Significance
Multiple myeloma (MM) and triple-negative breast cancer (TNBC) are dependent on 26S proteasome for malignancy. We have previously shown that the proteasome-regulating kinase DYRK2 is a viable target for both MM and TNBC. Here we identified a specific DYRK2 inhibitor, LDN192960, which alleviates both MM and TNBC progression via mechanisms including partial inhibition of proteasome activity. At this time we report a single drug target for 2 diverse cancers and highlight the importance of identifying proteasome regulators.
Keywords: DYRK, multiple myeloma, triple-negative breast cancer, kinase inhibitor, proteasome inhibitor
Abstract
Dependence on the 26S proteasome is an Achilles’ heel for triple-negative breast cancer (TNBC) and multiple myeloma (MM). The therapeutic proteasome inhibitor, bortezomib, successfully targets MM but often leads to drug-resistant disease relapse and fails in breast cancer. Here we show that a 26S proteasome-regulating kinase, DYRK2, is a therapeutic target for both MM and TNBC. Genome editing or small-molecule mediated inhibition of DYRK2 significantly reduces 26S proteasome activity, bypasses bortezomib resistance, and dramatically delays in vivo tumor growth in MM and TNBC thereby promoting survival. We further characterized the ability of LDN192960, a potent and selective DYRK2-inhibitor, to alleviate tumor burden in vivo. The drug docks into the active site of DYRK2 and partially inhibits all 3 core peptidase activities of the proteasome. Our results suggest that targeting 26S proteasome regulators will pave the way for therapeutic strategies in MM and TNBC.
Multiple myeloma (MM) and triple-negative breast cancer (TNBC) are diverse forms of neoplasia with a combined predicted incidence of >70,000 new cases in the United States with >16,000 deaths in 2018 (American Cancer Society 2018 Facts and Figures). MM arises from clonal proliferation of malignant plasma cells (1, 2), whereas TNBC is a highly metastatic form of breast cancer resistant to most hormone therapies due to a lack of estrogen, progesterone, and HER2 receptors (3). Although with no apparent similarities in physiological manifestations or current pharmacological interventions, both MM and TNBC are surprisingly dependent on the 26S proteasome function for survival and progression of disease (2, 4). The 26S proteasome is an essential protein complex that degrades the majority of cellular proteins in eukaryotes (5). The 20S core particle harbors the intrinsic chymotryptic (β5), tryptic (β2), and caspase-like (β1) peptidase activities, whereas the remaining subunits (RPT1–6, RPN1–3, 5–13, and 15) constitute a 19S regulatory particle that caps the 20S core and plays a role in ubiquitylated-substrate recruitment, unfolding, and translocation (5, 6).
The malignant plasma cells in MM are Ig secreting factories (4) and require the proteasome to play a vital role in ER-associated elimination of misfolded proteins for survival and progression (2, 7). FDA-approved drugs bortezomib, carfilzomib, and ixazomib directly bind to and inhibit the 20S core peptidase active site and alleviate MM progression thereby improving life expectancy of patients (2, 8), albeit with reported side effects (9). However, the 20S core subunits frequently accumulate mutations and/or increase copy number (10); when this happens, these patients may develop bortezomib resistance (11). There are limited options when patients exhibit relapsed or refractory MM coupled with proteasome-inhibitor resistance (12).
TNBC, on the other hand, exhibits a high rate of disease relapse with a marked dependence on the 26S proteasome function (4), but unlike MM, the 26S proteasome in TNBC functions to systematically degrade proapoptotic factors leading to neoplastic survival and malignant progression (4). Despite the proteasome dependence, proteasome inhibitors have shown modest efficacy in breast cancer and other solid tumors (13, 14) either due to poor drug penetration into the solid tumors (4) or insufficient potency of the drugs to inhibit all 3 core peptidases of the 20S proteasome (15).
Due to the dependence on 26S proteasome function in both forms of neoplasia and the shortcomings of current proteasome inhibitors, we need novel strategies to target the Achilles’ heel of MM and TNBC. The 26S proteasome has >300 conserved phosphorylation sites on its various subunits, yet very few kinases or phosphatases that regulate the phosphorylation state of these sites have been reported thus far (16–18). Our laboratory recently identified dual-specificity tyrosine phosphorylation-regulated kinase 2 or DYRK2 as a bona fide proteasome regulating kinase that phosphorylates Thr25 on RPT3 and leads to enhanced proteasome activity (17). Inhibition of DYRK2-mediated phosphorylation of RPT3 causes a dramatic reduction in all 3 peptidase activities of the 26S proteasome leading to a marked reduction in the rate of protein degradation, thereby impeding cell cycle progression thus reducing tumor growth (17, 19). As a proof of concept, we recently reported that the natural product curcumin, derived from turmeric, is a highly potent and selective inhibitor of DYRK2 that sensitized both MM and TNBC cell lines via partial inhibition of proteasome activity (19). Furthermore, bortezomib-resistant MM cells were equally sensitive to DYRK2 inhibition as compared to standard MM lines, thus suggesting that inhibiting DYRK2 is a viable therapeutic option for drug-resistant MM patients (19).
In the current study, we report a highly potent and selective small-molecule inhibitor of DYRK2, LDN192960, which alleviates neoplastic progression in both MM and TNBC. Our results establish that inhibition of DYRK2 is a therapeutic strategy to target dual 26S proteasome-adapted MM and TNBC progression leading to impediment of malignancy and potential improvement of patient survival.
Results
DYRK2 and Proteasome Subunits Are Up-Regulated in TNBC.
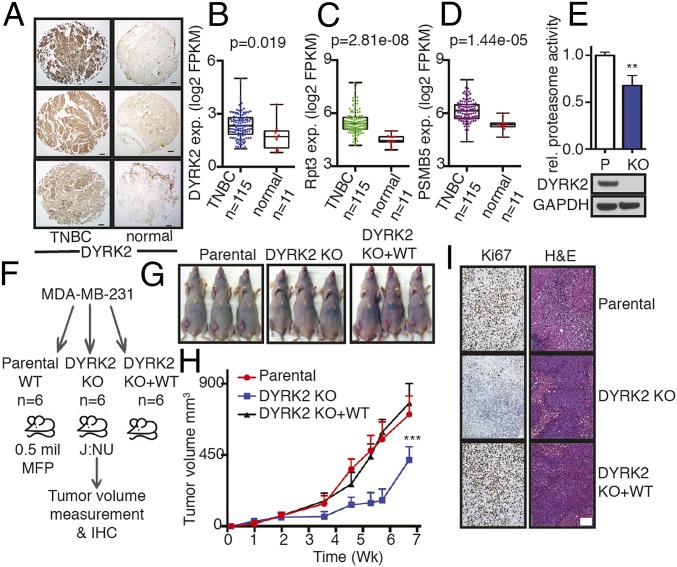
To establish DYRK2 as a viable target for TNBC treatment, we examined the differential expression status of DYRK2 in TNBC patient tissues. Immunohistochemistry of DYRK2 on patient-derived TNBC tumors showed a higher expression of DYRK2 in malignancy relative to adjacent normal breast tissues (Fig. 1A). TNBC is known to be highly heterogeneous, so a bioinformatic approach was undertaken to more comprehensively evaluate the expression status of DYRK2; the DYRK2 substrate RPT3; and the proteasome core subunit, PSMB5, in TNBC diseased states. PSMB5, the core 20S proteasome subunit β5, harbors the chymotryptic-like activity of the proteasome and is known to be overexpressed and/or mutated in malignancy (10, 20–22). We mined the TCGA Cancer Genome Atlas (https://www.cancer.gov/); downloaded the gene expression data of all cancer types and matched normal tissue controls (where available); and compared DYRK2, RPT3, and PSMB5 gene expression (SI Appendix, Fig. S1). As reported previously, DYRK2 expression was largely cancer type specific (17), although in the majority of cancers, we find DYRK2 to be overexpressed compared to normal controls (SI Appendix, Fig. S1 and Table S1). Specifically, in the 1,204 breast cancer patients, DYRK2 was overexpressed relative to matched normal controls (SI Appendix, Fig. S1). Within the breast cancer database, we identified 115 TNBC patients annotated with 11 matched normal tissue controls wherein DYRK2 expression was significantly up-regulated (*P < 0.05) (Fig. 1B). Both RPT3 and PSMB5 were also significantly overexpressed (***P < 0.001) (Fig. 1 C and D).
Fig. 1.
DYRK2 and proteasome are up-regulated in TNBC and promote tumor progression. (A) DYRK2 IHC of TNBC and adjacent normal breast tissue sections from patients. (Scale bar, 100 μm.) (B) DYRK2, (C) RPT3, and (D) PSMB5 differential gene expression in human TNBC and matched normal tissue as available from TCGA (TNBC vs. normal tissue, P value derived from a semipaired modification to the Student’s t test) (see also SI Appendix, Fig. S1). (E) Proteasome was affinity-purified from 1 mg of cell lysate of parental or DYRK2 KO MDA-MB-468 Rpn11-TBHA cells. Proteasome activity was measured with Suc-LLVY-AMC. **P < 0.01 (parental vs. DYRK2 KO, unpaired Student's t test, mean ± SD from n = 3 independent experiments). Immunoblotting of the cell lysates were carried out with indicated antibodies. (F) Experimental flow for TNBC xenograft study in G–I. (G and H) MDA-MB-231 parental or DYRK2 KO or DYRK2 KO + WT rescue cells were injected into the mammary fat pad of J:NU nude mice. Tumor volume was measured twice a week (n = 6 mice per condition), and growth curves were plotted. ***P < 0.001 (compared to parental group, 2-way ANOVA, mean ± SD with Tukey’s multiple comparison). (I) Histological examination of consecutive sections of the tumors (from G and H) with Ki67 and hematoxylin/eosin staining. (Scale bar, 100 μm.)
The proteasome was isolated from MDA-MB-468 parental cells or DYRK2-depleted cells using overexpressed TEV-Biotin-HA tagged RPN11 (RPN11-TBHA) as bait. The proteasome activity was measured on the pull-downs using the Suc-LLVY-AMC substrate peptide. DYRK2 knock-out (KO) cells had 30% reduced proteasome activity (Fig. 1E).
To establish the role of DYRK2 in TNBC, we generated DYRK2-depleted MDA-MB-231 cells and developed a mammary fat pad-derived breast cancer xenograft model in athymic nude mice J:NU (Fig. 1F). DYRK2 depletion led to significant tumor burden reduction in the mice xenografts which was completely restored in mice bearing DYRK2-depleted cells reintroduced with wild-type DYRK2 (Fig. 1 G and H). In fact, the loss of proliferating cells in the tumor as measured by Ki67 staining was also rescued in the DYRK2-reintroduced tumors (Fig. 1I).
These data suggest that DYRK2 inhibition could indeed be a promising mechanism for inhibiting the TNBC proteasome.
DYRK2 Promotes MM Progression.
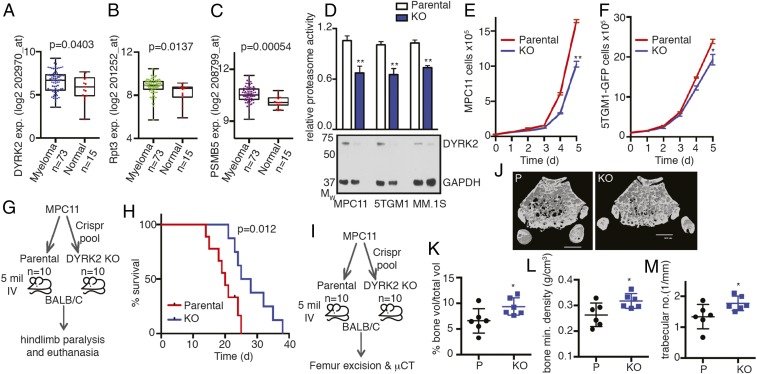
We examined whether MM also exhibited a similar overexpression pattern for DYRK2 and proteasome subunits as in TNBC (Fig. 1 B–D). We queried a publicly available dataset, GSE6477 (https://www.ncbi.nlm.nih.gov/geo/, GSE6477) (23, 24), for the differential expression of DYRK2, RPT3, and PSMB5 between normal and newly diagnosed MM disease states. Similar to TNBC, we found significant overexpression of all 3 genes (Fig. 2 A–C) suggesting the DYRK2 is indeed a potential target in MM. We depleted DYRK2 in MM cell lines using Crispr/Cas9. DYRK2 depletion was ascertained by anti-DYRK2 immunoblotting with GAPDH as a control (Fig. 2D). To further quantify the proteasome activity, cell lysates from parental and DYRK2-depleted MM cells were assayed with the fluorogenic peptide substrate, Suc-LLVY-AMC. DYRK2-depleted cells exhibited a 30 to 40% decrease in proteasome activity in both murine (MPC11 and 5TGM1-GFP) and human (MM.1S) MM cells (Fig. 2D).
Fig. 2.
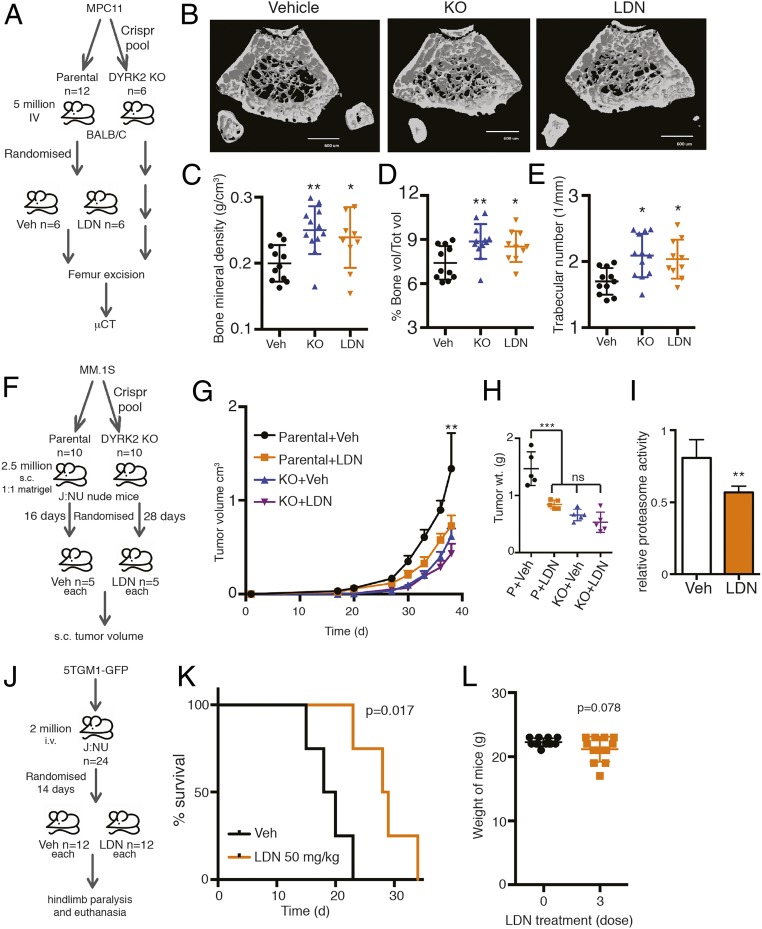
DYRK2 promotes myeloma progression and myeloma-mediated bone degeneration. (A) DYRK2, (B) RPT3, and (C) PSMB5 differential gene expression analyses were performed on the MM dataset with the accession number GSE6477 available at the Gene Expression Omnibus (myeloma vs. normal tissue, P value derived from empirical Bayes estimation on linear models of gene expression in limma package) (see also SI Appendix, Fig. S3). (D) Proteasome activity in total cell lysates from indicated MM parental and DYRK2 KO cells was measured with Suc-LLVY-AMC and normalized to total protein content. **P < 0.01 (parental vs. DYRK2 KO, unpaired Student's t test, mean ± SD from n = 3 independent experiments). Immunoblotting of the cell lysates was carried out with indicated antibodies. (E and F) Growth curves of myeloma cell lines MPC11 and 5TGM1-GFP (parental and DYRK2 KO). *P < 0.05, **P < 0.01 (2-way ANOVA, mean ± SD from n = 3 independent experiments). (G) Experimental flow for myeloma allograft survival study in H. (H) MPC11 parental or DYRK2 KO cells were injected i.v. into BALB/c mice (n = 10 per condition). Moribund mice with complete hindlimb paralysis were killed, and the Kaplan–Meier curve was derived (P value derived from survival curve comparison using Mantel–Cox Log-rank test). (I) Experimental flow for myeloma allograft study in J–M. (J) MPC11 parental or DYRK2 KO cells were injected i.v. into BALB/c mice (n = 6 per condition). Three weeks postinjection, mice were killed, and μCT imaging was carried out on formalin-fixed femur bone. Representative μCT image is shown. (Scale bar, 600 μm.) (K–M) Post-μCT the percentage of proximal femur trabecular bone volume over total volume (K), proximal femur bone mineral density (L), and proximal femur trabecular number (M) were quantified. *P < 0.05 (parental vs. DYRK2 KO, unpaired Student's t test, mean ± SD from n = 6 mice).
Next, we carried out proliferation assays to compare the rate of proliferation between parental and genome-edited MPC11 and 5TGM1-GFP cells. Consistent with our previous data, upon DYRK2 depletion, the rate of proliferation of both cell lines was significantly diminished over 5 d (Fig. 2 E and F).
DYRK2 promotes tumorigenesis in mouse xenograft models (17, 19). However, the exact role of DYRK2 in tumorigenesis in diverse cancer models is unclear (17, 25–29). To ascertain the role of DYRK2 in MM neoplastic progression and survival, we utilized a syngeneic MM model. MPC11 (Merwin plasma cell tumor 11) cells were derived from a plasmacytoma from the BALB/c strain of mice (30). We injected either parental or DYRK2-depleted MPC11 via tail vein into BALB/c mice of either sex, 10 mice for each cell strain. The main disease manifestation of MM has been termed CRAB for hypercalcemia, renal dysfunction, anemia, and bone degeneration (31, 32). MPC11-BALB/c syngeneic allografts are an established model for MM exhibiting bone degeneration 14 to 21 d postinjection (33). In our study, disease progression in mice was observed by movement difficulties leading to hindlimb paralysis. Moribund mice exhibiting complete hindlimb paralysis were killed, and Kaplan–Meier survival curves were generated to compare parental and DYRK2-depleted allograft-bearing mice. BALB/c mice with DYRK2-depleted MPC11 cells had a prolonged (>30%) delay in terminal MM disease progression (Fig. 2 G and H).
To study the effect of DYRK2 depletion on MM-mediated bone degeneration, MPC11 parental or DYRK2-depleted allograft-bearing BALB/c mice were killed 3 wk postinjection (Fig. 2I). Micro computed tomography (μCT) imaging was carried out to visualize cortical and trabecular bone structure on the formalin-fixed excised femurs. Quantitative analysis of the trabecular bone of the proximal femur region revealed that the averaged proximal femur trabecular parameters for DYRK2-depleted allograft-bearing mice were significantly higher in percent trabecular bone volume, with significantly higher bone mineral density and higher trabecular number than parental allografts (Fig. 2 J–M). Similar studies were carried out using the 5TGM1-GFP myeloma model (34). NSG mice were i.v. injected with either vector control or DYRK2-depleted 5TGM1-GFP cells (SI Appendix, Fig. S2 A and B). Mice were killed 3 wk postinjection, and femurs were excised. The femurs from mice bearing DYRK2-depleted cells had substantially fewer GFP+ foci (SI Appendix, Fig. S2 C and D), and femur cross section staining revealed significantly lower tartrate-resistant acid phosphatase activity signifying lower osteoclast activity (SI Appendix, Fig. S2E).
Thus, our data establish DYRK2 as a kinase that promotes MM cell proliferation and disease progression, which lead to accelerated malignancy and morbidity in vivo.
LDN192960 Is a Potent and Selective Inhibitor of DYRK2.
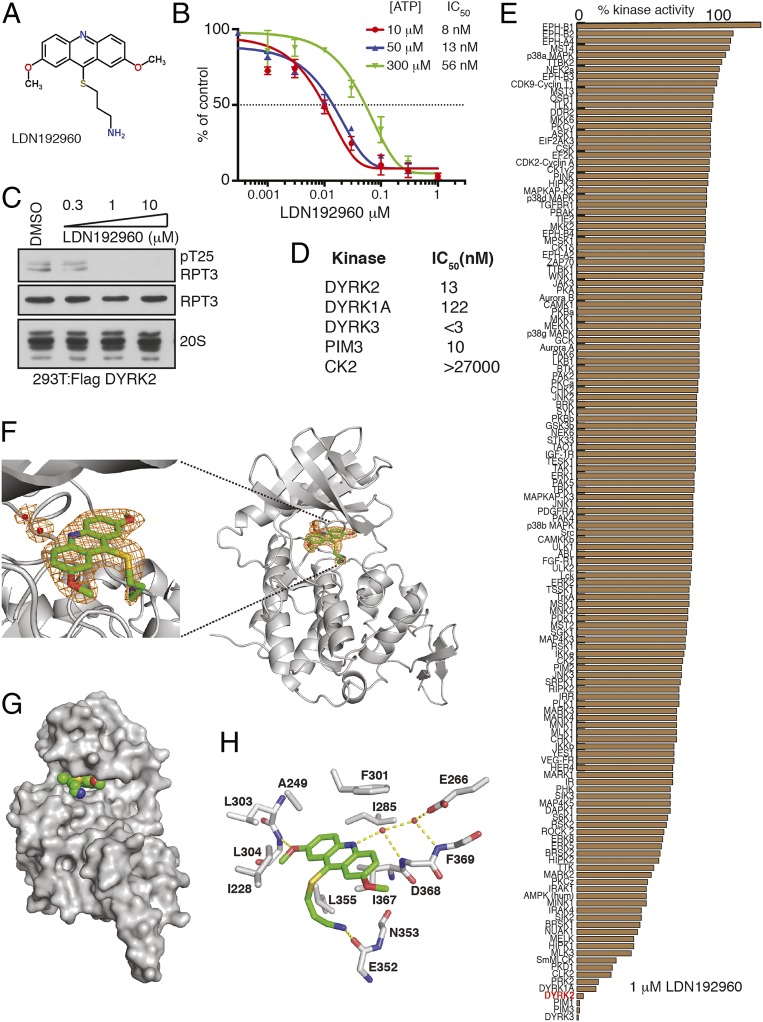
Because DYRK2 clearly plays an oncogenic role in TNBC and MM, we hypothesized that a small-molecule inhibitor for DYRK2 could potentially alleviate cancer progression. We recently reported that curcumin is a potent and selective inhibitor of DYRK2 (19). However, curcumin is highly hydrophobic, shows activity in vivo at high 300 mg/kg body weight, and in general is termed as an improbable drug (35). Hence, we identified LDN192960 (Fig. 3A) as a potential inhibitor of DYRK2. LDN192960 was developed initially as a Haspin inhibitor and was found to exhibit off-target effects on DYRK and PIM isoforms (36, 37). LDN192960 had an in vitro IC50 of 13 nM toward DYRK2 at 50 μM ATP and exhibits a 0.5- to 4-fold change in DYRK2 IC50 upon titration of 10 to 300 μM ATP (Fig. 3B). Kinetically, it exhibits a mixed mode of DYRK2 inhibition (SI Appendix, Fig. S4). To evaluate whether LDN192960 could suppress cellular DYRK2 activity, we treated HEK293T cells transiently overexpressing DYRK2-FLAG with increasing concentrations of LDN192960 and assessed RPT3 phosphorylation at Thr25, the major site of DYRK2 phosphorylation on the proteasome. We observed that LDN192960 treatment suppressed Thr25 RPT3 phosphorylation in a dose-dependent manner, with maximal effects observed at inhibitor concentrations of 1 to 10 μM (Fig. 3C). Furthermore, other than PIM and DYRK isoforms (Fig. 3D), LDN192960 does not inhibit any of the 130+ kinases tested to the same extent including other CMGC kinase family members that are closely related to the DYRKs (Fig. 3E).
Fig. 3.
LDN192960 is a potent and selective inhibitor of DYRK2. (A) Chemical structure of LDN192960. (B) In vitro IC50 of LDN192960 on purified DYRK2 over 3 different ATP concentrations. Results are means ± SD for triplicate reactions with similar results obtained in at least 1 other experiment (see also SI Appendix, Fig. S4). (C) HEK293T cells transiently expressing DYRK2 were treated with the indicated concentrations of LDN192960 over 2 h. Cells were lysed, and immunoblotting was carried out with the indicated antibodies. (D) Table listing IC50 values for indicated kinases for LDN192960. (E) Kinase profiling of LDN192960 at 1 μM was carried out against the panel of 140 kinases at the International Centre for Protein Kinase Profiling (http://www.kinase-screen.mrc.ac.uk/). (F) A composite omit map is contoured at 1.5 σ and shown as an orange mesh, revealing the presence of LDN192960 and 2 water molecules (red spheres). DYRK2 is shown as ribbons and colored in gray. PDB ID 6K0J. (G) LDN192960 occupies the ATP-binding pocket of DYRK2. DYRK2 is shown in a surface representation, and LDN192960 atoms are shown as spheres. (H) Detailed interactions between DYRK2 and LDN192960. Hydrogen bonds are shown as dashed lines (see also SI Appendix, Fig. S5 and Table S2).
To elucidate how LDN192960 specifically inhibits DYRK2, we crystallized DYRK2 in complex with LDN192960 and determined the structure at 2.35 Å (Fig. 3F and SI Appendix, Table S1). LDN192960 binds to the ATP-binding pocket of DYRK2 (Fig. 3G). It is sandwiched between several hydrophobic DYRK2 residues, including Ala249, Ile285, Phe301, Leu355, and Ile367 (Fig. 3H). One of the methoxy groups of LDN192960 forms a hydrogen bond with the main chain amide group Leu304. Two water molecules are also seen in the pocket and mediate a network of hydrogen bonds between LDN192960 and DYRK2 residues Glu266, Asp368, and Phe369. The amino side chain of LDN192960 extends out and forms a hydrogen bond with the main chain carbonyl group of Glu352 (Fig. 3H).
Together, these biochemical and structural data suggest that LDN192960 targets the ATP binding active site of DYRK2 and specifically inhibits DYRK2 kinase activity in vitro and in cells.
LDN192960 Impedes 26S Proteasome Activity in Cells and Induces Cytotoxicity.
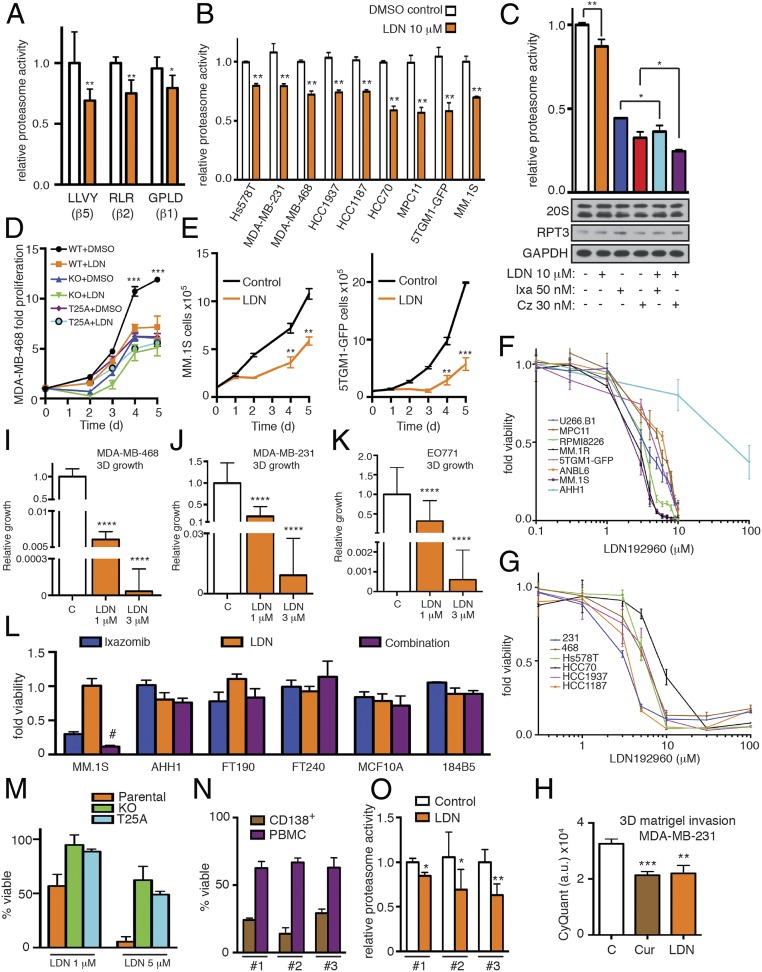
LDN192960 treatment results in significant reduction of all 3 (β1, β2, and β5) peptidase activities of the proteasome as seen by using specific fluorogenic peptide substrates (Fig. 4A). Moreover, MM and TNBC cells treated with LDN192960 exhibit a 20 to 40% inhibition of proteasome activity (Fig. 4B). Furthermore, a combination of LDN192960 with either ixazomib or carfilzomib exhibited a modest additive effect toward the inhibition of proteasome activity (Fig. 4C). Interestingly, 1 μM LDN192960 significantly affected the proliferation of MDA-MB-468 parental cells unlike the MDA-MB-468 DYRK2 KO or RPT3 Thr25Ala knock-in cells where 1 μM LDN192960 had a modest effect on proliferation (Fig. 4D). Similarly, LDN192960 inhibited cell proliferation in both MM cells tested (Fig. 4E). LDN192960 exhibits cytotoxicity to all MM cells tested with an EC50 between 1 and 10 μM; however, noncancerous AHH1 cells exhibited resistance >30 μM EC50 to LDN192960-mediated cytotoxicity (Fig. 4F). Similarly, all TNBC cells tested exhibited an LDN192960 EC50 of 1 to 10 μM (Fig. 4G). Treatment with 3 μM LDN192960 largely reduced the ability of MDA-MB-231 cells to invade in 3D matrigel invasion chemotaxis assays (Fig. 4H) to a similar extent as curcumin (19). Interestingly, 1 to 3 μM LDN192960 treatment markedly suppressed the ability of TNBC cells to form anchorage-independent 3D growth (Fig. 4 I–K). Next, we wanted to determine if LDN192960-ixazomib mediated additive impairment of proteasome activity (Fig. 4C) could have an effect on cancer cell viability. Interestingly, a marked additive cytotoxicity was observed in proteasome addicted MM cell line MM.1S, whereas noncancerous cells AHH1, FT190, FT240, MCF10A, and 184B5 did not exhibit additive cytotoxicity upon treatment with LDN192960 and ixazomib (Fig. 4L). Furthermore, 1 and 5 μM LDN192960 induced more pronounced cytotoxicity in MDA-MB-468 parental cells as compared to MDA-MB-468 DYRK2 KO or RPT3 Thr25Ala knock-in cells (Fig. 4M).
Fig. 4.
LDN192960 perturbs proteasome activity, induces cell death, and impedes proliferation and invasion. (A) Proteasome activity in total cell lysates from MDA-MB-468 cells with or without 10 μM LDN192960 treatment for 2 h was measured with Suc-LLVY-AMC or Ac-RLR-AMC or Ac-GPLD-AMC. *P < 0.05, **P < 0.01 (compared to control treated, 2-tailed paired Student’s t test, mean ± SD from n = 3 biological replicates). (B) Proteasome activity in total cell lysates from the indicated cells with or without 10 μM LDN192960 treatment for 2 h was measured with Suc-LLVY-AMC and normalized to total protein content. **P < 0.01 (compared to control treated for each cell line, ordinary 1-way ANOVA, mean ± SD from n = 3 independent experiments). (C) MDA-MB-231 cells were pretreated with indicated drugs (Ixa, Ixazomib; Cz, Carfilzomib; LDN, LDN192960) for 1 h, and proteasome activity was measured in cell lysates using Suc-LLVY-AMC. **P < 0.01, *P < 0.05 (compared to control treated, ordinary 1-way ANOVA, mean ± SD from n = 3 independent experiments). Immunoblotting of the cell lysates was carried out with indicated antibodies. (D) Fold proliferation of MDA-MB-468 WT, DYRK2 KO, and RPT3 Thr25Ala knock-in cells in presence of DMSO control or 1 μM LDN192960. ***P < 0.001 (2-way ANOVA, mean ± SD from n = 3 biological replicates). (E) Growth curves of MM.1S and 5TGM1-GFP control and LDN192960-treated cells (MM.1S 1 μM and 5TGM1-GFP 3 μM). **P < 0.01, ***P < 0.001 (2-way ANOVA, mean ± SD from n = 3 independent experiments). (F and G) LDN192960 induces cytotoxicity in all myeloma (F) and TNBC (G) tested with EC50 between 6 and 12 μM. Noncancerous AHH-1 cells had >30 µM EC50 for LDN192960. LDN192960 treatment was carried out for 36 h for this experiment. (H) Bar graph depicting cell invasion in a Matrigel transwell migration assay using DMSO treated or 3 μM curcumin or LDN192960-treated MDA-MB-231 cells. Data were acquired 18 h after seeding in upper chamber of 8 μm pore size transwells. Cells that invaded the Matrigel were quantified based on DNA content using CyQuant dye and data represented as arbitrary units (a.u.). **P < 0.01, ***P < 0.001 (compared to DMSO treated, 2-way ANOVA, mean ± SD from n = 2 independent experiments with triplicates in each). (I–K) Growth in 3D culture. MDA-MB-468 (I), MDA-MB-231 (J), and EO771 (K) cells were cultured in 1% methylcellulose for 2 to 4 wk in the presence of DMSO or LDN192960 at 1 or 3 μM. Areas of cell growth were quantified by analysis of images (5 per well). ****P < 0.0001 (compared to DMSO treated, 2-way ANOVA, mean ± SD from n = 2 independent experiments with triplicates in each). (L) Proteasome addicted MM.1S cells and noncancerous AHH1, FT190, FT240, MCF10A, and 184B5 cells were treated with ixazomib alone (MCF10A, 184B5, and MM.1S = 25 nM; FT190, FT240, and AHH1 = 50 nM) or with LDN192960 alone (FT190, FT240, MCF10A, and 184B5 = 5 μM; AHH1 = 10 μM; MM.1S = 3 μM) or the combination of ixazomib and LDN192960 for 24 h, and cell viability was analyzed by CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay kit. Viability of DMSO-treated cells was utilized as control. Data are represented as fold viability of DMSO-treated control for each cell line (# indicates statistical significance compared to each single drug treatment; 2-way ANOVA with multiple comparison, Fisher’s LSD test) (see also SI Appendix, Fig. S6). (M) MDA-MB-468 parental, DYRK2 KO, and RPT3 Thr25Ala knock-in cells were treated with or without LDN192960 at the indicated concentrations for 48 h. Cell viability was ascertained with CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay kit. Data were represented as percent viable compared to DMSO-treated cells. (N) Purified primary patient CD138+ myeloma and PBM cells were treated with 3 μM of LDN192960 or DMSO control for 24 h, and cell viability was ascertained with CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay kit. Data were represented as percent viable compared to DMSO-treated cells (PBMC from ATCC was used as control for patient 1 due to nonavailability of peripheral blood). (O) Proteasome activity in lysates from primary patient CD138+ myeloma cells treated with DMSO or 10 μM LDN192960 for 2 h was measured with Suc-LLVY-AMC. *P < 0.05 (compared to control treated, 2-tailed paired Student’s t test, mean ± SD from n = 3 independent replicates).
We next queried whether LDN192960 could in fact inhibit DYRK2-mediated proteasome function in primary patient-derived CD138+ MM cells and whether LDN192960-mediated cytotoxicity is specific for CD138+ MM cells compared to noncancerous peripheral blood mononuclear cells (PBMC). CD138+ MM cells were purified from the bone marrow aspirates of MM patients, and PBMCs were either purified from patient peripheral blood or purchased from ATCC. Postpurification, equal numbers of CD138+ MM cells were treated with DMSO or LDN192960. Interestingly, CD138+ cells were relatively more sensitive to 3 μM LDN192960 treatment for 24 h compared to the PBMC counterparts (Fig. 4N). Furthermore, upon treatment with 10 μM LDN192960 for 2 h, the CD138+ cell lysate exhibited a significant reduction of proteasome activity as compared to the DMSO control lysate (Fig. 4O).
Thus, LDN192960 impairs major hallmarks of cancer cells via partial inhibition of proteasome activity.
LDN192960 Delays MM Progression and Impedes Myeloma-Mediated Bone Degeneration.
We have shown that mice bearing MPC11 DYRK2 KO cells develop a much slower MM burden and consequently present better bone health relative to the parental counterparts (Fig. 2 F–I). To determine if LDN192960 treatment could inhibit bone degeneration in mice bearing parental MPC11 similar to the DYRK2 KO phenotype, we generated the syngeneic MPC11-BALB/c mouse allograft model. Parental cell-bearing mice were randomized into 2 groups for thrice weekly treatment with either PBS vehicle or 50 mg/kg LDN192960 (Fig. 5A). MPC11-DYRK2 KO–bearing mice were generated in parallel for comparison (no treatment). All mice were killed after 3 wk of treatment. The femurs were excised, formalin fixed, and imaged by μCT to visualize cortical and trabecular bone structure. Compared to vehicle-treated controls, both LDN192960-treated and DYRK2 KO-bearing mice exhibited comparable higher bone trabecular network (Fig. 5B). At the trabecular region, both the LDN192960-treated group and the DYRK2 KO-bearing group showed significantly higher trabecular bone volume fraction, bone mineral density, and trabecular number than the vehicle-treated cohort (Fig. 5 C–E).
Fig. 5.
LDN192960 impedes MM progression and delays myeloma-mediated bone degeneration. (A and B) MPC11 parental or genome edited (DYRK2 KO) cells were injected i.v. into inbred BALB/c mice (n = 12 for parental and n = 6 for DYRK2 KO). Two weeks postinjection, parental cell-bearing mice were randomized into 2 groups of n = 6 and treated with vehicle PBS or LDN192960 50 mg/kg thrice weekly. Two weeks posttreatment, the mice were killed, and formalin-fixed femur bones were imaged using μCT. Representative μCT image is shown. (Scale bar, 600 μm.) (C–E) Post-μCT the bone mineral density (C), the percentage of trabecular bone volume over total volume (D), and the trabecular number (E) were quantified for distal and proximal femurs. *P < 0.05, **P < 0.01 (compared to vehicle treated, 1-way ANOVA, mean ± SD, multiple comparisons with Fisher’s LSD test, from n = 6 mice each). (F) Experimental flow for MM.1S myeloma xenograft study in G. (G) MM.1S parental or genome-edited (DYRK2 KO) cells were injected s.c. into J:NU nude mice. Palpable tumor-bearing mice were randomized (16 d for parental and 28 d for DYRK2 KO) into 2 equal groups each and treated with vehicle control or LDN192960 3 times a week by i.p. injection, and tumor volume was measure twice a week. **P < 0.01 (compared to parental vehicle-treated group, 2-way ANOVA, mean ± SD from n = 5 mice each). (H) Post-42 d of injection, tumors were resected, and tumor weight was measured. ***P < 0.001, ns, not significant (compared to vehicle treated, ordinary 1-way ANOVA, mean ± SD from n = 5 mice each). (I) Proteasome activity in whole tumor lysates from parental vehicle or LDN192960-treated tumor-bearing mice was measured with Suc-LLVY-AMC. **P < 0.01 (compared to control treated, 2-tailed paired Student’s t test, mean ± SD from n = 3 different tumors each). (J) Experimental flow for 5TGM1-GFP myeloma allograft study in K. (K) 5TGM1-GFP cells were injected i.v. into J:NU mice and treated with 50 mg/kg LDN192960 or vehicle 14 d postinjection (n = 12 per condition). Moribund mice with complete hindlimb paralysis were killed, and Kaplan–Meier curve was derived (P value derived from survival curve comparison using Mantel–Cox Log-rank test). (L) Weight of mice before and after 3× 50 mg/kg LDN1929060 treatment (P value derived from Student’s t test, mean ± SD from n = 12 mice).
We know from our previous studies that DYRK2 KO cells do not exhibit further reduction of proteasome activity upon treatment with DYRK2 inhibitor (19). However, we understand that LDN192960 also inhibits PIM kinases, which have been known to be targets in MM (38, 39); hence, we wanted to ascertain the relative contributions of DYRK2 versus other kinases toward the in vivo effects observed with LDN192960 treatment. We generated an s.c. xenograft model by injecting MM.1S cells (parental or DYRK2 KO) into nude J:NU mice and investigated whether LDN192960 treatment could further reduce MM tumors in the absence of DYRK2 (Fig. 5F). Upon palpable tumor formation, parental and DYRK2 KO-bearing mice were randomized each into 2 groups and treated with vehicle or LDN192960 50 mg/kg. There was a dramatic reduction of tumor volume in parental xenografts upon LDN192960 treatment (Fig. 5 G and H) and a clear reduction of 26S proteasome activity in the tumor lysate as well (Fig. 5I). In DYRK2 KO-bearing xenografts, there was a modest but nonsignificant tumor reduction in volume and weight with LDN192960 treatment (Fig. 5 G and H). In addition, LDN192960 treatment improved survival of mice allografted with 5TGM1-GFP cells (Fig. 5 J and K) with mean survival 28.5 d as compared to 19 d for vehicle treated. Furthermore, there was no significant decrease in body weights of mice observed after 3× 50 mg/kg doses of LDN192960 (Fig. 5L).
Thus, our data indicate that LDN192960 can effectively impede MM cell growth in vivo via DYRK2 inhibition.
LDN192960 Inhibits Bortezomib-Resistant MM.
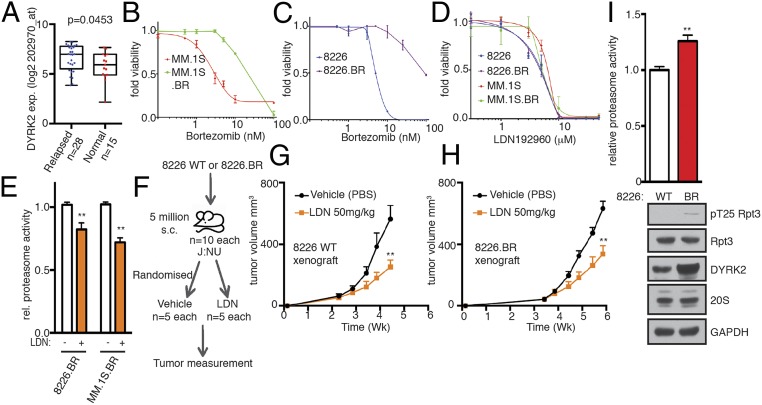
Bortezomib resistance is a major therapeutic impediment for MM patients (2). Various studies have reported diverse reasons for why malignant plasma cells develop bortezomib resistance. Plasma cells have been reported to develop bortezomib resistance by reducing misfolded protein levels and ER stress during differentiation, thereby uncoupling from the dependence on the proteasome (40). Other groups point to overexpression, polyploidy, and bortezomib-docking site mutations of the proteasome to be key for developing bortezomib resistance (10, 20–22). Interestingly, there was a modest yet statistically significant overexpression of DYRK2 in the relapsed MM patient sample dataset compared with newly diagnosed MM controls (Fig. 6A) along with RPT3 and PSMB5 which were overexpressed as well in the same GSE6477 dataset (SI Appendix, Fig. S3). Because DYRK2 is an upstream regulator of the 26S proteasome, we hypothesized that it may very well serve as a therapeutic target even in relapsed MM. To test this, we used bortezomib-resistant cell lines that were generated by adaptation to continuous proteasome inhibition (19, 41). RPMI8226.BR and MM.1S.BR cells exhibited >10- to 50-fold resistance to bortezomib compared with the respective bortezomib-sensitive parental RPMI8226 and MM.1S cells (Fig. 6 B and C). However, the EC50 for LDN192960 for either parental or bortezomib-resistant lines of the MM cells were identical (Fig. 6D), suggesting that LDN192960-mediated cytotoxicity is independent of bortezomib resistance. Furthermore, LDN192960 perturbed the proteasome activity of both 8226.BR and MM.1S.BR cells by 30 to 40% (Fig. 6E), similar to standard MM cells (Fig. 4B).
Fig. 6.
LDN192960 bypasses bortezomib resistance. (A) DYRK2 differential gene expression in human relapsed MM specimens and human normal tissue as available from a public database GSE6477 (relapsed vs. normal tissue, P value derived from empirical Bayes estimation on linear models of gene expression in limma package) (see also SI Appendix, Fig. S3). (B and C) Bortezomib EC50 for parental MM.1S and bortezomib-resistant MM.1S.BR cells (B) and for parental RPMI8226 and bortezomib-resistant 8226.BR cells (C). (D) LDN192960 EC50 for parental MM.1S, parental RPMI8226, MM.1S.BR, and 8226.BR cells. (E) Proteasome activity in total cell lysates from the indicated bortezomib-resistant cells with or without 10 μM LDN192960 treatment for 2 h was measured with Suc-LLVY-AMC and normalized to total protein content. **P < 0.01 (compared to control treated for each cell line, ordinary 1-way ANOVA, mean ± SD from n = 3 independent experiments). (F) Experimental flow for myeloma xenograft study in G and H. (G and H) The 8226 parental (G) or 8226.BR (H) cells were injected s.c. into J:NU nude mice. Palpable tumor-bearing mice were randomized (16 d for parental and 23 d for 8226.BR) into 2 equal groups each and treated with vehicle control or LDN192960 3 times a week by i.p. injection, and tumor volume was measured twice a week (n = 5 per condition). **P < 0.01 (compared to vehicle treated, 2-way ANOVA, mean ± SD, from n = 5 mice). (I) Proteasome activity in total cell lysates from 8226 WT or 8226.BR was measured with Suc-LLVY-AMC. **P < 0.01 (8226 WT vs. 8226.BR, unpaired Student’s t test, mean ± SD from n = 3 independent experiments). Immunoblotting of the cell lysates was carried out with indicated antibodies.
To further confirm the antitumor efficacy of LDN192960 in targeting bortezomib-resistant tumors, parental and bortezomib-resistant RPMI8226 cells were injected s.c. in J:NU mice (Fig. 6F). Mice with palpable tumors were randomized into 2 groups and injected intraperitoneally thrice weekly with either vehicle control or 50 mg/kg LDN192960. Indeed, LDN192960 treatment significantly reduced tumor burden in bortezomib-resistant RPMI8226.BR lines after 2 wk of treatment comparable to the level in parental RPMI8226 (Fig. 6 G and H). Interestingly, 8226.BR cells had a significantly higher proteasome activity than 8226 WT cells without any change in 20S or 19S RPT3 protein levels (Fig. 6I). However, 8226.BR cells had markedly higher DYRK2 and pT25 RPT3 signals which possibly contribute to the higher proteasome activity (Fig. 6I).
Thus, our data suggest that LDN192960 induces cytotoxicity in bortezomib-resistant MM, both in cells and in vivo.
LDN192960 Markedly Reduces TNBC Tumor Burden.
Recent work has shown that simultaneous inhibition of both β5 chymotryptic-like and β2 tryptic-like subunits of the proteasome is essential to sensitize TNBC (15). Since LDN192960-mediated DYRK2 inhibition perturbed all 3 core peptidase activities of the proteasome (Fig. 4A) and genetic depletion of DYRK2 led to reduction of TNBC tumors in mice (Fig. 1 G–I), we wanted to test the antitumor efficacy of the drug at targeting the solid tumor in vivo.
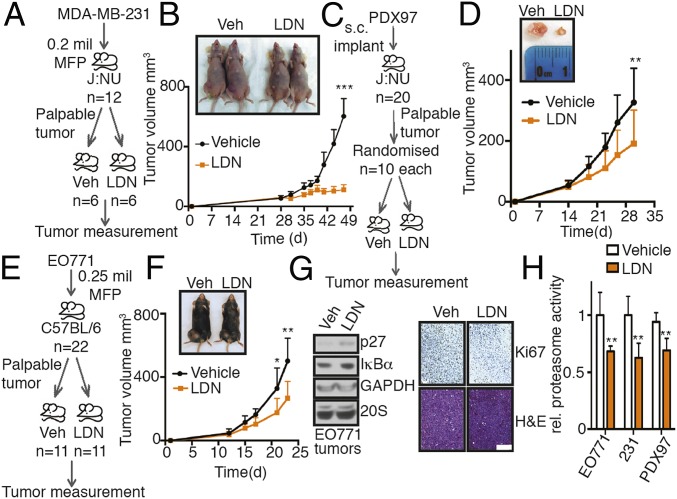
We evaluated the effect of LDN192960 in 3 different TNBC allo/xenograft models. Parental MDA-MB-231 mice with palpable mammary tumors were randomized into 2 groups and treated with either vehicle control or 50 mg/kg LDN192960 (Fig. 7A). Interestingly, there was a very dramatic tumor reduction after 2 wk of LDN192960 treatment, manifested by slower tumor growth (Fig. 7B) and reduced proteasome activity in the resected tumor lysates (Fig. 7H). To test the efficacy of LDN192960 in a primary TNBC patient-derived sample, we developed a patient-derived xenograft (PDX) model from fresh frozen tissues acquired from the University of California San Diego (UCSD) Moores Cancer Center Biorepository (Fig. 7C). PDX97 was derived from the primary tumor with grade 3 invasive triple-negative ductal carcinoma. Treatment of 50 mg/kg LDN192960 significantly reduced the tumor burden of J:NU mice bearing PDX97 tumors (Fig. 7D), along with a reduced proteasome activity in the resected tumor lysates (Fig. 7H).
Fig. 7.
LDN192960 perturbs TNBC progression. (A) Experimental flow for TNBC xenograft study in B. (B) MDA-MB-231 parental cells were injected into the mammary fat pad of J:NU nude mice. Palpable tumor-bearing mice were randomized into 2 equal groups and treated with vehicle control or LDN192960 3 times a week by i.p. injection, and tumor volume was measured twice a week (n = 6 per condition). ***P < 0.001 (compared to vehicle treated, 2-way ANOVA, mean ± SD, from n = 6 mice). (C) Experimental flow for TNBC PDX study in D. (D) Patient-derived PDX97 tumor specimens were surgically implanted s.c. into J:NU mice. Palpable tumor-bearing mice were randomized into 2 equal groups and were treated with vehicle control or LDN192960 as in B (n = 10 per condition). **P < 0.01 (compared to vehicle treated, 2-way ANOVA, mean ± SD, from n = 10 mice). (E) Experimental flow for TNBC allograft study in F. (F) EO771 parental cells were injected into the mammary fat pad of C57BL/6 immunocompetent mice. Palpable tumor-bearing mice were randomized into 2 equal groups and were treated with vehicle control or LDN192960 as in B (n = 11 per condition). **P < 0.01, *P < 0.05 (compared to vehicle treated, 2-way ANOVA, mean ± SD, from n = 11 mice). (G) Immunoblotting with indicated antibodies in tumor lysates from F and histological examination of consecutive sections of the fixed tumors from F with Ki67 and hematoxylin/eosin staining were carried out. (Scale bar, 100 μm.) (H) Proteasome activity in whole tumor lysates from vehicle or LDN192960-treated tumor-bearing mice from B, D, and F were measured with Suc-LLVY-AMC. **P < 0.01 (compared to control treated, 2-way ANOVA, mean ± SD from n = 3 different tumors each).
To further confirm this in a syngeneic TNBC model, we employed the basal-like TNBC cell line EO771, which was derived from a spontaneously developed medullary breast adenocarcinoma in C57BL/6 mice (42). We injected EO771 cells into the mammary fat pad of virgin female C57BL/6J mice and randomized palpable tumor-bearing mice into 2 groups followed by treatment with LDN192960 or DMSO as previously described (Fig. 7E). After 2 wk of treatment, the tumor growth rate of the LDN192960-treated cohort was significantly slower than vehicle-treated (Fig. 7F) with a marked reduction in Ki67 stained proliferating cells (Fig. 7G). We also observed a modest increase in proteasome substrates p27 and IκBα protein levels in LDN192960-treated tumor lysates (Fig. 7G) and a reduced proteasome activity in the tumor lysates (Fig. 7H).
Thus, LDN192960-mediated inhibition of DYRK2 significantly alleviates tumor burden in standard and PDX TNBC models, and partial inhibition of the proteasome contributes to this anti-TNBC tumor activity.
Discussion
The ubiquitin–proteasome system has long been the focus for development of clinical therapeutics in cancer, including but not limited to targeting deubiquitinases (43), manipulating E3 ligases (44), and inhibiting 19S subunits (45) and the 20S core (11) of the proteasome. Our work opens a possibility of inhibiting kinases playing a vital role in molecular regulation of the 26S proteasome. With over 300 phosphorylation sites on a 4.5-MDa complex with nearly 40 subunits, identifying kinases and phosphatases regulating the 26S proteasome could be the next novel paradigm for development of alternate mechanisms of targeting proteasome-inhibitor resistant or relapsed neoplasias.
Our current work proposes an alternate mechanism of targeting proteasome-addicted malignancies via perturbing upstream regulators of the 26S proteasome. We established DYRK2 as a bona fide proteasome kinase and showed that DYRK2 inhibition leads to impediment of the cell cycle via accumulation of proteasome substrates and proapoptotic factors leading to tumor regression (17, 19). In this study, we report that DYRK2 is overexpressed and promotes both TNBC (Fig. 1) and MM (Fig. 2) progression. We further report LDN192960 as a potent and selective small-molecule inhibitor of DYRK2 (Fig. 3) that induced cytotoxicity and growth inhibition in MM and TNBC cells with relatively modest effects in noncancerous lines (Fig. 4). LDN192960 restricted MM-mediated bone degeneration (Fig. 5) and bypassed bortezomib resistance in MM (Fig. 6). LDN192960 further reduced tumor burden in TNBC mouse allo/xenografts, and LDN192960-treated tumors exhibited partial inhibition of proteasome activity and a modest accumulation of proteasome substrates p27 and IκBα (Fig. 7).
Targeting the proteasome has been a major success story in MM therapeutics with multiple FDA-approved drugs currently used as front-line defenses (8). However, high incidence of proteasome-inhibitor resistance leaves patients with limited clinical options (11). The average 5-y survival rate for MM patients is only 50% (NCI), which highlights the need for novel MM therapeutics. Our work provides inroads into pharmacologically alleviating MM progression via inhibition of DYRK2. DYRK2 is overexpressed in both newly diagnosed MM and relapsed MM (Figs. 2A and 6A and SI Appendix, Fig. S3A), and inhibiting DYRK2 delays myeloma progress, even in bortezomib-resistant cases (Fig. 6). In addition, DYRK2 depletion/inhibition halts bone degeneration and promotes overall survival (Figs. 2 and 5 and SI Appendix, Fig. S2).
Recent work has shown that simultaneous inhibition of both β5 chymotryptic-like and β2 tryptic-like subunits of the proteasome is essential to sensitize TNBC (15). Since DYRK2 has higher expression in TNBC (Fig. 1 A and B) and can indirectly regulate all 3 peptidase activities of the proteasome (Fig. 4A), inhibiting the kinase could provide a novel mechanism of proteasome-directed therapeutics which has largely failed to date in TNBC clinical trials (13). The success of this concept was clearly demonstrated by the dramatic reduction in tumor volume upon genetic depletion (Fig. 1 G–I) or pharmacological inhibition of DYRK2 in TNBC allo/xenograft models (Fig. 7).
We further show that DYRK2 can be readily inhibited in vivo as the DYRK2 inhibitor, LDN192960, directly phenocopies DYRK2 genetic depletion and alleviates MM (Figs. 2 J–M and 5 B–E) and TNBC (Figs. 1 G–I and 7) progression. LDN192960 is an acridine-orange derivative (36, 37) and largely functions via specific inhibition of DYRK2 in vitro (Fig. 3), in cells (Figs. 4 A–C and O and 6E), and in vivo (Figs. 5I and 7H). LDN192960 has a 13-nM IC50 at 50 μM ATP in vitro and binds to the ATP binding site of DYRK2 via multiple hydrophobic interactions (Fig. 3 B and F–H) exhibiting a mixed mechanism of DYRK2 inhibition (SI Appendix, Fig. S4) that requires further biochemical dissection beyond the scope of this manuscript. In particular, Ile285 and Ile367 are involved in making strong hydrophobic/van der Waals interactions with LDN192960 and play crucial roles in determining the binding specificity because both residues are replaced by valines in DYRK1 (Val222 and Val306) (SI Appendix, Fig. S5). Indeed, LDN192960 showed a 9- to 10-fold selectivity toward DYRK2 compared to DYRK1A (Fig. 3D). The drug did exhibit in vitro off-target effects on PIM isoforms and DYRK3 (Fig. 3 D and E), both of which have been reported to be potentially oncogenic (38, 39, 46). However, tumors derived from DYRK2 KO cells did not show a significant reduction upon LDN192960 treatment (Fig. 5 G–I). DYRK2 phosphorylates a single site, Thr25, on the N terminus of RPT3 (Fig. 3C), and ablating this phosphorylation perturbs 20 to 40% proteasome activity in both cells (Figs. 1E, 4 A–C and O, and 6E) and tumors (Figs. 5I and 7H). LDN192960 did not affect the rate of proliferation (Fig. 4D) or percent viability of cells (Fig. 4M) harboring either DYRK2 depletion or RPT3 Thr25Ala knock-in as compared to parental lines. Interestingly, LDN192960 treatment led to a significant accumulation (****P < 0.001) of asynchronous cells into the G2-M stage of the cell cycle consistent with our previous studies with DYRK2 KO cells (SI Appendix, Fig. S6). Furthermore, the additive effect of LDN192960 and the proteasome inhibitors carfilzomib or Ixazomib on proteasome activity in MDA-MB-468 cells (Fig. 4C) was not seen in the cell viabilities of noncancerous myeloid (AHH1), fallopian tube epithelial (FT190 and FT240), or mammary (MCF10A and 184B5) lines (Fig. 4L). Interestingly, CD138+ primary patient-derived myeloma cells were more sensitive to LDN192960 than matched normal PBMCs isolated from peripheral blood (Fig. 4N). Together, these data suggest a significant portion of LDN192960’s anticancer activity is through the DYRK2-proteasome inhibition especially in proteasome addicted cancers. However, higher concentrations of LDN192960 do affect the viability of all cells tested including DYRK2 KO/RPT3 KI (Fig. 4 F and M); hence, we cannot completely rule out potential off-target effects on other DYRK2 substrates or other DYRK/PIM isoforms contributing to antitumor effects exhibited by higher concentrations of LDN192960. Thus, LDN192960 has a more pronounced cytotoxic effect toward cancer cells, relative to noncancerous ones (Figs. 4 F, L and N and 6 B–D), and dramatically perturbs 3D invasion through Matrigel matrix (Fig. 4H) and anchorage-independent 3D growth (Fig. 4 I–K) suggesting a potential therapeutic window for specific cancer targeting.
It is quite interesting how phosphorylation of a single site (Thr25) on RPT3 by DYRK2 could perturb 20 to 40% of the 26S proteasome activity. Cryo-EM structures reveal that the 19S proteasome-regulatory particle is conformationally dynamic (47, 48), and binding of the ubiquitinated substrate induces broad rearrangement in the regulatory particle that aligns multiple subunits including the deubiquitinase RPN11 and hexameric-ATPase ring with the entrance to the 20S core (48). This rearrangement is concomitant upon a 25° rotation of RPN2 about the RPT3-RPT6 coiled-coil region and allows the proteasome to enter a substrate-induced degradation mode (48). Since pThr25 is at the N terminus of the coiled coil, we speculate that it may facilitate these molecular motions by providing interactions between RPN2 and RPT3 that favor structural rearrangement toward the degradation-competent mode of the proteasome.
The role of DYRK2 in cancer has been controversial especially in breast and liver cancers (25–29). We do appreciate that DYRK2 function could be cancer type and cell type specific, yet in the context of breast and liver hepatocellular carcinoma cohorts, we saw strong DYRK2 overexpression (SI Appendix, Fig. S1 and Table S1). In fact, with a few exceptions, DYRK2 is significantly overexpressed (*P < 0.05) in many of the cancer types listed in TCGA as compared to matched normal tissue controls (SI Appendix, Fig. S1 and Table S1). In addition, IHC staining in patient TNBC samples with adjacent normal tissue showed a stronger IHC signal for DYRK2 in diseased state compared to normal (Fig. 1A). Various pan-DYRK inhibitors also exhibit antitumor activity (19, 49, 50), which further corroborates the role of DYRK2 as tumor promoting.
In fact, both DYRK2 protein expression and proteasome activity were significantly increased in bortezomib-resistant myeloma 8226.BR compared to the parental 8226.WT without any increase in proteasome levels (Fig. 6I). The stoichiometry of pThr25 signal is quite low (17); however, higher DYRK2 levels in 8226.BR cells led to a stronger endogenous pThr25 signal in these cells further suggesting an interesting correlation between DYRK2 and proteasome activation in neoplasia. Further work is needed to establish whether DYRK2 is a key player driving bortezomib resistance disease recurrence.
In the United States alone, nearly 4,000 TNBC and over 12,000 MM (the majority with bortezomib resistance) patient deaths were projected by the American Cancer Society 2018 Facts and Figures. Development of DYRK2 inhibitors with high in vivo potency and selectivity could indeed pave the way for a new option in proteasome-based therapeutics. Targeting DYRK2 could potentially be beneficial in treating other 26S proteasome-addicted neoplasia like mantle cell lymphoma where a partial inhibition of the 3 core peptidase activities of the proteasome could specifically tip the scales for cancer and thereby alleviate tumor burden in patients often with relapsed–refractory disease.
Materials and Methods
For details of general methods, proteasome assays, cell-based assays, mouse experiments, CD138+ myeloma and PBMC purification, histology, crystallography, kinase specificity screening, bioinformatic analyses, statistics and data representation, key resources table, and other methods, please refer to SI Appendix. Crystallography coordinates are deposited in the Protein Data Bank (PDB; ID 6K0J), and all data in the manuscript are available freely to readers. Bone marrow aspirates and matched peripheral blood samples were obtained from Health Insurance Portability and Accountability Act (HIPAA)-compliant deidentified consenting patients in accordance with Institutional Review Board approved protocols at University of California San Diego (UCSD).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Health DK018849-41 (to J.E.D.), DK018024-43 (to J.E.D.), CA184898 (to J.P.M.), CA121941 (to J.P.M.), GM074024 (to J.P.M.), CA194107 (to J.P.M.), CA210004 (to J.P.M.), National Library of Medicine training grant T15LM011271 (to O.C.), National Cancer Institute Training Grant CA009523 (to J.E.M.), the Mary Kay Ash Breast Cancer Grant 047.16 (to J.E.D.), UCSD-Moores Cancer Centre translational and clinical pilot project Award (to J.E.D. and C.C.), Cancer Research UK C52419/A22869 (to L.d.l.V.), National Key Research and Development Program of China 2017YFA0505200 (to J.X. and X.L.) and 2016YFC0906000 (to J.X.), and the National Natural Science Foundation of China Grant 91853202, 21625201, 21661140001, and 21521003 (to X.L.), the Beijing Outstanding Young Scientist Program BJJWZYJH01201910001001 (to X.L.), and the Beijing Municipal Science and Technology Project Grant Z171100000417001 (to X.L.). We thank Drs. Carolyn Worby and Kun-Liang Guan for valuable input, Drs. Alexandra Newton (UCSD), Michael Karin (UCSD), Richard Klemke (UCSD), and Robert Rottapel (University of Toronto) for providing cell lines, Alara Sedef Tuncer for experimental help, and Vibhav Nadkarni for scientific illustrations. We thank the Kinase-Screen team at International Centre for Protein Kinase Profiling, Dundee, UK. We also thank Dr. Richard Schwab, Sharmeela Kaushal, and Joseph Maroge at UCSD Moores Cancer Center for providing patient-derived samples.
Footnotes
The authors declare no competing interest.
Data deposition: Crystallography, atomic coordinates, and factors have been deposited in the Protein Data Bank (PDB ID 6K0J).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912033116/-/DCSupplemental.
References
- 1.Wright J. H., A case of multiple myeloma. J. Boston Soc. Med. Sci. 4, 195–204.5 (1900). [PMC free article] [PubMed] [Google Scholar]
- 2.Goldberg A. L., Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 199, 583–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchini G., Balko J. M., Mayer I. A., Sanders M. E., Gianni L., Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 13, 674–690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrocca F., et al. , A genome-wide siRNA screen identifies proteasome addiction as a vulnerability of basal-like triple-negative breast cancer cells. Cancer Cell 24, 182–196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coux O., Tanaka K., Goldberg A. L., Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Besche H. C., Peth A., Goldberg A. L., Getting to first base in proteasome assembly. Cell 138, 25–28 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touzeau C., Maciag P., Amiot M., Moreau P., Targeting Bcl-2 for the treatment of multiple myeloma. Leukemia 32, 1899–1907 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Kunacheewa C., Orlowski R. Z., New drugs in multiple myeloma. Annu. Rev. Med. 70, 521–547 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Dimopoulos M. A., et al. , Carfilzomib or bortezomib in relapsed or refractory multiple myeloma (ENDEAVOR): An interim overall survival analysis of an open-label, randomised, phase 3 trial. Lancet Oncol. 18, 1327–1337 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Oerlemans R., et al. , Molecular basis of bortezomib resistance: Proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood 112, 2489–2499 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Orlowski R. Z., Kuhn D. J., Proteasome inhibitors in cancer therapy: Lessons from the first decade. Clin. Cancer Res. 14, 1649–1657 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Kumar S. K., et al. , Natural history of relapsed myeloma, refractory to immunomodulatory drugs and proteasome inhibitors: A multicenter IMWG study. Leukemia 31, 2443–2448 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Engel R. H., et al. , A phase II study of single agent bortezomib in patients with metastatic breast cancer: A single institution experience. Cancer Invest. 25, 733–737 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Smith D. C., et al. , Phase 1 study of ixazomib, an investigational proteasome inhibitor, in advanced non-hematologic malignancies. Invest. New Drugs 33, 652–663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weyburne E. S., et al. , Inhibition of the proteasome β2 site sensitizes triple-negative breast cancer cells to β5 inhibitors and suppresses Nrf1 activation. Cell Chem. Biol. 24, 218–230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo X., et al. , UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proc. Natl. Acad. Sci. U.S.A. 108, 18649–18654 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo X., et al. , Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat. Cell Biol. 18, 202–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.VerPlank J. J. S., Lokireddy S., Zhao J., Goldberg A. L., 26S Proteasomes are rapidly activated by diverse hormones and physiological states that raise cAMP and cause Rpn6 phosphorylation. Proc. Natl Acad. Sci. USA 116, 4228–4237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee S., et al. , Ancient drug curcumin impedes 26S proteasome activity by direct inhibition of dual-specificity tyrosine-regulated kinase 2. Proc. Natl. Acad. Sci. U.S.A. 115, 8155–8160 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balsas P., Galán-Malo P., Marzo I., Naval J., Bortezomib resistance in a myeloma cell line is associated to PSMβ5 overexpression and polyploidy. Leuk. Res. 36, 212–218 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Wei W., et al. , PSMB5 is associated with proliferation and drug resistance in triple-negative breast cancer. Int. J. Biol. Markers 33, 102–108 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Barrio S., et al. , Spectrum and functional validation of PSMB5 mutations in multiple myeloma. Leukemia 33, 447–456 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Chng W. J., et al. , Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 67, 2982–2989 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Tiedemann R. E., et al. , Kinome-wide RNAi studies in human multiple myeloma identify vulnerable kinase targets, including a lymphoid-restricted kinase, GRK6. Blood 115, 1594–1604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imawari Y., et al. , Downregulation of dual-specificity tyrosine-regulated kinase 2 promotes tumor cell proliferation and invasion by enhancing cyclin-dependent kinase 14 expression in breast cancer. Cancer Sci. 109, 363–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mimoto R., Imawari Y., Hirooka S., Takeyama H., Yoshida K., Impairment of DYRK2 augments stem-like traits by promoting KLF4 expression in breast cancer. Oncogene 36, 1862–1872 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Mimoto R., Nihira N. T., Hirooka S., Takeyama H., Yoshida K., Diminished DYRK2 sensitizes hormone receptor-positive breast cancer to everolimus by the escape from degrading mTOR. Cancer Lett. 384, 27–38 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Mimoto R., et al. , DYRK2 controls the epithelial-mesenchymal transition in breast cancer by degrading Snail. Cancer Lett. 339, 214–225 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Yokoyama-Mashima S., et al. , Forced expression of DYRK2 exerts anti-tumor effects via apoptotic induction in liver cancer. Cancer Lett. 451, 100–109 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Laskov R., Scharff M. D., Synthesis, assembly, and secretion of gamma globulin by mouse myeloma cells. I. Adaptation of the Merwin plasma cell tumor-11 to culture, cloning, and characterization of gamma globulin subunits. J. Exp. Med. 131, 515–541 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson K. C., Progress and paradigms in multiple myeloma. Clin. Cancer Res. 22, 5419–5427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landgren O., Rajkumar S. V., New developments in diagnosis, prognosis, and assessment of response in multiple myeloma. Clin. Cancer Res. 22, 5428–5433 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferguson V. L., et al. , Effect of MPC-11 myeloma and MPC-11 + IL-1 receptor antagonist treatment on mouse bone properties. Bone 30, 109–116 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Oyajobi B. O., et al. , Detection of myeloma in skeleton of mice by whole-body optical fluorescence imaging. Mol. Cancer Ther. 6, 1701–1708 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson K. M., et al. , The essential medicinal chemistry of curcumin. J. Med. Chem. 60, 1620–1637 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuny G. D., et al. , Structure-activity relationship study of acridine analogs as haspin and DYRK2 kinase inhibitors. Bioorg. Med. Chem. Lett. 20, 3491–3494 (2010). [PMC free article] [PubMed] [Google Scholar]
- 37.Cuny G. D., et al. , Structure-activity relationship study of beta-carboline derivatives as haspin kinase inhibitors. Bioorg. Med. Chem. Lett. 22, 2015–2019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiasa M., et al. , Pim-2 kinase is an important target of treatment for tumor progression and bone loss in myeloma. Leukemia 29, 207–217 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Wang H. L., et al. , Discovery of ( R)-8-(6-Methyl-4-oxo-1,4,5,6-tetrahydropyrrolo[3,4- b]pyrrol-2-yl)-3-(1-methylcyclopropyl)-2-((1-methylcyclopropyl)amino)quinazolin-4(3 H)-one, a potent and selective Pim-1/2 kinase inhibitor for hematological malignancies. J. Med. Chem. 62, 1523–1540 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Leung-Hagesteijn C., et al. , Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell 24, 289–304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salem K., McCormick M. L., Wendlandt E., Zhan F., Goel A., Copper-zinc superoxide dismutase-mediated redox regulation of bortezomib resistance in multiple myeloma. Redox Biol. 4, 23–33 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugiura K., Stock C. C., Studies in a tumor spectrum. I. Comparison of the action of methylbis (2-chloroethyl)amine and 3-bis(2-chloroethyl)aminomethyl-4-methoxymethyl -5-hydroxy-6-methylpyridine on the growth of a variety of mouse and rat tumors. Cancer 5, 382–402 (1952). [DOI] [PubMed] [Google Scholar]
- 43.Pinto-Fernandez A., Kessler B. M., DUBbing cancer: Deubiquitylating enzymes involved in epigenetics, DNA damage and the cell cycle as therapeutic targets. Front. Genet. 7, 133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto K. M., et al. , Protacs: Chimeric molecules that target proteins to the Skp1-cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U.S.A. 98, 8554–8559 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., et al. , Capzimin is a potent and specific inhibitor of proteasome isopeptidase Rpn11. Nat. Chem. Biol. 13, 486–493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wippich F., et al. , Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Dong Y., et al. , Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565, 49–55 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matyskiela M. E., Lander G. C., Martin A., Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 20, 781–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uhl K. L., Schultz C. R., Geerts D., Bachmann A. S., Harmine, a dual-specificity tyrosine phosphorylation-regulated kinase (DYRK) inhibitor induces caspase-mediated apoptosis in neuroblastoma. Cancer Cell Int. 18, 82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li C., et al. , Anticancer activities of harmine by inducing a pro-death autophagy and apoptosis in human gastric cancer cells. Phytomedicine 28, 10–18 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.