Abstract
Background
Neural tube defects (NTDs) are severe common birth defects that result from a failure in neural tube closure (NTC). Our previous study has shown that decreased histone methylation altered the regulation of genes linked to NTC. However, the effect of alterations in histone acetylation in human fetuses with NTDs, which are another functional posttranslation modification, remains elusive. Thus, we aimed to identify acetylation sites and changes in histone in patients with NTDs.
Methods
First, we identified histone acetylation sites between control human embryonic brain tissue and NTDs using Nano‐HPLC‐MS/MS. Next, we evaluated the level of histone acetylation both groups via western blotting (WB). Finally, we used LC‐ESI‐MS and WB to compare whether histone H4 acetylation was different in NTDs.
Results
A total of 43 histone acetylation sites were identified in human embryonic brain tissue, which included 16 novel sites. Furthermore, we found an increased histone acetylation and H4K5ac in tissue with NTDs.
Conclusion
Our result present a comprehensive map of histone H4 modifications in the human fetal brain. Furthermore, we provide experimental evidence supporting a relationship between histone H4K5ac and NTDs. This offers a new insight into the pathological role of histone modifications in human NTDs.
Keywords: histone acetylation, human fetal brain, mass spectrometry, neural tube defects
A total of 43 histone acetylation sites were identified in human embryonic brain tissue, which included 16 novel sites. Furthermore, we found increased histone acetylation and H4K5ac in tissue with NTDs.
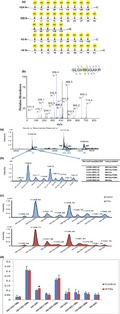
1. INTRODUCTION
Neural tube defects (NTDs) form a group of severe congenital malformations, including anencephaly, spina bifida, and encephalocele (Lei, Jingru, Yanjun, Hui, & Aiguo, 2017). The incidence of NTDs is approximately 1 in 1,000, and is estimated reach to 4–10 in 1,000 in northern China (Gu et al., 2007; Put, Straaten, Trijbels, & Blom, 2001). The etiology of NTDs is largely unknown; however, it is considered to be multifactorial, involving multiple interacting genes and environmental factors. The mechanisms by which environmental factors affect the process of neural tube closure and their critical interaction with genetic factors remain unknown.
Previous research has indicated that aberrant histone acetylation may cause NTDs. Shyamasundar et al have found that hyperglycemia alters epigenetic mechanisms, including histone methylation and histone acetylation, in mouse embryonic neural stem cells (NSCs). This, in turn, results in altered developmental control gene expression and may form the basis for NTDs (Shyamasundar et al., 2013). Furthermore, Yang et al. have shown that high glucose increases histone H3 acetylation, which leads to NTDs. An additional study has reported that high glucose‐induced oxidative stress represses sirtuin deacetylase expression and increases histone H3 acetylation, leading to NTDs (Yu, Wu, & Yang, 2016). Additional studies have shown that acetylation of histone H3 at lysine 27 (H3K27ac) at a specific differentiation gene enhancer region is necessary for the conversion of embryonic stem cells into NSCs (Rada‐Iglesias et al., 2011). Balmer has differentiated human embryonic stem cells to neuroectodermal precursors as a model to investigate the mechanisms of action of the histone deacetylase inhibitor, Tricostation A. This study reported that Tricostation A affects the differentiation of embryonic stem cells into NSCs (Balmer et al., 2012). In addition, previous animal studies have shown that injection of the histone deacetylase inhibitor, valproic acid, or Tricostatin A induces NTDs (Copp & Greene, 2010; Murko et al., 2013). Moreover, genetic studies have found that a loss of histone acetyltransferase EP300, CREBBP, and CITED2 function may cause the occurrence of NTDs (Copp & Greene, 2010). A case–control study, including 297 spina bifida cases and 300 controls, found 37 SNPs within EP300 and CREBBP were associated with the occurrence of spina bifida (Lu et al., 2010). Taken together, these reports suggest that histone acetylation plays an important role in neural tube development. Therefore, we sought to assess histone acetylation sites and their differential expression using mass spectrometry (MS) in human NTDs.
In this study, we used the Nano‐HPLC‐MS/MS (Q Exactive HF MS) to examine histone H4 acetylation in the brains of four healthy fetuses and four fetuses with NTDs and compare acetylation sites. Furthermore, we developed a rapid MS method (LC‐ESI‐MS) to compare the histone H4 acetylation expression between NTDs and healthy control brains. Our results showed that the histone H4 acetylation expression increased in NTDs. This indicated that abnormal modification of histone H4 acetylation is a potential pathogenic mechanism in NTDs.
2. MATERIALS AND METHODS
2.1. Ethical compliance
The investigation was approved by the Committee of Medical Ethics of the Capital Institute of Pediatrics. Written informed consent was obtained from all mothers who participated in this study.
2.2. Subjects
Tissue with NTDs (spina bifida) and healthy control tissue were selected from Shanxi Province in northern China. Brain tissue from four fetuses with spina bifida and four healthy control fetuses (gestational age, ~20 weeks) was analyzed (Table 1). All cases were from medical abortions diagnosed with NTDs using B‐mode ultrasound. Pathological diagnosis of NTDs was performed by experienced pathologists according to the International Classification of Disease. Age‐ and sex‐matched healthy control fetuses that were aborted for nonmedical reasons were also enrolled from this region. Any fetuses displaying pathological malformations or intrauterine growth retardation were excluded from the control group.
Table 1.
Clinical information of the eight individual fetuses
| No. | Source of brain tissue | Hcy level (pmol/mg) | folate level(ng/mg) | Gender | Age |
|---|---|---|---|---|---|
| 1 | Normal control | 3.361722 | 0.1109 | Male | 19 weeks |
| 2 | Normal control | 2.166592 | 0.0462 | Male | 20 weeks |
| 3 | Normal control | 4.092798 | 0.1093 | Female | 21 weeks |
| 4 | Normal control | 2.060746 | 0.0459 | Female | 20 weeks |
| 5 | Open lumbosacral spina bifida | 34.246276 | 0.1647 | Male | 24 weeks |
| 6 | Open thoracolumbar spine | 30.88228 | 0.1792 | Male | 20 weeks |
| 7 | Closed lumbosacral spina bifida | 57.575095 | 0.1436 | Female | 20 weeks |
| 8 | Open lumbar spina bifida | 42.48655 | 0.0698 | Female | 26 weeks |
2.3. Histone extraction
Histones were extracted from brain samples using acid extraction (Hake, Shechter, Dormann, & Allis, 2007). Briefly, tissue was homogenized in 10 ml lysis buffer containing 10 mM Tris‐Cl (pH 8.0), 1 mM KCl, 1.5 mM MgCl2, and 1 mM dithiothreitol (DTT). Protease and phosphatase inhibitors were added immediately before histone extraction. Nuclei were isolated by centrifugation (1,500 g for 10 min). Next, 0.2 N sulfuric acid (H2SO4) was added overnight at 4°C to isolate the histones. Following this, the supernatant was precipitated with trichloroacetic acid. Next, the histone‐containing pellets were washed with ice acetone and dissolved in distilled water. The extracted histones were stored at −80°C before analysis, as reported previously (Zhang et al., 2013).
2.4. In‐solution protein digestion
As described previously (Zhang et al., 2013), 40μg of extracted histones were digested. Briefly, disulfide bonds were reduced with 10mM (final concentration) DTT for 1hr at 37°C, followed by alkylation via 40mM (final concentration) iodoacetamide for 1hr at room temperature in the dark. The alkylation reaction was quenched with 40mM DTT for 30 min. After urea dilution to <1 M with 25mM NH4HCO3, sequence‐grade trypsin (Pierce, 90,057) was added at a ratio of 1:40 (enzyme: total protein), and proteins were digested at 37°C for 4 hr. Tryptic digestion was quenched by adding 1.0% trifluoroacetic acid. Finally, the solution was centrifuged at 13,000 g for 10 min to remove any insoluble material. The supernatant was collected and stored in 80°C for subsequent analysis.
2.5. Nano‐HPLC‐MS/MS
The digested samples were loaded onto 15‐cm C18 columns (particle size, 2 µm; diameter, 75 µm) using the autosampler connected to an UltiMate 3000RSLCnano system (Dionex). The peptides were eluted with a linear gradient of buffer B (0.1% formic acid in 95% ACN, v/v) from 4% to 45% in 120 min, followed by a steep increase to 90% in 5 min at a flow rate of 300 nl/min. The eluted peptides were ionized and sprayed into a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) using a Nanospray Flex source in positive mode. A full mass scan (m/z 350–2000) was used at a resolution of 120,000. Twenty of the most intense ions were isolated for MS/MS analysis. The automatic gain control (AGC) target was set at 1 × 106 ions, and the maximum ion injection time (IT) was 50 ms. Source ionization parameters were optimized with the spray voltage at 2.5 kV. Other parameters were as follows: capillary temperature, 320°C; S‐Lens RF level, 50.
The MS raw data were searched using Proteome Discoverer (version 2.1.0.81) against the database of human histones, downloaded from Uniprot (http://www.uniprot.org, October 2015). The precursor mass tolerance and fragment mass tolerance are 20 ppm and 0.05 Da, respectively. Peptides were generated from the trypsin (semi) digest with up to four missed cleavage sites. Fixed modification was carbamidomethyl (C) and variable modifications were acetyl (K) (42.011 Da) and oxidation (M). Peptide spectral matches (PSMs) were validated using a percolator based on q‐values at a 1% false discovery rate (FDR). Only proteins with a minimum of two quantifiable peptides were included in our final dataset.
2.6. LC‐ESI‐MS analysis of histone H4
LC‐ESI‐MS analysis of the extracted histone proteins was performed using an Agilent series 1,200 pump connected to the Agilent 6,530 Q‐TOF MS with an Agilent Jet stream electrospray ionization in positive ion mode. The LC condition was as follows: mobile phase, 3% ACN/0.1% aqueous formic acid; isobaric elution was conducted by hold at 3% ACN/0.1% aqueous formic acid for 1 hr. The flow rate was 0.2 ml/min. The column was a TSK‐GEL G2000SWXL, 7.8 × 300 mm, 5‐m column with a guard column of the same packing. The injection volume was 10 μl. The ESI system employed a 3.5‐kV spray voltage (positive polarity). The drying gas (nitrogen) temperature was set at 325°C, drying gas flow at 12 L/min, nebulizer pressure at 50 psi, and Fragmentor voltage at 175 V. The mass chromatograms were recorded in total ion current (TIC) within 500 and 2,000 m/z. The deconvoluted ESI mass spectra of histones were obtained using Mass Hunter 1.0. The peak averaged mass spectra were reconstructed and the mass of the histones and their isoforms were calculated. The relative abundance of each histone isoform was derived and transformed into relative percentage amount. t test was used to compare frequencies between categorical variables and groups. Differences were considered statistically significant if p < .05.
2.7. Western blotting
Extracted histone samples (5 µg) was separated on a 12% SDS‐PAGE (Invitrogen, NW04120BOX) and transferred to nitrocellulose membranes via electrophoretic migration. The membrane was blocked in 5% milk/TBST (0.9%NaCl, 10mM Tris‐HCl, 0.05% Tween 20 pH 7.5) and incubated with primary antibodies: Kac (1:1,000; CST, 9,441); H3 (1:100,000; Abcam, ab213257); H4K5ac (1:2000; Abcam, ab124636); H4 (1:2000; Abcam, ab222763) overnight at 4ºC. After washing three times with TBST, the membrane was immersed in 5% milk/TBST containing horseradish peroxidase (HRP)‐conjugated secondary antibody (1:5,000; Zhongshan Jinqiao, ZB5301) for 1 h at room temperature. The immune complexes were detected using ECL (Pierce, 32,209). Results were presented as mean ± standard deviation (SD) from at least three independent experiments. Data analysis was performed using t test. Differences were considered significant if p < .05.
3. RESULTS
3.1. Sample preparation and histone extraction
To explore the distribution of histone acetylation in the human fetal brain, samples were analyzed using Q Exactive HF. We matched all case and control samples for age and gender. Detailed clinical information is shown in Table 1. Coomassie blue‐stained SDS gel results (Figure 1) indicated that the five major components (H1, H2a, H2b, H3, and H4) of core histone could be separated clearly from the histone extract.
Figure 1.
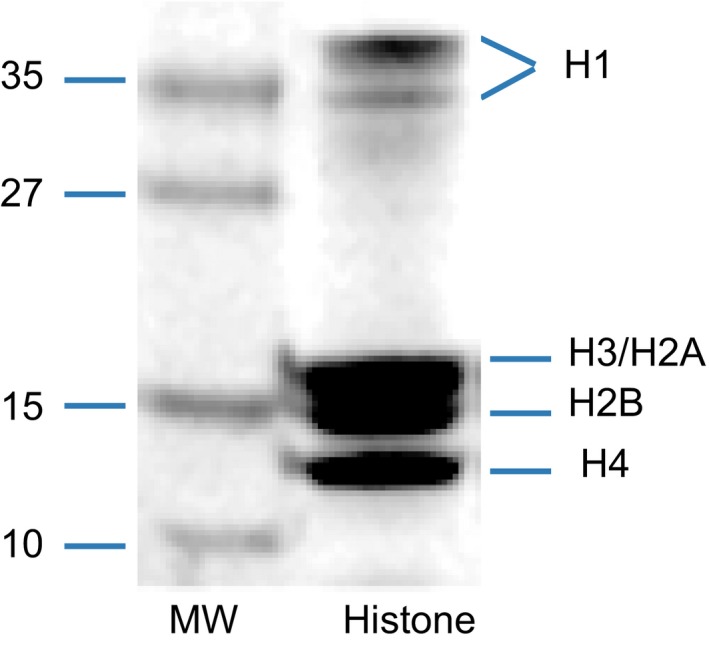
Coomassie Blue‐stained SDS gel with the extracted histone mixture for MS. The locations of the histone five major components (H1, H2a, H2b, H3, and H4) were noted. MW means molecular weight
3.2. Mass spectrometry mapping of histone acetylation in the human fetal brain
In this study, a total of 43 acetylation sites were identified in healthy control human fetal brains, including K74,K75,K95,K99,K118,K119,K124,K126,K128 in H2a; K5,K11,K12,K15,K16,K20,K23,K27,K28,K34,K43,K46,K57,K108,K116 in H2b; K9,K14,K18,K23,K27,K36,K79,K115,K122 in H3; and K5,K8,K12,K16,K20,K44,K59,K77,K79,K91 in histone H4 (Figure 2a). The spectra (Figure 2b) show the representative lysine acetylation human brain H4K12 peptide GLGKacGGAKR identified by MS/MS. The series of b‐ and y‐type acetylation fragment ions provided confident sequence information and indicated an unambiguous acetylation site. Among these sites, 27 acetylation sites had been detected in humans in previous reports (Horikoshi, 2013; Kimura, 2013). However, 16 acetylation sites, including H2aK74, H2aK75, H2aK99, H2aK118, H2aK124, H2aK126, H2aK128, H2bK16, H2bK23, H2bK27, H2bK28, H2bK34, H2bK43, H2bK57, H4K44, and H4K59 were reported for the first time. All identified histone peptides of two groups were listed in Table S1 and S2. All peptides contained histone modifications that were detected on histone H4 in the four healthy fetal brains was listed in Table 2. Taken together, these data identified a novel specific histone acetylation map in the human fetal brain.
Figure 2.
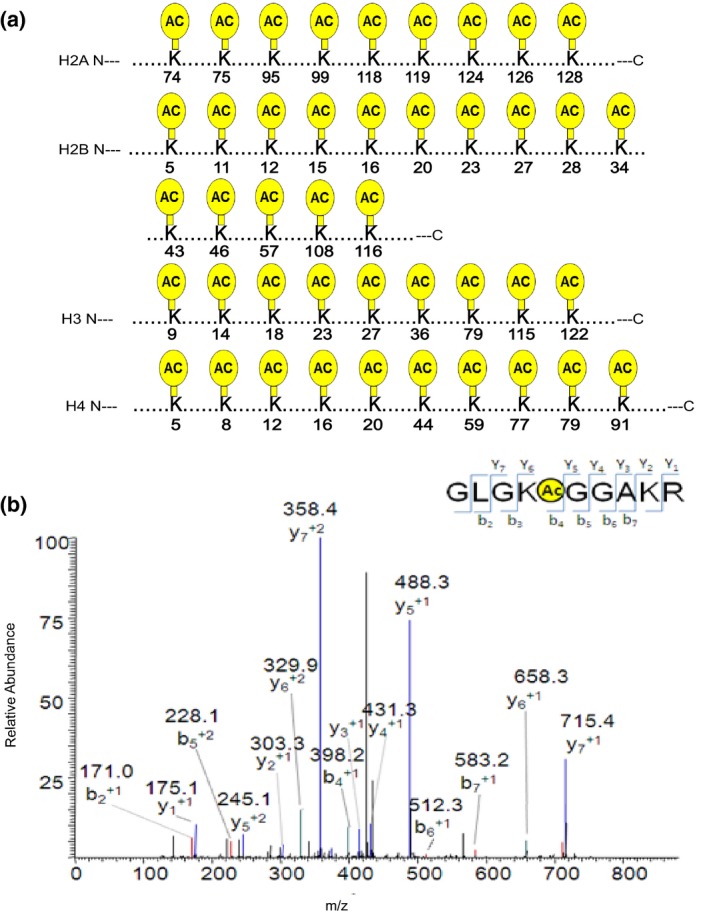
Schematic illustration of acetylation sites of histone lysine residues in human brain samples identified using HPLC‐MS/MS. (a) sites of histone acetylation detected in normal fetal brains. The yellow circle shape depicts acetylation sites in core histones (H3, H4, H2a, and H2b). (b) MS/MS spectra of a tryptic peptide ion histone H4K12 acetylated peptide (GLGKacGGAKR) in normal fetal brains. The x and y axes represent m/z and relative ion intensity, respectively. A series of b‐ and y‐type fragment ions are evident which not only provide reliable sequence information, but also indicate an unambiguous acetylation modification
Table 2.
Summary of acetylation peptides of histone H4 identified by Nano‐HPLC‐MS/MS in normal fetal brains
| Modification Site | Peptide sequence and modification |
MH + a [Da] |
chargeb | DeltaCnc | XCorrd | Confidencee |
|---|---|---|---|---|---|---|
| K5 | MSGRGKAcetylGGKGLGKGGAK | 1,787.913622 | 2 | 0 | 1.927143455 | High |
| GKAcetylGGKGLGKGGAK | 1,240.702806 | 2 | 0 | 3.515420198 | High | |
| KAcetylGGKGLGKGGAKR | 1,381.795702 | 2 | 0 | 2.113160849 | High | |
| GKAcetylGGKGLGKGGAKR | 1,438.815599 | 2 | 0 | 2.742053986 | High | |
| MSGRGKAcetylGGKGLGK | 1,490.771044 | 2 | 0 | 2.032351017 | High | |
| K8 | GKGGKAcetylGLGKGGAKR | 1,438.814745 | 2 | 0 | 4.259084702 | High |
| GKGGKAcetylGLGKGGAK | 1,240.702806 | 2 | 0 | 3.515420198 | High | |
| KGGKAcetylGLGKGGAKR | 1,381.795702 | 2 | 0 | 2.113160849 | High | |
| GGKAcetylGLGKGGAKR | 1,211.686449 | 2 | 0 | 2.340593338 | High | |
| RGKGGKAcetylGLGKGGAK | 1,600.866625 | 2 | 0 | 1.983496785 | High | |
| K12 | GKGGKGLGKAcetylGGAK | 1,240.702806 | 2 | 0 | 3.515420198 | High |
| KGGKGLGKAcetylGGAKR | 1,381.795702 | 2 | 0 | 2.113160849 | High | |
| GKGGKGLGKAcetylGGAKR | 1,438.815372 | 3 | 0 | 4.971486568 | High | |
| GGKGLGKAcetylGGAKR | 1,211.679857 | 2 | 0 | 3.015662432 | High | |
| GRGKGGKGLGKAcetylGGAK | 1,585.838169 | 3 | 0.0272 | 2.856269836 | High | |
| GKGLGKAcetylGGAKRHR | 1,391.808519 | 2 | 0 | 2.102574348 | High | |
| MSGRGKGGKGLGKAcetylGGA | 1,647.836595 | 2 | 0 | 2.614582539 | High | |
|
K16 |
MSGRGKGGKGLGKGGAKAcetyl | 1,789.964403 | 2 | 0 | 1.995563865 | High |
| GRGKGGKGLGKGGAKAcetyl | 1,483.818394 | 3 | 0.0496 | 2.297608614 | High | |
| GKGLGKGGAKAcetylRHR | 1,463.801561 | 2 | 0 | 2.216907024 | High | |
| KGGKGLGKGGAKAcetylR | 1,381.795702 | 2 | 0 | 2.113160849 | High | |
| GKGGKGLGKGGAKAcetylR | 1,438.815372 | 3 | 0 | 4.971486568 | High | |
| GGKGLGKGGAKAcetylR | 1,211.684374 | 2 | 0 | 2.888950586 | High | |
| K20 | KAcetyVLRDNIQGITKPAIR | 1,878.137106 | 4 | 0 | 5.521019936 | High |
| K44 | VKAcetylRISGLIYEETRGVLKVFLENVIR | 3,203.867927 | 5 | 0 | 2.860794067 | High |
| K59 | GVLKAcetylVFLENVIR | 1,428.857714 | 2 | 0 | 2.620787621 | High |
| VKRISGLIYEETRGVLKAcetylVFLENVIR | 3,203.867317 | 5 | 0 | 3.749208689 | High | |
| K77 | AVTYTEHAKAcetylRKTVTAMDVVYALK | 2,665.357151 | 3 | 0 | 2.517678738 | High |
| AVTYTEHAKAcetylRKTVTAMDVVYALKR | 2,995.549595 | 3 | 0 | 2.991867781 | High | |
| TYTEHAKAcetylRKAcetylTVTAMDVVYALKR | 2,623.424717 | 3 | 0 | 2.651541948 | High | |
| YTEHAKAcetylRKTVTAMDVVYALKR | 2,622.4202 | 3 | 0.0169 | 2.322541475 | High | |
| HAKAcetylRKTVTAMDVVYALKR | 2,199.261814 | 3 | 0 | 2.458812237 | High | |
| LENVIRDAVTYTEHAKAcetylR | 2,071.078647 | 3 | 0.0211 | 2.78817749 | High | |
| VTYTEHAKAcetylRKTVTAMDVVYALK | 2,622.408481 | 3 | 0 | 3.066841602 | High | |
| EHAKAcetylRKTVTAMDVVYALKR | 2,300.218845 | 3 | 0 | 2.742209196 | High | |
| AKAcetylRKTVTAMDVVYALKR | 2,048.173632 | 2 | 0 | 1.932291985 | High | |
| DAVTYTEHAKAcetylRKTVTAMDVVYALKR | 2,950.511143 | 3 | 0 | 3.67817688 | High | |
|
K79 |
TYTEHAKRKAcetylTVTAMDVVYALKR | 2,623.424717 | 3 | 0 | 2.651541948 | High |
| AKRKAcetylTVTAMDVVYALKR | 2,048.173632 | 2 | 0 | 1.932291985 | High | |
| KRKAcetylTVTAMDVVYALK | 2,009.098911 | 3 | 0 | 2.698174 | High | |
| KAcetylTVTAMDVVYALKR | 1,636.905369 | 3 | 0 | 2.493818045 | High | |
| VTYTEHAKRKAcetylTVTAMDVVYALK | 2,622.408481 | 3 | 0 | 3.066841602 | High | |
| DAVTYTEHAKRKAcetylTVTAMDVVYALKR | 2,950.605442 | 3 | 0 | 3.611026525 | High | |
| AVTYTEHAKRKAcetylTVTAMDVVYALKR | 2,995.549595 | 3 | 0 | 3.535665035 | High | |
| HAKRKAcetylTVTAMDVVYALKR | 2,199.292393 | 3 | 0 | 3.398030996 | High | |
| EHAKRKAcetylTVTAMDVVYALKR | 2,300.218845 | 3 | 0.0435 | 2.856972218 | High | |
| AVTYTEHAKRKAcetylTVTAMDVVYALK | 2,665.357151 | 3 | 0 | 2.517678738 | High | |
| K91 | TVTAMDVVYALKAcetylR | 1,536.852953 | 2 | 0 | 3.142895699 | High |
| KTVTAMDVVYALKAcetylR | 1,636.912327 | 3 | 0 | 4.236666203 | High |
Displays the protonated monoisotopic mass of the peptides. It is the measured mass.
ion charge; Displays the charge state of the peptide.
DeltaCn value displays the normalized score difference between the currently selected PSM and the highest‐scoring PSM for that spectrum. Credible criteria < 0.05.
Scores the number of fragment ions that are common to two different peptides with the same precursor mass and calculates the cross‐correlation score for all candidate peptides queried from the database.
Confidence: Indicates a confidence level associated with the peptide sequence at the top level. The false discovery rate (FDR) is a statistical value that estimates the number of false positive identifications among all identifications found by a peptide identification search. Specifies the target false discovery rate for peptide matches of high confidence. Peptide matches that pass the filter associated with the strict FDR (0.01) indicates a high confidence.
To further assess the histone acetylation alterations associated with NTDs, we compared histone acetylation sites between the NTD and healthy control samples. These results revealed no emerging or missing acetylation sites in NTD samples compared with healthy controls. The acetylation distribution of all fetal brain tissues is shown in Table 3. This suggests there is no difference in detectable acetylated histone sites between the NTD and control samples.
Table 3.
Acetylation distribution of eight fetal brain tissues
| Protein name | Modification site | Normal control | NTDs | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| H2a | K74 | ● | ● | ● | ○ | ● | ○ | ● | ● |
| K75 | ○ | ● | ○ | ○ | ● | ○ | ● | ● | |
| K95 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K99 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K118 | ○ | ○ | ● | ● | ○ | ○ | ○ | ● | |
| K119 | ● | ○ | ● | ○ | ● | ○ | ● | ● | |
| K124 | ● | ○ | ● | ○ | ● | ● | ● | ● | |
| K126 | ● | ○ | ● | ○ | ● | ○ | ○ | ● | |
| K128 | ● | ● | ○ | ○ | ○ | ● | ○ | ○ | |
| H2b | K5 | ● | ● | ● | ● | ● | ● | ● | ● |
| K11 | ● | ● | ● | ● | ○ | ● | ● | ● | |
| K12 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K15 | ● | ● | ● | ● | ● | ● | ○ | ● | |
| K16 | ● | ● | ● | ● | ● | ● | ○ | ● | |
| K20 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K23 | ○ | ● | ○ | ● | ● | ○ | ○ | ○ | |
| K27 | ○ | ● | ○ | ○ | ● | ○ | ○ | ○ | |
| K28 | ● | ● | ○ | ● | ● | ● | ○ | ○ | |
| K34 | ● | ● | ○ | ● | ● | ● | ● | ● | |
| K43 | ● | ● | ● | ● | ○ | ● | ● | ● | |
| K46 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K57 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K108 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K116 | ● | ● | ● | ● | ○ | ● | ○ | ● | |
| H3 | K9 | ○ | ● | ○ | ● | ○ | ○ | ○ | ● |
| K14 | ● | ○ | ○ | ● | ○ | ○ | ● | ● | |
| K18 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K23 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K27 | ● | ● | ● | ● | ● | ○ | ● | ● | |
| K36 | ● | ● | ● | ● | ● | ○ | ● | ● | |
| K79 | ○ | ● | ● | ○ | ○ | ○ | ● | ● | |
| K115 | ● | ○ | ● | ○ | ○ | ● | ○ | ● | |
| K122 | ● | ● | ● | ● | ○ | ○ | ○ | ● | |
| H4 | K5 | ● | ● | ● | ● | ○ | ○ | ● | ● |
| K8 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K12 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K16 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K20 | ○ | ○ | ● | ○ | ○ | ○ | ○ | ● | |
| K44 | ○ | ○ | ● | ● | ○ | ○ | ○ | ● | |
| K59 | ● | ● | ● | ● | ● | ● | ○ | ● | |
| K77 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K79 | ● | ● | ● | ● | ● | ● | ● | ● | |
| K91 | ● | ● | ● | ● | ● | ● | ○ | ● | |
●:detected in samples.
○: not detected in samples.
3.3. The histone acetylation in human fetal brain
Although the histone acetylation sites were same in NTDs and control samples, we could not exclude any quantitative change between groups. Therefore, we performed western blotting using a specific Kac antibody and found a significantly increased expression in the NTD samples when compared with normal samples (Figure 3).
Figure 3.
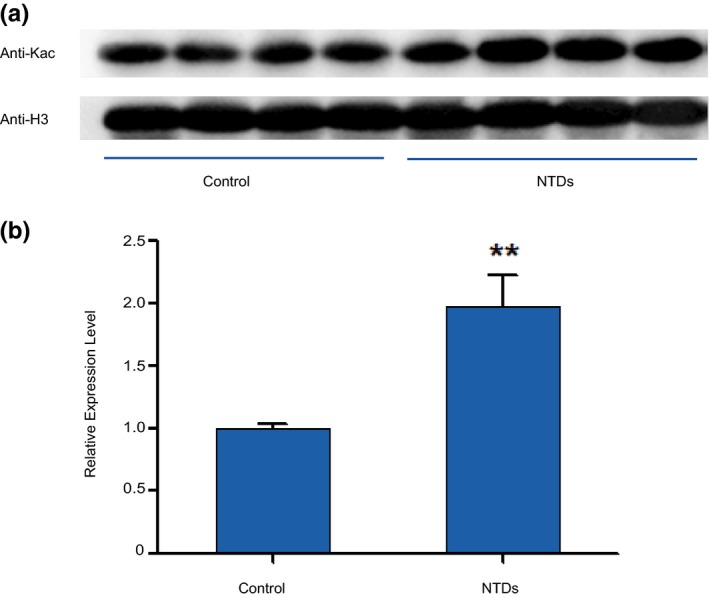
Differential levels of histone acetylation in control and NTD samples. (a) Western blot of Kac in the normal and NTD (spina bifida) samples. (b) The quantitative analysis of western blot showed that the levels of acetylation expression in NTDs are higher than in controls. The differentiation have a statistically significant (p < .01 compared with the control group, t test)
3.4. LC‐ESI‐MS analysis of posttranslational modifications of human brain histones
To evaluate the levels of acetylation in the four histone components between NTD and control samples, we next developed a rapid MS method (LC‐ESI‐MS). The extracted histones were separated using a molecular sieve chromatography column, which showed four major peaks, including H3, H4, H2a, and H2b (Figure 4a). The assignments were based on the masses predicted from known amino acid sequences and allowing for posttranslational modifications (Bonenfant, Coulot, Towbin, Schindler, & Oostrum, 2006; Zhang et al., 2004). Compared with the traditional method using a C4 or C18 column, the elution pattern was consistent and reproducible with similar results obtained for other human cell lines, such as Jurkat cells or bovine thymus. Extracted histone mixtures containing H2a, H2b, H3, and H4 were clearly separated (Figure 4a). The peak marked with 11,306.93 was identified as histone H4 (Figure 4a) by the deconvoluted ESI mass spectra analyzed by an on‐line ion trap with Mag Tran 1.0 software. Furthermore, Histone H4 with a series of acetylation sites (Figure 4b) was identified by mass analysis. Each histone protein modification was indicated by derived molecular mass calculated by software, as reported in Figure 4b, together with their percentage relative abundance. We detected the percentage relative abundance of histone H4 acetylation in NTDs and healthy controls samples (Figure 4c). The relative abundance of all samples was shown in Table 4. Among the nine types that were identified (H4+1AC+1Me, H4+1AC+2Me, H4+2AC, H4+2AC+1Me, H4+2AC+2Me, H4+3AC, H4+3AC+1Me, H4+3AC+2Me, H4+4AC), there was a significant difference in H4+2AC relative abundance between healthy controls and NTD samples. These results showed that the level of H4 with two acetylation was significantly increased in NTD samples compared with healthy controls (Figure 4d).
Figure 4.
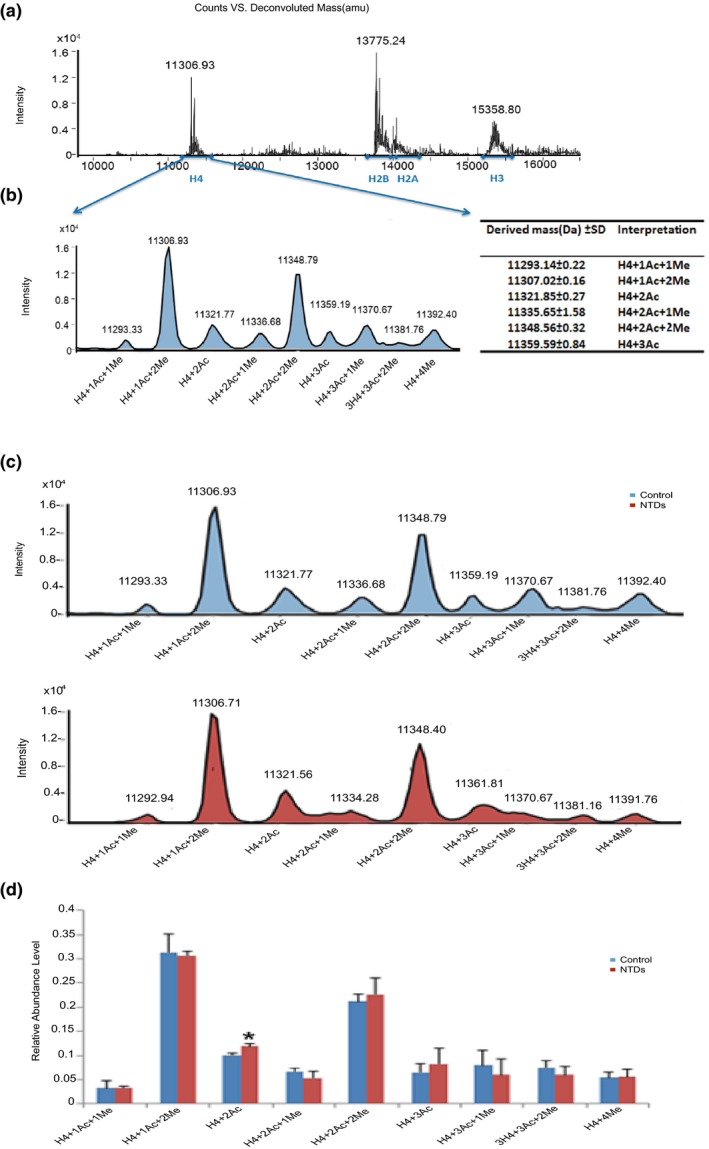
Histone modification profiles in control and NTD samples. (a) Deconvoluted ESI mass spectra of histones in Normal control fetal brains. The four peaks correspond to H4, H2b, H2a, and H3, respectively. They are marked with arrows in the spectrum. The peak of 11,306.93 Da corresponds to histone H4. (b) Reconstructed peak averaged mass spectra and the histone H4 isoforms with different acetylation was interpreted by the derived mass. H4+1AC+1Me on behalf of the H4 which have a acetylation site and a methylation site; H4+1AC+2Me on behalf the H4 which have a acetylation site and two methylation sites; H4+2AC on behalf the H4 which have two acetylation sites; H4+2AC+1Me on behalf the H4 which have two acetylation sites and a methylation site; H4+2AC+2Me on behalf the H4 which have two acetylation sites and two methylation sites; H4+3AC on behalf the H4 which have three acetylation sites. (c) Reconstructed peak averaged mass spectra of histone H4 isoforms in control and NTD samples. (d) Histograms of the quantitative analysis of histone acetylation modification along with their percentage relative abundance. (p < .05 compared with the control group, t test)
Table 4.
The relative abundance of peptides detected from the four normal fetal brains and four NTDs
| Noa | H4 + 1AC + 1Me | H4 + 1AC + 2Me | H4 + 2AC | H4 + 2AC + 1Me | H4 + 2AC + 2Me | H4 + 3AC | H4 + 3AC + 1Me | H4 + 3AC + 2Me | H4 + 4AC |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 902 | 3,031 | 1,286 | 1,096 | 2,424 | 1,427 | 571 | 888 | 1,028 |
| 2 | 315 | 2,986 | 768 | 529 | 1813 | 636 | 307 | 412 | 329 |
| 3 | 288 | 3,348 | 1,193 | 818 | 2,625 | 675 | 1,159 | 1,043 | 769 |
| 4 | 403 | 3,437 | 1,263 | 779 | 2,256 | 702 | 1,247 | 1,029 | 678 |
| 5 | 366 | 3,100 | 1,230 | 290 | 2,679 | 1,184 | 244 | 483 | 483 |
| 6 | 362 | 3,015 | 1,248 | 723 | 1875 | 532 | 932 | 910 | 757 |
| 7 | 249 | 2,664 | 906 | 403 | 1702 | 424 | 785 | 570 | 579 |
| 8 | 301 | 2,907 | 1,179 | 614 | 2,419 | 1,044 | 314 | 348 | 334 |
The number is the same as the clinical information number of the eight individual fetus.
3.5. H4K5ac expression validation
Based on previously reported alterations in H4K5ac in a mouse embryo model of maternal diabetes with NTDs (Salbaum & Kappen, 2012), we sought to validate the alterations in H4K5ac in NTDs and healthy controls in this study. Therefore, we performed western blotting using a specific anti‐H4K5ac antibody. These results indicated that H4K5ac expression was detected in all healthy control samples, but were significantly higher in the NTD brain samples (Figure 5).
Figure 5.
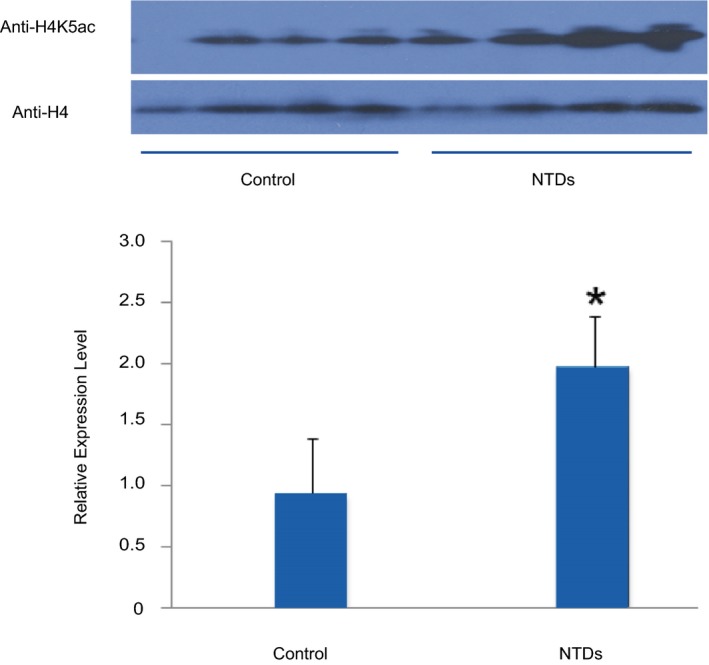
Histone acetylation comparation of H4K5ac in control and NTD samples. (a) Western blot of H4K5ac in the normal and NTD (spina bifida) samples. (b) Histograms of the quantitative analysis of H4K5ac expression using western blot (p < .05 compared with the control group, t test)
In summary, histone H4K5ac levels in brain samples with NTDs were significantly when compared with healthy controls. These results indicated quantitative difference in histone H4 acetylation in human NTDs.
4. DISCUSSION
Histone posttranslational modifications play an important role in the dynamics of chromatin structure and function. However, no comprehensive analysis of histone acetylation has been reported at the protein level in the developing human brain. In this study, we used a strategy combining mass spectrometric techniques (nano‐HPLC‐MS/MS and LC‐ESI‐MS), molecular sieve separation, and enzymatic digestions to characterize histone acetylation in human fetal samples. Taken together, we obtained a comprehensive overview of the human brain histone acetylation, in particular associated with histone H4 acetylation. Using two MS methods, we concluded that there is no difference in detectable of histone acetylation sites between the NTDs and control samples. But the acetylation level of histone H4 in NTDs increased. Here, we showed that the quantity of histone H4 acetylation was important for normal brain development; an increase in histone H4 acetylation was linked to NTDs.
Histone acetylation regulates many cellular processes, such as gene expression. Acetylation marks within the histone N terminal tails and globular domains can selectively make the structure of certain chromatin regions from tightly dispersed, facilitating the binding of transcription factors to DNA, thereby promoting the transcription of certain genes and enhancing the expression level of genes. Acetylated lysine within the H4 tails (K5, K8, and K12 of H4) and globular domains (K56 of H3 and K91 of H4) appear to serve important, albeit redundant, roles in chromatin assembly (Ma, Wu, Altheim, Schultz, & Grunstein, 1998; Ye et al., 2005). Another role for histone acetylation is the regulation of chromatin folding. Acetylation of the histone H4 tail plays a prominent role in determining the ability of chromatin to fold into higher‐order structures (Chang & Takada, 2016; Luger, Mader, Richmond, Sargent, & Richmond, 1997). Acetylation promotes a more open chromatin configuration by decreasing the interactions between nucleosomes. This releases the histone tails from linker DNA and reduces fiber–fiber interactions. In addition, histone acetylation regulates the formation of heterochromatin; acetylation of H4 lysine 16 is important to regulate chromatin accessibility(Zhang, Erler, & Langowski, 2017) and spread heterochromatin components, whereas acetylation of this site serves as a barrier to this spreading (Liou, Tanny, Kruger, Walz, & Moazed, 2005). Finally, histone acetylation is critical for gene transcription (Chen, Zhao, & Zhao, 2015; Chen et al., 2019). Acetylation promotes transcription by favoring a more open chromatin conformation that permits binding of the transcriptional machinery and by directly serving as recognition sites for factors that promote transcription (Shahbazian & Grunstein, 2007; Wilson & Merkenschlager, 2006).
Histone acetylation has a close association with the occurrence of NTDs. It is crucial in gene transcription and plays a key role in late brain development (Antonio & Silvia, 2013). Furthermore, it is dynamically regulated by several classes of histone deacetylases (HDACs) and families of histone acetyltransferases (HATs), such as P300, CBP, and GCN5 (Oc, 2012). These act on targeted regions of chromatin to regulate specific gene transcription. One study reported that Mark2 depletion reduces Dvl gene expression and interrupts neural stem cell (NSCs) growth and differentiation in mouse cell line, which are likely to be mediated through a decrease in class IIa HDAC phosphorylation and reduced H3K4ac and H3K27ac occupancies at the Dvl1/2 promoters(Chen et al., 2017). High glucose is a significant risk factor for NTDs. Bai et al. have reported that maternal diabetes leads to an increase in H4K5/K8/K12/K16 acetylation levels, and that the CBP selective inhibitor, C646, could efficiently prevent an increase or emergence of histone H4 acetylation and neuroepithelial cell proliferation (Bai et al., 2018). Furthermore, a deletion in the neuronal Nap1l2 (nucleosome assembly protein 1‐like 2) gene in mice leads to an increase in acetylated histone H3K9/14 at the Cdkn1c transcription site and causes NTDs (Attia, Rachez, De Pauw, Avner, & Rogner, 2007). Taken together with our results, the alterations in histone acetylation influence the gene transcription in a genome‐wide profile, and histone acetylation is associated with the occurrence of NTDs. Moreover, gene‐specific changes in histone H4 acetylation patterns may be a key early step in the pathological processes triggered by status epileptics (Huang, Doherty, & Dingledine, 2002). Previous research (Cho, Kim, Kim, & Sun, 2011) has found that the level of histone H4 acetylation is highly dynamic and tightly linked to neuronal type and differentiation stage. Furthermore, histone H4 acetylation contributes to the action of IGF‐I on gene expression in the mammalian central nerve system (Sun & D’Ercole, 2006). Histone modifications have complex interactions. H4K5ac results in decreased arginine methylation by PRMT1, PRMT3, and PRMT8 (Fulton, Zhang, He, Ho, & Zheng, 2017). Taken together, this indicates that inordinate histone H4 acetylation may be involved in etiology of NTDs.
5. CONCLUSION
In summary, this study presents a comprehensive map of histone H4 modifications in the human fetal brain and provides experimental evidence supporting a relationship between histone H4 acetylation and NTDs. Combined with our previous work that shows that aberrant histone methylation is involved in NTDs (Zhang et al., 2013), these results confirm that aberrant histone modification during early pregnancy is associated with the occurrence of NTDs.
CONFLICT OF INTEREST
All authors declared that no competing interests existed.
Supporting information
ACKNOWLEDGMENTS
We are grateful to all participating hospitals for their assistance in sample collection and recording of clinical information. We thank all of the women who participated for their cooperation. This study is supported by Beijing Natural Science Foundation (7182024) and Research Foundation of Capital Institute of Pediatrics (No. PY‐2018‐02). We thank Amy Birch, PhD, from Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.
Li D, Wan C, Bai B, Cao H, Liu C, Zhang Q. Identification of histone acetylation markers in human fetal brains and increased H4K5ac expression in neural tube defects. Mol Genet Genomic Med. 2019;7:e1002 10.1002/mgg3.1002
Contributor Information
Changyun Liu, Email: changyun1@163.com.
Qin Zhang, Email: maureenzq@hotmail.com.
REFERENCES
- Antonio, C. , & Silvia, S. (2013). Histone acetylation in neurodevelopment. Current Pharmaceutical Design, 19, 5043–5050. 10.2174/1381612811319280003 [DOI] [PubMed] [Google Scholar]
- Attia, M. , Rachez, C. , De Pauw, A. , Avner, P. , & Rogner, U. C. (2007). Nap1l2 promotes histone acetylation activity during neuronal differentiation. Molecular and Cellular Biology, 27(17), 6093–6102. 10.1128/MCB.00789-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, B. , Zhang, Q. , Wan, C. , Li, D. , Zhang, T. , & Li, H. (2018). CBP/p300 inhibitor C646 prevents high glucose exposure induced neuroepithelial cell proliferation. Birth Defects Research, 110(14), 1118–1128. 10.1002/bdr2.1360 [DOI] [PubMed] [Google Scholar]
- Balmer, N. V. , Weng, M. K. , Zimmer, B. , Ivanova, V. N. , Chambers, S. M. , Nikolaeva, E. , … Leist, M. (2012). Epigenetic changes and disturbed neural development in a human embryonic stem cell‐based model relating to the fetal valproate syndrome. Human Molecular Genetics, 21(18), 4104–4114. 10.1093/hmg/dds239 [DOI] [PubMed] [Google Scholar]
- Bonenfant, D. , Coulot, M. , Towbin, H. , Schindler, P. , & van Oostrum, J. (2006). Characterization of histone H2A and H2B variants and their post‐translational modifications by mass spectrometry. Molecular & Cellular Proteomics, 5(3), 541–552. M500288-MCP200 [DOI] [PubMed] [Google Scholar]
- Chang, L. , & Takada, S. (2016). Histone acetylation dependent energy landscapes in tri‐nucleosome revealed by residue‐resolved molecular simulations. Scientific Reports, 6(1), 34441 10.1038/srep34441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. P. , Zhao, Y. T. , & Zhao, T. C. (2015). Histone deacetylases and mechanisms of regulation of gene expression. Critical Reviews in Oncogenesis, 20(1‐2), 35–47. 10.1615/CritRevOncog.2015012997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. F. , Lin, Y. T. , Gallegos, D. A. , Hazlett, M. F. , Gomez‐Schiavon, M. , Yang, M. G. , … West, A. E. (2019). Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity‐inducible genes. Cell Reports, 26(5), 1174–1188.e5. 10.1016/j.celrep.2019.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S. , Zhang, Q. , Bai, B. , Ouyang, S. , Bao, Y. , Li, H. , & Zhang, T. (2017). MARK2/Par1b insufficiency attenuates DVL gene transcription via histone deacetylation in lumbosacral spina bifida. Molecular Neurobiology, 54(8), 6304–6316. 10.1007/s12035-016-0164-0 [DOI] [PubMed] [Google Scholar]
- Cho, B. , Kim, H. J. , Kim, H. , & Sun, W. (2011). Changes in the histone acetylation patterns during the development of the nervous system. Experimental Neurobiology, 20(2), 81–84. 10.5607/en.2011.20.2.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copp, A. J. , & Greene, N. D. (2010). Genetics and development of neural tube defects. The Journal of Pathology, 220 (2), 217–230. 10.1002/path.2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton, M. D. , Zhang, J. , He, M. , Ho, M. C. , & Zheng, Y. G. (2017). Intricate effects of alpha‐amino and lysine modifications on arginine methylation of the N‐terminal tail of histone H4. Biochemistry, 56(28), 3539–3548. 10.1021/acs.biochem.7b00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, X. , Lin, L. , Zheng, X. , Zhang, T. , Song, X. , Wang, J. , … Liu, J. (2007). High prevalence of NTDs in Shanxi Province: A combined epidemiological approach. Birth Defects Research Part A: Clinical and Molecular Teratology, 79(10), 702–707. 10.1002/bdra.20397 [DOI] [PubMed] [Google Scholar]
- Horikoshi, M. (2013). Histone acetylation: From code to web and router via intrinsically disordered regions. Current Pharmaceutical Design, 19(28), 5019–5042. [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Doherty, J. J. , & Dingledine, R. (2002). Altered histone acetylation at glutamate receptor 2 and brain‐derived neurotrophic factor genes is an early event triggered by status epilepticus. Journal of Neuroscience, 22(19), 8422–8428. 22/19/8422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, H. (2013). Histone modifications for human epigenome analysis. Journal of Human Genetics, 58(7), 439–445. 10.1038/jhg.2013.66 [DOI] [PubMed] [Google Scholar]
- Lei, J. , Jingru, Y. , Yanjun, X. , Hui, L. , & Aiguo, R. (2017). Prevalence of neural tube defects and the impact of prenatal diagnosis in three districts of Beijing, China. Paediatric and Perinatal Epidemiology, 31(4), 293–300. 10.1111/ppe.12354 [DOI] [PubMed] [Google Scholar]
- Liou, G. G. , Tanny, J. C. , Kruger, R. G. , Walz, T. , & Moazed, D. (2005). Assembly of the SIR complex and its regulation by O‐acetyl‐ADP‐ribose, a product of NAD‐dependent histone deacetylation. Cell, 121(4), 515–527. S0092-8674(05)00354-5 [DOI] [PubMed] [Google Scholar]
- Lu, W. , Guzman, A. R. , Yang, W. , Chapa, C. J. , Shaw, G. M. , Greene, R. M. , … Zhu, H. (2010). Genes encoding critical transcriptional activators for murine neural tube development and human spina bifida: A case‐control study. BMC Medical Genetics, 11(1), 141 10.1186/1471-2350-11-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger, K. , Mader, A. W. , Richmond, R. K. , Sargent, D. F. , & Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 389(6648), 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Ma, X. J. , Wu, J. , Altheim, B. A. , Schultz, M. C. , & Grunstein, M. (1998). Deposition‐related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proceedings of the National Academy of Sciences, 95(12), 6693–6698. 10.1073/pnas.95.12.6693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murko, C. , Lagger, S. , Steiner, M. , Seiser, C. , Schoefer, C. , & Pusch, O. (2013). Histone deacetylase inhibitor trichostatin A induces neural tube defects and promotes neural crest specification in the chicken neural tube. Differentiation, 85(1‐2), 55–66. 10.1016/j.diff.2012.12.001 [DOI] [PubMed] [Google Scholar]
- Oc, A. (2012). Expansion of the lysine acylation landscape. Angewandte Chemie (International edition in English), 51(16), 3755–3756. 1002/anie.201200316 [DOI] [PubMed] [Google Scholar]
- Rada‐Iglesias, A. , Bajpai, R. , Swigut, T. , Brugmann, S. A. , Flynn, R. A. , & Wysocka, J. (2011). A unique chromatin signature uncovers early developmental enhancers in humans. Nature, 470(7333), 279–283. 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salbaum, J. M. , & Kappen, C. (2012). Responses of the embryonic epigenome to maternal diabetes. Birth Defects Research Part A: Clinical and Molecular Teratology, 94(10), 770–781. 10.1002/bdra.23035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian, M. D. , & Grunstein, M. (2007). Functions of site‐specific histone acetylation and deacetylation. Annual Review of Biochemistry, 76(1), 75–100. 10.1146/annurev.biochem.76.052705.162114 [DOI] [PubMed] [Google Scholar]
- Shechter, D. , Dormann, H. L. , Allis, C. D. , & Hake, S. B. (2007). Extraction, purification and analysis of histones. Nature Protocols, 2(6), 1445–1457. 10.1038/nprot.2007.202 [DOI] [PubMed] [Google Scholar]
- Shyamasundar, S. , Jadhav, S. P. , Bay, B. H. , Tay, S. S. , Kumar, S. D. , Rangasamy, D. , & Dheen, S. T. (2013). Analysis of epigenetic factors in mouse embryonic neural stem cells exposed to hyperglycemia. PLoS ONE, 8(6), e65945 10.1371/journal.pone.0065945 PONE‐D‐13‐05290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, L. Y. , & D’Ercole, A. J. (2006). Insulin‐like growth factor‐I stimulates histone H3 and H4 acetylation in the brain in vivo. Endocrinology, 147(11), 5480–5490. 10.1210/en.2006-0586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Put, N. M. , van Straaten, H. W. , Trijbels, F. J. , & Blom, H. J. (2001). Folate, homocysteine and neural tube defects: An overview. Experimental Biology and Medicine, 226(4), 243–270. 10.1177/153537020122600402 [DOI] [PubMed] [Google Scholar]
- Wilson, C. B. , & Merkenschlager, M. (2006). Chromatin structure and gene regulation in T cell development and function. Current Opinion in Immunology, 18(2), 143–151. 10.1016/j.coi.2006.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Ai, X. , Eugeni, E. E. , Zhang, L. , Carpenter, L. R. , Jelinek, M. A. , … Parthun, M. R. (2005). Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Molecular Cell, 18(1), 123–130. S1097-2765(05)01148-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Wu, Y. , & Yang, P. (2016). High glucose‐induced oxidative stress represses sirtuin deacetylase expression and increases histone acetylation leading to neural tube defects. Journal of Neurochemistry, 137(3), 371–383. 10.1111/jnc.13587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Freitas, M. A. , Wickham, J. , Parthun, M. R. , Klisovic, M. I. , Marcucci, G. , & Byrd, J. C. (2004). Differential expression of histone post‐translational modifications in acute myeloid and chronic lymphocytic leukemia determined by high‐pressure liquid chromatography and mass spectrometry. Journal of the American Society for Mass Spectrometry, 15(1), 77–86. S1044030503007165 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Xue, P. , Li, H. , Bao, Y. , Wu, L. , Chang, S. , … Zhang, T. (2013). Histone modification mapping in human brain reveals aberrant expression of histone H3 lysine 79 dimethylation in neural tube defects. Neurobiology of Diseases, 54, 404–413. 10.1016/j.nbd.2013.01.014 [DOI] [PubMed] [Google Scholar]
- Zhang, R. , Erler, J. , & Langowski, J. (2017). Histone acetylation regulates chromatin accessibility: Role of H4K16 in inter‐nucleosome interaction. Biophysical Journal, 112(3), 450–459. 10.1016/j.bpj.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


