Abstract
Faulty or damaged mRNAs are detected by the cell when translating ribosomes stall during elongation and trigger pathways of mRNA decay, nascent protein degradation, and ribosome recycling. The most common mRNA defect in eukaryotes is probably inappropriate poly-adenylation at near-cognate sites within the coding region. How ribosomes stall selectively when they encounter poly(A) is unclear. Here, we use biochemical and structural approaches in mammalian systems to show that poly-lysine, encoded by poly(A), favors a peptidyl-tRNA conformation sub-optimal for peptide bond formation. This conformation partially slows elongation, permitting poly(A) mRNA in the ribosome’s decoding center to adopt an rRNA-stabilized single-stranded helix. The reconfigured decoding center clashes with incoming aminoacyl-tRNA, thereby precluding elongation. Thus, coincidence detection of poly-lysine in the exit tunnel and poly(A) in the decoding center allows ribosomes to detect aberrant mRNAs selectively, stall elongation, and trigger downstream quality control pathways essential for cellular homeostasis.
The accumulation of aberrant proteins can disrupt intracellular homeostasis, cause protein misfolding stress and is implicated in a wide range of diseases ranging from diabetes to neurodegeneration1–3. To minimize aberrant protein production, cells have evolved numerous quality control pathways to selectively identify misfolded or misprocessed proteins and route them for degradation1,2. Although many of these quality control pathways query the protein directly, others are aimed at detecting defective mRNAs that are likely to produce defective proteins4,5. The critical decision point in all quality control pathways is the initial recognition step that triggers the downstream degradation reactions6.
Many of the pathways that recognize defective mRNAs rely on their translation7–9. One important cue used by the cell is stalling of the ribosome during elongation. The stall can be caused for many reasons including excessive mRNA secondary structure10, runs of rare or difficult-to-decode codons11, mRNA truncation12,13, and a poly(A) sequence14–16. The stalled ribosome triggers the ‘no-go’ or ‘non-stop’ pathways of mRNA decay 10,13,17,18, nascent protein degradation via ribosome-associated quality control (RQC) 12,19–23, and ribosome recycling 12,24,25 to maintain protein homeostasis26–30.
There are at least two mechanisms of detecting a stalled ribosome depending on the context. Stalling at the end of a truncated mRNA results in an empty ribosomal A-site that is efficiently recognized by the ribosome rescue factor Pelota (Dom34 in yeast) in complex with the GTPase Hbs124,25,31,32. A second mechanism involves detecting the indirect consequences of a stalled ribosome: collision with the elongating ribosome behind it. Experimental conditions predicted to incur collisions were correlated with markers of downstream quality control including endonucleolytic mRNA cleavage and ribosome ubiquitination33. The collided di-ribosome complex was found to attain a distinctive configuration34,35 that was shown in vitro and in cells to be recognized specifically by the ubiquitin ligase ZNF598 (Hel2 in yeast)34 to initiate downstream quality control pathways14,15,34–38,
Although there are many ways to induce stalling experimentally, the most common reason in cells is thought to be inappropriate poly-adenylation at near-cognate sites within the coding region 17,39–43. The absence of an in-frame stop codon in these mRNAs allows ribosomes to translate into the poly(A) tail. Estimates of premature polyadenylation range up to ~10-20% of mRNAs17, suggesting that this might be a major source of ribosome stalling in cells. Consistent with this idea, crosslinking experiments show that ZNF598 is strongly enriched on ribosomes specifically containing the lysyl-tRNA that decodes the AAA codon41. Despite the prevalence of this pathway, it remains unclear how ribosomes stall selectively on poly(A).
The two candidate contributors to poly(A)-mediated stalling are the nascent polypeptide and the mRNA. Early studies observed that poly-basic sequences inside the ribosome exit tunnel can slow translation in vitro44, and can initiate downstream quality control in yeast45. Later work indicated that at least some of the effects in yeast may be due to particularly inefficient decoding of certain codons, most notably arginine-encoding CGA, as a major contributor to stalling11,46. In mammalian cells, poly-arginine and AAG-encoded poly-lysine do not induce stalling as effectively as poly(A)-encoded poly-lysine, arguing for a strong contribution from the mRNA sequence14–16. Consistent with this idea, experiments in a reconstituted bacterial translation system have shown that iterated AAA codons are very poorly decoded relative to iterated AAG codons47.
These collected observations indicate that both the polypeptide and mRNA might contribute to stalling, but the molecular mechanisms behind their effects on elongation are not understood. Any proposed mechanism must necessarily explain how the cell ignores short poly(A) sequences of 3 or 4 codons, which occur within normal coding regions, while stalling selectively on longer poly(A) sequences characteristic of aberrantly processed mRNAs. This is the problem we set out to address in this study using a combination of biochemical, structural, and cellular assays.
Results
Reconstitution of poly(A)-triggered stalling in vitro
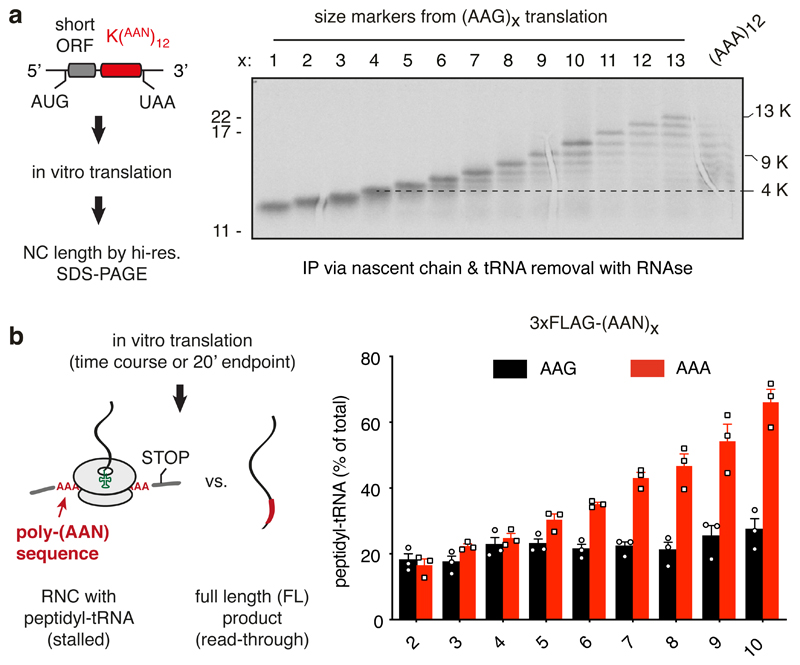
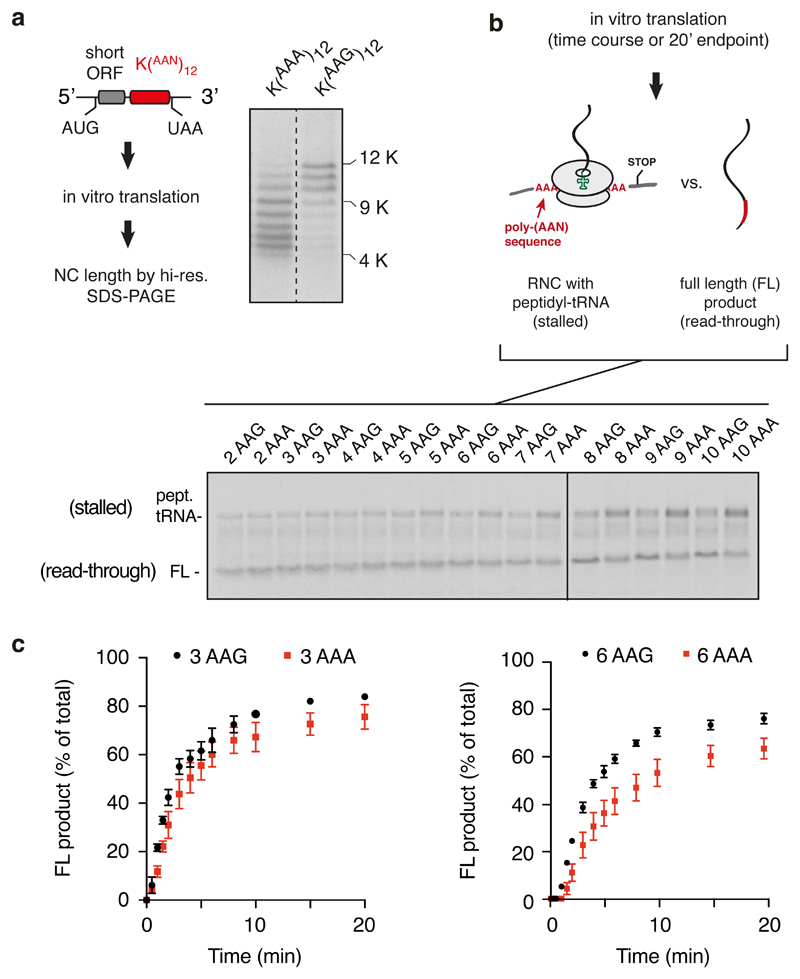
We sought to reconstitute in vitro the two key physiologically relevant features of poly(A)-mediated stalling: specificity for poly(A) over iterated AAG codons, and a length threshold above the longest poly(A) sequences within normal coding regions. We designed a synthetic mRNA containing a short open reading frame followed by in-frame iterated AAA or AAG codons preceding stop codons in all three frames (Fig. 1a, diagram). The short polypeptide facilitated single amino acid resolution on SDS-PAGE of the 35S-labelled translation products (Fig. 1a). Using translation extracts prepared from human cultured cells, we found that the majority of ribosomes stall after translating between 5-9 AAA codons (Fig. 1a; Extended Data Fig. 1a). Very little stalling was seen earlier than 5 AAA codons, and most ribosomes had stalled within 10 AAA codons. By contrast, ribosomes elongate more effectively through lysine-encoding AAG codons, with most ribosomes synthesising at least 10 lysine residues.
Fig. 1. Reconstitution of ribosome stalling on poly(A) mRNA in vitro.
(A) Analysis of nascent chain products resulting from in vitro translation of iterated AAG or AAA lysine codons in human cell lysate. Uncropped gel image is available online (Source Data). (B) Analysis of iterated AAG versus AAA codons for stalling in rabbit reticulocyte lysate. The translation reaction was performed for 20 min after which the proportion of stalled products was assessed by the relative amounts of peptidyl-tRNA versus full length polypeptide. The mean ± SEM (n=3) is plotted together with individual data points from independent experiments. The ‘background’ of ~20% peptidyl-tRNA even in the absence of stalling is due to failed termination at the stop codon, which is located within a few nucleotides of the 3’ end of the mRNA. Later in vitro stalling experiments with a longer 3’UTR that protrudes outside the mRNA channel (e.g., Fig. 6a) showed improved termination efficiency (~95%). An overly short 3’UTR presumably makes the mRNA more flexible in the mRNA channel and less able to recruit eRF1.
Similar codon- and length-dependence was observed in rabbit reticulocyte lysate translation extracts (Fig. 1b). In these experiments, we directly monitored stalling by analyzing tRNA attached versus the terminated product in constructs containing different numbers of AAA versus AAG codons. As shown in Extended Data Fig. 1b and quantified in Fig. 1b, no differences in read-through to the stop codon were seen for 4 or fewer iterated codons, with 6 codons showing the first reliable difference. Synchronised time courses of read-through (Extended Data Fig. 1c) showed essentially identical kinetics and efficiency of (AAA)3 versus (AAG)3, but a detectable difference between (AAA)6 versus (AAG)6. Thus, the in vitro translation system displays specificity of stalling on poly(A) over AAG-encoded poly-lysine and shows a length threshold beyond the longest poly(A) sequence found in any human open reading frame. These results suggest that the in vitro assembled poly(A)-stalled ribosome-nascent chain (RNC) complex is a physiologically relevant intermediate.
Cryo-EM structure of a poly(A)-stalled ribosome
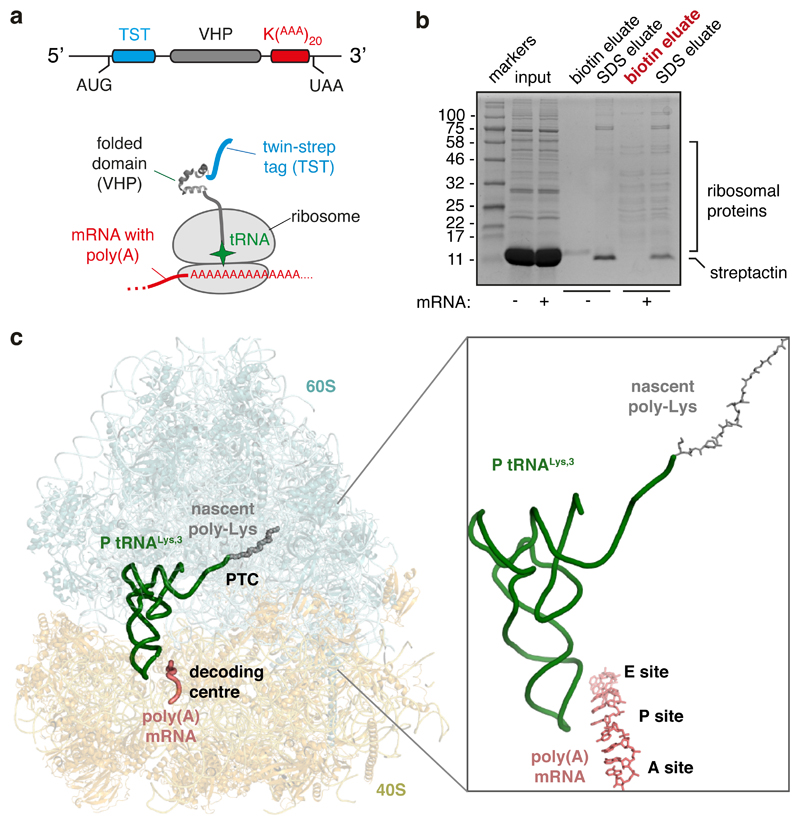
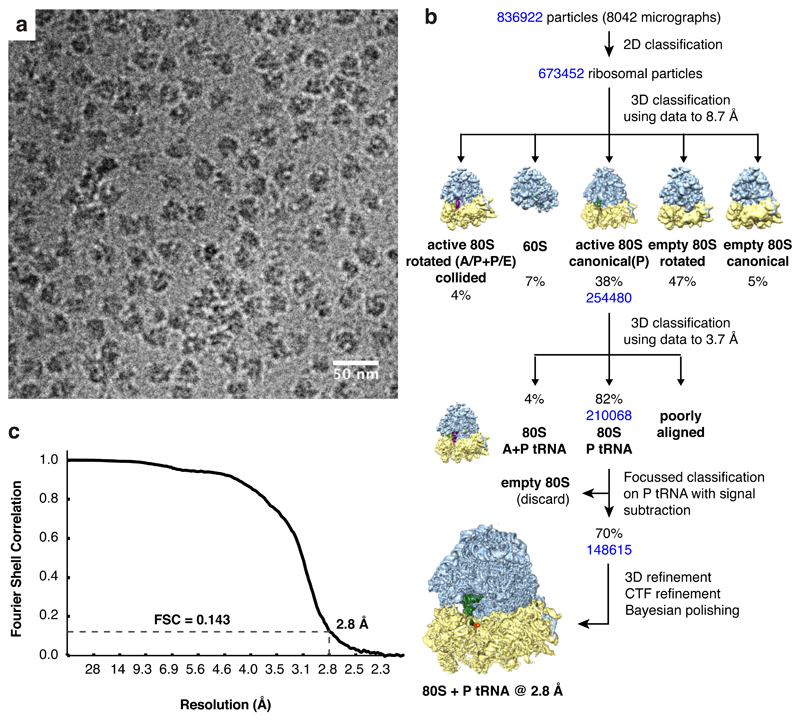
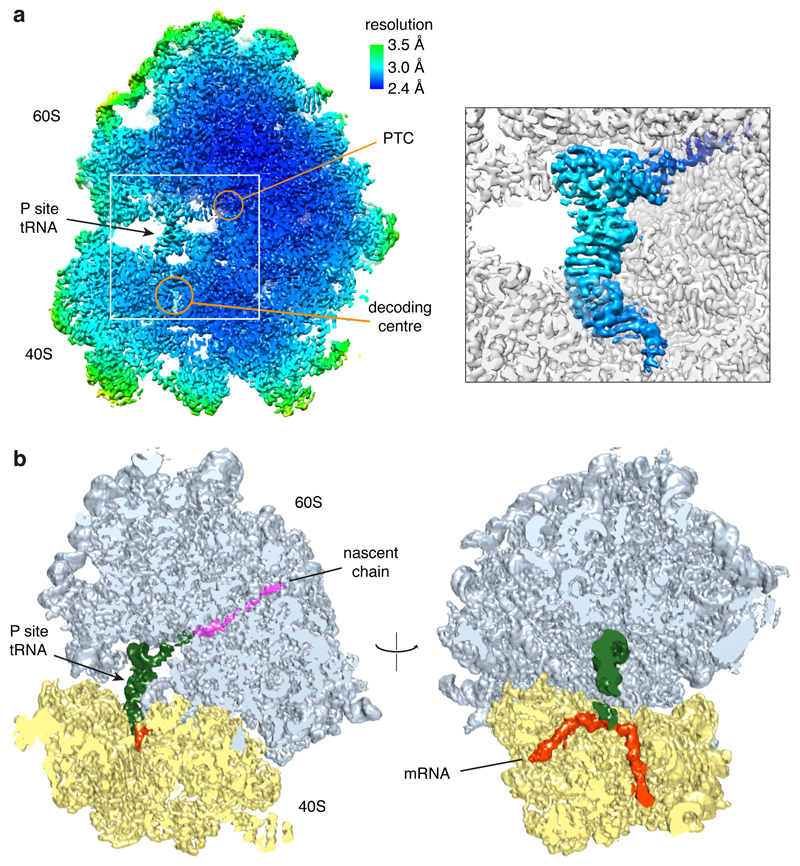
Using the rabbit reticulocyte lysate translation system and the construct shown in Fig. 2a, we affinity-purified poly(A)-stalled RNCs (Fig. 2b) and used single particle electron cryomicroscopy (cryo-EM) to analyze the ribosomal particles (Extended Data Fig. 2). Of the translating ribosomes, the majority (~90%) were observed in the canonical (non-rotated) state with P-site tRNA and the remainder were seen in the rotated state with hybrid tRNAs. This latter population proved to be trailing ribosomes within collided di-ribosome complexes as evidenced by additional EM density on the A-site side. Earlier experiments have shown that only ~10% of mRNAs undergo two rounds of initiation in this in vitro system34, explaining this di-ribosome population. We therefore excluded the trailing ribosomes from our analysis and reconstructed a structure from the majority population of poly(A)-stalled non-rotated ribosomes at an overall resolution of 2.8 Å (Fig. 2c; Extended Data Figs. 2, 3).
Fig. 2. Structure of the ribosome stalled during poly(A) translation.
(A) Schematic of the construct and ribosome-nascent chain complex (RNC) used for structure determination. (B) Characterization of the RNC purification strategy for structure determination. Rabbit reticulocyte lysate translation reactions containing or lacking the mRNA depicted in panel A were subjected to affinity purification via the N-terminal Twin-Strep-tag and eluted sequentially with biotin and SDS. Aliquots of the total translation reaction (input) and 32-fold excess of each eluate were analyzed by SDS-PAGE and staining with Coomassie blue. The sample subjected to cryo-EM analysis is indicated in red. (C) Overview of the poly(A)-stalled RNC structure with key elements indicated in the inset.
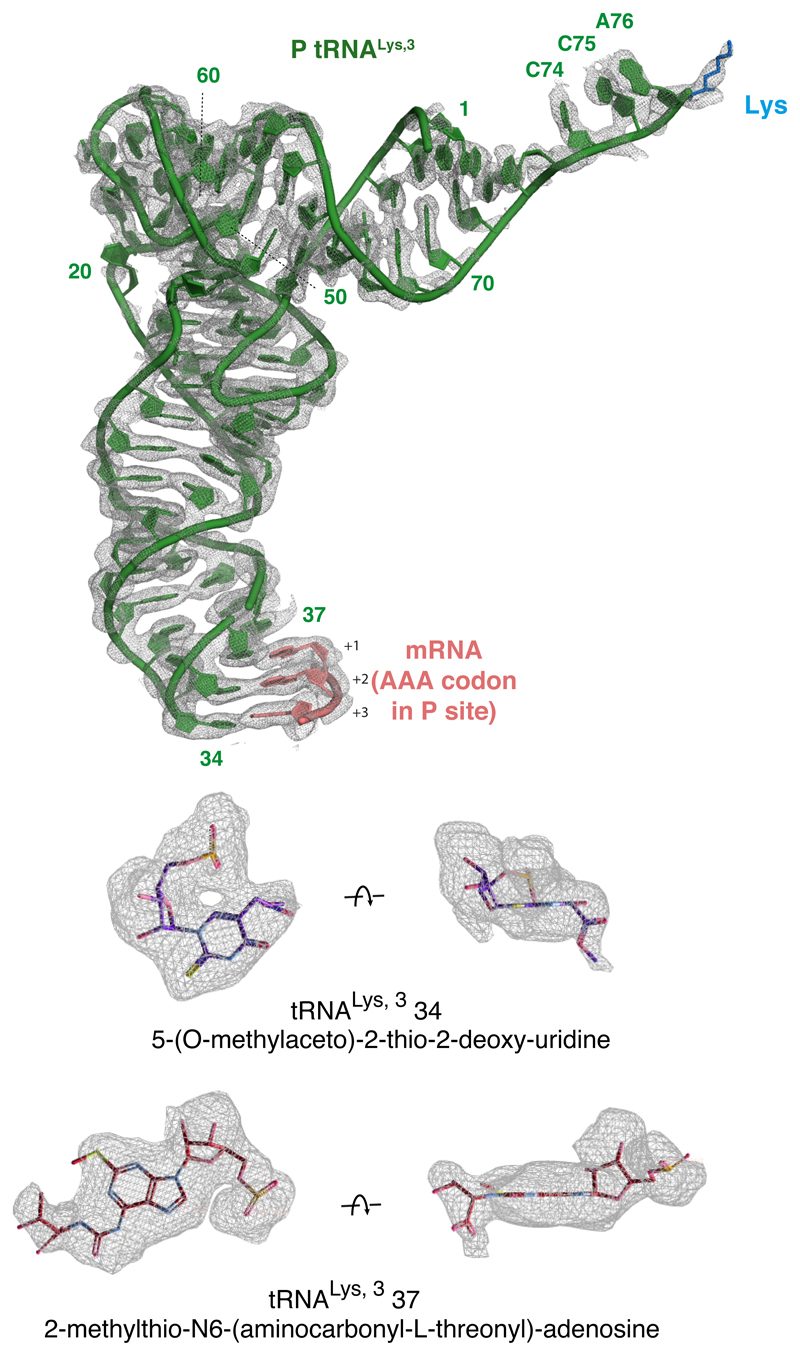
Based on biochemical analysis of stalling (Fig. 1), the particles used for reconstruction should contain nascent chains of different lengths due to stalling at different positions along the poly(A) sequence. Nevertheless, nearly all RNCs will contain at least five lysine residues at the C-terminus of the nascent chain and poly(A) throughout the mRNA channel, thereby improving homogeneity in these regions of the reconstruction. The structure shows P-site tRNALys,3 (which decodes the AAA codon) linked to a nascent chain (Fig. 2c; Extended Data Fig. 4). The A and E sites are unoccupied and weak density is seen for the recently described ‘Z-site’ tRNA48. Density for the nascent polypeptide is observed from the peptidyl-transferase center (PTC) through most of the ribosome exit tunnel, and density for the mRNA is seen through most of the mRNA channel (Extended Data Fig. 3). Local resolution approached 2.4 Å at the core, including at the PTC and the decoding center (Extended Data Fig. 3). This permitted the building and interpretation of atomic models at these key functional regions. As described in turn, the decoding center conformation impedes tRNA delivery and the nascent peptide geometry at the PTC impedes peptidyl transfer.
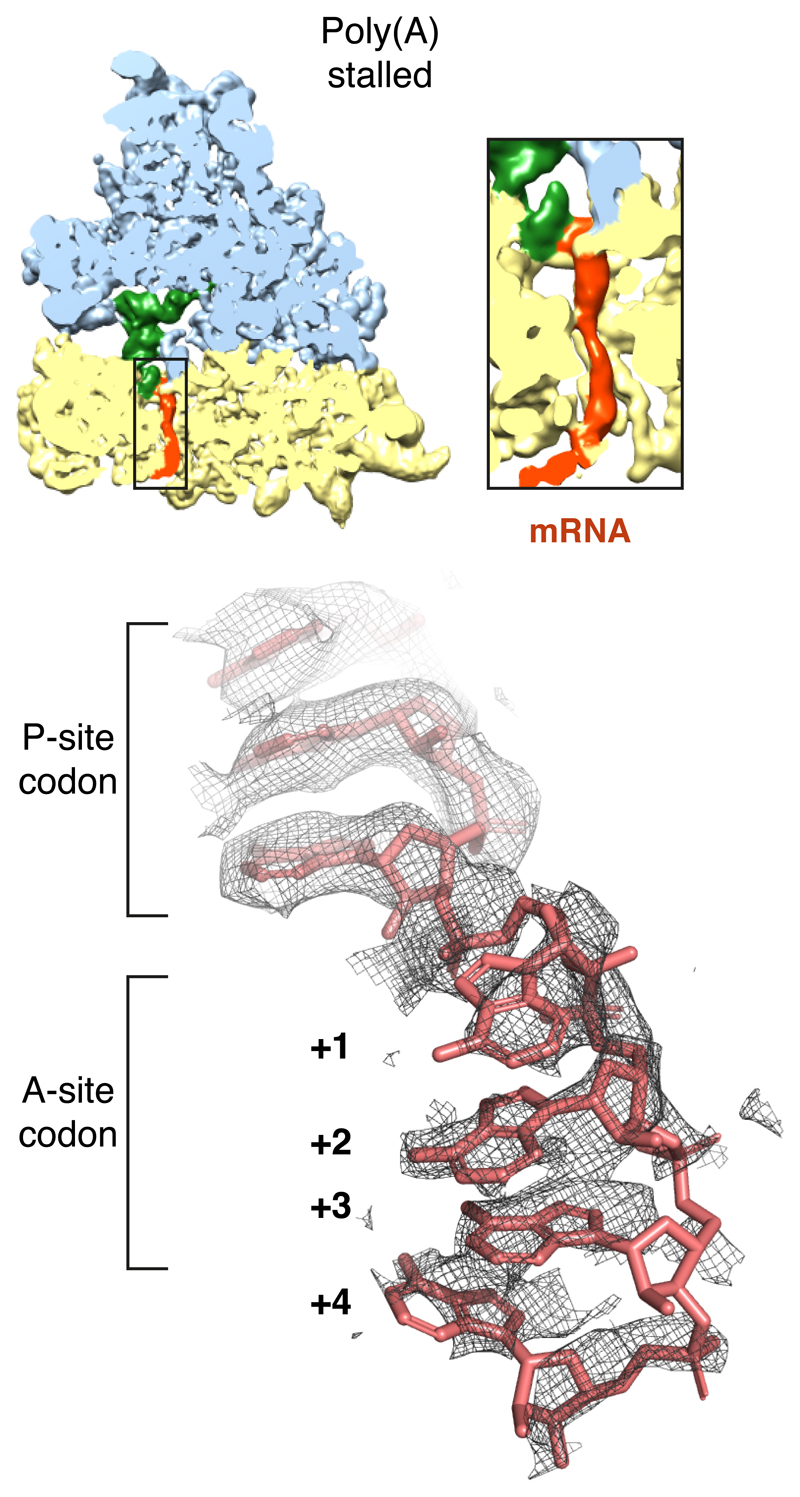
Decoding center rearrangement induced by poly(A)
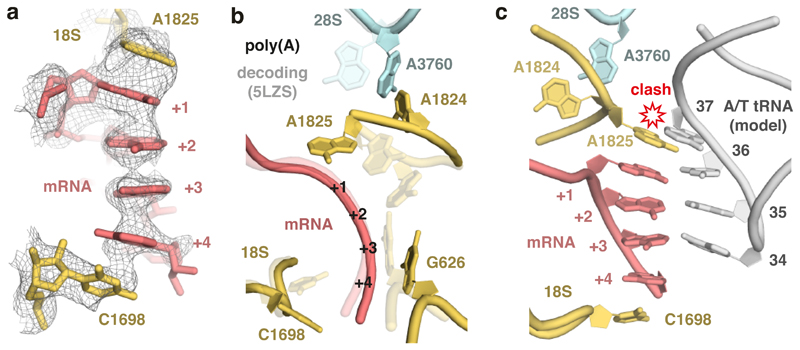
Because poly-lysine encoded by poly(A) stalls ribosomes more potently than poly-lysine encoded by AAG codons (Fig. 1a; Extended Data Fig. 1)14–16, we suspected that the mRNA might contribute to stalling. Inspection of the decoding center suggested a mechanism for poly(A) specificity. Four adenosine bases of the mRNA (+1 to +4) stack to form a single-stranded helix (Extended Data Fig. 5) that is stabilized by stacking interactions with the universally conserved 18S rRNA bases A1825 and C1698 (Fig. 3a). A1825 ordinarily occupies a ‘flipped-in’ position but flips ‘out’ and participates in the decoding of both sense and stop codons32,49. The flipped-out A1825 and C1698 nucleotides in the poly(A)-stalled ribosome stack on the poly(A) helix and differ from the positions seen during decoding (Fig. 3b) or termination32.
Fig. 3. Poly(A)-induced decoding center rearrangement.
(A) The ribosome A-site containing the poly(A) single-stranded helix (pink), capped by stacking interactions with 18S rRNA bases (yellow), is shown fitted within the EM density map (mesh). (B) Comparison of the decoding center configurations in the poly(A)-stalled ribosome (solid model) and the ribosome trapped during decoding (PDB 5LZS; transparent model). The mRNA bases and tRNA (in the case of decoding) are omitted for clarity. (C) Superimposed models of the poly(A)-stalled ribosome and the A/T tRNA (grey) positioned as it would be during decoding (PDB 5LZS). Base 37 of the tRNA clashes with 18S rRNA base A1825, which cannot move to its ‘flipped in’ position because A3760 of 28S rRNA occupies this space. Base modifications would be present at position 37 (and 34) when the incoming tRNA is the cognate Lys-tRNALys,3, but this is not shown for clarity.
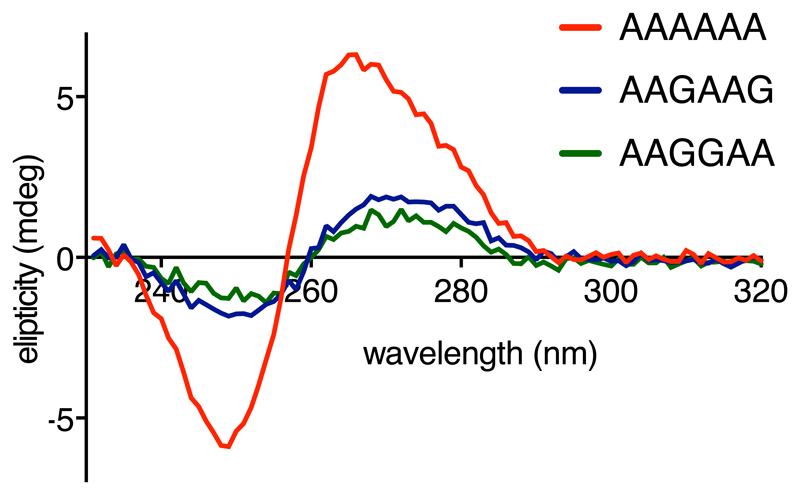
The rearranged decoding center in the poly(A)-stalled ribosome is incompatible with delivery of the tRNA•eEF1A•GTP ternary complex due to a clash between tRNA base 37 and A1825 (Fig. 3c). This clash would be particularly severe for Lys-tRNALys,3 due to a bulky 2-methylthio-N6-(aminocarbonyl-L-threonyl) modification on A37 (Extended Data Fig. 4). Notably, A1825 is disfavored from reverting to its ‘in’ position to relieve this clash for two reasons. First, its stacking interaction with the poly(A) helix extends and stabilizes this helix, favoring the ‘out’ position. Second, the universally conserved A3760 of 28S rRNA has moved relative to its usual position and now occupies the ‘in’ position of A1825 (Fig. 3b). Thus, the poly(A)-stalled ribosome contains a rearranged decoding center that is incompatible with Lys-tRNALys,3 delivery (Fig. 3c), providing one reason elongation is strongly impeded. Importantly, iterated AAG codons, which are less effective than poly(A) at triggering stalling (Fig. 1), are also less favorable for helix formation as measured by circular dichroism of oligonucleotides (Extended Data Fig. 6).
Suboptimal geometry of the nascent polypeptide at the PTC
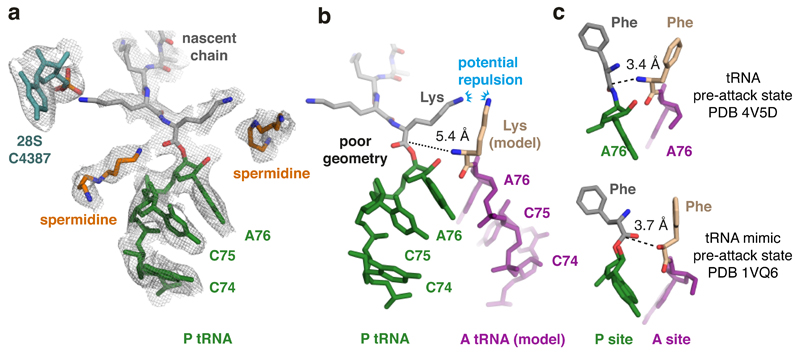
Although decoding center rearrangement explains how poly(A) mRNA contributes to stalling, it remained unclear why ribosomes do not stall at the first occurrence of AAAA in the A-site. An important clue to resolving this conundrum came from inspection of the PTC, which provided a second reason for impaired elongation. The most proximal lysine attached to A76 of the P-site tRNA is oriented with its side chain pointing toward the PTC’s A-site and its backbone carbonyl pointing in the opposite direction (Fig. 4a). This unusual orientation, not observed in an elongation-competent RNC structure (Extended Data Fig. 7), seems to be favored because the preceding lysine residue points in the opposite direction where it interacts with the backbone oxygen of 28S rRNA base C4387.
Fig. 4. Peptidyl-tRNA is mis-positioned in the poly(A) stalled ribosome.
(A) The P-site tRNA (green), attached nascent chain with the first three lysines (grey), and 28S rRNA residue C4387, which interacts with the penultimate Lys side chain, are shown fitted within the EM density map (mesh). Putative spermidine molecules (orange), hundreds of which are thought to bind ribosomes and facilitate translation50,51, were modeled into otherwise unaccounted density. (B) Superimposed models of the peptidyl-tRNA in the poly(A)-stalled ribosome with the A-site tRNA positioned for peptidyl transfer (PDB 4V5D). The amino acid in this structure (phenylalanine) was replaced with lysine to model the situation during poly(A) translation. The proximal lysine of the nascent chain faces the lysine of the accommodating A-site tRNA, resulting in potential charge repulsion between their respective epsilon amines. The backbone geometry of the nascent chain attachment to A76 is suboptimal for peptidyl transfer, as highlighted by the 5.4 Å distance between the α-amino group of the aminoacyl tRNA and the incorrectly oriented backbone carbonyl of the peptidyl tRNA. (C) Shown are models depicting the P- and A-site amino acids attached to the P- and A-site nucleotide (A76) for the indicated structures. Note that in both pre-attack structures, the attacking atom is within 4 Å of the P-site target bond, unlike the 5.4 Å distance in the poly(A)-stalled ribosome.
The consequence of this altered geometry would be two-fold: First, the P-site lysine side chain would form repulsive charge interactions with the lysine side chain of an incoming lysyl-tRNA at the A-site (Fig. 4b), potentially reducing the efficiency of accommodation. Second, the backbone carbon of the ester bond that is attacked by the α-amine of aminoacyl-tRNA is displaced further from the A-site relative to the position observed in mammalian elongation-competent RNCs (Extended Data Fig. 7) and bacterial peptidyl-transfer intermediates (Fig. 4c, compare to Fig. 4b). The altered backbone geometry and the ~5.4 Å distance between the α-amine and the ester bond, ordinarily less than 4 Å49, are likely to reduce the efficiency of peptide bond formation.
The PTC also contained two regions of unaccounted density that we have provisionally assigned as spermidine molecules. This assignment is plausible because spermidine is an abundant (~1-3 mM) cytosolic polyamine, fits the observed density, and is known to bind ribosomes at hundreds of sites50. Whether these putative spermidine molecules directly contribute to ribosome stalling or represent incidental interactions captured in our structure is currently unknown. The role of polyamines in any specific step of translation has been challenging to investigate because changes in polyamine levels potently impact multiple aspects of ribosome function in vitro and in cells50–52.
Poly-lysine in the exit tunnel impairs peptide bond formation
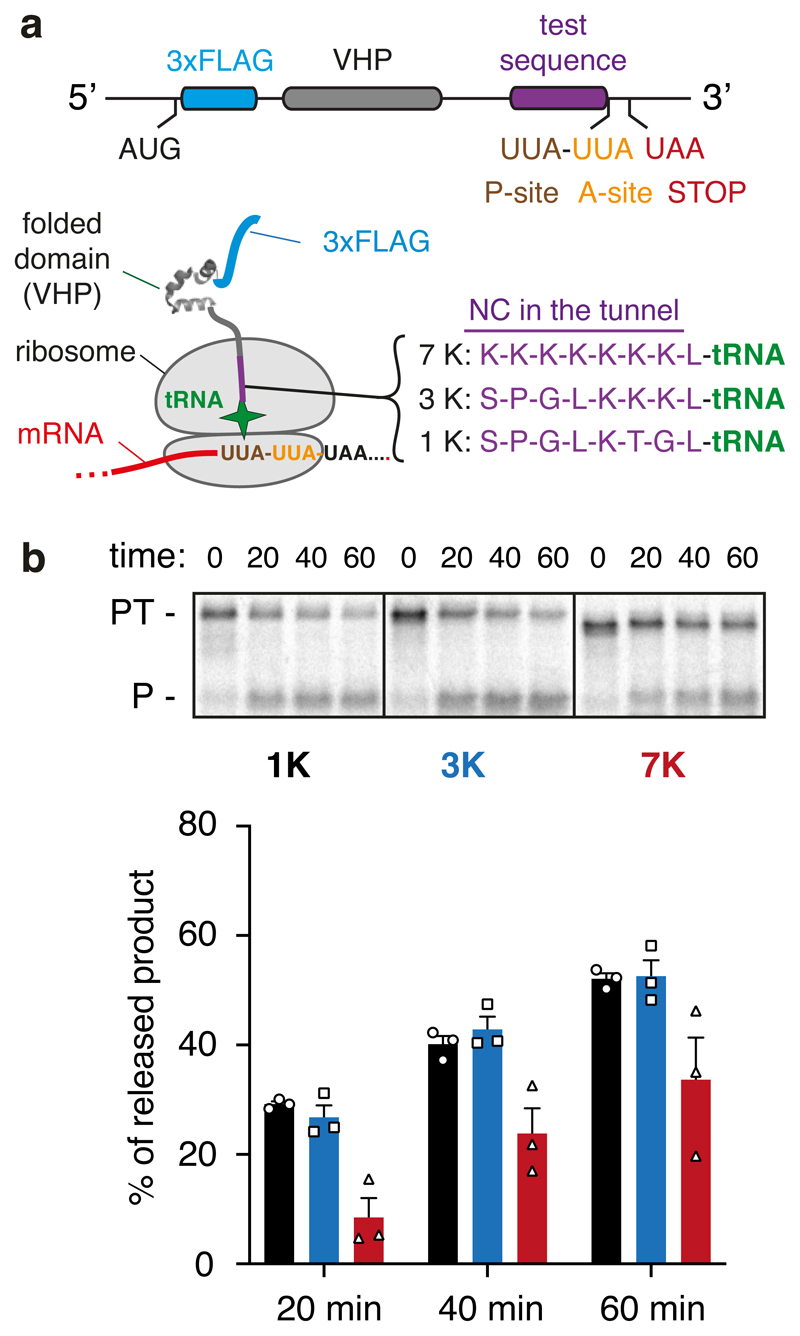
Our structural observations at the PTC suggested the possibility that peptide bond formation may be impaired when poly-lysine resides inside the ribosomal exit tunnel. To test this idea experimentally, we measured the puromycin reactivity of matched RNCs containing 1, 3 or 7 lysines (encoded by AAG codons) positioned in the proximal region of the exit tunnel (Fig. 5a). Puromycin mimics the 3’ end of an aminoacylated tRNA, binds to the A-site, and reacts with the P-site peptidyl-tRNA in a reaction identical to peptidyl-transfer. If nascent chain geometry is suboptimal for peptide bond formation, this should be reflected as reduced reactivity to puromycin. Importantly, the puromycin reaction exclusively reports on geometry at the PTC independent of any effects on elongation via changes at the decoding center.
Fig. 5. Analysis of PTC geometry by puromycin reactivity.
(A) Experimental strategy. A construct was designed to cause ribosome stalling at the second of two rare UUA leucine codons (see Methods). This RNC is known to be functional because translation resumes when liver tRNA is added53. RNCs were produced using this strategy in which one of three test sequences (7K, 3K, and 1K, as indicated) are positioned inside the ribosome tunnel. The lysine residues were encoded with AAG codons to minimize stalling before the desired UUA codon. After the translation reaction to synchronise ribosomes at the rare codon stall, the sample was moved to ice and the salt concentration was increased to 500 mM to prevent splitting of ribosomal subunits by rescue factors. Puromycin was then added to 2 μM final concentration and polypeptide release from tRNA was judged by SDS-PAGE. (B) Stalled RNCs containing the 1K, 3K, and 7K test sequences were evaluated for their reactivity to puromycin over a 60 min time course as a measure of peptidyl-transfer capacity. A representative experiment out of three is shown in the top panel, with the bottom panel showing a graph depicting the mean ± SEM (n=3) and individual data points from independent experiments. The positions of peptidyl-tRNA (PT) and puromycin-released peptide (P) are indicated to the left of the gel. Uncropped gel image is available online (Source Data).
To prepare the RNCs for this experiment, we exploited the fact that rabbit reticulocyte lysate has exceptionally low abundance of tRNAs for many codons that do not exist in hemoglobins. Earlier studies had established that the ribosome can be induced to stop at two sequential rare leucine codons (UUA) and re-started upon addition of total liver tRNA53. Because a single UUA does not stall ribosomes, we can deduce that the ribosome is stalled in a reversible manner with the second UUA codon in an A-site, awaiting the appropriate aminoacyl-tRNA. Importantly, any ribosomes that failed to stall would terminate, resulting in a polypeptide without an attached tRNA. Thus, peptidyl-tRNA at the start of the experiment is an indicator of a temporarily paused but functional elongation intermediate; its conversion to free nascent polypeptide by puromycin is an indicator of peptidyl-transfer.
Using this strategy, we found that elongation intermediates containing 7 exit tunnel lysine residues are substantially less reactive to puromycin than intermediates containing 1 or 3 lysine residues (Fig. 5b). A low concentration of puromycin (2 µM) was used to ensure that the rate of peptide release was sufficiently slow to allow accurate measurements to be made. Because all three samples contain leucyl-tRNA in the P-site and identical mRNA in the A site and beyond, the difference can be attributed to the preceding polypeptide sequence inside the exit tunnel. We conclude that 7 lysine residues inside the exit tunnel, similar to the sample analyzed by cryo-EM, is sufficient to impair peptide bond formation consistent with the altered geometry seen in the structure. Even though only the final three lysines were sufficiently static to be modelled accurately in the EM map, the preceding lysine residues are nevertheless important for impairing peptidyl transfer, presumably by constraining the final residues in a suboptimal geometry. We believe this explains earlier observations that basic residues inside the exit tunnel slow translation44.
Coincidence detection of poly-lysine and polyl(A) triggers stalling
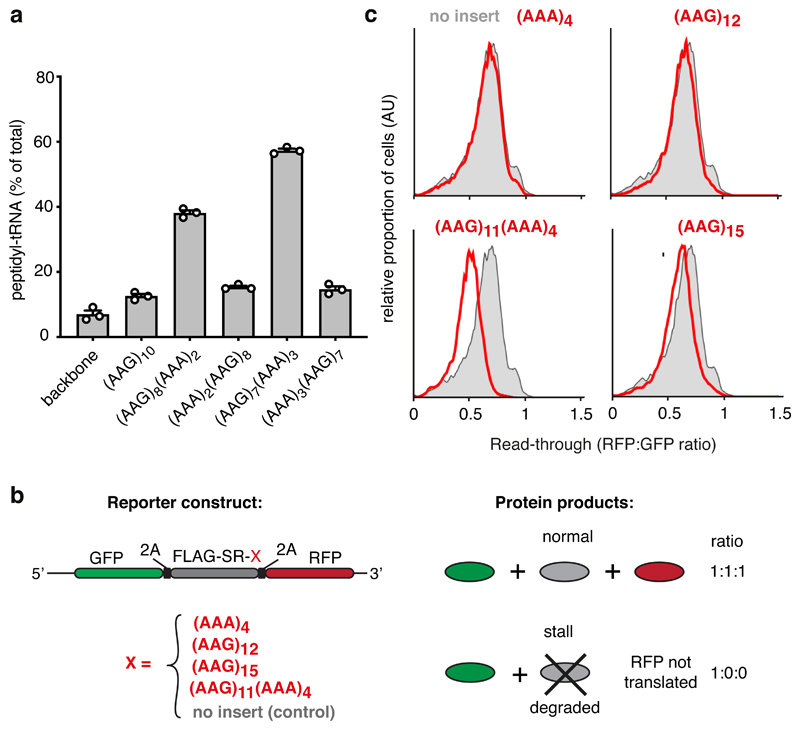
Based on our structural and biochemical observations, we postulated that poly-lysine mediated slowing via the PTC facilitates poly(A)-dependent rearrangement of the decoding center to mediate stalling. Such a ‘coincidence detection’ model would explain why stalling on poly(A) only occurs after several lysine residues have entered the ribosome exit tunnel, thereby avoiding stalls on short poly(A) sequences. To test the model, we analyzed ribosome stalling by poly-lysine encoded by (AAG)7(AAA)3, a sequence that would position seven lysine residues in the exit tunnel before decoding of (AAA)3 begins. Stalling efficiency with (AAG)7(AAA)3 was ~60% [comparable to (AAA)10] under conditions where (AAG)10 shows only background levels of stalling (Fig. 6a). In contrast, the reversed (AAA)3(AAG)7 sequence was ineffective at stalling, indicating that (AAA)3 cannot stall ribosomes in the absence of preceding lysine residues. Furthermore, the final (AAG)3 codons in this sequence, which are preceded by seven lysine residues already in the tunnel, does not trigger stalling.
Fig. 6. Coincidence detection of the nascent chain and mRNA mediate stalling.
(A) Iterated combinations of AAG and AAA codons inserted near the end of an open reading frame were analyzed for stalling by in vitro translation. The proportion of stalled product was assessed by the relative amount of peptidyl-tRNA versus full length terminated polypeptide and plotted as % peptidyl-tRNA. A construct lacking the AAG/AAA insert is shown for comparison (‘backbone’). The mean ± SEM (n=3) together with individual data points from independent experiments are plotted. (B) Schematic diagram of the dual-color reporter construct for detection of terminal stalling by flow cytometry. “2A” indicates the viral P2A sequence which causes skipping of peptide bond formation without interrupting elongation. If a test sequence inserted downstream of the FLAG-SR region causes terminal stalling, production of GFP is unaffected while RFP is not produced. Thus, the RFP:GFP ratio will be less than 1 and serves as a quantitative measure of terminal stalling. (C) Iterated combinations of AAG and AAA codons were analyzed for stalling in HEK293T cells using a dual color reporter in which the test sequence of interest (red text) is inserted between an N-terminal GFP and C-terminal RFP. Shown are histograms from flow cytometry analysis of the RFP:GFP ratio as a measure of ribosome read-through of the test sequence (red traces) relative to that seen in the absence of an insert (grey trace). Gating strategy for the histogram is shown in Source Data online.
Stalling was also induced by (AAG)8(AAA)2, with the reverse (AAA)2(AAG)8 having little effect (Fig. 6a), illustrating that even two AAA (but not AAG) codons in the mRNA channel are sufficient to stall ribosomes when preceded by poly-lysine. Yet, (AAA)4 does not detectably stall translation on its own (Fig. 1b), arguing that two lysine residues in the P-site of the PTC combined with (AAA)2 at the decoding center are not sufficient for stalling even though these are the minimal elements observed in the poly(A)-stalled structure. The experiment systematically increasing the length of poly(A) (Fig. 1b) instead shows that stalling efficacy of (AAA) codons in the mRNA channel increases progressively with the number of lysine residues in the exit tunnel. This result further argues that the observed configuration of the proximal two lysine residues (Fig. 4a) is progressively favored by weak exit tunnel interactions of upstream lysine residues. Although these upstream interactions are not sufficiently static to be seen in our structure, their effect on peptide bond formation (Fig. 5b) and on mRNA-mediated stalling (Fig. 6a) can be detected functionally.
Using a cell-based assay for ribosome stalling (Fig. 6b), we found that (AAA)4, which is insufficient to cause stalling, does so when preceded by (AAG)11 (Fig. 6c). The importance of (AAA)4 in stalling by (AAG)11(AAA)4 is evidenced by the nearly complete read-through of (AAG)12 or (AAG)15. Thus, synergy between lysine residues in the exit tunnel and poly(A) in the mRNA channel is seen in cells, although the threshold lengths for stalling are somewhat longer than the in vitro system (e.g., Fig. 6a). The different thresholds can be rationalised by the ~5-fold faster translation rate in cells, which allows ~5-fold less time per elongation cycle for decoding center rearrangement. Conversely, an artificially long time spent in the non-rotated state would favor decoding center rearrangement when the A-site contains (AAAA). Protracted waiting times would explain why decoding of even the second and third AAA codons (but not AAG codons) is highly inefficient in a fully reconstituted in vitro system47. Presumably, relatively dilute translation factors in cell-free translation reactions cause ribosomes to spend more time waiting for the next aminoacyl-tRNA, favoring the intramolecular (and hence, dilution-independent) rearrangement of the decoding center. Thus, our coincidence detection model in which stalling by (AAAA) in the decoding center depends on slowed elongation now rationalises otherwise inconsistent observations across experimental systems.
Discussion
Based on the findings in this study, we propose a model (Fig. 7) that explains why ribosomes elongate through short poly(A) stretches found in normal open reading frames but stall on long poly(A) stretches of an improperly processed mRNA. When poly(A) first enters the A-site, it likely samples the helical state very rapidly. Because decoding of (AAA)(ANN) is generally highly efficient, we infer that decoding center rearrangement is slower than tRNA•eEF1A•GTP engagement under normal conditions. For this reason, the ribosome elongates through the next few AAA codons until the lysine side chains of the nascent polypeptide begin making interactions with the proximal part of the exit tunnel.
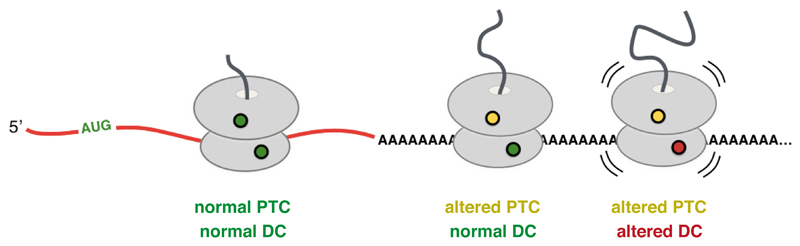
Fig. 7. Coincidence detection model for ribosome stalling on poly(A).
Translation of the first few AAA codons progressively slows elongation due to lysine interactions within the exit tunnel favoring a peptidyl-tRNA geometry that is suboptimal for peptide bond formation. In the context of slow peptide bond formation, the decoding center has greater opportunities for rearrangement, which further slows elongation and can cause the ribosome to stall. PTC and DC refer to peptidyl-transferase center and decoding center, respectively.
These interactions progressively stabilize the C-terminal region of the nascent polypeptide in a geometry unfavorable for peptidyl-transfer. In this geometry, the lysine side chain of the last amino acid points into the A site, causing steric and repulsive interactions with an incoming lysyl-tRNA. Interestingly, charge repulsion by long side chains has been observed to impede peptidyl-transfer in another context54. Here, the lysine or arginine side chain in the penultimate position of the nascent polypeptide can impede an incoming basic amino acid, an effect exaggerated by macrolide antibiotic binding in the exit tunnel. Inefficient peptidyl-transfer would increase the probability of tRNA dissociation from the A-site.
The additional time spent in the non-rotated, empty A-site configuration is permissive for rRNA engagement of the poly(A) helix, the presumed rate-limiting step in stalling. Poly(A) favors single-stranded helix formation due to stronger stacking interactions allowed by favorable electrostatics relative to other bases55. Furthermore, guanines disrupt poly(A) helices (Extended Data Fig. 6)56, explaining why iterated AAG codons are less effective stall sequences. Thus, coincidence detection of poly-lysine in the exit tunnel of the large subunit and poly(A) in the decoding center of the small subunit, over 90 Å away, allows ribosomes to selectively stall on poly(A) and avoid stalling within normal coding regions.
Once the decoding center has rearranged, the ribosome slows substantially. Even when a tRNA displaces this state, it may base pair incorrectly with the +2 to +4 bases to cause frameshifting as seen in functional assays14. Furthermore, accommodation and peptidyl-transfer are still impeded, providing opportunities for the rearranged state to form again. Severe ribosome slowing on poly(A) allows time for a trailing ribosome to catch up and collide. The collided ribosome is then recognised selectively by the ubiquitin ligase ZNF598 (Hel2 in yeast)34, which ubiquitinates the 40S subunit to initiate downstream recycling and quality control pathways34,35. In the absence of ZNF598, these downstream steps are not initiated, allowing the ribosome to elongate eventually through poly(A), albeit very slowly and with reduced reading frame fidelity14.
In addition to addressing the problem of ribosome stalling on poly(A), our study represents the first example to our knowledge of stalling due to mRNA interactions at the decoding center. All earlier ribosome stalling sequences operate only via PTC rearrangement induced by nascent chain interactions with the ribosome exit tunnel57. Our findings illustrate how the plasticity of the decoding center has also been exploited to selectively recognize particular mRNA sequences and trigger acute stalling. Because decoding center rearrangement takes time, mRNA recognition can be made contingent on elongation being modulated by the polypeptide chain. Thus, there may be other examples in which a modest slowing of elongation by the nascent polypeptide colludes with decoding center rearrangement to trigger stalling.
In addition to the nascent polypeptide57, elongation kinetics can be modulated by cellular stress58,59, protein folding60, and polypeptide-binding factors61, raising the possibility that stalling and its downstream consequences can be regulated by these parameters. For example, stalling at otherwise innocuous AAAA sequences within normal open reading frames might be triggered selectively in the context of slow elongation during some but not other situations. The human transcriptome contains a variety of short poly(A) sequences that are candidates for stall-mediated regulation as has been speculated16. Such regulation might be particularly prominent in the malaria parasite Plasmodium falciparum, which has numerous transcripts with poly(A) in their coding regions62 and whose abundances change markedly during its complex life cycle. More generally, the combination of the A site mRNA element and a conformational hindrance in the PTC might be more broadly used to modulate translation in other contexts63. Thus, variants of the coincidence detection mechanism described here could be used by cells as a novel mechanism to regulate translation and mRNA stability in response to changing cellular conditions.
Methods
Constructs
Templates for in vitro transcription and translation experiments in Fig 1a and Extended Data Fig. 1a used a gene block encoding 1xFLAG followed by 33 amino acids. Experiments in Fig. 1b and Extended Data Fig. 1b,1c employed a longer gene block encoding 3xFLAG, 52 amino acids, and three rare TTA codons used for pausing the ribosome in synchronised time courses (Extended Data Fig. 1c). These templates were used for PCR with a forward primer containing the SP6 promoter and reverse primer encoding the desired polybasic test sequence followed by stop codons in all three reading frames. PCR employed Phusion high fidelity polymerase, after which the PCR products were purified for use in transcription and translation reactions as previously described64. The in vitro translation experiments in Fig. 6a used an SP64-based plasmid encoding an N-terminal 3xFLAG tag, the autonomously folding villin headpiece (VHP) domain, and the unstructured cytosolic fragment of Sec61β65. This base plasmid was used to insert a stretch of 10 lysine codons (AAG and/or AAA as indicated in the figure) 30 codons downstream of the VHP domain and 30 codons upstream of the stop codon. The resulting plasmids were used as templates for PCR and in vitro transcription as described previously64. Experiments in Fig. 5 used the same base plasmid, except that custom reverse primers were used to generate constructs encoding the desired C-terminal test sequences. For cryo-EM sample preparation, we used an SP64-based plasmid encoding the twin-strep-tag (TST), the VHP domain, and the cytosolic domain of Sec61β followed by twenty AAA codons [K(AAA)20]. This plasmid was used as the template for PCR and in vitro transcription as described above. Constructs for in vivo based experiments presented in Fig. 6c were generated using previously described dual-fluorescent reporters encoding GFP-P2A-FLAG-[test sequence]-P2A-RFP14, where the test sequences are indicated in the figure.
Mammalian In vitro translations
Most in vitro translation reactions were in a rabbit reticulocyte lysate (RRL) system for 20 min at 32°C as described previously64. For the time course experiments in Extended Data Fig. 1c, we used RRL containing only its endogenous tRNA (i.e., not supplemented with liver tRNA). RRL is severely lacking in the tRNA used to decode the UUA codon53, resulting in the ribosome stalling at the (UUA)3 sequence just preceding the test sequence. Translation reactions were allowed to proceed for 10 minutes, then pactamycin was added to 0.2 µM final concentration to inhibit further initiation. At the same time, total pig liver tRNA was added to 50 µg/ml final concentration to restart translation elongation. Samples were collected at the indicated time points directly into SDS-PAGE sample buffer in order to immediately quench the reactions. The experiment in Fig. 5 testing puromycin sensitivity of various RNCs was also performed using RRL lacking liver tRNA. After a 20 min translation reaction at 32°C, samples were moved to ice and KAc was added to a final concentration of 500 mM. This prevents engagement of ribosome splitting factors, allowing the assay of puromycin reactivity without the confounding effects of ribosome splitting. Puromycin was then added to 2 µM final concentration and samples were moved to 32°C. Aliquots were removed at various time points and transferred directly into SDS-PAGE sample buffer for analysis by gel electrophoresis. Experiments in Fig. 1a and Extended Data Fig. 1a were performed in lysates generated from HEK 293T cells. In brief, translation-competent lysate was generated from four 15 cm plates of cells at around ~80% confluency. After two washes with ice cold PBS, cells were scraped and spun for 5 min at 1,000 rpm at 4°C. An additional wash followed by a spin was performed and cells were resuspended in 1 ml of hypotonic buffer (10 mM HEPES pH 7.6, 10 mM K(OAc), 1.5 mM Mg(OAc)2, 2 mM DTT). After swelling for 30 min on ice, cells were ruptured by passage through a pre-chilled 26G needle attached to a 2 ml syringe. Nuclei, cellular debris, and large organelles were sedimented by 15 min centrifugation at 15,000 g at 4°C in a tabletop microcentrifuge. The supernatant was collected and dialysed against 500 ml of dialysis buffer (10 mM HEPES pH 7.6, 90 mM K(OAc), 1.5 mM Mg(OAc)2, 1 mM DTT) for 2h at 4°C using a 0.5-3 ml 10,000 MWCO dialysis cassette. The dialysed lysate was spun for 15 min at 15,000 g at 4°C in a tabletop microcentrifuge to remove any precipitates. The lysate was supplemented with one-tenth the volume of 10x translation buffer [20 mM K(OAc), 15 mM MgCl2, 3 mM DTT, 130 mM HEPES pH 7.6, 4 mM spermidine, 0.4 mg/ml creatine kinase, 1 mg/ml pig liver tRNA, 120 mM creatine phosphate, 10 mM ATP, 10 mM GTP, 400 μM each of all amino acids except methionine]. The sample was then mixed well and spun again for 15 min at 15,000 g at 4°C to remove any aggregates. The resulting supernatant was aliquoted and flash frozen in liquid nitrogen. A typical translation reaction contained 12 μl of HEK lysate as prepared above, 1 μl of 35S Methionine (500 μCi/ml), 1 μl (~100 ng) of mRNA generated as described above, but further purified using RNeasy RNA isolation kit (Qiagen) and water to 20 μl. Translations proceeded for 30 min at 37°C. To purify nascent polypeptides, reaction mixtures were added to 450 μl of prechilled IP buffer [50 mM HEPES pH 7.6, 100 mM NaCl, 10 mM Mg(OAc)2, 0.5% Triton X-100] and 3.5 μl of anti-FLAG M2 affinity resin (Sigma). IPs were incubated for 1.5 h at 4°C with end-over-end rolling. After 4 washes with IP buffer, the resin was incubated with 10 μl of 1 x RNC buffer [50 mM HEPES ph 7.6, 100 mM K(OAc), 5 mM Mg(OAc)2] containing 0.2 mg/ml of RNAse A for 15 min at 37°C to digest the attached tRNA. All samples were analyzed directly by SDS-PAGE (except where digestion with RNAse A was performed as indicated on the individual figures) using 18% (Fig 1a and Extended Data Fig. 1a), 15% (Extended Data Fig. 1b, 1c), or 12% (Fig. 5b) Bis-Tris pH 6.5 gels run in MES-SDS running buffer. This gel system preserves the aminoacyl-tRNA species during electrophoresis. Quantification was performed by phosphorimaging followed by densitometry of the bands using Fiji software. Graphs were produced using GraphPad Prism. In each case, an arithmetic mean obtained from two (time course experiments) or three (remaining experiments) experiments was plotted with error bars corresponding to standard error of the mean (SEM) calculated using the GraphPad software.
Analysis of stalling in cells by flow cytometry
HEK 293T and HEK TRex cells were from Invitrogen and cultured in DMEM (Gibco) containing 10% Fetal Calf Serum (FCS). The cells were routinely tested for mycoplasma and found to be negative. Cells were not authenticated. For the analysis of stalling on polybasic sequences, cells were first plated on 24-well plate and transfected 24 h later with dual-fluorescent reporter constructs using TransIt 293 reagent (Mirus) according to manufacturer’s protocol. After 20 h of expression, cells were washed with PBS and trypsinised. After inhibiting trypsin, cells were collected into 1.5 ml tubes, spun at 5,000 rpm for 3 min at 4°C, washed with 1 ml of ice cold PBS, spun again and resuspended in 500 µl of ice cold PBS. Flow cytometry data was collected on LSRII instrument (Becton Dickinson) and analyzed by Flow Jo software. In each case, around 20,000 GFP-positive events were analyzed.
Circular dichroism (CD) measurements
CD measurements were performed as described before56. Briefly, RNA oligonucleotides (AAAAAA, AAGAAG and AAGGAA) were synthesized by Integrated DNA Technologies and dissolved in RNAse-free water to a final concentration of 250 μM, diluted to 25 μM in CD buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 1 mM Mg(OAc)2, 1 mM TCEP), and analyzed in a J-815 (Jasco) instrument at 340–200 nm at 0.5-nm intervals. The spectra were obtained at 50 nm min−1 with standard (100 mdeg) sensitivity. For each RNA, nine individual spectra were measured and averaged. A baseline of CD buffer alone was subtracted from RNA data. Plots were generated using Graphpad Prism.
Cryo-EM specimen and grid preparation
To prepare the poly(A)-stalled RNC, a 4 ml RRL-based translation reaction programmed with TST-VHPβ-K(AAA)20 mRNA was incubated for 30 min at 32°C. 40 µl of Strep-Tactin High Performance Sepharose beads (GE Healthcare) were directly incubated with the reaction mixture for 1.5 h at 4°C with end-over-end rolling. The resin was washed 5 times with 1x Ribosome-Nascent Chain (RNC) buffer (50 mM HEPES ph 7.6, 100 mM K(OAc), 5 mM Mg(OAc)2), then the RNCs were eluted with 80 µl of 1xRNC buffer containing 50 mM biotin for 1 h at 4°C. The eluate was adjusted to a ribosome concentration of ~100 nM (absorbance at 260 nm of 4.3) and was applied at 4°C at 100% relative humidity to glow-discharged UltrAuFoil R2/2 200 mesh grids coated with a 60 Å layer of amorphous carbon prepared in-house. After incubation for 30 s and blotting for 5 s, the grid was plunge-frozen into liquid ethane using a Vitrobot Mk III (FEI) and stored in liquid N2.
Data Collection
The poly(A)-stalled ribosome dataset was recorded on an FEI Falcon III camera in integrated mode using the M06 300 kV Titan Krios G3 microscope at the UK national electron bio-imaging center (eBIC) equipped with an X-FEG source and using EPU software (FEI). The dataset contained 8960 movies (19 frames; a dose of 2.2 e- frame-1 Å-2; 0.47s exposure; 75,000X magnification), resulting in a pixel size of 1.085 Å (refer to Table 1 for data statistics).
Table 1. Cryo-EM data collection, refinement and validation statistics.
| Poly(A)-stalled ribosome (EMD-10181, PDB 6SGC) |
|
|---|---|
| Data collection and processing | |
| Magnification | 75,000 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 41.8 |
| Defocus range (μm) | -1.5 to -2.7 |
| Pixel size (Å) | 1.085 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 673,452 |
| Final particle images (no.) | 148,615 |
| Map resolution (Å) | 2.80 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 2.4-3.5 |
| Refinement | |
| Initial model used (PDB code) | 5LZS |
| Model resolution (Å) | 2.80 |
| FSC threshold | 0.143 |
| Model resolution range (Å) | 2.4-3.5 |
| Map sharpening B factor (Å2) | -20 |
| Model composition | |
| Nonhydrogen atoms | 219,039 |
| Protein residues | 11,945 |
| Nucleic acid residues | 5,730 |
| Metals (Mg2+/Zn2+) | 275/8 |
| Ligands | 2 |
| B factors (Å2) | |
| Protein | 49.9 |
| Nucleotide | 64.5 |
| Ligand | 29.2 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.500 |
| Validation | |
| MolProbity score | 1.65 |
| Clashscore | 7.01 |
| Poor rotamers (%) | 0.01 |
| Ramachandran plot | |
| Favored (%) | 96.15 |
| Allowed (%) | 3.78 |
| Disallowed (%) | 0.07 |
Data Processing
Data processing for the poly(A)-stalled ribosome (Extended Data Fig. 2) was performed in RELION-3.066. Movies were aligned as 9 x 9 patches using MotionCorr267 with dose-weighting. Contrast transfer function (CTF) was estimated using CTFFIND-4.168 and 8042 micrographs with good CTF (and corresponding to a CTF figure of merit > 0.2 and CTF maximum resolution better than 6 Å) were selected for further processing. 839,989 particles were picked using an 80S ribosome 3D reference and extracted in a 512 pixel box, which was then downscaled into a 128 pixel box (4.34 Å/pixel). Particle sorting and two-dimensional classification were performed to yield 673,452 ribosome particles. Initial three-dimensional refinement was carried out using a 70 Å lowpass-filtered map of a rabbit ribosome as reference to yield a starting ribosome map with an estimated angular accuracy of 0.71°. The data was 3D-classified without alignments to isolate active 80S ribosomes in the canonical state with a P-site tRNA (38%, 254480 particles) and this subset was refined to an angular accuracy of 0.59° and re-extracted in a 400 pixel box (1.39 Å/pixel). To identify the subset of poly(A)-stalled ribosomes accurately, we performed another round of masked 3D classification without alignments (which retained 210068 particles or 82% of particles) and additionally performed focused classification with partial signal subtraction (FCwSS) on this subset. Briefly, PDB 5LZS32 was docked into the density of our starting ribosome map using UCSF Chimera and all ribosome signal outside a generous mask around the P/P tRNA was subtracted. Masked 3D classification into 5 classes was performed without image alignment for 25 iterations and a regularisation parameter (T) of 10. The procedure yielded a 70% class of 148615 particles with strong density for the P-site tRNA, mRNA and the most proximal lysine of the nascent chain. Subtracted ribosome density was restored and these particles were refined to a final angular accuracy of 0.35° to yield a 3.2 Å map. These particles were finally re-extracted again in a 512 pixel box (1.085 Å/pixel) and the newly-developed CTF refinement and bayesian polishing procedures were performed as described66. The final round of 3D refinement (angular accuracy of 0.24°) and post-processing using a generous solvent mask and a user-provided sharpening B-factor of -20 yielded a 2.8 Å map of the poly(A)-stalled ribosome (Extended Data Fig. 3).
Model Building
To build a model for the poly(A)-stalled ribosome, PDB 5LZS32 was first docked into the sharpened density for the stalled ribosome and the A/T and E/E tRNA and eEF1 chains were deleted. The mRNA sequence was mutated to (A)10. The P-site Lys-tRNALys,3 from a previously published crystal structure69 was rigid body-fitted into the P-site tRNA density as well as into the ‘Z-site’ tRNA48 density of a blurred map in Coot and real-space-refined. The L1 stalk and uL1 protein models were also added. Density for the most proximal three lysine residues of the nascent chain were sufficiently clear to allow placement of side chains. Poly-Lys (with only Cβ stubs) was modelled in the other positions in an extended conformation with Ramachandran restraints in Coot and had good geometry and torsion angles. The bond between the 3’O of A76 and the carbonyl C of the attached lysine was added as a custom bond geometric restraint via an additional parameter input file during refinement in phenix.real_space_refine. Two regions of unaccounted density near the PTC were fitted with spermidine molecules (ligand code SPD). We cannot be certain these densities represent spermidine, but it seems plausible because spermidine is an abundant cytosolic polyamine required for translation51,52, fits the observed density, and is well established to bind ribosomes in hundreds of sites50. The overall model was adjusted manually in Coot to conform with the density using suitably blurred maps (with B-factors between 0 and -200), saved in mmCIF/PDBx format and real space-refined using phenix.real_space_refine70. Model statistics (Table 1) were generated automatically using Molprobity via the Phenix GUI71. All reported resolutions are based on the Fourier shell correlation (FSC) 0.143 criterion72.
Molecular Graphics
Structural figures were generated using Pymol (Schrödinger, LLC), UCSF Chimera73 or Coot74. The FSC curve in Extended Data Fig. 2 was generated in Microsoft Excel and annotated in Adobe Illustrator CC 2019.
Extended Data
Extended Data Fig. 1. Additional characterization of ribosome stalling in vitro.
(A) A second example of nascent chain products resulting from in vitro translation of iterated AAG or AAA lysine codons in human cell lysate, as in Fig. 1A. The positions of nascent chain products containing 4, 9, or 12 lysines are indicated. (B) Analysis of iterated AAG versus AAA codons for stalling in rabbit reticulocyte lysate. The translation reaction was performed for 20 min after which the proportion of stalled products was assessed by the relative amounts of peptidyl-tRNA versus full length polypeptide. The ‘background’ of ~20% peptidyl-tRNA even in the absence of stalling is due to failed termination at the stop codon, which is located within a few nucleotides of the 3’ end of the mRNA. Later in vitro stalling experiments with a longer 3’UTR that protrudes outside the mRNA channel showed improved termination efficiency (~95%). An overly short 3′UTR presumably makes the mRNA more flexible in the mRNA channel and less able to recruit eRF1. Multiple experiments such as this one were quantified to produce the graph shown in Fig. 1B. (C) Time course of the appearance of full length (FL) product for constructs containing the indicated number of iterated AAG or AAA codons. Translation was synchronised by first pausing the ribosome at a run of rare leucine codons just preceding the poly-basic encoding sequence, then restarting translation at time 0 by addition of tRNA. The mean ± SEM for each time point calculated from two experiments are plotted.
Extended Data Fig. 2. Cryo-EM analysis of ribosomes stalled on poly(A).
(A) Representative micrograph of poly(A)-stalled ribosomes used for single particle analysis. (B) Data processing scheme used for structure determination in Relion 3.0. 3D classification reveals that ~90% of active ribosomes are in the canonical state with P/P tRNA while ~10% are seen in the rotated state with A/P and P/E hybrid state tRNAs. The majority of the rotated state ribosomes also contain density for a preceding ribosome and therefore represent ribosomes that have collided with a poly(A)-stalled ribosome. (C) Fourier shell correlation (FSC) curve of the final map illustrating an overall resolution of 2.8 Å.
Extended Data Fig. 3. Characterization of cryo-EM map.
(A) Local resolution of the poly(A)-stalled ribosome sliced through the centre. The positions of key elements are indicated. PTC: peptidyl-transferase centre. Inset (right) highlights the high local resolution at the PTC and decoding centre (B) Slices through the density map at the plane of the polypeptide exit tunnel (left) and mRNA channel (right). Continuous nascent chain density corresponding to a mixture of poly-Lys lengths and Cα positions is contoured at a different level to the rest of the map and is shown in magenta, and mRNA density is shown in red. The P site tRNA is green, 40S subunit in yellow, and 60S subunit in light blue.
Extended Data Fig. 4. Experimental EM density for P-site Lys-tRNALys,3.
Map-to-model fits for the P-site Lys-tRNALys,3 with the AAA codon of the mRNA in the P-site and the first nascent chain side chain (Lysine). Base modifications at positions 34 and 37 of the tRNA are shown within the EM density.
Extended Data Fig. 5. Views of the mRNA density in the EM map of the poly(A)-stalled ribosome.
The density map is sliced through the ribosome in a plane that reveals the decoding centre and shows the mRNA within the small subunit. The large and small subunits (blue and yellow, respectively), P-site tRNA (green) and mRNA (red) are colored. The inset shows a zoomed in region of the mRNA channel, illustrating that the poly(A) mRNA is ordered through most of the channel. The bottom panel shows the mRNA density in the P- and A-sites in the final refined and sharpened map. The mRNA is well ordered in the P-site due to base-pairing with the P-site tRNA, and ordered in the A-site due to stabilizing interactions with rRNA as shown in Fig. 3.
Extended Data Fig. 6. Guanosine interrupts the intrinsic helical propensity of poly(A).
Circular dichroism (CD) spectra of AAAAAA (red), AAGAAG (blue) and AAGGAA (green) RNA oligonucleotides are plotted. These spectra are averaged from 9 independent measurements performed on the same samples. The AAAAAA oligo displays a CD signature characteristic for the helical conformation of poly(A), as described previously 52. Introduction of guanosines significantly disrupts this helical structure.
Extended Data Fig. 7. Comparison of peptidyl-tRNA geometry in different mammalian RNC structures.
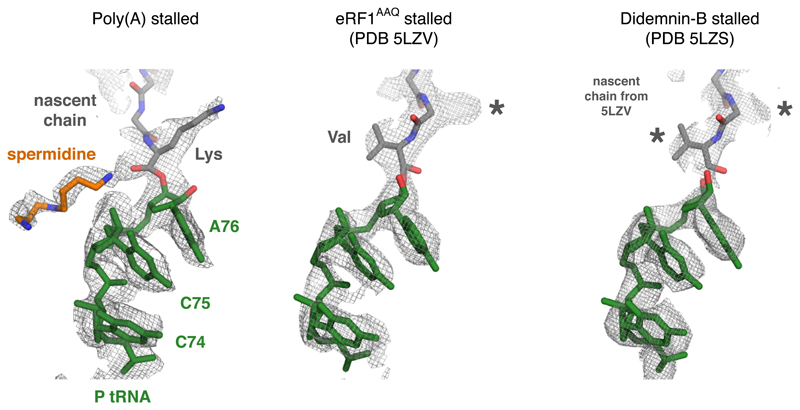
Shown are the EM density maps for the peptidyl-tRNA region at the PTC for the indicated structures. The fitted models are shown for the poly(A)-stalled ribosome and the RNC stalled at the stop codon with a dominant-negative eRF1AAQ mutant (PDB code 5LZV). The 5LZV RNC is in a geometry competent for peptidyl-transfer (or in this case, peptide release by eRF1). The structure from the didemnin-B stalled RNCs contains a mixture of nascent chains stalled at different positions. Thus, the nascent chain density represents an average of a variety of peptidyl-tRNAs. Note that the nascent chain model from 5LZV fits well into the density map, indicating that the majority of peptidyl-tRNAs assume this configuration during active elongation. The geometry for the poly(A) peptidyl-tRNA is unambiguously different from this optimal geometry. Lys and Val refer to the lysine and valine side chains of modeled nascent chains. The asterisks indicate density for side chains that are not shown.
Acknowledgments
We thank T. Tang and L. Passmore for useful discussions, help with CD measurements, and sharing data prior to publication; J. Grimmett and T. Darling for advice, data storage and high performance computing; M. Daly, G. Cannone, and J. Brown for technical support; S. Scheres, T. Nakane and P. Emsley for advice; the MRC Laboratory of Molecular Biology Electron Microscopy Facility for access and support of electron microscopy, sample preparation and data collection; Diamond for access and support of the Cryo-EM facilities at the UK national electron bio-imaging center (eBIC) (proposal EM17434-53, funded by the Wellcome Trust, MRC and BBSRC); Z. Yang for data collection support at eBIC; Hegde and Ramakrishnan lab members for useful discussions. This work was supported by the UK Medical Research Council (MC_UP_A022_1007 to RSH and MC_U105184332 to VR), a Wellcome Trust Senior Investigator award (WT096570), the Agouron Institute, and the Louis-Jeantet Foundation (VR), Bio-X graduate fellowship to JC and NIH grants GM51266 and GM113078 to JDP.
Footnotes
Reporting summary statement - Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The poly(A)-stalled ribosome map has been deposited to the EMDB with accession codes EMDB 10181. Atomic coordinates have been deposited to the Protein Data Bank under accession code PDB 6SGC. A re-refined version of 5LZV that includes the ester bond between the P-site tRNA and the attached Valine of the nascent chain with the correct bond length (used in Extended Data Fig. 7b, 7c) is available upon request. Source data for Fig. 1a, 5a, 6c, and Extended Data Fig. 1a, 1b are available with the paper online. All other data are available upon request.
Author contributions: V.C. generated the EM structures, built and interpreted molecular models, and wrote the first draft of the manuscript; S.J. prepared and characterized samples for structure determination, performed biochemical and cell assays of stalling, and interpreted these data; J.C. and J.D.P. provided supporting data that corroborated the stalling model; A.B. and S.S. produced an initial stalled ribosome structure that seeded the project; V.R. provided overall project guidance and helped interpret the structure; R.S.H. conceived the project, provided overall project guidance, helped interpret the findings, and wrote later drafts of the manuscript; all authors contributed to manuscript editing.
Competing Interests Statement: The authors declare no competing interests.
References
- 1.Wolff S, Weissman JS, Dillin A. Differential scales of protein quality control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science (80-. ) 2016;353 doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 3.Labbadia J, Morimoto RI. The Biology of Proteostasis in Aging and Disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol. 2019;20:406–420. doi: 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hoof A, Wagner EJ. A brief survey of mRNA surveillance. Trends Biochem Sci. 2011;36:585–592. doi: 10.1016/j.tibs.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shao S, Hegde RS. Target Selection during Protein Quality Control. Trends Biochem Sci. 2016;41:124–137. doi: 10.1016/j.tibs.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013;29:691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inada T. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim Biophys Acta - Gene Regul Mech. 2013;1829:634–642. doi: 10.1016/j.bbagrm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–4. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letzring DP, Wolf AS, Brule CE, Grayhack EJ. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA. 2013;19:1208–17. doi: 10.1261/rna.039446.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao S, Von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuboi T, et al. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3’ end of aberrant mRNA. Mol Cell. 2012;46:518–29. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 14.Juszkiewicz S, Hegde RS. Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol Cell. 2017;65:743–750.e4. doi: 10.1016/j.molcel.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sundaramoorthy E, et al. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol Cell. 2017;65:751–760.e4. doi: 10.1016/j.molcel.2016.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arthur LL, et al. Translational control by lysine-encoding A-rich sequences. Sci Adv. 2015;1:e1500154. doi: 10.1126/sciadv.1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frischmeyer PA, et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science (80-. ) 2002;295:2258–2261. doi: 10.1126/science.1067338. [DOI] [PubMed] [Google Scholar]
- 18.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–4. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 19.Bengtson MH, Joazeiro CaP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–24. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Defenouillere Q, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–54. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–17. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yonashiro R, et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. Elife. 2016;5:1–16. doi: 10.7554/eLife.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izawa T, Park S-H, Zhao L, Hartl FU, Neupert W. Cytosolic Protein Vms1 Links Ribosome Quality Control to Mitochondrial and Cellular Homeostasis. Cell. 2017;171:890–903.e18. doi: 10.1016/j.cell.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Izawa T, et al. Roles of Dom34:Hbs1 in Nonstop Protein Clearance from Translocators for Normal Organelle Protein Influx. Cell Rep. 2012;2:447–453. doi: 10.1016/j.celrep.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Choe Y-J. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature. 2016;531:191–5. doi: 10.1038/nature16973. [DOI] [PubMed] [Google Scholar]
- 30.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–54. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Becker T, et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–20. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 32.Shao S, et al. Decoding Mammalian Ribosome-mRNA States by Translational GTPase Complexes. Cell. 2016;167:1229–1240.e15. doi: 10.1016/j.cell.2016.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simms CL, Yan LL, Zaher HS. Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol Cell. 2017;68:361–373.e5. doi: 10.1016/j.molcel.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juszkiewicz S, et al. ZNF598 Is a Quality Control Sensor of Collided Ribosomes. Mol Cell. 2018;72:469–481.e7. doi: 10.1016/j.molcel.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeuchi K, et al. Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 2019;38:e100276. doi: 10.15252/embj.2018100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuo Y, et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00188-1. 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitron CS, Park JH, Brandman O. Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA. 2017;23:798–810. doi: 10.1261/rna.060897.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saito K, Horikawa W, Ito K. Inhibiting K63 polyubiquitination abolishes no-go type stalled translation surveillance in Saccharomyces cerevisiae. PLoS Genet. 2015;11:e1005197. doi: 10.1371/journal.pgen.1005197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozsolak F, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–29. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–31. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garzia A, et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun. 2017;8 doi: 10.1038/ncomms16056. 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graber JH, Cantor CR, Mohr SC, Smith TF. Genomic detection of new yeast pre-mRNA 3’-end-processing signals. Nucleic Acids Res. 1999;27:888–94. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guydosh NR, Green R. Translation of poly(A) tails leads to precise mRNA cleavage. RNA. 2017;23:749–761. doi: 10.1261/rna.060418.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J, Deutsch C. Electrostatics in the Ribosomal Tunnel Modulate Chain Elongation Rates. J Mol Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem. 2009;284:10343–10352. doi: 10.1074/jbc.M808840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamble CE, Brule CE, Dean KM, Fields S, Grayhack EJ. Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell. 2016;166:679–690. doi: 10.1016/j.cell.2016.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koutmou KS, et al. Ribosomes slide on lysine-encoding homopolymeric A stretches. Elife. 2015;4 doi: 10.7554/eLife.05534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown A, Baird MR, Yip MC, Murray J, Shao S. Structures of translationally inactive mammalian ribosomes. Elife. 2018;7 doi: 10.7554/eLife.40486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 50.Dever TE, Ivanov IP. Roles of polyamines in translation. J Biol Chem. 2018;293:18719–18729. doi: 10.1074/jbc.TM118.003338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandal S, Mandal A, Johansson HE, Orjalo AV, Park MH. Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc Natl Acad Sci. 2013;110:2169–2174. doi: 10.1073/pnas.1219002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atkins JF, Lewis JB, Anderson CW, Gesteland RF. Enhanced differential synthesis of proteins in a mammalian cell-free system by addition of polyamines. J Biol Chem. 1975;250:5688–95. [PubMed] [Google Scholar]
- 53.Feng Q, Shao S. In vitro reconstitution of translational arrest pathways. Methods. 2018;137:20–36. doi: 10.1016/j.ymeth.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 54.Sothiselvam S, et al. Binding of Macrolide Antibiotics Leads to Ribosomal Selection against Specific Substrates Based on Their Charge and Size. Cell Rep. 2016;16:1789–99. doi: 10.1016/j.celrep.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mignon P, Loverix S, Steyaert J, Geerlings P. Influence of the - interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res. 2005;33:1779–1789. doi: 10.1093/nar/gki317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang TTL, Stowell JAW, Hill CH, Passmore LA. The intrinsic structure of poly(A) RNA determines the specificity of Pan2 and Caf1 deadenylases. Nat Struct Mol Biol. 2019 doi: 10.1038/s41594-019-0227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson DN, Arenz S, Beckmann R. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr Opin Struct Biol. 2016;37:123–133. doi: 10.1016/j.sbi.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Shalgi R, et al. Widespread Regulation of Translation by Elongation Pausing in Heat Shock. Mol Cell. 2013;49:439–452. doi: 10.1016/j.molcel.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu B, Han Y, Qian S-B. Cotranslational Response to Proteotoxic Stress by Elongation Pausing of Ribosomes. Mol Cell. 2013;49:453–463. doi: 10.1016/j.molcel.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cymer F, von Heijne G. Cotranslational folding of membrane proteins probed by arrest-peptide–mediated force measurements. Proc Natl Acad Sci. 2013;110:14640–14645. doi: 10.1073/pnas.1306787110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butkus ME, Prundeanu LB, Oliver DB. Translocon "pulling" of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J Bacteriol. 2003;185:6719–22. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Agirrezabala X, et al. Ribosome rearrangements at the onset of translational bypassing. Sci Adv. 2017;3:e1700147. doi: 10.1126/sciadv.1700147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma A, Mariappan M, Appathurai S, Hegde RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 2010;619:339–63. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell. 2013;50:637–48. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–21. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bénas P, et al. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA. 2000;6:1347–55. doi: 10.1017/s1355838200000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Afonine PV, et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr Sect D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen VB, et al. MolProbity : all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 73.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 74.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]