Abstract
Relationship between Toll-like receptor-2 (TLR2) and cancer risk has been illustrated in some studies, but their conclusions are inconsistent. Therefore, we designed this meta-analysis to explore a more accurate conclusion of whether TLR2 affects cancer risks. Articles were retrieved from various literature databases according to the criteria. We used STATA to calculate the odds ratio (OR) and 95% confidence interval (95% CI) to evaluate the relationship between certain polymorphism of TLR2 and cancer risk. Finally, 47 case–control studies met the criteria, comprising 15851 cases and 21182 controls. In the overall analysis, people are more likely to get cancer because of -196 to -174del in TLR2 in all five genetic models, B vs. A (OR = 1.468, 95% Cl = 1.129–1.91, P=0.005); BB vs. AA (OR = 1.716, 95% Cl = 1.178–2.5, P=0.005); BA vs. AA (OR = 1.408, 95% Cl = 1.092–1.816, P=0.008); BB+BA vs. AA (OR = 1.449, 95% Cl = 1.107–1.897, P=0.007); BB vs. BA+AA (OR = 1.517, 95% Cl = 1.092–2.107, P=0.013). Meanwhile, rs4696480 could significantly increase the risk of cancer in Caucasians, furthermore, rs3804099 significantly decreased cancer risk in overall analysis, but more subjects are necessary to confirm the results. All in all, this meta-analysis revealed that not only -196 to -174del increased the risk of among overall cancers, Caucasians are more likely to get cancer because of rs4696480, while rs3804099 polymorphism could reduce the risk of cancer in some genetic models. There is no direct evidence showing that rs5743708, rs3804100 and rs1898830 are related to cancer.
Keywords: Cancer risk, Meta-analysis, TLR2, Toll-like receptor 2
Introduction
Cancer prevalence increases rapidly and becomes a major threat to human health in today’s world. As we all know, genes are inextricably linked to the development of cancer. In many cancer studies, such as gastric cancer [1], colorectal cancer, breast cancer [2], cervical cancer [3], Toll-like receptor (TLR)-2 (TLR2) has been determined as a pathogenic factor involved in tumorigenesis. The TLR2 gene located on human chromosome 4q32, includes one coding exon and two non-coding exons [4]. TLRs are mainly expressed in immune-related cells and immune-related epithelial cells, their role in tissue resistance to microbes is achieved by identifying conserved bacterial molecules [5]. Therefore some researchers believe that TLR2 play a significant role in the innate immune response through releasing pro-inflammatory cytokines [6].
-196 to -174del is a 22-bp deletion in TLR2 gene, which has been shown to cause a decrease in the transcriptional activity of the TLR2 gene [7]. However, in the past few years, there are inconsistent conclusions about the relationship between -196 to -174del and cancer risk. One paper noted that -196 to -174del in association with Helicobacter pylori significantly increased the risk of gastric cancer in patients [1]. But Hishida et al. [8] suggested that -196 to -174del had no relationship with gastric cancer. About reproductive tumors, some literatures suggested that -196 to -174del is not associated with breast cancer [9] and cervical cancer [3], but on the contrary, Theodoropoulos et al. [10] think that -196 to -174del may produce a significant increase in the risk of breast cancer. Mandal et al. [11] revealed that -196 to -174del polymorphism in TLR2 gene seems to be associated with the upgraded prostate cancer risk, while Singh et al. [12] drew out that -196 to -174del showed a three- to five-folds risk of bladder cancer comparison with people without this mutation.
For rs3804099 (c.597T>C) and rs3804100 (c.1350T>C), Etokebe et al. [13] and Semlali et al. [14] found no association between these two SNPs and breast cancer; Tongtawee et al. [15] demonstrated that rs3804099 and rs3804100 had no relationship with gastric cancer. However, the study of Xie et al. [16] found that the risk of hepatocellular carcinoma in TLR2 rs3804099 and rs3804100 carriers was reduced. For rs4696480 (g.6686T>A), de Barros Gallo et al. [17] thought that rs4696480 was associated with oral cancer in Caucasians, but Semlali et al. [18] found no difference in rs4696480 expression between the breast cancer patients and the controls in Asians.
Therefore, considering the limitations of individual study sample sizes and the contradictions of their conclusions, we designed this meta-analysis to study the relationship between TLR2 polymorphisms. (rs3804099, rs3804100, rs4696480, rs5743708 (c.2258G> A), rs1898830 (g.8013A> G) and -196 to -174del) and cancer risk.
Materials and methods
Database searching
Up to October 2019, PubMed, Embase, Google Scholar, Web of Science, Wanfang database and CNKI database were used by two investigators for article identification. We used the following strategy for the searching of relevant citations: (TLR2 OR (Toll-like receptors-2) OR CD282) AND (cancer OR tumor OR carcinoma OR neoplasms OR malignancy) AND (polymorphism OR mutation OR variant OR SNP OR genotype). Since the present study is a meta-analysis, no institutional review board approval and patient consent were required.
Inclusion and exclusion criteria
Articles included in our research must meet the following conditions: (1) study the relationship between cancer risk and TLR2 polymorphism; (2) provide sufficient data for extraction and calculation; (3) subjects are human patients; (4) the case–control study included control group and cancer patients case group. When duplicate data appeared in different publications, only the latest publication data were used. If the study did not meet the above criteria, it was excluded.
Data extraction and quality assessment
We extracted data from these articles, such as cancer type, first author, ethnicity, source of control, publication year, number of cases and controls, etc. Any differences were resolved through group discussions until all consensus was reached. We used Newcastle–Ottawa Scale (NOS) to evaluate the quality of the article (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). We carefully recorded seven aspects including ‘adequacy of case definition’, ‘representativeness of the cases’, ‘selection of controls’, ‘definition of controls’, ‘comparability cases/controls’, ‘ascertainment of exposure’ and ‘ascertainment of exposure’ to evaluate.
Statistical analysis
The STATA software was used for meta-statistical analysis. The relationship between the TLR2 rs3804099, rs3804100, rs4696480, rs5743708, rs1898830, -196 to -174del and cancer risk was assessed using pooled odds ratios (ORs) with 95% confidence intervals (95% CIs) under dominant, recessive, homozygous codominance, heterozygous codominance, and allelic control genetic models. Heterogeneity was estimated using Q test and I2 statistics [19]. When heterogeneity existed (P<0.1), random-effects model was applied, otherwise, fixed-effect model was used [20]. The Hardy–Weinberg equilibrium (HWE) of the control group was calculated using the chi-square test. In addition, we performed a stratified analysis based on cancer type, race, source of control and quality score. The sensitivity analysis was used to evaluate the stability of the overall analysis and the publication bias was evaluated by Egger’s test and Begg’s funnel plot [21].
False-positive report probability analysis and trial sequential analysis
We also used the false-positive report probability (FPRP) to evaluate the results; 0.2 was set as thePRP threshold and assigned a prior probability of 0.25 to detect the OR of 0.67/1.50 (protective/risk effects). The significant result with the FPRP values less than 0.2 were considered a worthy finding [22,23]. Trial sequential analysis (TSA) was conducted with the guideline of a former publication [24,25]. We set a significance of 5% for type I error, as well as a 30% significance of type II error, to calculate the required sample size, and built the TSA monitoring boundaries.
In silico analysis
For evaluating the linkage disequilibrium (LD) between different polymorphisms, we downloaded the dataset including the polymorphisms information of TLR2 gene from the 1000 Genomes Project, which contained the distribution of gene polymorphisms among CHB (Han Chinese in Beijing, China), CHS (southern Han Chinese, China), CEU (Utah residents with Northern and Western European ancestry from the CEPH collection), JPT (Japanese in Tokyo, Japan) and YRI (Yoruba in Ibadan, Nigeria), ESN (Esan in Nigeria) patients, and we used Haplpoview software to visualize the association between different polymorphisms, the relationship between them were assessed by r2 statistics. We also performed the expression quantitative trait loci (eQTL) analysis using GTEx portal website (http://www.gtexportal.org/home/) to predict potential associations between the SNPs and gene expression levels [26,27].
Results
Search results
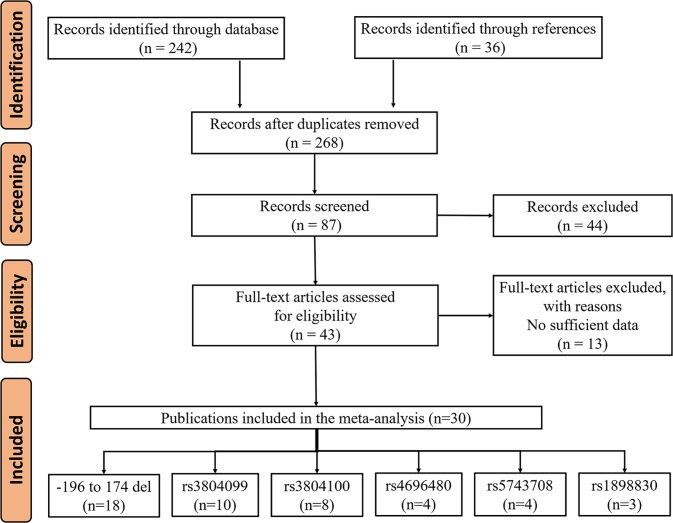
We used online databases to find 242 articles, and found another 36 articles by reviewing the references. After removing the duplicates, we found a total of 268 records in the database. We first screened the duplicate articles and then screened 43 of the high-quality articles on the NOS (Supplementary Table S1). Of the 43 articles selected, 13 were rejected for insufficient data. At last, 30 articles met the criteria, including 47 case–control studies. The flowchart of our study selection is shown in Figure 1. This meta-analysis collected individuals with different genetic backgrounds (e.g. Asians, Africans and Caucasians). The detailed characteristics of these publications are provided in Table 1.
Figure 1. Flowchart of enrolled studies selection procedure.
Table 1. Characteristics of the enrolled studies on TLR2 polymorphism and cancer.
| First author | Year | Ethnicity | Genotyping method | Source of control | Cancer type | Cases | Control | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | BA | BB | Total | A% | B% | AA | BA | BB | Total | A% | B% | HWE | ||||||
| (-196 to -174del) | ||||||||||||||||||
| Tahara et al. | 2007 | Asian | AS-PCR | PB | Gastric cancer | 126 | 112 | 51 | 289 | 63.0% | 37.0% | 73 | 65 | 8 | 146 | 72.3% | 27.7% | Y |
| Pandey et al. | 2009 | Asian | PCR | PB | Cervical cancer | 102 | 43 | 5 | 150 | 82.3% | 17.7% | 114 | 35 | 1 | 150 | 87.7% | 12.3% | Y |
| Hishida et al. | 2010 | Asian | PCR | HB | Gastric cancer | 243 | 267 | 73 | 583 | 64.6% | 35.4% | 722 | 730 | 184 | 1636 | 66.4% | 33.6% | Y |
| Srivastava et al. | 2010 | Asian | PCR-RFLP | PB | Gallbladder cancer | 132 | 94 | 6 | 232 | 77.2% | 22.8% | 163 | 87 | 4 | 254 | 81.3% | 18.7% | N |
| Zeng et al. | 2011a | Asian | DHPLC | HB | Gastric cancer | 119 | 110 | 19 | 248 | 70.2% | 29.8% | 187 | 246 | 63 | 496 | 62.5% | 37.5% | Y |
| Nischalk et al. | 2011 | Caucasian | PCR | PB | Hepatocellular carcinoma | 115 | 63 | 11 | 189 | 77.5% | 22.5% | 248 | 92 | 7 | 347 | 84.7% | 15.3% | Y |
| Oliveira et al. | 2012 | Caucasian | PCR-RFLP | PB | Gastric cancer | 116 | 50 | 8 | 174 | 81.0% | 19.0% | 189 | 34 | 2 | 225 | 91.6% | 8.4% | Y |
| Mandal et al. | 2012 | Asian | PCR | PB | Prostate cancer | 135 | 54 | 6 | 195 | 83.1% | 16.9% | 193 | 52 | 5 | 250 | 87.6% | 12.4% | Y |
| Theodoropoulos et al. | 2012 | Caucasian | PCR | PB | Breast cancer | 120 | 113 | 28 | 261 | 67.6% | 32.4% | 432 | 46 | 2 | 480 | 94.8% | 5.2% | Y |
| Singh et al. | 2013 | Asian | PCR | PB | Bladder cancer | 110 | 79 | 11 | 200 | 74.8% | 25.3% | 119 | 73 | 8 | 200 | 77.8% | 22.3% | Y |
| Bi et al. | 2014 | Asian | PCR | PB | Cervical cancer | 40 | 47 | 15 | 102 | 62.3% | 37.7% | 36 | 50 | 14 | 100 | 61.0% | 39.0% | Y |
| Castano-Rodriguez et al. | 2014 | Asian | MassARRAY | HB | Gastric cancer | 7 | 44 | 35 | 86 | 33.7% | 66.3% | 19 | 95 | 106 | 220 | 30.2% | 69.8% | Y |
| Zidi et al. | 2014 | African | PCR | HB | Cervical cancer | 89 | 20 | 13 | 122 | 81.1% | 18.9% | 196 | 37 | 27 | 260 | 82.5% | 17.5% | N |
| Devi et al. | 2015 | Asian | PCR | PB | Breast cancer | 251 | 191 | 20 | 462 | 75.0% | 25.0% | 491 | 246 | 33 | 770 | 79.7% | 20.3% | Y |
| Proenca et al. | 2015 | African | PCR | PB | Colorectal cancer | 144 | 39 | 5 | 188 | 87.0% | 13.0% | 200 | 36 | 4 | 240 | 90.8% | 9.2% | Y |
| Zidi et al. | 2015 | African | PCR | PB | Cervical cancer | 93 | 26 | 11 | 130 | 81.5% | 18.5% | 196 | 37 | 27 | 260 | 82.5% | 17.5% | N |
| AL-Harras et al. | 2016 | African | PCR-RFLP | PB | Breast cancer | 44 | 22 | 6 | 72 | 76.4% | 23.6% | 61 | 33 | 6 | 100 | 77.5% | 22.5% | Y |
| Huang et al. | 2018 | Asian | PCR | PB | Gastric cancer | 105 | 124 | 31 | 260 | 64.2% | 35.8% | 132 | 113 | 15 | 260 | 72.5% | 27.5% | Y |
| rs3804099 | ||||||||||||||||||
| Etokebe et al. | 2009 | Caucasian | TaqMan | PB | Breast cancer | 29 | 44 | 16 | 89 | 57.3% | 42.7% | 26 | 48 | 15 | 89 | 56.2% | 43.8% | Y |
| Slattery et al. | 2012 | Caucasian | GoldenGate | PB | Colon cancer | 1255 | 300 | 1555 | - | - | 1531 | 425 | 1956 | - | - | - | ||
| Xie et al. | 2012 | Asian | SNaPshot | HB | Hepatocellular carcinoma | 19 | 71 | 121 | 211 | 25.8% | 74.2% | 15 | 117 | 100 | 232 | 31.7% | 68.3% | N |
| Miedema et al. | 2012 | Caucasian | AS-PCR | HB | Lymphoblastic leukemia | 51 | 94 | 37 | 182 | 53.8% | 46.2% | 48 | 102 | 28 | 178 | 55.6% | 44.4% | N |
| Slattery et al. | 2012 | Caucasian | GoldenGate | PB | Rectal cancer | 238 | 372 | 144 | 754 | 56.2% | 43.8% | 299 | 477 | 183 | 959 | 56.0% | 44.0% | Y |
| Zeljic et al. | 2013 | Caucasian | TaqMan | PB | Oral cancer | 29 | 39 | 25 | 93 | 52.2% | 47.8% | 37 | 67 | 0 | 104 | 67.8% | 32.2% | N |
| Semlali et al. | 2017 | Asian | TaqMan | PB | Breast cancer | 35 | 58 | 32 | 125 | 51.2% | 48.8% | 33 | 71 | 42 | 146 | 46.9% | 53.1% | Y |
| Semlali et al. | 2018 | Asian | TaqMan | PB | Colon cancer | 42 | 50 | 19 | 111 | 60.4% | 39.6% | 28 | 47 | 27 | 102 | 50.5% | 49.5% | Y |
| Tongtawee et al. | 2018 | Asian | TaqMan | HB | Gastric cancer | 62 | 13 | 13 | 88 | 77.8% | 22.2% | 194 | 56 | 62 | 312 | 71.2% | 28.8% | N |
| Zeng et al. | 2011b | Asian | PCR-RFLP | HB | Gastric cancer | 132 | 99 | 17 | 248 | 73.2% | 26.8% | 216 | 231 | 49 | 496 | 66.8% | 33.2% | Y |
| rs3804100 | ||||||||||||||||||
| Purdu et al. | 2008 | Caucasian | TaqMan | PB | Non-Hodgkin lymphoma | 1658 | 272 | 12 | 1942 | 92.4% | 7.6% | 1556 | 233 | 9 | 1798 | 93.0% | 7.0% | Y |
| Etokebe et al. | 2009 | Caucasian | TaqMan | PB | Breast cancer | 76 | 13 | 0 | 89 | 92.7% | 7.3% | 84 | 11 | 0 | 95 | 94.2% | 5.8% | Y |
| Xie et al. | 2012 | Asian | SNaPshot | HB | Hepatocellular carcinoma | 14 | 67 | 130 | 211 | 22.5% | 77.5% | 11 | 110 | 111 | 232 | 28.4% | 71.6% | N |
| Miedema et al. | 2012 | Caucasian | AS-PCR | HB | Lymphoblastic leukemia | 170 | 18 | 1 | 189 | 94.7% | 5.3% | 165 | 18 | 0 | 183 | 95.1% | 4.9% | Y |
| Castano-Rodriguez et al. | 2014 | Asian | MassARRAY | HB | Gastric cancer | 47 | 34 | 4 | 85 | 75.3% | 24.7% | 122 | 76 | 14 | 212 | 75.5% | 24.5% | Y |
| Semlali et al. | 2017 | Asian | TaqMan | PB | Breast cancer | 99 | 24 | 1 | 124 | 89.5% | 10.5% | 115 | 27 | 4 | 146 | 88.0% | 12.0% | Y |
| Semlali et al. | 2018 | Asian | TaqMan | PB | Colon cancer | 99 | 13 | 2 | 114 | 92.5% | 7.5% | 82 | 19 | 2 | 103 | 88.8% | 11.2% | Y |
| Tongtawee et al. | 2018 | Asian | TaqMan | HB | Gastric cancer | 66 | 22 | 0 | 88 | 87.5% | 12.5% | 230 | 70 | 12 | 312 | 84.9% | 15.1% | N |
| rs4696480 | ||||||||||||||||||
| Miedema et al. | 2012 | Caucasian | AS-PCR | HB | Hepatocellular carcinoma | 42 | 99 | 44 | 185 | 49.5% | 50.5% | 60 | 83 | 38 | 181 | 56.1% | 43.9% | Y |
| Gallo et al. | 2017 | Caucasian | TaqMan | PB | Oral cancer | 12 | 39 | 24 | 75 | 42.0% | 58.0% | 31 | 34 | 24 | 89 | 53.9% | 46.1% | N |
| Semlali et al. | 2017 | Asian | TaqMan | PB | Breast cancer | 46 | 51 | 29 | 126 | 56.7% | 43.3% | 50 | 63 | 25 | 138 | 59.1% | 40.9% | Y |
| Semlali et al. | 2018 | Asian | TaqMan | PB | Colon cancer | 30 | 49 | 27 | 106 | 51.4% | 48.6% | 26 | 41 | 25 | 92 | 50.5% | 49.5% | Y |
| rs5743708 | ||||||||||||||||||
| Nischalk et al. | 2011 | Caucasian | PCR | PB | Hepatocellular carcinoma | 174 | 15 | 0 | 189 | 96.0% | 4.0% | 319 | 28 | 0 | 347 | 96.0% | 4.0% | Y |
| Slattery et al. | 2012 | Caucasian | GoldenGate | PB | Rectal cancer | 727 | 27 | 754 | - | - | 913 | 46 | 959 | - | - | |||
| Slattery et al. | 2012 | Caucasian | GoldenGate | PB | Colon cancer | 1467 | 88 | 1555 | - | - | 1864 | 92 | 1956 | - | - | |||
| Kina et al. | 2018 | Caucasian | PCR | PB | Glioma | 32 | 18 | 70 | 120 | 34.2% | 65.8% | 184 | 35 | 6 | 225 | 89.6% | 10.4% | N |
| rs1898830 | ||||||||||||||||||
| Xie et al. | 2012 | Asian | SNPshot | HB | Hepatocellular carcinoma | 47 | 92 | 72 | 211 | 44.1% | 55.9% | 34 | 118 | 80 | 232 | 40.1% | 59.9% | Y |
| Slattery et al. | 2012 | Caucasian | GoldenGate | PB | Rectal cancer | 305 | 363 | 86 | 754 | 64.5% | 35.5% | 410 | 437 | 111 | 958 | 65.6% | 34.4% | Y |
| Slattery et al. | 2012 | Caucasian | GoldenGate | PB | Colon cancer | 705 | 674 | 176 | 1555 | 67.0% | 33.0% | 896 | 833 | 227 | 1956 | 67.1% | 32.9% | Y |
Abbreviations: H-B, hospital based; P-B, population based. P>0.05 means conformed to HWE.
Meta-analysis results
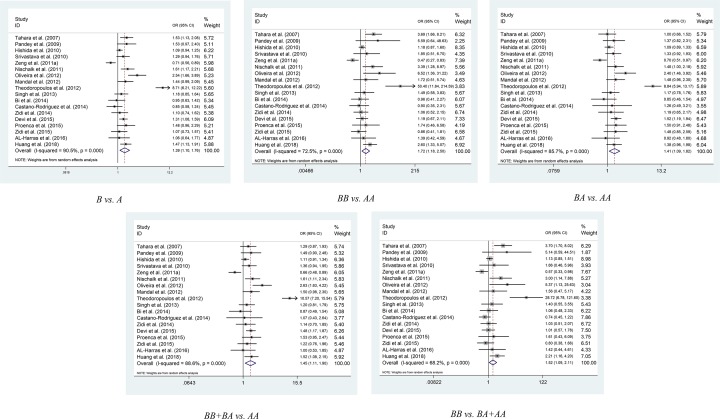
The results of pooled analysis for TLR2 polymorphism and cancer susceptibility are provided in Table 2. For -196 to -174del, we collected 18 articles containing 3943 cases and 4574 controls [1–3,6,8–12,28–36]. In the overall analysis, -196 to -174del significantly increased the risk of cancer [B vs. A (OR = 1.468, 95% Cl = 1.129–1.91, P=0.005); BB vs. AA (OR = 1.716, 95% Cl = 1.178–2.5, P=0.005); BA vs. AA (OR = 1.408, 95% Cl = 1.092–1.816, P=0.008); BB+BA vs. AA (OR = 1.449, 95% Cl = 1.107–1.897, P=0.007); BB vs. BA+AA (OR = 1.517, 95% Cl = 1.092–2.107, P=0.013)] (Figure 2). Among the subgroup of Caucasians, -196 to -174del produces a significant increase in the risk of cancer, too [B vs. A (OR = 3.291, 95% Cl = 1.139–9.51, P=0.028); BB vs. AA (OR = 9.878, 95% Cl = 1.83–53.322, P=0.008); BA vs. AA (OR = 3.156, 95% Cl = 1.034–9.634, P=0.044); BB+BA vs. AA (OR = 3.555, 95% Cl = 1.098–11.51, P=0.034); BB vs. BA+AA (OR = 7.294, 95% Cl = 1.752-30.369, P=0.006)]. During the subgroup analysis of HB, -196 to -174del was found to be associated with cancer [B vs. A (OR = 1.576, 95% Cl = 1.193–2.08, P<0.001); BB vs. AA (OR = 2.274, 95% Cl = 1.43–3.616, P<0.001); BA vs. AA (OR = 1.543, 95% Cl = 1.143–2.081, P<0.001); BB+BA vs. AA (OR = 1.624, 95% Cl = 1.186–2.223, P<0.001); BB vs. BA+AA (OR = 2.011, 95% Cl = 1.317–3.07, P=0.001)]. In addition, in the subgroup analysis of Asians, the models of BB+BA vs. AA (OR = 1.203, 95% Cl = 1.015–1.427, P=0.033) and B vs. A (OR = 1.169, 95% Cl = 1.005–1.361, P=0.043) suggested that -196 to -174del increased the risk of cancer. Meanwhile, when -196 to -174del conformed to HWE in the control group, analysis of all models showed that the deletion of these 22 genes increased the risk of cancer (Supplementary Table S2). By the way, the BA vs. AA model in the N subgroup suggested that -196 to-174del was related to the cancer risk (OR = 1.335, 95% Cl = 1.015–1.757, P=0.039).
Table 2. Results of pooled analysis for TLR2 polymorphism and cancer susceptibility.
| Comparison | Subgroup | n | Cases | Controls | PH | PZ | HR (95% CI) |
|---|---|---|---|---|---|---|---|
| (-196 to -174del) | |||||||
| B vs. A | Overall | 18 | 3943 | 6394 | <0.001 | 0.005* | 1.468 (1.129–1.91) |
| BB vs. AA | Overall | 18 | 3943 | 6394 | <0.001 | 0.005* | 1.716 (1.178–2.5) |
| BA vs. AA | Overall | 18 | 3943 | 6394 | <0.001 | 0.008* | 1.408 (1.092–1.816) |
| BB+BA vs. AA | Overall | 18 | 3943 | 6394 | <0.001 | 0.007* | 1.449 (1.107–1.897) |
| BB vs. BA+ AA | Overall | 18 | 3943 | 6394 | <0.001 | 0.013* | 1.517 (1.092–2.107) |
| B vs. A | Asian | 11 | 2807 | 4482 | <0.001 | 0.043* | 1.169 (1.005–1.361) |
| BB vs. AA | Asian | 11 | 2807 | 4482 | 0.003 | 0.098 | 1.373 (0.943–2) |
| BA vs. AA | Asian | 11 | 2807 | 4482 | 0.039 | 0.054 | 1.168 (0.997–1.367) |
| BB+BA vs. AA | Asian | 11 | 2807 | 4482 | 0.008 | 0.033* | 1.203 (1.015–1.427) |
| BB vs. BA+ AA | Asian | 11 | 2807 | 4482 | 0.005 | 0.177 | 1.256 (0.902–1.748) |
| B vs. A | Caucasian | 3 | 624 | 1052 | <0.001 | 0.028* | 3.291 (1.139–9.51) |
| BB vs. AA | Caucasian | 3 | 624 | 1052 | 0.007 | 0.008* | 9.878 (1.83–53.322) |
| BA vs. AA | Caucasian | 3 | 624 | 1052 | <0.001 | 0.044* | 3.156 (1.034–9.634) |
| BB+BA vs. AA | Caucasian | 3 | 624 | 1052 | <0.001 | 0.034* | 3.555 (1.098–11.51) |
| BB vs. BA+ AA | Caucasian | 3 | 624 | 1052 | 0.029 | 0.006* | 7.294 (1.752–30.369) |
| B vs. A | African | 4 | 512 | 860 | 0.653 | 0.159 | 1.163 (0.943–1.436) |
| BB vs. AA | African | 4 | 512 | 860 | 0.796 | 0.746 | 1.076 (0.693–1.67) |
| BA vs. AA | African | 4 | 512 | 860 | 0.652 | 0.075 | 1.296 (0.974–1.724) |
| BB+BA vs. AA | African | 4 | 512 | 860 | 0.72 | 0.106 | 1.232 (0.956–1.586) |
| BB vs. BA+AA | African | 4 | 512 | 860 | 0.755 | 0.897 | 1.029 (0.666–1.59) |
| B vs. A | PB | 14 | 2904 | 3782 | <0.001 | 0.001* | 1.576 (1.193–2.08) |
| BB vs. AA | PB | 14 | 2904 | 3782 | <0.001 | 0.001* | 2.274 (1.43–3.616) |
| BA vs. AA | PB | 14 | 2904 | 3782 | <0.001 | 0.005* | 1.543 (1.143–2.081) |
| BB+BA vs. AA | PB | 14 | 2904 | 3782 | <0.001 | 0.002* | 1.624 (1.186–2.223) |
| BB vs. BA+AA | PB | 14 | 2904 | 3782 | 0.001 | 0.001* | 2.011 (1.317–3.07) |
| B vs. A | HB | 4 | 1039 | 2612 | 0.016 | 0.502 | 0.92 (0.721–1.173) |
| BB vs. AA | HB | 4 | 1039 | 2612 | 0.048 | 0.552 | 0.866 (0.54–1.39) |
| BA vs. AA | HB | 4 | 1039 | 2612 | 0.122 | 0.841 | 0.984 (0.837–1.156) |
| BB+BA vs. AA | HB | 4 | 1039 | 2612 | 0.038 | 0.716 | 0.942 (0.684–1.298) |
| BB vs. BA+AA | HB | 4 | 1039 | 2612 | 0.121 | 0.43 | 0.917 (0.739–1.138) |
| B vs. A | Gastric cancer | 6 | 1640 | 2983 | <0.001 | 0.194 | 1.22 (0.904–1.647) |
| BB vs. AA | Gastric cancer | 6 | 1640 | 2983 | <0.001 | 0.176 | 1.565 (0.818–2.995) |
| BA vs. AA | Gastric cancer | 6 | 1640 | 2983 | 0.002 | 0.309 | 1.171 (0.864–1.586) |
| BB+BA vs. AA | Gastric cancer | 6 | 1640 | 2983 | <0.001 | 0.216 | 1.246 (0.879–1.764) |
| BB vs. BA+AA | Gastric cancer | 6 | 1640 | 2983 | <0.001 | 0.223 | 1.401 (0.814–2.411) |
| B vs. A | Breast cancer | 3 | 795 | 1350 | <0.001 | 0.212 | 2.31 (0.621–8.593) |
| BB vs. AA | Breast cancer | 3 | 795 | 1350 | <0.001 | 0.2 | 4.049 (0.478–34.306) |
| BA vs. AA | Breast cancer | 3 | 795 | 1350 | <0.001 | 0.197 | 2.347 (0.642–8.58) |
| BB+BA vs. AA | Breast cancer | 3 | 795 | 1350 | <0.001 | 0.2 | 2.52 (0.613–10.36) |
| BB vs. BA+AA | Breast cancer | 3 | 795 | 1350 | <0.001 | 0.233 | 3.176 (0.476–21.196) |
| B vs. A | Cervical cancer | 4 | 504 | 770 | 0.474 | 0.269 | 1.121 (0.916–1.372) |
| BB vs. AA | Cervical cancer | 4 | 504 | 770 | 0.453 | 0.782 | 1.061 (0.696–1.618) |
| BA vs. AA | Cervical cancer | 4 | 504 | 770 | 0.554 | 0.177 | 1.215 (0.916–1.613) |
| BB+BA vs. AA | Cervical cancer | 4 | 504 | 770 | 0.586 | 0.207 | 1.177 (0.914–1.515) |
| BB vs. BA+AA | Cervical cancer | 4 | 504 | 770 | 0.456 | 0.848 | 1.041 (0.692–1.566) |
| B vs. A | Y | 15 | 3459 | 5620 | <0.001 | 0.008* | 1.447 (1.103–1.897) |
| BB vs. AA | Y | 15 | 3459 | 5620 | <0.001 | 0.004* | 1.915 (1.227–2.991) |
| BA vs. AA | Y | 15 | 3459 | 5620 | <0.001 | 0.02* | 1.422 (1.057–1.915) |
| BB+BA vs. AA | Y | 15 | 3459 | 5620 | <0.001 | 0.013* | 1.494 (1.088–2.052) |
| BB vs. BA+AA | Y | 15 | 3459 | 5620 | <0.001 | 0.009* | 1.673 (1.137–2.461) |
| B vs. A | N | 3 | 484 | 774 | 0.709 | 0.14 | 1.168 (0.951–1.434) |
| BB vs. AA | N | 3 | 484 | 774 | 0.597 | 0.84 | 1.05 (0.655–1.681) |
| BA vs. AA | N | 3 | 484 | 774 | 0.872 | 0.039* | 1.335 (1.015–1.757) |
| BB+BA vs. AA | N | 3 | 484 | 774 | 0.839 | 0.07 | 1.258 (0.981–1.613) |
| BB vs. BA+AA | N | 3 | 484 | 774 | 0.615 | 0.959 | 0.988 (0.62–1.575) |
| rs3804099 | |||||||
| B vs. A | Overall | 9 | 1901 | 2618 | 0.001 | 0.723 | 0.967 (0.806–1.162) |
| BB vs. AA | Overall | 9 | 1901 | 2618 | 0.029 | 0.29 | 0.84 (0.609–1.16) |
| BA vs. AA | Overall | 9 | 1901 | 2618 | 0.643 | 0.008* | 0.827 (0.717–0.952) |
| BB+BA vs. AA | Overall | 9 | 1901 | 2618 | 0.446 | 0.016* | 0.85 (0.744–0.97) |
| BB vs. BA+AA | Overall | 10 | 3456 | 4574 | 0.001 | 0.946 | 0.991 (0.768–1.28) |
| B vs. A | Asian | 5 | 783 | 1288 | 0.013 | 0.177 | 0.838 (0.648–1.083) |
| BB vs. AA | Asian | 5 | 783 | 1288 | 0.721 | 0.005* | 0.65 (0.482–0.877) |
| BA vs. AA | Asian | 5 | 783 | 1288 | 0.892 | 0.001* | 0.69 (0.55–0.867) |
| BB+BA vs. AA | Asian | 5 | 783 | 1288 | 0.994 | <0.001 | 0.684 (0.555–0.843) |
| BB vs. BA+AA | Asian | 5 | 783 | 1288 | 0.005 | 0.559 | 0.869 (0.542–1.393) |
| B vs. A | Caucasian | 4 | 1118 | 1330 | 0.025 | 0.3 | 1.147 (0.885–1.486) |
| BB vs. AA | Caucasian | 4 | 1118 | 1330 | 0.024 | 0.455 | 1.283 (0.667–2.47) |
| BA vs. AA | Caucasian | 4 | 1118 | 1330 | 0.819 | 0.425 | 0.929 (0.774–1.114) |
| BB+BA vs. AA | Caucasian | 4 | 1118 | 1330 | 0.87 | 0.866 | 0.985 (0.829–1.171) |
| BB vs. BA+AA | Caucasian | 5 | 2673 | 3286 | 0.01 | 0.647 | 1.082 (0.771–1.518) |
| B vs. A | Breast cancer | 2 | 214 | 235 | 0.647 | 0.364 | 0.885 (0.68–1.152) |
| BB vs. AA | Breast cancer | 2 | 214 | 235 | 0.611 | 0.399 | 0.796 (0.47–1.351) |
| BA vs. AA | Breast cancer | 2 | 214 | 235 | 0.887 | 0.302 | 0.792 (0.509–1.233) |
| BB+BA vs. AA | Breast cancer | 2 | 214 | 235 | 0.765 | 0.276 | 0.793 (0.523–1.203) |
| BB vs. BA+AA | Breast cancer | 2 | 214 | 235 | 0.621 | 0.713 | 0.921 (0.592–1.432) |
| B vs. A | Gastric Cancer | 2 | 336 | 808 | 0.831 | 0.002* | 0.728 (0.594–0.893) |
| BB vs. AA | Gastric Cancer | 2 | 336 | 808 | 0.75 | 0.026* | 0.605 (0.389–0.942) |
| BA vs. AA | Gastric Cancer | 2 | 336 | 808 | 0.926 | 0.018* | 0.706 (0.529–0.942) |
| BB+BA vs. AA | Gastric Cancer | 2 | 336 | 808 | 0.956 | 0.004* | 0.681 (0.524–0.886) |
| BB vs. BA+AA | Gastric Cancer | 2 | 336 | 808 | 0.928 | 0.083 | 0.683 (0.444–1.051) |
| BB vs. BA+ AA | Colon Cancer | 2 | 1666 | 2058 | 0.243 | 0.034* | 0.841 (0.716–0.987) |
| B vs. A | PB | 5 | 1172 | 1400 | 0.004 | 0.985 | 0.997 (0.759–1.311) |
| BB vs. AA | PB | 5 | 1172 | 1400 | 0.01 | 0.762 | 0.912 (0.502–1.658) |
| BA vs. AA | PB | 5 | 1172 | 1400 | 0.764 | 0.252 | 0.901 (0.754–1.077) |
| BB+BA vs. AA | PB | 5 | 1172 | 1400 | 0.468 | 0.385 | 0.928 (0.785–1.098) |
| BB vs. BA+AA | PB | 6 | 2727 | 3356 | 0.021 | 0.549 | 0.915 (0.683–1.225) |
| B vs. A | HB | 4 | 729 | 1218 | 0.007 | 0.658 | 0.934 (0.691–1.263) |
| BB vs. AA | HB | 4 | 729 | 1218 | 0.29 | 0.155 | 0.794 (0.577–1.091) |
| BA vs. AA | HB | 4 | 729 | 1218 | 0.624 | 0.005* | 0.713 (0.564–0.902) |
| BB+BA vs. AA | HB | 4 | 729 | 1218 | 0.679 | 0.005* | 0.734 (0.591–0.912) |
| BB vs. BA+AA | HB | 4 | 729 | 1218 | 0.012 | 0.782 | 1.073 (0.65–1.772) |
| B vs. A | Y | 5 | 1327 | 1792 | 0.13 | 0.036* | 0.895 (0.807–0.993) |
| BB vs. AA | Y | 5 | 1327 | 1792 | 0.233 | 0.087 | 0.828 (0.668–1.028) |
| BA vs. AA | Y | 5 | 1327 | 1792 | 0.484 | 0.058 | 0.856 (0.729–1.005) |
| BB+BA vs. AA | Y | 5 | 1327 | 1792 | 0.258 | 0.028* | 0.844 (0.725–0.982) |
| BB vs. BA+ AA | Y | 5 | 1327 | 1792 | 0.437 | 0.265 | 0.898 (0.742–1.086) |
| B vs. A | N | 4 | 574 | 826 | 0.004 | 0.37 | 1.179 (0.823–1.688) |
| BB vs. AA | N | 4 | 574 | 826 | 0.008 | 0.596 | 1.262 (0.534–2.98) |
| BA vs. AA | N | 4 | 574 | 826 | 0.628 | 0.042* | 0.73 (0.54–0.988) |
| BB+BA vs. AA | N | 4 | 574 | 826 | 0.469 | 0.315 | 0.87 (0.663–1.142) |
| BB vs. BA+AA | N | 4 | 574 | 826 | 0.002 | 0.242 | 1.564 (0.739–3.308) |
| rs3804100 | |||||||
| B vs. A | Overall | 8 | 2842 | 3081 | 0.422 | 0.254 | 1.076 (0.949–1.219) |
| BB vs. AA | Overall | 8 | 2842 | 3081 | 0.682 | 0.412 | 0.823 (0.516–1.311) |
| BA vs. AA | Overall | 8 | 2842 | 3081 | 0.487 | 0.603 | 1.041 (0.896–1.209) |
| BB+BA vs. AA | Overall | 8 | 2842 | 3081 | 0.758 | 0.641 | 1.035 (0.894–1.199) |
| BB vs. BA+AA | Overall | 8 | 2842 | 3081 | 0.243 | 0.061 | 1.343 (0.987–1.827) |
| B vs. A | Asian | 5 | 622 | 1005 | 0.152 | 0.71 | 1.037 (0.856–1.257) |
| BB vs. AA | Asian | 5 | 622 | 1005 | 0.66 | 0.153 | 0.655 (0.366–1.17) |
| BA vs. AA | Asian | 5 | 622 | 1005 | 0.276 | 0.543 | 0.917 (0.692–1.213) |
| BB+BA vs. AA | Asian | 5 | 622 | 1005 | 0.688 | 0.391 | 0.888 (0.677–1.165) |
| BB vs. BA+AA | Asian | 5 | 622 | 1005 | 0.105 | 0.079 | 1.346 (0.966–1.875) |
| B vs. A | Caucasian | 3 | 2220 | 2076 | 0.937 | 0.237 | 1.105 (0.937–1.304) |
| BB vs. AA | Caucasian | 3 | 2220 | 2076 | 0.618 | 0.494 | 1.337 (0.582–3.075) |
| BA vs. AA | Caucasian | 3 | 2220 | 2076 | 0.87 | 0.317 | 1.095 (0.917–1.308) |
| BB+BA vs. AA | Caucasian | 3 | 2220 | 2076 | 0.908 | 0.268 | 1.104 (0.927–1.315) |
| BB vs. BA+AA | Caucasian | 3 | 2220 | 2076 | 0.612 | 0.51 | 1.323 (0.576–3.039) |
| B vs. A | PB | 4 | 2269 | 2142 | 0.365 | 0.555 | 1.049 (0.896–1.228) |
| BB vs. AA | PB | 4 | 2269 | 2142 | 0.471 | 0.91 | 0.959 (0.465–1.977) |
| BA vs. AA | PB | 4 | 2269 | 2142 | 0.402 | 0.495 | 1.061 (0.894–1.26) |
| BB+BA vs. AA | PB | 4 | 2269 | 2142 | 0.384 | 0.514 | 1.057 (0.894–1.251) |
| BB vs. BA+ AA | PB | 4 | 2269 | 2142 | 0.479 | 0.911 | 0.96 (0.466–1.978) |
| B vs. A | HB | 4 | 573 | 939 | 0.308 | 0.266 | 1.124 (0.915–1.381) |
| BB vs. AA | HB | 4 | 573 | 939 | 0.512 | 0.336 | 0.74 (0.4–1.368) |
| BA vs. AA | HB | 4 | 573 | 939 | 0.346 | 0.872 | 0.975 (0.715–1.329) |
| BB+BA vs. AA | HB | 4 | 573 | 939 | 0.83 | 0.829 | 0.967 (0.715–1.308) |
| BB vs. BA+AA | HB | 4 | 573 | 939 | 0.146 | 0.033* | 1.449 (1.031–2.036) |
| B vs. A | Breast cancer | 2 | 213 | 241 | 0.429 | 0.886 | 0.968 (0.617–1.517) |
| BA vs. AA | Breast cancer | 2 | 213 | 241 | 0.663 | 0.662 | 1.118 (0.679–1.839) |
| BB+BA vs. AA | Breast cancer | 2 | 213 | 241 | 0.533 | 0.867 | 1.042 (0.641–1.695) |
| B vs. A | Gastric cancer | 2 | 173 | 524 | 0.493 | 0.598 | 0.918 (0.669–1.261) |
| BB vs. AA | Gastric cancer | 2 | 173 | 524 | 0.259 | 0.168 | 0.481 (0.17–1.362) |
| BA vs. AA | Gastric cancer | 2 | 173 | 524 | 0.88 | 0.531 | 1.129 (0.772–1.652) |
| BB+BA vs. AA | Gastric cancer | 2 | 173 | 524 | 0.675 | 0.927 | 1.018 (0.703–1.473) |
| BB vs. BA+AA | Gastric cancer | 2 | 173 | 524 | 0.27 | 0.142 | 0.463 (0.165–1.295) |
| B vs. A | Y | 6 | 2543 | 2537 | 0.666 | 0.546 | 1.045 (0.905–1.207) |
| BB vs. AA | Y | 6 | 2543 | 2537 | 0.706 | 0.824 | 0.935 (0.516–1.695) |
| BA vs. AA | Y | 6 | 2543 | 2537 | 0.683 | 0.436 | 1.065 (0.909–1.248) |
| BB+BA vs. AA | Y | 6 | 2543 | 2537 | 0.688 | 0.467 | 1.059 (0.907–1.237) |
| BB vs. BA+AA | Y | 6 | 2543 | 2537 | 0.693 | 0.771 | 0.916 (0.508–1.653) |
| B vs. A | N | 2 | 299 | 544 | 0.075 | 0.741 | 1.091 (0.652–1.824) |
| BB vs. AA | N | 2 | 299 | 544 | 0.188 | 0.308 | 0.674 (0.316–1.439) |
| BA vs. AA | N | 2 | 299 | 544 | 0.108 | 0.507 | 0.855 (0.537–1.36) |
| BB+BA vs. AA | N | 2 | 299 | 544 | 0.563 | 0.499 | 0.855 (0.543–1.346) |
| BB vs. BA+AA | N | 2 | 299 | 544 | 0.073 | 0.789 | 0.716 (0.062–8.24) |
| rs4696480 | |||||||
| B vs. A | Overall | 4 | 492 | 500 | 0.323 | 0.03* | 1.216 (1.019–1.452) |
| BB vs. AA | Overall | 4 | 492 | 500 | 0.344 | 0.032* | 1.463 (1.034–2.069) |
| BA vs. AA | Overall | 4 | 492 | 500 | 0.059 | 0.167 | 1.407 (0.867–2.281) |
| BB+BA vs. AA | Overall | 4 | 492 | 500 | 0.076 | 0.115 | 1.415 (0.919–2.179) |
| BB vs. BA+AA | Overall | 4 | 492 | 500 | 0.836 | 0.296 | 1.169 (0.872–1.568) |
| B vs. A | Asian | 2 | 232 | 230 | 0.628 | 0.772 | 1.039 (0.801–1.348) |
| BB vs. AA | Asian | 2 | 232 | 230 | 0.563 | 0.692 | 1.106 (0.671–1.824) |
| BA vs. AA | Asian | 2 | 232 | 230 | 0.711 | 0.77 | 0.939 (0.616–1.433) |
| BB+BA vs. AA | Asian | 2 | 232 | 230 | 0.981 | 0.968 | 0.992 (0.672–1.465) |
| BB vs. BA+AA | Asian | 2 | 232 | 230 | 0.382 | 0.596 | 1.125 (0.728–1.738) |
| B vs. A | Caucasian | 2 | 260 | 270 | 0.424 | 0.007* | 1.393 (1.094–1.775) |
| BB vs. AA | Caucasian | 2 | 260 | 270 | 0.406 | 0.009* | 1.903 (1.171–3.091) |
| BA vs. AA | Caucasian | 2 | 260 | 270 | 0.252 | 0.001* | 1.984 (1.307–3.012) |
| BB+BA vs. AA | Caucasian | 2 | 260 | 270 | 0.261 | 0.001* | 1.95 (1.317–2.887) |
| BB vs. BA+AA | Caucasian | 2 | 0.848 | 0.351 | 1.208 (0.812–1.798) | ||
| B vs. A | PB | 3 | 307 | 319 | 0.21 | 0.176 | 1.167 (0.933–1.458) |
| BB vs. AA | PB | 3 | 307 | 319 | 0.217 | 0.152 | 1.369 (0.891–2.105) |
| BA vs. AA | PB | 3 | 307 | 319 | 0.044 | 0.421 | 1.322 (0.67–2.611) |
| BB+BA vs. AA | PB | 3 | 307 | 319 | 0.056 | 0.349 | 1.336 (0.729–2.449) |
| BB vs. BA+AA | PB | 3 | 307 | 319 | 0.652 | 0.408 | 1.167 (0.809–1.681) |
| B vs. A | Y | 3 | 417 | 411 | 0.463 | 0.158 | 1.15 (0.947–1.396) |
| BB vs. AA | Y | 3 | 417 | 411 | 0.502 | 0.163 | 1.31 (0.897–1.916) |
| BA vs. AA | Y | 3 | 417 | 411 | 0.183 | 0.238 | 1.211 (0.881–1.665) |
| BB+BA vs. AA | Y | 3 | 417 | 411 | 0.227 | 0.158 | 1.239 (0.921–1.666) |
| BB vs. BA+AA | Y | 3 | 427 | 411 | 0.677 | 0.412 | 1.146 (0.827–1.588) |
| rs5743708 | |||||||
| B vs. A | Overall | 2 | 309 | 572 | <0.001 | 0.321 | 4.076 (0.255–65.24) |
| BA vs. AA | Overall | 2 | 309 | 572 | 0.022 | 0.338 | 1.697 (0.575–5.011) |
| BB+BA vs. AA | Overall | 4 | 2618 | 3487 | <0.001 | 0.312 | 1.651 (1.348–2.022) |
| rs1898830 | |||||||
| B vs. A | Overall | 3 | 2520 | 3146 | 0.391 | 0.939 | 1.003 (0.928–1.085) |
| BB vs. AA | Overall | 3 | 2520 | 3146 | 0.323 | 0.646 | 0.961 (0.809–1.14) |
| BA vs. AA | Overall | 3 | 2520 | 3146 | 0.056 | 0.806 | 0.971 (0.768–1.227) |
| BB+BA vs. AA | Overall | 3 | 2520 | 3146 | 0.075 | 0.813 | 0.975 (0.791–1.202) |
| BB vs. BA+AA | Overall | 3 | 2520 | 3146 | 0.998 | 0.77 | 0.977 (0.835–1.143) |
| B vs. A | Caucasian | 2 | 2309 | 2914 | 0.623 | 0.655 | 1.019 (0.939–1.106) |
| BB vs. AA | Caucasian | 2 | 2309 | 2914 | 0.779 | 0.972 | 1.003 (0.837–1.202) |
| BA vs. AA | Caucasian | 2 | 2309 | 2914 | 0.515 | 0.355 | 1.056 (0.941–1.187) |
| BB+BA vs. AA | Caucasian | 2 | 2309 | 2914 | 0.518 | 0.433 | 1.045 (0.936–1.167) |
| BB vs. BA+AA | Caucasian | 2 | 2309 | 2914 | 0.955 | 0.777 | 0.975 (0.822–1.158) |
| B vs. A | PB | 2 | 2309 | 2914 | 0.623 | 0.655 | 1.019 (0.939–1.106) |
| BB vs. AA | PB | 2 | 2309 | 2914 | 0.779 | 0.972 | 1.003 (0.837–1.202) |
| BA vs. AA | PB | 2 | 2309 | 2914 | 0.515 | 0.355 | 1.056 (0.941–1.187) |
| BB+BA vs. AA | PB | 2 | 2309 | 2914 | 0.518 | 0.433 | 1.045 (0.936–1.167) |
| BB vs. BA+AA | PB | 2 | 2309 | 2914 | 0.955 | 0.777 | 0.975 (0.822–1.158) |
Abbreviations: n, polymorphisms did not conform to HWE in the control group; P-B, population based; PH, P-value of Q test for heterogeneity test; PZ, means statistically significant (P<0.05); Y, polymorphisms conformed to HWE in the control group.
* P-value less than 0.05 was considered as statistically significant.
Figure 2. Meta-analysis of the association between TLR2 -196 to -174 del polymorphism and cancer risk.
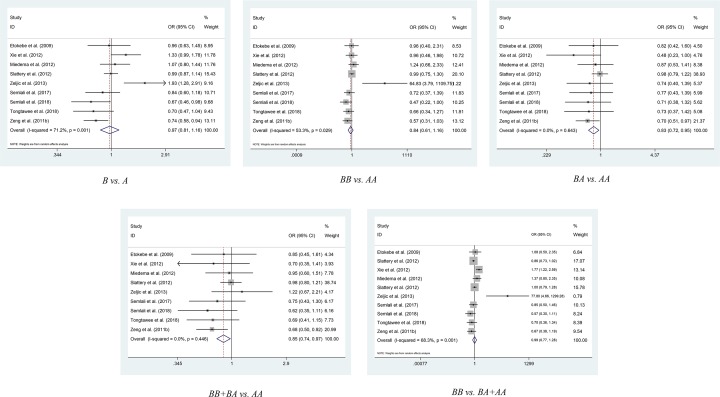
There are nine studies on rs3804099 polymorphism including a total of 3456 cases and 4574 controls [13–16,18,37–40]. According to overall analysis, rs3804099 significantly decreased cancer risk [BA vs. AA (OR = 0.827, 95% Cl = 0.717–0.952, P=0.008), BB+BA vs. AA (OR = 0.85, 95% Cl = 0.744–0.97, P=0.016)] (Figure 3). About Asians, rs3804099 polymorphism reduced the risk of cancer in the model of BA vs. AA (OR = 0.69, 95% Cl = 0.55–0.867, P=0.001) and BB vs. AA (OR = 0.65, 95% Cl = 0.482–0.877, P=0.005). In the subgroup of gastric cancer patients, we found that rs3804099 polymorphism reduced the risk of cancer [B vs. A (OR = 0.728, 95% Cl = 0.594–0.893, P=0.002), BB vs. AA (OR = 0.605, 95% Cl = 0.389–0.942, P=0.026), BA vs. AA (OR = 0.706, 95% Cl = 0.529–0.942, P=0.018), BB+BA vs. AA (OR = 0.681, 95% Cl = 0.524–0.886, P=0.004)] and the model of BB vs. BA+AA is not associated with reduced risk of gastric cancer. Part of the model in the hospital-based analysis was associated with reduced cancer risk [BA vs. AA (OR = 0.713, 95% Cl = 0.564–0.902, P=0.005), BB+BA vs. AA (OR = 0.734, 95% Cl = 0.591–0.912, P=0.005)].
Figure 3. Meta-analysis of the association between TLR2 rs3804009 del polymorphism and cancer risk.
There are four studies on rs4696480 polymorphism including a total of 492 cases and 500 controls [14,17,18,38]. In some models of the overall analysis, rs4696480 significantly increased cancer risk [B vs. A (OR = 1.216, 95% Cl = 1.019–1.452, P=0.03); BB vs. AA (OR = 1.463, 95% Cl = 1.034–2.069, P=0.032)]. It is worth mentioning that rs4696480 makes Caucasians more susceptible to cancer [B vs. A (OR = 1.393, 95% Cl = 1.094–1.775, P=0.007), BB vs. AA (OR = 1.903, 95% Cl = 1.171–3.091, P=0.009), BA vs. AA (OR = 1.984, 95% Cl = 1.307–3.012, P=0.001), BB+BA vs. AA (OR = 1.95, 95% Cl = 1.317–2.887, P=0.001)]. Thus, we can conclude that a subgroup analysis by ethnicity suggests that rs4696480 is related to cancer risk in Caucasians, but not in other ethnic groups (Table 2 and Supplementary Figure S1).
For rs3804100 polymorphism, we collected eight publications which contained 2842 cases and 3081 controls [1,13–16,18,38,41]. But only in hospital-based analysis we found the model of BB vs. BA+AA (OR = 1.449, 95% Cl = 1.031–2.036, P=0.033) added to the risk of cancer. None of the other models showed any association between rs3804100 and cancer risk, either in the analysis of overall group or in other subgroups (Table 2 and Supplementary Figure S2).
As for rs5743708 [6,37,42] and rs1898830 [16,37], they were found to have no significant correlation with cancer, either in overall analysis or in other subgroup analysis (Table 2 and Supplementary Figures S3 and S4).
Sensitivity analysis and publication bias
By the way, we removed individual study one by one when conducted the sensitivity analysis. We did not observe any significant changes in the OR and corresponding 95% CI values, so the stability of our results was confirmed. All the details of sensitivity analysis are shown in the Supplementary Table S2 and Figure S5.
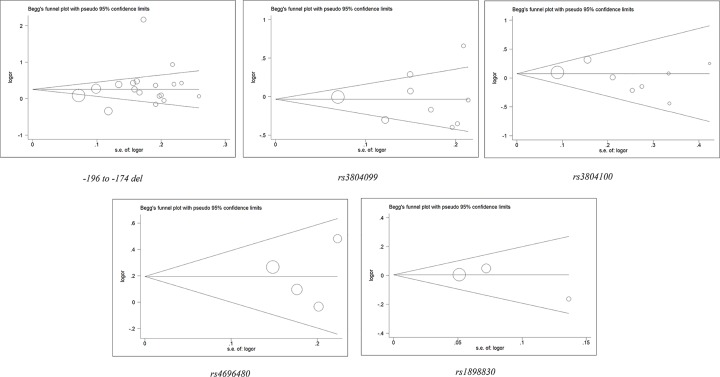
We used the Begg’s test to evaluate publication bias for selected literature. These funnel plots in Figure 4 showed the relationship between the cancer risk and the TLR2 polymorphism in this meta-analysis. Among the various polymorphic sites, the funnel plots were symmetrically distributed. This showed that there was no publication bias. The Egger’s test further analyzed the publication bias, and showed that no significant evidence of publication bias was observed in our study (P=0.937 for SNP rs4696480; P=0.291 for - 196 to - 174del polymorphism; P=0.991 for SNP rs3804099) (Supplementary Table S3).
Figure 4. Begg’s funnel plot for TLR2 polymorphisms and overall cancer publication bias (B vs. A).
For Begg’s funnel plot, the x-axis is log (OR), and the y-axis is natural logarithm of OR. The horizontal line in the figure represents the overall estimated log (OR). The two diagonal lines indicate the pseudo 95% confidence limits of the effect estimate.
Results of FPRP and TSA
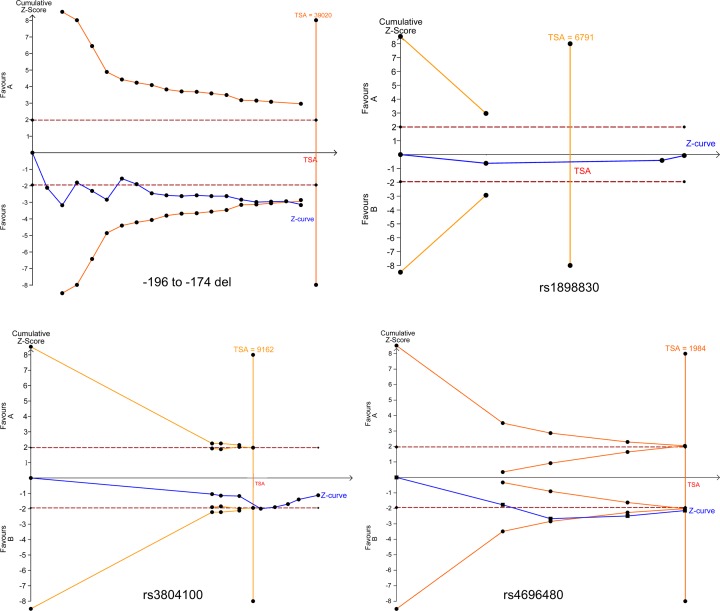
The FPRP values for positive findings at different prior probability levels are shown in Table 3. For -196 to -174del variant, almost all the statistical power high than 0.2, for the FPRP values, under the prior probability of 0.25, the FPRP values for each group is less than 0.2, except the five genetic models about Caucasian subgroup. Which means that the results on Caucasian subgroup are not stable, more studies are needed to illustrate the results. For the other positive results on rs3804099, rs3804100 and rs4696480, almost all the statistical power was higher than 0.5, and under the prior probability of 0.25, the FPRP values for each group is less than 0.2, which means that the results are reliable. The results of TSA are shown in Figure 5, we analyzed the required sample size of each polymorphism. The required sample size of -196 to -174del variant is approximately 39020, although the sample size in the current study did not meet the required number, we observed that the cumulative z-curve crossed the trial sequential monitoring boundary and the traditional significant boundary (Z = 1.96, α = 0.05), which means that our conclusions were robust with the sufficient evidence. For rs3804100 (required sample size: 9162) and rs4696480 (required sample size: 1984), we observed that the cumulative z-curve crossed the trial sequential monitoring boundary and the traditional significant boundary, and meet the required number. The TSA result about rs1898830 showed that the mutant allele performed the similar impact on cancer risk compare with the wild allele, no more samples are needed to confirm the result (Figure 5). However, The TSA results of rs3804099 and rs5743708 indicated that more objects are need to drag out the robust conclusion (Supplementary Figure S6).
Table 3. FPRP values for associations between the risk of cancer and the frequency of genotypes.
| Comparison | Subgroup | n | PZ | OR (95% CI) | Statistical power | ||||
|---|---|---|---|---|---|---|---|---|---|
| 0.25 | 0.1 | 0.01 | 0.001 | ||||||
| (-196 to -174del) | |||||||||
| B vs. A | Overall | 18 | 0.005* | 1.468 (1.129–1.91) | 0.564 | 0.022† | 0.064† | 0.427 | 0.883 |
| BB vs. AA | Overall | 18 | 0.005* | 1.716 (1.178–2.5) | 0.237 | 0.054† | 0.146† | 0.652 | 0.950 |
| BA vs. AA | Overall | 18 | 0.008* | 1.408 (1.092–1.816) | 0.683 | 0.035† | 0.099† | 0.547 | 0.924 |
| BB+BA vs. AA | Overall | 18 | 0.007* | 1.449 (1.107–1.897) | 0.597 | 0.034† | 0.096† | 0.539 | 0.922 |
| BB vs. BA+ AA | Overall | 18 | 0.013* | 1.517 (1.092–2.107) | 0.468 | 0.073† | 0.192† | 0.723 | 0.963 |
| B vs. A | Asian | 11 | 0.043* | 1.169 (1.005–1.361) | 0.999 | 0.117† | 0.285 | 0.814 | 0.978 |
| BB+BA vs. AA | Asian | 11 | 0.033* | 1.203 (1.015–1.427) | 0.994 | 0.106† | 0.262 | 0.796 | 0.975 |
| B vs. A | Caucasian | 3 | 0.028* | 3.291 (1.139–9.51) | 0.073 | 0.532 | 0.773 | 0.974 | 0.997 |
| BB vs. AA | Caucasian | 3 | 0.008* | 9.878 (1.83–53.322) | 0.014 | 0.621 | 0.831 | 0.982 | 0.998 |
| BA vs. AA | Caucasian | 3 | 0.044* | 3.156 (1.034–9.634) | 0.096 | 0.577 | 0.804 | 0.978 | 0.998 |
| BB+BA vs. AA | Caucasian | 3 | 0.034* | 3.555 (1.098–11.51) | 0.075 | 0.579 | 0.805 | 0.978 | 0.998 |
| BB vs. BA+ AA | Caucasian | 3 | 0.006* | 7.294 (1.752–30.369) | 0.015 | 0.561 | 0.793 | 0.977 | 0.998 |
| B vs. A | PB | 14 | 0.001* | 1.576 (1.193–2.08) | 0.364 | 0.011† | 0.031† | 0.263 | 0.783 |
| BB vs. AA | PB | 14 | 0.001* | 2.274 (1.43–3.616) | 0.040 | 0.039† | 0.108† | 0.571 | 0.931 |
| BA vs. AA | PB | 14 | 0.005* | 1.543 (1.143–2.081) | 0.427 | 0.031† | 0.086† | 0.510 | 0.913 |
| BB+BA vs. AA | PB | 14 | 0.002* | 1.624 (1.186–2.223) | 0.310 | 0.023† | 0.067† | 0.441 | 0.888 |
| BB vs. BA+ AA | PB | 14 | 0.001* | 2.011 (1.317–3.07) | 0.087 | 0.040† | 0.111† | 0.578 | 0.933 |
| B vs. A | Y | 15 | 0.008* | 1.447 (1.103–1.897) | 0.603 | 0.036† | 0.101† | 0.551 | 0.925 |
| BB vs. AA | Y | 15 | 0.004* | 1.915 (1.227–2.991) | 0.141 | 0.083† | 0.214 | 0.750 | 0.968 |
| BA vs. AA | Y | 15 | 0.02* | 1.422 (1.057–1.915) | 0.637 | 0.088† | 0.224 | 0.760 | 0.970 |
| BB+BA vs. AA | Y | 15 | 0.013* | 1.494 (1.088–2.052) | 0.510 | 0.072† | 0.189 | 0.719 | 0.963 |
| BB vs. BA+ AA | Y | 15 | 0.009* | 1.673 (1.137–2.461) | 0.290 | 0.085† | 0.218 | 0.754 | 0.969 |
| BA vs. AA | N | 3 | 0.039* | 1.335 (1.015–1.757) | 0.797 | 0.129† | 0.307 | 0.830 | 0.980 |
| rs3804099 | |||||||||
| BA vs. AA | Overall | 9 | 0.008* | 0.827 (0.717–0.952) | 0.999 | 0.024† | 0.069† | 0.448 | 0.891 |
| BB+BA vs. AA | Overall | 9 | 0.016* | 0.85 (0.744–0.97) | 1.000 | 0.045† | 0.125† | 0.611 | 0.941 |
| BB vs. AA | Asian | 5 | 0.005* | 0.65 (0.482–0.877) | 0.434 | 0.032† | 0.091† | 0.524 | 0.917 |
| BA vs. AA | Asian | 5 | 0.001* | 0.69 (0.55–0.867) | 0.287 | 0.064† | 0.170† | 0.692 | 0.958 |
| B vs. A | Gastric cancer | 2 | 0.002* | 0.728 (0.594–0.893) | 0.801 | 0.009† | 0.025† | 0.223 | 0.743 |
| BB vs. AA | Gastric cancer | 2 | 0.026* | 0.605 (0.389–0.942) | 0.334 | 0.190† | 0.413 | 0.886 | 0.987 |
| BA vs. AA | Gastric cancer | 2 | 0.018* | 0.706 (0.529–0.942) | 0.652 | 0.076† | 0.199† | 0.732 | 0.965 |
| BB+BA vs. AA | Gastric cancer | 2 | 0.004* | 0.681 (0.524–0.886) | 0.563 | 0.022† | 0.063† | 0.426 | 0.882 |
| BB vs. BA+ AA | Colon cancer | 2 | 0.034* | 0.841 (0.716-0.987) | 0.998 | 0.093† | 0.235 | 0.771 | 0.971 |
| BA vs. AA | HB | 4 | 0.005* | 0.713 (0.564–0.902) | 0.712 | 0.020† | 0.057† | 0.400 | 0.871 |
| BB+BA vs. AA | HB | 4 | 0.005* | 0.734 (0.591–0.912) | 0.807 | 0.019† | 0.055† | 0.391 | 0.867 |
| B vs. A | Y | 5 | 0.036* | 0.895 (0.807–0.993) | 1.000 | 0.098† | 0.247 | 0.783 | 0.973 |
| BB+BA vs. AA | Y | 5 | 0.028* | 0.844 (0.725–0.982) | 0.999 | 0.078† | 0.202 | 0.736 | 0.966 |
| BA vs. AA | N | 4 | 0.042* | 0.73 (0.54–0.988) | 0.722 | 0.147† | 0.341 | 0.851 | 0.983 |
| rs3804100 | |||||||||
| BB vs. BA+ AA | HB | 4 | 0.033* | 1.449 (1.031–2.036) | 0.579 | 0.144† | 0.336 | 0.848 | 0.983 |
| rs4696480 | |||||||||
| B vs. A | Overall | 4 | 0.03* | 1.216 (1.019–1.452) | 0.990 | 0.085† | 0.218 | 0.754 | 0.969 |
| BB vs. AA | Overall | 4 | 0.032* | 1.463 (1.034–2.069) | 0.556 | 0.145† | 0.337 | 0.848 | 0.983 |
| B vs. A | Caucasian | 2 | 0.007* | 1.393 (1.094–1.775) | 0.725 | 0.029† | 0.084† | 0.501 | 0.910 |
| BB vs. AA | Caucasian | 2 | 0.009* | 1.903 (1.171–3.091) | 0.168 | 0.143† | 0.333 | 0.846 | 0.982 |
| BA vs. AA | Caucasian | 2 | 0.001* | 1.984 (1.307–3.012) | 0.095 | 0.040† | 0.110† | 0.576 | 0.932 |
| BB+BA vs. AA | Caucasian | 2 | 0.001* | 1.95 (1.317–2.887) | 0.095 | 0.026† | 0.075† | 0.470 | 0.899 |
Statistical power was calculated using the number of observations in the subgroup and the OR and P values in this table. Abbreviations: CI, confidence interval; H-B, hospital based; HWE (Y), polymorphisms conformed to HWE in the control group.
*P-value less than 0.05 was considered as statistically significant.
†The significant result with the FPRP values less than 0.2 was considered a worthy finding.
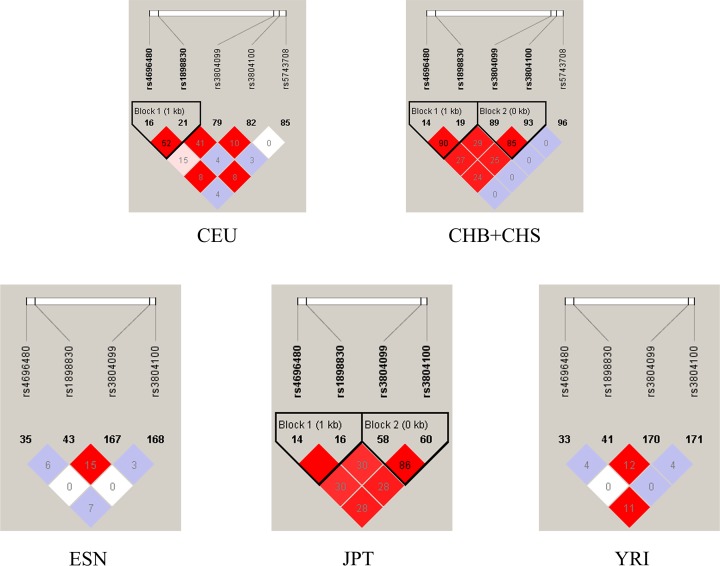
Figure 6. LD analyses for TLR2 polymorphisms in populations from 1000 genomes Phase 3.
The number of each cell represents r2 and white color cells show no LD between polymorphisms.
Figure 5. TSA for TLR2 polymorphism under the allele contrast model (B vs. A).
LD analyses and in-silico analysis of TLR2 expression
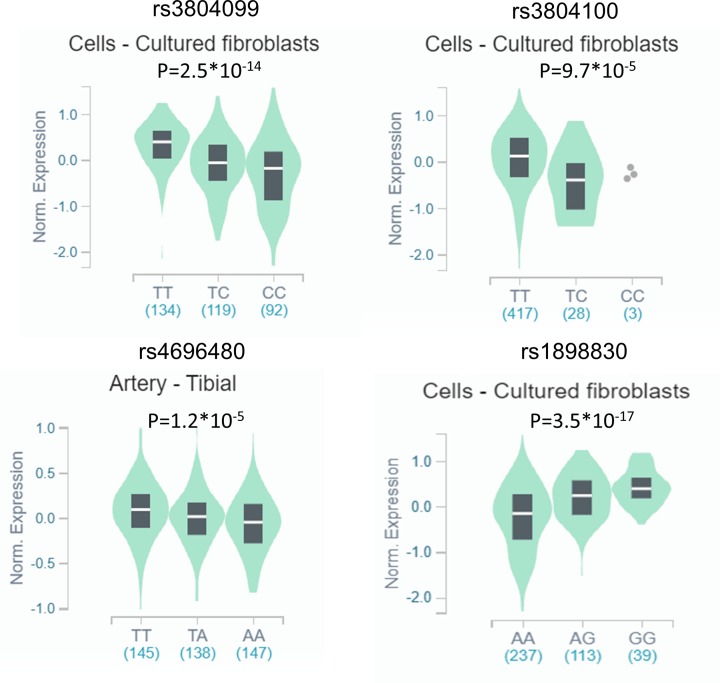
LD analysis was conducted to evaluate the presence of bins in different TLR2 polymorphisms, aiming to understand the internal linkages, the results of which are shown in Figure 6. Highlighted, there is significant LD between rs4696480 and rs1898830 in CEU, CHB and CHS, and JPT populations (CEU: r2 = 0.52; CHB and CHS: r2 = 0.90; JPT: r2 = 1.0). The LD between rs3804099 and rs3804100 is also remarkable in CHB and CHS and JPT populations (CHB and CHS: r2 = 0.85; JPT: r2 = 0.86) (Supplementary Table S4). According to the result on GTEx portal data, we found that the mutant allele leads to an increase expression of TLR2 mRNA in rs1898830 (P=3.5*10−17), while the mutant allele of rs3804099 (P=2.5*10−14), rs3804100 (P=9.7*10−5) and rs4696480 (P=1.2*10−5) lead to a decreased expression of TLR2 (Figure 7).
Figure 7. In-silico analysis of TLR2 expression concerned to its polymorphisms.
Discussion
TLRs are expressed in mast cells and several other cell types, which could recognize microbial components and trigger inflammatory response. TLR2 is type I transmembrane transporter which plays an important role in immune inflammatory response [43], and have been shown to influence host defense and disease progression [44]. There have been four previous meta-analyses on TLR2. But two of the studies were limited to gastric cancer [45,46]. One of these articles suggested that - 196 to - 174del was associated with the rise of cancer risk and the rs3804099 can decrease cancer risk [47]. Another article suggested that -196 to -174del had no relationship with cervical cancer [48]. For assessing the real influence of TLR2 on cancer risk, we collected more samples than before. And our meta-analysis combines many types of cancers to study the relationship between TLR2 polymorphism and cancer risk as comprehensively as possible.
For -196 to -174del, it is a 22-bp deletion at the promoter region of TLR2 gene. Transcriptional reduction in the TLR2 gene due to this substitution may significantly alter the function of the promoter [49]. Chen et al.’s meta-analysis [45] thought that this polymorphism is not associated with gastric cancer. Yang et al. [48] published a meta-analysis in 2018 suggesting that -196 to -174del had nothing to do with cervical cancer. And in our calculations, we revealed that the deletion of these 22 genes does increase the risk of cancer, especially among Caucasians. However, the subgroup calculations of gastric, breast and cervical cancers had no obvious significance.
Synonymous mutations are associated with disease, such as rs3804099 and rs3804100 of TLR2 [16]. We found that rs3804099 is protective against gastric cancer which is consistent with Wang et al. [47]. As for rs3804100, unfortunately, we only came to the conclusions related to cancer in the subgroup of hospital-based. This conclusion is extremely contingent because of the small number of samples and the limitations of the source of the sample. Taking into account the vast majority of calculations and references, we reserve the conclusion that rs3804100 is not related to cancer. And we are the first meta-analysis involving rs4696480. The overall analysis of B vs. A and BB vs. AA shown that rs4696480 has increased the risk of cancer. At the same time, the calculation results also show that its influence on cancer is particularly obvious among the Caucasian population.
Although our conclusions about -196 to -174del, rs3804099 and rs3804100 are consistent with the previous two meta-analyses, we included more case–control studies, so our meta-analysis is more convincing. And we also clearly observe that ‘ethnic’ factors are critical in assessing the role of TLR2 in cancer risk. The calculation of -196 to -174del and rs4696480 both found that Caucasians make a significant increase in the cancer risk. And in the model of BB vs. AA and BA vs. AA, rs3804099 deduce the cancer risk in Asians. Furthermore, as the results showing -196 to -174del and rs4696480 are associated with the tumorigenesis, so that these polymorphisms could be a potential biomarker to remind people with the polymorphism pay more attention to the occurrence of cancer, and solve the problem as soon as possible. In the current study, we also evaluated the LD between different polymorphisms of TLR2, we found that there are significantly LD among rs4696480 and rs1898830, rs3804099 and rs3804100. Based on the results, it could guide the researchers to put these polymorphisms together when assess their effect on cancer risks or other bioscience mechanisms. At the same time, we should also be aware of some of the limitations of our article. First of all, based on the results of TSA, we found that the sample size of -196 to -174 del, rs3804100 and rs4696480 is enough to generate the reliable conclusion in the current study, however, larger number of patients are needed to confirm the effect of rs3804099, rs1898830 and rs5743708 to cancer risks. Second, we lack in-depth studies of the effects of environment, lifestyle, bacterial infections and other factors of cancer risk.
Conclusion
Our meta-analysis suggested that -196 to -174del increased the risk of cancer; rs4696480 increases the risk of cancer in Caucasians; rs3804099 reduced the risk of cancer, especially gastric cancer. While there is no direct evidence showing that rs5743708,3804100 and rs1898830 are related to cancer.
Supplementary Material
Abbreviations
- CEU
Utah residents with Northern and Western European ancestry from the CEPH collection
- CHB
Han Chinese in Beijing, China
- CHS
Southern Han Chinese, China
- FPRP
false-positive report probability
- HWE
Hardy–Weinberg equilibrium
- JPT
Japanese in Tokyo, Japan
- LD
linkage disequilibrium
- NOS
Newcastle–Ottawa Scale
- OR
odds ratio
- TLR2
Toll-like receptor-2
- TSA
trial sequential analysis
- 95% CI
95% confidence interval
Contributor Information
Si-Min Wang, Email: wsm_cmu@163.com.
Li Zuo, Email: zuoli1978@hotmail.com.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
Conception and design: S.-L.G. and Y.-D.C. Collection and assembly of data: C.Y., J.C. and L.-F.Z. Data analysis and interpretation: S.-L.G., Y.-D.C. and L.Z. Manuscript writing: S.-L.G., YD.C. and S.-M.W. Final approval of manuscript: all authors.
References
- 1.Castano-Rodriguez N., Kaakoush N.O., Pardo A.L., Goh K.L., Fock K.M. and Mitchell H.M. (2014) Genetic polymorphisms in the Toll-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer. Hum. Immunol. 75, 808–815 10.1016/j.humimm.2014.06.001 [DOI] [PubMed] [Google Scholar]
- 2.Devi K.R., Chenkual S., Majumdar G., Ahmed J., Kaur T., Zonunmawia J.C. et al. (2015) TLR2∆22 (-196-174) significantly increases the risk of breast cancer in females carrying proline allele at codon 72 of TP53 gene: a case-control study from four ethnic groups of North Eastern region of India. Tumour Biol. 36, 9995–10002 10.1007/s13277-015-3795-2 [DOI] [PubMed] [Google Scholar]
- 3.Bi X., Yu X., Wang N., Yin F. and Wang Y. (2014) Study on TLRs polymorphisms and cervical cancer susceptibility. Prog. Obstet. Gynecol. 23, 520–523 [Google Scholar]
- 4.Haehnel V., Schwarzfischer L., Fenton M.J. and Rehli M. (2002) Transcriptional regulation of the human Toll-Like Receptor 2 gene in monocytes and macrophages. J. Immunol. 168, 5629–5637 10.4049/jimmunol.168.11.5629 [DOI] [PubMed] [Google Scholar]
- 5.Yu L. and Chen S. (2008) Toll-like receptors expressed in tumor cells: targets for therapy. Cancer Immunol. Immunother. 57, 1271–1278 10.1007/s00262-008-0459-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nischalke H.D., Coenen M., Berger C., Aldenhoff K., Muller T., Berg T. et al. (2011) The toll-like receptor 2 (TLR2) -196 to -174 del/ins polymorphism affects viral loads and susceptibility to hepatocellular carcinoma in chronic hepatitis C. Int. J. Cancer 130, 1470–1475 10.1002/ijc.26143 [DOI] [PubMed] [Google Scholar]
- 7.Noguchi E., Nishimura F., Fukai H., Kim J., Ichikawa K., Shibasaki M. et al. (2004) An association study of asthma and total serum immunoglobin E levels for Toll-like receptor polymorphisms in a Japanese population. Clin. Exp. Allergy 34, 177–183 10.1111/j.1365-2222.2004.01839.x [DOI] [PubMed] [Google Scholar]
- 8.Hishida A., Matsuo K., Goto Y., Naito M., Wakai K., Tajima K. et al. (2010) No associations of Toll-like receptor 2 (TLR2) -196 to -174del polymorphism with the risk of Helicobacter pylori seropositivity, gastric atrophy, and gastric cancer in Japanese. Gastric Cancer 13, 251–257 10.1007/s10120-010-0567-y [DOI] [PubMed] [Google Scholar]
- 9.Al-Harras M.F., Houssen M.E., Shaker M.E., Farag K., Farouk O., Monir R. et al. (2016) Polymorphisms of glutathione S-transferase pi 1 and toll-like receptors 2 and 9: association with breast cancer susceptibility. Oncol. Lett. 11, 2182–2188 10.3892/ol.2016.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theodoropoulos G.E., Saridakis V., Karantanos T., Michalopoulos N.V., Zagouri F., Kontogianni P. et al. (2012) Toll-like receptors gene polymorphisms may confer increased susceptibility to breast cancer development. Breast 21, 534–538 10.1016/j.breast.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 11.Mandal R.K., George G.P. and Mittal R.D. (2012) Association of Toll-like receptor (TLR) 2, 3 and 9 genes polymorphism with prostate cancer risk in North Indian population. Mol. Biol. Rep. 39, 7263–7269 10.1007/s11033-012-1556-5 [DOI] [PubMed] [Google Scholar]
- 12.Singh V., Srivastava N., Kapoor R. and Mittal R.D. (2013) Single-nucleotide polymorphisms in genes encoding toll-like receptor -2, -3, -4, and -9 in a case-control study with bladder cancer susceptibility in a North Indian population. Arch. Med. Res. 44, 54–61 10.1016/j.arcmed.2012.10.008 [DOI] [PubMed] [Google Scholar]
- 13.Etokebe G.E., Knezević J.K., Petričević B., Pavelić J., Vrbanec D. and Dembić Z. (2009) Single-nucleotide polymorphisms in genes encoding toll-like receptor -2, -3, -4, and -9 in case-control study with breast cancer. Genet. Test Mol. Biomarkers 13, 729–734 10.1089/gtmb.2009.0045 [DOI] [PubMed] [Google Scholar]
- 14.Semlali A., Almutairi M., Parine N.R., Al Amri A., Shaik J.P., Al Naeem A. et al. (2017) No genetic relationship between TLR2 rs4696480, rs3804100, and rs3804099 gene polymorphisms and female breast cancer in Saudi populations. Onco Targets Ther. 10, 2325–2333 10.2147/OTT.S121618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tongtawee T., Simawaranon T., Wattanawongdon W., Dechsukhum C. and Leeanansaksiri W. (2018) Toll-like receptor 2 and 4 polymorphisms associated with Helicobacter pylori susceptibility and gastric cancer. Turk. J. Gastroenterol. 30, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie J., Shi M., Song Y., Shen B., Deng X., Jin J. et al. (2012) The association between Toll-like receptor 2 single-nucleotide polymorphisms and hepatocellular carcinoma susceptibility. BMC Cancer 12, 57 10.1186/1471-2407-12-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Barros Gallo C., Marichalar-Mendia X., Setien-Olarra A., Acha-Sagredo A., Bediaga N.G., Gainza-Cirauqui M.L. et al. (2017) Toll-like receptor 2 rs4696480 polymorphism and risk of oral cancer and oral potentially malignant disorder. Arch. Oral Biol. 82, 109–114 10.1016/j.archoralbio.2017.06.003 [DOI] [PubMed] [Google Scholar]
- 18.Semlali A., Parine N.R., Al-Numair N.S., Almutairi M., Hawsawi Y.M., Amri A.A. et al. (2018) Potential role of Toll-like receptor 2 expression and polymorphisms in colon cancer susceptibility in the Saudi Arabian population. Onco Targets Ther. 11, 8127–8141 10.2147/OTT.S168478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins J.P. and Thompson S.G. (2002) Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 20.Higgins J.P., Thompson S.G., Deeks J.J. et al. (2003) Measuring inconsistency in meta-analyses. BMJ 327, 557–560 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egger M., Smith G.D. and Schneider M. (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L. and Rothman N. (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 96, 434–442 10.1093/jnci/djh075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J., Wang M.Y., Qiu L.X., Zhu M.L., Shi T.Y., Zhou X.Y. et al. (2013) Genetic variations of mTORC1 genes and risk of gastric cancer in an Eastern Chinese population. Mol. Carcinog. 52, E70–E79 10.1002/mc.22013 [DOI] [PubMed] [Google Scholar]
- 24.Meng J., Wang S., Zhang M., Fan S., Zhang L. and Liang C. (2018) TP73 G4C14-A4T14 polymorphism and cancer susceptibility: evidence from 36 case-control studies. Biosci. Rep. 38, 10.1042/BSR20181452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao F., Pu J., Wen Q., Huang Q., Zhang Q., Huang B. et al. (2017) Association between the ERCC2 Asp312Asn polymorphism and risk of cancer. Oncotarget 8, 48488–48506 10.18632/oncotarget.17290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhuo Z.J., Liu W., Zhang J., Zhu J., Zhang R., Tang J. et al. (2018) Functional polymorphisms at ERCC1/XPF genes confer neuroblastoma risk in Chinese children. EBioMedicine 30, 113–119 10.1016/j.ebiom.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan S., Meng J., Zhang L., Zhang X. and Liang C. (2019) CAV1 polymorphisms rs1049334, rs1049337, rs7804372 might be the potential risk in tumorigenicity of urinary cancer: a systematic review and meta-analysis. Pathol. Res. Pract. 215, 151–158 10.1016/j.prp.2018.11.009 [DOI] [PubMed] [Google Scholar]
- 28.Tahara T., Arisawa T., Wang F., Shibata T., Nakamura M., Sakata M. et al. (2007) Toll-like receptor 2 -196 to 174del polymorphism influences the susceptibility of Japanese people to gastric cancer. Cancer Sci. 98, 1790–1794 10.1111/j.1349-7006.2007.00590.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandey S., Mittal R.D., Srivastava M., Srivastava K., Singh S., Srivastava S. et al. (2009) Impact of Toll-like receptors [TLR] 2 (-196 to -174 del) and TLR 4 (Asp299Gly, Thr399Ile) in cervical cancer susceptibility in North Indian women. Gynecol. Oncol. 114, 501–505 10.1016/j.ygyno.2009.05.032 [DOI] [PubMed] [Google Scholar]
- 30.Srivastava K., Srivastava A., Kumar A. and Mittal B. (2010) Significant association between toll-like receptor gene polymorphisms and gallbladder cancer. Liver Int. 30, 1067–1072 10.1111/j.1478-3231.2010.02268.x [DOI] [PubMed] [Google Scholar]
- 31.Zeng H.M., Pan K.F., Zhang Y., Zhang L., Ma J.L., Zhou T. et al. (2011) Genetic variants of toll-like receptor 2 and 5, Helicobacter pylori infection, and risk of gastric cancer and its precursors in a chinese population. Cancer Epidemiol. Biomarkers Prev. 20, 2594–2602 10.1158/1055-9965.EPI-11-0702 [DOI] [PubMed] [Google Scholar]
- 32.de Oliveira J.G. and Silva A.E. (2012) Polymorphisms of the TLR2 and TLR4 genes are associated with risk of gastric cancer in a Brazilian population. World J. Gastroenterol. 18, 1235–1242 10.3748/wjg.v18.i11.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zidi S., Verdi H., Yilmaz-Yalcin Y., Yazici A.C., Gazouani E., Mezlini A. et al. (2014) Involvement of Toll-like receptors in cervical cancer susceptibility among Tunisian women. Bull. Cancer 101, E31–5 10.1684/bdc.2014.2037 [DOI] [PubMed] [Google Scholar]
- 34.Zidi S., Sghaier I., Gazouani E., Mezlini A. and Yacoubi-Loueslati B. (2016) Evaluation of Toll-Like Receptors 2/3/4/9 gene polymorphisms in cervical cancer evolution. Pathol. Oncol. Res. 22, 323–330 10.1007/s12253-015-0009-6 [DOI] [PubMed] [Google Scholar]
- 35.Huang J., Hang J.J., Qin X.R., Huang J. and Wang X.Y. (2018) Interaction of H. pylori with toll-like receptor 2 -196 to -174 ins/del polymorphism is associated with gastric cancer susceptibility in southern China. Int. J. Clin. Oncol. 24, 494–500 [DOI] [PubMed] [Google Scholar]
- 36.Proença M.A., de Oliveira J.G., Cadamuro A.C., Succi M., Netinho J.G., Goloni-Bertolo E.M. et al. (2015) TLR2 and TLR4 polymorphisms influence mRNA and protein expression in colorectal cancer. World J. Gastroenterol. 21, 7730–7741 10.3748/wjg.v21.i25.7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slattery M.L., Herrick J.S., Bondurant K.L. and Wolff R.K. (2012) Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int. J. Cancer 130, 2974–2980 10.1002/ijc.26314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miedema K.G., Tissing W.J., Te Poele E.M., Kamps W.A., Alizadeh B.Z., Kerkhof M. et al. (2012) Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia 26, 1203–1210 10.1038/leu.2011.341 [DOI] [PubMed] [Google Scholar]
- 39.Zeng H., Zhang Y., Zhang L. et al. (2011) The correlation between polymorphisms of Toll-like receptor 2 and Toll-like receptor 9 and susceptibility to gastric cancer. Chin. J. Prev. Med. 45, 588–592 [PubMed] [Google Scholar]
- 40.Zeljic K., Supic G., Jovic N., Kozomara R., Brankovic-Magic M., Obrenovic M. et al. (2014) Association of TLR2, TLR3, TLR4 and CD14 genes polymorphisms with oral cancer risk and survival. Oral Dis. 20, 416–424 10.1111/odi.12144 [DOI] [PubMed] [Google Scholar]
- 41.Purdue M.P., Lan Q., Wang S.S., Kricker A., Menashe I., Zheng T.-Z. et al. (2008) A pooled investigation of Toll-like receptor gene variants and risk of non-Hodgkin lymphoma. Carcinogenesis 30, 275–281 10.1093/carcin/bgn262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kina I., Sultuybek G.K., Soydas T., Yenmis G., Biceroglu H., Dirican A. et al. (2018) Variations in Toll-like receptor and nuclear factor-kappa B genes and the risk of glioma. Br. J. Neurosurg. 33, 165–170 [DOI] [PubMed] [Google Scholar]
- 43.Akira S., Takeda K. and Kaisho T. (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 10.1038/90609 [DOI] [PubMed] [Google Scholar]
- 44.Takeda K., Kaisho T. and Akira S. (2003) Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 10.1146/annurev.immunol.21.120601.141126 [DOI] [PubMed] [Google Scholar]
- 45.Chen J., Hu S., Liang S., Chen Q., Yang Q., Zheng W. et al. (2013) Associations between the four Toll-Like receptor polymorphisms and the risk of gastric cancer: a meta-analysis. Cancer Biother. Radiopharm. 28, 674–681 10.1089/cbr.2012.1395 [DOI] [PubMed] [Google Scholar]
- 46.Cheng C., Lingyan W., Yi H., Cheng Z., Huadan Y., Xuting X. et al. (2014) Association between TLR2, MTR, MTRR, XPC, TP73, TP53 genetic polymorphisms and gastric cancer: a meta-analysis. Clin Res Hepatol Gastroenterol. 38, 346–359 10.1016/j.clinre.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Liu L., Liu Y. and Zhang K. (2013) TLR-2 gene polymorphisms and susceptibility to cancer: evidence from meta-analysis. Genet. Test Mol. Biomarkers 17, 864–872 10.1089/gtmb.2013.0246 [DOI] [PubMed] [Google Scholar]
- 48.Yang S., Liu L., Xu D. and Li X. (2018) The relationship of the TLR9 and TLR2 genetic polymorphisms with cervical cancer risk: a meta-analysis of case-control studies. Pathol. Oncol. Res. 10.1007/s12253-018-0465-x [DOI] [PubMed] [Google Scholar]
- 49.Slpponen P., Kekki M., Haapakoski J., Ihamäki T. and Siurala M. (1985) Gastric cancer risk in chronic gastritis: statistical calculations of cross-selectional data. Int. J. Cancer 35, 173–177 10.1002/ijc.2910350206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.