Abstract
Purpose
Colorectal cancer (CRC) stem cells are tumorigenic, capable of self-renewal, and resistant to therapy. Although the expression pattern and functions of micro RNA (miR)-194 in CRC cells have been widely investigated, little is known about its role in CRC stem cells. Therefore, the aim of this study was to investigate the potential role of miR-194 in CRC stem cells.
Materials and methods
CRC stem cells were isolated from the SW620 colon cancer cell line using microbeads. The expression levels of miR-194 and slingshot 2 (SSH2) in CRC stem cells were detected by RT-PCR and Western blot. A luciferase reporter assay was performed to confirm that miR-194 directly targets SSH2. Proliferation of CRC stem cells was examined by colony formation and MTT assays. Apoptosis in CRC stem cells was detected by cell cycle and apoptosis assays. The role of miR-194 in tumor growth was determined in vivo.
Results
Cells positive for CD44 and CD133 accounted for approximately 88.7% of the isolated population after microbead isolation. We reveal for the first time that miR-194 expression is decreased in CRC stem cells. Specifically, miR-194 is involved in inhibiting the proliferation of CRC stem cells and promoting CRC stem cell apoptosis by directly targeting SSH2. Furthermore, overexpression of miR-194 resulted in blocking the G1/S transition, the induction of cellular apoptotic process, thereby suppressing the malignant behaviors of CRC stem cells.
Conclusion
This study represents a novel characterization of miR-194 function in CRC stem cells, which may aid in the development of promising therapeutic strategies targeting CRC.
Keywords: colorectal cancer, miR-194, SSH2, proliferation, stem cell
Introduction
Colorectal cancer (CRC) is ranked the third most common type of primary malignant tumor in adults, and is the fourth most common cause of tumor-related mortality worldwide.1 Numerous recent studies have indicated that CRC contains a critical subpopulation of cells with stem cell-like properties, ie, they are tumorigenic, capable of self-renewal, and resistant to therapy.2,3 Accumulating evidence has shown that CRC stem cells play crucial roles in CRC development and may be a promising new target for therapeutic intervention;4 however, it is still unclear how CRC stem cells regulate the development and progression of CRC. In this study, therefore, we focused on the precise biological role of CRC stem cells isolated and cultured from the CRC cell line, SW620.
MicroRNAs (miRNAs) constitute a large class of small, single-stranded, noncoding RNAs that posttranscriptionally regulate target mRNA expression.5 Thousands of miRNAs have been recognized as participating in varied physiological and pathological processes associated with metabolism, signal transduction, cell adhesion, cell movement, cell differentiation, and apoptosis.6 Accordingly, miRNAs have been found to act both as oncogenes and tumor suppressors, depending on the target gene and cancer type.7,8
The expression of miR-194 is markedly reduced in various cancers, including gastric,9 liver,10 and lung cancers,11 as well as CRC,12 suggesting that miR-194 possesses tumor-suppressive properties. Although the expression pattern and function of miR-194 in these cancer cells have been widely investigated, little is known about its role in stem cells, particularly CRC stem cells. Notably, our microarray results showed that miR-194 was the most downregulated miRNA in CRC stem cells compared with that in non-stem cells, suggesting that miR-194 may play an important regulatory role in CRC stem cells.13
Slingshot (SSH) phosphatases belong to a gene family of three members (SSH1, SSH2 and SSH3). Slingshot phosphatases and LIM-kinases are involved in activation/stabilization of filament–actin through cofilin de/phosphorylation.14 Of several phosphatases that have been reported to activate cofilin by dephosphorylation, the slingshot family of phosphatases (SSH1, SSH2 and SSH3) has emerged as the primary mediator in restoring cofilin’s actin depolymerizing activity and inducing cell motility.
The aim of the present study was to investigate the role of miR-194 in CRC stem cells and identify putative target genes. The potential mechanisms through which miR-194 regulates the biological behaviors of CRC stem cells were also investigated. Our findings will contribute to elucidating the function of miR-194 in CRC stem cells and may aid the development of therapeutic strategies for CRC.
Materials And Methods
Cell Culture
The human CRC cell line SW620 and HEK 293T cells were obtained from the Chinese Academy of Sciences, Shanghai, China. All the cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) and incubated at 37 °C in 95% air/5% CO2. Microbeads (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) were used to isolate CD44+/CD133+ and CD44−/CD133− cells from the SW620 cell line. After isolation, CD44+/CD133+ cells were grown in serum-free DMEM/F12 medium (Gibco) supplemented with 10 ng/mL epidermal growth factor (EGF) and 10 ng/mL basic EGF (bEGF; Sigma–Aldrich, St Louis, MO, USA). Isolated CD44−/CD133− cells were cultured in DMEM/F12 medium supplemented only with 10% FBS. All the cells were maintained in the same above-mentioned environment during the experiments.
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted from cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The concentration and purity of each extracted RNA sample were measured using a Thermo Scientific NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). cDNA was reverse-transcribed from RNA samples using the stem-loop reverse transcript primer method. Real-time PCR was carried out using SYBR Premix EX Taq (Takara, Dalian, Liaoning, China) in an Applied Biosystems 7500 quantitative Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The sequences of the primers used are as follows: miR-194: 5′-TGTAACAGCAACTCCA-3′ (forward), 5′-GTGCAGGGTCCGAGGT-3′ (reverse); SSH2: 5′-CAAGAATCAGCCACAGACGG-3′ (forward), 5′-TAGGACGATGCTTTCTTCAG-3′ (reverse); GAPDH: 5′-AGAGGCAGGGATGATGTTCTG-3′ (forward), 5′-GACTCATGACCACAGTCCATGC-3′ (reverse). Relative expression levels were calculated using the 2−ΔΔCT method.
Western Blot Analysis
Total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer containing phenylmethylsulfonyl fluoride (PMSF; Santa Cruz Biotechnology, Santa Cruz, CA, USA). The bicinchoninic acid (BCA) protein assay kit (Beyotime Institute of Biotechnology, Haimen, Jiangsu, China) was used to determine protein concentrations. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated overnight at 4 °C with anti-SSH2 (1:500, Santa Cruz Biotechnology) and anti-β-actin (1:1000, Santa Cruz Biotechnology) primary antibodies, followed by incubation with a horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (IgG, 1:3000 dilution; Santa Cruz Biotechnology). Signals were detected using a chemiluminescence detection kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA). β-Actin was used as an internal reference and the experiments were performed in triplicate.
Cell Transfection
miR-194 mimics and the negative control were synthesized by GenePharma (Shanghai, China). CRC stem cells were cultured in 6-well plates at 2 × 105 cells per well and incubated overnight until reaching 60% confluence. Transfections were conducted using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The cells were used for subsequent analyses 48 h after transfection.
Vector Construction and Luciferase Reporter Assay
A gene fragment containing the three miR-194 binding sites in present in SSH2 was designed and cloned into the pGL3-Control vector (Promega, Madison, WI, USA) to form the 3′-UTR of the luciferase reporter vector (SSH2-3′-UTR-Wt). Point mutations were then introduced into the SSH2-3′-UTR-Wt sequence to generate SSH2-3′-UTR-Mut. The control Renilla luciferase-encoding plasmid (pRL-TK; Promega), SSH2-3′-UTR-Wt or SSH2-3′-UTR-Mut, and miR-194 or negative control (NC) were co-transfected into HEK 293T cells using Lipofectamine 2000 Reagent (Invitrogen). Luciferase activity was assayed 48 h after transfection using the Dual-Luciferase reporter assay (Promega). Relative luciferase activity was expressed as the ratio of firefly to Renilla luciferase activity.15
Colony Formation Assay
A total of 500 cells infected with miR-194-expressing recombinant lentivirus (Hanbio, Shanghai, China) were seeded in each well of a 6-well plate. After 14 days of culture, the colonies were fixed in methanol for 10 min and then stained with a 1% crystal violet solution (Beyotime Institute of Biotechnology) for 20 min for imaging.
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2H-Tetrazolium Bromide (MTT) Assay
Cells transfected with miRNA were plated at 2000 cells per well in 96-well plates. Then, MTT (50 mg per well, Sigma–Aldrich) was added at different time points and cultured for an additional 4 h. The cells were lysed for 15 min and the plates gently shaken for 5 min. Dimethyl sulfoxide (DMSO; Sigma–Aldrich) was used to terminate the reaction and the absorbance was then assayed at 490 nm.16
Cell Cycle Assay
For cell cycle assay, cells were fixed in the presence of 70% ethanol at 4°C overnight. After washing with phosphate-buffered saline (PBS), these cells were incubated in PBS containing 20 μg/mL of propidium iodide (Sigma–Aldrich), 200 μg/mL of RNase A, and 0.1% Triton X-100 (BD Biosciences, San Jose, CA, USA) at 37°C for 30 mins. Cell nuclei (1 × 106 cells) were stained with propidium iodide (Sigma–Aldrich). A FACSCalibur flow cytometer (BD Biosciences) was used to quantify the percentage of cells in the G0/G1, S, and G2/M phases of the cell cycle.
Apoptosis Assay
Cells were washed with ice-cold PBS, trypsinized, and resuspended in 1× binding buffer at 1×106 cells/mL. After gentle vortex, the cells were stained with fluorescein isothiocyanate (FITC) using an FITC–Annexin V Apoptosis Detection Kit (Sigma–Aldrich) followed by a 15 min incubation at room temperature in the dark according to the manufacture’s protocol. A FACSCalibur flow cytometer (BD Biosciences) was used to detect the rate of apoptosis. The experiment was performed in triplicate.
In Vivo Study
Cells infected with miR-194-expressing lentivirus or the negative control were used for in vivo analysis. Four-week-old BALB/c nude mice were obtained from the Animal Experimental Center of Fudan University, and provided food ad libitum. In total, 5 × 105 cells were implanted subcutaneously into the shoulder area of each animal. Tumors that developed were measured with a slide caliper every 7 days until 35 days post injection. Tumor volumes were calculated according to the following formula: volume (mm3)=length×width2/2. All the mice were euthanized 35 days after injection and the subcutaneous tumors were weighed. The animal study protocol was approved by the ethics committee of Fudan University. All the experiments were conducted strictly in accordance with a protocol approved by the Administrative Panel on Laboratory Animal Care of Fudan University.
Statistical Analysis
Experimental data were expressed as means ± standard deviation (SD). The t-test or one-way analysis of variance (ANOVA) was used to determine significance. Values were considered significantly different at P<0.05. All statistical analyses were performed using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA).
Results
Isolation and Identification of CRC Stem Cells
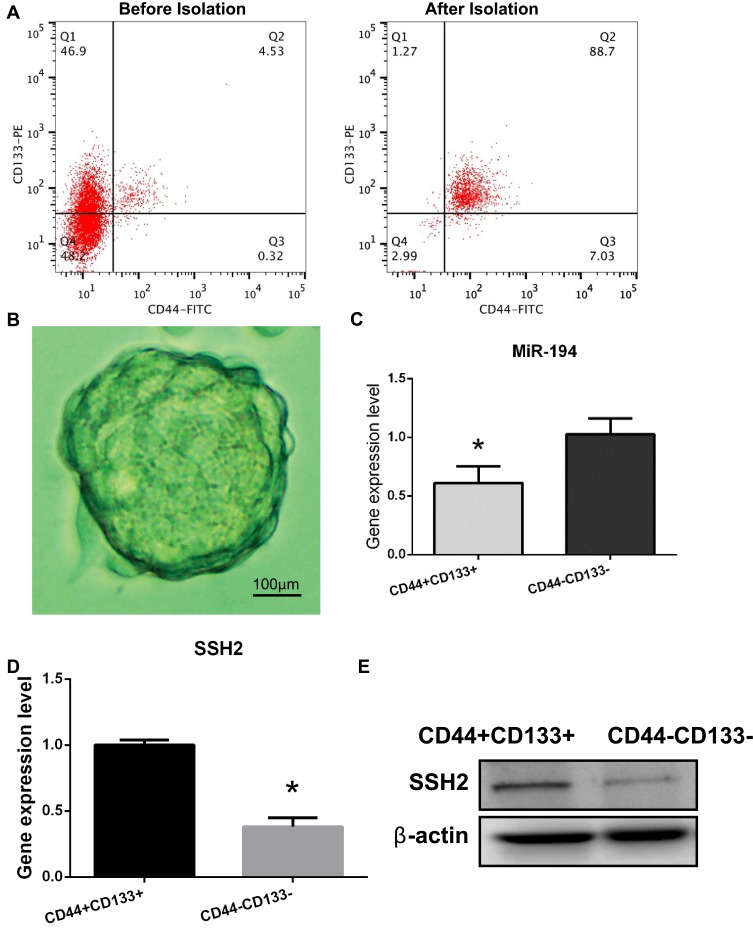
Previous studies have identified CD44 and CD133 as stem cell markers.13 Therefore, CD44+/CD133+ cells were isolated from the SW620 cell line using microbeads and then evaluated by flow cytometry (Figure 1A). Cells positive for CD44 and CD133 accounted for approximately 88.7% of the isolated population after microbead isolation. After 10 days of culture, sphere formation could be observed under an inverted microscope (Figure 1B). These results imply that the CD44+/CD133+ cells possess stem cell characteristics.
Figure 1.
Isolation and identification of colorectal cancer (CRC) stem cells in the SW620 cell line and determination of miR-194 and slingshot 2 (SSH2) expression levels in CRC stem cells and non-stem cells. (A) The proportions of CD44+ and CD133+ cells were analyzed by flow cytometry before and after isolation with microbeads. Before isolation, CD44+/CD133+ cells accounted for 4.53% of the total number of cells. After isolation, CD44+/CD133+ cells accounted for 88.7% of the total number of cells. (B) CRC stem cells were cultured in serum-free medium and sphere formation was evaluated under a microscope after 10 days. Scale bar = 100 µm. (C) RT-qPCR results showed that the expression levels of miR-194 were reduced (*P<0.05 according to the two-tailed t test). (D)The expression levels of SSH2 were increased in CRC stem cells compared with those in CRC non-stem cells (*P<0.05 according to the two-tailed t test). (E) The SSH2 protein expression levels were increased in CRC stem cells compared with those in CRC non-stem cells.
Expression Levels of miR-194 and SSH2 in CRC Stem and Non-Stem Cells
Differential miRNA expression between CRC stem and non-stem cells was previously determined by miRNA microarray.13 Of 1711 human miRNAs evaluated, 31 were found to be significantly downregulated in CRC stem cells. Because miR-194 was found to be the most significantly downregulated miRNA in CRC stem cells, this miRNA was selected for further studies. The RT-qPCR results confirmed that miR-194 expression was reduced in CRC stem cells compared with that in CRC non-stem cells (Figure 1C), Next, the mRNA expression levels of SSH2 were quantified in CD44+/CD133+ cells and CD44−/CD133− cells. The results showed that SSH2 expression was significantly upregulated in CD44+/CD133+ cells compared with that in CD44−/CD133− cells (Figure 1D) (P<0.05). Analysis of SSH2 protein levels by Western blot yielded a similar result (Figure 1E). Combined, these data indicated that the expression of SSH2 was upregulated in CRC stem cells, while that of miR-194 was downregulated.
miR-194 Directly Regulates SSH2 Expression in CRC Stem Cells
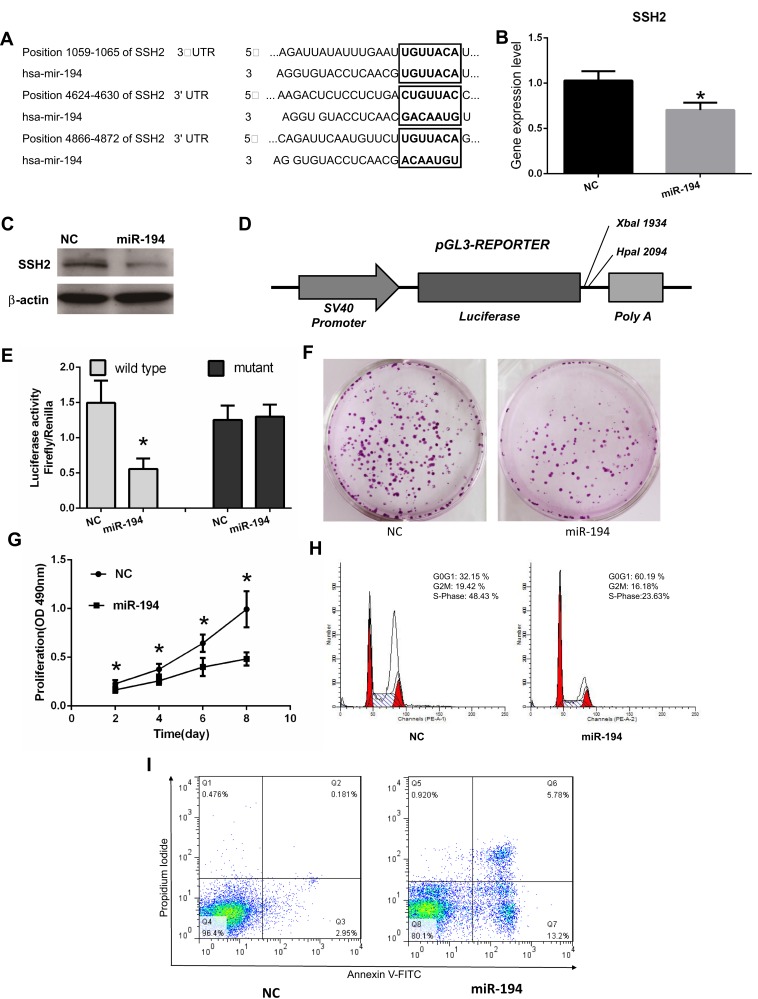
Bioinformatics databases (TargetScan, PicTar, and RNAhybrid) were used to predict conserved miRNA-194 target genes. Because SSH2 harbors three highly conserved miR-194 binding sites at positions 1059–1065, 4624–4630, and 4866–4872 in its 3′-UTR, SSH2 was predicted to be a target for miR-194 (Figure 2A). To confirm whether miR-194 directly targets SSH2, miR-194 mimics were employed to assess the effect of miR-194 on SSH2 expression. For this, CRC stem cells were transfected with either miR-194 or the NC. Western blot and RT-qPCR results indicated that SSH2 expression was reduced in miR-194-transfected cells compared with that in cells transfected with miR-194-NC (Figure 2B and C). This suggests that miR-194 regulates the expression of SSH2 in CRC stem cells.
Figure 2.
miR-194 inhibits cell proliferation, blocks the G1/S transition, and promotes apoptosis in colorectal cancer (CRC) stem cells by directly targeting the slingshot 2 (SSH2) gene. (A) The 3′-UTR of SSH2 contains three binding sites for miR-194. (B) NC represents CRC stem cells transfected with miR-194-NC; miR-194 represents CRC stem cells transfected with miR-194. The SSH2 mRNA expression levels, determined via quantitative RT-PCR, were reduced in CRC stem cells transfected with miR-194 compared with those in CRC stem cells transfected with miR-194-NC (*P<0.05). (C) The SSH2 protein expression level, determined via Western blot, was reduced in CRC stem cells transfected with miR-194 compared with that in CRC stem cells transfected with miRNA-194-NC. (D) Schematic presentation of luciferase reporter vector, this containing either the wild-type or mutant 3′-UTR of SSH2 were co-transfected into HEK 293T cells with miR-194-NC or miR-194. (E) Compared with co-transfection with miR-194-NC, co-transfection with miR-194 led to a significant decrease in the luciferase activity of the pGL3-SSH2-3′ UTR-Wt reporter (P<0.05 according to the two-tailed t test), but not in that of the pGL3-SSH2-3′-UTR-Mut reporter. (F) Colony formation assay demonstrating that CRC stem cells transfected with miR-194-NC formed a substantially higher number of colonies than CRC stem cells transfected with miR-194(P<0.05 according to the two-tailed t test). (G) MTT assay showing that CRC stem cell proliferation was significantly decreased in miR-194 transfected cells compared with that in cells transfected with miRNA-194-NC (P<0.05 according to ANOVA). (H) miR-194 blocks the G1/S transition in CRC stem cells. Cells were transfected with miR-194 or miR-194-NC and subjected to flow cytometry for cell-cycle analysis. The results showed that, compared with CRC stem cells transfected with miRNA-194-NC, the proportion of miRNA-194-transfected CRC stem cells at the G1 phase was increased, whereas the proportion at the S phase was markedly reduced(P<0.05 according to the two-tailed t test). (I) The effect of miR-194 and miR-194-NC on CRC stem cell apoptosis was investigated by Annexin V–FITC/PI staining. A greater percentage of apoptotic cells were observed among miR-194-transfected cells than among those transfected with miRNA-194-NC(P<0.05 according to the two-tailed t-test).
Next, a luciferase reporter assay was performed to elucidate the molecular mechanisms through which miR-194 negatively regulates SSH2 expression (Figure 2D). A gene fragment containing the three miR-194 binding sites in SSH2 was cloned into the SSH2-3′-UTR-Wt reporter plasmid, and the resulting plasmid was co-transfected into CRC stem cells with either miR-194 or miR-194-NC. As expected, co-transfection of SSH2-3′-UTR-Wt with miR-194, but not miR-194-NC, led to a marked downregulation of luciferase reporter activity (Figure 2E). These findings suggested that miR-194 inhibits SSH2 expression. Point mutations were subsequently introduced into the miR-194 binding sites in SSH2-3′-UTR-Mut to disrupt the predicted miR-194 binding sites. Co-transfection of miR-194 and SSH2-3′-UTR-Mut did not substantially alter the luciferase activity (Figure 2E). The above results indicate that miR-194 inhibits SSH2 expression by directly binding to the putative binding sites present in the 3′-UTR of SSH2.
miR-194 Inhibited Cell Proliferation, Blocked the G1/S Transition, and Promoted Apoptosis in CRC Stem Cells
The functional effect of miR-194 on the behaviors of CRC stem cells was assessed by determining the effects of miR-194 overexpression on cell proliferation, apoptosis, and cell cycle progression. To ascertain whether miR-194 upregulation affects the proliferative ability of CRC stem cells, we conducted plate colony formation assays and MTT assays. The colonies were significantly lower in the miR-194 group (186±63) as compared with that in the miR-194-NC group (52±16, Figure 2F, P<0.05). As shown in Figure 2G, the proliferation of colorectal cancer stem cells was significantly reduced in the miR-194 group compared with that in the miR-194-NC group (P<0.05). These data suggested that miR-194 functions as a repressor in regulating CRC stem cell proliferation.
A cell cycle assay was performed to examine the effect of miR-194 overexpression on CRC stem cell apoptosis. As shown in Figure 2H, the percentage of G0-G1 phase cells in the miR-194 group was 60.19%±5.35, whereas it was only 32.15%±4.87 in the miR-194-NC group (Fig. 5A b and B b, P < 0.01). The percentage of cells reentering into S phase in the miR-194 group (23.63%±4.11) was significantly less than that in the miR-194-NC group (48.43%±5.35, P < 0.01). These results indicate that high miR-194 concentrations induce cell cycle arrest at the G1 phase, thereby blocking the G1/S transition.
To further investigate the effects of miR-194 overexpression on CRC stem cell apoptosis, an apoptosis assay was conducted. As shown in Figure 2I, the apoptosis rate of colorectal cancer stem cells in the miR-194 group (5.78%±1.02)was increased as compared with that in the miR-194-NC group (0.18%±0.10, P<0.01). These data clearly indicate that miR-194 induces apoptosis in CRC stem cells.
miR-194 Overexpression Reduces Tumor Size
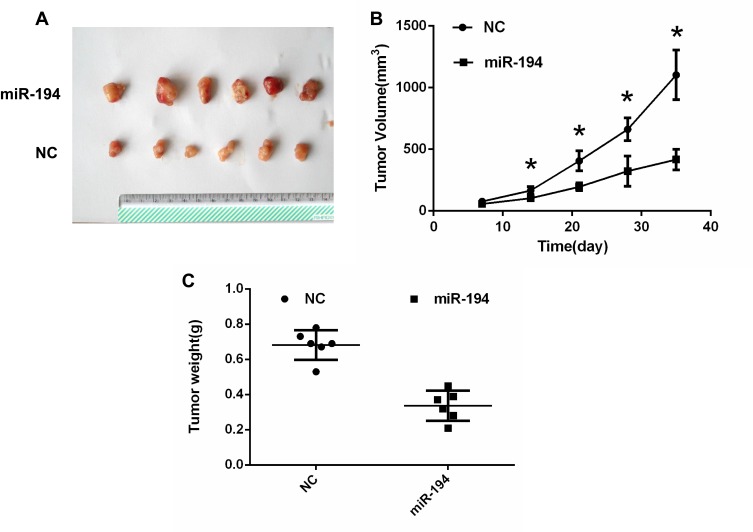
To further determine the role of miR-194 in tumor growth, a xenograft tumor model was established in nude mice (Figure 3A). As illustrated in Figure 3B, tumor volumes were smaller in the miR-194 group than in the miR-194-NC group at 14,21,28,35 days after inoculation. In addition, when the mice were euthanized at 35 days after inoculation, tumors were found to weigh less in the miR-194 group than in the miR-194-NC group (Figure 3C). These findings suggest that overexpression of miR-194 suppresses CRC stem cell proliferation in vivo.
Figure 3.
Colorectal cancer (CRC) stem cells transfected with miR-194 or miR-194-NC were implanted subcutaneously into the shoulder area of mice. (A) A xenograft tumor model was established in nude mice. At 14 days, The tumors were removed from the animal models and displayed. (B)Tumor growth with implantation of CRC stem cells transfected with miR-194 was considerably slower than that with implantation of CRC stem cells transfected with miR-194-NC at 14,21,28,35 days after inoculation (*P < 0.05 according to ANOVA; n = 6). (C) At 5 weeks after implantation, tumor weight with implantation of CRC stem cells transfected with miR-194 was lower than that with implantation of CRC stem cells transfected with miR-194-NC (P<0.05 according to the two-tailed t-test; n = 6).
Discussion
Cancer stem cells constitute a small subpopulation of cancer cells present in various types of tumors. These cells exhibit stem cell-like properties, including the potential for self-renewal and differentiation into multiple lineages, and are resistant to radiation and chemotherapy. Numerous studies investigating CRC have focused on modulating miR-194 function. However, it is still unknown whether miR-194 expression is closely associated with CRC stem cells – the “seed cells” in CRC.
Cancer stem cells isolated from various tumors are normally identified as such through the expression of specific surface epitopes. CRC stem cells have been reported to express specific surface markers, namely, CD24, CD44, CD133, Lgr5, and CD166.4,17–19 In this study, the stem cell phenotype of the CD44+/CD133+ cells identified by flow cytometry was verified by formation of stem cell spheres after 10 days of serum-free culture.
Several recent studies have demonstrated that microRNAs are important regulatory factors in cancer stem cells, controlling the growth and migratory, invasive, and metastatic capacity of tumors. Wang et al found that miR-20a and miR-106a were significantly upregulated in glioma stem cells isolated from both primary human glioma specimens and a glioblastoma cell line. miR-20a and 106a have been shown to enhance the invasive ability of glioma stem cells through regulation of the TIMP metallopeptidase inhibitor 2 (TIMP2) gene, suggesting that both miRNAs represent potential therapeutic targets.20 miR-135a can inhibit the tumorigenic potential of medulloblastoma stem cells by directly targeting the Rac/Cdc42 guanine nucleotide exchange factor 6 (ARHGEF6) gene.21 Takahashi et al identified miR-27b, which was strongly downregulated in breast cancer stem cells, as a potential tumor suppressor. Surprisingly, knockdown of miRNA-27b resulted in cancer stem cell generation.22 These reports showed that microRNAs have important biological functions in cancer stem cells.
A previous study suggested that miR-194 is significantly dysregulated in CRC patients,23 while another study showed that miR-194 is highly induced during epithelial maturation in the small intestine.24 Additionally, Zhao et al suggested that overexpression of miR-194 inhibits cell proliferation, induces G0/G1 or S phase cell cycle arrest, and increases apoptosis and dysregulated cell invasion and migration. Furthermore, overexpression of miR-194 was shown to suppress xenografted CRC growth, while loss of miR-194 in APC Min/+ mouse colons promoted de novo tumor formation.25 Although miR-194 dysregulation has been implicated in various human tumors, the precise function of miR-194 in CRC stem cells remains largely unknown. In this study, we found that miR-194 expression was decreased in CRC stem cells. We conducted MTT assays and plate colony formation assays to ascertain whether miR-194 functions as a suppressor in regulating CRC stem cell proliferation, as well as cell cycle and the apoptosis assays to investigate the effect of miR-194 upregulation on CRC stem cell apoptosis. The results clearly indicate that miR-194 can induce apoptosis in CRC stem cells. Furthermore, we also showed that overexpression of miR-194 induces cellular apoptotic processes, thereby regulating the malignant behaviors of CRC stem cells. Additionally, we confirmed in vivo that miR-194 overexpression leads to reduced tumor size in nude mice, suggesting that miR-194 can suppress tumor growth. Our data indicate that miR-194 plays a vital role in protecting against the malignant behaviors of CRC stem cells.
SSH2 belongs to a gene family composed of three members (SSH1, SSH2, and SSH3) that have been shown to control essential cellular processes, including invasion, migration, and motility. Members of the SSH phosphatase family have been reported to act downstream of the α6β4 integrin/Rac1 signaling pathway,14 and be negatively regulated by phosphorylation of their serine residues. SSH2 family members are required for the effective migration and invasion of tumor cells through maintenance of the unidirectionality of membrane protrusions.26 The results of a previous study implied that SSH2 knockdown in the UOK257 renal cancer cell line preferentially induces caspase 3/7 activity, which is closely associated with apoptotic processes in tumor cells.27 However, no study has provided a detailed evaluation of the antiapoptotic function of SSH2 in tumor stem cells. In this study, we found a negative correlation between SSH2 and miR-194 expression in CRC stem cells. Furthermore, the expression level of SSH2, as determined by quantitative RT-PCR and Western blot, was downregulated after miR-194 transfection. However, further studies are needed to confirm whether miR-194-mediated regulation of SSH2 is direct or indirect. Among the numerous potential miR-194 gene targets identified in bioinformatics databases, we found that SSH2 contains three binding sites for miR-194, strongly suggesting that this microRNA interacts directly with SSH2. To test this assumption, we performed a luciferase reporter assay; as expected, co-transfection of miR-194 and SSH2-3′-UTR-Wt resulted in a marked downregulation of luciferase reporter activity, whereas co-transfection of miR-194-NC and SSH2-3′-UTR-Wt or miR-194 and SSH2-3′-UTR-Mut had no significant effect on luciferase activity. Our present data indicate that miR-194 can suppress the malignant behaviors of CRC stem cells by directly suppressing SSH2 transcription. However, most of our results were from cell experiments in vitro, we just found the phenomenon of overexpression of miR-194 reducing tumor size in vivo. The mechanism is still not very clear, this needs further research.
Conclusion
In summary, we have shown, for the first time, that miR-194 expression is decreased in CRC stem cells. Our results indicate that miR-194 inhibits CRC stem cell proliferation and promotes apoptosis by directly targeting SSH2. Furthermore, overexpression of miR-194 was shown to result in the induction of apoptosis, thereby suppressing the malignant behaviors of CRC stem cells. Therefore, this study reports a novel characterization of miR-194 in CRC stem cells, which may aid in the development of promising therapeutic strategies targeting CRC.
Acknowledgment
This work was supported by grants from the National Natural Science Foundation for Young Scholars (81201618), Shanghai Science and Technology Commission Medical Guide Project (134119a1400), and “Belt and Road” Young Scientist Communication International Cooperation Project (17410742100).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30(3):363–371. doi: 10.1002/stem.1031 [DOI] [PubMed] [Google Scholar]
- 4.Elsaba TM, Martinez-Pomares L, Robins AR, et al. The stem cell marker CD133 associates with enhanced colony formation and cell motility in colorectal cancer. PLoS One. 2010;5(5):e10714. doi: 10.1371/journal.pone.0010714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 7.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi: 10.1038/nrg2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10(6):389–402. doi: 10.1038/nrc2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song Y, Zhao F, Wang Z, et al. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann Surg Oncol. 2012;19:S509–S517. doi: 10.1245/s10434-011-1999-2 [DOI] [PubMed] [Google Scholar]
- 10.Krützfeldt J, Rösch N, Hausser J, Manoharan M, Zavolan M, Stoffel M. MicroRNA-194 is a target of transcription factor 1 (Tcf1, HNF1α) in adult liver and controls expression of frizzled-6. Hepatology. 2012;55(1):98–107. doi: 10.1002/hep.24658 [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Liu T, Fang O, Leach LJ, Hu X, Luo Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27(kip1). Oncogene. 2014;33(12):1506–1514. doi: 10.1038/onc.2013.108 [DOI] [PubMed] [Google Scholar]
- 12.Chiang Y, Song Y, Wang Z, et al. microRNA-192, −194 and −215 are frequently downregulated in colorectal cancer. Exp Ther Med. 2012;3(3):560–566. doi: 10.3892/etm.2011.436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Xiang J, Chen Z, et al. miRNA expression profile of colon cancer stem cells compared to non-stem cells using the SW1116 cell line. Oncol Rep. 2012;28(6):2115–2124. doi: 10.3892/or.2012.2054 [DOI] [PubMed] [Google Scholar]
- 14.Kligys K, Claiborne JN, DeBiase PJ, et al. The slingshot family of phosphatases mediates Rac1 regulation of cofilin phosphorylation, laminin-332 organization, and motility behavior of keratinocytes. J Biol Chem. 2007;282(44):32520–32528. doi: 10.1074/jbc.M707041200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma J, Yao Y, Wang P, et al. MiR-181a regulates blood-tumor barrier permeability by targeting Krüppel-like factor 6. J Cereb Blood Flow Metab. 2014;34(11):1826–1836. doi: 10.1038/jcbfm.2014.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS Lett. 2015;589(2):207–214. doi: 10.1016/j.febslet.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 17.Wang C, Xie J, Guo J, Manning HC, Gore JC, Guo N. Evaluation of CD44 and CD133 as cancer stem cell markers for colorectal cancer. Oncol Rep. 2012;28(4):1301–1308. doi: 10.3892/or.2012.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF. Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci U S A. 2010;107(8):3722–3727. doi: 10.1073/pnas.0915135107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang R, Jian X, Zhao J, Bai J. Knockdown of miR-27a sensitizes colorectal cancer stem cells to TRAIL by promoting the formation of Apaf-1-caspase-9 complex. Oncotarget. 2017;8(28):45213–45223. doi: 10.18632/oncotarget.16779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Wang B, Shi Y, et al. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34(11):1407–1419. doi: 10.1038/onc.2014.75 [DOI] [PubMed] [Google Scholar]
- 21.Hemmesi K, Squadrito ML, Mestdagh P, et al. miR-135a inhibits cancer stem cell-driven medulloblastoma development by directly repressing arhgef6 expression. Stem Cells. 2015;33(5):1377–1389. doi: 10.1002/stem.1958 [DOI] [PubMed] [Google Scholar]
- 22.Takahashi RU, Miyazaki H, Takeshita F, et al. Loss of microRNA-27b contributes to breast cancer stem cell generation by activating ENPP1. Nat Commun. 2015;6:7318. doi: 10.1038/ncomms8318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B, Shen ZL, Gao ZD, et al. MiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle. 2015;14(7):1046–1058. doi: 10.1080/15384101.2015.1007767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hino K, Tsuchiya K, Fukao T, et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14(7):1433–1442. doi: 10.1261/rna.810208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao HJ, Ren LL, Wang ZH, et al. MiR-194 deregulation contributes to colorectal carcinogenesis via targeting AKT2 pathway. Theranostics. 2014;4(12):1193–1208. doi: 10.7150/thno.8712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108(2):233–246. doi: 10.1016/S0092-8674(01)00638-9 [DOI] [PubMed] [Google Scholar]
- 27.Lu X, Boora U, Seabra L, et al. Knockdown of Slingshot 2 (SSH2) serine phosphatase induces Caspase3 activation in human carcinoma cell lines with the loss of the Birt-Hogg-Dubé tumour suppressor gene (FLCN). Oncogene. 2014;33(8):956–965. doi: 10.1038/onc.2013.27 [DOI] [PubMed] [Google Scholar]