Abstract
Mammalian mRNA biogenesis is dependent on specific recognition of a hexanucleotide AAUAAA motif in the polyadenylation signals (PAS) of pre-mRNA transcripts by the cleavage and polyadenylation specificity factor (CPSF) complex. Here we report a 3.1 Å resolution cryo-electron microscopy structure of a core CPSF module bound to the PAS hexamer motif. The structure reveals molecular interactions responsible for base-specific recognition, providing a rationale for mechanistic differences between mammalian and yeast 3’ polyadenylation.
In mammalian cells, a central hexamer A(A/U)UAAA motif is a key cis-acting element within the polyadenylation signal (PAS) required for 3’ cleavage and polyadenylation of pre-mRNA transcripts1–3. The PAS hexamer motif is specifically recognized by the Cleavage and Polyadenylation Specificity Factor (CPSF), a multi-subunit complex responsible for polyadenylation site definition, pre-mRNA cleavage and polyA-polymerase recruitment, and for coupling 3’ end formation with transcriptional termination4–9. A core CPSF assembly composed of proteins CPSF160, WDR33, CPSF30 and Fip1 binds the A(A/U)UAAA motif, via direct interactions with CPSF30 and WDR33, and is sufficient to recruit polyA-polymerase in vitro10,11. Despite the critical function of CPSF in mammalian mRNA biogenesis, the molecular interactions that underpin its specificity for the PAS hexamer motif are poorly understood.
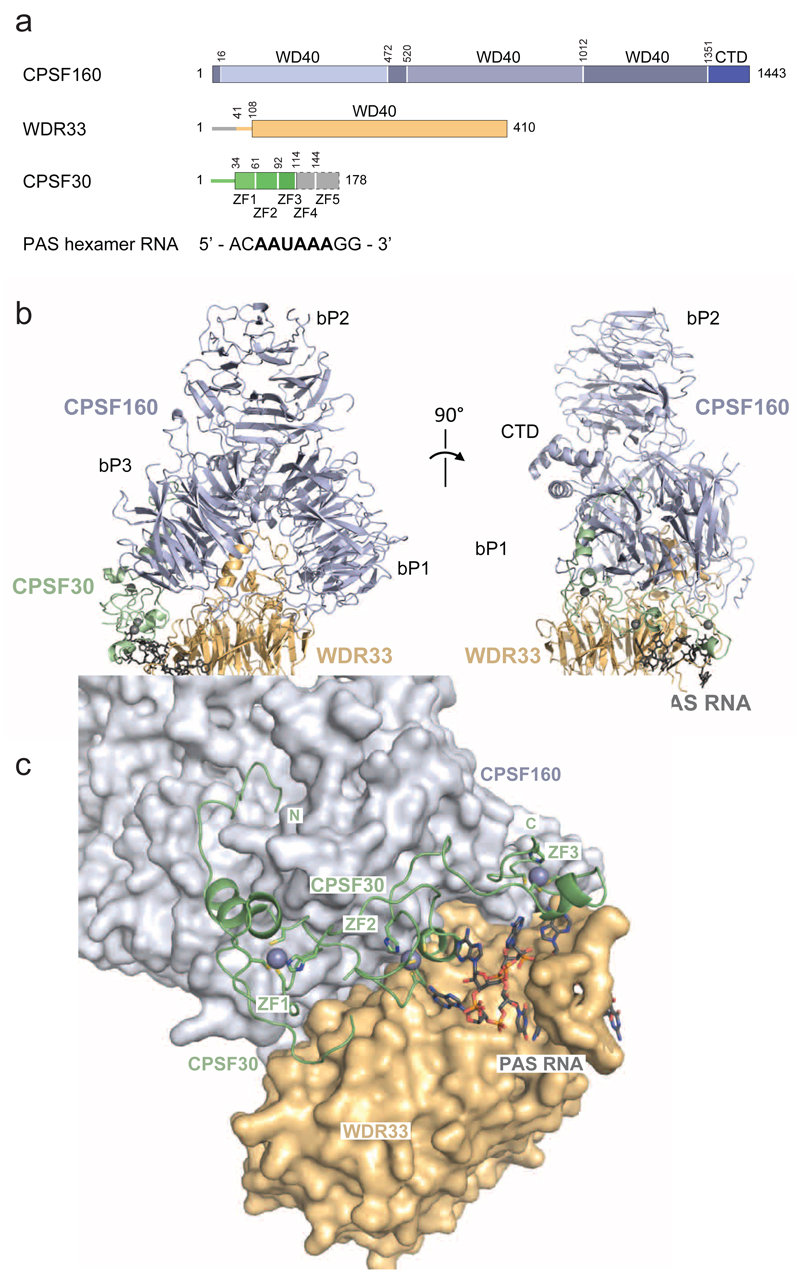
To address this question, we reconstituted by coexpression in baculovirus-infected insect cells a core human CPSF polyadenylation module consisting of full-length CPSF160, an N-terminal fragment of WDR33 containing the WD40 domain, a truncated CPSF30 fragment containing zinc-finger (ZF) domains ZF1-ZF5 and a conserved fragment of Fip1 corresponding to residues Gly130-Lys195Fip1, bound to a 10-nucleotide RNA containing the AAUAAA hexamer (Supplementary Fig. 1a). Using cryo-electron microscopy (cryo-EM) and single-particle analysis, we determined the structure of the core CPSF-AAUAAA complex at an overall resolution of 3.1 Å, as indicated by the Fourier shell correlation (FSC) plot (Supplementary Fig. 1b-g). The structure comprises full-length CPSF160 (residues Met1-Phe1443CPSF160), WDR33 (Leu41-Lys410WDR33), domains ZF1-3 of CPSF30 (Met1-Ile115CPSF30) and an RNA fragment containing the AAUAAA hexamer and two additional 3’-terminal nucleotides (Fig. 1a). No density could be observed for Fip1 and CPSF30 domains ZF4 and ZF5, suggesting that these domains form a flexible module. This is in good agreement with previous interaction studies showing that yeast Fip1 is an intrinsically disordered protein that interacts with Yth1 (the yeast CPSF30 ortholog) via its ZF4 and ZF5 domains12.
Figure 1. Molecular architecture of CPSF160-WDR33-CPSF30 in complex with the AAUAAA PAS hexamer motif.
(a) Schematic diagram depicting CPSF160, WDR33 and CPSF30 constructs used for complex reconstitution (not to scale). N- and C-terminal regions for which no electron density was observed are depicted in grey. CPSF160 beta-propeller domains (bP1-3) and C-terminal domain (CTD) are highlighted with different shades of blue. CPSF160 regions forming beta-propeller domain 3 (bP3) are non-contiguous in the primary structure of the protein. (b) Front (left) and side (right) views of the CPSF160-WDR33-CPSF30-AAUAAA hexamer motif complex shown in cartoon (proteins) and stick (RNA) format. CPSF160 beta-propeller domains and C-terminal domain are indicated as bP1 to bP3 and CTD, respectively. Zinc atoms are displayed as grey spheres. (c) Zoomed-in view of CPSF30 binding to a cleft formed by CPSF160 C-terminal beta-propeller (bP3) and WDR33. CPSF30 is represented as cartoon, PAS RNA as sticks and CPSF160-WDR33 as surface. Zinc atoms are displayed as grey spheres.
Overall, the structure of the human CPSF160-WDR33-CPSF30 complex is highly similar to a recently determined structure of the core polyadenylation module of the yeast CPF complex comprising Cft1 (the yeast CPSF160 ortholog), Pfs2 (WDR33) and Yth1 (CPSF30)13 (Fig. 1b, Supplementary Figure 2a). The tandem ZF1-3 domains of CPSF30 line a cleft at the CPSF160-WDR33 interface near the N-terminal extension of WDR33 (Fig. 1c). The N-terminal region of CPSF30 upstream of the ZF1 domain extends over the surface of the CPSF160 beta-propeller domain bP3, inserting its N-terminus into the central cavity of bP3. These findings are in good agreement with recent biochemical characterization of subunit interactions within the human CPSF core module and an X-ray crystallographic structure of the CPSF160-WDR33 heterodimer14.
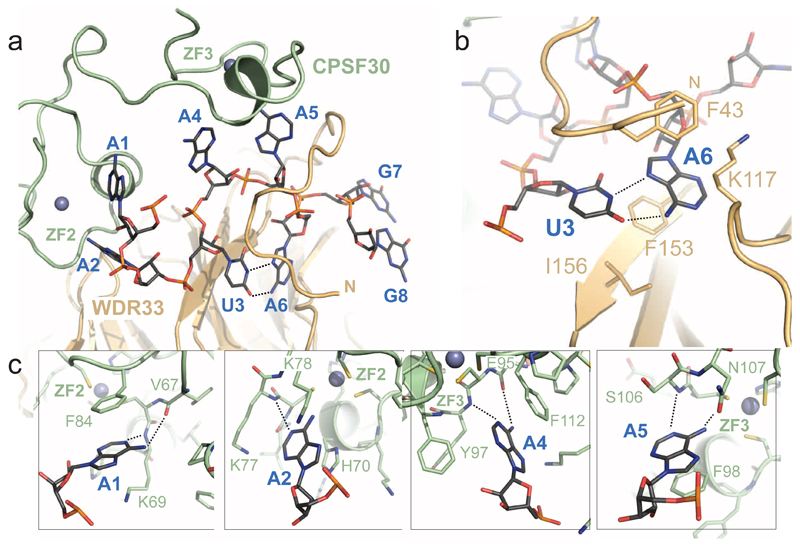
The ribose-phosphate backbone of the AAUAAA hexamer adopts an S-shaped conformation (Fig. 2a, Supplementary Figure 2b). The RNA is clamped at the CPSF30-WDR33 interface, establishing an intricate network of interactions that involve both the RNA backbone and the nucleobases (Supplementary Figure 3). The N-terminal region of WDR33 comprising residues Leu41-Lys55WDR33, folds over the RNA, contacting nucleotides U3, A5 and A6 and thereby locking the RNA in place (Fig. 2a). The basic residues Lys46WDR33, Arg47WDR33, Arg49WDR33 and Arg54WDR33 contact the ribose-phosphate backbone of the RNA (Supplementary Figure 3). The centerpiece of the PAS hexamer is a Hoogsteen base pair formed between nucleotides U3 and A6 (Fig. 2b), sandwiched by π-π stacking interactions with Phe153WDR33 and Phe43WDR33, both of which are invariant in mammalian WDR33 orthologs. Nucleotides A1-A2 are bound by the CPSF30 ZF2 domain, while nucleotides A4-A5 are bound by ZF3. The two segments of the AAUAAA hexamer are bound by the ZF domains in a nearly identical manner (Supplementary Fig. 2c). Altogether, these structural insights rationalize previous findings that implicated CPSF30 ZF2-3 domains and WDR33 N-terminus in specific recognition of the AAUAAA motif10,11,14
Figure 2. AAUAAA motif recognition by CPSF30 and WDR33.
(a) Overall view of PAS hexamer RNA recognition by CPSF30 ZNF2 and ZNF3 domains and WDR33. CPSF30 and WDR33 are shown in cartoon format the PAS RNA as sticks. Hoogsteen base-pair hydrogen bonds between nucleobases of U3 and A6 are indicated as dotted lines. (b) Zoomed-in view of the U3-A6 Hoogsteen base-pair sandwiched between WDR33 F43 and F153/I156 residues. Hydrogen bonds are indicated as dotted lines. (c) Recognition of nucleotides A1, A2, A4 and A5 by CPSF30 ZNF2 and ZNF3 domains.
Each of the A1, A2, A4 and A5 bases occupies a pocket in CPSF30 in which the base stacks with a strictly conserved aromatic residue: A1 with Phe84CPSF30, A2 with His70CPSF30, A4 with Phe112CPSF30, and A5 with Phe98CPSF30 (Fig. 2c). The nucleobases of A1, A4 and A5 are further recognized via their Watson-Crick edges by sequence-specific hydrogen bonding interactions with their N1 and 6-amino groups (Fig. 2c). A1 is contacted by the main-chain amide of Lys69CPSF30 and carbonyl of Val67CPSF30; A4 by the main-chain amide of Tyr97CPSF30 and carbonyl of Glu95CPSF30; A5 is contacted by the main-chain amide of Ser106CPSF30 and side-chain carbonyl of Asn107CPSF30. By contrast, the interaction with the nucleobase of A2 is less specific since a single hydrogen bond is established between the main-chain amide of Lys77CPSF30 and the N1 group of the adenine base. Finally, the U3-A6 base pair does not appear to be recognized by base-specific interactions (Fig. 2b,c).
AAUAAA and AUUAAA are the two most common hexamer motifs found within the polyadenylation signals of mammalian mRNAs15–17. The observed base-specific recognition of adenosines at positions 1, 4 and 5 within the hexamer is consistent with their invariance in the consensus sequence. Conversely, the lack of base-specific recognition at position 2 is reflected by transcriptome-wide studies of mammalian mRNA polyadenylation showing that the second position within the PAS hexamer is the most variable15–17. Notably, the structural insights are also in good agreement with the observation that although the identity of each base within the PAS hexamer consensus strongly affects the efficiency of RNA cleavage and polyadenylation in a HeLa cell nuclear extract, the second position within the hexamer is the most permissive to base substitutions18. Hoogsteen base-pairing between nucleotides U3 and A6 explains their invariance in the PAS hexamer motif even in the absence of base-specific interactions with WDR33; although other isosteric pyrimidine-purine combinations could potentially fit in the U3-A6 binding pocket in WDR33, these base-pair combinations are energetically unfavorable due to non-optimal hydrogen bonding interactions, which likely results in substantially reduced affinity.
Whereas the AAUAAA hexamer is strongly conserved in mammalian pre-mRNA transcripts, the polyadenylation signals of yeast pre-mRNAs are less well-defined and typically contain degenerate A-rich motifs termed positioning elements19. Both structural superposition and sequence alignment of human CPSF30 and yeast Yth1 indicate that the A1, A4 and A5 recognition pockets are conserved in the Yth1 ZF2 and ZF3 domains (Supplementary Fig. 4a,b), implying that Yth1 recognizes adenosine nucleotides in positioning elements. In contrast to higher eukaryotic WDR33 orthologs, yeast Pfs2 lacks an aromatic residue equivalent to Phe43WDR33 in its N-terminal region (Supplementary Fig. 4c). As a result, the U3-A6 binding pocket is not conserved in Pfs2, possibly explaining why yeast positioning elements are degenerate and do not conform to a well-defined consensus sequence.
In conclusion, structural insights presented in this study reveal the molecular interaction framework that underpins polyadenylation site definition in mammalian pre-mRNA 3’ end processing. While this manuscript was in preparation, another cryo-EM structure of the human CPSF160-WDR33-CPSF30 complex bound to the AAUAAA PAS motif was reported20, with similar conclusions. Despite emerging commonalities in the molecular architectures of yeast and mammalian cleavage and polyadenylation complexes, their structural features responsible for RNA binding have diverged to recognize distinct polyadenylation signals. Ongoing studies of yeast CPF and mammalian CPSF complexes will shed light on the molecular mechanisms that couple polyadenylation signal recognition to the nuclease and polyA polymerase enzymatic activities, as well as to transcriptional termination, and provide insights into their regulation.
Online Methods
Protein expression and purification
Human CPSF subunit constructs were cloned into MacroLab vectors developed by Scott Gradia (University of California, Berkeley)21. His6-tagged full-length CPSF160 (Uniprot Q10570) was inserted into the 438B vector (Addgene #55219); His6-(StrepII)2-WDR33 (Uniprot Q9C0J8-1, residues 1-410) was inserted into a modified version of the 438B vector; untagged CPSF30 (Uniprot O95639-3, residues 1-178), was inserted into the 438A vector (Addgene #55218); StrepII-GFP-tagged Fip1 (Uniprot Q6UN15-4, residues 130-195) was inserted into the 438Rgfp vector (Addgene #55211) by ligation-independent cloning. The four constructs were combined in a single plasmid using the MacroBac ligation-independent cloning system 21. Recombinant baculovirus was generated using the Bac-to-Bac system (Invitrogen) according to standard protocols and used to infect Sf9 cells at a density of 1.0 × 106 ml−1. Cells were harvested 72 hr post infection, resuspended in 20 mM Tris-Cl pH 7.5, 200 mM NaCl, 10% glycerol and 0.05% Tween20, supplemented with Protease Inhibitor Cocktail (GE Healthcare), and lysed by sonication. The complex was purified on Ni-NTA resin (Qiagen), followed by purification on Streptactin superflow resin (IBA). Expression tags were removed by overnight incubation with TEV protease. Synthetic 10-mer PAS RNA (5’-ACAAUAAAGG-3’) was added in 1.2-fold molar excess and the resulting protein-RNA complex was further purified by size exclusion chromatography on a Superdex-200 column (GE Healthcare) in 20 mM HEPES pH 7.5, 140 mM KCl and 1 mM DTT. Peak fractions (as indicated in Supplementary figure 1A) were collected and flash-frozen in liquid nitrogen.
Cryo-EM sample preparationand data collection
The CPSF-PAS RNA complex was vitrified on Quantifoil Cu 400 mesh R1.2/1.3 grids previously glow-discharged for 15 seconds on each side. 4 μl of sample at 0.3 mg/ml was deposited on the EM grid, incubated for 15 seconds, blotted for 7 seconds and plunge-frozen in liquid ethane-propane mixture using a FEI Vitrobot Mark IV at 22 °C and 100% humidity. Data collection was performed on a FEI Titan Krios microscope operated at 300 kV and equipped with a Gatan K2 Summit detector and a Gatan Quantum GIF LS energy filter. Image acquisition and initial processing was managed by Focus22. Micrographs with a pixel size of 1.058 Å were collected in super-resolution mode and consisted of 50 frames for a total exposure time of 10 seconds and a total dose of 80 e- / Å2. The defocus ranged between -2.2 and -1 μm.
Image processing
Motion correction and averaging were performed with MotionCor223. Final micrographs are dose-weighted sums of the full stack removing the first frame. Contrast transfer function (CTF) parameters were determined with CTFFIND424. Subsequent processing was performed in RELION-225. A dataset of 1070 images was selected with a CTF resolution estimate between 2.6 and 3.0 Å. 1600 particles were manually picked from a random subset of micrographs and the resulting 2D class averages used as input for the RELION-2 Autopick function on binned micrographs, obtaining a total 265,000 particles. After selection of 263,000 particles with best Autopick figure of merit, the particles were subjected to two rounds of 2D classification, selecting particles from 72 and 27 classes, respectively. The final dataset contained 137,000 particles representing 51% of the picked particle dataset. Aligned particles were then extracted from unbinned micrographs and an initial reference-free model was generated using RELION-2 (Initial model function). 3D classification of the dataset into 3 classes yielded models with similar features, number of particles and final resolution, suggesting structural homogeneity. Therefore, the whole dataset was directly subjected to 3D refine alignment. After post-processing in RELION-2 the final map reached a resolution of 3.1 Å. The resolution was measured by Fourier shell correlation between reconstructions from two independently refined half-datasets (gold-standard FSC in RELION-2). Local resolution was calculated within RELION-2 with ResMap26 from two data-independent reconstructions.
Model building
Model building was performed in Coot27 using the CPSF160-WDR33 heterodimer as starting model (PDB 6F9N) and refined using phenix.real_space_refine28. Figures were prepared with Chimera29 and PyMol (https://www.pymol.org)
Supplementary Material
Acknowledgements
We are indebted to H. Stahlberg for access to the Bio EM Lab at the C-CINA Center for Cellular Imaging and Nanoanalytics at the University of Basel. We thank L. Kovacik and M. Chani for technical assistance with cryo-EM image acquisition. We are grateful to J. Novacek and M. Peterek (Core Facility for Cryo-electron Microscopy and Tomography, CEITEC, Masaryk University, Brno, Czech Republic) for initial sample analysis. We are grateful to A. Scaiola at the ScopeM Scientific Center for Optical and Electron Microscopy at ETH Zurich for assistance with sample preparation and preliminary sample analysis. This work was supported by the European Research Council (ERC) Starting Grant ANTIVIRNA (Grant no. ERC-StG-337284). M.F. was supported by a Long-Term Fellowship from the European Molecular Biology Organization (EMBO ALTF-343-2013). R.A. acknowledges support from the European Union 7th Framework Program (PROSPECTS, HEALTH-F4-2008-201648), the European Research Council (ERC Advanced Grants no. 233226 and no. 670821), and the Innovative Medicines Initiative Joint Undertaking (ULTRA-DD, grant no. 115766). M.J. is International Research Scholar of the Howard Hughes Medical Institute and Vallee Scholar of the Bert L & N Kuggie Vallee Foundation.
Footnotes
Data availability
The cryo-EM density map has been deposited in the Electron Microscopy Data Bank under accession code EMD-4225. The corresponding atomic model has been deposited in the Protein Data Bank under accession code 6FBS.
Author Contributions
M.C. and M.J. designed experiments. M.C. prepared all samples, and determined the cryo-EM structure of the CPSF160-WDR33-CPSF30-PAS RNA complex. L.M.M. assisted with cryo-EM sample preparation. M.F. assisted with cryo-EM data analysis and data interpretation, supervised by R.A. M.C., M.F. and M.J. wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Proudfoot NJ, Brownlee GG. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–4. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 2.Shi Y, Manley JL. The end of the message: multiple protein-RNA interactions define the mRNA polyadenylation site. Genes Dev. 2015;29:889–97. doi: 10.1101/gad.261974.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA. 2012;3:385–96. doi: 10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienroth S, Wahle E, Suter-Crazzolara C, Keller W. Purification of the cleavage and polyadenylation factor involved in the 3'-processing of messenger RNA precursors. J Biol Chem. 1991;266:19768–76. [PubMed] [Google Scholar]
- 5.Chan S, Choi EA, Shi Y. Pre-mRNA 3'-end processing complex assembly and function. Wiley Interdiscip Rev RNA. 2011;2:321–35. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller W, Bienroth S, Lang KM, Christofori G. Cleavage and polyadenylation factor CPF specifically interacts with the pre-mRNA 3' processing signal AAUAAA. EMBO J. 1991;10:4241–9. doi: 10.1002/j.1460-2075.1991.tb05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murthy KG, Manley JL. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–11. [PubMed] [Google Scholar]
- 8.Shi Y, et al. Molecular architecture of the human pre-mRNA 3' processing complex. Mol Cell. 2009;33:365–76. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiang K, Tong L, Manley JL. Delineating the structural blueprint of the pre-mRNA 3'-end processing machinery. Mol Cell Biol. 2014;34:1894–910. doi: 10.1128/MCB.00084-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan SL, et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3' processing. Genes Dev. 2014;28:2370–80. doi: 10.1101/gad.250993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schonemann L, et al. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 2014;28:2381–93. doi: 10.1101/gad.250985.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barabino SM, Ohnacker M, Keller W. Distinct roles of two Yth1p domains in 3'-end cleavage and polyadenylation of yeast pre-mRNAs. EMBO J. 2000;19:3778–87. doi: 10.1093/emboj/19.14.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casanal A, et al. Architecture of eukaryotic mRNA 3'-end processing machinery. Science. 2017;358:1056–1059. doi: 10.1126/science.aao6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clerici M, Faini M, Muckenfuss LM, Aebersold R, Jinek M. Structural basis of AAUAAA polyadenylation signal recognition by the human CPSF complex. eLife. 2018 doi: 10.1038/s41594-017-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–83. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruber AJ, et al. A comprehensive analysis of 3' end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016;26:1145–59. doi: 10.1101/gr.202432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu J, Lutz CS, Wilusz J, Tian B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–93. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Z, Sherman F. 3'-end-forming signals of yeast mRNA. Mol Cell Biol. 1995;15:5983–90. doi: 10.1128/mcb.15.11.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y, et al. Molecular basis for the recognition of the human AAUAAA polyadenylation signal. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1718723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gradia SD, et al. MacroBac: New Technologies for Robust and Efficient Large-Scale Production of Recombinant Multiprotein Complexes. Methods Enzymol. 2017;592:1–26. doi: 10.1016/bs.mie.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biyani N, et al. Focus: The interface between data collection and data processing in cryo-EM. J Struct Biol. 2017;198:124–133. doi: 10.1016/j.jsb.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–30. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Afonine PV, Headd JJ, Terwilliger TC, Adams PD. New tool: phenix.real_space_refine. Computational Crystallography Newsletter. 2013;4:43–44. [Google Scholar]
- 29.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.