Significance
Ebp1 deletion causes developmental defects with massive cell death and cessation of cell proliferation through dysregulation of epigenetic gene silencing unit, a Suv39H1/DNMT1 axis. Our study indicates that EBP1 regulates global gene expression via at least 2 mechnaisms. First, EBP1 acts as a transcriptional repressor for DNMT1 gene expression, allowing the escape of the repressive chromatin state. Second, EBP1 binds to the promoter region of the target gene inhibiting the association of DNMT1 with the downstream gene such as Survivin, regulating gene expression. Hence, these findings provide a molecular mechanism of how EBP1 functions in cell survival and transcriptional regulation by modulating epigenetic regulators during development.
Keywords: Ebp1, epigenetic control, transcriptional repression, DNA methylation, cell death
Abstract
ErbB3-binding protein 1 (EBP1) is implicated in diverse cellular functions, including apoptosis, cell proliferation, and differentiation. Here, by generating genetic inactivation of Ebp1 mice, we identified the physiological roles of EBP1 in vivo. Loss of Ebp1 in mice caused aberrant organogenesis, including brain malformation, and death between E13.5 and 15.5 owing to severe hemorrhages, with massive apoptosis and cessation of cell proliferation. Specific ablation of Ebp1 in neurons caused structural abnormalities of brain with neuron loss in [Nestin-Cre; Ebp1flox/flox] mice. Notably, global methylation increased with high levels of the gene-silencing unit Suv39H1/DNMT1 in Ebp1-deficient mice. EBP1 repressed the transcription of Dnmt1 by binding to its promoter region and interrupted DNMT1-mediated methylation at its target gene, Survivin promoter region. Reinstatement of EBP1 into embryo brain relived gene repression and rescued neuron death. Our findings uncover an essential role for EBP1 in embryonic development and implicate its function in transcriptional regulation.
The ErbB3-binding protein 1, Ebp1 gene (Pa2g4) (chromosome 12q13.2 in humans; chromosome 10 D3 in mouse) is comprised of 10 exons and encodes 2 alternatively spliced EBP1 isoforms, p48 and p42. Pa2g4 is over 8.4 kb in length and is located ∼1.3 kb downstream of Erbb3, encoding the heregulin (human epidermal growth factor) receptor. EBP1 was isolated owing to its interaction with ERBB3 (1); the 2 EBP1 isoforms are expressed in all tissues and cells, including cells that do not express the ERBB3 receptor, and only p42 EBP1 that is 54 amino acids shorter than p48 at its N terminus, but not p48, binds to ERBB3 (2–4). Recent studies demonstrated that the long form, the p48 protein, suppresses apoptosis and promotes cell proliferation, acting as an oncoprotein through Akt activation and p53 degradation (5, 6), whereas p42 EBP1, known to be a potent tumor suppressor (7–10), elicits inhibition of PI3K activity via p85-subunit degradation (11). During early brain development, only p48 EBP1 is expressed and contributes to enhance neurite growth (12) and axon regeneration (13). However, the physiological functions of EBP1 in vivo remain unclear.
Mammalian cell growth and development are regulated by specific patterns of gene expression that are controlled by 2 major epigenetic systems: histone methylation and DNA methylation. For example, the methylated lysines (K) 4, 36, and 79 on Histone H3 are found in active chromatin, whereas trimethylation of K9 and K27 on Histone H3 are generally associated with repressive chromatin structure that represses gene expression. Histone H3-K9 (H3K9) methylation has shown to be a prerequisite for DNA methylation (14). Histone methyltransferase Suv39H1, specific for K9 of H3 and SUV39H1-mediated H3K9 trimethylation, appears more closely related with DNA methylation, recruiting DNMT1, a major DNA methyltransferase, reinforcing the stability of heterochromatin and subsequent gene silencing (15, 16). DNMT1 is essential for embryonic development, inactivation of heterochromatin structure, and tissue specification (17, 18). Although the role of DNA methylation and histone methylation in gene silencing is well established, the precise mechanisms by which signals for specific gene expression, mediated by these factors, remain to be elucidated and may be induced by cellular stress such as cell cycle arrest and apoptosis. In mammalian cells, EBP1 inhibits apoptosis and accelerates cell proliferation, and both of the 2 isoforms of EBP1 differentially modulate transcriptional activity in different types of cancer cells (4, 19). Considering that EBP1 associates with multiple transcriptional regulatory proteins and possesses evolutionarily conserved lysine-rich RNA/DNA binding domain, it might be possible that EBP1 contributes to the transcriptional regulation by modulating epigenetic control.
Here, we report in vivo evidence for an essential role of EBP1 of epigenetic regulation during embryonic development. Genetic ablation of Ebp1 causes embryonic lethality with massive cell death with dysregulation of transcriptional repression unit, SUV39H1/DNMT1. Ebp1(−/−) mice demonstrated up-regulation of Suv39H1-dependent H3K9 trimethylation and activation of DNMT1, displaying markedly increased global DNA methylation. EBP1 directly binds to the promoter region of DNNT1 and represses its transcriptional expression. On the other hand, EBP1 interferes with DNMT1-mediated DNA methylation on its target gene, Survivin promoter. Reintroduction of AAV2-EBP1 significantly reduced neuronal death and relived gene repression in the embryonic brain slices. Taken together, our findings suggest the molecular mechanism of how EBP1 controls the gene-silencing unit to govern the gene expression during development.
Results
Genetic Deletion of Ebp1 in Mice Causes Embryonic Lethality with Developmental Defects.
We targeted Ebp1 in the mouse by replacing exons 6 to 10 coupled with the insertion of IRES-eGFP cassette downstream of the Ebp1 STOP codon and NeoR gene (Fig. 1A). Following PCR and Southern blot analysis, 4 recombinant embryonic stem (ES) clones were chosen for blastocyst injection (SI Appendix, Fig. S1A). Eleven highly chimeric male mice were generated and mated with C57BL/6 female mice to generate Ebp1(+/−) mice (SI Appendix, Fig. S1B). Targeting of the gene resulted in the complete abrogation of Ebp1 gene expression (SI Appendix, Fig. S1 C and D). Unlike the only previous report of Ebp1-deficient mice carrying a gene trap insertion that showed that Ebp1 knockout mice were viable, but exhibited transient growth retardation (20), in our study, no homozygous mutant pups were obtained among 468 pups from the Ebp1(+/−) intercrosses, suggesting embryonic lethality. Heterozygous mice were viable, but the ratio between wild-type (WT) and Ebp1 mutant mice was only about 1:1.7 (36.8% and 63.2%, respectively) (SI Appendix, Fig. S1E). In addition, heterozygous mice appeared to be smaller (∼28%, 1 wk), but they were able to gain body weight comparable to that of WT mice after 2 mo (SI Appendix, Fig. S1F).
Fig. 1.
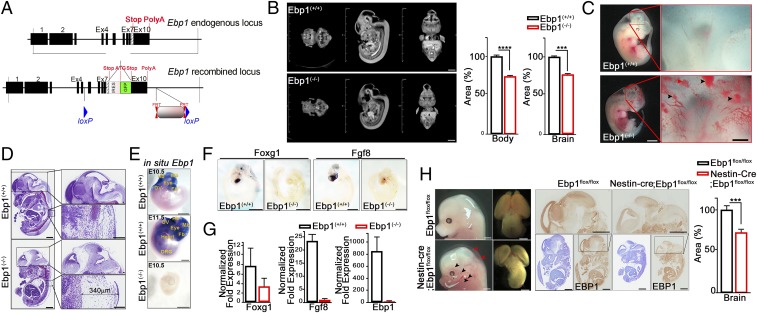
The loss of Ebp1 causes embryonic lethality and developmental defects. (A) Generation of Ebp1 knockout mice. Schematic representation of the targeting strategy. (B) Surface rendering and sagittal cross-section of the mouse at E13.5 images were acquired by micro-CT. Data are shown as mean ± SEM; ***P < 0.0005, ****P < 0.0001 versus the Ebp1(+/+). (Scale bar: 1.0 mm.) (C) Gross morphology of Ebp1 wild-type (Ebp1(+/+)) and knockout (Ebp1(−/−)) mouse littermates at E11. Block arrows indicate bleeding. (Scale bar: 500 μm.) (D) Embryo sagittal sections were stained using cresyl violet. (Scale bar: 100 μm.) (E) Whole-mount RNA in situ hybridization assay for Ebp1 expression from E10.5 and E11.5 embryos. (Scale bar: 100 μm.) (F) Whole-mount RNA in situ hybridization assay for Foxg1 and Fgf8 expression from E10.5 embryos. (Scale bar: 100 μm.) (G) Quantitative RT-PCR analysis of regulator of brain development from E13.5 Ebp1(+/+) or Ebp1(−/−) brain. The relative fold changes were quantified and are shown in the bar graphs. (H) Gross morphology of Ebpflox/flox and [Nestin-Cre; Ebp1flox/flox] mouse littermate at E14.5. Ebp1(−/−) embryos showed bleeding (black arrows) and edema (red arrows) in the brain. (Scale bar: 1 mm.) The paraffin-embedded sections were immunostained with anti-EBP1 antibody at E14.5 (Top). Embryo coronal sections were stained with cresyl violet (Bottom). The bar graph shows the percentage of the brain area. Data are shown as mean ± SEM; ***P < 0.0001. (Scale bar: 1 mm.)
Embryos collected from timed mating were examined and genotyped. From embryonic day (E) 9.5 to 11.5, we found live embryos of all expected genotypes in a normal Mendelian distribution. Between E13.5 and E15.5, we found much lower numbers of Ebp1(−/−) embryos, and these were consistently smaller compared to their WT and heterozygote littermates. At E17.5, Ebp1(−/−) embryos were most underrepresented and, when present, were either resorbing or had unrecognizable organ features. Therefore, Ebp1 knockout resulted in embryonic lethality between E13.5 and E15.5 (SI Appendix, Fig. S1G). Microcomputed tomographic (micro-CT) imaging provided an anatomical image with much smaller body and whole brain volume (26% and 24%, respectively) in Ebp1(−/−) embryos (E13.5) compared to those of the WT, although their somite numbers were similar (50 to 53) (Fig. 1B).
Among the distinct developmental abnormalities, Ebp1(−/−) embryos exhibited prominently dilated vessels and hemorrhages at E11.5 and edemas at E13.5, throughout the entire body, particularly in the brain (Fig. 1C and SI Appendix, Fig. S1H), and displayed dilated cartilage primordium and deficient brain organogenesis (Fig. 1D), suggesting growth retardation and possible defects in vessel development with malformation of the brain (Fig. 1 B–D and SI Appendix, Fig. S1I). Whole-mount staining of E10.5 Ebp1(−/−) embryos for the blood vessel differentiation marker, CD31, was profoundly weaker and revealed large vessels, especially in the brain, that were more dilated compared to those in the WT embryo, which revealed normal vessel branching (SI Appendix, Fig. S1 J and K). Quantitative (q) RT-PCR using E13.5 yolk sac revealed a series of angiogenic markers, including CD31, endoglin, Gata4, Vegf, Sox18, and Sox17, whose levels were notably reduced following the diminution of Ebp1 expression (SI Appendix, Fig. S1L). In situ hybridization of E10.5 to E11.5 embryos of WT mice displayed Ebp1 normally expressed in most brain regions at E10.5 and encompassed entire organs and tissues at E11.5. In contrast, Ebp1(−/−) mutants (E10.5) completely lost the expression of Ebp1 (Fig. 1E) throughout the entire embryo. Certain probes, critical for early brain development, were not found in Ebp1(−/−) embryos compared to those in WT embryos. Notably, in E10.5 Ebp1(−/−) embryos, Foxg1, a neuroectoderm marker normally expressed in the telencephalic region (21), and transcription factor Fgf 8, critical for the isthmic organizer (IsO) at the midbrain–hindbrain boundary (22) and which regulates vasculogenesis and angiogenesis (23), were not detected (Fig. 1F), indicating the presence of defects in the brain organization and malformation in the forebrain and mid/hindbrain of Ebp1(−/−) mutants. This was supported by qRT-PCR of RNA isolated from the brain of WT and Ebp1(−/−) mutants (E11.5) that showed marked reduction of Foxg1 and Fgf 8 levels (Fig. 1G).
To clarify the role of EBP1 in the brain development, we next genetically ablated Ebp1 in the brain using Ebp1 conditional knockout mice (Ebp1flox/flox) mice crossing with a Nestin-Cre driver (24, 25), as EBP1 is predominantly expressed in neurons among brain cells (SI Appendix, Fig. S2A) (13). We analyzed Ebp1 inactivation using Southern blot or an anti-Ebp1 antibody to detect exons 6 to 10, which was deleted on Cre recombination of the Ebp1flox allele (SI Appendix, Fig. S2B; see Materials and Methods). Ebp1 was absent at E13.5 to postnatal day (P) 7 in the entire brain regions, including the hippocampus and cortical region from [Nestin-Cre; Ebp1flox/flox], but not control mice or Ebp1flox/flox mice (SI Appendix, Fig. S2 C–F). The homozygous mutant [Nestin-Cre; Ebp1flox/flox] was viable; however, histological analysis of [Nestin-Cre; Ebp1flox/flox] mutant embryos showed smaller brains as well as severe edema and bleedings with structural abnormalities compared to that of control Ebp1flox/flox mice, consistent with the findings in the brain of Ebp1(−/−) embryos (Fig. 1H). Taken together, these observations reveal that EBP1 is required for proper brain development.
Loss of Ebp1 Leads to Failure in the Regulation of Apoptosis and Cell Proliferation.
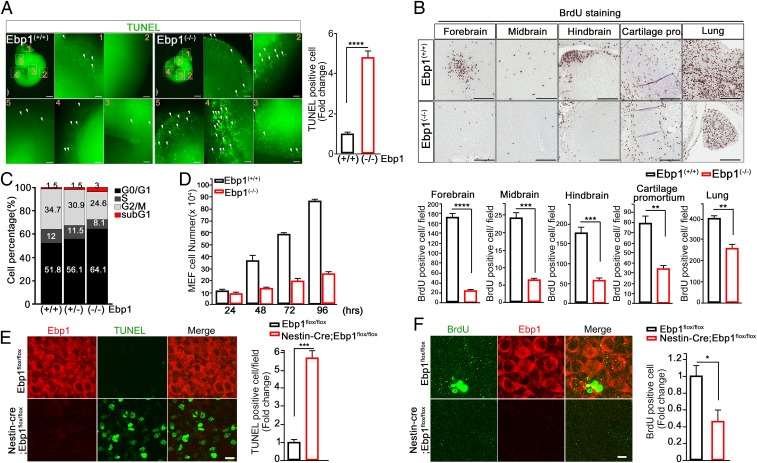
As we previously reported that EBP1 prevented apoptotic cell death and enhanced cell proliferation (3, 4), we wondered whether the impairment of development in Ebp1-deficient mice was caused by cell death and/or defects in proliferation. Whole-mount staining of Ebp1(−/−) embryos (E11.5) showed a noticeable degree of apoptosis throughout all of the tissues, revealing a marked increase in the number of terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL)-positive cells (Fig. 2A). In contrast, a relative number of proliferating cells shown by BrdU incorporation and proliferating cell nuclear antigen (PCNA)-positive cells were dramatically reduced in Ebp1(−/−) embryos compared to that in WT embryos, including all of the brain regions and cartilage primordium (Fig. 2B and SI Appendix, Fig. S3 A and B). In Ebp1(−/−) mouse embryo fibroblasts (MEFs), Annexin V staining displayed massive apoptotic death (SI Appendix, Fig. S3C), and this was supported by the up-regulation of proapoptotic Bax and down-regulation of anti-apoptotic Bcl2 expression at both RNA and protein levels in the absence of EBP1 (SI Appendix, Fig. S3D).
Fig. 2.
Ebp1 deficiency leads to marked cell death and failure in cell proliferation. (A) Whole-mount TUNEL staining of E11.5 of Ebp1(+/+) and Ebp1(−/−) brain reveals apoptotic cells (Left). White arrows indicate the TUNEL-positive signals. Quantification of TUNEL-positive signal is shown as a bar graph (Right). Data represent the mean ± SEM of 3 independent experiments. ****P < 0.0001 versus Ebp1 wild type. (Scale bar: 500 μm and 100 μm [magnification].) (B) The paraffin section was stained with anti-BrdU antibody (brown). (Scale bar: 100 µm.) Quantification of BrdU-positive signal is shown as a bar graph (Bottom). Each value represents the mean ± SEM of triplicate measurements. ****P < 0.0001, ***P < 0.0005, and **P < 0.005. (C) MEF cells (passage 2) were stained with propidium (PI), and cell-cycle profiles were determined by FACS. (D) MEF cell proliferation was determined by toluidine blue cell (24 to 96 h) between the wild-type and knockout MEF cells. (E) The embryo brain slice was fixed at DIV14 and stained with TUNEL. Quantification of TUNEL-positive signal is shown as a bar graph (Right). Data represent the mean ± SEM of 3 independent experiments. ***P < 0.0005 versus Ebp1flox/flox mouse. (Scale bar: 50 μm.) (F) The embryo brain slice was treated with BrdU at DIV13. The slice was stained with anti-BrdU antibody. (Scale bar: 50 µm.) Quantification of BrdU-positive signal is shown as a bar graph. Each value represents the mean ± SEM of triplicate measurements. *P < 0.05.
Cell cycle analysis with MEFs demonstrated a more than 2-fold increase in the population of sub G1 and growth arrest in the G1 phase in Ebp1(−/−) MEFs (Fig. 2C) as indicated by an increased p21Waf1 expression and decrease in expression of G1/S transition markers, cyclin E1 and CDK2 (SI Appendix, Fig. S3E). Moreover, we observed a failure in proliferation at early passages (P2–P3) in Ebp1(−/−) MEFs, whereas WT MEFs were found to grow faster at early passages and showed an immortal phenotype at later passages (Fig. 2D and SI Appendix, Fig. S3F), further corroborating the importance of EBP1 in cell proliferation.
To more specifically access the contribution of EBP1 in the brain, we performed whole-brain slice culture analysis of [Nestin-Cre; Ebp1flox/flox] mutant embryos (E14.5). This whole-brain slice displayed massive cell death as detected by TUNEL reactivity and a much lower proliferation rate as shown by BrdU-positive cells in [Nestin-Cre; Ebp1flox/flox] mutant embryos compared with control embryo brain slice (Fig. 2 E and F). Thus, our findings suggest that the developmental failure observed in Ebp1 deficiency is at least in part due to the role of EBP1 in the prevention of cell death and regulation of cell proliferation.
Ebp1 Deletion Reconfigures Epigenetic System-Related Gene Expression, Causing Dysregulation of H3K9 Trimethylation and DNA Methylation.
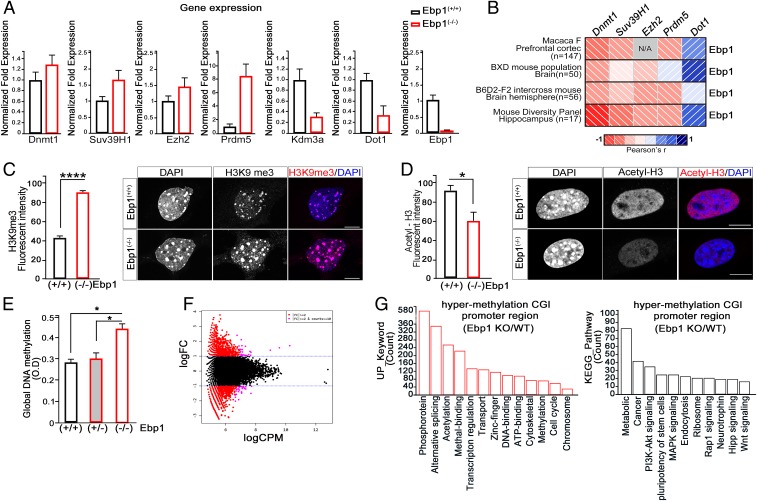
To explore the molecular consequences of EBP1 loss, we carried out microarray-based transcriptome profiling. Microarray analysis of the Ebp1(−/−) mouse embryo revealed a list of genes whose expression was significantly different compared to that in WT. Approximately 5,015 genes with >2-fold changes (P < 0.05), of which 2,382 genes were up-regulated, and 2,633 genes were down-regulated (SI Appendix, Fig. S4A). Among the differentially expressed gene profiles, surprisingly, a group of genes whose expression was closely associated with the epigenetic systems such as DNA methylation and histone modification was considerably altered (SI Appendix, Fig. S4B). Importantly, the H3K9 methylation-related genes affecting mammalian gene expression were found to be up-regulated (SI Appendix, Fig. S4C). Accordingly, qRT-PCR of a group of epigenetic factors involved in H3K9 methylation from RNA isolated from Ebp1(+/+) WT and Ebp1(−/−) mutant mice at E13.5 showed a remarkable increase in levels of the following: DNA (cytosine-5)-methyltransferase (Dnmt1), histone-lysine N-methyltransferase Su(var)3–9 homolog 1 (Suv39H1;H3K9), enhancer of Zeste 2 homolog 2 (Ezh2), and PR domain zinc finger protein 5 (Prdm5), generally associated with gene repression. On the other hand, lysine demethylase 3A (Kdm3a) and histone demethylase were down-regulated. Likewise, the levels of disruptor of telomeric silencing 1 (Dot1) that acts as H3K79 methyltransferases found in active chromatin were relatively low in Ebp1(−/−) mice (Fig. 3A). The correlation analysis of diverse brain transcriptomes including monkey and a different genetic reference population of mice brain strongly supported our finding that Ebp1 expression is negatively correlated with genes associated with transcriptional repression such as Dmnt1, Suv39H1, Ezh1, or Prdm5, whereas positively correlated with the transcriptional activation-related gene, Dot1 (Fig. 3B and Dataset S1). Taken together, Ebp1 loss might cause inappropriate gene silencing during development, leading to up-regulation of DNA and/or histone methylation.
Fig. 3.
Ebp1 loss is associated with up-regulation of gene-silencing related genes. (A) Quantitative RT-PCR analysis of methylation related genes from E13.5 Ebp1(+/+) or Ebp1(−/−) yolk sacs. The relative fold changes were quantified and shown in the bar graphs. The mRNA levels were normalized to the levels of GAPDH. (B) Correlation matrices showing Pearson’s r between Ebp1 and the other genes (i.e., Dnmt1, Suv39h1, Ezh2, Prdm5, and Dot1) in the various species as indicated. See Dataset S1. (C and D) Fixed MEF cells (passage 2) were stained with anti-H3K9me3 (C) and anti-acetylation H3 antibody (D). The intensity of methylation and acetylation of H3 are shown as a bar graph. (Scale bar: 10 µm.) Data represent the mean ± SEM of 3 independent experiments. *P < 0.01, ****P < 0.0001 versus wild type. (E) gDNA was extracted from E13.5 embryonic brain. Global DNA methylation levels were measured and are shown as bar graphs. Data represent the mean ± SEM of 3 independent experiments. *P < 0.05. (F and G) MBD sequencing was performed in E15.5 Ebp1(+/+) and Ebp1(−/−) embryonic brain. (F) Smear plot of MBD-seq data showing the overall average (x axis) versus log2 fold change in methylation levels. Differentially methylated genes are shown in red, and nonsignificant changes are shown in black. (G) Hypermethylated CGI promoter in Ebp1(−/−)/Ebp1(+/+) mouse was subjected to GO-based classification using the DAVID gene-term classification tool. Annotation clusters with enrichment score >2.0 and P value <0.05 were selected as enriched functional categories.
H3K9 trimethylation is a prerequisite for DNA methylation, and we found that consistent with the increased expression levels of histone-lysine N-methyltransferases, Suv39H1 methyltransferase activity was also found to increase, revealing higher levels of H3K9 trimethylation in Ebp1(−/−) MEFs compared with that in WT (SI Appendix, Fig. S4D). The depletion of Ebp1 by siRNA exhibited increased levels of H3K9 trimethylation along with increased Suv39H1 levels (SI Appendix, Fig. S4E). Particularly in Ebp1(−/−) MEFs, enriched H3K9 trimethylation was visualized at DAPI-dense heterochromatin. Conversely, H3 acetylation was much less seen in Ebp1(−/−) MEFs but was enriched at euchromatin regions in Ebp1(+/+) MEFs, implying that a lack of EBP1 caged heterochromatin status (Fig. 3 C and D).
Next, we determined whether Suv39H1-mediated H3K9 trimethylation in terms of Ebp1 loss could be related to DNA methylation. Indeed, genomic DNA methylation levels markedly increased in E13.5 embryos of Ebp1(−/−) compared to that in Ebp1+/− or WT mice (Fig. 3E). Global DNA methylation pattern analysis by methyl-CpG-binding domain protein (MBD)-based enrichment coupled with next-generation sequencing (NGS) (MBD-seq) revealed profound differences in the methylation of either the promoter or nonpromoter regions of CpG islands (CGIs) between Ebp1(−/−) and Ebp1(+/+) mice (Fig. 3F and SI Appendix, Fig. S4F). Intriguingly, DAVID Gene Ontology (GO) term analysis of genes showed that hypermethylated genes (enrichment score: 3.77) in Ebp1(−/−) mice were associated with transcriptional regulation, chromosome modification, DNA binding, and growth/developmental signaling pathways, including PI3-kinase, MAPK, Hipp, and Wnt signaling (Fig. 3G and SI Appendix, Fig. S4G). Therefore, our findings suggest that the loss of Ebp1 causes high levels of DNA methylation and H3K9 trimethylation, which are the hallmarks of gene silencing, reflecting genes that are developmentally important were repressed in Ebp1(−/−) mice.
EBP1 Controls Transcriptional Regulation through Repression of Dmnt1 Transcription.
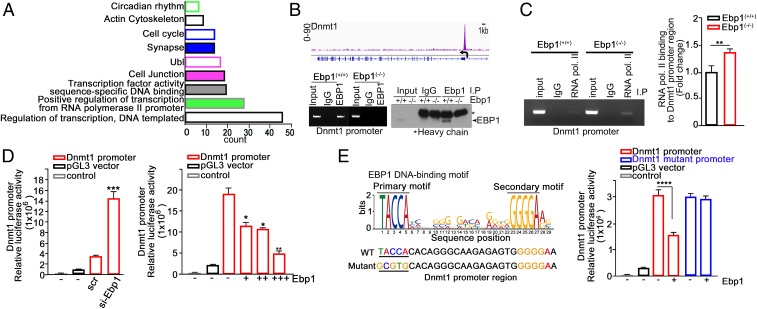
To gain insights into the molecular mechanism underlying EBP1-dependent transcriptional regulation by DNA methylation, we aimed to identify direct target of EBP1 using chromatin immunoprecipitation analyses coupled with sequencing (ChIP-seq). Using model-based analysis of ChIP-seq (MACS), the resulting peaks highlighted 331 putative EBP1-bound regions and identified Ebp1-ChIP-seq targets that were enriched in proximal promoter regions (∼88%) of all of the chromosomes (SI Appendix, Fig. S5 A and B and Dataset S2). GO analysis revealed a strong enrichment of EBP1-associated genes in transcriptional control, including DNA binding, RNA polymerase, synapse, and cell cycle (Fig. 4A). Among these genes, whose expression was significantly altered upon loss of Ebp1 according to our microarray analysis (P < 0.05), 31 down-regulated and 36 up-regulated genes showed an association with EBP1. EBP1-associated genes that were down-regulated upon Ebp1 loss were involved in transcriptional regulation and cell survival. For instance, EBP1 binds to the promoter region of activating transcription factor (ATF7), which inhibits apoptosis (26) and BCL6, which regulates lymphocyte function and survival by suppressing p53 (27). In contrast, up-regulated genes are mostly involved in gene silencing and cell death such as DNMT1 or Trp53, a tumor suppressor gene that triggers cell cycle arrest and apoptosis (SI Appendix, Fig. S5 C and D).
Fig. 4.
EBP1 binds to the Dnmt1 promoter region and represses Dnmt1 expression. (A) MEF cells were used for ChIP-seq analysis with the anti-EBP1 antibody. Gene Ontology function enrichment was analyzed using DAVID. P values <0.05 were selected as categories. (B) Overview of the Ebp1-binding site in the Dnmt1 gene (Top). The chromatin was immunoprecipitated using anti-Ebp1 and assayed by PCR with a primer specific for the Dnmt1 proximal promoter region. MEF cells (passage 2) were subjected to chromatin immunoprecipitation with anti-EBP1 antibody and verified by immunoblot (Bottom). (C) ChIP assay was used to measure the binding of RNA polymerase II to Dnmt1 promoter region in Ebp1(+/+) and Ebp1(−/−) MEF cells. Quantification of the binding of RNA polymerase II to Dnmt1 promoter region is shown as a bar graph (Right). Data represent the mean ± SEM of 3 independent experiments. **P = 0.0027 (D) The 293T cells were cotransfected with the pGL3-Dnmt1 promoter and scramble RNA or si-Ebp1 (Left). The 293T cells were cotransfected with pGL3-Dnmt1 promoter along with control or increasing concentration of Ebp1 plasmid (Right). Data represent the mean ± SEM of 3 independent experiments. *P < 0.05, **P < 0.005, ***P < 0.0005 versus control. (E) Identification of a binding motif for the EBP1 and mutation in the predicted EBP1 binding site in the Dnmt1 promoter (Left). Cells were cotransfected with pGL3-Dnmt1 promoter (WT or mutant) along with control or Ebp1 plasmid (Right). Control: nontransfected cell. Data represent the mean ± SEM of 3 independent experiments. ****P < 0.0001.
As we found Dnmt1 genes for the potent target of EBP1 in ChIP-seq analysis, we confirmed by ChIP assays that EBP1 bound to the promoter region (−500 to +200) of Dnmt1 (Fig. 4B). In the presence of EBP1, RNA polymerase II barely bound to the Dnmt1 promoter region, whereas its promoter binding increased upon EBP1 deficiency (Fig. 4C). Employing Dnmt1 promoter luciferase reporter plasmid (containing −500 to +200), we showed that the depletion of Ebp1 by siRNA enhanced the promoter activity, whereas increased expression of EBP1 correspondingly suppressed the activity (Fig. 4D). Computational analysis of ChIP-seq data predicted a putative EBP1 binding sequences (P value = 3.7e-9) within the EBP1 binding region (−500 to +200). To ensure the functional association of EBP1 in Dnmt1 gene regulation, we constructed a mutated Dnmt1 promoter luciferase reporter (containing the −500 to +200), in which the putative EBP1 binding sequence (around −106 [TACCA]) was mutated by randomized sequence (GCGTG). As expected, EBP1 suppressed wild-type promoter activity. However, EBP1 overexpression in the presence of mutant promoter did not interfere with the promoter activity, suggesting that this sequence is crucial for EBP1 to meditate transcriptional repression (Fig. 4E). In addition, a high level of DNMT1 was observed in Ebp1(−/−) MEFs, and this was dramatically abolished upon reconstitution of Ebp1 into Ebp1(−/−) MEFs compared to that of Ebp1(+/+) MEF cells (SI Appendix, Fig. S5E). Thus, our data indicate that EBP1 acts as a critical transcriptional repressor during embryonic development that regulates the transcriptional suppression of Dnmt1 thereby maintaining open chromatin of genes related to normal development.
EBP1 Impairs DNMT1-Mediated Promoter Methylation.
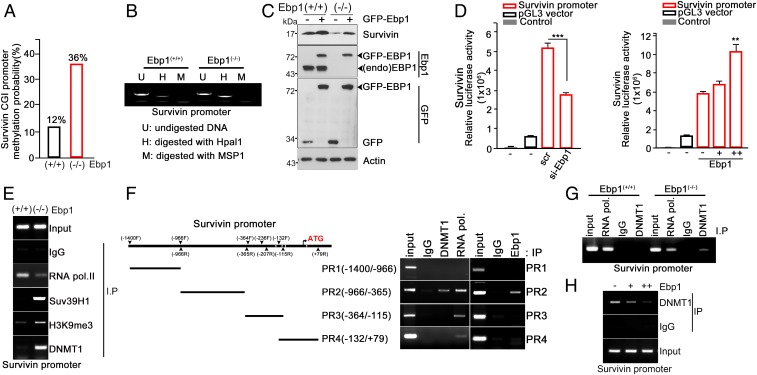
DNMT1 represses gene expression by methylation onto promoter of its target genes. Thus, it was reasonable to hypothesize that failure in proper transcriptional regulation of Dnmt1 upon loss of Ebp1 resulted in dysregulation of DNMT1 target genes. To validate our hypothesis we chose one of well-known Dmnt1 repressive gene, Survivin (28). Importantly, transcription and protein levels of Survivin were evidently impaired in Ebp1(−/−) mouse embryos (E13.5) (SI Appendix, Fig. S6 A and B) and its promoter region was highly methylated upon loss of EBP1 (MBD-seq) (Fig. 5A and Dataset S3). To determine whether loss of Ebp1 could lead to Survivin promoter methylation and subsequent gene silencing, we performed methylation-sensitive PCR, and CpG methylation was evidently observed in the Survivin promoter in the absence of Ebp1 (Fig. 5B). Forced expression of EBP1 in Ebp1(−/−) MEFs restored Survivin expression and even enhanced Survivin expression in the Ebp1(+/+) MEFs (Fig. 5C). Additionally, knockdown of EBP1 reduced Survivin promoter activity, whereas overexpression of EBP1 enhanced Survivin promoter activity, as shown by a luciferase assay (Fig. 5D). Therefore, Ebp1 deficiency resulted in the suppression of Survivin expression through promoter methylation.
Fig. 5.
EBP1 suppresses DNMT1-mediated Survivin gene repression. (A) MBD sequencing was performed in E15.5 Ebp1(+/+) and Ebp1(−/−) embryonic brain. The bar graph shows the methylation probability of the Survivin CGI promoter region. See Dataset S3. (B) Genomic DNA (gDNA) was extracted from Ebp1(+/+) and Ebp1(−/−) MEF cells. The extracted gDNA was subjected to methylation-specific PCR (MSP). (C) MEF with the indicated genotypes was transfected with control or GFP-Ebp1. Survivin expression levels were determined by immunoblotting. Actin served as the loading control. (D) Si-scramble RNA and si-Ebp1 were transfected into 293T cells (Left). The 293T cells were cotransfected with pGL3-Survivin promoter along with control or increasing the amount of Ebp1 plasmid (Right). Control: nontransfected cell. Data represent the mean ± SEM of 3 independent experiments. **P < 0.005, ***P < 0.0005 versus control. (E) MEF cells were subjected to ChIP assay with anti-IgG, RNA polymerase II, Suv39H1, H3K9me3, and DNMT1 antibody. The purified DNA was subjected to PCR using Survivin promoter primers. (F) ChIP assay was performed using anti-IgG, DNMT1, RNA polymerase II, and EBP1 antibody. PCR was carried out with the indicated primer sets (Right). (G) MEF cells were subjected to ChIP assay with anti-IgG, RNA polymerase II, and DNMT1 antibody. The purified DNA was subjected to PCR using Survivin promoter primers. (H) Ebp1(−/−) MEF cells were transfected with control or increasing concentration of Ebp1 plasmid. MEF cells were subjected to ChIP assay with anti-IgG and DNMT1 antibody.
To further define how loss of Ebp1 caused hypermethylation of Survivin promoter in Ebp1(−/−) mice brain, we performed a ChIP assay using indicated antibodies with Ebp1(+/+) and Ebp1(−/−) MEFs. In the absence of Ebp1, Suv39H1 and H3K9 trimethylation, as well as DNMT1, were found to be associated with the Survivin promoter, while RNA polymerase II was notably weakly associated in this promoter compared to the WT (Fig. 5E). More importantly, EBP1 bound the −966/−365 region of the Survivin promoter, where DNMT1 and RNA polymerase II also occupied the Survivin promoter (Fig. 5F) as Survivin promoter possessed the similar EBP1 binding motif to that in the Dnmt1 promoter. DNMT1 was found to bind more strongly to this region in Ebp1(−/−) MEF as compared to WT-MEF (Fig. 5G), and increased EBP1 expression correspondingly disrupted DNMT1 binding to the Survivin promoter (Fig. 5H). This finding implies that EBP1 interferes with DNMT1 recruitment onto the Survivin promoter, subsequently prohibiting DNA methylation. In addition to Survivin, various Dnmt1 target genes were shown to have relatively high methylation on their promoters in the absence of EBP1 as compared with WT (MBD-seq) (SI Appendix, Fig. S6C). Especially, among DNMT1 target genes, Kcna2 and Klf13, which are developmentally essential (29, 30) and possess EBP1 binding motif, were highly methylated upon loss of EBP1. Accordingly, these genes were transcriptionally suppressed (SI Appendix, Fig. S6D) and CpG methylation in their promoter region was abundant in the absence of Ebp1 compared with WT (SI Appendix, Fig. S6E), supporting the notion that EBP1 could alleviate DNMT1 binding at its target promoter. Thus, EBP1 not only transcriptionally represses the expression of DNMT1 but also disrupts its functions at the target gene, probably promoting the necessary gene expression during embryonic development.
Reinstatement of EBP1 Restored Neuron Loss and Attenuated H3K9 Trimethylation.
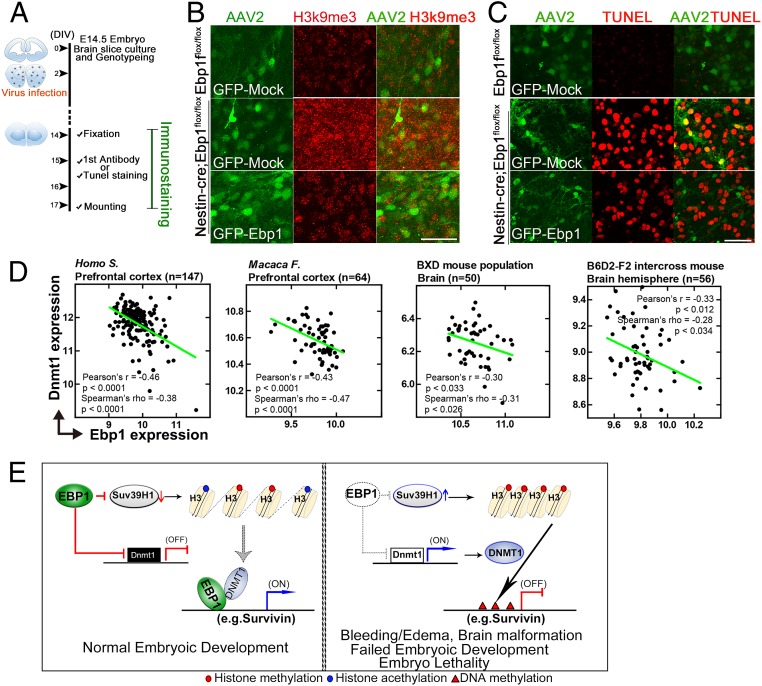
The defects found in embryo brain of [Nestin-Cre; Ebp1flox/flox] mice were all phenocopies of embryo brain of Ebp1(−/−) mice, implying that the developmental defect of brain with abundant cell death observed in [Nestin-Cre; Ebp1flox/flox] mice should be mainly caused by transcriptional repression due to loss of Ebp1. Therefore we hypothesized that reintroduction of EBP1 in the embryo neuron of [Nestin-Cre; Ebp1flox/flox] mice might rescue gene silencing and abundant cell death observed in [Nestin-Cre; Ebp1flox/flox] mice. Using the adeno-associated virus (AAV) 2 delivery system, AAV2-GFP-EBP1 was infected into neurons in the ex vivo slice culture of [Nestin-Cre; Ebp1flox/flox] embryo (E14.5) brain (Fig. 6A). Compared to control embryo brain slices (E14.5), [Nestin-Cre; Ebp1flox/flox] embryo brain slices displayed notably enriched H3K9 trimethylation as well as high TUNEL reactivity compared to control mice. However, reintroduction of EBP1 using AAV2-GFP-EBP1 robustly ameliorated H3K9 trimethylation and prevented cell death at the basal level (Fig. 6 B and C). This constitutes direct evidence that EBP1 is important for preventing cell death through regulation of epigenetic control in brain development.
Fig. 6.
Genetic ablation of Ebp1 in the CNS impairs brain development with neuronal death and reinstatement of Ebp1 reverses the effects of Ebp1 loss. (A–C) Embryo brain slice cultures were prepared from E14.5. The slices were infected with AAV2-control or AAV2-Ebp1 at the DIV2 and cultured for an additional 12 d (A). The slice was stained with H3K9me3 (red, B) and the embryo brain slices were stained with TUNEL (red, C). (Scale bar: 50 μm.) Images shown here are representative of at least 3 independent experiments. (D) Scatterplots of covariation analysis between Ebp1 and Dnmt1 expression from various species as indicated. See Dataset S4. (E) Schematic illustration of the proposed molecular mechanisms of Ebp1 in embryonic development in mice.
Remarkably, our covariation analysis between Ebp1 and Dnmt1 expression in the brain of human, monkey, and mouse revealed that Ebp1 expression is inversely correlated with Dnmt1 expression in various species (Fig. 6D and Dataset S4), implying that conceivably this event could occur in not only mouse but also other species, including human brain, corroborating the hypothesis that physiological function of EBP1 could alleviate aberrant gene repression that can be caused by altered expression of DNMT1 during brain development.
Discussion
In the current study, we showed that embryos of Ebp1(−/−) mice exhibited developmental abnormalities with massive cell death. These defects could be due in part to deficits in cell death and failure during cell cycle progression (Fig. 2). Particularly, we suggest that an important role for EBP1 in this development is the repression of the gene-silencing unit Suv39H1/DNMT1, leading to target gene expression. In Ebp1-deficient mice, we observed an unusual high level of histone/DNA methylation-related proteins and global transcriptional repression during development (Figs. 3 and 4). Reinstatement of EBP1 expression rescued neuronal death and relived the gene repression in the embryonic brain slices (Fig. 6 B and C). This study, therefore, sheds light on in vivo functions of EBP1, which will help elucidate the epigenetic underpinnings of embryonic development and advance our understanding of how the epigenetic gene-silencing unit is molecularly regulated for maintaining proper gene expression.
Despite independent studies from others and our demonstrating that EBP1 regulates diverse cellular events including proliferation, survival, and differentiation in the past 20 y, the precise molecular mechanisms of how EBP1 contributes to the multiple levels of cellular events are not well defined and few studies are available to provide the in vivo evidence of EBP1 functions due to lack of mammalian model system. In fly, overexpression of EBP1 disrupts muscle progenitors, leading to neurogenic-like states. EBP1 protein is robustly expressed in myoblasts during development and regenerative myogenesis, controlling the balance between proliferation and differentiation in a chicken embryo model (31). In Xenopus, loss of Ebp1 leads to down-regulation of the neural border zone, neural crest, and cranial placode genes by interacting with transcription factor, Six1 (32). The only reported Ebp1 knockout mice study from Zhang et al. (20) showed that homozygous Ebp1(−/−) mice were viable and 30% smaller than wild-type littermates with transient growth retardation, which shows phenotypes resembling our heterozygous Ebp1(+/−) mouse. Presumably, the discrepancy of Ebp1 knockout mice phenotypes might be due to the way in which the Ebp1 gene was disrupted. While Zhang et al. (20) used gene trap insertion at intron 2 of the Ebp1 gene, we chose conditional deletion of the Ebp1 exons 6 to 10, avoiding targeting of the Ebp1 exon 1, because the short distance between Ebp1 and Erbb3 genes (∼1.3 kb) could lead to the deregulation of ErbB3 expression, and affect ErbB3 putative regulatory elements present in 3′ of the ErbB3 gene. Nevertheless, no other reports of in vivo functions of EBP1 in knockout mice exist until now to our knowledge. Therefore, the genetic disruption of Ebp1 in a mice model would be useful as a model to study the precise roles of EBP1 and underlying molecular mechanisms.
It is of interest that the neuron loss phenotype of the homozygous Ebp1(−/−) embryo and [Nestin-Cre; Ebp1flox/flox] mutant embryos resembles that of disruption of the tropomyosin-related kinase (Trk) receptor family, reflecting p48EBP1 as a downstream mediator of NGF/BDNF-Trk signaling (12). For example, homozygous null mice of Nrtk1, encoding TrkA, exhibited abundant neuronal loss in dorsal root and sympathetic ganglia (33), and germ line mutation of the kinase domain of Nrtk2 died within the first 2 wk after birth, with neuron loss in both the central and peripheral nervous systems (34). Moreover, the disruption of the Nrtk2 gene led to developmental defects in cardiac vascularization (35). Furthermore, lack of Bdnf also led to excessive neuronal death and died during the second postnatal week (36). In addition to neurotrophin-Trk receptor signaling, the alteration of most of the growth factors and their receptor signaling which are known to be essential for development, including heregulin and ErbB2/3/4 signaling (37) and Fgf8 (38) and Fgfr1 and Fgfr2 (39, 40) signaling, causes embryonic lethality with massive cell death and/or anomalies, shown in homozygous Ebp1(−/−) embryos. Therefore we cannot rule out systemic and/or spatiotemporal regulation of EBP1 function by other growth factor signaling during development.
Embryonic development including brain is regulated by spatiotemporal coordination of specific patterns of gene expression. Identification of the molecular pathway that directs chromatin structure and gene expression is essential for understanding development and has important relevance for determining mechanisms of developmental disorders. Interestingly, we found that loss of Ebp1 caused enriched H3K9 trimethylation and impoverished H3 acetylation in Ebp1(−/−) MEFs, caging the heterochromatin status. Moreover, we identified that EBP1 directly binds to the specific promoter region of Dnmt1 and represses its expression, consequently alleviating DNA methylation of the DNMT1 target gene, for example, Survivin, up-regulating its gene expression in development, whereas DNMT1, Suv39H1, and K9 methylated H3 are in a complex on the Survivin promoter in Ebp1(−/−) MEFs (Fig. 5). Survivin is prominently expressed and prevents apoptosis during early embryonic development (41) and vascular tube formation and new vessel formation are accompanied by increased Survivin levels (42, 43). Besides Survivin, Kcna2 and Klf13, which possess the EBP1 binding motif, were highly methylated compared with other DNMT1 target genes that do not possess the EBP1 binding motif upon Ebp1 loss (SI Appendix, Fig. S6C), suggesting that probably EBP1 could inhibit DNMT1 binding at its target promoter. Hence, it might be plausible that EBP1 plays a critical role in manipulating proper gene expression controlling both DNA and histone methylation.
Since the most obvious defect observed in the Ebp1(−/−) embryo is brain malformation and since EBP1 expression is predominant in neurons among cell types in the central nervous system (SI Appendix, Fig. S2A) (13), we genetically ablated Ebp1 in neurons in the central nervous system using a Nestin-Cre driver. As expected, [Nestin-Cre; Ebp1flox/flox] mutant embryos displayed developmental abnormalities with massive neuronal death (Fig. 2) as well as accumulation of K9 methylated H3 compared to that of control Ebp1flox/flox mice (Fig. 6 B and C). Importantly, AAV2-GFP-EBP1 expression in brain slice rescued the defects in [Nestin-Cre; Ebp1flox/flox] mutant embryos, protecting neuron death and reversed closed chromatin to open chromatin. Moreover, covariation analysis with brain transcriptomes showed not only in mice but also in human brain that Ebp1 and Dmnt1 expression was negatively correlated (Fig. 6D). Thus, we propose EBP1 as a potent regulator of gene expression that may coordinate the regulation of multiple genes by modulating epigenetic regulators during development (Fig. 6E) and further investigation of EBP1 on developmental disorders in human involved in epigenetic diseases may inform therapeutic strategies to restore balancing of gene expression.
Materials and Methods
Animals.
Ebp1(−/−) or Ebp1flox/flox mice were generated from the geneOway (France). All experimental protocols were approved by the Institutional Animal Care for Ethics and Use Committee of Sunkyunkwan University (SUSM, SKKUIACUC 2018-11-14-2), and the study followed institutional and National Institutes of Health guidelines for laboratory animal care. Details of the materials and methods are presented in SI Appendix, SI Materials and Methods.
Statistical Analyses.
Graphs and associated statistical analyses were generated using GraphPad Prism (GraphPad, La Jolla, CA). The data were generated by performing the experiments at least 3 times. All data are presented as mean ± SEM. The statistical significance of the 2 groups was assessed by an unpaired t test.
Data Availability.
The data discussed in the paper are in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government [Ministry of Science, Information and Communication Technology and Future Planning (MSIP)] (2016R1A5A2945889), NRF-2017R1A2B4001846, and by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea (HI17C0227).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916306116/-/DCSupplemental.
References
- 1.Yoo J. Y., et al. , Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br. J. Cancer 82, 683–690 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lessor T. J., Yoo J. Y., Xia X., Woodford N., Hamburger A. W., Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J. Cell. Physiol. 183, 321–329 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Ahn J. Y., et al. , Nuclear Akt associates with PKC-phosphorylated Ebp1, preventing DNA fragmentation by inhibition of caspase-activated DNase. EMBO J. 25, 2083–2095 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Z., Ahn J. Y., Liu X., Ye K., Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc. Natl. Acad. Sci. U.S.A. 103, 10917–10922 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim C. K., et al. , Negative regulation of p53 by the long isoform of ErbB3 binding protein Ebp1 in brain tumors. Cancer Res. 70, 9730–9741 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Kim C. K., et al. , Long isoform of ErbB3 binding protein, p48, mediates protein kinase B/Akt-dependent HDM2 stabilization and nuclear localization. Exp. Cell Res. 318, 136–143 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Xia X., Cheng A., Lessor T., Zhang Y., Hamburger A. W., Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell. Physiol. 187, 209–217 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Woodford N., Xia X., Hamburger A. W., Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Res. 31, 2168–2177 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y., et al. , Suppression of salivary adenoid cystic carcinoma growth and metastasis by ErbB3 binding protein Ebp1 gene transfer. Int. J. Cancer 120, 1909–1913 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., et al. , The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc. Natl. Acad. Sci. U.S.A. 102, 9890–9895 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko H. R., et al. , P42 Ebp1 regulates the proteasomal degradation of the p85 regulatory subunit of PI3K by recruiting a chaperone-E3 ligase complex HSP70/CHIP. Cell Death Dis. 5, e1131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon I. S., Ahn J. Y., p48 Ebp1 acts as a downstream mediator of Trk signaling in neurons, contributing neuronal differentiation. Neurochem. Int. 58, 215–223 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Ko H. R., et al. , Neuron-specific expression of p48 Ebp1 during murine brain development and its contribution to CNS axon regeneration. BMB Rep. 50, 126–131 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehnertz B., et al. , Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192–1200 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Estève P. O., et al. , Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 20, 3089–3103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes S., Pathways to silencing unite. Nat. Cell Biol. 8, 315 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Qin W., Leonhardt H., Spada F., Usp7 and Uhrf1 control ubiquitination and stability of the maintenance DNA methyltransferase Dnmt1. J. Cell. Biochem. 112, 439–444 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Leng F., et al. , Methylated DNMT1 and E2F1 are targeted for proteolysis by L3MBTL3 and CRL4DCAF5 ubiquitin ligase. Nat. Commun. 9, 1641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko H. R., et al. , P42 Ebp1 functions as a tumor suppressor in non-small cell lung cancer. BMB Rep. 48, 159–165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., et al. , Alterations in cell growth and signaling in ErbB3 binding protein-1 (Ebp1) deficient mice. BMC Cell Biol. 9, 69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martynoga B., Morrison H., Price D. J., Mason J. O., Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113–127 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Crossley P. H., Martin G. R., The mouse Fgf8 gene encodes a family of polypeptides and is expressed in regions that direct outgrowth and patterning in the developing embryo. Development 121, 439–451 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Mattila M. M., et al. , FGF-8b increases angiogenic capacity and tumor growth of androgen-regulated S115 breast cancer cells. Oncogene 20, 2791–2804 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Betz U. A., Vosshenrich C. A., Rajewsky K., Müller W., Bypass of lethality with mosaic mice generated by Cre-loxP-mediated recombination. Curr. Biol. 6, 1307–1316 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Tronche F., et al. , Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Persengiev S. P., Devireddy L. R., Green M. R., Inhibition of apoptosis by ATFx: A novel role for a member of the ATF/CREB family of mammalian bZIP transcription factors. Genes Dev. 16, 1806–1814 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phan R. T., Dalla-Favera R., The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432, 635–639 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Estève P. O., Chin H. G., Pradhan S., Human maintenance DNA (cytosine-5)-methyltransferase and p53 modulate expression of p53-repressed promoters. Proc. Natl. Acad. Sci. U.S.A. 102, 1000–1005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbins C. A., Tempel B. L., Kv1.1 and Kv1.2: Similar channels, different seizure models. Epilepsia 53 (suppl. 1), 134–141 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Martin K. M., Metcalfe J. C., Kemp P. R., Expression of Klf9 and Klf13 in mouse development. Mech. Dev. 103, 149–151 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Figeac N., Serralbo O., Marcelle C., Zammit P. S., ErbB3 binding protein-1 (Ebp1) controls proliferation and myogenic differentiation of muscle stem cells. Dev. Biol. 386, 135–151 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Neilson K. M., et al. , Pa2G4 is a novel Six1 co-factor that is required for neural crest and otic development. Dev. Biol. 421, 171–182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smeyne R. J., et al. , Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature 368, 246–249 (1994). [DOI] [PubMed] [Google Scholar]
- 34.Klein R., et al. , Targeted disruption of the trkB neurotrophin receptor gene results in nervous system lesions and neonatal death. Cell 75, 113–122 (1993). [PubMed] [Google Scholar]
- 35.Wagner N., et al. , Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms’ tumor transcription factor Wt1. Genes Dev. 19, 2631–2642 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ernfors P., Lee K. F., Jaenisch R., Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature 368, 147–150 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Erickson S. L., et al. , ErbB3 is required for normal cerebellar and cardiac development: A comparison with ErbB2-and heregulin-deficient mice. Development 124, 4999–5011 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Sun X., Meyers E. N., Lewandoski M., Martin G. R., Targeted disruption of Fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes Dev. 13, 1834–1846 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaguchi T. P., Harpal K., Henkemeyer M., Rossant J., fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 8, 3032–3044 (1994). [DOI] [PubMed] [Google Scholar]
- 40.Arman E., Haffner-Krausz R., Chen Y., Heath J. K., Lonai P., Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. U.S.A. 95, 5082–5087 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawamura K., et al. , Survivin acts as an antiapoptotic factor during the development of mouse preimplantation embryos. Dev. Biol. 256, 331–341 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Mesri M., et al. , Suppression of vascular endothelial growth factor-mediated endothelial cell protection by survivin targeting. Am. J. Pathol. 158, 1757–1765 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zwerts F., et al. , Lack of endothelial cell survivin causes embryonic defects in angiogenesis, cardiogenesis, and neural tube closure. Blood 109, 4742–4752 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in the paper are in SI Appendix.