Abstract
Introduction
Methylenetetrahydrofolate reductase (MTHFR) is essential in mediating folate metabolism, and thus plays an important role in diabetes and diabetic complications. MTHFR C677T (rs1801133 C>T) polymorphism has been proposed to be linked with type 2 diabetes mellitus (T2DM) susceptibility. However, the conclusions are inconsistent. Therefore, we rechecked their linkage aiming to obtain a more reliable estimation by performing an updated meta‐analysis.
Methods
We searched electronic databases PubMed, EMBASE, CNKI, and Wanfang to obtain studies updated to October 2019.
Results
After carefully screening, we finally incorporated 68 studies with 10,812 cases and 8,745 controls. The genotype frequency of C677T polymorphism was analyzed pooled to generate odds ratios (ORs) and 95% confidence intervals (CIs). Pooled results presented that MTHFR C677T polymorphism was significantly associated with T2DM under homozygous (OR = 1.64, 95% CI = 1.39–1.94), heterozygous (OR = 1.38, 95% CI = 1.20–1.59), recessive (OR = 1.41, 95% CI = 1.23–1.61), dominant (OR = 1.47, 95% CI = 1.27–1.70), and allele (OR = 1.37, 95% CI = 1.23–1.52) genetic models. Stratified analysis demonstrated that C677T genotype was associated with T2DM in Asian populations, but not Caucasian and African populations.
Conclusion
Our results indicated that MTHFR C677T polymorphism confers to T2DM, especially in Asian populations. Much more large‐scale case–control studies are needed to strengthen such conclusion in the future.
Keywords: C677T, meta‐analysis, MTHFR, polymorphism, T2DM
Pooled results presented that MTHFR C677T polymorphism was significantly associated with type 2 diabetes mellitus (T2DM) under homozygous (odds ratio [OR] = 1.61, 95% confidence interval [CI] = 1.37–1.90), heterozygous (OR = 1.32, 95% CI = 1.15–1.51), recessive (OR = 1.39, 95% CI = 1.22–1.58), dominant (OR = 1.40, 95% CI = 1.22–1.60), and allele (OR = 1.32, 95% CI = 1.20–1.45) genetic models. Stratified analysis demonstrated that C677T genotype was associated with T2DM in Asian populations, but not Caucasian and African populations. Our results indicated that MTHFR C677T polymorphism confers to T2DM, especially in Asian populations.
1. INTRODUCTION
Type 2 diabetes mellitus (T2DM) is a major public health problem that not only affects individual life quality, but also increases social economic burden (DeFronzo et al., 2015). The frequency of T2DM in China is increasing quickly, with estimation of about 380 million T2DM patients by 2025 (van Dieren, Beulens, van der Schouw, Grobbee, & Neal, 2010). The etiology of T2DM remains partly elucidated. Evidences suggest that T2DM is a complex disease caused by the combinations of environmental and genetic risk factors (Wareham, Franks, & Harding, 2002; Zeggini et al., 2007).
Previous reports showed that individuals with insufficient intake of folic acid were more likely to have T2DM. Folate is a methyl group donor in the synthesis of intracellular methylation reactions and de novo deoxynucleoside (Blount et al., 1997). When folate deficiency, the DNA stability will be impaired (Duthie, 1999). Methylenetetrahydrofolate reductase (MTHFR) is a folate‐metabolizing enzyme that participates in folic acid circulation and DNA synthesis (Friso et al., 2002). MTHFR catalyzes the irreversible reduction of 5,10‐methylenetetrahydrofolate to 5‐methyltetrahydrofolate (Niclot et al., 2006). Dysfunction or low activity of MTHFR may decrease the level of methyl pool; consequently, it inhibits the successful deoxynucleoside synthesis and intracellular methylation reactions (Rozen, 1997).
The human gene MTHFR (OMIM number: 607093) is located on chromosome 1p36.3. Of all the identified SNPs, C677T (Ala222Val, rs1801133 C>T) is one of the most investigated genetic variations (Adinolfi et al., 2005; Liew & Gupta, 2015). The C677T polymorphism is a C to T transition at base pair 677, which results in the amino acid transition from Ala to Val. Such amino acid transition significantly decreases the activity of MTHFR (Weisberg, Tran, Christensen, Sibani, & Rozen, 1998). Recent data suggested that there exist an association between C677T and the susceptibility of T2DM. However, the role of C667T in risk of T2DM was discrepant. Several meta‐analyses that were conducted to solve this conflicting role somehow failed. To get a precise estimation, we re‐analyzed the role of MTHFR C677T on T2DM via including larger eligible investigations.
2. MATERIALS AND METHODS
2.1. Literature search
We carried out a comprehensive literature search in the following databases: PubMed, EMBASE, CNKI, and Wanfang. The searching was updated to October 2019 without any language limitations. The combination of the following search terms was adopted: ‘MTHFR or methylenetetrahydrofolate reductase’, and ‘polymorphism or polymorphisms or SNP or single nucleotide polymorphism or variant’ and ‘diabetes or mellitus or diabetes mellitus or T2DM’. To expand the included studies, we also retrieved eligible references from the selected studies. The GenBank reference sequence and version number for the gene is: MTHFR (NM_005957.5).
2.2. Inclusion/exclusion criteria
We set the following criteria when performing the selection work: (a) evaluating the association of MTHFR C677T polymorphism with T2DM risk; (b) case–control design; (c) odds ratios (ORs) and their 95% confidence intervals (CIs) were able to obtain; and (d) reports Hardy–Weinberg equilibrium (HWE). Exclusion criteria were as follows: (a) reviews or meta‐analyses; (b) case‐only studies or case reports; and (c) duplicate publications.
2.3. Data extraction
We arranged three authors to handle data extraction: two authors to extract data independently and one author to resolve the disagreement. The following data were selectively extracted from each study: first author's surname, year of publication, country, ethnicity, genotyping methods, and genotypic distribution. The stratification analysis was conducted by ethnicity (Asians, Caucasians, and Africans) and HWE (HWE <0.05 and HWE >0.05).
2.4. Statistical methods
STATA 11.0 software (Stata Corporation) was adopted to conduct the current meta‐analysis. We first used Chi‐square test to check whether the genotype frequency of C677T among the controls was in HWE. After that, we determined the relationship between MTHFR C677T polymorphism and T2DM risk by calculating pooled ORs with the corresponding 95% CIs. We totally used five genetic models: homozygote model (TT vs. CC), heterozygote model (CT vs. CC), recessive model (TT vs. CT/CC), dominant model (CT/TT vs. CC), and allele model (T vs. C) to detect such relationship. Stratification analyses were also taken by ethnicity (Asian, Caucasian, and African) and HWE (HWE <0.05 and HWE >0.05), aiming to detect the source of heterogeneity. We carried out Chi‐square‐based Q statistic test and inconsistency index statistics (I 2) to calculate heterogeneity between study results. If the studies were homogeneous (with phet < .10 or I 2 > 50%), the random‐effects model (the DerSimonian and Laird method) was chosen. Otherwise, ORs were calculated using the fixed‐effects model (the Mantel‐Haenszel method). Sensitivity analysis was performed to assess the strength of the conclusion, by sequentially excluding each study at a time. Begg's funnel plot and Egger's linear regression test were conducted to assess publication bias. We also conducted quality assessment to detect the quality of each study using the quality assessment criteria (He et al., 2014). All the statistics were two‐sided. p < .05 was considered as significant.
3. RESULTS
3.1. Study search
General process of publication selection was graphically shown in Figure 1. Initial retrieval from PubMed and EMBASE databases got a total of 78 and 45 potentially relevant published records, respectively. We also obtained 18 articles from Chinese databases CNKI and Wanfang. After titles and abstracts screening, 81 nonrelevant records were excluded. The remaining 60 articles and eight additional articles identified from retrieved studies were included in the final meta‐analysis (Al‐Harbi et al., 2015; Al‐Salihi, Ajeena, Al‐Kashwan, & Al‐Lebban, 2016; Zidan, El Mougy, Moustafa, El attar, & Mohamed, 2019; Benrahma et al., 2012; Bluthner et al., 1999; Cao, Huang, Mao, & Gao, 2005; Chang et al., 2011; Chen, Ning, Zhu, Li, & Shi, 2004; Chen et al., 2010; P. Chen, Pan, Sun, Bai, & Fu, 2008; Dai & Yu, 2012; El Hajj Chehadeh et al., 2016; Eroglu et al., 2007; Errera et al., 2006; Fekih‐Mrissa et al., 2017; Fujita et al., 1999; Guo, Pan, Chu, Guo, & Sun, 2005; Guo et al., 2002; Hu, Zhang, Fang, Qin, & Liu, 2009; Hu, Gan, Li, & Bi, 2001; Jimenez‐Ramirez et al., 2017; Ksiazek, Bednarek‐Skublewska, & Buraczynska, 2004; Lin, Wang, & Liu, 2009; Liu et al., 2014; Luo, Yan, Li, Cheng, & Song, 2007; Luo, Yan, Ma, Cheng, & Song, 2008; Mao, Gao, Qin, & Shi, 2004; Mehri et al., 2010; Mei, Chen, & Zheng, 2012; Mtiraoui et al., 2007; Neugebauer, Baba, & Watanabe, 1998; Nithya et al., 2017; Odawara & Yamashita, 1999; Pirozzi et al., 2018; Qiu, 2009; Rahimi et al., 2009; Ramanathan, Harichandana, Kannan, Elumalai, & Sfd, 2019; Raza, Abbas, Siddiqi, & Mahdi, 2017; Settin, El‐Baz, Ismaeel, Tolba, & Allah, 2015; Shang, Wang, & Liu, 2017; Shi, He, Cheng, Wang, & Liu, 2006; J. Shi, Li, Yu, Chen, & Tao, 2002; Shpichinetsky et al., 2000; Soares et al., 2008; J. Sun, Xu, Xue, Zhu, & Lu, 2005; Sun, Xu, & Zhu, 2001; Sun, Xu, Zhu, & Lu, 2004; Sun, Xu, Lu, & Zhu, 2009; Sun, Chen, et al., 2004; Sun, Wang, Shi, & Yang, 2013; Wang et al., 2017; Wang, Hu, Xiao, & Wan, 2014; Wang, Wang, & Li, 2018; Wang, Wang, Xue, Chen, & Zou, 2001; Wang, Wang, Xue, Cheng, et al., 2001; Wen, Lu, Li, Wu, & Zhang, 2008; Wirta et al., 1998; Xiao, Hu, Shan, Guan, & Ren, 2006; Xu, Zhang, Shan, & Ma, 2003; Yang, Lu, & Pan, 2001; Yilmaz, Agachan, Ergen, Karaalib, & Isbir, 2004; Yoshioka et al., 2004; Yue, Liu, Kang, Hu, & Qiu, 2006; Zhang, Li, Liu, & Hu, 2007; Zhang, Xiang, Weng, & Li, 2002; Zhang & Liu, 2009; Zhi et al., 2016; Zhou, Li, & Zhang, 2004).
Figure 1.
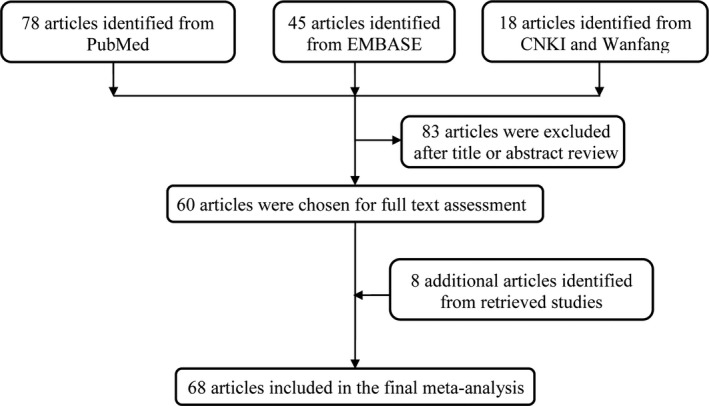
Search flow diagram
3.2. Study characteristics
The study characteristics of the final selected studies were presented in Table 1. A total of 68 studies with 10,812 cases and 8,745 controls were included in our final meta‐analysis. Among these eligible studies, 52 were done on Asians, 11 studies were done on Caucasians, and five studies were done on Africans. As to the HWE, genotype distribution in the controls of 52 studies was agreed with the HWE, and 16 studies were not. We also classified the studies into low‐quality studies (48 studies) and high‐quality studies (20 studies) by quality score.
Table 1.
Characteristics of studies included in the current meta‐analysis
| Surname | Year | Country | Ethnicity | Genotype method | Case | Control | HWE | Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | Total | CC | CT | TT | Total | |||||||
| Neugebauer | 1998 | Japan | Asian | PCR‐RFLP | 24 | 31 | 12 | 67 | 86 | 43 | 17 | 146 | 0.003 | 6 |
| Wirta V | 1998 | Finland | Caucasian | PCR‐RFLP | 46 | 30 | 8 | 84 | 60 | 48 | 7 | 115 | 0.520 | 8 |
| Bluthner | 1999 | Germany/Poland | Caucasian | PCR‐RFLP | 74 | 50 | 23 | 147 | 67 | 68 | 15 | 150 | 0.708 | 6 |
| Fujita | 1999 | Japan | Asian | PCR‐RFLP | 31 | 57 | 17 | 105 | 20 | 39 | 9 | 68 | 0.142 | 7 |
| Odawara | 1999 | Japan | Asian | PCR‐RFLP | 52 | 65 | 26 | 143 | 38 | 68 | 25 | 131 | 0.578 | 7 |
| Shpichinetsky | 2000 | Israel | Caucasian | PCR‐RFLP | 23 | 22 | 10 | 55 | 21 | 16 | 6 | 43 | 0.316 | 3 |
| Hu S | 2001 | China | Asian | PCR‐RFLP | 49 | 48 | 16 | 113 | 30 | 24 | 1 | 55 | 0.121 | 5 |
| Sun J | 2001 | China | Asian | PCR‐RFLP | 32 | 33 | 20 | 85 | 10 | 16 | 31 | 57 | 0.008 | 4 |
| Wang L | 2001 | China | Asian | PCR‐RFLP | 52 | 68 | 41 | 161 | 37 | 36 | 12 | 85 | 0.502 | 5 |
| Wang L | 2001 | China | Asian | PCR‐RFLP | 65 | 75 | 39 | 179 | 37 | 38 | 10 | 85 | 0.959 | 7 |
| Yang G | 2001 | China | Asian | PCR‐RFLP | 17 | 27 | 23 | 67 | 26 | 28 | 8 | 62 | 0.914 | 6 |
| Guo Q | 2002 | China | Asian | PCR‐RFLP | 12 | 19 | 22 | 53 | 12 | 11 | 5 | 28 | 0.391 | 7 |
| Shi J | 2002 | China | Asian | PCR‐RFLP | 12 | 31 | 7 | 50 | 22 | 29 | 5 | 56 | 0.291 | 5 |
| Zhang G | 2002 | China | Asian | PCR‐RFLP | 56 | 108 | 34 | 198 | 40 | 49 | 11 | 100 | 0.484 | 7 |
| Xu J | 2003 | China | Asian | PCR‐RFLP | 39 | 54 | 30 | 123 | 20 | 25 | 7 | 52 | 0.853 | 8 |
| Chen A | 2004 | China | Asian | PCR‐RFLP | 24 | 45 | 22 | 91 | 21 | 9 | 5 | 35 | 0.038 | 7 |
| Ksiazek P | 2004 | Poland | Caucasian | PCR‐RFLP | 159 | 123 | 44 | 326 | 71 | 83 | 16 | 170 | 0.237 | 10 |
| Mao L | 2004 | China | Asian | PCR‐RFLP | 35 | 37 | 11 | 83 | 26 | 18 | 3 | 47 | 0.960 | 8 |
| Sun J | 2004 | China | Asian | PCR‐RFLP | 102 | 76 | 42 | 220 | 74 | 34 | 22 | 130 | <0.001 | 9 |
| Sun L | 2004 | China | Asian | PCR‐RFLP | 27 | 52 | 27 | 106 | 29 | 18 | 3 | 50 | 0.925 | 7 |
| Yilmaz H | 2004 | Turkey | Caucasian | PCR‐RFLP | 121 | 98 | 30 | 249 | 101 | 93 | 20 | 214 | 0.831 | 8 |
| Yoshioka K | 2004 | Japan | Asian | PCR‐RFLP | 21 | 13 | 6 | 40 | 71 | 107 | 29 | 207 | 0.260 | 9 |
| Zhou J | 2004 | China | Asian | PCR‐RFLP | 16 | 78 | 45 | 139 | 8 | 31 | 30 | 69 | 0.998 | 8 |
| Cao H | 2005 | China | Asian | PCR‐RFLP | 14 | 20 | 6 | 40 | 26 | 18 | 3 | 47 | 0.960 | 7 |
| Guo L | 2005 | China | Asian | PCR‐RFLP | 60 | 51 | 50 | 161 | 58 | 34 | 35 | 127 | <0.001 | 8 |
| Sun J | 2005 | China | Asian | PCR‐RFLP | 101 | 78 | 49 | 228 | 63 | 31 | 20 | 114 | <0.001 | 10 |
| Errera FI | 2006 | Brazil | Caucasian | PCR‐RFLP | 44 | 41 | 10 | 95 | 36 | 57 | 14 | 107 | 0.244 | 9 |
| Shi C | 2006 | China | Asian | PCR‐RFLP | 108 | 60 | 18 | 186 | 68 | 34 | 7 | 109 | 0.338 | 8 |
| Xiao Y | 2006 | China | Asian | PCR‐RFLP | 16 | 53 | 4 | 73 | 47 | 25 | 1 | 73 | 0.245 | 7 |
| Yue H | 2006 | China | Asian | PCR‐RFLP | 66 | 131 | 55 | 252 | 17 | 11 | 2 | 30 | 0.903 | 8 |
| Eroglu Z | 2007 | Turkey | Caucasian | PCR‐RFLP | 51 | 45 | 7 | 103 | 63 | 58 | 7 | 128 | 0.171 | 7 |
| Luo D | 2007 | China | Asian | PCR‐RFLP | 65 | 102 | 26 | 193 | 42 | 35 | 14 | 91 | 0.151 | 7 |
| Mtiraoui N | 2007 | Tunisia | Caucasian | PCR‐RFLP | 163 | 135 | 62 | 360 | 270 | 94 | 36 | 400 | <0.001 | 12 |
| Zhang C | 2007 | China | Asian | PCR‐RFLP | 28 | 29 | 19 | 76 | 34 | 19 | 12 | 65 | 0.006 | 8 |
| Chen P | 2008 | China | Asian | PCR‐RFLP | 19 | 70 | 27 | 116 | 14 | 73 | 37 | 124 | 0.014 | 9 |
| Luo D | 2008 | China | Asian | PCR‐RFLP | 59 | 63 | 19 | 141 | 43 | 31 | 11 | 85 | 0.166 | 8 |
| Soares AL | 2008 | Brazil | Caucasian | PCR‐RFLP | 15 | 8 | 2 | 25 | 9 | 5 | 2 | 16 | 0.363 | 3 |
| Wen J | 2008 | China | Asian | PCR‐RFLP | 22 | 50 | 23 | 95 | 27 | 25 | 5 | 57 | 0.816 | 6 |
| Hu L | 2009 | China | Asian | PCR‐RFLP | 47 | 63 | 49 | 159 | 26 | 17 | 9 | 52 | 0.053 | 7 |
| Lin R | 2009 | China | Asian | PCR‐RFLP | 56 | 36 | 47 | 139 | 93 | 22 | 24 | 139 | <0.001 | 10 |
| Qiu Y | 2009 | China | Asian | PCR‐RFLP | 83 | 68 | 48 | 199 | 53 | 29 | 18 | 100 | <0.001 | 9 |
| Rahimi Z | 2009 | Iran | Asian | PCR‐RFLP | 33 | 27 | 5 | 65 | 33 | 22 | 4 | 59 | 0.898 | 5 |
| Sun J | 2009 | China | Asian | PCR‐RFLP | 94 | 73 | 48 | 215 | 78 | 38 | 26 | 142 | <0.001 | 10 |
| Zhang Q | 2009 | China | Asian | PCR‐RFLP | 66 | 94 | 66 | 226 | 26 | 17 | 9 | 52 | 0.053 | 8 |
| Chen A | 2010 | China | Asian | PCR‐RFLP | 57 | 74 | 27 | 158 | 34 | 17 | 4 | 55 | 0.373 | 8 |
| Mehri S | 2010 | Tunisia | African | PCR‐RFLP | 50 | 49 | 16 | 115 | 66 | 38 | 12 | 116 | 0.078 | 8 |
| Chang YH | 2011 | China | Asian | PCR | 1 | 25 | 30 | 56 | 3 | 23 | 36 | 62 | 0.781 | 6 |
| Houda Benrahma | 2012 | Morocco | African | PCR‐RFLP | 160 | 97 | 25 | 282 | 114 | 122 | 26 | 262 | 0.420 | 10 |
| Dai H | 2012 | China | Asian | PCR‐RFLP | 51 | 54 | 15 | 120 | 31 | 27 | 2 | 60 | 0.176 | 8 |
| Mei Q | 2012 | China | Asian | PCR‐RFLP | 17 | 51 | 23 | 91 | 17 | 70 | 37 | 124 | 0.076 | 8 |
| Sun L | 2013 | China | Asian | PCR‐RFLP | 180 | 243 | 48 | 471 | 30 | 42 | 6 | 78 | 0.094 | 11 |
| Liu K | 2014 | China | Asian | PCR‐RFLP | 103 | 54 | 6 | 163 | 54 | 23 | 0 | 77 | 0.123 | 8 |
| Han Wang | 2014 | China | Asian | TaqMan | 234 | 293 | 66 | 593 | 298 | 312 | 70 | 680 | 0.377 | 12 |
| Al‐Harbi EM | 2015 | Bahrain | Asian | PCR‐RFLP | 116 | 43 | 12 | 171 | 135 | 47 | 6 | 188 | 0.449 | 10 |
| Ahmad Settin | 2015 | Egypt | African | PCR‐RFLP | 111 | 65 | 27 | 203 | 156 | 135 | 20 | 311 | 0.195 | 11 |
| Al‐Salihi NJ | 2016 | Iraqi | Asian | PCR‐RFLP | 28 | 28 | 5 | 61 | 12 | 10 | 0 | 22 | 0.167 | 4 |
| El Hajj Chehadeh SW | 2016 | United Arab Emirates | Asian | TaqMan | 155 | 49 | 5 | 209 | 132 | 27 | 10 | 169 | <0.001 | 10 |
| Xueyuan Zhi | 2016 | China | Asian | TaqMan | 28 | 86 | 66 | 180 | 76 | 172 | 102 | 350 | 0.826 | 11 |
| Fekih‐Mrissa N | 2017 | Tunisia | African | PCR‐RFLP | 56 | 102 | 2 | 160 | 124 | 68 | 8 | 200 | 0.726 | 11 |
| Jimenez‐Ramirez FJ | 2017 | Puerto Rico | Caucasian | PCR‐RFLP | 72 | 8 | 9 | 89 | 184 | 159 | 57 | 400 | 0.020 | 10 |
| K Nithya | 2017 | India | Asian | PCR‐RFLP | 173 | 25 | 2 | 200 | 94 | 6 | 0 | 100 | 0.757 | 10 |
| Raza ST | 2017 | India | Asian | PCR‐RFLP | 152 | 162 | 65 | 379 | 102 | 52 | 26 | 180 | <0.001 | 11 |
| Shang G | 2017 | China | Asian | PCR‐RFLP | 84 | 106 | 36 | 226 | 66 | 91 | 37 | 194 | 0.573 | 11 |
| Wang D | 2017 | China | Asian | PCR‐RFLP | 69 | 72 | 21 | 162 | 162 | 127 | 13 | 302 | 0.052 | 10 |
| Pirozzi FF | 2018 | Brazil | Caucasian | PCR‐RFLP | 17 | 22 | 8 | 47 | 30 | 38 | 9 | 77 | 0.560 | 7 |
| Wang J | 2018 | China | Asian | PCR‐RFLP | 176 | 101 | 103 | 380 | 183 | 70 | 53 | 306 | <0.001 | 11 |
| Ramanathan G | 2019 | India | Asian | PCR‐RFLP | 72 | 71 | 2 | 145 | 81 | 19 | 0 | 100 | 0.293 | 10 |
| Zidan | 2019 | Egypt | African | PCR‐RFLP | 30 | 51 | 39 | 120 | 54 | 6 | 0 | 60 | 0.683 | 9 |
Abbreviations: HWE, Hardy–Weinberg equilibrium; PCR‐RFLP, polymerase chain reaction‐restriction fragment length polymorphism.
3.3. Meta‐analysis results
Table 2 and Figure 2 illustrated the main results of the current meta‐analysis. We adopted five genetic models to assess the association between MTHFR C677T and T2DM: homozygote model TT versus CC, heterozygous model CT versus CC, recessive model TT versus CT/CC, dominant model CT/TT versus CC, and allele model T versus C. There was a significant association between MTHFR C677T polymorphism and T2DM under homozygous (OR = 1.64, 95% CI = 1.39–1.94), heterozygous (OR = 1.38, 95% CI = 1.20–1.59), recessive (OR = 1.41, 95% CI = 1.23–1.61), dominant (OR = 1.47, 95% CI = 1.27–1.70), and allele (OR = 1.37, 95% CI = 1.23–1.52) genetic models in a random‐effects model.
Table 2.
Meta‐analysis of the association between MTHFR C677T polymorphism and T2DM susceptibility
| Variables | No. of studies | Homozygous | Heterozygous | Recessive | Dominant | Allele | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TT versus CC | CT versus CC | TT versus CT/CC | CT/TT versus CC | T versus C | |||||||
| OR (95% CI) | phet | OR (95% CI) | phet | OR (95% CI) | phet | OR (95% CI) | phet | OR (95% CI) | phet | ||
| All | 68 | 1.64 (1.39–1.94) | <.001 | 1.38 (1.20–1.59) | <.001 | 1.41 (1.23–1.61) | <0.001 | 1.47 (1.27–1.70) | <.001 | 1.37 (1.23–1.52) | <.001 |
| Ethnicity | |||||||||||
| Asian | 52 | 1.78 (1.48–2.15) | <.001 | 1.51 (1.33–1.70) | <.001 | 1.43 (1.23–1.67) | <0.001 | 1.60 (1.40–1.82) | <.001 | 1.44 (1.29–1.59) | <.001 |
| Caucasian | 11 | 1.20 (0.81–1.79) | .007 | 0.79 (0.52–1.21) | <.001 | 1.43 (1.14–1.79) | 0.457 | 0.87 (0.57–1.32) | <.001 | 0.97 (0.72–1.32) | <.001 |
| African | 5 | 1.70 (0.63–4.57) | <.001 | 1.88(0.75–4.74) | <.001 | 1.45 (0.62–3.39) | 0.002 | 2.15 (0.86–5.42) | <.001 | 1.92 (0.98–3.73) | <.001 |
| HWE | |||||||||||
| >0.05 | 52 | 1.76 (1.45–2.13) | <.001 | 1.34 (1.14–1.57) | <.001 | 1.50 (1.29–1.75) | 0.006 | 1.48 (1.26–1.73) | <.001 | 1.39 (1.24–1.56) | <.001 |
| ≤0.05 | 16 | 1.38 (0.99–1.92) | <.001 | 1.48 (1.11–1.99) | <.001 | 1.19 (0.92–1.55) | <0.001 | 1.44 (1.07–1.95) | <.001 | 1.29 (1.01–1.65) | <.001 |
| Quality score | |||||||||||
| >9 | 20 | 1.46 (1.12–1.89) | <.001 | 1.29 (0.99–1.69) | <.001 | 1.37 (1.12–1.68) | 0.003 | 1.36 (1.05–1.75) | <.001 | 1.30 (1.09–1.56) | <.001 |
| ≤9 | 48 | 1.78 (1.43–2.23) | <.001 | 1.42 (1.21–1.67) | <.001 | 1.45 (1.21–1.73) | 0.001 | 1.53 (1.29–1.82) | <.001 | 1.40 (1.23–1.60) | <.001 |
The GenBank reference sequence and version number for the gene is: MTHFR (NM_005957.5). Values were in bold if 95% CIs excluded 1 or p values less than .05.
Abbreviations: CI, confidence interval; Het, heterogeneity; HWE, Hardy–Weinberg equilibrium; OR, odds ratio; T2DM, type 2 diabetes mellitus.
Figure 2.
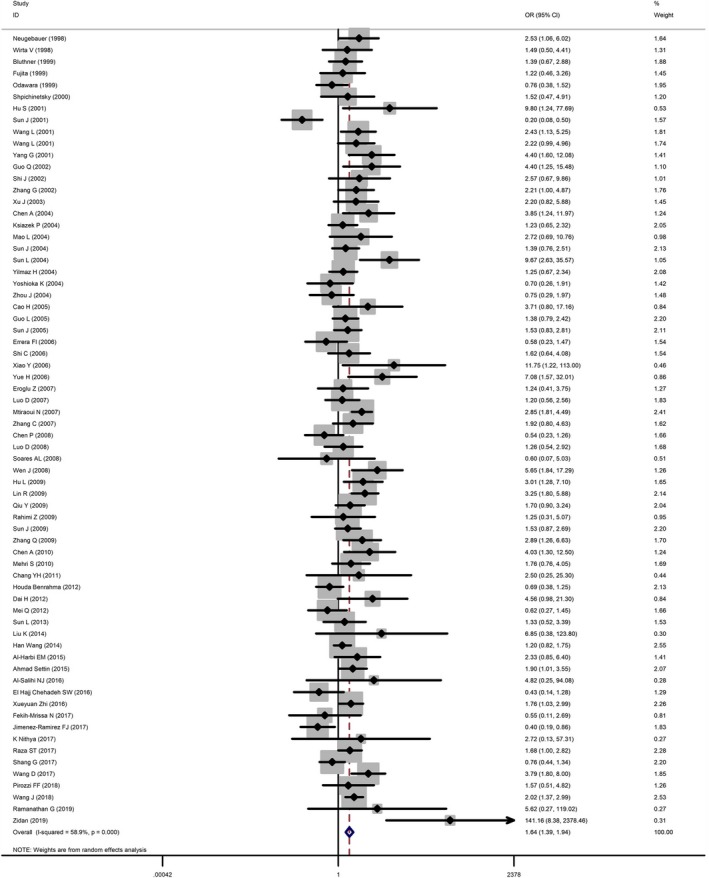
Forest plot of association between MTHFR C677T polymorphism and T2DM under homozygous model. The horizontal lines represent the study‐specific ORs and 95% CIs, respectively. The diamond represents the pooled results of OR and 95% CI. CI, confidence interval; OR, odds ratio; T2DM, type 2 diabetes mellitus
In the subgroup analysis based on ethnicity, we divided the included studies into three ethnicities: Asian, Caucasian, and African. We found significant association between MTHFR C677T genotype and T2DM in Asian populations, under each genetic models homozygous (OR = 1.78, 95% CI = 1.48–2.15), heterozygous (OR = 1.51, 95% CI = 1.33–1.70), recessive (OR = 1.43, 95% CI = 1.23–1.67), dominant (OR = 1.60, 95% CI = 1.40–1.82), and allele (OR = 1.44, 95% CI = 1.29–1.59). However, no relationship was found between MTHFR C677T genotype and T2DM in Caucasian and African, except for the recessive model in Caucasian group (OR = 1.43, 95% CI = 1.14–1.79). When stratified by HWE, significant association was also observed in the subgroup of HWE >0.05, under homozygous (OR = 1.76, 95% CI = 1.45–2.13), heterozygous (OR = 1.34, 95% CI = 1.14–1.57), recessive (OR = 1.50, 95% CI = 1.29–1.75), dominant (OR = 1.48, 95% CI = 1.26–1.73), and allele (OR = 1.39, 95% CI = 1.24–1.56) genetic models. Significant association was detected in heterozygous (OR = 1.48, 95% CI = 1.11–1.99), dominant (OR = 1.44, 95% CI = 1.07–1.95), and allele (OR = 1.29, 95% CI = 1.01–1.65) in the subgroup of HWE < 0.05. A subgroup analysis stratified by quality score was also conducted. Significant association was detected in homozygous (OR = 1.46, 95% CI = 1.12–1.89), recessive (OR = 1.37, 95% CI = 1.12–1.68), dominant (OR = 1.36, 95% CI = 1.05–1.75), and allele (OR = 1.30, 95% CI = 1.09–1.56) genetic models, in the subgroup of quality score > 9. Significant association was also detected in homozygous (OR = 1.78, 95% CI = 1.43–2.23), heterozygous (OR = 1.42, 95% CI = 1.21–1.67), recessive (OR = 1.45, 95% CI = 1.21–1.73), dominant (OR = 1.53, 95% CI = 1.29–1.82), and allele (OR = 1.40, 95% CI = 1.23–1.60) genetic models, in the subgroup of quality score ≤9.
3.4. Heterogeneity and sensitivity analysis
As shown in Table 1, substantial heterogeneities could be found among all the genetic models (p < .001) for the MTHFR C677T. Therefore, the random‐effect model was used to calculate the pooled ORs and 95% CIs for all the models.
Sensitivity analysis using sequential leave‐one‐out strategy was carried out to explore the influence of a single study on the pooled ORs. The omission of each study did not impact the recalculated ORs, indicating the credibility and reliability of our results (Figure 3).
Figure 3.
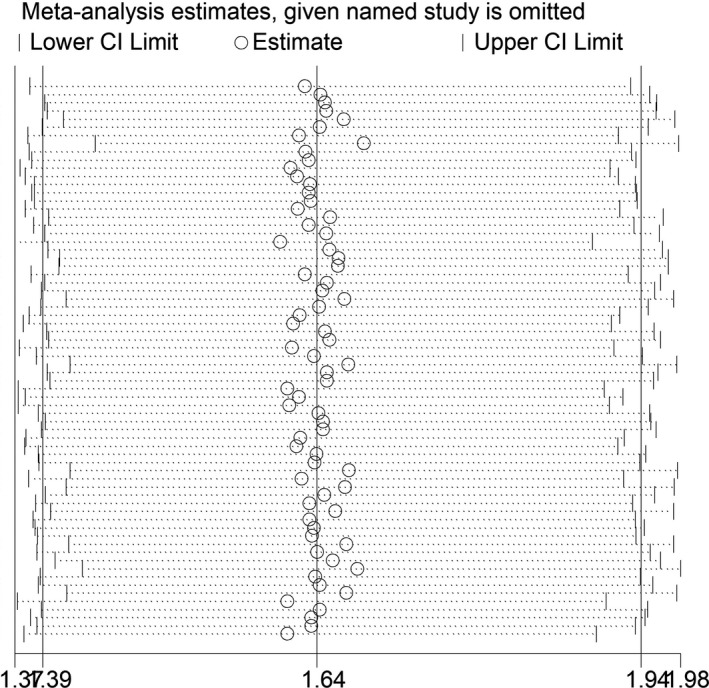
Sensitivity analysis of the association between MTHFR C677T polymorphism and T2DM. Each point represents the recalculated OR after deleting a separate study. OR, odds ratio; T2DM, type 2 diabetes mellitus
3.5. Publication bias
Begg's funnel plot and quantitative Egger's test were adopted to test the publication bias of the current meta‐analysis. As indicated by the symmetrical shape of the Begg's funnel plots, no significant publication bias was observed (Figure 4). Moreover, Egger's test also suggested the nonexistence of publication bias among the studies (data not shown).
Figure 4.
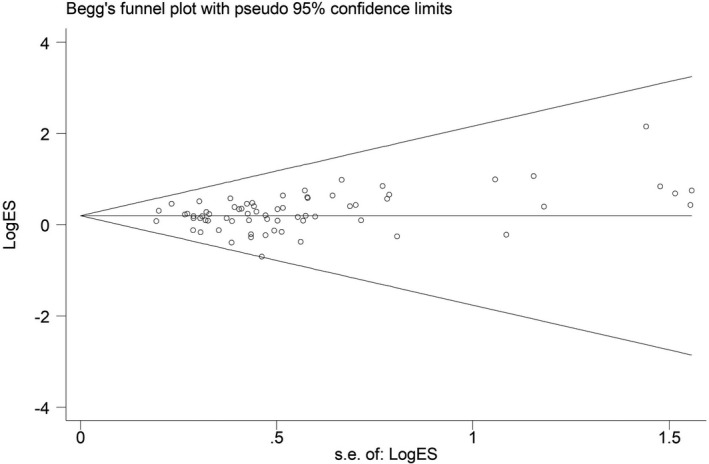
Funnel plot analysis for assessing publication bias for MTHFR C677T polymorphism under homozygous model. Each point represents a separate study for the indicated association
4. DISCUSSION
To our knowledge, the current meta‐analysis represents the largest and most comprehensive one regarding the relationship between MTHFR C677T and T2DM so far. Our analysis provided strong evidence that MTHFR C677T was significantly associated with T2DM, especially in Asians. Sensitivity analysis indicated that there was no significant change in the overall results by removing one study in each turn. Publication bias analysis also showed that the results are convincible.
MTHFR C677T is a functional genetic variation that leads to amino acid substitution from alanine to valine (Ueland, Hustad, Schneede, Refsum, & Vollset, 2001). Such amino acid shift was illustrated to compromise the enzyme activity to nearly 50%, compared to the wild‐type MTHFR enzyme (Weisberg et al., 1998). Although the relationship between T2DM susceptibility and MTHFR C677T genotype has been largely investigated, contradictory conclusions still remain. In 2006, no evidence of association was found by F.I.V. Errera et al. between the 677TT genotype of MTHFR and T2DM, in Brazilian populations (Errera et al., 2006). In a study conducted in China in 2014, Wang et al. found that C677T in the MTHFR may influence the risk of T2DM (Wang et al., 2014). Recently, in a case–control study conducted in the population of Brazilian with 47 T2DM cases and 78 controls by Flavio Fontes Pirozzi et al. (Pirozzi et al., 2018), no correlation was found between the MTHFR C677T in the development of T2DM.
Due to the divergent results among single‐country studies, several systematic meta‐analyses have been undertaken to determine conclusively whether MTHFR C677T is associated with the risk of T2DM. In 2013, Chinese academics Zhong, Rodriguez, Yang, and Li (2013) conducted a meta‐analysis regarding MTHFR C677T and T2DM. Their meta‐analysis included 4,855 T2DM patients and 5,242 controls. However, they failed to obtain clear evidence of a significant association of MTHFR C677T and T2DM across all 39 studies conducted in 15 countries. They also failed to provide compelling evidence of an association specifically for African, Asian, or Caucasian populations. Interestingly, Khalid et al. (Al‐Rubeaan et al., 2013) observed that there was a significant relationship between MTHFR C677T polymorphism and T2DM in Arab population, in 2013. In 2014, Zhu et al. (2014) conducted an updated meta‐analysis in Chinese population aiming to better identify the role of C677T polymorphism in T2DM. They included 29 studies with 4,656 T2DM patients and 2,127 controls. They detected a significant relationship between MTHFR C677T polymorphism and T2DM in the Chinese Han population.
Genotype frequencies at the C677T locus of MTHFR vary widely by ethnicity (Errera et al., 2006; Yilmaz et al., 2004), raising the possibility that any association between this SNP and the risk of T2DM may likewise depend on ethnicity. Thus, we further put our focus on ethnic stratification analysis based on the groups that emerged from our literature searches: African, Asian, and Caucasian. Our analysis provided strong evidence that MTHFR C677T was significantly associated with T2DM in Asians, but not in Caucasians or Africans. Thus, it is necessary to identify the role of C677T in different ethnicities.
Several weaknesses should be pointed out before interpreting our conclusion. First, selection bias could not be avoided as only the articles in English and Chinese were analyzed. Other studies written in other languages were unable to include. Second, analyzing one SNP in MTHFR was far more enough, as the development of T2DM was associated with multiple SNPs in multiple genes. Third, we also failed to determine the role of other potential influential factors in the initiation of T2DM. These potential influential factors such as life style, environment exposures, and gene–environment interactions were reported to be associated with T2DM. Fourth, it is inevitable to avoid several shortages such as misclassified genotypes, unwell‐matched sources of controls, and inconsistent qualities of the included studies, due to the retrospective nature of meta‐analysis. Finally, between‐study heterogeneity was found in all comparisons, which may compromise the reliability of conclusion.
5. CONCLUSION
In all, this meta‐analysis provides a precise conclusion that MTHFR C677T polymorphism was significantly associated with T2DM, especially in Asian populations. Further well‐designed, large‐scale, and in‐depth studies are warranted to check such relationship.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
Y.M. and X.L. conceived the study design and wrote the paper. K.M., L.Z., M.L., and M.Z. performed the selection, collected the data, and performed the statistical analysis. M.G. and G.Q. were responsible for the quality control of data. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
This study was supported by grants from School fund of Guilin Medical University (No. 2017KY0487).
Meng Y, Liu X, Ma K, et al. Association of MTHFR C677T polymorphism and type 2 diabetes mellitus (T2DM) susceptibility. Mol Genet Genomic Med. 2019;7:e1020 10.1002/mgg3.1020
REFERENCES
- Adinolfi, L. E. , Ingrosso, D. , Cesaro, G. , Cimmino, A. , D'Antò, M. , Capasso, R. , … Ruggiero, G. (2005). Hyperhomocysteinemia and the MTHFR C677T polymorphism promote steatosis and fibrosis in chronic hepatitis C patients. Hepatology, 41(5), 995–1003. 10.1002/hep.20664 [DOI] [PubMed] [Google Scholar]
- Al‐Harbi, E. M. , Farid, E. M. , Gumaa, K. A. , Darwish, A. H. , Alenizi, M. , & Singh, J. (2015). Genetic combination of angiotensin‐converting enzyme with methylene tetrahydrofolate reductase polymorphisms and the risk of type 2 diabetes mellitus in Bahrain. Journal of the Renin‐Angiotensin‐Aldosterone System, 16(1), 172–177. 10.1177/1470320313478286 [DOI] [PubMed] [Google Scholar]
- Al‐Rubeaan, K. , Siddiqui, K. , Saeb, A. T. , Nazir, N. , Al‐Naqeb, D. , & Al‐Qasim, S. (2013). ACE I/D and MTHFR C677T polymorphisms are significantly associated with type 2 diabetes in Arab ethnicity: A meta‐analysis. Gene, 520(2), 166–177. 10.1016/j.gene.2013.02.017 [DOI] [PubMed] [Google Scholar]
- Al‐Salihi, N. J. , Ajeena, I. M. , Al‐Kashwan, T. A. , & Al‐Lebban, Z. (2016). The association of methylenetetrahydrofolate reductase (MTHFR)/C677T polymorphisms with the development of peripheral neuropathy in type 2 diabetes mellitus. Medical Journal of Babylon, 13(2), 452–458. [Google Scholar]
- Benrahma, H. , Abidi, O. , Melouk, L. , Ajjemami, M. , Rouba, H. , Chadli, A. , … Barakat, A. (2012). Association of the C677T polymorphism in the human methylenetetrahydrofolate reductase (MTHFR) gene with the genetic predisposition for type 2 diabetes mellitus in a Moroccan population. Genetic Testing and Molecular Biomarkers, 16(5), 383–387. 10.1089/gtmb.2011.0179 [DOI] [PubMed] [Google Scholar]
- Blount, B. C. , Mack, M. M. , Wehr, C. M. , MacGregor, J. T. , Hiatt, R. A. , Wang, G. , … Ames, B. N. (1997). Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: Implications for cancer and neuronal damage. Proceedings of the National Academy of Sciences of the United States of America, 94(7), 3290–3295. 10.1073/pnas.94.7.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluthner, M. , Bruntgens, A. , Schmidt, S. , Strojek, K. , Grzeszczak, W. , & Ritz, E. (1999). Association of methylenetetrahydrofolate reductase gene polymorphism and diabetic nephropathy in type 2 diabetes? Nephrology, Dialysis, Transplantation, 14(1), 56–57. 10.1093/ndt/14.1.56 [DOI] [PubMed] [Google Scholar]
- Cao, H. , Huang, D. , Mao, L. , & Gao, Y. (2005). Association of homocysteine, methylenetetrahydrofolate reductase gene polymorphism with nephropathy in type 2 diabetes mellitus. ACTA Universitatis Medicinalis Nanjing (Natural Science), 25(4), 249–251. [Google Scholar]
- Chang, Y. H. , Fu, W. M. , Wu, Y. H. , Yeh, C. J. , Huang, C. N. , & Shiau, M. Y. (2011). Prevalence of methylenetetrahydrofolate reductase C677T and A1298C polymorphisms in Taiwanese patients with Type 2 diabetic mellitus. Clinical Biochemistry, 44(17–18), 1370–1374. 10.1016/j.clinbiochem.2011.09.020 [DOI] [PubMed] [Google Scholar]
- Chen, A. , Ning, Y. , Zhu, X. , Li, L. , & Shi, H. (2004). Study on the relationship between gene polymorphisms of N5,10‐methylenetetrahydrofolate reductase and nephropathy in type 2 diabetes mellitus in Gansu Han Chinese of China. Chinese Journal of Prevention and Control of Chronic Non‐communication Diseases, 12(5), 195–197. [Google Scholar]
- Chen, A.‐R. , Zhang, H.‐G. , Wang, Z.‐P. , Fu, S.‐J. , Yang, P.‐Q. , Ren, J.‐G. , … Tian, L.‐H. (2010). C‐reactive protein, vitamin B12 and C677T polymorphism of N‐5,10‐methylenetetrahydrofolate reductase gene are related to insulin resistance and risk factors for metabolic syndrome in Chinese population. Clinical and Investigative Medicine, 33, 290–297. 10.25011/cim.v33i5.14354 [DOI] [PubMed] [Google Scholar]
- Chen, P. , Pan, Y. , Sun, D. , Bai, J. , & Fu, S. (2008). The relationship between methylenetetrahydrofolate reductase gene C677T polymorphism and type 2 diabetes mellitus. Journal of Qiqihar Medical College, 29, 672–673. [Google Scholar]
- Dai, H. , & Yu, Z. (2012). An association study of MTHFR and eNOS genes polymorphism with diabetic nephropathy. Chinese Journal of Trauma and Disability Medicine, 20, 4–6. [Google Scholar]
- DeFronzo, R. A. , Ferrannini, E. , Groop, L. , Henry, R. R. , Herman, W. H. , Holst, J. J. , … Weiss, R. (2015). Type 2 diabetes mellitus. Nature Reviews Disease Primers, 1, 15019 10.1038/nrdp.2015.19 [DOI] [PubMed] [Google Scholar]
- Duthie, S. J. (1999). Folic acid deficiency and cancer: Mechanisms of DNA instability. British Medical Bulletin, 55(3), 578–592. 10.1258/0007142991902646 [DOI] [PubMed] [Google Scholar]
- El Hajj Chehadeh, S. W. , Jelinek, H. F. , Al Mahmeed, W. A. , Tay, G. K. , Odama, U. O. , Elghazali, G. E. , & Al Safar, H. S. (2016). Relationship between MTHFR C677T and A1298C gene polymorphisms and complications of type 2 diabetes mellitus in an Emirati population. Meta Gene, 9, 70–75. 10.1016/j.mgene.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu, Z. , Erdogan, M. , Tetik, A. , Karadeniz, M. , Cetinalp, S. , Kosova, B. , … Yılmaz, C. (2007). The relationship of the methylenetetrahydrofolate reductase C677T gene polymorphism in Turkish type 2 diabetic patients with and without nephropathy. Diabetes/Metabolism Research and Reviews, 23(8), 621–624. 10.1002/dmrr.735 [DOI] [PubMed] [Google Scholar]
- Errera, F. , Silva, M. , Yeh, E. , Maranduba, C. , Folco, B. , Takahashi, W. , … Passos‐Bueno, M. R. (2006). Effect of polymorphisms of the MTHFR and APOE genes on susceptibility to diabetes and severity of diabetic retinopathy in Brazilian patients. Brazilian Journal of Medical and Biological Research, 39(7), 883–888. 10.1590/S0100-879X2006000700005 [DOI] [PubMed] [Google Scholar]
- Fekih‐Mrissa, N. , Mrad, M. , Ibrahim, H. , Akremi, I. , Sayeh, A. , Jaidane, A. , … Gritli, N. (2017). Methylenetetrahydrofolate reductase (MTHFR) (C677T and A1298C) polymorphisms and vascular complications in patients with type 2 diabetes. Canadian Journal of Diabetes, 41(4), 366–371. 10.1016/j.jcjd.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Friso, S. , Choi, S.‐W. , Girelli, D. , Mason, J. B. , Dolnikowski, G. G. , Bagley, P. J. , … Selhub, J. (2002). A common mutation in the 5,10‐methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proceedings of the National Academy of Sciences of the United States of America, 99(8), 5606–5611. 10.1073/pnas.062066299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, H. , Narita, T. , Meguro, H. , Ishii, T. , Hanyu, O. , Suzuki, K. , … Ito, S. (1999). No association between MTHFR gene polymorphism and diabetic nephropathy in Japanese type II diabetic patients with proliferative diabetic retinopathy. Journal of Diabetes and Its Complications, 13(5–6), 284–287. 10.1016/S1056-8727(99)00057-4 [DOI] [PubMed] [Google Scholar]
- Guo, L. , Pan, Q. , Chu, M. , Guo, F. , & Sun, M. (2005). Relationship between genetic polymorphisms of methylenetetrahydrofolate reductase and macrovascular diseases in type 2 diabetes. Journal of Clinical Internal Medicine, 22, 468–470. [Google Scholar]
- Guo, Q. , Lu, J. , Qin, H. , Sheng, C. , Yin, S. , & Pan, C. (2002). Changes of the plasma homocysteine and its mechanism in type 2 diabetes with microangiopathy. Chinese Journal of Diabetes, 10(1), 32–36. [Google Scholar]
- He, J. , Liao, X.‐Y. , Zhu, J.‐H. , Xue, W.‐Q. , Shen, G.‐P. , Huang, S.‐Y. , … Jia, W.‐H. (2014). Association of MTHFR C677T and A1298C polymorphisms with non‐Hodgkin lymphoma susceptibility: Evidence from a meta‐analysis. Scientific Reports, 4, 6159 10.1038/srep06159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, L. , Zhang, Q. , Fang, M. , Qin, J. , & Liu, J. (2009). Association of non‐alcoholic fatty liver with plasma homocysteine and methylenetetrahydrofolate reductase gene polymorphism in patients of type 2 diabetes mellitus in Shanxi. Chinese Journal of General Practitioners, 08, 385–388. [Google Scholar]
- Hu, S. , Gan, P. , Li, J. , & Bi, H. (2001). The relationship between the mutation of methylenetetrahydrofolate reductase gene 677C–>T and the diabetic microangiopathy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 18(2), 118–121. [PubMed] [Google Scholar]
- Jimenez‐Ramirez, F. J. , Castro, L. M. , Ortiz, C. , Concepcion, J. , Renta, J. Y. , Morales‐Borges, R. H. , … Duconge, J. (2017). Role of treatment‐modifying MTHFR677C>T and 1298A>C polymorphisms in metformin‐treated Puerto Rican patients with type‐2 diabetes mellitus and peripheral neuropathy. Drug Metabolism and Personalized Therapy, 32(1), 23–32. 10.1515/dmpt-2016-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek, P. , Bednarek‐Skublewska, A. , & Buraczynska, M. (2004). The C677T methylenetetrahydrofolate reductase gene mutation and nephropathy in type 2 diabetes mellitus. Medical Science Monitor, 10(2), BR47‐51. [PubMed] [Google Scholar]
- Liew, S. C. , & Gupta, E. D. (2015). Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: Epidemiology, metabolism and the associated diseases. European Journal of Medical Genetics, 58(1), 1–10. 10.1016/j.ejmg.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Lin, R. , Wang, C. , & Liu, X. (2009). Matched case–control study on the association between polymorphism of methylenetetrahydrofolate reductase (MTHFR) gene and diabetic nephropathy in type 2 diabetic patients. Modern Preventive Medicine, 36(20), 3801–3804. [Google Scholar]
- Liu, K. , Pang, D. , Zou, C. , Wu, S. , Ping, Z. , & Zeng, Q. (2014). Relationship of C677T polymorphisms of MTHFR gene with diabetic nephropathy in southeast Guangxi. International Journal of Laboratory Medicine, 35(13), 1670–1672. [Google Scholar]
- Luo, D. , Yan, S. , Li, J. , Cheng, Y. , & Song, Y. (2007). The association between methylenetetrahydrofolate reductase gene C677T polymorphism and coronary heart disease in type 2 diabetes mellitus. Chinese Journal of Clinical Laboratory Science, 25, 114–116. [Google Scholar]
- Luo, D. , Yan, S. , Ma, H. , Cheng, Y. , & Song, Y. (2008). Relationship between homocysteine methylene tetrahyrdrofolate reductase polymorphism and coronary heart disease in Chinese type 2 diabetes mellitus. Journal of China‐Japan Friendship Hospital, 22, 24–27. [Google Scholar]
- Mao, L. , Gao, Y. , Qin, W. , & Shi, H. (2004). The association of methylenetetrahydrofolate reductase gene polymorphism with cerebral infarction in type 2 diabetes mellitus. Acta Academiae Medicinae Nantong, 24(2), 146–147, 150. [Google Scholar]
- Mehri, S. , Koubaa, N. , Nakbi, A. , Hammami, S. , Chaaba, R. , Mahjoub, S. , … Hammami, M. (2010). Relationship between genetic polymorphisms of angiotensin‐converting enzyme and methylenetetrahydrofolate reductase as risk factors for type 2 diabetes in Tunisian patients. Clinical Biochemistry, 43(3), 259–266. 10.1016/j.clinbiochem.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Mei, Q. , Chen, P. , & Zheng, L. (2012). Correlation study between gene polymorphism of methylene tetrahydrofolate reductase and type 2 diabetes. China Medical Herald, 9, 162–163. [Google Scholar]
- Mtiraoui, N. , Ezzidi, I. , Chaieb, M. , Marmouche, H. , Aouni, Z. , Chaieb, A. , … Almawi, W. Y. (2007). MTHFR C677T and A1298C gene polymorphisms and hyperhomocysteinemia as risk factors of diabetic nephropathy in type 2 diabetes patients. Diabetes Research and Clinical Practice, 75(1), 99–106. 10.1016/j.diabres.2006.05.018 [DOI] [PubMed] [Google Scholar]
- Neugebauer, S. , Baba, T. , & Watanabe, T. (1998). Methylenetetrahydrofolate reductase gene polymorphism as a risk factor for diabetic nephropathy in NIDDM patients. The Lancet, 352(9126), 454 10.1016/S0140-6736(05)79188-1 [DOI] [PubMed] [Google Scholar]
- Niclot, S. , Pruvot, Q. , Besson, C. , Savoy, D. , Macintyre, E. , Salles, G. , … Baudry‐Bluteau, D. (2006). Implication of the folate‐methionine metabolism pathways in susceptibility to follicular lymphomas. Blood, 108(1), 278–285. 10.1182/blood-2005-04-1567 [DOI] [PubMed] [Google Scholar]
- Nithya, K. , Isabel, W. , Angeline, T. , Priscilla, A. S. , Shakila, H. , & Asirvatham, A. J. (2017). MTHFR C677T gene polymorphism in Type 2 diabetes mellitus patients with and without vascular complications: A case‐control study. Meta Gene, 14, 79–84. 10.1016/j.mgene.2017.08.005 [DOI] [Google Scholar]
- Odawara, M. , & Yamashita, K. (1999). A common mutation of the methylenetetrahydrofolate reductase gene as a risk factor for diabetic nephropathy. Diabetologia, 42(5), 631–632. 10.1007/s001250051206 [DOI] [PubMed] [Google Scholar]
- Pirozzi, F. F. , Belini Junior, E. , Okumura, J. V. , Salvarani, M. , Bonini‐Domingos, C. R. , & Ruiz, M. A. (2018). The relationship between of ACE I/D and the MTHFR C677T polymorphisms in the pathophysiology of type 2 diabetes mellitus in a population of Brazilian obese patients. Archives of Endocrinology and Metabolism, 62(1), 21–26. 10.20945/2359-3997000000005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Y. (2009). C677T polymorphism of MTHFR gene in type 2 diabetes with carotid atherosclerosis. Zhe Jiang Medical Journal, 31, 426–428. [Google Scholar]
- Rahimi, Z. , Nomani, H. , Mozafari, H. , Vaisi‐Raygani, A. , Madani, H. , Malek‐Khosravi, S. , & Parsian, A. (2009). Factor V G1691A, prothrombin G20210A and methylenetetrahydrofolate reductase polymorphism C677T are not associated with coronary artery disease and type 2 diabetes mellitus in western Iran. Blood Coagulation & Fibrinolysis, 20(4), 252–256. 10.1097/MBC.0b013e3283255487 [DOI] [PubMed] [Google Scholar]
- Ramanathan, G. , Harichandana, B. , Kannan, S. , Elumalai, R. , & Sfd, P. (2019). Association between end‐stage diabetic nephropathy and MTHFR (C677T and A1298C) gene polymorphisms. Nephrology (Carlton), 24(2), 155–159. 10.1111/nep.13208 [DOI] [PubMed] [Google Scholar]
- Raza, S. T. , Abbas, S. , Siddiqi, Z. , & Mahdi, F. (2017). Association between ACE (rs4646994), FABP2 (rs1799883), MTHFR (rs1801133), FTO (rs9939609) genes polymorphism and type 2 diabetes with dyslipidemia. International Journal of Molecular and Cellular Medicine, 6(2), 121–130. 10.22088/acadpub.BUMS.6.2.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, R. (1997). Genetic predisposition to hyperhomocysteinemia: Deficiency of methylenetetrahydrofolate reductase (MTHFR). Thrombosis and Haemostasis, 78(1), 523–526. 10.1055/s-0038-1657581 [DOI] [PubMed] [Google Scholar]
- Settin, A. , El‐Baz, R. , Ismaeel, A. , Tolba, W. , & Allah, W. A. (2015). Association of ACE and MTHFR genetic polymorphisms with type 2 diabetes mellitus: Susceptibility and complications. Journal of the Renin‐Angiotensin‐Aldosterone System, 16(4), 838–843. 10.1177/1470320313516172 [DOI] [PubMed] [Google Scholar]
- Shang, G. , Wang, H. , & Liu, J. (2017). Relationship between methylenetetrahydrofolate reductase gene polymorphism and type 2 diabetes mellitus. Journal of Medical Research, 46(9), 72–75. [Google Scholar]
- Shi, C. , He, Y. , Cheng, G. , Wang, W. , & Liu, J. (2006). Detection of the 677 C‐T variant of MTHFR gene in Chinese diabetic patients with fluorescent MGB probe real‐time PCR. Chinese Journal of Diabetes, 14, 258–260. [Google Scholar]
- Shi, J. , Li, B. , Yu, Y. , Chen, Y. , & Tao, R. (2002). The relationship between the polymorphism of MTHFR gene and type 2 diabetes mellitus. Journal of Jilin University (Medicine Edition), 28, 371–374. [Google Scholar]
- Shpichinetsky, V. , Raz, I. , Friedlander, Y. , Goldschmidt, N. , Wexler, I. D. , Ben‐Yehuda, A. , & Friedman, G. (2000). The association between two common mutations C677T and A1298C in human methylenetetrahydrofolate reductase gene and the risk for diabetic nephropathy in type II diabetic patients. Journal of Nutrition, 130(10), 2493–2497. 10.1093/jn/130.10.2493 [DOI] [PubMed] [Google Scholar]
- Soares, A. L. , Fernandes, A. P. , Cardoso, J. E. , Sousa, M. O. , Lasmar, M. C. , Novelli, B. A. , … Carvalho, M. G. (2008). Plasma total homocysteine levels and methylenetetrahydrofolate reductase gene polymorphism in patients with type 2 diabetes mellitus. Pathophysiology of Haemostasis and Thrombosis, 36(5), 275–281. 10.1159/000252825 [DOI] [PubMed] [Google Scholar]
- Sun, J. Z. , Xu, Y. , Lu, H. , & Zhu, Y. (2009). Polymorphism of the methylenetetrahydrofolate reductase gene association with homocysteine and ischemic stroke in type 2 diabetes. Neurol India, 57(5), 589–593. 10.4103/0028-3886.57808 [DOI] [PubMed] [Google Scholar]
- Sun, J. , Xu, Y. , Xue, J. , Zhu, Y. , & Lu, H. (2005). Methylenetetrahydrofolate reductase polymorphism associated with susceptibility to coronary heart disease in Chinese type 2 diabetic patients. Molecular and Cellular Endocrinology, 229(1–2), 95–101. 10.1016/j.mce.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Sun, J. , Xu, Y. , & Zhu, Y. (2001). The association of methylenetetrahydrofolate reductase gene polymorphism with nephropathy in type 2 diabetes mellitus in Chinese. Chinese Journal of Internal Medicine, 40(8), 529–532. [PubMed] [Google Scholar]
- Sun, J. , Xu, Y. , Zhu, Y. , & Lu, H. (2004). Genetic polymorphism of methylenetetrahydrofolate reductase as a risk factor for diabetic nephropathy in Chinese type 2 diabetic patients. Diabetes Research and Clinical Practice, 64(3), 185–190. 10.1016/j.diabres.2003.10.022 [DOI] [PubMed] [Google Scholar]
- Sun, L. , Chen, L. , Ren, J. , Wang, D. , Zheng, X. , & Xu, L. (2004). Relationship of plasma homocysteine and gene polymorphism of homocysteine metabolism related enzyme with diabetic peripheral neuropathy. Chinese Journal of Endocrinology and Metabolism, 20(6), 536–537. [Google Scholar]
- Sun, L. , Wang, S. , Shi, X. , & Yang, Z. (2013). Interactions between APOE and MTHFR mutations is associated with the risk for type 2 diabetic nephropathy. Journal of Medical Molecular Biology, 10(2), 95–99. [Google Scholar]
- Ueland, P. M. , Hustad, S. , Schneede, J. , Refsum, H. , & Vollset, S. E. (2001). Biological and clinical implications of the MTHFR C677T polymorphism. Trends in Pharmacological Sciences, 22(4), 195–201. 10.1016/S0165-6147(00)01675-8 [DOI] [PubMed] [Google Scholar]
- van Dieren, S. , Beulens, J. W. , van der Schouw, Y. T. , Grobbee, D. E. , & Neal, B. (2010). The global burden of diabetes and its complications: An emerging pandemic. European Journal of Cardiovascular Prevention and Rehabilitation, 17(Suppl 1), S3–S8. 10.1097/01.hjr.0000368191.86614.5a [DOI] [PubMed] [Google Scholar]
- Wang, D. , Bai, L. , Zhai, Q. , Li, Y. , Cao, M. , Hai, J. , & Zhang, Q. (2017). Association of MTHFR C677T and A1298C polymorphisms with the development of type 2 diabetic nephropathy and their interaction with environmental factors. International Journal of Clinical and Experimental Pathology, 10(3), 3778–3785. [Google Scholar]
- Wang, H. , Hu, C. , Xiao, S. H. , & Wan, B. (2014). Association of tagging SNPs in the MTHFR gene with risk of type 2 diabetes mellitus and serum homocysteine levels in a Chinese population. Disease Markers, 2014, 725731 10.1155/2014/725731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Wang, F. , & Li, W. (2018). The significance of C677T mutation in type 2 diabetes mellitus combined with macrovascular complications. China Medical Herald, 15(14), 102–105. [Google Scholar]
- Wang, L. , Wang, J. , Xue, Y. , Chen, Y. , & Zou, H. (2001). Relationship between methylenetetrahydrofolate reductase gene polymorphism and diabetic nephropathy. Zhonghua Yi Xue Yi Chuan Xue Za Zhi, 18(4), 276–278. [PubMed] [Google Scholar]
- Wang, L. , Wang, J. , Xue, Y. , Cheng, Y. , Zhou, H. , & Qiu, F. (2001). Relation between methylenetetrahydrofolate reductase gene polymorphism and diabetic retinopathy. Chinese Journal of Ocular Fundus Diseases, 17(3), 198–200. [Google Scholar]
- Wareham, N. J. , Franks, P. W. , & Harding, A. H. (2002). Establishing the role of gene‐environment interactions in the etiology of type 2 diabetes. Endocrinology and Metabolism Clinics of North America, 31(3), 553–566. 10.1016/S0889-8529(02)00007-5 [DOI] [PubMed] [Google Scholar]
- Weisberg, I. , Tran, P. , Christensen, B. , Sibani, S. , & Rozen, R. (1998). A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Molecular Genetics and Metabolism, 64(3), 169–172. 10.1006/mgme.1998.2714 [DOI] [PubMed] [Google Scholar]
- Wen, J. , Lu, Z. , Li, X. , Wu, D. , & Zhang, Y. (2008). Correlation of MTHFR gene polymorphism and plasma homocysteine with microalbuminuria in type 2 diabetes. Shanghai Medical Journal, 31, 47–51. [Google Scholar]
- Wirta, V. , Huang, X.‐H. , Wirta, O. , Rantalaiho, V. , Pasternack, A. , Jokela, H. , … Lehtimäki, T. (1998). Mutation C677T of methylenetetrahydrofolate reductase gene is not associated with coronary artery disease, but possibly with albuminuria, in type 2 diabetic patients. Clinical Chemistry and Laboratory Medicine, 36(8), 625–628. 10.1515/CCLM.1998.109 [DOI] [PubMed] [Google Scholar]
- Xiao, Y. , Hu, Z. , Shan, K. , Guan, Z. , & Ren, X. (2006). An Investigation on the relationship between the polymorphism of MTHFR gene and type 2 diabetic cardiovascular disease. Journal of Guiyang Medical College, 31, 317–319. [Google Scholar]
- Xu, J. , Zhang, J. , Shan, B. , & Ma, H. (2003). Relationship between methylenetetrahydrofolate reductase gene polymorphism and diabetic nephropathy in type 2 diabetes mellitus in the Hans of Hebei Province. Clinical Focus, 18(14), 787–789. [Google Scholar]
- Yang, G. , Lu, J. , & Pan, C. (2001). Study on the relationship between N5,10‐methylenetetrahydrofolate reductase gene polymorphism and the susceptibility to microangiopathy in type 2 diabetes mellitus. Chinese Journal of Endocrinology and Metabolism, 17(4), 224–227. [Google Scholar]
- Yilmaz, H. , Agachan, B. , Ergen, A. , Karaalib, Z. E. , & Isbir, T. (2004). Methylene tetrahydrofolate reductase C677T mutation and left ventricular hypertrophy in Turkish patients with type II diabetes mellitus. Journal of Biochemistry and Molecular Biology, 37(2), 234–238. 10.5483/BMBRep.2004.37.2.234 [DOI] [PubMed] [Google Scholar]
- Yoshioka, K. , Yoshida, T. , Umekawa, T. , Kogure, A. , Takakura, Y. , Toda, H. , & Yoshikawa, T. (2004). Methylenetetrahydrofolate reductase gene polymorphism is not related to diabetic nephropathy in Japanese Type 2 diabetic patients. Diabetic Medicine, 21(9), 1051–1052. 10.1111/j.1464-5491.2004.01192.x [DOI] [PubMed] [Google Scholar]
- Yue, H. , Liu, J. , Kang, W. , Hu, L. , & Qiu, J. (2006). Relationship between plasma level of homocysteine and urine microalbumin in incipient type 2 diabetic nephropathy. Chinese Journal of General Pratitioners, 5, 725–729. [Google Scholar]
- Zeggini, E. , Weedon, M. N. , Lindgren, C. M. , Frayling, T. M. , Elliott, K. S. , Lango, H. , … Hattersley, A. T. (2007). Replication of genome‐wide association signals in UK samples reveals risk loci for type 2 diabetes. Science, 316(5829), 1336–1341. 10.1126/science.1142364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Li, Z. , Liu, G. , & Hu, R. (2007). MTHFR, eNOS gene polymorphisms connecting research of the patients with T2DM complicating cerebral infarction. Journal of Clinical Internal Medine, 24, 458–460. [Google Scholar]
- Zhang, G. , Xiang, K. , Weng, Q. , & Li, J. (2002). Association between 677C/T polymorphism of methylenetetrahydrofolate reductase gene and type 2 diabetes with macrovascular complications in Shanghai. Chinese Journal of Endocrinology and Metabolism, 18, 362–365. [Google Scholar]
- Zhang, Q. , & Liu, J. (2009). The relationship of carotid intima‐media thickness with C677T polymorphism of MTHFR and plasma homocysteine level in type 2 diabetes. Chinese Journal of Diabetes, 17, 356–358. [Google Scholar]
- Zhi, X. , Yang, B. , Fan, S. , Li, Y. , He, M. , Wang, D. A. , … Sun, G. (2016). Additive interaction of MTHFR C677T and MTRR A66G polymorphisms with being overweight/obesity on the risk of type 2 diabetes. International Journal of Environmental Research and Public Health, 13(12), 10.3390/ijerph13121243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, J. H. , Rodriguez, A. C. , Yang, N. N. , & Li, L. Q. (2013). Methylenetetrahydrofolate reductase gene polymorphism and risk of type 2 diabetes mellitus. PLoS ONE, 8(9), e74521 10.1371/journal.pone.0074521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J. , Li, X. , & Zhang, J. (2004). Relationship between the C677T polymorphism in the methylenetetrahydrofolate reductase gene and cerebral infarction complicated type 2 diabetes. Journal of Apoplexy and Nervous Diseases, 21, 136–138. [Google Scholar]
- Zhu, B. , Wu, X. , Zhi, X. , Liu, L. , Zheng, Q. , & Sun, G. (2014). Methylenetetrahydrofolate reductase C677T polymorphism and type 2 diabetes mellitus in Chinese population: A meta‐analysis of 29 case‐control studies. PLoS ONE, 9(7), e102443 10.1371/journal.pone.0102443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidan, A. R. , El Mougy, H. M. , Moustafa, H. S. , El attar, S. , & Mohamed, E. F. (2019). Methylenetetrahydrofolate reductase C677T gene polymorphism and diabetic nephropathy susceptibility in patients with type 2 diabetes mellitus. The Scientific Journal of Al‐Azhar Medical Faculty, Girls, 3, 14–22. 10.4103/sjamf.sjamf_38_18 [DOI] [Google Scholar]


