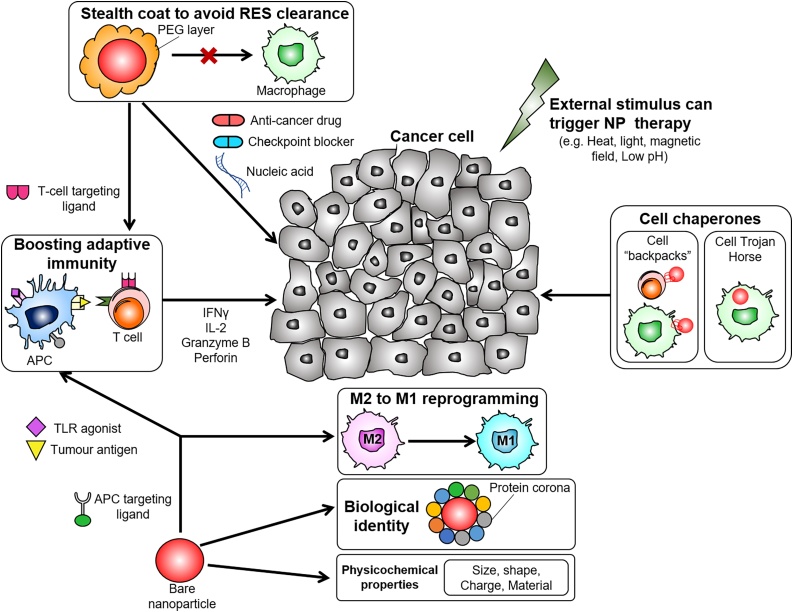
MiRNAs play a significant role in immune system modulation [188]. As a result, inhibiting or upregulating specific miRNAs can potentially modulate the immune response to tumours, for example by controlling macrophage polarization or by regulating their activities in the tumour micro-environment (TME) [189,190]. For instance, upregulation of specific miRNAs, such as let-7c [191,192], and miR-146a [193,194] has been shown to drive immunosuppressive and pro-tumour activity of (pro-tumour) “M2”-type tumour-associated macrophages (TAMs). In contrast, (anti-tumour) “M1”-type macrophages, characterized by tumoricidal activity, can be induced by increased expression of a.o. miR-125b [192,195], and miR-155 [[196], [197], [198]]. As immunomodulation is often constrained by poor specificity, NP-based specific delivery of immunomodulatory agents has the potential to improve on-target delivery while minimizing off-target effects (Fig. 6) [199]. Although numerous studies have demonstrated the anti-tumour efficacy of NP-based immunomodulation, targeting macrophages, T cells, and dendritic cells (DCs) [200,201], there is a startling lack of studies on immunomodulation through miRNA-loaded NPs. Importantly, the design of miRNA-based NP platforms for immunomodulation may build upon existing RNA-based NP technologies or immune system targeting strategies, briefly summarized below [8,[202], [203], [204]].
Fig. 6.
NPs can be applied in cancer immunotherapy to i) enhance the activity of T cells, ii) induce macrophage reprogramming towards the tumoricidal M1 phenotype, iii) target immune cells and exploit their tumour infiltration ability to penetrate inside the tumour mass, where release can be triggered by either internal or external stimuli. Image modified with permission from Gun, S. Y. et al., Redox biology, p.101174 (2019) (published under Creative Commons attribution Licence CC-BY).
Researchers have used monocytes/macrophages as enablers of NP-based anti-cancer therapy, by virtue of their ability to naturally home to tumours. Choi et al. show that macrophages that phagocytized gold nanoshells (Au-NS) can accumulate into breast tumour spheroids, resulting in cancer cell death through photo-induced ablation [205,206]. Anselmo et al. and Doshi et al. have attached a therapeutic cargo to monocytes in the form of a “cellular backpack” and demonstrate that monocyte-associated “backpacks” could accumulate to greater degree in inflamed organs, as compared to a “free backpacks” [207,208]. Alternatively, Song et al. demonstrate the TAM-specific delivery of siRNA targeted against vascular endothelial growth factor (VEGF) and placental growth factor (PIGF), that support the pro-tumour activity of TAMs [209]. Mice being administered with siRNA-loaded NPs showed decreased TAM accumulation in the tumour microenvironment and striking reductions in tumour growth compared to control mice.
Similarly, T cells, which display cytotoxic activity against tumour cells, can be exploited to facilitate intratumoral delivery of NPs [210,211]. For example, Siriwon et al. show that CAR T cells can effectively deliver and release an antagonist against T cell activity deep within the tumour microenvironment, thus supporting effector T cell cytotoxicity [212]. NPs can also be used to deliver cytotoxic T lymphocyte molecule-4 (CTLA-4) siRNA into tumour-infiltrating T cells (TILs), to increase the proliferation and the anti-tumour activity of effector T cells [213]. Smith et al. have utilized NPs to deliver leukemia-specific CAR genes into T cells, achieving long term disease remission [214]. This strategy could potentially be used to achieve a sufficient pool of therapeutic T cells to elicit “on-demand” cancer immunity [214].
NPs-based immunotherapy can also be achieved by targeting dendritic cells (DCs). DCs present epitopes of tumour-associated antigens (TAAs) to T cells, activating T cells for their attack of TAA-presenting tumour cells [215]. Self-assembled DNA–RNA nanocapsules have been develoded to that deliver tumour neoantigens to DCs, increasing the CD8+ T cells and reducing the growth of colorectal tumours [8]. Warashina et al. have developed a lipid NP for DC-specific delivery of siRNA against a suppressor of cytokine signaling 1 (SOCS1). Treated DCs displayed a marked increase in cytokine production with consequent reduction of tumour growth in vivo [216]. Kranz et al. have further exploited the antiviral defence mechanisms of DCs, using DC-targeting RNA-lipoplexes to trigger the release of pro-inflammatory interferon-α [217].
7.2. Neurodegenerative disorders
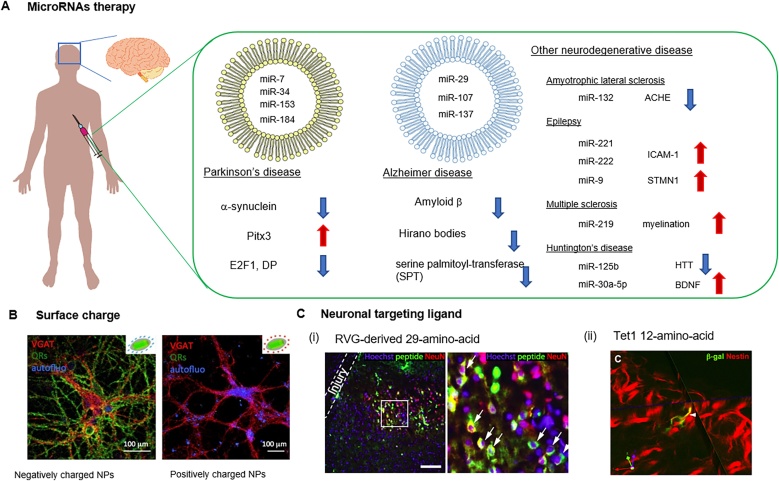
Neurodegenerative disorders, such as Parkinson’s disease (PD), Alzheimer’s disease (AD), amyotrophic lateral sclerosis (ALS), multiple sclerosis (MS), and Huntington’s disease (HD), are characterized by, among other features, the progressive loss of neuronal synapses, leading to neuronal death. Recent studies have reported that abnormal miRNA expression contributes to the degradation of neurons [218], suggesting that neurodegenerative diseases can be classified as an RNA disorder [219]. miRNA-loaded NPs that regulate RNA transcripts may thus represent a form of novel therapeutics for treating neurodegenerative diseases (Fig. 7) [220].
Fig. 7.
(A) Representative potential microRNA-therapies for treating neurodegenerative diseases. Targeted NPs release miRNA to neurons by: (B) surface charge (reproduced with permission from Dante, S. et al., ACS Nano 11, 6630–6640 (2017), or (C) surface functionalization with specific neuronal binding ligand such as (i) RVG-derived 29-amino-acid and (ii) Tet1, 12-amino-acid (reproduced with permission Kumar, P. et al. ACS Nano 2016, 10, 8 7926–7933 and Kwon, E. J. et al., Biomaterials 31, 2417–2424 (2010), respectively).
Comparative studies of miRNA profiles in post-mortem brain tissue from PD patients and healthy controls have shown altered miRNA levels in PD patients. For instance, miR-7, miR-153, and miR-433 drive the over expression of α-synuclein, which has been associated with aberrant soluble oligomeric conformations in in vitro experiments [221,222]. Dysregulation of miR-34 has been associated with the pathogenesis of PD along with altered mitochondria activity and oxidative stress [223]. In addition, several studies show reduced expression of miR-133 in PD patients, which regulates dopaminergic neuron development, differentiation and maturation [224]. miR-133 also further regulates Pitx3, a transcription factor involved in dopaminergic neuron differentiation [225].
Additionally, many miRNAs are abnormally regulated in the brains of AD patients. For instance, decreased expressions of miR-29, miR-189, and miR-9 are found in the AD brain, as evidenced by genome-sequencing and miRNA profiling. Decreased expressions of miR-298, miR-328 and miR-195 have been found in AD mouse models [226,227]. Recent studies have shown the relationship between miRNA dysregulation and extracellular amyloid β plaque (Aβ) production and aggregation which gives rise to neuro-degeneration [228]. For example, decreased expression of miR-107 may contribute toward faster disease progression by regulating beta-site amyloid precursor protein (APP) cleaving enzyme 1 (BACE1) [229]. miR-106a and miR-520c have been found to suppress the expression of APP in vitro [230]. miR-17 and miR-20a, along with miR-147, miR-153, miR-644, and miR-655 as well as miR-323-3p, have been found to directly target APP [231]. Additional studies have identified dysregulated miRNAs that contribute to AD pathogenesis through atypical regulation of numerous proteins, which is involved in disease progression. Downregulated miR-103 or miR-107 expression can also lead to increased cofilin 1 (CFL1) levels in AD brains, which may associate with the formation of intracellular Hirano bodies [232]. Other studies show that decreased expressions of miR-9, miR-137, miR-181c, and miR-29b-1 in AD brain can upregulate serine palmitoyltransferase (SPT), long chain subunit 1 (SPTLC1) and 2 (SPTLC2), which are the rate-limiting enzymes in the synthesis of ceramides [233].
The dysregulation of miRNA also plays an essential role in other neurodegenerative diseases. Many studies have identified ALS-relevant transcripts and pathways that are targeted by dysregulated miRNAs. For instance, miR-132 regulates acetyl-cholinesterase and polypyrimidine tract binding protein 2 (PTBP2), which influences alternative splicing of tau in ALS [234]. Wang et al. showed that miR-219 and miRNA-338 downregulation is observed in murine CNS, regulating oligodendrocyte differentiation and myelination [235]. Several miRNAs may also play a role in HD, particularly by targeting the Huntingtin gene (Htt). Sinha et al. show that miR-125b, miR-214, miR-150, miR-146a have the potential to reduce Htt aggregates [236]. The encapsulation of these miRNA in NPs may further represent a feasible strategy for more efficient delivery, thus contributing to improved therapeutic outcomes.
To date, delivery of miRNAs to the brain and to the spinal cord mainly relies on recombinant adeno-associated virus (rAAV) [237]. Miyazaki et al. showed that miR-196a delivery using AAV vector could enhance the decay of mRNA androgen receptor by silencing Elav-like family member 2 (CELF2) in mice [238]. They found that the early delivery of miR-196a by an AAV vector could ameliorate the spinal and bulbar muscular atrophy phenotype in mice. Another study showed that miR-132 delivery using AAV9 vector could recover motor function, thus regulating muscle locomotion as well as survival rate of HD model mice [239].
As AAV vectors have a limited capacity for encapsulation (4.5–5 kb) due to size limitations, non-viral vectors may substantially increase miRNA delivery efficiency to the brain, improving neurodegenerative disease treatment. Drug delivery to the brain mediated by NPs can also allow blood-brain barrier (BBB) crossing and selective targeting of neurons. For example, heat generation from iron oxide NPs has been shown to enhance BBB bypass of NPs [240]. Dante et al. showed that negatively charged (—COOH) NPs could effectively localize at the membrane of neurons and influence neural excitability [241,242]. Ligand-based targeting has been also considered for the improved delivery of NPs to brain and neurons. Kumar et al. reported that a short peptide-derived rabies virus glycoprotein (RVG) localize on neuronal cells in vitro through specific binding to the acetylcholine receptor [243]. Kwon et al. proposed a non-viral delivery vehicle to adult neural stem cells using a 12-amino-acid, Tet1, which could bind specifically to neuronal cells [244]. Therefore, NPs represent potential therapeutic tools in neurodegenerative disorders by virtue of their ability to deliver miRNA treatments to deeper regions of the brain.
8. Conclusions
As important regulators of cell behavior in normal and pathological conditions, miRNAs hold immense potential for clinical applications, ranging from cancer therapy to TE approaches for the treatment of bone and cardiac defects, to neurological disorders, as described in the above paragraphs [3]. Moreover, because miRNAs inhibit/promote the expression of a multitude of genes compared with mRNA and siRNA, miRNA therapy potentially carries distinct advantages for disease treatment and regenerative medicine [25,26]. Efficient and safe delivery of miRNAs is of paramount importance for their exploitation in the clinics, as naked miRNAs are susceptible to rapid degradation and traditional transfection reagents are not suitable for in-human applications [8].
Encapsulation of miRNAs into NPs can overcome these delivery challenges, resulting in enhanced targeting efficacy and reduced off-target effects of the encapsulated payload. Importantly, the design of NPs with genetic cargo has primarily focused on siRNA, DNA, plasmids or oligonucleotides, and only few have detailed work with miRNA. In this work, we discussed the advantages of nanomedicines for miRNA delivery, reviewed the recent applications of NPs and miRNAs carriers, and highlighted potential new fields of use, which can benefit from miRNA-based therapies. The development of miRNA-loaded NPs is nascent and, often, authors describe methods for preparing NPs for miRNA delivery while referring to previous protocols, which are originally developed for drugs or other types of RNA or DNA cargo. Few studies present a thorough comparison between miRNA-loaded NPs and NPs with other RNA (for example siRNA-loaded NPs) [245]. Even so, these comparative studies only investigated the physicochemical properties of the NPs or their uptake by cells in vitro and it remains to be validated if these findings are also relevant in vivo. Indeed, as crucial biochemical differences exist between miRNAs and other nucleic acids, we should be cautious to extrapolate or predict outcomes for miRNA-loaded NPs based on the findings of other DNA/RNA-associated NPs. Nonetheless, our understanding of miRNA-loaded NPs is only beginning, and the future holds several therapeutic possibilities for miRNA-loaded NPs.
Acknowledgements
This work was supported by the MIT-POLITO grant BIOMODE – Compagnia di San Paolo (M.C., R.K., V.C.), the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (BIORECAR, grant agreement No. 772168, V.C.), National Research Foundation, Prime Minister’s Office, Singapore, under its CREATE program for the Singapore-MIT Alliance for Research and Technology BioSystems and Micromechanics IRG (S.L., R.K.), a core grant to Singapore Immunology Network (SIgN) from Agency for Science, Technology and Research (A*STAR) (G.A.), National Science Foundation, Science and Technology Center on Emergent Behaviors of Integrated Cellular Systems (CBET-0939511) (T.O., R.K.) and from the US National Cancer Institute (U01 CA214381-01) (R.K.). C.M. acknowledges funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 658665. M.C. and C.P acknowledge support from the joint “Doctorate of Bioengineering and Medical-Surgical Sciences” of University of Turin and Politecnico di Torino.
References
- 1.Wery M., Kwapisz M., Morillon A. Noncoding RNAs in gene regulation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:728–738. doi: 10.1002/wsbm.148. [DOI] [PubMed] [Google Scholar]
- 2.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J., Zhang Z. miRNA regulatory variation in human evolution. Trends Genet. 2013;29:116–124. doi: 10.1016/j.tig.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H., Shyh Chang N., Segrè A.V., Shinoda G., Shah S.P., Einhorn W.S., Takeuchi A., Engreitz J.M., Hagan J.P., Kharas M.G., Urbach A., Thornton J.E., Triboulet R., Gregory R.I., Altshuler D., Daley G.Q. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong S., Yang B., Guo H., Kang F. MicroRNAs regulate osteogenesis and chondrogenesis. Biochem. Biophys. Res. Commun. 2012;418:587–591. doi: 10.1016/j.bbrc.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 6.Luo X., Xu C., Xiao J., Shan H., Yang B., Pan Z., Lin H., Chu W., Wang Z., Lu Y. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J. Cell. Sci. 2011;124:3187. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 7.Lin L., Li N., Li D., Zhang Q., Wang C., Wang P., Tao W., Ding G., Wang Z., Zhang P., Zhuang S.-M., Zhou W., Cao X., Hou J., Liu X., Zheng L. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma. J. Biol. Chem. 2011;286:36677–36685. doi: 10.1074/jbc.M111.270561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu G., Mei L., Vishwasrao H.D., Jacobson O., Wang Z., Liu Y., Yung B.C., Fu X., Jin A., Niu G., Wang Q., Zhang F., Shroff H., Chen X. Intertwining DNA-RNA nanocapsules loaded with tumor neoantigens as synergistic nanovaccines for cancer immunotherapy. Nat. Commun. 2017;8:1482. doi: 10.1038/s41467-017-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu G., Yao W., Xiao W., Li H., Xu H., Lang B. MicroRNA-34a functions as an anti-metastatic microRNA and suppresses angiogenesis in bladder cancer by directly targeting CD44. J. Exp. Clin. Cancer Res. 2012;33 doi: 10.1186/s13046-014-0115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan L., Yu J.T., Hu N., Tan L. Non-coding RNAs in Alzheimer’s disease. Mol. Neurobiol. 2013;47:382–393. doi: 10.1007/s12035-012-8359-5. [DOI] [PubMed] [Google Scholar]
- 11.Martins M., Rosa A., Guedes L.C., Fonseca B.V., Gotovac K., Violante S., Mestre T., Coelho M., RosaMá M.M., Martin E.R., Vance J.M., Outeiro T.F., Wang L., Borovecki F., Ferreira J.J., Oliveira S.A. Convergence of mirna expression profiling, α-synuclein interacton and GWAS in Parkinson’s disease. PLoS One. 2011;6:e25443. doi: 10.1371/journal.pone.0025443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji X., Takahashi R., Hiura Y., Hirokawa G., Fukushima Y., Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin. Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 13.Dong S., Cheng Y., Yang J., Li J., Liu X., Wang X., Wang D., Krall T.J., Delphin E.S., Zhang C. MicroRNA expression signature and the role of MicroRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Rooij E., Sutherland L.B., Liu N., Williams A.H., Mcanally J., Gerard R.D., Richardson J.A., Olson E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demirtas D.A., Demirtas A.O., Biskin A., Demirtas M.A. Micro-RNA in health and disease. Int. J. Cardiol. Res. 2018;5:116–123. [Google Scholar]
- 16.Quiat D., Olson E.N. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J. Clin. Invest. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanczyk J., Leslie Pedrioli D.M., Brentano F., Sanchez-Pernaute O., Kolling C., Gay R.E., Detmar M., Gay S., Kyburz D. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008;58:1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 19.Ignacio C., Hicks S.D., Burke P., Lewis L., Szombathyne-Meszaros Z., Middleton F.A. Alterations in serum microRNA in humans with alcohol use disorders impact cell proliferation and cell death pathways and predict structural and functional changes in brain. BMC Neurosci. 2015;16:55. doi: 10.1186/s12868-015-0195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu D.C., Li Q.G., Ding X.W., Ding Y.T., Yu D.C., Li Q.G., Ding X.W., Ding Y.T. Circulating microRNAs: potential biomarkers for cancer. Int. J. Mol. Sci. 2011;12:2055–2063. doi: 10.3390/ijms12032055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y., Murray-Stewart T., Wang Y., Yu F., Li J., Marton L.J., Casero R.A.B., Oupický D. Self-immolative nanoparticles for simultaneous delivery of microRNA and targeting of polyamine metabolism in combination cancer therapy. J. Control. Release. 2016;246:110–119. doi: 10.1016/j.jconrel.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santos-Carballal B., Aaldering L.J., Ritzefeld M., Pereira S., Sewald N., Moerschbacher B.M., Götte M., Goycoolea F.M. Physicochemical and biological characterization of chitosan-microRNA nanocomplexes for gene delivery to MCF-7 breast cancer cells. Sci. Rep. 2015;5 doi: 10.1038/srep13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu F., Liao J.Z., Xiang G.Y., Zhao P.X., Ye F., Zhao Q., He X.X. MiR-101 and doxorubicin codelivered by liposomes suppressing malignant properties of hepatocellular carcinoma. Cancer Med. 2017;6:651–661. doi: 10.1002/cam4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco E., Shen H., Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015;33:941–951. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee R.C., Feinbaum R.L., Ambrost V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 28.Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 29.Lewis B.P., Burge C.B., Bartell D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denli A.M., Tops B.B.J., Plasterk R.H.A., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 32.Schwarz D.S., Hutvágner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:119–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 33.Lim L.P., Lau C.L., Nelson, Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 34.Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science (80-.) 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 36.Valencia-Sanchez M.A., Liu J., Hannon G.J., Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 37.Montgomery R.L., Hullinger T.G., Semus H.M., Dickinson B.A., Seto A.G., Lynch J.M., Stack C., Latimer P.A., Olson E.N., van Rooij E. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grueter C.E., Van Rooij E., Johnson B.A., Deleon S.M., Sutherland L.B., Qi X., Gautron L., Elmquist J.K., Bassel-Duby R., Olson E.N. A cardiac MicroRNA governs systemic energy homeostasis by regulation of MED13. Cell. 2012;149:671–683. doi: 10.1016/j.cell.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trang P., Wiggins J.F., Daige C.L., Cho C., Omotola M., Brown D., Weidhaas J.B., Bader A.G., Slack F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouchie A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013;31:577. doi: 10.1038/nbt0713-577. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y., Gao D.Y., Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira D.M., Rodrigues P.M., Borralho P.M., Rodrigues C.M.P. Delivering the promise of miRNA cancer therapeutics. Drug Discov. Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Gray G.D., Basu S., Wickstrom E. Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3’-alkylamino oligodeoxynudeotides, 2’-o-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochem. Pharmacol. 1997;53:1465–1476. doi: 10.1016/s0006-2952(97)82440-9. [DOI] [PubMed] [Google Scholar]
- 44.van Dongen S., Abreu-goodger C., Enright A.J. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loinger A., Shemla D.Y., Simon D.I., Margalit H., Biham O. Competition between small RNAs: a quantitative view. Biophys. J. 2012;102:1712–1721. doi: 10.1016/j.bpj.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu B., Zhao X., Lee L.J., Lee R.J. Targeted delivery systems for oligonucleotide therapeutics. AAPS J. 2009;11:195–203. doi: 10.1208/s12248-009-9096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberbauer R., Schreiner G.F., Meyer T.W. Renal uptake of an 18-mer phosphorothioate oligonucleotide. Kidney Int. 1995:1226–1232. doi: 10.1038/ki.1995.406. [DOI] [PubMed] [Google Scholar]
- 48.Czauderna F., Fechtner M., Dames S., Aygün H., Klippel A., Pronk G.J., Giese K., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raemdonck K., Vandenbroucke R.E., Demeester J., Sanders N.N., De Smedt S.C. Maintaining the silence: reflections on long-term RNAi. Drug Discov. Today. 2008;13:917–931. doi: 10.1016/j.drudis.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Y., Leisegang M.S., Carmichael G., Yong Jin H., Gonzalez-Martin A., Miletic A.V., Lai M., Knight S., Sabouri-Ghomi M., Head S.R., Macauley M.S., Rickert R.C., Xiao C. Transfection of microRNA mimics should be used with caution. Front. Genet. 2015;6 doi: 10.3389/fgene.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson D.W., Bracken C.P., Szubert J.M., Goodall G.J. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS One. 2013;8:55214. doi: 10.1371/journal.pone.0055214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X., He X., Liu Y., Zhang H., Chen H., Guo S., Liang Y. MiR-101-3p inhibits the growth and metastasis of non-small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT-1. Biomed. Pharmacother. 2017;93:1065–1073. doi: 10.1016/j.biopha.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Dang T.M., Wong W.C., Ong S.M., Li P., Lum J., Chen J., Poidinger M., Zolezzi F., Wong S.C. MicroRNA expression profiling of human blood monocyte subsets highlights functional differences. Immunology. 2015;145:404–416. doi: 10.1111/imm.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visvanathan J., Lee S., Lee B., Lee J.W., Lee S.K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Solingen C., Seghers L., Bijkerk R., Duijs J.M., Roeten M.K., van Oeveren‐Rietdijk A.M., Baelde H.J., Monge M., Vos J.B., de Boer H.C., Quax P.H. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J. Cell. Mol. Med. 2009;13:1577–1585. doi: 10.1111/j.1582-4934.2008.00613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X., Gu S., Chen B.F., Shen W.L., Yin Z., Xu G.W., Hu J.J., Zhu T., Li G., Wan C., Ouyang H.W., Lee T.L., Chan W.Y. Nanoparticle delivery of stable miR-199a-5p agomir improves the osteogenesis of human mesenchymal stem cells via the HIF1a pathway. Biomaterials. 2015;53:239–250. doi: 10.1016/j.biomaterials.2015.02.071. [DOI] [PubMed] [Google Scholar]
- 57.Lennox K.A., Behlke M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 58.Velpurisiva P., Gad A., Piel B., Jadia R., Rai P. Nanoparticle design strategies for effective cancer immunotherapy. J. Biomed. 2017;2:64–77. doi: 10.7150/jbm.18877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pecot C.V., Calin G.A., Coleman R.L., Lopez-Berestein G., Sood A.K. RNA interference in the clinic: challenges and future directions. Nat. Rev. Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cun D., Jensen D.K., Maltesen M.J., Bunker M., Whiteside P., Scurr D., Foged C., Nielsen H.M., Krohn D., Jonas M., Bunker M., Whiteside P., Scurr D., Foged C., Mørck H. High loading efficiency and sustained release of siRNA encapsulated in PLGA nanoparticles: quality by design optimization and characterization. Eur. J. Pharm. Biopharm. 2011;77:26–35. doi: 10.1016/j.ejpb.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 61.Decuzzi P., Godin B., Tanaka T., Lee S.Y., Chiappini C., Liu X., Ferrari M. Size and shape effects in the biodistribution of intravascularly injected particles. J. Control. Release. 2009;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 62.Kim S.H., Mok H., Jeong J.H., Kim S.W., Park T.G. Comparative evaluation of target-specific GFP gene silencing efficiencies for antisense ODN, synthetic siRNA, and siRNA plasmid complexed with PEI-PEG-FOL conjugate. Bioconjug. Chem. 2006;17:241–244. doi: 10.1021/bc050289f. [DOI] [PubMed] [Google Scholar]
- 63.Liang G., Zhu Y., Jing A., Wang J., Hu F., Feng W., Xiao Z., Chen B. Cationic microRNA-delivering nanocarriers for efficient treatment of colon carcinoma in xenograft model. Gene Ther. 2016;23:829–838. doi: 10.1038/gt.2016.60. [DOI] [PubMed] [Google Scholar]
- 64.Lee T.J., Yoo J.Y., Shu D., Li H., Zhang J., Yu J.G., Jaime-Ramirez A.C., Acunzo M., Romano G., Cui R., Sun H.L., Luo Z., Old M., Kaur B., Guo P., Croce C.M. RNA nanoparticle-based targeted therapy for glioblastoma through inhibition of oncogenic miR-21. Mol. Ther. 2017;25:1544–1555. doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caracciolo G., Caputo D., Pozzi D., Colapicchioni V., Coppola R. Size and charge of nanoparticles following incubation with human plasma of healthy and pancreatic cancer patients. Colloids Surf. B Biointerfaces. 2014;123:673–678. doi: 10.1016/j.colsurfb.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 66.Parodi A., Corbo C., Cevenini A., Molinaro R., Palomba R., Pandolfi L., Agostini M., Salvatore F., Tasciotti E. Enabling cytoplasmic delivery and organelle targeting by surface modification of nanocarriers. Nanomedicine. 2015;10:1923–1940. doi: 10.2217/nnm.15.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang L., Guo S. Nanoparticles escaping RES and endosome: challenges for siRNA delivery for cancer therapy. J. Nanomater. 2011;2011:12. [Google Scholar]
- 68.Barbero F., Russo L., Vitali M., Piella J., Salvo I., Borrajo M.L., Busquets-fité M., Grandori R., Bastús N.G., Casals E. Formation of the protein corona: the interface between nanoparticles and the immune system. Semin. Immunol. 2017;34:52–60. doi: 10.1016/j.smim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 69.Nierenberg D., Khaled A.R., Flores O. Formation of a protein corona influences the biological identity of nanomaterials. Rep. Pract. Oncol. Radiother. 2018;23:300–308. doi: 10.1016/j.rpor.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arvizo R.R., Miranda O.R., Moyano D.F., Walden C.A., Giri K., Bhattacharya R., Robertson J.D., Rotello V.M., Reid J.M., Mukherjee P. Modulating pharmacokinetics, tumor uptake and biodistribution by engineered nanoparticles. PLoS One. 2011;6:e24374. doi: 10.1371/journal.pone.0024374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He C., Hu Y., Yin L., Tang C., Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 72.Aggarwal P., Hall J.B., McLeland C.B., Dobrovolskaia M.A., McNeil S.E. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv. Drug Deliv. Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parodi A., Quattrocchi N., van de Ven A.L., Chiappini C., Evangelopoulos M., Martinez J.O., Brown B.S., Khaled S.Z., Yazdi I.K., Enzo M.V., Isenhart L., Ferrari M., Tasciotti E. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat. Nanotechnol. 2013;8:61–68. doi: 10.1038/nnano.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu C.M.J., Zhang L., Aryal S., Cheung C., Fang R.H., Zhang L. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10980–10985. doi: 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cosco D., Cilurzo F., Maiuolo J., Federico C., Di Martino M.T., Cristiano M.C., Tassone P., Fresta M., Paolino D. Delivery of miR-34a by chitosan/PLGA nanoplexes for the anticancer treatment of multiple myeloma. Sci. Rep. 2015;5:1–11. doi: 10.1038/srep17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saraiva C., Talhada D., Rai A., Ferreira R., Ferreira L., Bernardino L., Ruscher K. MicroRNA-124-loaded nanoparticles increase survival and neuronal differentiation of neural stem cells in vitro but do not contribute to stroke outcome in vivo. PLoS One. 2018;13:e0193609. doi: 10.1371/journal.pone.0193609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cai C., Xie Y., Wu L., Chen X., Liu H., Zhou Y., Zou H., Liu D., Zhao Y., Kong X., Liu P. PLGA-based dual targeted nanoparticles enhance miRNA transfection efficiency in hepatic carcinoma. Sci. Rep. 2017;7 doi: 10.1038/srep46250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ekin A., Karatas O.F., Culha M., Ozen M. Designing a gold nanoparticle-based nanocarrier for microRNA transfection into the prostate and breast cancer cells. J. Gene Med. 2014;16:331–335. doi: 10.1002/jgm.2810. [DOI] [PubMed] [Google Scholar]
- 79.Zhang T., Xue X., He D., Hsieh J.T. A prostate cancer-targeted polyarginine-disulfide linked PEI nanocarrier for delivery of microRNA. Cancer Lett. 2015;365:156–165. doi: 10.1016/j.canlet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 80.Du J., Lane L.A., Nie S. Stimuli-responsive nanoparticles for targeting the tumor microenvironment. J. Control. Release. 2015;219:205–214. doi: 10.1016/j.jconrel.2015.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rao J.P., Geckeler K.E. Polymer nanoparticles: preparation techniques and size-control parameters. Prog. Polym. Sci. 2011;36:887–913. [Google Scholar]
- 82.Lamprecht A., Ubrich N., Hombreiro Pérez M., Lehr C.M., Hoffman M., Maincent P. Influences of process parameters on nanoparticle preparation performed by a double emulsion pressure homogenization technique. Int. J. Pharm. 2000;196:177–182. doi: 10.1016/s0378-5173(99)00422-6. [DOI] [PubMed] [Google Scholar]
- 83.Lee Y.S., Johnson P.J., Robbins P.T., Bridson R.H. Production of nanoparticles-in-microparticles by a double emulsion method: a comprehensive study. Eur. J. Pharm. Biopharm. 2013;83:168–173. doi: 10.1016/j.ejpb.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 84.Mohamed A., Kunda N.K., Ross K., Hutcheon G.A., Saleem I.Y. Polymeric nanoparticles for the delivery of miRNA to treat Chronic Obstructive Pulmonary Disease (COPD) Eur. J. Pharm. Biopharm. 2019;136:1–8. doi: 10.1016/j.ejpb.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 85.Yang J., Liu H., Zhang X. Design, preparation and application of nucleic acid delivery carriers. Biotechnol. Adv. 2014;32:804–817. doi: 10.1016/j.biotechadv.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 86.Bhargava-Shah A., Foygel K., Devulapally R., Paulmurugan R. Orlistat and antisense-miRNA-loaded PLGA-PEG nanoparticles for enhanced triple negative breast cancer therapy. Nanomedicine. 2016;11:235–247. doi: 10.2217/nnm.15.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Su X., Kuang L., Battle C., Shaner T., Mitchell B.S., Fink M.J., Jayawickramarajah J. Mild two-step method to construct DNA-conjugated silicon nanoparticles: scaffolds for the detection of microRNA-21. Bioconjugate. 2014;25:1739–1743. doi: 10.1021/bc5004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Degliangeli F., Kshirsagar P., Brunetti V., Pompa P.P., Fiammengo R. Absolute and direct microRNA quantification using DNA–gold nanoparticle probes. J. Am. Chem. Soc. 2014;136:2264–2267. doi: 10.1021/ja412152x. [DOI] [PubMed] [Google Scholar]
- 89.Yang S.W., Vosch T. Rapid detection of microRNA by a silver nanocluster DNA probe. Anal. Chem. 2011;83:6935–6939. doi: 10.1021/ac201903n. [DOI] [PubMed] [Google Scholar]
- 90.Koo H., Lee S., Hee Na J., Hwa Kim S., Kwang Hahn S., Choi K., Chan Kwon I., Young Jeong S., Kim K. Bioorthogonal copper-free click chemistry in vivo for tumor-targeted delivery of nanoparticles. Angew. Chem. Int. Ed. 2012;124:12006–12010. doi: 10.1002/anie.201206703. [DOI] [PubMed] [Google Scholar]
- 91.Roode L.E., Brighton H., Bo T., Perry J.L., Parrott M.C., Kersey F., Luft J.C., Bear J.E., DeSimone J.M., Davis I.J. Subtumoral analysis of PRINT nanoparticle distribution reveals targeting variation based on cellular and particle properties. Nanomed. Nanotechnol. Biol. Med. 2016;12:1053–1062. doi: 10.1016/j.nano.2015.12.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sambade M., Deal A., Schorzman A., Luft J.C., Bowerman C., Chu K., Karginova O., Van Swearingen A., Zamboni W., DeSimone J., Anders C.K. Efficacy and pharmacokinetics of a modified acid-labile docetaxel-PRINT® nanoparticle formulation against non-small-cell lung cancer brain metastases. Nanomedicine. 2016;11:1947–1955. doi: 10.2217/nnm-2016-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herranz-Blanco B., Ginestar E., Zhang H., Hirvonen J., Santos H.A. Microfluidics platform for glass capillaries and its application in droplet and nanoparticle fabrication. Int. J. Pharm. 2017;516:100–105. doi: 10.1016/j.ijpharm.2016.11.024. [DOI] [PubMed] [Google Scholar]
- 94.Karnik R., Gu F.X., Bose S., Basto P., Cannizzaro C., Langer R., Farokhzad O.C. Microfluidic synthesis of polymeric nanoparticles. ASME 2008 6th Int. Conf. Nanochannels, Microchannels, Minichannels. 2008:1921–1922. [Google Scholar]
- 95.Chen D., Love K.T., Chen Y., Eltoukhy A.A., Kastrup C., Sahay G., Jeon A., Dong Y., Whitehead K.A., Anderson D.G. Rapid discovery of potent siRNA-containing lipid nanoparticles enabled by controlled microfluidic formulation. J. Am. Chem. Soc. 2012;134:6948–6951. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 96.Desai N. Challenges in development of nanoparticle-based therapeutics. AAPS J. 2012;14:282–295. doi: 10.1208/s12248-012-9339-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Emerich D.F., Thanos C.G. The pinpoint promise of nanoparticle-based drug delivery and molecular diagnosis. Biomol. Eng. 2006;23:171–184. doi: 10.1016/j.bioeng.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 98.Griset A.P., Walpole J., Liu R., Gaffey A., Colson Y.L., Grinstaff M.W. Expansile nanoparticles: synthesis, characterization, and in vivo efficacy of an acid-responsive polymeric drug delivery system. J. Am. Chem. Soc. 2009;131:2469–2471. doi: 10.1021/ja807416t. [DOI] [PubMed] [Google Scholar]
- 99.Ghorbani M., Hamishehkar H., Arsalani N., Akbar A. A novel dual-responsive core-crosslinked magnetic-gold nanogel for triggered drug release. Mater. Sci. Eng. C. 2016;68:436–444. doi: 10.1016/j.msec.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 100.Lee H.J., Namgung R., Kim W.J., Kim J.I., Park I.K. Targeted delivery of microRNA-145 to metastatic breast cancer by peptide conjugated branched PEI gene carrier. Macromol. Res. 2013;21:1201–1209. [Google Scholar]
- 101.Ibrahim A.F., Weirauch U., Thomas M., Grun̈weller A., Hartmann R.K., Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinom. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 102.Liu Q., Li R.T., Qian H.Q., Wei J., Xie L., Shen J., Yang M., Qian X.P., Yu L.X., Jiang X.Q., Liu B.R. Targeted delivery of miR-200c/DOC to inhibit cancer stem cells and cancer cells by the gelatinases-stimuli nanoparticles. Biomaterials. 2013;34:7191–7203. doi: 10.1016/j.biomaterials.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 103.Chiou G.Y., Cherng J.Y., Hsu H.S., Wang M.L., Tsai C.M., Lu K.H., Chien Y., Hung S.C., Chen Y.W., Wong C.I., Tseng L.M., Huang P.I., Yu C.C., Hsu W.H., Chiou S.H. Cationic polyurethanes-short branch PEI-mediated delivery of Mir145 inhibited epithelial–mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. J. Control. Release. 2012;159:240–250. doi: 10.1016/j.jconrel.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 104.Yang Y.P., Chien Y., Chiou G.Y., Cherng J.Y., Wang M.L., Lo W.-L., Chang Y.L., Huang P.I., Chen Y.W., Shih Y.H., Chen M.T., Chiou S.H. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials. 2012;33:1462–1476. doi: 10.1016/j.biomaterials.2011.10.071. [DOI] [PubMed] [Google Scholar]
- 105.Venkiteswaran S., Thomas T., Thomas T.J. Selectivity of polyethyleneimines on DNA nanoparticle preparation and gene transport. ChemistrySelect. 2016;1:1144–1150. [Google Scholar]
- 106.Bieber T., Elsässer H.-P. Preparation of a low molecular weight polyethylenimine for efficient cell transfection. Biotechniques. 2001;30:74–81. doi: 10.2144/01301st03. [DOI] [PubMed] [Google Scholar]
- 107.Sun C., Tang T., Uludağ H., Cuervo J.E. Molecular dynamics simulations of DNA/PEI complexes: effect of PEI branching and protonation state. Biophys. J. 2011;100:2754–2763. doi: 10.1016/j.bpj.2011.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Höbel S., Aigner A. Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013;5:484–501. doi: 10.1002/wnan.1228. [DOI] [PubMed] [Google Scholar]
- 109.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 110.Jokerst J.V., Lobovkina T., Zare R.N., Gambhir S.S. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond.) 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Danhier F., Ansorena E., Silva J.M., Coco R., Le Breton A., Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J. Control. Release. 2012;161:505–522. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 112.Wong S.Y., Pelet J.M., Putnam D. Polymer systems for gene delivery—past, present, and future. Prog. Polym. Sci. 2007;32:799–837. [Google Scholar]
- 113.Serrano M.C., Pagani R., Vallet-Reg M., Penã J., Izquierdo I., Portoles M.T. In vitro biocompatibility assessment of poly (ε-caprolactone) films using L929 mouse fibroblasts. Biomaterials. 2004;25:5603–5611. doi: 10.1016/j.biomaterials.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 114.Grizzi I., Garreau H., Li S., Vert M. Hydrolytic degradation of devices based on poly (dl-lactic acid) size-dependence. Biomaterials. 1995;16:305–311. doi: 10.1016/0142-9612(95)93258-f. [DOI] [PubMed] [Google Scholar]
- 115.Patil Y., Panyam J. Polymeric nanoparticles for siRNA delivery and gene silencing. Int. J. Pharm. 2009;367:195–203. doi: 10.1016/j.ijpharm.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yuan X., Shah B.A., Kotadia N.K., Li J., Gu H., Wu Z. The development and mechanism studies of cationic chitosan-modified biodegradable PLGA nanoparticles for efficient siRNA drug delivery. Pharm. Res. 2010;27:1285–1295. doi: 10.1007/s11095-010-0103-0. [DOI] [PubMed] [Google Scholar]
- 117.Wang S., Zhang J., Wang Y., Chen M. Hyaluronic acid-coated PEI-PLGA nanoparticles mediated co-delivery of doxorubicin and miR-542-3p for triple negative breast cancer therapy. Nanomed. Nanotechnol. Biol. Med. 2016;12:411–420. doi: 10.1016/j.nano.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 118.Grossen P., Witzigmann D., Sieber S., Huwyler J. PEG-PCL-based nanomedicines: a biodegradable drug delivery system and its application. J. Control. Release. 2017;260:46–60. doi: 10.1016/j.jconrel.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 119.Mattu C., Pabari R.M., Boffito M., Sartori S., Ciardelli G., Ramtoola Z. Comparative evaluation of novel biodegradable nanoparticles for the drug targeting to breast cancer cells. Eur. J. Pharm. Biopharm. 2013;85(3):463–472. doi: 10.1016/j.ejpb.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 120.Prabaharan M., Mano J.F. Chitosan-based particles as controlled drug delivery systems. Drug Deliv. 2004;12:41–57. doi: 10.1080/10717540590889781. [DOI] [PubMed] [Google Scholar]
- 121.Hu L., Sun Y., Wu Y. Advances in chitosan-based drug delivery vehicles. Nanoscale. 2013;5:3103. doi: 10.1039/c3nr00338h. [DOI] [PubMed] [Google Scholar]
- 122.Bernkop-Schnürch A., Dünnhaupt S. Chitosan-based drug delivery systems. Eur. J. Pharm. Biopharm. 2012;81:463–469. doi: 10.1016/j.ejpb.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 123.Costa D.F., Torchilin V.P. Micelle-like nanoparticles as siRNA and miRNA carriers for cancer therapy. Biomed. Microdev. 2018;20 doi: 10.1007/s10544-018-0298-0. [DOI] [PubMed] [Google Scholar]
- 124.Bravo-Anaya L.M., Fernández-Solís K.G., Rosselgong J., Nano-Rodríguez J.L.E., Carvajal F., Rinaudo M. Chitosan-DNA polyelectrolyte complex: influence of chitosan characteristics and mechanism of complex formation. Int. J. Biol. Macromol. 2019;126:1037–1049. doi: 10.1016/j.ijbiomac.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 125.Layek B., Lipp L., Singh J. Cell penetrating peptide conjugated chitosan for enhanced delivery of nucleic acid. Int. J. Mol. Sci. 2015;16:28912–28930. doi: 10.3390/ijms161226142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nydert P., Dragomir A., Hjelte L. Chitosan as a carrier for non-viral gene transfer in a cystic-fibrosis cell line. Biotechnol. Appl. Biochem. 2008;51:153–157. doi: 10.1042/BA20070197. [DOI] [PubMed] [Google Scholar]
- 127.Cosco D., Federico C., Maiuolo J., Bulotta S., Molinaro R., Paolino D., Tassone P., Fresta M. Physicochemical features and transfection properties of chitosan/poloxamer 188/poly(d,l-lactide-co-glycolide) nanoplexes. Int. J. Nanomed. 2014;9:2359–2372. doi: 10.2147/IJN.S58362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sonia T.A., Sharma C.P. Chitosan and its derivatives for drug delivery perspective. In: Jayakumar R., Muzzarelli R.A.A., Prabaharan M., editors. Chitosan Biomater. I. Springer; Berlin, Heidelberg: 2011. pp. 23–53. [Google Scholar]
- 129.Layek B., Singh J. N-Hexanoyl, N-octanoyl and N-decanoyl chitosans: Binding affinity, cell uptake, and transfection. Carbohydr. Polym. 2012;89:403–410. doi: 10.1016/j.carbpol.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 130.Chae S.Y., Son S., Lee M., Jang M.K., Nah J.W. Deoxycholic acid-conjugated chitosan oligosaccharide nanoparticles for efficient gene carrier. J. Control. Release. 2005;109:330–344. doi: 10.1016/j.jconrel.2005.09.040. [DOI] [PubMed] [Google Scholar]
- 131.Mandke R., Singh J. Effect of acyl chain length and unsaturation on physicochemical properties and transfection efficiency of N-acyl-substituted low-molecular-weight chitosan. J. Pharm. Sci. 2012;101:268–282. doi: 10.1002/jps.22767. [DOI] [PubMed] [Google Scholar]
- 132.Dicker K.T., Gurski L.A., Pradhan-Bhatt S., Witt R.L., Farach-Carson M.C., Jia X. Hyaluronan: a simple polysaccharide with diverse biological functions. Acta Biomater. 2014;10:1558–1570. doi: 10.1016/j.actbio.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cadete A., Alonso M.J. Targeting cancer with hyaluronic acid-based nanocarriers: recent advances and translational perspectives. Nanomedicine. 2016;11:2341–2357. doi: 10.2217/nnm-2016-0117. [DOI] [PubMed] [Google Scholar]
- 134.Ganesh S., Iyer A.K., Morrissey D.V., Amiji M.M. Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials. 2013;34:3489–3502. doi: 10.1016/j.biomaterials.2013.01.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Landesman-Milo D., Goldsmith M., Ben-Arye S.L., Witenberg B., Brown E., Leibovitch S., Azriel S., Tabak S., Morad V., Peer D. Hyaluronan grafted lipid-based nanoparticles as RNAi carriers for cancer cells. Cancer Lett. 2013;334:221–227. doi: 10.1016/j.canlet.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 136.Sun X., Zhang N. Cationic polymer optimization for efficient gene delivery. Mini Rev. Med. Chem. 2010;10:108–125. doi: 10.2174/138955710791185109. [DOI] [PubMed] [Google Scholar]
- 137.Cohen Z.R., Ramishetti S., Peshes-Yaloz N., Goldsmith M., Wohl A., Zibly Z., Peer D. Localized RNAi therapeutics of chemoresistant grade IV glioma using hyaluronan-grafted lipid-based nanoparticles. ACS Nano. 2015;9:1581–1591. doi: 10.1021/nn506248s. [DOI] [PubMed] [Google Scholar]
- 138.Haussecker D. Current issues of RNAi therapeutics delivery and development. J. Control. Release. 2014;195:49–54. doi: 10.1016/j.jconrel.2014.07.056. [DOI] [PubMed] [Google Scholar]
- 139.Lee M.S., Lee J.E., Byun E., Kim N.W., Lee K., Lee H., Sim S.J., Lee D.S., Jeong J.H. Target-specific delivery of siRNA by stabilized calcium phosphate nanoparticles using dopa–hyaluronic acid conjugate. J. Control. Release. 2014;192:122–130. doi: 10.1016/j.jconrel.2014.06.049. [DOI] [PubMed] [Google Scholar]
- 140.Sekhon B.S., Kamboj S.R. Inorganic nanomedicine-part 1. Nanomed. Nanotechnol. Biol. Med. 2010;6:516–522. doi: 10.1016/j.nano.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 141.Sekhon B.S., Kamboj S.R. Inorganic nanomedicine-part 2. Nanomed. Nanotechnol. Biol. Med. 2010;6:612–618. doi: 10.1016/j.nano.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 142.Mulens V., del P. Morales M., Barber D.F. Development of magnetic nanoparticles for cancer gene therapy: a comprehensive review. ISRN Nanomater. 2013;2013:1–14. [Google Scholar]
- 143.Schade A., Delyagina E., Scharfenberg D., Skorska A., Lux C., David R., Steinhoff G. Innovative strategy for microRNA delivery in human mesenchymal stem cells via magnetic nanoparticles. Int. J. Mol. Sci. 2013;14:10710–10726. doi: 10.3390/ijms140610710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yin P.T., Shah B.P., Lee K.B. Combined magnetic nanoparticle‐based microRNA and hyperthermia therapy to enhance apoptosis in brain cancer cells. Small. 2014;10:4106–4112. doi: 10.1002/smll.201400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cha W., Fan R., Miao Y., Zhou Y., Qin C., Shan X., Wan X., Li J. Mesoporous silica nanoparticles as carriers for intracellular delivery of nucleic acids and subsequent therapeutic applications. Molecules. 2017;22 doi: 10.3390/molecules22050782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bertucci A., Prasetyanto E.A., Septiadi D., Manicardi A., Brognara E., Gambari R., Corradini R., De Cola L. Combined delivery of temozolomide and anti‐miR221 PNA using mesoporous silica nanoparticles induces apoptosis in resistant glioma cells. Small. 2015;11:5687–5695. doi: 10.1002/smll.201500540. [DOI] [PubMed] [Google Scholar]
- 147.Tivnan A., Orr W.S., Gubala V., Nooney R., Williams D.E., McDonagh C., Prenter S., Harvey H., Domingo-Fernández R., Bray I.M., Piskareva O., Ng C.Y., Lode H.N., Davidoff A.M., Stallings R.L. Inhibition of neuroblastoma tumor growth by targeted delivery of MicroRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS One. 2012;7:e38129. doi: 10.1371/journal.pone.0038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Banik M., Basu T. Calcium phosphate nanoparticles: a study of their synthesis, characterization and mode of interaction with salmon testis DNA. Dalton Trans. 2014;43:3244–3259. doi: 10.1039/c3dt52522h. [DOI] [PubMed] [Google Scholar]
- 149.Jung H., Kim S.A., Yang Y.G., Yoo H., Lim S.J., Mok H. Long chain microRNA conjugates in calcium phosphate nanoparticles for efficient formulation and delivery. Arch. Pharm. Res. 2015;38:705–715. doi: 10.1007/s12272-014-0451-0. [DOI] [PubMed] [Google Scholar]
- 150.Mieszawska A.J., Mulder W.J.M., Fayad Z.A., Cormode D.P. Multifunctional gold nanoparticles for diagnosis and therapy of disease. Mol. Pharm. 2013;10:831–847. doi: 10.1021/mp3005885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Connor E.E., Mwamuka J., Gole A., Murphy C.J., Wyatt M.D. Gold nanoparticles are taken up by human cells but do not cause acute cytotoxicity. Small. 2005;1:325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- 152.Jain P.K., Lee K.S., El-Sayed I.H., El-Sayed M.A. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. J. Phys. Chem. B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 153.Ghosh R., Singh L.C., Shohet J.M., Gunaratne P.H. A gold nanoparticle platform for the delivery of functional microRNAs into cancer cells. Biomaterials. 2013;34:807–816. doi: 10.1016/j.biomaterials.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 154.Couvreur P., Barret G. Critical review in therapeutic drug carrier System. J. Pediatr. Surg. 2005;5:1–20. [PubMed] [Google Scholar]
- 155.Zhao Y., Huang L. Adv. Genet. Academic Press; 2014. Lipid nanoparticles for gene delivery; pp. 13–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 157.Cheng X., Lee R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016;99:129–137. doi: 10.1016/j.addr.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 158.Hsu S.H., Yu B., Wang X., Lu Y., Schmidt C.R., Lee R.J., Lee L.J., Jacob S.T., Ghoshal K. Cationic lipid nanoparticles for therapeutic delivery of siRNA and miRNA to murine liver tumor. Nanomed. Nanotechnol. Biol. Med. 2013;9:1169–1180. doi: 10.1016/j.nano.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bejerano T., Etzion S., Elyagon S., Etzion Y., Cohen S. Nanoparticle delivery of miRNA-21 mimic to cardiac macrophages improves myocardial remodeling after myocardial infarction. Nano Lett. 2018;18:5885–5891. doi: 10.1021/acs.nanolett.8b02578. [DOI] [PubMed] [Google Scholar]
- 160.Zhou Y.L., Yang Q.Q., Yan Y.Y., Zhu C., Zhang L., Tang J.B. Localized delivery of miRNAs targets cyclooxygenases and reduces flexor tendon adhesions. Acta Biomater. 2018;70:237–248. doi: 10.1016/j.actbio.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 161.Zhang X., Li Y., Chen Y.E., Chen J., Ma P.X. Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects. Nat. Commun. 2016;7:1–15. doi: 10.1038/ncomms10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang J., Brown M.E., Zhang H., Martinez M., Zhao Z., Bhutani S., Yin S., Trac D., Xi J.J., Davis M.E. High-throughput screening identifies microRNAs that target Nox2 and improve function after acute myocardial infarction. Am. J. Physiol. Circ. Physiol. 2017;312:H1002–H1012. doi: 10.1152/ajpheart.00685.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kouri F.M., Hurley L.A., Daniel W.L., Day E.S., Hua Y., Hao L., Peng C.Y., Merkel T.J., Queisser M.A., Ritner C., Zhang H. miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev. 2015;29:732–745. doi: 10.1101/gad.257394.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Costa P.M., Cardoso A.L., Custódia C., Cunha P., Pereira De Almeida L., Pedroso De Lima M.C. MiRNA-21 silencing mediated by tumor-targeted nanoparticles combined with sunitinib: a new multimodal gene therapy approach for glioblastoma. J. Control. Release. 2015;207:31–39. doi: 10.1016/j.jconrel.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 165.Cui X., Sun Y., Shen M., Song K., Yin X., Di W., Duan Y. Enhanced chemotherapeutic efficacy of paclitaxel nanoparticles Co-delivered with MicroRNA-7 by inhibiting paclitaxel-induced EGFR/ERK pathway activation for ovarian cancer therapy. ACS Appl. Mater. Interfaces. 2018;10:7821–7831. doi: 10.1021/acsami.7b19183. [DOI] [PubMed] [Google Scholar]
- 166.Paoletti C., Divieto C., Chiono V. Impact of biomaterials on differentiation and reprogramming approaches for the generation of functional cardiomyocytes. Cells. 2018;7:114. doi: 10.3390/cells7090114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang L.L., Liu Y., Chung J.J., Wang T., Gaffey A.C., Lu M., Cavanaugh C.A., Zhou S., Kanade R., Atluri P., Morrisey E.E., Burdick J.A. Sustained miRNA delivery from an injectable hydrogel promotes cardiomyocyte proliferation and functional regeneration after ischaemic injury. Nat. Biomed. Eng. 2017;1:983–992. doi: 10.1038/s41551-017-0157-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Jayawardena T.M., Egemnazarov B., Finch E.A., Zhang L., Payne J.A., Pandya K., Zhang Z., Rosenberg P., Mirotsou M., Dzau V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Di Mauro V., Iafisco M., Salvarani N., Vacchiano M., Carullo P., Ramírez-Rodríguez G.B., Patrício T., Tampieri A., Miragoli M., Catalucci D. Bioinspired negatively charged calcium phosphate nanocarriers for cardiac delivery of MicroRNAs. Nanomedicine. 2016;11:891–906. doi: 10.2217/nnm.16.26. [DOI] [PubMed] [Google Scholar]
- 170.Barnett R.E., Conklin D.J., Ryan L., Keskey R.C., Ramjee V., Sepulveda E.A., Srivastava S., Bhatnagar A., Cheadle W.G. Anti-inflammatory effects of miR-21 in the macrophage response to peritonitis. J. Leukoc. Biol. 2016;99(2):361–371. doi: 10.1189/jlb.4A1014-489R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nakasa T., Yoshizuka M., Andry Usman M., Elbadry Mahmoud E., Ochi M. MicroRNAs and bone regeneration. Curr. Genomics. 2015;16:441–452. doi: 10.2174/1389202916666150817213630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhao L., Wu J., Xu M., Liu W., Song J., Yan S., Gao L., Zhang Y. Induction of osteogenic differentiation of stem cells via a lyophilized microRNA reverse transfection formulation on a tissue culture plate. Int. J. Nanomed. 2013;8:1595. doi: 10.2147/IJN.S43244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang Z., Wu G., Wei M., Liu Q., Zhou J., Qin T., Feng X., Liu H., Feng Z., Zhao Y. Improving the osteogenesis of human bone marrow mesenchymal stem cell sheets by microRNA-21-loaded chitosan/hyaluronic acid nanoparticles via reverse transfection. Int. J. Nanomed. 2016;11:2091. doi: 10.2147/IJN.S104851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Oliveto S., Mancino M., Manfrini N., Biffo S. Role of microRNAs in translation regulation and cancer. World J. Biol. Chem. 2017;8:45. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Conde J., Edelman E.R., Artzi N. Target-responsive DNA/RNA nanomaterials for microRNA sensing and inhibition: The jack-of-all-trades in cancer nanotheranostics? Adv. Drug Deliv. Rev. 2015;81:169–183. doi: 10.1016/j.addr.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 176.Jin Y.Y., Andrade J., Wickstrom E. Non-specific blocking of MIR-17-5p guide strand in triple negative breast cancer cells by amplifying passenger strand activity. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0142574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Aaldering L.J., Tayeb H., Krishnan S., Fletcher S., Wilton S.D., Veedu R.N. Smart functional nucleic acid chimeras: enabling tissue specific RNA targeting therapy. RNA Biol. 2015;12:412–425. doi: 10.1080/15476286.2015.1017234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Li N., Zhao X., Wang L., Zhang S., Cui M., He J. miR-494 suppresses tumor growth of epithelial ovarian carcinoma by targeting IGF1R. Tumor Biol. 2016;37:7767–7776. doi: 10.1007/s13277-015-4603-8. [DOI] [PubMed] [Google Scholar]
- 179.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C., Vonderheide R.H., Pittet M.J., Jain R.K., Zou W., Howcroft T.K., Woodhouse E.C., Weinberg R.A., Krummel M.F. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Valkenburg K.C., De Groot A.E., Pienta K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018;15:366–381. doi: 10.1038/s41571-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Deng X., Cao M., Zhang J., Hu K., Yin Z., Zhou Z., Xiao X., Yang Y., Sheng W., Wu Y., Zeng Y. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 182.Xu S., Nasr S.H., Chen D., Zhang X., Sun L., Huang X., Qian C. MiRNA extraction from cell-free biofluid using protein corona formed around carboxyl magnetic nanoparticles. ACS Biomater. Sci. Eng. 2017;4:654–662. doi: 10.1021/acsbiomaterials.7b00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Agnihotri S.A., Mallikarjuna N.N., Aminabhavi T.M. Recent advances on chitosan-based micro- and nanoparticles in drug delivery. J. Control. Release. 2004;100:5–28. doi: 10.1016/j.jconrel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 184.Trivedi M., Singh A., Talekar M., Pawar G., Shah P., Amiji M. MicroRNA-34a encapsulated in hyaluronic acid nanoparticles induces epigenetic changes with altered mitochondrial bioenergetics and apoptosis in non-small-cell lung cancer cells. Sci. Rep. 2017;7:1–17. doi: 10.1038/s41598-017-02816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Poliseno L., Pandolfi P.P. PTEN ceRNA networks in human cancer. Methods. 2015;77:41–50. doi: 10.1016/j.ymeth.2015.01.013. [DOI] [PubMed] [Google Scholar]
- 186.Saadatpour L., Fadaee E., Fadaei S., Mansour R.N., Mohammadi M., Mousavi S.M., Goodarzi M., Verdi J., Mirzaei H. Glioblastoma: exosome and microRNA as novel diagnosis biomarkers. Cancer Gene Ther. 2016;23:415. doi: 10.1038/cgt.2016.48. [DOI] [PubMed] [Google Scholar]
- 187.Wang G., Wang J.J., Tang H.M., To S.S.T. Targeting strategies on miRNA-21 and PDCD4 for glioblastoma. Arch. Biochem. Biophys. 2015;580:64–74. doi: 10.1016/j.abb.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 188.Yang Y. Cancer immunotherapy: harnessing the immune system to battle cancer. J. Clin. Invest. 2015;125:3335–3337. doi: 10.1172/JCI83871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Self-Fordham J.B., Naqvi A.R., Uttamani J.R., Kulkarni V., Nares S. MicroRNA: dynamic regulators of macrophage polarization and plasticity. Front. Immunol. 2017;8:1–18. doi: 10.3389/fimmu.2017.01062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Essandoh K., Li Y., Huo J., Fan G.C. MiRNA-mediated macrophage polarization and its potential role in the regulation of inflammatory response. Shock. 2016;46:122–131. doi: 10.1097/SHK.0000000000000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ge Y., Zhang L., Nikolova M., Reva B., Fuchs E. Strand-specific in vivo screen of cancer-associated miRNAs unveils a role for miR-21 in SCC progression. Nat. Cell Biol. 2015;18:111–121. doi: 10.1038/ncb3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Murphy A.J., Guyre P.M., Pioli P.A. Estradiol suppresses NF-κB activation through coordinated regulation of let-7a and miR-125b in primary human macrophages. J. Immunol. 2010;184:5029–5037. doi: 10.4049/jimmunol.0903463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF- B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Huang C., Liu X.J., QunZhou, Xie J., Ma T.T., Meng X.M., Li J. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. Int. Immunopharmacol. 2016;32:46–54. doi: 10.1016/j.intimp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 195.Chaudhuri A.A., So A.Y.L., Sinha N., Gibson W.S.J., Taganov K.D., O’Connell R.M., Baltimore D. Mir-125b potentiates macrophage activation. J. Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Meghna Talekar A.O. Macrophage polarization and the effect of MicroRNA-155 administered in water-in-oil-in-water multiple emulsion formulations. J. Clin. Cell. Immunol. 2015;6(2):1–7. [Google Scholar]
- 197.Martinez-Nunez R.T., Louafi F., Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor α1 (IL13Rα1) J. Biol. Chem. 2011;286:1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.O’Connell R.M., Taganov K.D., Boldin M.P., Cheng G., Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gun S.Y., Lee S.W.L., Sieow J.L., Wong S.C. Targeting immune cells for cancer therapy. Redox Biol. 2019:101174. doi: 10.1016/j.redox.2019.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Shao K., Singha S., Clemente-Casares X., Tsai S., Yang Y., Santamaria P. Nanoparticle-based immunotherapy for cancer. J. Am. Chem. Soc. 2018;11:40. doi: 10.1021/nn5062029. [DOI] [PubMed] [Google Scholar]
- 201.Fang R.H., Zhang L. Nanoparticle-based modulation of the immune system. Annu. Rev. Chem. Biomol. Eng. 2016;7:305–326. doi: 10.1146/annurev-chembioeng-080615-034446. [DOI] [PubMed] [Google Scholar]
- 202.Silva J.M., Zupancic E., Vandermeulen G., Oliveira V.G., Salgado A., Videira M., Gaspar M., Graca L., Préat V., Florindo H.F. In vivo delivery of peptides and Toll-like receptor ligands by mannose-functionalized polymeric nanoparticles induces prophylactic and therapeutic anti-tumor immune responses in a melanoma model. J. Control. Release. 2015;198:91–103. doi: 10.1016/j.jconrel.2014.11.033. [DOI] [PubMed] [Google Scholar]
- 203.Dolina J.S., Sung S.S.J., Novobrantseva T.I., Nguyen T.M., Hahn Y.S. Lipidoid nanoparticles containing PD-L1 siRNA delivered in vivo enter Kupffer cells and enhance NK and CD8+ T cell-mediated hepatic antiviral immunity. Mol. Ther. - Nucleic Acids. 2013;2:e72. doi: 10.1038/mtna.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Heo M.B., Lim Y.T. Programmed nanoparticles for combined immunomodulation, antigen presentation and tracking of immunotherapeutic cells. Biomaterials. 2014;35:590–600. doi: 10.1016/j.biomaterials.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 205.Choi M.R., Stanton-maxey K.J., Stanley J.K., Levin C.S., Bardhan R., Akin D., Badve S., Sturgis J., Robinson J.P., Bashir R., Halas N.J., Clare S.E. A cellular Trojan horse for delivery of therapeutic nanoparticles into tumors. Nano Lett. 2007;7:3759–3765. doi: 10.1021/nl072209h. [DOI] [PubMed] [Google Scholar]
- 206.Choi M.R., Bardhan R., Stanton-Maxey K.J., Badve S., Nakshatri H., Stantz K.M., Cao N., Halas N.J., Clare S.E. Delivery of nanoparticles to brain metastases of breast cancer using a cellular Trojan horse. Cancer Nanotechnol. 2012;3:47–54. doi: 10.1007/s12645-012-0029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Anselmo A.C., Gilbert J.B., Kumar S., Gupta V., Cohen R.E., Rubner M.F., Mitragotri S. Monocyte-mediated delivery of polymeric backpacks to inflamed tissues: a generalized strategy to deliver drugs to treat inflammation. J. Control. Release. 2015;199:29–36. doi: 10.1016/j.jconrel.2014.11.027. [DOI] [PubMed] [Google Scholar]
- 208.Doshi N., Swiston A.J., Gilbert J.B., Alcaraz M.L., Cohen R.E., Rubner M.F., Mitragotri S. Cell-based drug delivery devices using phagocytosis-resistant backpacks. Adv. Healthc. Mater. 2011;23:105–109. doi: 10.1002/adma.201004074. [DOI] [PubMed] [Google Scholar]
- 209.Song Y., Tang C., Yin C. Combination antitumor immunotherapy with VEGF and PIGF siRNA via systemic delivery of multi-functionalized nanoparticles to tumor-associated macrophages and breast cancer cells. Biomaterials. 2018;185:117–132. doi: 10.1016/j.biomaterials.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 210.Steinfeld U., Pauli C., Kaltz N., Bergemann C., Lee H. T lymphocytes as potential therapeutic drug carrier for cancer treatment. Int. J. Pharm. 2006;311:229–236. doi: 10.1016/j.ijpharm.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 211.Zupke O., Distler E., Jurchott A., Paiphansiri U., Dass M., Thomas S., Hartwig F., Theobald M., Landfester K., Mailander V., Herr W. Nanoparticles and antigen-specific T-cell therapeutics : a comprehensive study on uptake and release. Nanomedicine. 2015;10:1063–1076. doi: 10.2217/nnm.14.160. Sections View Article. [DOI] [PubMed] [Google Scholar]
- 212.Siriwon N., Kim Y.J., Siegler E., Chen X., Rohrs J.A., Liu Y., Wang P. CAR-T cells surface-engineered with drug-encapsulated nanoparticles can ameliorate intratumoral T-cell hypofunction. Cancer Immunol. Res. 2018;6:812–825. doi: 10.1158/2326-6066.CIR-17-0502. [DOI] [PubMed] [Google Scholar]
- 213.Li S.Y., Liu Y., Xu C.F., Shen S., Sun R., Du X.J., Xia J.X., Zhu Y.H., Wang J. Restoring anti-tumor functions of T cells via nanoparticle-mediated immune checkpoint modulation. J. Control. Release. 2016;231:17–28. doi: 10.1016/j.jconrel.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 214.Smith T.T., Stephan S.B., Moffett H.F., Mcknight L.E., Reiman D., Bonagofski E., Wohlfahrt M.E., Pillai S.P.S., Stephan M.T. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat. Nanotechnol. 2017;12:813–820. doi: 10.1038/nnano.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Quezada S.A., Simpson T.R., Peggs K.S., Merghoub T., Vider Jelena, Fan X., Blasberg R., Yagita H., Muranski P., Antony P.A., Restifo N.P., Allison J.P. Tumor-reactive CD4+ T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J. Exp. Med. 2010;207:637–650. doi: 10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Warashina S., Nakamura T., Sato Y., Fujiwara Y., Hyodo M., Hatakeyama H., Harashima H. A lipid nanoparticle for the efficient delivery of siRNA to dendritic cells. J. Control. Release. 2016;225:183–191. doi: 10.1016/j.jconrel.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 217.Kranz L.M., Diken M., Haas H., Kreiter S., Loquai C., Reuter K.C., Meng M., Fritz D., Vascotto F., Hefesha H., Grunwitz C., Vormehr M., Hüsemann Y., Selmi A., Kuhn A.N., Buck J., Derhovanessian E., Rae R., Attig S., Diekmann J., Jabulowsky R.A., Heesch S., Hassel J., Langguth P., Grabbe S., Huber C., Türeci Ö., Sahin U. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534:396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 218.Bilen J., Liu N., Bonini N.M. A new role for microRNA pathways: modulation of degeneration induced by pathogenic human disease proteins. Cell Cycle. 2006;5:2835–2838. doi: 10.4161/cc.5.24.3579. [DOI] [PubMed] [Google Scholar]
- 219.Johnson R., Noble W., Tartaglia G.G., Buckley N.J. Neurodegeneration as an RNA disorder. Prog. Neurobiol. 2012;99:293–315. doi: 10.1016/j.pneurobio.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Karnati H.K., Panigrahi M.K., Gutti R.K., Greig N.H., Tamargo I.A. miRNAs: key players in neurodegenerative disorders and epilepsy. J. Alzheimers Dis. 2015;48:563–580. doi: 10.3233/JAD-150395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Doxakis E. Post-transcriptional regulation of α-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 2010;285:12726–12734. doi: 10.1074/jbc.M109.086827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang G., van der Walt J.M., Mayhew G., Li Y.J., Züchner S., Scott W.K., Martin E.R., Vance J.M. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of α-synuclein. Am. J. Hum. Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Miñones-Moyano E., Porta S., Escaramís G., Rabionet R., Iraola S., Kagerbauer B., Espinosa-Parrilla Y., Ferrer I., Estivill X., Martí E. MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 224.Kim J., Inoue K., Ishii J., Vanti W.B., Voronov S.V., Murchison E., Hannon G., Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science (80-.) 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.de Mena L., Coto E., Cardo L.F., Díaz M., Blázquez M., Ribacoba R., Salvador C., Pastor P., Samaranch L., Moris G., Menéndez M., Corao A.I., Alvarez V. Analysis of the Micro-RNA-133 and PITX3 genes in Parkinson’s disease. Am. J. Med. Genet. Part B: Neuropsychiatr. Genet. 2010;153:1234–1239. doi: 10.1002/ajmg.b.31086. [DOI] [PubMed] [Google Scholar]
- 226.Boissonneault V., Plante I., Rivest S., Provost P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J. Biol. Chem. 2009;284:1971–1981. doi: 10.1074/jbc.M807530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhu H.C., Wang L.M., Wang M., Song B., Tan S., Teng J.F., Duan D.X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res. Bull. 2012;88:596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 228.Hébert S.S., De Strooper B. miRNAs in neurodegeneration. Science (80-.) 2007;317:1179–1180. doi: 10.1126/science.1148530. [DOI] [PubMed] [Google Scholar]
- 229.Wang W.X., Rajeev B.W., Stromberg A.J., Ren N., Tang G., Huang Q., Rigoutsos I., Nelson P.T. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hébert S.S., De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 231.Delay C., Calon F., Mathews P., Hébert S.S. Alzheimer-specific variants in the 3’UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 2011;6:70. doi: 10.1186/1750-1326-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yao J., Hennessey T., Flynt A., Lai E., Beal M.F., Lin M.T. MicroRNA-related cofilin abnormality in Alzheimer’s disease. PLoS One. 2010;5:e15546. doi: 10.1371/journal.pone.0015546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Geekiyanage H., Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in Sporadic Alzheimer’s disease. J. Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Smith P.Y., Delay C., Girard J., Papon M.A., Planel E., Sergeant N., Buée L., Hébert S.S. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet. 2011;20:4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- 235.Wang H., Moyano A.L., Ma Z., Deng Y., Lin Y., Zhao C., Zhang L., Jiang M., He X., Ma Z., Lu F., Xin M., Zhou W., Yoon S.O., Bongarzone E.R., Lu Q.R. miR-219 cooperates with miR-338 in myelination and promotes myelin repair in the CNS. Dev. Cell. 2017;40 doi: 10.1016/j.devcel.2017.03.001. 566–582.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Sinha M., Ghose J., Bhattarcharyya N.P. Micro RNA -214,-150,-146a and-125b target Huntingtin gene. RNA Biol. 2011;8:1005–1021. doi: 10.4161/rna.8.6.16035. [DOI] [PubMed] [Google Scholar]
- 237.Colella P., Ronzitti G., Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. - Methods Clin. Dev. 2018;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Miyazaki Y., Adachi H., Katsuno M., Minamiyama M., Jiang Y.M., Huang Z., Doi H., Matsumoto S., Kondo N., Iida M., Tohnai G. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat. Med. 2012;18:1136. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 239.Fukuoka M., Takahashi M., Fujita H., Chiyo T., Popiel H.A., Watanabe S., Furuya H., Murata M., Wada K., Okada T., Nagai Y., Hohjoh H. Supplemental treatment for Huntington’s disease with miR-132 that is deficient in Huntington’s disease brain. Mol. Ther. - Nucleic Acids. 2018;11:79–90. doi: 10.1016/j.omtn.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dobson J. Remote control of cellular behaviour with magnetic nanoparticles. Nat. Nanotechnol. 2008;3:139. doi: 10.1038/nnano.2008.39. [DOI] [PubMed] [Google Scholar]
- 241.Dante S., Petrelli A., Petrini E.M., Marotta R., Maccione A., Alabastri A., Quarta A., De Donato F., Ravasenga T., Sathya A., Cingolani R., Proietti Zaccaria R., Berdondini L., Barberis A., Pellegrino T. Selective targeting of neurons with inorganic nanoparticles: revealing the crucial role of nanoparticle surface charge. ACS Nano. 2017;11:6630–6640. doi: 10.1021/acsnano.7b00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Walters R., Medintz I.L., Delehanty J.B., Stewart M.H., Susumu K., Huston A.L., Dawson P.E., Dawson G. The role of negative charge in the delivery of quantum dots to neurons. ASN Neuro. 2015;7 doi: 10.1177/1759091415592389. 1759091415592389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kumar P., Wu H., McBride J.L., Jung K.E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 244.Kwon E.J., Lasiene J., Jacobson B.E., Park I.-K., Horner P.J., Pun S.H. Targeted nonviral delivery vehicles to neural progenitor cells in the mouse subventricular zone. Biomaterials. 2010;31:2417–2424. doi: 10.1016/j.biomaterials.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Louw A.M., Kolar M.K., Novikova L.N., Kingham P.J., Wiberg M., Kjems J., Novikov L.N. Chitosan polyplex mediated delivery of miRNA-124 reduces activation of microglial cells in vitro and in rat models of spinal cord injury. Nanomed. Nanotechnol. Biol. Med. 2016;12:643–653. doi: 10.1016/j.nano.2015.10.011. [DOI] [PubMed] [Google Scholar]