Significance
Early diagnosis and characterization of the severity of CFTR mutations carried in cystic fibrosis (CF) impacts life expectancy and quality of life for patients. We demonstrate a testing platform that combines analysis of perspiration samples by desorption electrospray ionization mass spectrometry and a machine-learning method of gradient boosted decision trees, with an accuracy for the correct identification of CF cases of 98 ± 2%. Our sampling method is minimally invasive; it only requires swiping a standard microscope slide across the patient’s forehead, with no sample processing. The whole collection and testing process takes less than 2 min, which suggests a faster alternative with comparable accuracy to the conventional sweat chloride test, which takes up to 3 h.
Keywords: desorption electrospray ionization, mass spectrometry, machine learning, cystic fibrosis
Abstract
The gold standard for cystic fibrosis (CF) diagnosis is the determination of chloride concentration in sweat. Current testing methodology takes up to 3 h to complete and has recognized shortcomings on its diagnostic accuracy. We present an alternative method for the identification of CF by combining desorption electrospray ionization mass spectrometry and a machine-learning algorithm based on gradient boosted decision trees to analyze perspiration samples. This process takes as little as 2 min, and we determined its accuracy to be 98 ± 2% by cross-validation on analyzing 277 perspiration samples. With the introduction of statistical bootstrap, our method can provide a confidence estimate of our prediction, which helps diagnosis decision-making. We also identified important peaks by the feature selection algorithm and assigned the chemical structure of the metabolites by high-resolution and/or tandem mass spectrometry. We inspected the correlation between mild and severe CFTR gene mutation types and lipid profiles, suggesting a possible way to realize personalized medicine with this noninvasive, fast, and accurate method.
Cystic fibrosis (CF) is the most common life-threatening autosomal recessive disease in the United States and is the result of mutations in the CF transmembrane conductance regulator (CFTR) gene (1, 2). Although there is no cure, CFTR modulator drugs and intensive therapy regimens can improve the outlook for people with CF, if diagnosed early (3). The data accumulated from several longitudinal studies (4–6) demonstrated the benefits of early diagnosis prompted by newborn screening (NBS), which is now universally available in the United States. However, NBS only identifies newborns at risk for CF, and the benefits can only be fully realized when the appropriate confirmatory diagnostic testing is in place. Despite its initial description more than 60 y ago, the sweat chloride test by cholinergic pilocarpine iontophoresis remains to this date as the clinical standard for CF diagnosis (7).
The sweat chloride test (8) includes first stimulation of a high rate of sweat secretion in a small area of skin by pilocarpine iontophoresis, collection of sweat, and determination of chloride concentration in the sweat collected (9). The testing routine, particularly when working with newborns, entails great technical skill, sophisticated equipment, and cumbersome sweat collection devices. It is also recognized that newborns are an especially challenging population to test as the failure rate for sample collection is not negligible (10). In addition, current testing methodology takes up to 3 h to complete, including time spent analyzing chloride content, which is prone to technical errors (10). Perhaps more troublesome is that it is now clearly recognized that the test has a wide diagnostic range with many individuals falling into intermediate value categories, and even a few of those affected may have completely normal values (11–14). These recognized drawbacks motivate the development of a new diagnostic test for CF as an alternative to the current sweat chloride test.
Various research efforts have been employed on developing metabolomics for CF diagnosis. Metabolites from different sources of biological samples, for example, blood (15, 16), sweat (17–20), breath (21–23), sputum (24, 25), and stool (26, 27), have been analyzed for biomarkers that could be useful in CF diagnosis. Particularly, Esteves et al. (18) pressed silica plates onto human skin, extracted what was transferred to the silica plates with methanol:water (1:1), and then employed electrospray ionization mass spectrometry for metabolite identification. Macedo et al. (17) used standard pilocapine-stimulated sweat collection, which was subjected to capillary electrophoresis for biomarker discovery. Those efforts can be summarized as a bottom-up approach that first identifies metabolic changes and then develops diagnostic assays.
We present here a different approach that gives diagnostic predictions without necessarily knowing the specific metabolites, by combining desorption electrospray ionization (DESI) mass spectrometry (28) and machine learning. In 2010, Eberlin et al. made the first medical applications of DESI by grading the degree of malignancy in brain tumors (29). Since then, there have been many DESI studies for chemical imaging and diagnosis of tissue samples (30–32). The present work builds on what Zhou and Zare did in 2017 to classify genders, ethnicities, and ages of individuals from a DESI MS analysis of sweat (33).
As a second important improvement, we sought the simplest and least invasive sampling method possible that at the same time will lead to the highest rate of success in obtaining a valid sample. To accomplish this, we selected a standard microscope glass slide as the collection device and simply swiped it across the forehead or nose of a subject to collect the sample. The swiping process involves applying gentle pressure to the microscope slide as it is moved across the forehead of the patient, a task that takes about 5 s to perform. Then, the samples collected in this manner are the product of perspiration present in the skin surface rather than stimulated sweat. This introduces an important feature of our method as it obviates the need for active sweat gland stimulation and eliminates the known influence of the secretory rate achieved on the concentrations of analytes in sweat. Next, we applied DESI directly to the glass slide without any extra sample preparation steps. We employed a machine-learning model of gradient boosted decision trees (GBDT) (34) to recognize the pattern of the metabolites in the sample and searched for the distinction of CF versus healthy controls. The reason why we choose GBDT method is that it not only exhibits high classification performance but also provides an explainable model. The tree model makes predictions based on a series of tests of the intensities of peaks. Although molecules from exogenous sources such as lotions or creams are picked up by the mass spectrometer, the machine learning algorithm tends to pick the features that exist in all subjects of a class (i.e., CF or healthy control). Therefore, exogenous sourced molecules are usually not selected because their large variance across different subjects causes them to be rejected by the machine learning algorithm.
In addition, statistical bootstrap (35) was used to provide an estimate of the uncertainty in the prediction, which provides further information for healthcare providers to decide whether additional diagnostic testing (e.g., further gene sequencing, CFTR functional assays (7)) is necessary. In addition, we selected important mass peaks by the feature selection algorithm to gain insight as to which metabolites and lipids contribute to the prediction of disease state. The concept of important features selected by the algorithm are different from the traditional definition of biomarkers, but they are related in the following 2 ways: 1) if an important feature has been identified as a known biomarker, it will add credibility to the model and 2) if an important feature is previously unknown, it may suggest a biomarker candidate for future study. Finally, given the large number of CFTR mutations with variable spectrum of dysfunction associated (36), we examined the correlation between gene mutations and patterns in lipid composition.
Results
Mass Spectrometric Analysis of Perspiration Samples.
DESI mass spectrometry was applied on 277 perspiration samples at negative ion mode. On the surface of each sample, 30 spots are randomly selected to be analyzed by DESI; their mass spectra are then averaged. The spots are chosen randomly from the glass area seen to contain the perspiration sample. In DESI-MS imaging of tissues, each pixel or spot is different because they are composed of different cells. However, each spot is taken from the perspiration sample obtained from the skin surface and is believed to represent a homogeneous distribution of perspiration molecules. Therefore, we believe it is better to analyze the average rather than individual spots. It is not necessary to remove the background because the algorithm tends to choose the feature that is different between the CF and healthy control samples. The background peaks that are contained in both samples will not be used during classification. Of the 277 samples, 57 were collected from CF patients, while 220 came from healthy controls. The CF and healthy controls have similar age brackets (SI Appendix, Fig. S1).
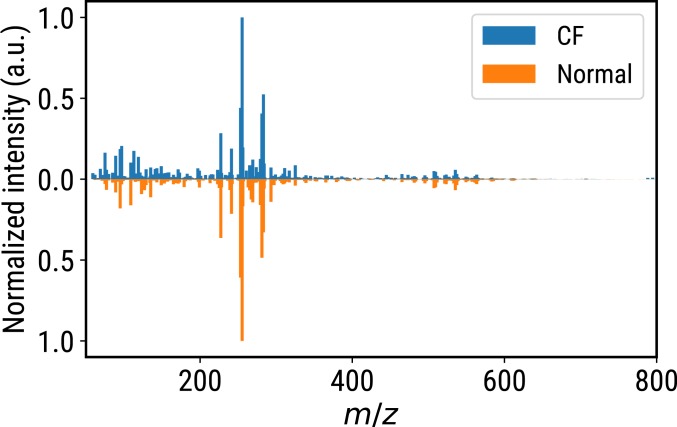
Fig. 1 shows the average mass spectra for CF patients and healthy controls at negative ion mode. SI Appendix, Fig. S2, shows the same spectra in an overlaid manner. We introduce the terms m/z (the mass-to-charge ratio of an ion analyzed by the mass spectrometer) and peak (molecules with a specific m/z appear as a peak in the mass spectrum). Most of the ions detected in the region m/z = 200 to 350 are fatty acids, while the ions in the region m/z = 450 to 600 are mostly fatty acid dimers or diacylglycerols. A visual examination reveals that there is little difference between CF and healthy control samples in the m/z = 200 to 350 region.
Fig. 1.
The average mass spectra for CF patients and healthy controls.
Predictive Diagnosis with Machine Learning.
Approximately 1,000 distinct peaks were extracted from the whole set of samples. GBDT (34) was applied to the samples to classify them between CF and healthy states. GBDT is a tree-based algorithm that uses a sequence of rules to evaluate the chance of the sample being in a specific category. Given the limited size of samples, 6-fold rotational cross-validation was employed to evaluate the performance of the model. The dataset was randomly split into 6 parts; in each of the 6 rounds, 1 part was chosen to be the test set, and all other parts were used as the training set. The goal of cross-validation is to test the ability of the model to predict unseen data. With the evaluation of 6-fold cross-validation (SI Appendix, Table S1), the accuracy of the model was determined to be 98 ± 2%. With the reported sweat chloride test performance metrics for the diagnosis of CF of an accuracy of 98%, specificity of 92.8%, and sensitivity of 100% (24), our method for analyzing perspiration reaches a comparable performance but with the added advantage of it being a highly feasible and faster alternative.
Table 1 presents the values of all of the metrics. The definitions of recall (sensitivity), precision, and specificity are
where denotes true negative, denotes true positive, denotes false negative, and denotes false positive. These metrics measure the performance of the model under different conditions. AUC is the area under the receiver operating characteristics curve (SI Appendix, Fig. S3), which illustrates the diagnostic ability of the model as its discrimination threshold is varied.
Table 1.
The average performance of the prediction model
| Metric | Value |
| Accuracy | 98 ± 2% |
| Recall | 96 ± 7% |
| Precision | 94 ± 7% |
| Specificity | 98 ± 2% |
Uncertainty of Diagnosis.
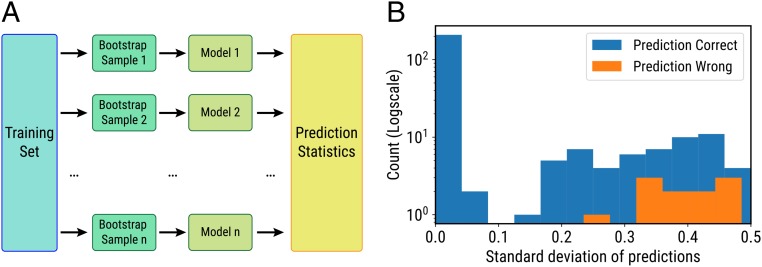
Considering the needs in clinical practice, healthcare providers are often faced with a necessity to make decisions based on the level of confidence that a given test result has as a predictor. Further diagnostic tests may have to be performed if the prediction has a high uncertainty. We borrowed the idea of bootstrapping, which relies on random sampling with replacement, to construct the confidence interval of the prediction (35). The dataset generated was separated into a training set and a test set with a ratio of 5:1. Twenty bootstrap sample sets were then created from random sampling with replacement such that each sample set had the same number of samples as the original training set. In the next step, 20 unique models were created by training solely on the corresponding bootstrap sample set, and 20 predictions can be obtained for each sample in the test set. The statistics of prediction can then be constructed from those predictions. Fig. 2A shows a graphical illustration of this method.
Fig. 2.
(A) Graphical illustration of bootstrap methods. (B) The empirical distribution of the SD of correct and wrong predictions. The y axis is the number of samples for which the prediction is correct or wrong.
Intuitively, the percentage of correct predictions will increase as the uncertainty of prediction decreases. Fig. 2B shows the empirical distribution of the SD of correct and wrong predictions. The SD is a measure of uncertainty. As the SD increases, the percentage of correct predictions decreases.
Further, we performed an experiment to simulate real diagnostic practice. We established a threshold of 0.1, which means the prediction will be marked as “confident” only if the SD of the prediction is lower than the threshold. Over a similar 6-fold cross-validation process as previously described, we have on average 76% of the samples marked as confident, and the prediction accuracy of the confident samples is 100% throughout the cross-validation process.
Feature Selection and Identification.
The model described above is able to determine whether a sample comes from a CF patient or a healthy control without explicitly knowing what biomarker or set of biomarkers are contributing to the distinction. However, it is still desired to identify the important chemical species in the classification procedure. An important feature, which is identified as a known biomarker, will help rationalize the decision-making process. Moreover, an important feature that is previously unknown will lead to possible metabolite discovery with relevance to the disease state that ultimately could serve as a biomarker of the degree of function or dysfunction present at the individual level.
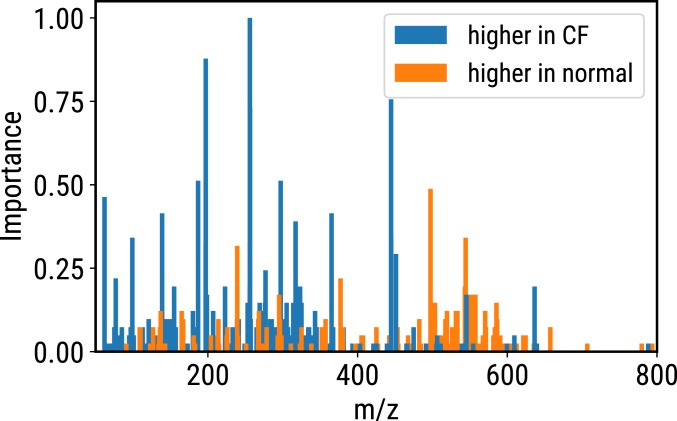
The GBDT algorithm is capable of evaluating the importance of a feature by measuring the number of times when using this feature as a branching point for the tree. The more the feature is used, the higher the importance. Fig. 3 shows a bar plot of the peak importance calculated by the GBDT model. Over the 1,222 peaks extracted from the samples, 280 peaks (SI Appendix, Table S2) are selected to be important features to distinguish between healthy controls and CF patients. The important features are mostly lipids because the perspiration samples we collected consist of mainly lipids.
Fig. 3.
The importance calculated by GBDT model depending on the number of times the feature is used in decision making. Blue bars show peaks with higher intensity in CF patients, while orange bars show peaks with higher intensity in healthy normal.
The intensity of a peak in a mass spectrum correlates with the concentration of 1 or more chemical species in the sample. The chemical species that are represented by the important peaks were tentatively identified using high-resolution and/or tandem mass spectrometry (SI Appendix, Figs. S6–S33). SI Appendix, Table S3, lists several important peaks and their identifications. Many of those identified molecules are found to be biologically relevant. For example, m/z = 305.2480 (error 0.0 ppm) and its tandem mass spectrum matched those of Mead acid FA(20:3). Here 20 stands for 20 carbons in the fatty acid molecule, while 3 means there are 3 carbon–carbon double bonds. The proposed chemical structure of FA(20:3) is shown in SI Appendix, Fig. S5A. This molecule was found to have a higher mean concentration in CF samples, which agrees with the result of increased Mead acid level in blood and tissues of CF patients (37). Another peak selected to be important by GBDT is m/z = 255.2328, which is identified to be palmitic acid FA(16:0) based on high-accuracy mass measurements (mass error 1.57 ppm) and tandem mass spectrometry. The proposed chemical structure of FA(20:3) is shown in SI Appendix, Fig. S5B. Previous studies show that palmitate has a higher concentration in the plasma of CF patients (38), and CF patients absorb more of this fatty acid than healthy controls (39). Those studies confirm our result, which shows that the concentration of FA(16:0) is higher in CF patients. Although chemical identification is not necessary, the feature selection adds credibility to the model.
Correlation Between Gene Mutations and Lipid Profiles.
A large number of mutations are known to be associated with CF (40). This contributes to the known variability seen from patient to patient. Different CFTR mutations usually vary in the severity of the defect produced and have different cellular mechanisms (36). The genetic mutation of each CF patient that contributed a sample is presented in SI Appendix, Table S4. Following the categorization scheme proposed by McKone et al. (41), the mutations were classified into 2 categories, mild and severe, for practical considerations. For each peak, a test was performed on the null hypothesis of the distributions of that specific peak intensity are the same among mild and strong mutations. SI Appendix, Table S5, shows the peaks with P values less than 0.05. Also, statistical feature selection is performed with least absolute shrinkage and selection operator (Lasso) (42), whose test accuracy is 77 ± 9% on a 6-fold cross-validation. The peaks with nonzero coefficients are recognized as important features, which are shown in SI Appendix, Table S6. Interestingly, the peak at m/z = 609.5109 is believed to be significantly different by hypothesis testing, as well as an important feature by Lasso. This peak is tentatively identified as triacylglycerol TG(34:0) by a high-accuracy mass measurement (mass error 1.48 ppm). A previous study on lipid profiles in CF demonstrated that CF patients in all age groups had higher triacylglycerol (43). As a further step, our results show that TG levels are different between mild and severe CFTR mutations, and there is a correlation between CFTR gene mutations and lipid profiles.
Discussion
We present here a method for simple, fast, and accurate CF testing, which uses DESI mass spectrometry to analyze the chemical composition of perspiration and machine learning to recognize underlying patterns that distinguish between healthy controls and CF patients. Compared with other methods including electrospray ionization mass spectrometry and capillary electrophoresis–mass spectrometry, DESI enables the analysis of a sample surface without sample processing, which reduces the time needed, as well as the variation induced in sample preparation steps. The advantages introduced by our simple sampling procedures constitute an unquestionable improvement over current gold standard CF diagnostic methodology. In addition, we employed statistical bootstrap to estimate the confidence level of the prediction given by the algorithm, which provides further information for healthcare providers to make decisions in practice. Although metabolite identification is not necessary for diagnosis, we have pinpointed the important chemical features in the prediction process and tentatively identified some of them by high-accuracy and/or tandem mass spectrometry. The discovery of biologically related molecules adds credibility to the model. In the final part, we correlated the CFTR gene mutations with the change of lipid profiles in perspiration, which suggests an alternative to gene sequencing for building a patient model for personalized medicine. Future improvement to this work can be achieved by enrolling more patients to increase the sample size of a genetically diverse CF population, as well as further identification of the important metabolites present in CF patients. In addition, we believe our method opens the possibility of using the peaks identified to monitor the change in CFTR function introduced by novel CFTR modulators being introduced to the clinic.
Materials and Methods
Sample Collection.
All experiments involving human subjects were approved by Stanford University’s Institutional Review Board, which required adults to give an oral consent form and that all data collected would be anonymized. Perspiration samples are collected by gently swiping a standard microscope glass slide across the forehead of each participant. It is possible that some dead skin is also collected in this procedure, but it is not readily dissolved by the droplet spray, so we refer to the samples analyzed as perspiration. Samples were stored at room temperature until batch processing. The interval between collection and mass spectrometric analysis ranges from 1 to 8 wk. The lipid profile changes during storage, but the formulation of our algorithm tends to use features that do not vary much with time for classification, as both CF and healthy control samples were stored for comparable time periods. The rationale behind our claim is if a feature varies strongly with time, then the difference between CF and normal samples with respect to this feature will disappear, and it will be difficult to use this feature to separate CF and normal samples.
Mass Spectrometry Analysis.
The DESI method was employed for sweat sample analysis. A laboratory-built DESI source with an x–y stage was set up in front of an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific). The mass spectrum was collected under negative ion mode with m/z of 50 to 1,000. The DESI source used methanol–water (9:1 vol/vol) as the solvent with a flow rate of 4 μL/min. The nitrogen gas pressure was set to 120 psi. In all experiments, the Orbitrap ion analyzer was calibrated and operated under a resolution of 60,000 to ensure accurate measurements. On the surface of each sample, 30 spots were randomly selected to be analyzed by DESI.
Data Analysis.
The Thermo proprietary raw files were converted to mzML file formats and then read in Python with the mzml package (44). The 30 spectra of each sample were averaged. A handwritten peak finding algorithm was employed to convert the continuous spectrum to sparse peak profiles. A total of 1,222 peaks are chosen such that each peak appears in at least 60 samples. Each sample was then vectorized by the peak values with a resolution of 0.1 m/z, which means that each sample is converted to a 1,222-dimensional vector based on the intensities of the specific peaks. Six-fold rotational cross-validation was employed to evaluate the performance of a model. The dataset was randomly split into 6 parts; in each of the 6 rounds, 1 part was chosen to be the test set, and all other parts are used as the training set. The mechanism of the GBDT model is that it only uses the important features to make a prediction. Therefore, the metrics we reported are based only on the selected features.
Different classification algorithms of logistic regression with l1 regularization (Lasso), support vector machines, random forests, and gradient boosted decision trees (GBDT) are tested based on the performance of cross-validation. The number of trees in the GBDT model is 100. The algorithms are implemented in open-source packages of scikit-learn (45) and lightgbm (34).
Chemical Identification.
The important features selected by the GBDT algorithm are correlated with specific peaks in the mass spectrum. The chemicals represented by the peaks are identified by searching the database of Lipid MAPS (https://www.lipidmaps.org), METLIN (https://metlin.scripps.edu), and MassBank (www.massbank.jp), according to their high-resolution and high-accuracy m/z values. Further, we performed collision-induced dissociation (CID) tandem mass spectrometry to obtain the fragmentation profile of a specific peak. The raw data can be found at https://osf.io/j59h2/?view_only=c0212307a2714d909559550a65db0213. The CID spectra corresponding to the chemical identification are provided as SI Appendix, Figs. S6–S33. The fragmentation profiles are compared with the standard from the database, if available. Given the complicated biological matrix (the perspiration sample), not all fragmented ions can be matched with the standard samples. Therefore, we claim tentative identification of the chemicals. In addition, the exact position and stereochemistry of the unsaturated bonds are not able to be determined with tandem mass spectrometry. However, given the source of the samples is human beings, we can claim the most probable metabolites are the human-sourced ones.
Supplementary Material
Acknowledgments
Z.Z. is grateful for the support of a Stanford Graduate Fellowship. This work was supported by National Science Foundation under the Data-Driven Discovery Science in Chemistry (D3SC) for Early Concept Grants for Exploratory Research (EAGER Grant CHE-1734082) and the Ross Mosier CF Laboratories at Stanford.
Footnotes
The authors declare no competing interest.
Data deposition: Raw data can be accessed through the Open Science Framework at https://osf.io/j59h2/?view_only=c0212307a2714d909559550a65db0213, DOI:10.17605/OSF.IO/J59H2.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909630116/-/DCSupplemental.
References
- 1.Ratjen F., et al. , Cystic fibrosis. Nat. Rev. Dis. Primers 1, 15010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quinton P. M., Cystic fibrosis: A disease in electrolyte transport. FASEB J. 4, 2709–2717 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Kerem E., Mutation specific therapy in CF. Paediatr. Respir. Rev. 7 (suppl. 1), S166–S169 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Farrell P. M., et al. ; Wisconsin Cystic Fibrosis Neonatal Screening Study Group , Nutritional benefits of neonatal screening for cystic fibrosis. N. Engl. J. Med. 337, 963–969 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Grosse S. D., et al. ; CDC , Newborn screening for cystic fibrosis: Evaluation of benefits and risks and recommendations for state newborn screening programs. MMWR Recomm. Rep. 53, 1–36 (2004). [PubMed] [Google Scholar]
- 6.Grosse S. D., Rosenfeld M., Devine O. J., Lai H. J., Farrell P. M., Potential impact of newborn screening for cystic fibrosis on child survival: A systematic review and analysis. J. Pediatr. 149, 362–366 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Farrell P. M., et al. , Diagnosis of cystic fibrosis: Consensus guidelines from the cystic fibrosis foundation. J. Pediatr. 181, S4–S15.e1 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Gibson L. E., Cooke R. E., A test for concentration of electrolytes in sweat in cystic fibrosis of the pancreas utilizing pilocarpine by iontophoresis. Pediatrics 23, 545–549 (1959). [PubMed] [Google Scholar]
- 9.LeGrys V. A., et al. , Sweat testing: Sample collection and quantitative chloride analysis; approved guideline. Clin. Lab Stand Ins. 29 (suppl. 27), C34-A2 (2009). [Google Scholar]
- 10.Legrys V. A., McColley S. A., Li Z., Farrell P. M., The need for quality improvement in sweat testing infants after newborn screening for cystic fibrosis. J. Pediatr. 157, 1035–1037 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munck A., et al. ; ECFS Neonatal Screening Working Group , Cystic Fibrosis Screen Positive, Inconclusive Diagnosis (CFSPID): A new designation and management recommendations for infants with an inconclusive diagnosis following newborn screening. J. Cyst. Fibros. 14, 706–713 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lebecque P., et al. , Mutations of the cystic fibrosis gene and intermediate sweat chloride levels in children. Am. J. Respir. Crit. Care Med. 165, 757–761 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Zirbes J., Koepke R., Kharrazi M., Milla C., Sweat chloride (SC) concentration and CFTR mutation class among infants identified by newborn screening (NBS) in California (CA). J. Cyst. Fibros. 9 (suppl. 1), S7 (2010). [Google Scholar]
- 14.Kharrazi M., et al. ; California Cystic Fibrosis Newborn Screening Consortium , Newborn screening for cystic fibrosis in California. Pediatrics 136, 1062–1072 (2015). [DOI] [PubMed] [Google Scholar]
- 15.DiBattista A., et al. , Metabolic signatures of cystic fibrosis identified in dried blood spots for newborn screening without carrier identification. J. Proteome Res. 18, 841–854 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Shoki A. H., Mayer-Hamblett N., Wilcox P. G., Sin D. D., Quon B. S., Systematic review of blood biomarkers in cystic fibrosis pulmonary exacerbations. Chest 144, 1659–1670 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Macedo A. N., et al. , The sweat metabolome of screen-positive cystic fibrosis infants: Revealing mechanisms beyond impaired chloride transport. ACS Cent. Sci. 3, 904–913 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esteves C. Z., et al. , Skin biomarkers for cystic fibrosis: A potential non-invasive approach for patient screening. Front Pediatr. 5, 290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinton P., et al. , β-adrenergic sweat secretion as a diagnostic test for cystic fibrosis. Am. J. Respir. Crit. Care Med. 186, 732–739 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Mattar A. C. V., Leone C., Rodrigues J. C., Adde F. V., Sweat conductivity: An accurate diagnostic test for cystic fibrosis? J. Cyst. Fibros. 13, 528–533 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Aurora P., et al. ; London Cystic Fibrosis Collaboration , Multiple-breath washout as a marker of lung disease in preschool children with cystic fibrosis. Am. J. Respir. Crit. Care Med. 171, 249–256 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Tate S., MacGregor G., Davis M., Innes J. A., Greening A. P., Airways in cystic fibrosis are acidified: Detection by exhaled breath condensate. Thorax 57, 926–929 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robroeks C. M. H. H. T., et al. , Metabolomics of volatile organic compounds in cystic fibrosis patients and controls. Pediatr. Res. 68, 75–80 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Warwick W. J., et al. , Evaluation of a cystic fibrosis screening system incorporating a miniature sweat stimulator and disposable chloride sensor. Clin. Chem. 32, 850–853 (1986). [PubMed] [Google Scholar]
- 25.Yang J., Eiserich J. P., Cross C. E., Morrissey B. M., Hammock B. D., Metabolomic profiling of regulatory lipid mediators in sputum from adult cystic fibrosis patients. Free Radic. Biol. Med. 53, 160–171 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy J. L., Wootton S. A., Bond S. A., Jackson A. A., Energy content of stools in normal healthy controls and patients with cystic fibrosis. Arch. Dis. Child. 66, 495–500 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaakoush N. O., Pickford R., Jaffe A., Ooi C. Y., Is there a role for stool metabolomics in cystic fibrosis? Pediatr. Int. (Roma) 58, 808–811 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Takáts Z., Wiseman J. M., Gologan B., Cooks R. G., Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306, 471–473 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Eberlin L. S., et al. , Discrimination of human astrocytoma subtypes by lipid analysis using desorption electrospray ionization imaging mass spectrometry. Angew. Chem. Int. Ed. Engl. 49, 5953–5956 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ifa D. R., Eberlin L. S., Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin. Chem. 62, 111–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarmusch A. K., et al. , Lipid and metabolite profiles of human brain tumors by desorption electrospray ionization-MS. Proc. Natl. Acad. Sci. U.S.A. 113, 1486–1491 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jarmusch A. K., et al. , Differential lipid profiles of normal human brain matter and gliomas by positive and negative mode desorption electrospray ionization—Mass spectrometry imaging. PLoS One 11, e0163180 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Z., Zare R. N., Personal information from latent fingerprints using desorption electrospray ionization mass spectrometry and machine learning. Anal. Chem. 89, 1369–1372 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Ke G., et al. , “Lightgbm: A highly efficient gradient boosting decision tree” in Advances in Neural Information Processing Systems, Guyon I., et al., Eds. (Neural Information Processing Systems Foundation, La Jolla, CA, 2017), pp. 3146–3154. [Google Scholar]
- 35.Efron B., Tibshirani R., Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1, 54–75 (1986). [Google Scholar]
- 36.Milla C. E., Moss R. B., Recent advances in cystic fibrosis. Curr. Opin. Pediatr. 27, 317–324 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Seegmiller A. C., Abnormal unsaturated fatty acid metabolism in cystic fibrosis: Biochemical mechanisms and clinical implications. Int. J. Mol. Sci. 15, 16083–16099 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrell P. M., Mischler E. H., Engle M. J., Brown D. J., Lau S. M., Fatty acid abnormalities in cystic fibrosis. Pediatr. Res. 19, 104–109 (1985). [DOI] [PubMed] [Google Scholar]
- 39.Murphy J. L., Jones A. E., Stolinski M., Wootton S. A., Gastrointestinal handling of [1-13C]palmitic acid in healthy controls and patients with cystic fibrosis. Arch. Dis. Child. 76, 425–427 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cystic Fibrosis Mutation Database http://www.genet.sickkids.on.ca/cftr/Home.html. Accessed 2 March 2019.
- 41.McKone E. F., Emerson S. S., Edwards K. L., Aitken M. L., Effect of genotype on phenotype and mortality in cystic fibrosis: A retrospective cohort study. Lancet 361, 1671–1676 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Tibshirani R., Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58, 267–288 (1994). [Google Scholar]
- 43.Figueroa V., Milla C., Parks E. J., Schwarzenberg S. J., Moran A., Abnormal lipid concentrations in cystic fibrosis. Am. J. Clin. Nutr. 75, 1005–1011 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Kösters M., et al. , pymzML v2.0: Introducing a highly compressed and seekable gzip format. Bioinformatics 34, 2513–2514 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Pedregosa F., et al. , Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.