Abstract
Background
Familial nonmedullary thyroid cancer (FNMTC) accounts for approximately 3%–9% of all thyroid cancers; however, the mechanisms underlying FNMTC remain unclear. Environmental and genetic (especially genetic mutation) factors may play important roles in FNMTC etiology, development, and pathogenesis.
Methods
Three affected members, including two first‐degree relatives, and three healthy members of a family with FNMTC were studied. We performed whole‐exome and targeted gene sequencing to identify gene mutations that may be associated with FNMTC pathogenesis. The results were analyzed using Exome Aggregation Consortium data and the Genome Aggregation Database and further validated using Sanger sequencing.
Results
Of 28 pivotal genes with rare nonsynonymous mutations found, 7 were identified as novel candidate FNMTC pathogenic genes (ANO7, CAV2, KANK1, PIK3CB, PKD1L1, PTPRF, and RHBDD2). Among them, three genes (PIK3CB, CAV2, and KANK1) are reportedly involved in tumorigenesis through the PI3K/Akt signaling pathway.
Conclusion
We identified seven pathogenic genes in affected members of a family with FNMTC. The PI3K/Akt signaling pathway is thought to be closely related to the development of FNMTC, and three of the susceptibility genes identified herein are associated with this pathway. These findings expand our understanding of FNMTC pathogenesis and underscore PI3K/Akt pathology as a potential therapy target.
Keywords: familial nonmedullary thyroid cancer, genetic mutation, Sanger sequencing, targeted gene sequencing, whole‐exome sequencing
We identified 7 pathogenic genes in affected members of a family with FNMTC. The PI3K/Akt signaling pathway is thought to be closely related to the development of FNMTC, and three of the susceptibility genes identified herein are associated with this pathway. These findings expand our understanding of FNMTC pathogenesis and underscore PI3K/Akt pathology as a potential therapy target.
1. INTRODUCTION
Thyroid cancer is the most common endocrine tumor. There are more than 800,000 cases diagnosed each year in the United States, and the incidence continues to rise (Miller et al., 2016). Nonmedullary thyroid cancer (NMTC) originates from follicular cells and encompasses papillary thyroid cancer (PTC), follicular thyroid cancer, and anaplastic thyroid carcinoma. NMTC accounts for approximately 90% of all thyroid cancers (Zhang & Xing, 2016). Only a small subset of NMTC, mainly PTC, shows family aggregation, which accounts for 3%–9% of all thyroid cancers (Klubo‐Gwiezdzinska et al., 2017).
Familial NMTC (FNMTC) is defined as a family of two or more first‐degree relatives diagnosed as having thyroid cancer derived from the thyroid follicular epithelium cell and excludes a history of exposure to radiation. FNMTC can be divided into two groups on the basis of its clinical manifestations. The first group is called syndromic FNMTC and is characterized by the familial syndrome, including familial adenomatous polyposis, Gardner's syndrome, Cowden's disease, Carney complex type 1, Werner syndrome, papillary renal neoplasia, etc. The second group, termed nonsyndromic FNMTC, is mainly characterized by NMTC (Bano & Hodgson, 2016; Yu et al., 2015). Recently, researchers have found that FNMTC has higher recurrence, greater invasiveness, and worse prognosis than sporadic NMTC (SNMTC). In addition, the age of FNMTC onset is earlier than SNMTC, which generally means that patients undergo more surgical procedures or other active treatments (Tavarelli et al., 2015; Wang et al., 2015; Zhang, Yang, Meng, Chen, & Pang, 2016). At present, the genetics and pathogenesis of SNMTC are relatively clear, whereas for FNMTC, they remain ambiguous. FNMTC was first reported in 1955 by Robinson and Orr in several monozygotic twins who were diagnosed as having thyroid cancer (Robinson & Orr, 1955). Current research suggests that FNMTC is an autosomal dominant genetic disease with incomplete penetrance and variable expressivity. Despite some reports, the specific pathogenic genes and mechanisms underlying FNMTC are not yet clear. Therefore, finding the pathogenic FNMTC susceptibility gene mutations would be of great significance and would enable a more in‐depth assessment of the risk of FNMTC among members of a family with the disease.
Here we report a study on a pedigree containing three PTC‐affected members. We conducted whole‐exome sequencing for these three affected family members as well as for three healthy family members. We identified gene mutation sites that were common to the affected relatives but were not found in the healthy relatives. Using Sanger sequencing, we verified the mutations in these candidate pathogenic FNMTC susceptibility genes.
1.1. Case Report
One kindred with FNMTC (patient II.2) was referred to our institution for further investigation of the patient's clinical and genetic characteristics. Patient II.2 was examined and was diagnosed as having PTC. Patient I.1 was the proband's father, who was diagnosed as having PTC. Two other relatives (III.1 and III.2) were also diagnosed as having PTC on the basis of ultrasonographic examinations. Other family members were negative for the presence of thyroid nodules or of thyroid cancer as assessed by ultrasonography. The family had not been exposed to radioactive materials, and they did not have a history of other primary cancers (Figure 1).
Figure 1.
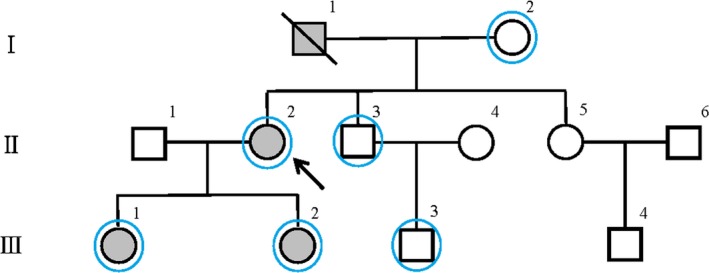
Pedigree of the family with familial non‐medullary thyroid cancer (FNMTC). Filled and unfilled symbols represent NMTC‐affected and NMTC‐unaffected individuals, respectively. Circles and squares represent females and males, respectively. The arrow indicates the proband; the slash, a deceased member. Blue circles represent family members who participated in the whole‐exome sequencing
2. METHODS
2.1. Editorial policies and ethical considerations
This study was approved by the Ethics Committee of Anhui Medical University. The peripheral blood of the proband and his family members were obtained at the Department of Otorhinolaryngology, Head and Neck Surgery of the First Affiliated Hospital of Anhui Medical University. Each person volunteered to participate in this study and signed an informed consent form to be included in the present study.
2.2. Genetic studies
Peripheral blood was collected from the proband, the two other affected relatives, and three healthy relatives. DNA was extracted from the peripheral blood using standard procedures and following manufacturer's instructions (Vazyme Biotech Co. Ltd). High‐throughput sequencing of DNA samples was performed after whole‐exome capture. The Exome Aggregation Consortium (ExAC) data and the Genome Aggregation Database (gnomAD) were used to filter the results obtained. The filter criteria were the allele frequency of the variant in the East Asian populations of the ExAC database (ExAC EAS) of <0.05 and of the gnomAD database (gnomAD EAS) of <0.05 and excluded synonymous mutations or noncoding flanking regions. The remaining genes that have been found to be closely related to tumor development were screened out. Sanger sequencing was used for final validation.
2.3. Sanger sequencing
GenBank sequences of ANO7 (accession number: NM‐001001891.3), CAV2 (accession number: NM‐001233.5), KANK1 (accession number: NM‐015158.3), PIK3CB (accession number: NM‐006219.2), PKD1L1 (accession number: NM‐138295.4), PTPRF (accession number: NM‐002840.4), RHBDD2 (accession number: NM‐001040456.2) were referred to the reference sequence database in National Center for Biotechnology Information (NCBI). Sanger sequencing was used to verify genetic susceptibility mutations. The primers are listed in Table 1.
Table 1.
Primers of susceptibility mutations
| Gene | Forward primer (5'→3') | Reverse primer (5'→3') |
|---|---|---|
| ANO7 | TGCTCTGACTACGAGGACACT | ACCAAGGTGAGATGGGGGAC |
| CAV2 | CCGCTGTGATCCAATTATCC | CCAGACCTGGGGTCCAAGTA |
| KANK1 | AGCCACCATGCAGATGACAC | CAGGCGTTTCAGAGCAATGG |
| PIK3CB | TTGCCTTCTTTTGACCTATCTT | TTCTGAGCCCTTTTCTTTCTT |
| PTPRF | AGGAGCTTCAGGCTACTCTGT | TGGGAGTTGGGTACTCAGGA |
| RHBDD2 | TAGGCGGCTTATAACCTGGC | TGATGACACAGCCTCGAATG |
| PKD1L1 | GCGACTCACATTTTACTTCCA | CATCCCCTGTCCTTCCTT |
3. RESULTS
3.1. Identification of pathogenic FNMTC susceptibility gene mutations
We performed whole‐exome sequencing using the genomic DNA extracted from the peripheral blood from the three affected and three healthy members of a family who were diagnosed as having FNMTC to identify candidate pathogenic genes. The pedigree of the family with FNMTC is illustrated in Figure 1. After we conducted bioinformatics analyses, we identified 28 rare pivotal single‐nucleotide polymorphism (SNP) mutations, and among them, 7 FNMTC susceptibility candidate genes with SNP mutations: anoctamin 7 (ANO7, OMIM: 605096), caveolin 2 (CAV2, OMIM: 601048), KN motif and ankyrin repeat domains 1 (KANK1, OMIM: 607704), phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit beta (PIK3CB, OMIM: 602925), polycystin 1 like 1, transient receptor potential channel interacting (PKD1L1, OMIM: 609721), protein tyrosine phosphatase receptor type F (PTPRF, OMIM: 179590), and rhomboid domain containing 2 (RHBDD2, OMIM: 615203) (Figure 2, Tables 2 and 3). Sanger sequencing was subsequently performed to verify the mutations in these seven candidate genes (Figure 3).
Figure 2.
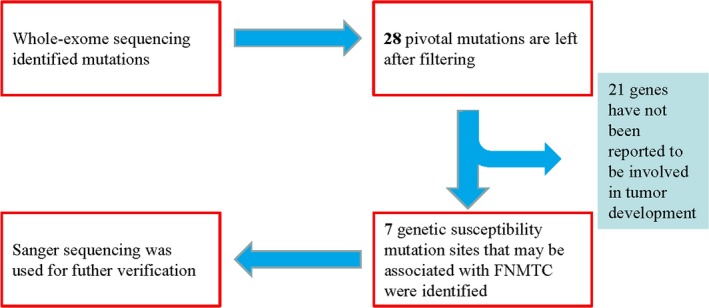
Flow chart for mutation screening. In total, 28 pivotal genes with mutations were selected from the whole‐exome sequencing. Among them, seven genes with nonsynonymous mutation sites were identified as potentially involved in tumorigenesis and development. Sanger sequencing was used for further verification of the mutations
Table 2.
Twenty‐eight pivotal genes with nonsynonymous mutations in the present family with FNMTC
| Gene | Cytoband | SNP annotation | ExAC ALL | ExAC EAS | gnomAD ALL | gnomAD EAS |
|---|---|---|---|---|---|---|
| AK2 | 1p35.1 | rs113711467 | 0.0097 | 0.0043 | 0.001 | 0 |
| ALDH9A1 | 1q24.1 | rs141131078 | 0.0003 | 0.0037 | — | 0.0018 |
| ANO7 | 2q37.3 | rs57677160 | 0.2022 | 0.0145 | 0.2013 | 0.0185 |
| C5orf49 | 5p15.31 | rs76872483 | — | 0.0429 | 0.0035 | 0.0347 |
| C9orf129 | 9q22.31 | rs62572859 | — | 0.041 | 0.1587 | 0.0316 |
| CAV2 | 7q31.2 | rs8940 | 0.1531 | 0.0079 | 0.1522 | 0.0105 |
| CEP162 | 6q14.2 | rs201104500 | 0.0003 | 0.0034 | 0.0003 | 0.0043 |
| CYS1 | 2p25.1 | — | 0.0013 | 0.0185 | 0.0005 | 0.0105 |
| KANK1 | 9p24.3 | rs28374506 | 0.0424 | 0.0087 | 0.0909 | 0.0086 |
| KRTAP10‐12 | 21q22.3 | rs61745911 | 0.0799 | 0.0266 | 0.1131 | 0.0309 |
| KRTAP10‐9 | 21q22.3 | rs62220926 | 0.0835 | 0.0083 | 0.1299 | 0.013 |
| MUC4 | 3q29 | — | — | 0 | — | 0.0016 |
| NUS1 | 6q22.1 | rs28362519 | 0.0007 | 0.0095 | 0.0005 | 0.008 |
| PCDHA8 | 5q31.3 | — | 0.0003 | 0.0032 | 0.0002 | 0.0037 |
| PIK3CB | 3q22.3 | rs375961764 | — | 0.0001 | — | 0 |
| PKD1L1 | 7p12.3 | rs17131915 | 0.0058 | 0.0062 | 0.0169 | 0.0074 |
| PTPRF | 1p34.2 | rs17849118 | 0.0003 | 0.0049 | — | 0.0012 |
| RHBDD2 | 7q11.23 | rs11547498 | 0.0004 | 0.0054 | 0.0002 | 0.0031 |
| RHCE | 1p36.11 | rs143715642 | 0.0005 | 0.0074 | 0.0005 | 0.0086 |
| SFTPC | 8p21.3 | rs75902455 | 0.0004 | 0.0051 | 0.0004 | 0.0062 |
| SH3TC2 | 5q32 | rs186864272 | 0.0001 | 0.0019 | 0.0001 | 0.0025 |
| SORBS3 | 8p21.3 | rs3758036 | 0.0035 | 0.0475 | 0.0022 | 0.0401 |
| SPAG11B | 8p23.1 | — | 0.0002 | 0.0002 | — | 0 |
| SZT2 | 1p34.2 | rs150966402 | 0.0014 | 0.0191 | . | 0.0234 |
| TMPRSS13 | 11q23.3 | — | 0.0001 | 0.0015 | — | 0.0018 |
| TTC22 | 1p32.3 | rs12144325 | 0.1191 | 0.0163 | 0.1073 | 0.013 |
| VLDLR | 9p24.2 | rs17848383 | 0.0002 | 0.0034 | 0.0001 | 0.0025 |
| ZDHHC11 | 5p15.33 | — | — | 0.0245 | — | 0.0171 |
Abbreviations: EAS, East Asian population; ExAC, Exome Aggregation Consortium; FNMTC, familial non‐medullary thyroid cancer; gnomAD, Genome Aggregation Database; SNP, single‐nucleotide polymorphism.
Table 3.
Seven genetic susceptibility genes identified from the 28 pivotal genes with mutations in this FNMTC family
| Gene | ANO7 | CAV2 | KANK1 | PIK3CB | PKD1L1 | PTPRF | RHBDD2 |
| Cytoband | 2q37.3 | 7q31.2 | 9p24.3 | 3q22.3 | 7p12.3 | 1p34.2 | 7q11.23 |
| Accession number | NM‐001001891.3 | NM‐001233.5 | NM‐015158.3 | NM‐006219.2 | NM‐138295.4 | NM‐002840.4 | NM‐001040456.2 |
| Type | Nonsynonymous SNV | Nonsynonymous SNV | Nonsynonymous SNV | Nonsynonymous SNV | Nonsynonymous SNV | Nonsynonymous SNV | Nonsynonymous SNV |
| SNP annotation | rs57677160 | rs8940 | rs28374506 | rs375961764 | rs17131915 | rs17849118 | rs11547498 |
| OMIM | 605,096 | 601,048 | 607,704 | 602,925 | 609,721 | 179,590 | 615,203 |
| ExAC ALL | 0.20 | 0.15 | 0.04 | — | 0.01 | 0.00 | 0.00 |
| ExAC EAS | 0.01 | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.01 |
| GnomAD ALL | 0.20 | 0.15 | 0.09 | — | 0.02 | — | 0.00 |
| GnomAD EAS | 0.02 | 0.01 | 0.01 | 0.00 | 0.01 | 0.00 | 0.00 |
| Tumor‐associated disease | Prostate cancer; breast cancer | Thyroid cancer; prostate cancer; breast cancer; renal cell carcinoma | Gastric cancer; renal cell carcinoma; peripheral nerve sheath tumor | Thyroid cancer; breast cancer; glioblastoma | Breast cancer | Gastric cancer; prostate cancer; lung cancer | Breast cancer; colorectal cancer |
| Gene Ontology annotation | Intracellular calcium activated chloride channel activity; phospholipid scramblase activity | Protein homodimerization activity; D1 dopamine receptor binding | Beta‐catenin binding | Transferase activity; transfers phosphorus‐containing groups; kinase activity | Calcium channel activity | Phosphatase activity | Serine‐type endopeptidase activity |
| Pathway | Ion channel transport; transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds | EGF/EGFR signaling pathway; syndecan−2‐mediated signaling events | — | Glioma and development dopamine D2 receptor transactivation of EGFR | — | Transmission across chemical synapses; adhesion | — |
Abbreviations: EAS, East Asian population; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; ExAC, Exome Aggregation Consortium; FNMTC, familial nonmedullary thyroid cancer; gnomAD, Genome Aggregation Database; SNP, single‐nucleotide polymorphism; SNV, single‐nucleotide variant.
Figure 3.
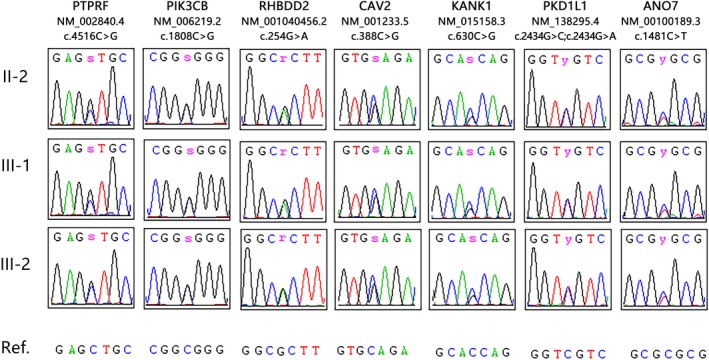
Sequence chromatograms for PTPRF (NM‐002840.4, c.4516C>G), PIK3CB (NM‐006219.2, c.1808C>G), RHBDD2 (NM‐001040456.2, c.254G>A), CAV2 (NM‐001233.5, c.388C>G), KANK1 (NM‐015158.3, c.630C>G), PKD1L1 (NM‐138295.4, c.2434G>C, A), ANO7 (NM‐001001891.3, c.1481C>T) in this family
4. DISCUSSION
Thyroid cancer is the fifth most common cancer in women in the United States, and the incidence worldwide continues to rise (Cabanillas, McFadden, & Durante, 2016). The prognosis is generally favorable for patients receiving an early diagnosis of PTC and follicular thyroid cancer. However, the risk of death increases significantly in clinical stages III and IV owing to metastasis, especially among patients with FNMTC. Therefore, understanding the mechanisms of FNMTC development and progression will likely improve the diagnosis, treatment, and prognosis of families with this disease. High‐throughput sequencing technologies can detect mutation sites in humans and have been widely used to study the molecular mechanisms of various types of cancers. In the present study, we identified 28 rare nonsynonymous SNPs and seven genes with rare nonsynonymous SNPs (ANO7, CAV2, KANK1, PIK3CB, PKD1L1, PTPRF, and RHBDD2) associated with cancer development in a pedigree with members diagnosed as having FNMTC. These seven genes have not previously been reported as being associated with FNMTC.
Among the seven candidate pathogenic FNMTC susceptibility genes, PIK3CB and CAV2 have been reported to be involved in the development of thyroid cancer (Aldred et al., 2004; Campos et al., 2014; Xing, 2010). PIK3CB encodes an isoform of the catalytic subunit of phosphoinositide 3‐kinase (PI3K). Recent studies have shown that the accumulation of genetic alterations in the PI3K/Akt pathway may increase the invasiveness of thyroid cancer, which may be one of the reasons why FNMTC is more aggressive than SNMTC. CAV2 is a major component of the inner surface of caveolae and is involved in essential cellular functions, including signal transduction, cellular growth control, and apoptosis. Caveolins are known to play an important role in tumor initiation and progression, potentially functioning as tumor suppressors. CAV1 is expressed in thyroid cancer and other tumors, whereas less is known about CAV2. Studies have found that CAV2 promotes the growth of renal cell carcinoma through the epidermal growth factor receptor (EGFR)/PI3K/Akt signaling pathway, and the expression of CAV2 is associated with poor prognosis in invasive breast cancer (Liu, Shangli, & Hu, 2018). Given the rare mutations observed in CAV2 in the present family with FNMTC and its role in the EGFR/PI3K/Akt signaling pathway, we speculate that CAV2 is likely to be a susceptibility gene in the pathogenesis of FNMTC. KANK1 is a member of the Kank family, which is involved in the regulation of actin polymerization and cell motility through PI3K/Akt signaling pathways (Kakinuma, Zhu, Wang, Roy, & Kiyama, 2009). In view of the biological function of KANK1 and the accumulation of genetic alterations in the PI3K/Akt signaling pathway, we speculate that KANK1 may also play an important role in FNMTC.
PKD1L1 encodes a member of the polycystin protein family. Polycystin‐1 and polycystin‐2 are the products of PKD1 and PKD2, genes that are mutated in most cases of autosomal dominant polycystic kidney disease (Yuasa et al., 2002). However, after additional inquiry into the medical history of the present family and after auxiliary clinical examination, no member of this family had experienced or had received a diagnosis of polycystic kidney disease.
Although the functions of ANO7, PTPRF, and RHBDD2 in thyroid cancer are still unclear, all three have been found to participate in and promote tumor development. ANO7 is a member of anoctamin family (Kaikkonen et al., 2018). The anoctamin family functions as ion channels and phospholipid scramblases. Although the role of ANO7 in thyroid cancer is unclear, ANO5 has been reported to be downregulated in thyroid cancer and may promote thyroid cancer cell migration and invasion by affecting the Janus kinase–signal transducer and activator of transcription (JAK/STAT3) signaling pathway (Chang et al., 2017). PTPRF is a member of the protein tyrosine phosphatase family. As a signaling molecule, members of this protein family can regulate a variety of cellular processes, such as cell growth, differentiation, mitotic cycle, and oncogenic transformation (Tian, Yang, Yang, Sun, & Liu, 2018). Mutations in PTPRF may affect cellular signal transduction, leading to a series of physiological and pathological changes. RHBDD2 is a member of the rhomboid family. RHBDD2 overexpression has been suggested to play a role in breast cancer tumor progression, facilitating the development of more aggressive phenotypes in at least a subset of breast carcinomas (Abba et al., 2009).
Among the seven genes analyzed in the present study, PIK3CB, CAV2, and KANK1 may be involved in the development of FNMTC through the PI3K/Akt signaling pathway, possibly by increasing the invasiveness of thyroid cancer cells. Thus, we speculate that the PI3K/Akt signaling pathway is likely to play an important role in the development of FNMTC and is likely to be related to the invasiveness of FNMTC.
5. CONCLUSION
In summary, our study identified 28 pivotal genes and seven pathogenic genes (ANO7, CAV2, KANK1, PIK3CB, PKD1L1, PTPRF, and RHBDD2) in a family with FNMTC. The PI3K/Akt signaling pathway may be closely related to the development of FNMTC in this family. These genes may help to diagnose FNMTC, elucidate its pathogenesis, and assess the degree of genetic risk among the various family members. However, owing to the limited sample size of the present study, the prevalence of alterations in these genes in FNMTC remains uncertain and requires future study with a large number of families.
CONFLICT OF INTEREST
The authors have no conflict of interest to disclose.
ACKNOWLEDGMENT
This work was funded by the Natural Science Foundation of Anhui Province (Grant Nos. 1808085MH252 and 1808085MH251).
Zhu J, Wu K, Lin Z, et al. Identification of susceptibility gene mutations associated with the pathogenesis of familial nonmedullary thyroid cancer. Mol Genet Genomic Med. 2019;7:e1015 10.1002/mgg3.1015
Junwei Zhu and Kaile Wu contributed equally to the study and are considered co–first authors.
REFERENCES
- Abba, M. C. , Lacunza, E. , Nunez, M. I. , Colussi, A. , Isla‐Larrain, M. , Segal‐Eiras, A. , … Aldaz, C. M. (2009). Rhomboid domain containing 2 (RHBDD2): A novel cancer‐related gene over‐expressed in breast cancer. Biochimica et Biophysica Acta, 1792(10), 988–997. 10.1016/j.bbadis.2009.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldred, M. A. , Huang, Y. , Liyanarachchi, S. , Pellegata, N. S. , Gimm, O. , Jhiang, S. , … Eng, C. (2004). Papillary and follicular thyroid carcinomas show distinctly different microarray expression profiles and can be distinguished by a minimum of five genes. Journal of Clinical Oncology, 22(17), 3531–3539. 10.1200/JCO.2004.08.127 [DOI] [PubMed] [Google Scholar]
- Bano, G. , & Hodgson, S. (2016). Diagnosis and Management of Hereditary Thyroid Cancer. Recent Results in Cancer Research, 205, 29–44. 10.1007/978-3-319-29998-3_3 [DOI] [PubMed] [Google Scholar]
- Cabanillas, M. E. , McFadden, D. G. , & Durante, C. (2016). Thyroid cancer. Lancet, 388(10061), 2783–2795. 10.1016/S0140-6736(16)30172-6 [DOI] [PubMed] [Google Scholar]
- Campos, M. , Kool, M. , Daminet, S. , Ducatelle, R. , Rutteman, G. , Kooistra, H. S. , … Mol, J. A. (2014). Upregulation of the PI3K/Akt pathway in the tumorigenesis of canine thyroid carcinoma. Journal of Veterinary Internal Medicine, 28(6), 1814–1823. 10.1111/jvim.12435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Z. , Cai, C. , Han, D. , Gao, Y. , Li, Q. , Feng, L. , … Wei, Q. (2017). Anoctamin5 regulates cell migration and invasion in thyroid cancer. International Journal of Oncology, 51(4), 1311–1319. 10.3892/ijo.2017.4113 [DOI] [PubMed] [Google Scholar]
- Kaikkonen, E. , Rantapero, T. , Zhang, Q. , Taimen, P. , Laitinen, V. , Kallajoki, M. , … Schleutker, J. (2018). ANO7 is associated with aggressive prostate cancer. International Journal of Cancer, 143(10), 2479–2487. 10.1002/ijc.31746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakinuma, N. , Zhu, Y. , Wang, Y. , Roy, B. C. , & Kiyama, R. (2009). Kank proteins: Structure, functions and diseases. Cellular and Molecular Life Sciences, 66(16), 2651–2659. 10.1007/s00018-009-0038-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klubo‐Gwiezdzinska, J. , Yang, L. , Merkel, R. , Patel, D. , Nilubol, N. , Merino, M. J. , … Kebebew, E. (2017). Results of Screening in Familial Non‐Medullary Thyroid Cancer. Thyroid, 27(8), 1017–1024. 10.1089/thy.2016.0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Shangli, Z. , & Hu, Z. (2018). CAV2 promotes the growth of renal cell carcinoma through the EGFR/PI3K/Akt pathway. Onco Targets Ther, 11, 6209–6216. 10.2147/OTT.S172803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. D. , Siegel, R. L. , Lin, C. C. , Mariotto, A. B. , Kramer, J. L. , Rowland, J. H. , … Jemal, A. (2016). Cancer treatment and survivorship statistics, 2016. CA: A Cancer Journal for Clinicians, 66(4), 271–289. 10.3322/caac.21349 [DOI] [PubMed] [Google Scholar]
- Robinson, D. W. , & Orr, T. G. (1955). Carcinoma of the thyroid and other diseases of the thyroid in identical twins. AMA Archives of Surgery, 70(6), 923–928. 10.1001/archsurg.1955.01270120131015 [DOI] [PubMed] [Google Scholar]
- Tavarelli, M. , Russo, M. , Terranova, R. , Scollo, C. , Spadaro, A. , Sapuppo, G. , … Pellegriti, G. (2015). Familial non‐medullary thyroid cancer represents an independent risk factor for increased cancer aggressiveness: A retrospective analysis of 74 families. Front Endocrinol (Lausanne), 6, 117 10.3389/fendo.2015.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X. , Yang, C. , Yang, L. , Sun, Q. , & Liu, N. (2018). PTPRF as a novel tumor suppressor through deactivation of ERK1/2 signaling in gastric adenocarcinoma. OncoTargets and Therapy, 11, 7795–7803. 10.2147/OTT.S178152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Cheng, W. , Li, J. , Su, A. , Wei, T. , Liu, F. , & Zhu, J. (2015). Endocrine tumours: Familial nonmedullary thyroid carcinoma is a more aggressive disease: A systematic review and meta‐analysis. European Journal of Endocrinology, 172(6), R253–R262. 10.1530/EJE-14-0960 [DOI] [PubMed] [Google Scholar]
- Xing, M. (2010). Genetic alterations in the phosphatidylinositol‐3 kinase/Akt pathway in thyroid cancer. Thyroid, 20(7), 697–706. 10.1089/thy.2010.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. , Dong, L. I. , Li, D. , Chuai, S. , Wu, Z. , Zheng, X. , … Gao, M. (2015). Targeted DNA sequencing detects mutations related to susceptibility among familial non‐medullary thyroid cancer. Scientific Reports, 5, 16129 10.1038/srep16129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa, T. , Venugopal, B. , Weremowicz, S. , Morton, C. C. , Guo, L. , & Zhou, J. (2002). The sequence, expression, and chromosomal localization of a novel polycystic kidney disease 1‐like gene, PKD1L1, in human. Genomics, 79(3), 376–386. 10.1006/geno.2002.6719 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Yang, S. , Meng, X. Y. , Chen, G. , & Pang, R. Z. (2016). Clinical analysis of familial nonmedullary thyroid carcinoma. World Journal of Surgery, 40(3), 570–573. 10.1007/s00268-015-3342-8 [DOI] [PubMed] [Google Scholar]
- Zhang, T. , & Xing, M. (2016). HABP2 G534E mutation in familial nonmedullary thyroid cancer. Journal of the National Cancer Institute, 108(6), v415 10.1093/jnci/djv415 [DOI] [PMC free article] [PubMed] [Google Scholar]


