Significance
While telomerase has been extensively studied, its core RNA component (TR) is extremely divergent in sequence and biogenesis pathways, which has hindered our understanding of the evolutionary pathway from the small ciliate TR to the more complex fungal and vertebrate TRs. Here we report a conserved structure of the authentic TR from Arabidopsis thaliana termed AtTR, which is different from an RNA previously described as the templating telomerase RNA, AtTER1. This breakthrough establishes the correct A. thaliana TR and enables identification of TR homologs across the plant kingdom. Structural and functional analyses of AtTR reveals chimeric structural features similar to those of TRs from ciliates and multicellular eukaryotes, supporting the idea that all TRs are homologous, sharing a common ancestor.
Keywords: ribonucleoprotein, reverse transcriptase, pseudoknot, telomeres
Abstract
Telomerase is essential for maintaining telomere integrity. Although telomerase function is widely conserved, the integral telomerase RNA (TR) that provides a template for telomeric DNA synthesis has diverged dramatically. Nevertheless, TR molecules retain 2 highly conserved structural domains critical for catalysis: a template-proximal pseudoknot (PK) structure and a downstream stem-loop structure. Here we introduce the authentic TR from the plant Arabidopsis thaliana, called AtTR, identified through next-generation sequencing of RNAs copurifying with Arabidopsis TERT. This RNA is distinct from the RNA previously described as the templating telomerase RNA, AtTER1. AtTR is a 268-nt Pol III transcript necessary for telomere maintenance in vivo and sufficient with TERT to reconstitute telomerase activity in vitro. Bioinformatics analysis identified 85 AtTR orthologs from 3 major clades of plants: angiosperms, gymnosperms, and lycophytes. Through phylogenetic comparisons, a secondary structure model conserved among plant TRs was inferred and verified using in vitro and in vivo chemical probing. The conserved plant TR structure contains a template-PK core domain enclosed by a P1 stem and a 3′ long-stem P4/5/6, both of which resemble a corresponding structural element in ciliate and vertebrate TRs. However, the plant TR contains additional stems and linkers within the template-PK core, allowing for expansion of PK structure from the simple PK in the smaller ciliate TR during evolution. Thus, the plant TR provides an evolutionary bridge that unites the disparate structures of previously characterized TRs from ciliates and vertebrates.
Many noncoding RNAs (ncRNAs) function as integral components of ribonucleoprotein (RNP) complex enzymes that govern such cellular processes as translation, RNA splicing, and telomere maintenance (1). The telomerase RNA (TR or TER) assembles with the telomerase reverse transcriptase (TERT) protein to form the catalytic core of an enzyme that maintains telomere function and genome integrity by continually adding telomeric DNA repeats onto chromosome ends (2). TR contains a template for the synthesis of G-rich telomere repeat arrays catalyzed by TERT. In addition, TR harbors highly conserved structural domains that serve as a scaffold for binding accessory proteins that facilitate RNP biogenesis, engagement with the chromosome terminus, and regulation of telomerase enzyme activity (3).
The essential role of telomerase in telomere maintenance is universally conserved across Eukarya, except for a small group of insect species that evolved a retrotransposon-mediated mechanism (4). Nevertheless, key aspects of the telomerase RNP have diverged dramatically, including the sequence and length of TR, the protein composition of the holoenzyme, and the mechanism of RNP maturation (5). For example, TR genes in ciliated protozoa encode relatively small RNAs (140 to 210 nt long) that are transcribed by RNA polymerase III (Pol III) (6, 7). The La-related protein P65 in Tetrahymena recognizes the 3′ poly-U tail of TR and bends the RNA to facilitate telomerase RNP assembly (8, 9). In contrast, fungi maintain much larger TR molecules (900 to 2,400 nt) that are transcribed by RNA polymerase II (Pol II) (3). The 3′ end maturation of fungal TRs requires components of the canonical snRNA biogenesis pathway and results in RNP assembly with Sm and Lsm proteins (10, 11). Like fungi, vertebrates also use Pol II to transcribe a TR with size ranging from 312 to 559 nt (12). However, vertebrate telomerase RNP processing and biogenesis proceeds via a small nucleolar RNA (snoRNA) maturation pathway (13). In vertebrates, a highly conserved structural motif in the 3′ H/ACA domain of TR binds the protein components of the H/ACA snoRNP (Dyskerin, NOP10, NHP2, and GAR1), which then protect the 3′ end of the mature TR from exonuclease degradation (14–16).
Within TR, 2 conserved domains are critical for telomerase catalysis (17). The first is the template-pseudoknot (PK) domain (T-PK) that bears a single-stranded template region typically corresponding to 1.5 to 2 copies of the telomeric repeat (3). The 5′ boundary of the TR template is defined by a template boundary element (TBE) that promotes polymerase fidelity by preventing incorporation of nontelomeric nucleotides into telomeric DNA (18–21). In addition to the template and TBE, the PK structure located downstream of the template is essential for TERT–TR interaction and enzyme activity (22, 23). The PK structures from vertebrates and yeast TRs are generally larger and more stable (12, 17), harboring longer helices than the PK structures of ciliate TR, which are relatively primitive and less stable (24, 25). Nuclear magnetic resonance studies of TR fragments reveal a unique triple-helix structure in the PK which plays an essential but poorly understood role in promoting telomerase activity (26). Another essential domain of TR, known as helix IV in ciliates and CR4/5 in vertebrates, can reconstitute telomerase activity in trans together with the T-PK domain (27–30). TRs from other groups of eukaryotes, including echinoderms and trypanosomes, also have a second structural domain called eCR4/5 that can bind independently to TERT in trans and is functionally equivalent to the vertebrate CR4/5. Therefore, the requirement of 2 conserved structural TR domains for telomerase activity is universally conserved among all major groups of eukaryotes, from Trypanosomes to vertebrates (31).
We previously described the identification of 2 telomerase-associated RNAs from Arabidopsis thaliana, termed AtTER1 and AtTER2 (32, 33). AtTER1 was proposed to serve as the template for telomeric DNA synthesis by telomerase (32); however, recent data have refuted the role of AtTER1 in telomere maintenance (34, 35). Moreover, Fajkus et al. (34) recently reported the identification of a novel telomerase RNA from A. thaliana, termed AtTR, that is required for telomere maintenance and is conserved across land plants. Here we present results of a next-generation sequencing analysis of TERT-associated RNAs, which independently led to the identification of AtTR as the bona fide RNA component for Arabidopsis telomerase. We show that AtTR is crucial for telomere maintenance in vivo and sufficient to reconstitute telomerase activity with A. thaliana TERT (AtTERT) protein in vitro. In addition, using phylogenetic sequence analysis of homologous TRs from the 3 distantly related plant lineages—including angiosperms, gymnosperms, and the early-branching lycophytes—we determine a conserved structural model for plant TRs that was verified using chemical probing and mutagenesis. Our findings provide an evolutionary bridge to unite the disparate structures of the previously characterized TRs from ciliates and vertebrates, as well as a new platform to explore the evolution of the telomerase RNP enzyme.
Results
AtTR Is the Predominant RNA Associated with Active Telomerase in Arabidopsis.
Prompted by collaborative work with the Beilstein laboratory, which indicated that AtTER1 was not the authentic TR component for A. thaliana telomerase (35), we developed an unbiased approach to identify ncRNAs associated with the AtTERT protein through RNA immunoprecipitation (RIP) analysis using anti-AtTERT antibody. RIP was performed under native conditions with mild salt and detergent concentrations to retain weak interactions. Next-generation sequencing of copurified RNAs identified 177 RNA sequences that were significantly enriched in the wild-type (WT) samples but not in the tert null samples (Fig. 1A). The previously reported telomerase RNA template AtTER1 and the TERT-associated RNA AtTER2 were not found among these AtTERT-associated RNAs. To address the possibility that AtTER1 was masked by other more abundant RNAs, we used more stringent conditions to purify active telomerase by size exclusion chromatography before RIP (Fig. 1B). Telomerase activity was detected by quantitative telomere repeat amplification protocol (qTRAP) with the peak activity in a fraction corresponding to an apparent molecular mass of ∼300 kDa (Fig. 1C). A scatterplot of RNAs purified and sequenced from fractions with peak telomerase activity revealed a single RNA that was enriched more than 100-fold above background (Fig. 1D). This is the same RNA independently reported by Fajkus et al. (34) and dubbed AtTR. Since AtTER1 overlaps with the 5′ region of RAD52 locus (36) (SI Appendix, Fig. S1A), we performed additional TERT RIP experiments to directly test whether RAD52 mRNA was present in the IP. While RAD52 mRNA could be amplified from the IP, an RNA corresponding to the previously described AtTER1 could not (SI Appendix, Fig. S1 B and C). These results are inconsistent with AtTER1 being a functional telomerase RNA and instead support the recent findings of Fajkus et al. (34) and Dew-Budd et al. (35) indicating that AtTER1 is not required for telomere maintenance.
Fig. 1.
A single RNA species is enriched in active telomerase complexes. (A) Scatterplot representing RNA targets enriched in a direct RIP-seq experiment. WT and tert null mutant samples are compared to identify potential AtTERT-associated RNAs, labeled as blue or red according to their relative enrichment in WT greater than 2-fold or 5-fold, respectively. (B) Schematic representation of the experimental design for identification of telomerase-associated RNAs. (C) Size exclusion chromatogram of A. thaliana protein lysate. The blue curve shows the elution profile, and the red bars show the relative telomerase activity from each fraction. (D) Scatterplot of RNAs copurified with the active A. thaliana telomerase complex. AtTR is the only RNA molecule significantly enriched in WT samples compared with tert mutants.
AtTR Is Required for Telomere Repeat Synthesis by A. thaliana Telomerase.
AtTR was originally described as a noncoding Pol III transcript involved in the stress response (37). AtTR bears a 9-nt sequence of 5′-CUAAACCCU-3′ complementary to the A. thaliana 7-nt telomeric DNA sequence (TTTAGGG)n (38). Mapping of its 5′ and 3′ ends by rapid amplification of cDNA ends revealed that AtTR is 268 nt long (SI Appendix, Fig. S2A). The size of endogenous AtTR was verified by Northern blot analysis (SI Appendix, Fig. S2B). Using direct terminator exonuclease treatment in combination with pyrophosphohydrolase, we found that AtTR bears a 5′ triphosphate structure (SI Appendix, Fig. S2C). AtTR is widely expressed but is most abundant in actively dividing cell culture. Notably, AtTR is also abundant in mature leaves, where AtTERT is conspicuously absent and telomerase activity is negligible (SI Appendix, Fig. S2D).
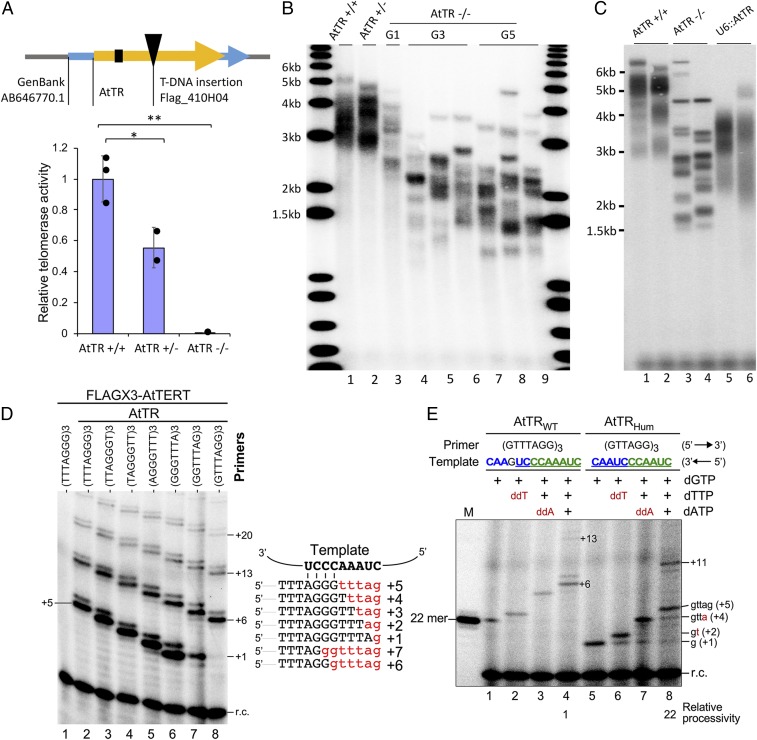
We used 2 genetic approaches to determine whether AtTR is required for telomerase activity and telomere maintenance in vivo. First, we found that a homozygous T-DNA insertion allele of AtTR (Flag_410H04) completely abolished AtTR RNA production as well as telomerase activity detected by qTRAP, while plants bearing a heterozygous mutation had ∼50% of the WT level of AtTR and 50% of the WT telomerase qTRAP activity (Fig. 2A and SI Appendix, Fig. S3 A and B). Terminal restriction fragment (TRF) analyses showed progressive shortening of the telomere tract in homozygous Flag_410H04 mutants over 5 generations (Fig. 2B), reminiscent of tert null mutants (39). Second, 2 independent CRISPR-mediated deletions that remove either a 49-nt sequence including the template or a 14 nt sequence downstream of the template disrupted telomere maintenance (SI Appendix, Fig. S4). We performed genetic complementation experiments on Flag_410H04 AtTR null mutants using an AtTR construct driven by the U6 promoter (U6::AtTR). Transformants with U6::AtTR expression exhibited restored telomerase activity and increased telomere length (Fig. 2C and SI Appendix, Fig. S3 C and D). These findings confirm that AtTR is necessary for both telomerase enzyme activity and telomere maintenance in A. thaliana.
Fig. 2.
AtTR is the RNA template for Arabidopsis telomerase. (A, Top) Schematic representation of the AtTR gene showing the template domain (black box) and the location of a T-DNA insertion. (A, Bottom) Relative telomerase activity of WT, heterozygous, and homozygous AtTR mutants determined by quantitative TRAP assay. (B) TRF analysis of telomere length in AtTR mutants across multiple generations. (C) TRF results for genetic complementation with AtTR driven by the U6 promoter. Third-generation AtTR−/− mutants were untransformed or transformed with U6::AtTR. (D) In vitro reconstitution of A. thaliana telomerase activity. Sequences of the putative template with the annealing position of 7 circular permuted telomeric DNA primers are shown on the right. The predicted primer-extended products are shown in red. A. thaliana telomerase is reconstituted in vitro from synthesized FLAGx3-AtTERT and 1.5 µM of T7 transcribed full-length AtTR (268 nt). The affinity-purified telomerase was assayed for activity in the presence of 32P-dGTP, dTTP, dATP, and 7 plant telomeric DNA primers with permuted sequences. A radiolabeled 18-mer recovery control (r.c.) was added before product purification and precipitation. Numbers to the right of the gel denote the number of nucleotides added to the primer. (E) Template-directed nucleotide addition by A. thaliana telomerase. Telomerase was reconstituted in vitro with AtTERT and either AtTRWT or AtTRHum. The reconstituted telomerase was assayed for activity in the presence of 32P-dGTP and different combinations of dTTP, dATP, ddTTP, or ddATP. A 21-nt plant telomeric DNA primer (GTTTAGG)3 was used for AtTR, and an 18-nt human telomeric DNA primer (GTTAGG)3 was used for the AtTRhum. A radiolabeled 18-mer recovery control was added before product purification and precipitation. Numbers and sequences of nucleotides added to the primers are indicated.
AtTR and AtTERT Reconstitute Active Telomerase In Vitro.
We next asked whether AtTR can assemble with AtTERT in vitro to reconstitute active telomerase. As shown in Fig. 2D, recombinant FLAGx3-AtTERT protein synthesized in rabbit reticulocyte lysate was assembled with T7 RNA polymerase transcribed AtTR in vitro and the reconstituted telomerase was immunopurified, followed by a direct primer extension assay (Fig. 2D). Seven A. thaliana telomeric DNA primers with permuted sequences of TTTAGGG bearing different 3′ terminal sequences were examined using in vitro reconstituted telomerase enzyme. The reaction with (GTTTAGG)3 generated a 7-nt ladder pattern of products with major bands at positions +6, +13, and +20 (Fig. 2D, lane 8), consistent with the 7-nt telomeric DNA repeats synthesized by A. thaliana telomerase. A. thaliana telomerase exhibited similar levels of activity with the different permuted telomeric DNA primers and generated the expected offset banding patterns (Fig. 2D, lanes 2 to 7), indicating correct primer-template alignment and specific use of the template. Importantly, the primer extension activity is AtTR-dependent, as no activity was detected in the absence of AtTR (Fig. 2D, lane 1).
To further examine the templating function of AtTR, we generated an AtTR template mutant (AtTRhum) with a template sequence similar to the human TR (hTR) template that allows the synthesis of 6-nt TTAGGG repeats. The telomeric TTAGGG repeats are ubiquitously conserved in most lineages of eukaryotes (3). The 9-nt AtTR template sequence 5′-CUAAACCCUGAACC-3′ for the synthesis of 7-nt repeats (TTTAGGG)n is flanked by a G residue at it 3′ boundary and potentially could be expanded to a longer 14-nt template by mutating the G residue to A. To convert the native A. thaliana template sequence to a human-like template, we simply deleted 1 A residue in the polymerization template sequence and the nonconserved G residue in the alignment sequence, which resulted in a 12-nt 5′-CUAACCCUAACC-3′ template for synthesizing TTAGGG repeats (Fig. 2E). As expected, the telomerase reconstituted from the AtTRhum template mutant generated the first major band at position +5(+gttag) and the second major band at +11, indicating the addition of a 6-nt DNA repeat using the human-like template (Fig. 2E, lane 8). Moreover, the inclusion of dideoxy-ribonucleotides, either ddTTP or ddATP, terminated the primer extension reaction at the expected positions on the template of the AtTRWT and AtTRhum (Fig. 2E, lanes 2–3 and 6–7). In addition, under processive conditions with all 3 nucleotides, the AtTRhum template with a long 6-nt alignment region led to a significantly higher processivity based on the ratio of +11/+5 products (Fig. 2E, lanes 4 and 8), consistent with a previous finding that longer templates correlate with higher repeat addition processivity (40). Taken together, these data demonstrate that the template sequence 43-CUAAACCCU-51 within AtTR is a bona fide template for telomeric DNA repeat synthesis by A. thaliana TERT.
Plant TRs Share a Conserved Secondary Structure.
To discern the structure of AtTR, we used phylogenetic comparative analysis to infer a secondary structure model from the sequence alignment of plant TR homologs identified from 3 major clades of land plant species: angiosperms, gymnosperms, and lycophytes (Fig. 3A). Orthologs of AtTR were identified by searching genomic sequence data predominantly from the National Center for Biotechnology using sequence homology search tools including BLAST, Fragrep2 (41), and Infernal (42). While the BLAST search was able to find TR homologs from closely related species, Fragrep2 allowed identification of TR homologs from more distantly related species by using position-specific weight matrix (PWM)-based searches with PWMs derived from multiple sequence alignments, as opposed to using the primary sequence as the search query. Collectively, we identified 85 AtTR orthologs, including 70 from angiosperms, 11 from gymnosperms, and 4 from lycophytes (SI Appendix, Table S1). To infer secondary structure, multiple sequence alignment analysis was performed with 16 representative TR sequences (12 angiosperms, 3 gymnosperms, and 1 lycophytes) selected from the 85 sequences to allow at least 1 representative from each individual order spanning 3 distinct clades (SI Appendix, Fig. S5). All TR sequences, including those from the basal groups, gymnosperms, and lycophytes, can be reliably aligned with the TR sequence from angiosperms, revealing universally conserved structural elements of plant TRs. From the alignment of 16 divergent plant TR sequences, universal or group-specific nucleotide covariations were identified to infer base-paired structural elements (Fig. 3 B–D and SI Appendix, Fig. S6). Comparison of TR secondary structures from 3 representative species—A. thaliana from angiosperms, Picea glauca (spruce) from gymnosperms and Selaginella kraussiana (spikemoss) from lycophytes—revealed 3 common structural features: a conserved T-PK core domain enclosed by stem P1c; a long stem comprising consecutive short base-paired regions termed P4, P5, and P6; and a long-range base-paired stem P1a formed between the extreme distal 5′ and 3′ sequences (Fig. 3 B–D).
Fig. 3.
Plant TRs share a conserved secondary structure. (A) Evolutionary relationship between major land plant clades. A single representative species of each order is included. An asterisk denotes the species with the secondary structure models shown in B, C, and D. Representative TR secondary structures determined by phylogenetic sequence analysis are shown for A. thaliana from angiosperms (B), P. glauca (spruce) from gymnosperms (C), and S. kraussiana (spike moss) from lycophytes (D). The characteristic TR PK is shaded in yellow. Universal covariations (green line), group-specific covariations (blue line), and plant invariant residues (red) are indicated and based on sequence alignment of 16 divergent plant species spanning 8 eudicots, 2 monocots, 2 early-branching angiosperms, 3 gymnosperms, and 1 lycophyte. The aligned sequences are shown in SI Appendix, Fig. S5. (E) In vitro chemical probing of AtTR secondary structure by SHAPE. Chemical reactivities per nucleotide are plotted on the AtTR secondary structure. (F) In vivo chemical probing of AtTR structure by DMS-MaPseq. Average mutation frequencies per nucleotide are plotted on the AtTR secondary structure.
The plant T-PK core domain resembles those from ciliate, fungal, and vertebrate TRs consisting of a template, a universal PK structure formed by stems P2 and P3, and a core-enclosing stem P1c (Fig. 3 B–D). However, the plant T-PK core domain contains additional plant-specific stems, namely P1.1 (in P. glauca and S. kraussiana), P2.1 (in A. thaliana and P. glauca), and P2.2 (in P. glauca and S. kraussiana) (Fig. 3 B–D). The P1.1 stem can be found in the invertebrate echinoderm and fungal TRs, and could potentially function as a TBE (17, 43). The P2.1 and P2.2 stems are not present in all plant TRs, suggesting that they are more adaptable and may be important for a function specific to some plant groups. One possible role for the variable P2.1 and P2.2 stems is to maintain the length of the linker between the template and the PK structure within the T-PK core domain.
In addition to the T-PK core domain, the plant TR contains a long helical structure with 3 consecutive short stems—P4, P5, and P6—located near the 3′ end between P1a and P1b (Fig. 3 B–D). The location and structure of the plant P4/P5/P6 stem resembles the vertebrate CR4/5 domain, echinoderm eCR4/5 domain, or ciliate helix IV, all of which are essential for telomerase activity (28, 29, 43). The 3-way junction formed among P1a, P1b, and P4/5/6 appears to be a conserved feature of plant TR (Fig. 3 B–D). This P1a-mediated 3-way junction is unique to plant TR and is not found in other known TRs.
This conserved secondary structure model of AtTR is supported by chemical modification probing analysis. Selective 2’-hydroxyl acylation analysis by primer extension (SHAPE) was used to examine the accessibility of each nucleotide in the in vitro folded RNA (44). N-methylisatoic anhydride modification of individual nucleotides was monitored, and SHAPE activity plotted on the structural model to identify unpaired residues (Fig. 3E and SI Appendix, Fig. S7). Consistent with our AtTR structural model, most unpaired nucleotides showed significant SHAPE activity.
We also probed the AtTR structure in vivo by dimethyl sulfide (DMS) footprinting and mutational profiling (DMS-MaPseq). DMS methylates the base-pairing faces of single-stranded, unprotected adenosines and cytidines. Such modifications cause the stalling of conventional reverse transcriptases during cDNA synthesis, allowing for footprinting studies. These modifications can also result in mismatches in cDNA when TGIRT reverse transcriptase is used (45). DMS modifications were analyzed by primer extension (DMS footprinting), while DMS-induced mutational rates per position were calculated by coupling TGITR cDNA synthesis with high throughput sequencing. DMS footprinting identified 38 accessible nucleotides that mapped to predicted single-stranded residues (SI Appendix, Fig. S8). Results of DMS MaPseq extended these findings and revealed a detailed map of nucleotide accessibility (Fig. 3F and SI Appendix, Fig. S9). Accessible nucleotides were concentrated in the predicted single-stranded regions within the T-PK and P1b-P1c linker. Taken together, these in vitro and in vivo structural probing results provide strong support for our AtTR secondary structure model.
In addition to inferring the conserved secondary structure, the multiple sequence alignment of the 16 representative plant TRs spanning land plant evolution revealed 5 highly conserved regions (CRs), CR1 to CR5, containing nucleotides that are invariant among these 16 distantly related species (SI Appendix, Fig. S5). Such remarkable conservation of nucleotide identity usually predicts essential functions of these regions, as evident in vertebrate TRs (12). CR1 corresponds to the template of AtTR. CR2 and CR3 form the universal P2 and P3 stems of the PK, while CR4 and CR5 form a P5 structural element that includes the short 3-bp P5 stem, an asymmetric internal loop and the upper part of stem P4 (Fig. 3 B–D). While lacking the P6.1 stem-loop, the universal P5 structural element of the plant TR resembles the CR4/5 domain conserved in vertebrate, fission yeast, and filamentous fungal TRs (17, 28). This highly conserved P5 stem may serve as a protein-binding site or play a crucial role in telomerase function.
The AtTR PK Domain Is Essential for Telomerase Function and Homologous to Human TR.
With a robust secondary structure model for AtTR, we sought to map the structural elements essential for telomerase activity. Full-length or truncated AtTR constructs were assembled with recombinant FLAGx3-AtTERT in vitro, and the immunopurified enzymes were analyzed for telomerase activity by direct primer extension. Analysis of 3 truncated AtTR fragments—11 to 179, 25 to 153, and 42 to 136 (Fig. 4A)—showed that AtTR-25 to 153 is the minimal PK fragment sufficient to reconstitute ∼40% of WT activity without the P4/5/6 domain (Fig. 4B, lanes 2 and 3). The core-enclosing P1c stem appeared to be important for telomerase function, as the AtTR-42–136 fragment with P1c removed was unable to reconstitute any significant activity (Fig. 4B, lane 4). Equivalent to the CR4/5 domain of human TR, the 3′ P1a/4/5/6 domain of AtTR can also function in trans as a separate RNA molecule to stimulate the reconstituted activity from the basal 40% to 66% of the WT level (Fig. 4C). A basal activity of telomerase reconstituted from the T-PK domain alone was previously reported with Trypanosome and Echinoderm TRs (31, 43), indicating an evolutionary transition of functional dependence for the 2 conserved TR domains.
Fig. 4.
Functional characterization of critical structural elements in AtTR. (A) Schematic of the AtTR secondary structure. The 5′ and 3′ residues of truncated AtTR fragments are denoted on the AtTR structure. The positions and identities of specific point mutations introduced are indicated. (B) Identification of a minimal PK fragment and (C) functional analysis of stem P1a/4/5/6. Full-length AtTR (AtTR-FL) and various AtTR truncated fragments were assembled with AtTERT in vitro and analyzed for activity by a primer extension assay. The number of nucleotides (+6, +13 or +20) added in each major band of product are indicated. The P1a/4/5/6 fragment was generated by deleting residues 25 to 153 from the AtTR-FL and replacing them with a GAAA tetraloop. The relative activities of the reactions are indicated under the gel. A recovery control (r.c.) is shown. (D) The functional requirement of invariant U residues in PK domain. (E) The effect of P1c linker length on template boundary definition. (F) Compensatory mutagenesis analysis of stem P5. (G) Compensatory mutagenesis analysis of stem P2.1. AtTR-FL constructs bearing specific point mutations are assembled with AtTERT in vitro and analyzed for telomerase activity. For analyzing template boundary definition with AtTR-38UU, the reconstituted enzyme was analyzed in the absence (−) or presence (+) of dCTP in addition to dGTP, dATP, and dTTP.
The PK structure of plant TRs highly resembles the PK structures in ciliate and vertebrate TRs with differences in size and complexity. In the human TR PK structure, the invariant U residues in the J2/3 upstream region (J2/3u) are essential for telomerase activity (46). To determine whether the invariant U residues in plant TR PK are functionally homologous to the human TR, we reconstituted telomerases with two AtTR mutants, U92C and UU94/95CC. The activity assays of the mutant enzyme showed no activity (Fig. 4D, lanes 2 and 3), indicating that these U residues in the AtTR PK domain are absolutely required for telomerase activity. Therefore, the T-PK domains of AtTR and hTR are both structurally and functionally homologous.
Another critical function provided by the T-PK domain is defining the functional template boundary through specific structural elements, that is, the P1 stem in vertebrate TR (20). The P1c stem in the T-PK domain of AtTR resembles the P1 stem in human TR and presumably functions as the template boundary element. To test this idea, we generated an AtTR mutant 38UU with 2 U residues inserted between the P1c stem and the template to increase the linker length, a critical determinant of the template boundary. In the WT AtTR template, a G residue immediately flanks the 5′ boundary and does not serve as a template even in the presence of dCTP substrate (Fig. 4E, lanes 1 and 2). However, in the presence of dCTP, the telomerase enzyme reconstituted with the AtTR mutant 38UU uses the G residue as a template beyond the template boundary (Fig. 4E, lanes 3 and 4). Thus, A. thaliana and human telomerases share a homologous mechanism for template boundary definition.
While the overall secondary structure of AtTR is well supported by covariation evidence and chemical probing data, we performed mutagenesis analysis to provide additional support for the highly conserved P5 stem and the plant-specific P2.1 stem (Fig. 4A). The 3-bp P5 stem is formed by 2 highly conserved regions, CR4 and CR5, with only limited covariation support for 1 of the 3 base pairs. We thus generated AtTR full-length constructs, P5-m1 and -m2, with 2 single point mutations, G194C and C239G, introduced to disrupt the invariant G:C base-pairing in the P5 stem, or a compensatory mutant P5-m3 with both point mutations to restore the base-pairing (Fig. 4A). The activity assay showed that P5-m1 and -m2 single point mutations abolished telomerase activity (Fig. 4F, lanes 2 and 3), while the compensatory mutation P5-m3 restored activity (Fig. 4F, lane 4), consistent with the essential base-paired structure of stem P5. A similar mutagenesis approach was used to confirm the base-paired structure and the functional importance of stem P2.1 (Fig. 4G). Taken together, these in vitro studies strongly support the robustness of the phylogenetic comparative analysis for inferring RNA secondary structure in plant TR.
Discussion
Telomerase emerged in early eukaryotes as a specialized reverse transcriptase with an integral RNA template to counteract the end-replication problem and maintain genomic integrity. While the catalytic TERT component of telomerase is conserved among eukaryotes, the TR component has diverged significantly during evolution. A missing piece in the evolutionary history of telomerase has been plant TR. Recent studies from the Fajkus (34) and Beilstein (35) laboratories indicated that the previously identified AtTER1 (32) was not the authentic TR in A. thaliana. The results from our independent study support this conclusion. We were unable to detect AtTER1 using two purification schemes, one designed to identify RNAs loosely associated with AtTERT, and a second more stringent approach to identify RNAs associated with partially purified, enzymatically active telomerase. The misidentification of AtTER1 in the previous study may have resulted from a primer extension strategy that used biased primers corresponding to predicted Arabidopsis TR template and that inadvertently recovered a low-abundance RNA molecule derived from the RAD52 locus that copurified with telomerase. Our next-generation sequencing approach also failed to recover AtTER2, a second telomerase-associated RNA proposed to negatively regulate enzyme activity in response to DNA damage (32, 33). Reevaluation of the AtTER2 locus in relation to telomerase and telomeres is now underway.
Nevertheless, the single RNA enriched by 100-fold in enzymatically active telomerase fractions from our more stringent purification scheme was AtTR, the same RNA molecule uncovered independently by the Fajkus laboratory using an in silico strategy to find plant TRs (34). To investigate the function of AtTR, we used a combination of Arabidopsis genetics and in vitro reconstitution experiments using a rigorous non-PCR assay of direct primer extension to test the authenticity of this putative telomerase RNA template. We determined that AtTR was not only required for telomere maintenance in vivo, but also possessed a functional template for telomeric DNA synthesis by AtTERT in vitro. Our observations agree with those of Fajkus et al. (34) and confirm that AtTR is the bona fide telomerase RNA subunit for A. thaliana.
AtTR was first described in 2012 by Wu et al. (37) as a root-specific, conserved Pol III-dependent ncRNA. The ATTR gene (GenBank accession no. AB646770.1) includes a U6-like type III promoter and poly(T) terminator. The promoter has a consensus cis upstream sequence element and a TATA box-like element 25 bp upstream of the transcription start site. The discovery of plant TRs as Pol III RNA transcripts leads to an interesting question: was the first TR a Pol II or Pol III transcript? TR was originally identified in ciliates as a small Pol III RNA transcript with sizes ranging from 140 to 210 nt (Fig. 5). RNA polymerase III is generally employed for transcribing small RNA such as 5S rRNA and tRNA due to its sequence-dependent termination at a U-rich termination site. A large RNA would encounter a high frequency of U-rich sequences and suffer premature termination with Pol III transcription, which is consistent with the small size of ciliate TR (7). Surprisingly, TRs identified later in vertebrates and fungi are larger Pol II transcripts with sizes of 312 to 559 nt and 920 to 2,425 nt, respectively (12, 17). While it seems reasonable to assume that the Pol III TR transcript is more ancestral, TRs from early branching flagellates, including Trypanosomes, are large Pol II transcripts ranging in size from 781 to 993 nt (Fig. 5). Discerning the origin of TR will require discovery of TRs from the early branching lineages of eukaryotes, a daunting task considering the extremely divergent nature of TR.
Fig. 5.
Evolution of TR PK structures. (Left) A simplified phylogenetic tree of major eukaryotic lineages. Branch length in the tree does not reflect evolutionary distance. The lineages with TR transcribed by Pol II (green) and Pol III (orange) are depicted. The size range of TRs from each group is indicated. (Right) The PK structures of TRs from the major groups of eukaryotes including ciliates, plants, fish, and mammals. Trypanosome TR does not have a PK structure in the template core domain (31). The P2 and P3 stems conserved from ciliates to mammals are shown in red, with highly conserved nucleotides explicitly denoted. The vertebrate-specific stem extension P2a is shown in blue, while the mammal-specific stem extension P2a.1 is shown in green. The lengths of joining sequences, J2/3 upstream (J2/3u) or downstream (J2/3d) regions, between stems P2 and P3 are indicated.
The conserved secondary structures of plant TRs presented in this study were determined using phylogenetic comparative analysis, a gold standard for inferring RNA secondary structures (12, 47). Moreover, the secondary structure of AtTR was verified by in vitro and in vivo chemical probing approaches under native conditions as well as mutagenesis analysis using an in vitro reconstitution system. In the AtTR structure, the most crucial structural element is the PK, which is conserved in all known TRs except Trypanosome (Fig. 5). Trypanosome TR contains 2 structural domains, the template core and eCR4/5, both of which are required for telomerase activity in vitro and can function in trans as 2 separate RNA fragments (31). However, the minimal template core domain of Trypanosome TR does not contain a PK, arguing that the critical TR PK was a later adaptation. Nevertheless, helix III of Trypanosome TR is potentially homologous to the PK forming helix III of Tetrahymena TR, as both helices are located between the template and the core enclosing helix, that is, helix I in Tetrahymena TR or the P1 stem in other TRs.
The PK structure of Tetrahymena TR only requires formation of a 4-bp stem between the loop sequence of helix III and an upstream complementary sequence (Fig. 5). This 4-bp stem is structurally equivalent to the vertebrate P2 stem, which is longer and contains 2 consecutive stems, P2a and P2b, and with an additional P2a.1 stem in the mammalian TR PK (Fig. 5). How this primitive ciliate TR PK evolved to the more complex vertebrate TR PK has been unclear. The structure of plant TR PK now provides an explanation for the structural transition from ciliate to vertebrate PK. Similar to ciliate PK, plant PK contains a short, unstable, 4-bp P2 stem and a longer 8- to 9-bp P3 stem. DMS chemical probing of the A. thaliana TR PK reveals mild modification of the P2 stem, consistent with a less stable helix (Fig. 3F). Notably, the ciliate and plant PK structures differ in the length of the joining sequences, J2/3 upstream (J2/3u) and J2/3 downstream (J2/3d) (Fig. 5). The length of J2/3u increases from 3 nt in Tetrahymena to 8 nt in plants, similar to the 8-nt J2b/3 in vertebrate TR PK (Fig. 5). The length of J2/3d sequence also increases from 4 nt in Tetrahymena to 14 nt in the A. thaliana PK. We propose that the longer J2/3d makes it possible to expand the short 4-bp P2 stem to a longer P2a/P2b stem in vertebrate PK during evolution. Notably, plant TR contains additional stems (P2.1 and P2.2) located between the template and the P2 stem (Fig. 5). These additional stems may reflect selective pressure to maintain the spatial constraints for the enzyme active site as the P2 stem expands during evolution. Therefore, the plant TR PK provides an evolutionary bridge for the structural transition from ciliate TR to vertebrate TR.
Materials and Methods
RIP-Seq.
Anti-AtTERT antibody was affinity-purified with an EpiMAX affinity purification kit (Abcam) following the manufacturer’s protocol. It was preincubated with protein A magnetic beads (Dynabeads) before IP experiments. For the direct RIP-seq, 1.2 g of WT (Col-0) and tert Arabidopsis flowers were ground in liquid nitrogen and homogenized in RIP buffer (100 mM Tris-OAC pH 7.5, 100 mM KGlu, 1 mM MgCl2, 0.5% Triton X-100, 0.1% Tween 20, 20 μL/mL Plant protease inhibitor mixture [Sigma-Aldrich], 1 μL/mL RNaseOUT [Thermo Fisher Scientific], and 2.5 mM DTT). After clearing by centrifugation, protein complexes were immunoprecipitated using preincubated anti-AtTERT magnetic beads for 2.5 h at 4 °C. After incubation, beads were washed 7 times with RIP buffer and then resuspended with 1 mL of TRIzol reagent (Invitrogen) to extract RNA. For RIP-seq after gel filtration, fractions with peak telomerase activity were incubated in TERT buffer (50 mM Tris-OAC pH 7.5, 150 mM KGlu, 5 mM MgCl2, 5% glycerol, 20 μL/mL Plant protease inhibitor mixture, 1 μL/mL RNaseOUT, 0.1 mM PMSF, and 1.5 mM DTT) with preincubated anti-AtTERT magnetic beads for 3 h at 4 °C. Beads were washed 7 times with RIP buffer, and the remaining RNA was extracted using Direct-zol RNA Kits (Zymo Research) including in-column DNase treatment. After rRNA depletion, construction of Illumina sequencing libraries was performed with the NEBNext Ultra Directional RNA Library Prep Kit, and libraries were sequenced on a 2 × 300 Illumina MiSeq platform by Texas A&M AgriLife Genomics and Bioinformatics Service.
Target-Specific DMS-Mapseq.
Target-specific DMS-MaPseq was performed as described previously (45, 48) with modifications. Total RNA was extracted from DMS- or mock-treated samples using RNA Clean & Concentrator-5 (Zymo Research) with in-column DNase digestion. RNA quality was analyzed on agarose gels, with 5 μg of high-quality RNA combined with gene-specific primers (5 pmol each) in a total volume of 11 μL. The mixture was heated to 75 °C for 3 min and annealed at 55 °C for 15 min. TGIRT reaction buffer including 4 μL of 5× First-Strand buffer (Thermo Fisher Scientific), 1 μL of 0.1 M DTT, 1 μL of RNaseOUT (Thermo Fisher Scientific) and 1 μL TGIRT-III (Ingex; catalog no. TGIRT50) was added, and the solution was incubated at room temperature for 30 min. Then 2 μL of 10 mM dNTP was added, and the well-mixed reaction was processed at 60 °C for 2.5 h. After RT, 1 μL of cDNA solution was directly added into a 50-μL PCR using Phusion High-Fidelity DNA Polymerase (New England BioLabs) to amply AtTR or ACT2 mRNA with an approximate product size of 260 bp. PCR products were gel-purified and quantified using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific). Without fragmentation, the cleaned PCR products were directly assembled into Illumina sequencing libraries using the NEBNext Ultra II DNA Library Prep Kit (New England BioLabs) with 25 ng input. One mock library and 2 DMS libraries were built for each genotype. Finally, the libraries were quantified using Agilent TapeStation, followed by sequencing on an 150 × 2 Illumina NextSeq 500 platform at Texas A&M University.
Telomerase Direct Primer Extension.
Activity of immunopurified telomerase was analyzed by direct primer extension assay as described previously (49). The telomerase enzyme on beads was assayed in a 10-µL reaction containing 1× telomerase reaction buffer (50 mM Tris⋅HCl pH 8.0, 50 mM NaCl, 0.5 mM MgCl2, 5 mM BME, and 1 mM spermidine), 1 µM DNA primer, and specified dNTPs or ddNTPs and 0.18 µM of 32P-dGTP (3,000 Ci/mmol, 10 mCi/mL; PerkinElmer). Reactions were incubated at 30 °C for 60 min and terminated by phenol/chloroform extraction, followed by ethanol precipitation. The 22-mer size marker was prepared in a 10-µL reaction containing (GGGTTTA)3 oligo, 1× terminal deoxynucleotidyl transferase (TdT) reaction buffer, 5 units of TdT (Affymetrix), and 0.1 µM of 32P-dGTP. The reaction was incubated at room temperature for 3 s and terminated by addition of 10 µL of 2× formamide loading buffer (10 mM Tris⋅HCl pH 8.0, 80% [vol/vol] formamide, 2 mM EDTA, 0.08% bromophenol blue, and 0.08% xylene cyanol). The DNA products were resolved on a 10% (wt/vol) polyacrylamide/8 M urea denaturing gel, dried, exposed to a phosphorstorage screen, and imaged on a Typhoon gel scanner (GE Healthcare).
Detailed descriptions of other materials and methods, including plant growth conditions, size exclusion chromatography, SHAPE, in vivo DMS modification, DMS footprinting, Northern blot analysis, TRF, TRAP, bioinformatics analysis, sequence alignment, and in vitro telomerase reconstitution are provided in SI Appendix, Materials and Methods. Primers are listed in SI Appendix, Table S2.
Data Availability.
Raw data of two independent RIP seq and DMS-MaPseq are deposited in Gene Expression Omnibus (GEO) under BioProject ID PRJNA588284. Nucleotide sequence data of plant TRs reported are available in the Third Party Annotation Section of the DDBJ/ENA/GenBank databases under the accession numbers TPA: BK011296–BK011375.
Supplementary Material
Acknowledgments
We thank Mark Beilstein for sharing unpublished data from his laboratory demonstrating that AtTER1 is not the authentic template for A. thaliana telomerase. We thank Anna Nelson-Dittrich for collaboration on initial RIP experiments and Andrew Nelson for helpful conversations related to RNA-seq analysis and initial phylogenetic analysis of AtTR. We also thank Jorge Cruz-Reyes, Zhiye Wang, and Andrew Hillhouse for expert technical advice and the Texas A&M Genomics and Bioinformatics Service and the Genomics Core at the Texas A&M Institute for Genome Sciences and Society for high-throughput sequencing. This work was supported by NIH Grants R01 GM094450 (to J.J.-L.C.) and R01 GM065383 (to D.E.S.) and NSF Grants MCB1616078 (to J.J.-L.C.) and MCB151787 (to D.E.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw data from 2 independent RIP-seq and DMS-MaPseq experiments have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (BioProject ID PRJNA588284.). TRs identified in this work have been deposited in GenBank (accession no. TPA: BK011296–BK011375).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915312116/-/DCSupplemental.
References
- 1.Wilusz J. E., Sunwoo H., Spector D. L., Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 23, 1494–1504 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shay J. W., Wright W. E., Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 20, 299–309 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Podlevsky J. D., Chen J. J., Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 13, 720–732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casacuberta E., Drosophila: Retrotransposons making up telomeres. Viruses 9, E192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan E. D., Collins K., Biogenesis of telomerase ribonucleoproteins. RNA 18, 1747–1759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greider C. W., Blackburn E. H., A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331–337 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Lingner J., Hendrick L. L., Cech T. R., Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 8, 1984–1998 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Singh M., et al. , Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol. Cell 47, 16–26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang J., et al. , Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350, aab4070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Box J. A., Bunch J. T., Tang W., Baumann P., Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature 456, 910–914 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Noël J. F., Larose S., Abou Elela S., Wellinger R. J., Budding yeast telomerase RNA transcription termination is dictated by the Nrd1/Nab3 non-coding RNA termination pathway. Nucleic Acids Res. 40, 5625–5636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J. L., Blasco M. A., Greider C. W., Secondary structure of vertebrate telomerase RNA. Cell 100, 503–514 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Mitchell J. R., Cheng J., Collins K., A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19, 567–576 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C., Meier U. T., Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 23, 1857–1867 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egan E. D., Collins K., Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol. Cell. Biol. 30, 2775–2786 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tseng C. K., Wang H. F., Schroeder M. R., Baumann P., The H/ACA complex disrupts triplex in hTR precursor to permit processing by RRP6 and PARN. Nat. Commun. 9, 5430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi X., et al. , The common ancestral core of vertebrate and fungal telomerase RNAs. Nucleic Acids Res. 41, 450–462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Autexier C., Greider C. W., Boundary elements of the Tetrahymena telomerase RNA template and alignment domains. Genes Dev. 9, 2227–2239 (1995). [DOI] [PubMed] [Google Scholar]
- 19.Tzfati Y., Fulton T. B., Roy J., Blackburn E. H., Template boundary in a yeast telomerase specified by RNA structure. Science 288, 863–867 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Chen J. L., Greider C. W., Template boundary definition in mammalian telomerase. Genes Dev. 17, 2747–2752 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansson L. I., et al. , Structural basis of template-boundary definition in Tetrahymena telomerase. Nat. Struct. Mol. Biol. 22, 883–888 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn E. H., Collins K., Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 3, a003558 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podlevsky J. D., Chen J. J., It all comes together at the ends: Telomerase structure, function, and biogenesis. Mutat. Res. 730, 3–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Autexier C., Greider C. W., Mutational analysis of the Tetrahymena telomerase RNA: Identification of residues affecting telomerase activity in vitro. Nucleic Acids Res. 26, 787–795 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilley D., Blackburn E. H., The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl. Acad. Sci. U.S.A. 96, 6621–6625 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Theimer C. A., Blois C. A., Feigon J., Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 17, 671–682 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Mitchell J. R., Collins K., Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 6, 361–371 (2000). [DOI] [PubMed] [Google Scholar]
- 28.Chen J. L., Opperman K. K., Greider C. W., A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 30, 592–597 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason D. X., Goneska E., Greider C. W., Stem-loop IV of tetrahymena telomerase RNA stimulates processivity in trans. Mol. Cell. Biol. 23, 5606–5613 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie M., et al. , Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J. Biol. Chem. 283, 2049–2059 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Podlevsky J. D., Li Y., Chen J. J., The functional requirement of two structural domains within telomerase RNA emerged early in eukaryotes. Nucleic Acids Res. 44, 9891–9901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cifuentes-Rojas C., Kannan K., Tseng L., Shippen D. E., Two RNA subunits and POT1a are components of Arabidopsis telomerase. Proc. Natl. Acad. Sci. U.S.A. 108, 73–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Cifuentes-Rojas C., et al. , An alternative telomerase RNA in Arabidopsis modulates enzyme activity in response to DNA damage. Genes Dev. 26, 2512–2523 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fajkus P., et al. , Telomerase RNAs in land plants. Nucleic Acids Res. 47, 9842–9856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dew-Budd K., Cheung J., Palos K., Forsythe E. S., Beilstein M. A., Evolutionary and biochemical analyses reveal conservation of the Brassicaceae telomerase ribonucleoprotein complex. bioRxiv 10.1101/760785, 760785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samach A., Melamed-Bessudo C., Avivi-Ragolski N., Pietrokovski S., Levy A. A., Identification of plant RAD52 homologs and characterization of the Arabidopsis thaliana RAD52-like genes. Plant Cell 23, 4266–4279 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J., et al. , A novel hypoxic stress-responsive long non-coding RNA transcribed by RNA polymerase III in Arabidopsis. RNA Biol. 9, 302–313 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Richards E. J., Ausubel F. M., Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell 53, 127–136 (1988). [DOI] [PubMed] [Google Scholar]
- 39.Riha K., McKnight T. D., Griffing L. R., Shippen D. E., Living with genome instability: Plant responses to telomere dysfunction. Science 291, 1797–1800 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Chen J. L., Greider C. W., Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 22, 304–314 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mosig A., Chen J. J.-L., Stadler P. F.. Homology search with fragmented nucleic acid sequence patterns. Lecture Notes in Comput. Sci. 4645, 335–345 (2007). [Google Scholar]
- 42.Nawrocki E. P., Eddy S. R., Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Podlevsky J. D., Li Y., Chen J. J., Structure and function of echinoderm telomerase RNA. RNA 22, 204–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkinson K. A., Merino E. J., Weeks K. M., Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Zubradt M., et al. , DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J. L., Greider C. W., Functional analysis of the pseudoknot structure in human telomerase RNA. Proc. Natl. Acad. Sci. U.S.A. 102, 8080–8085 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pace N. R., Smith D. K., Olsen G. J., James B. D., Phylogenetic comparative analysis and the secondary structure of ribonuclease P RNA–A review. Gene 82, 65–75 (1989). [DOI] [PubMed] [Google Scholar]
- 48.Ding Y., et al. , In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505, 696–700 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Chen Y., Podlevsky J. D., Logeswaran D., Chen J. J., A single nucleotide incorporation step limits human telomerase repeat addition activity. EMBO J. 37, e97953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of two independent RIP seq and DMS-MaPseq are deposited in Gene Expression Omnibus (GEO) under BioProject ID PRJNA588284. Nucleotide sequence data of plant TRs reported are available in the Third Party Annotation Section of the DDBJ/ENA/GenBank databases under the accession numbers TPA: BK011296–BK011375.