Significance
The conserved proteasome-binding protein PI31 serves as an adapter to couple proteasomes with cellular motors to mediate their transport to distal tips of neurons where protein breakdown occurs. We generated global and conditional PI31 knockout mouse strains and show that this protein is required for protein homeostasis, and that its conditional inactivation in neurons disrupts synaptic structures and long-term survival. This work establishes a critical role for PI31 and local protein degradation in the maintenance of neuronal architecture, circuitry, and function. Because mutations in the PI31 pathway cause neurodegenerative diseases in humans, reduced PI31 activity may contribute to the etiology of these diseases.
Keywords: ubiquitin−proteasome system/protein degradation, neurodegenerative diseases, axon/synapse, SCF complex/F-box protein, amyotrophic lateral sclerosis/ataxia
Abstract
Proteasome-mediated degradation of intracellular proteins is essential for cell function and survival. The proteasome-binding protein PI31 (Proteasomal Inhibitor of 31kD) promotes 26S assembly and functions as an adapter for proteasome transport in axons. As localized protein synthesis and degradation is especially critical in neurons, we generated a conditional loss of PI31 in spinal motor neurons (MNs) and cerebellar Purkinje cells (PCs). A cKO of PI31 in these neurons caused axon degeneration, neuronal loss, and progressive spinal and cerebellar neurological dysfunction. For both MNs and PCs, markers of proteotoxic stress preceded axonal degeneration and motor dysfunction, indicating a critical role for PI31 in neuronal homeostasis. The time course of the loss of MN and PC function in developing mouse central nervous system suggests a key role for PI31 in human neurodegenerative diseases.
The regulated breakdown of proteins is essential for cell function and survival. The ubiquitin−proteasome system (UPS) is responsible for the degradation of soluble intracellular proteins, whereas autophagy primarily serves to remove protein aggregates and damaged organelles (1–8). UPS-mediated protein degradation involves 2 major steps: First, a protein is tagged with a chain of ubiquitin molecules, and then polyubiquitinated substrates are cleaved into small peptides by the 26S proteasome (2, 9, 10). Although protein homeostasis is thought to be a critical feature of neuronal circuit function, the major emphasis has been the importance of local protein translation (11, 12). It has been suggested that local protein degradation in distal axons and dendrites is important for quality control of synaptic proteins, and also for plasticity and remodeling of synapses (13–20). However, in the absence of tools to selectively inhibit proteasomes at synapses, the precise role of local proteasome activity remains unclear.
Proteasome activity is regulated at multiple levels, including transcription of proteasomal proteins, assembly of subunits, posttranslational modifications, and the utilization of different regulatory particles (10, 21–23). One structurally conserved proteasome regulator is PI31 (proteasome inhibitor of 31kD). PI31 was originally discovered based on its ability to inhibit hydrolysis of small peptides by 20S particles in vitro (24, 25). Conversely, studies in Drosophila, yeast, and plants indicate that PI31 promotes protein breakdown in vivo (26–28). PI31 can be adenosine 5′-diphosphate-ribosylated by Tankyrase, and this modification promotes assembly of 26S proteasome (29). PI31 also binds directly to Fbxo7/PARK15, a conserved F-box protein that is the substrate recognition component of an SCF E3 ligase complex (26, 30–32). Unexpectedly, loss of Fbxo7 causes cleavage and inactivation of PI31, reduced proteasome activity, and neuronal degeneration (26, 33, 34). Significantly, impaired UPS function and mutations in Fbxo7/PARK15 are associated with neurodegenerative diseases (33–42).
We recently showed that, in addition to its effect on proteasome assembly, PI31 is an adapter for neuronal proteasome transport, suggesting a key role in protein homeostasis and synaptic function (43). To examine the physiological function of PI31, we generated global and conditional knockout mouse strains and investigated how loss of PI31 affects 2 major types of neurons—spinal motor neurons (MNs) and cerebellar Purkinje cells (PCs). Spinal MNs reside in the ventral horn of the spinal cord, while their long axons, that can extend over 1 m in humans, innervate effector muscles at a specialized synapse called the neuromuscular junction (NMJ) (44). PCs are the sole output neurons of the cerebellar cortex. Their dendrites receive inputs from cerebellar granule cell (GC) parallel fibers and inferior olivary nucleus climbing fibers, while their axons project through the inner granular layer (IGL) of the cerebellar cortex, where mature GCs and Golgi interneurons reside, to synapse onto deep cerebellar nuclei neurons (DCNn) in the deep cerebellar nuclei (DCN) (45, 46). Both of these neurons are also involved in a wide range of neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy in the case of spinal MNs, and ataxias, autism, and cerebellar essential tremor in the case of PCs (47–53).
Our study reveals that PI31 is an essential gene, as knockout embryos died at mid to late gestation, although the development and differentiation of many embryonic cell types appeared normal overall. Strikingly, in both MNs and PCs of the cerebellum, conditional loss of PI31 function impaired protein homeostasis in neuronal processes; disrupted the architecture of synapses, axons, and dendrites; and compromised motor function. Importantly, degeneration of neuronal synapses and processes became progressively more severe with age, culminating in neuronal loss. Inactivation of PI31 in MNs and PCs recapitulated the progressive neuropathology and motor dysfunction of previously described mouse models of ALS and ataxia, respectively, and was reminiscent of the severe behavioral and anatomical defects associated with human spinal MN and PC neurodegenerative diseases (54–58). Collectively, this work establishes a critical role for PI31 and protein degradation in the maintenance of neuronal architecture, circuitry, and function. Because mutations that impair PI31 function are thought to cause neurodegenerative diseases in humans, reduced PI31 activity may contribute to age-related neurodegenerative diseases.
Results
Generation of PI31-Null Mice.
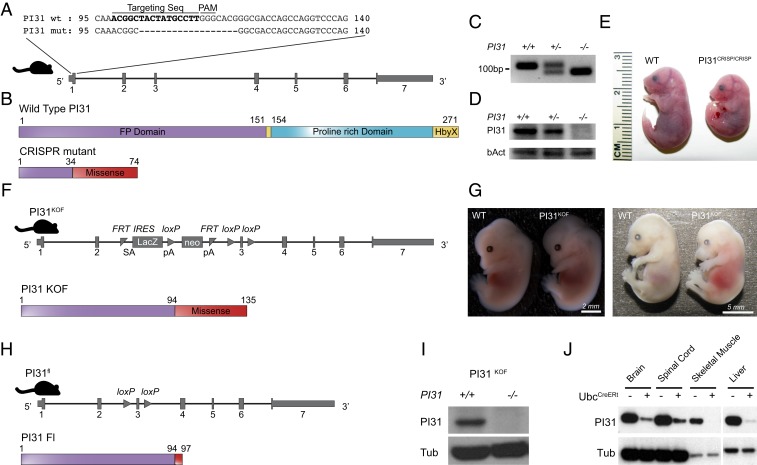
In order to examine the physiological role of PI31, we used 2 independent approaches to generate constitutive and conditional PI31 loss-of-function mouse mutants (Fig. 1). First, we used CRISPR/Cas9 technology to introduce a 16-base pair (bp) deletion in exon 1 of the mouse PI31 gene. This resulted in a frame shift and a premature stop codon (Fig. 1 A–C). Western blot analysis confirmed that no PI31 protein was detectable in PI31CRISP/CRISP embryos, indicating that this allele is a null mutant (Fig. 1D). PI31CRISP/CRISP embryos appeared normal overall prior to embryonic stage E13.5. However, at E15.5, small size differences became detectable, and, by E18.5, reduced growth was apparent (Fig. 1E). PI31CRISP/CRISP die perinatally, and we never observed any newborns. Histological analysis of these embryos indicated that many tissues and organs, such as heart, kidney, gut, muscle, and skin, developed normally overall and contained differentiated cells with appropriate morphology and patterning (SI Appendix, Fig. S1). We conclude that PI31 function, unlike complete inactivation of proteasomes, is not universally required for the growth, differentiation, and survival of many cell types (23).
Fig. 1.
Inactivation of PI31 results in embryonic lethality. Two independent approaches were used to generate constitutive and conditional PI31 loss-of-function mouse mutants. (A) CRISPR/Cas9 technology was used to introduce a 16-bp deletion in exon 1 of the mouse PI31 gene. Alignment of WT and PI31CRISP mutant DNA sequences illustrates the extent of this deletion; the sequence of the targeting guide RNA is depicted in bold. The deletion causes a frame shift and generates a premature stop codon. (B) Schematic presentation of WT PI31 and PI31CRISP protein domains. The mutant protein derived from the PI31CRISP allele is predicted to retain the first 34 amino acids (encoded by nucleotides 1 to 102 of the PI31 coding sequence), followed by 40 missense amino acids and a premature stop codon (FP domain is required for homodimerization and heterodimerization with other PI31 molecules and\or the E3 ligase FBXO7; proline-rich domain is proline-rich unstructured domain; HbyX domain is a domain commonly found in modulators of proteasome activity and considered to be important for binding the 20S proteasome particle). (C) PCR analysis of WT and PI31CRISP DNA confirms the presence of a deletion in the PI31CRISP mutant. (D) PI31 protein levels were examined by Western blot analysis of brain extracts from E13.5 embryos. PI31 protein was readily detected in WT embryos, but levels were reduced in PI31CRISP/+ and not detectable in PI31CRISP/CRISP embryos. (E) PI31CRISP/CRISP mutants are embryonic lethal. Representative pictures of WT and PI31CRISP/CRISP embryos at E18.5 indicate growth retardation and developmental delay. (F) Illustration of the gene architecture and predicted protein product in the KOF PI31KOF allele. A PI31 KO mouse mutant strain (PI31KOF) was generated from commercially available ES cells (Psmf1tm1a(EUCOMM)Hmgu). LacZ is translated from internal ribosome entry site (IRES), and this insertion is predicted to truncate PI31 by retaining the first 94aa encoded in exons 1 and 2, followed by 41 missense amino acids and a stop codon. (G) PI31KOF homozygotes are embryonic lethal. Representative pictures of WT and PI31KOF/KOF embryos at ages E13.5 and E15.5. (H) PI31KOF mice were bred with a strain expressing FLP1 recombinase in order to generate a PI31 allele where the third exon is flanked by 2 loxP sites (PI31fl and Psmf1tm1c(EUCOMM)Hmgu). After Cre recombination, the mutant protein derived from the PI31fl allele is predicted to retain the first 94 amino acids, followed by 3 mutant amino acids and a stop codon. (I and J) Validation of PI31 KO by Western blot. (I) No PI31 protein was detectable in MEFs derived from P31KOF/KOF embryos. (J) PI31f/f mice were bred with mice expressing a Tamoxifen-inducible Cre recombinase under control of the Ubc promoter. Reduced PI31 protein levels were measured in multiple tissues 2 wk after tamoxifen injection.
Because PI31 complete knockout mutants died at advanced embryonic stages, we generated conditional PI31 mutants in order to more carefully examine the role of PI31 in postembryonic cell types. For this purpose, we used the “knockout-first” (KOF) strategy which allows the production of reporter knockouts, conditional knockouts, and null alleles following exposure to the site-specific recombinases Cre and Flp (59, 60). A PI31 KOF (PI31KOF) mutant line was generated from commercially available embryonic stem (ES) cells (Psmf1tm1a(EUCOMM)Hmgu) as illustrated in Fig. 1F. The lacZ insertion introduces a strong splice acceptor, which is predicted to cause severe truncation of the PI31 protein and eliminate domains critical for function (Fig. 1F). Similar to PI31CRISP/CRISP mice, PI31KOF/KOF mice are recessive embryonic lethal, and, by E15.5, embryos were hemorrhagic and smaller in size (Fig. 1G). Western blot analysis confirmed that PI31 protein was not detectable in PI31KOF/KOF embryos (Fig. 1I). We conclude that both the PI31CRISP/CRISP and PI31KOF/KOF strains represent loss-of-function mutants in which expression of PI31 protein was successfully ablated, and that PI31 is an essential gene for mouse development. In order to generate conditional alleles for more in-depth analysis of PI31 function in specific postembryonic cell types, PI31KOF mice were bred with mice expressing FLP1 recombinase to generate a PI31 allele where the third exon is flanked by 2 loxP sites (PI31fl and Psmf1tm1c(EUCOMM)Hmgu) (Fig. 1H). PI31fl/fl mice express normal levels of PI31, are viable and fertile, and appear completely normal. However, introduction of various Cre drivers successfully inactivated PI31 and led to tissue-specific loss of PI31 protein, as indicated in Fig. 1J.
Inactivation of PI31 Causes Neurological Defects, Proteotoxic Stress, and Reduced Proteasome Assembly.
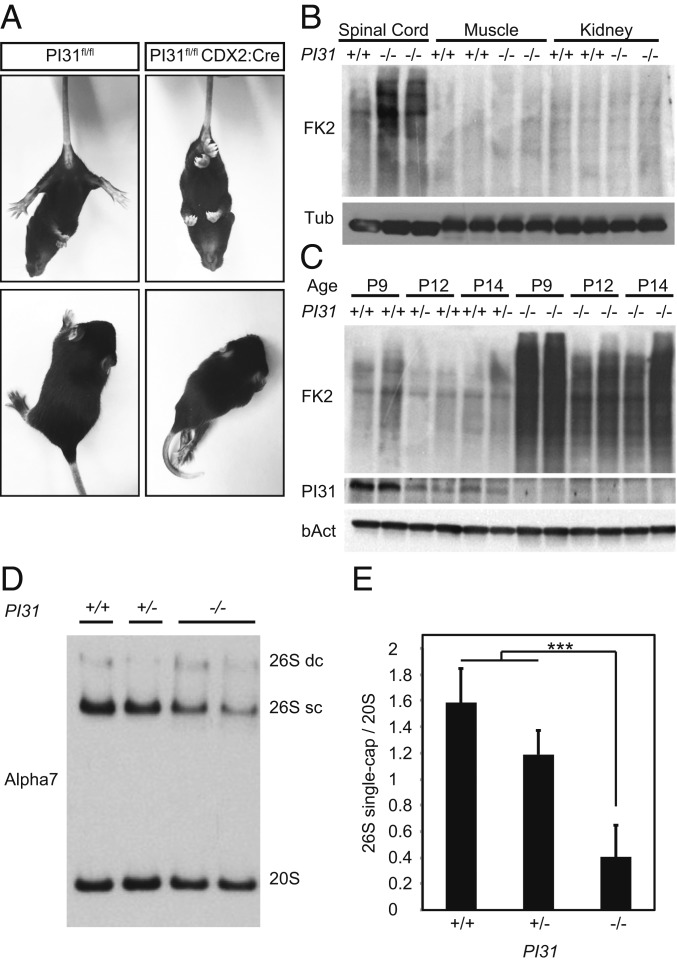
We used PI31fl/fl mice to investigate the physiological role of PI31 in different types of neurons. First, we crossed PI31fl/fl mice with CDX2-Cre mice to inactivate PI31 in the caudal part of the embryo. PI31fl/fl CDX2-Cre mice were viable, but, around P6, they began to develop progressive neuromotor phenotypes, characterized by spasticity, rigid muscle tone, strong tremor, and a severely impaired righting response (Fig. 2A and Movie S1). When these mice were picked up by their tails, they displayed hind leg clasping between episodes of tremor, a hallmark of neuromotor dysfunction (Fig. 2A). Mutant mice were only able to move using their front legs, since their hind limbs were hyperextended and paralyzed (Fig. 2A and Movie S1). These phenotypes became progressively more severe with age, and, by 3 to 4 wk, all mice died. We also noted that, although PI31 was deleted in all caudal tissues—including skin, muscle, and kidney—we did not observe any obvious phenotypes in these tissues.
Fig. 2.
Caudal inactivation of PI31 leads to severe motor defects and proteotoxic stress in the spinal cord. (A) Representative pictures of PI31fl/fl CDX2:Cre mice and a control littermate 14 d postnatal. (Upper) Hind feet clasping, and (Lower) hind limb paralysis in mutant mice. (B) Western blot analysis of spinal cord, muscle, and kidney of PI31fl/fl CDX2:Cre mice and their control littermates at P14. Blotting for polyubiquitinated proteins (FK2) indicates that spinal cord tissue was more sensitive to the loss of PI31 than muscle or kidney. (C) Western blot analysis of spinal cord extracts from PI31fl/fl CDX2:Cre mice and control littermates at indicated ages. Immunoblots for ubiquitin revealed a dramatic accumulation of polyubiquitinated proteins in PI31 mutants. PI31 expression in WT spinal cords was correlated with the accumulation of polyubiquitinated proteins in age-matched PI31 KOs. (D). MEF derived from PI31KOF embryos have decreased levels of 26S proteasomes. Western blot analysis following native gel electrophoresis of WT, heterozygous, and PI31 KO MEFs revealed a reduction in the levels of single-capped 26S proteasome. The 26S double-capped (26S dc) and single-capped (26S sc) and 20S proteasomes were detected with an anti-Alpha7 proteasome subunit antibody. (E) Quantification of proteasome profiles. Densitometry analysis of single-capped 26S and 20S proteasomes was performed, and the ratio of single-capped 26S to 20S proteasomes was calculated for each sample. Pooled data of densitometry analysis of 3 independent experiments show a reduction of single-capped 26S proteasomes in PI31 KO MEFs. Statistical analysis was performed using one-way ANOVA test; *** stands for P value = 0.001. Error bars indicate SD.
Next, we looked for evidence that protein homeostasis was disrupted in PI31fl/fl CDX2-Cre mice. For this purpose, we used the FK2 antibody to detect accumulation of poly-ubiquitin (poly-Ub) proteins, which serves as a readout for proteasome function (61). While no differences between control and PI31fl/fl CDX2-Cre were seen in protein extracted from muscle or kidney, we detected a clear and significant accumulation of poly-Ub proteins in spinal cord extracts from mutant mice (Fig. 2B). Highest levels of FK2 staining were seen in P9 mutant mice, at a time when motoric problems were apparent, but well before the peak of this phenotype and death of mutant animals (Fig. 2C). These observations indicate that inactivation of PI31 in the spinal cord leads to defects in the degradation of poly-Ub proteins, consistent with impaired proteasome function. It was previously shown that PI31 can promote the assembly of active 26S proteasomes from 19S and 20S particles (29). Therefore, we investigated whether loss of PI31 affects the proteasome profile. For this purpose, we used Western blot analysis of native gels to analyze extracts from mouse embryonic fibroblasts (MEFs) derived from PI31KOF/KOF embryos and controls (Fig. 2 D and E). This revealed a small but significant reduction in the levels of single-capped 26S proteasomes, without substantial change of individual subunits (SI Appendix, Fig. S2A). It is worth noting that PI31-null MEFs were viable for extended periods in culture, indicating that these biochemical changes and the modest accumulation of poly-Ub conjugates in mutant MEFs under normal growth conditions did not have profound biological consequences (SI Appendix, Fig. S2). We also saw induction of p62, which is a hallmark of proteotoxic stress (SI Appendix, Fig. S2B) (62, 63). Since p62 delivers poly-Ub aggregates to the autophagic machinery for degradation in lysosome, it is possible that this backup pathway can clear proteins escaping degradation in PI31-null MEFs sufficiently well enough to avoid overt biological consequences. We also treated MEFs with different concentrations of the proteasome inhibitors bortezomib and MG132 to look for a role of PI31 under conditions where protein breakdown is compromised (64). For both proteasome inhibitors, we observed that PI31-null MEFs survived less well (SI Appendix, Fig. S2 C and D). This reveals a modest but significant requirement for PI31 function in fibroblasts under stress conditions. Taken together, our results indicate that loss of PI31 causes decreased proteasome activity and reduces the breakdown of poly-Ub proteins, but that this has little biological consequence for many cell types. In contrast, neurons appear to be particularly sensitive to the loss of PI31 function.
Inactivation of PI31 in MNs Disrupts the NMJ and Models ALS Phenotypes.
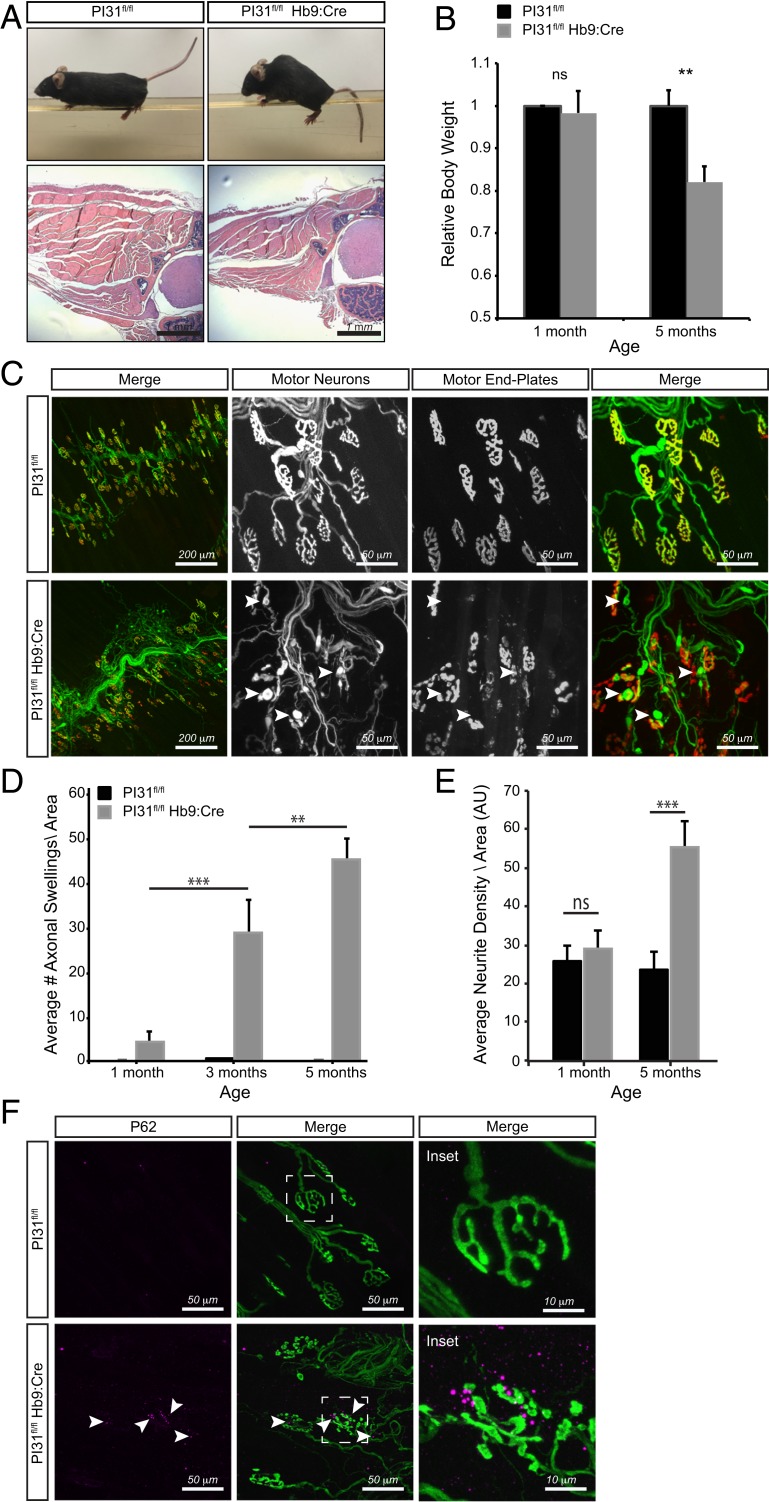
In order to examine a possible requirement of PI31 in neurons in more detail, we used several specific neuronal Cre lines. First, we used the Hb9-Cre to ablate PI31 function in MNs (65, 66). PI31fl/fl Hb9-Cre mice were initially viable but developed motor defects, kyphosis of the spine, and muscle atrophy by the age of 5 mo that became progressively more severe with age (Fig. 3 A and B). External inspection of these mice revealed severe kyphosis due to atrophy and weakness of paraspinal muscles, and also overt motoric defects that recapitulate phenotypes described for mouse ALS models (Fig. 3A and Movie S2) (53, 54, 67). Histological analysis of thoracic cross-sections of 5-mo-old PI31fl/fl Hb9cre mice revealed highly atrophied musculature in PI31 mutants (Fig. 3A) and concomitant reduction in body weight (Fig. 3B). Next, we examined the architecture of the NMJ by staining MN axons with β3-tubulin/synapsin, and the postsynaptic muscle end plate with α-bungarotoxin (Fig. 3C). Compared to age-matched controls, PI31 mutants displayed severe structural abnormalities, including presynaptic fragmentation of the NMJ, axonal tip swellings, and massive axonal sprouting that are all notable at the age of 5 mo (Fig. 3 C–E). These phenotypes recapitulate defects described for several other mouse ALS models (53, 67, 68). The swelling of axonal tips was apparent in PI31-null MNs as early as 1 mo after birth and became progressively more severe with age (Fig. 3D). Interestingly, we did not detect any overt morphological abnormalities, or accumulation of poly-Ub proteins and p62 granules and changes in other ALS related proteotoxic markers (FUS1 and TDP43) in MN cell bodies at this stage (SI Appendix, Fig. S5). Moreover, conditional inactivation of PI31 in muscle did not cause any striking anatomical or behavioral phenotypes, even in mice that were aged up to 8 mo. Finally, we also observed accumulation of p62-positive aggregates at the NMJ of PI31fl/fl Hb9-Cre mice, but not in control littermates, indicating that loss of PI31 disrupts proteostasis in axon terminals of MNs (Fig. 3F). Notably, these phenotypes are remarkably similar to those caused by conditional inactivation of Rpt3, a critical component of the proteasome regulatory particle that is required for the degradation of most proteasome substrates (54, 69).
Fig. 3.
Inactivation of PI31 in MNs causes progressive muscle denervation and atrophy. (A) PI31fl/fl Hb9:Cre mice develop motor defects, kyphosis of the spine, and muscle atrophy. Representative picture of PI31fl/fl Hb9:Cre mice and their WT controls at 6 mo of age. A representative hematoxylin/eosin staining of thoracic cross-sections of 5-mo-old PI31fl/fl Hb9cre mice and control littermates (PI31fl/fl without Cre) showing atrophied musculature of KO mice. (B) Relative body weight of 1- and 5-mo-old PI31fl/fl Hb9:Cre mice and their control littermates. PI31fl/fl Hb9:Cre mice suffer progressive weight loss. For 1MO, control n = 8 and KO n = 4; for 5MO, control n = 8 and KO n = 4. Weight of WT controls at each age was set to 1. Statistical analysis was performed with a 2-tailed paired t test; ** stands for P value < 0.01, ns stands for not significant. Error bars indicate SD. (C) Inactivation of PI31 in MNs disrupts the structure of the NMJ. Innervation of the triangularis sterni muscle was visualized by staining for β3-tubulin/synapsin (green), and postsynaptic muscle end plates were visualized with α-Bungarotoxin (red). (D and E) The 5-mo-old PI31fl/fl Hb9:Cre mutant mice have fragmented NMJs, axonal swellings (indicated with white arrowheads), and massive axonal sprouting. Loss of PI31 in MNs results in a progressive pathology, illustrated by the increase in axonal tip swellings and neurite sprouting with age. (D) A bar diagram of the average number of axonal swelling per area unit (AU = 850 µm2) at different ages as annotated. (E). Axonal sprouting was quantified by measuring neurite density per AU. Data are represented as bar diagrams of the average neurite density at different ages as annotated (see Material and Methods and SI Appendix, Figs. SI and S4 for more details); ** stands for P value < 0.01; *** stands for P value < 0.001. Error bars indicate SD. (F) P62 granules accumulate at the NMJ of PI31fl/fl Hb9:Cre, but not in control littermates. Representative image is of NMJ in the triangularis sterni muscle of PI31fl/fl and PI31fl/fl Hb9:Cre mice (4MO females); p62 granules are in magenta (marked with white arrowheads), and MNs are in green (stained by anti β3-tubullin and synapsin antibodies). Dashed white boxes indicate insets shown to the right.
Inactivation of PI31 in PCs Causes Progressive Motor Defects and Disrupts Protein Homeostasis.
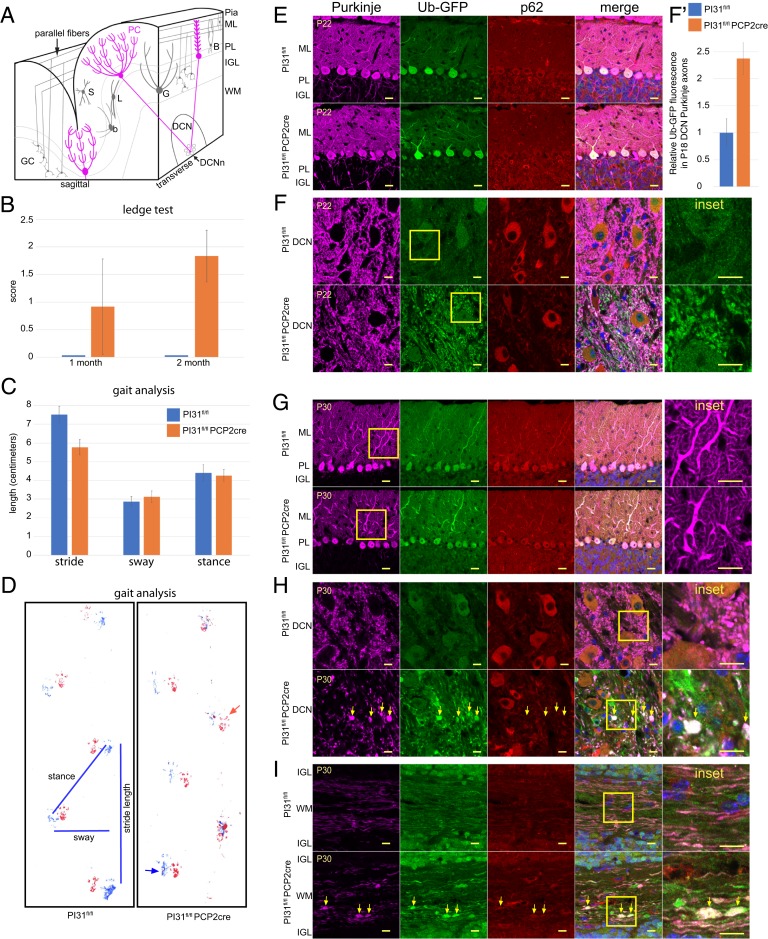
We also examined the role of PI31 in the cerebellum, as it provides an outstanding model for studying how proteotoxic stress impairs neuronal function, structure, and survival (70–78). PCs are very large neurons that are particularly sensitive to proteotoxic stress, which is thought to be a major factor contributing to cerebellar diseases (71, 79). As the PC is the sole output neuron of the cerebellar cortex, defects in PC function cause both motor and cognitive dysfunction. We used the PC-specific Cre driver Tg(Pcp2-Cre)GN135Gsat to examine how conditional loss of PI31 affects the health, structure, and viability of these cells as well as associated mouse behaviors. Behavioral analysis of PI31fl/fl Pcp2-Cre mice revealed a series of defects characteristic for impaired PC function, and these defects became progressively more severe with age. First, we used the ledge test and gait analysis to look for possible abnormalities in motor coordination in PI31fl/fl Pcp2-Cre mice (Fig. 4 B–D). At P21, PI31fl/fl Pcp2-Cre mice were indistinguishable from their PI31fl/fl littermates. However, by P30, PI31fl/fl Pcp2-Cre mice displayed an aberrant halting gait and frequently lost their balance (Fig. 4B). By P60, mutants had severely disrupted balance, frequently dragged their bodies along the ledge, and fell on their heads when lowering themselves into the cage. Locomotor defects were also seen with gait analysis in older mutant mice (Fig. 4C and Movie S3). Taken together, these behavioral abnormalities suggest defects in PC function.
Fig. 4.
Loss of PI31 in PCs leads to aberrant dendrite and axon morphology. (A) Schematic illustrating cerebellar anatomy. PCs (magenta) are the only output of the cerebellar cortex. Their dendrites in the molecular layer (ML) receive inputs from GC parallel fibers and climbing fibers from the inferior olivary nucleus. PC cell bodies form the PC layer (PL), while their axons project through the IGL where mature GCs and Golgi interneurons (G) are found, to the DCN, where they synapse with the DCNn. WM is white matter. (B–D) PI31fl/fl Pcp2-Cre mice display progressive behavioral defects. Up to P22, PI31fl/fl Pcp2-Cre mice were not distinguishable from PI31fl/fl mice in their righting reflex and visual observation of gait. (B) By P30, PI31fl/fl Pcp2-Cre mice began to show loss of balance in the ledge test, which became more severe by P60. The number of mice used for this experiment were: for P30, PI31fl/fl (n = 12) and PI31fl/fl Pcp2-Cre (n = 12), for P60, PI31fl/fl (n = 15) and PI31fl/fl Pcp2-Cre (n = 24). Error bars indicate SD. (C and D) At 10 mo, the gait of PI31fl/fl Pcp2-Cre mice was disturbed, and mutants frequently lost balance. Gait analysis showed a significant decrease in stride length (P = 0.00258 [2-tailed t test), PI31fl/fl [n4] and PI31fl/fl Pcp2-Cre [n4]). Error bars indicate SD. (D) An example of footprints used for gait analysis with measurements of length, sway, and stance. Hind paw prints are blue, and forepaw prints are red. The red arrow points to a common misplaced step for PI31fl/fl Pcp2-Cre mice, while the blue arrow indicates the hind paw touching the surface with splayed toes. (E–I) Cerebellum labeled for Calbindin (magenta) to mark PCs, Ub-GFP (green), p62 (red), and Hoechst 33342 (blue) for nuclei. Yellow box denotes Inset. PI31fl/fl and PI31fl/fl Pcp2-Cre denote PI31fl/fl Ub-GFP and PI31fl/fl Pcp2-Cre Ub-GFP mice, respectively. (E and F) Mature PCs at P22 appeared virtually normal in PI31fl/fl Pcp2-Cre mice when compared to PI31fl/fl mice. (E) PC cell bodies appeared normal, and their dendritic arbors were fully developed. (F) PI31fl/fl Pcp2-Cre mice Purkinje axons in the DCN appeared morphologically normal, but show an increase in Ub-GFP over their control PI31fl/fl siblings. (F′) Level of Ub-GFP increased in P18 PC axons in the DCN of PI31fl/fl Pcp2-Cre mice (significance p – 0.0037 [2-tailed t test], PI31fl/fl [n3] and PI31fl/fl Pcp2-Cre [n3]). Error bars indicate SD (SI Appendix, Fig. S6). (G and H) By P30, PI31fl/fl Pcp2-Cre mice PC primary and secondary dendrites appeared swollen, while their axon terminals in the DCN showed swellings with increased Ub-GFP (yellow arrows), indicating impaired protein breakdown. (I) Loss of PI31 caused swellings (yellow arrows) along the length of the axons in the white matter (P30). (E–H) Single confocal slices. (I) A maximum intensity projection of a confocal stack. (Scale bars: E and G, 20 µm; F, H, and I, 10 µm.)
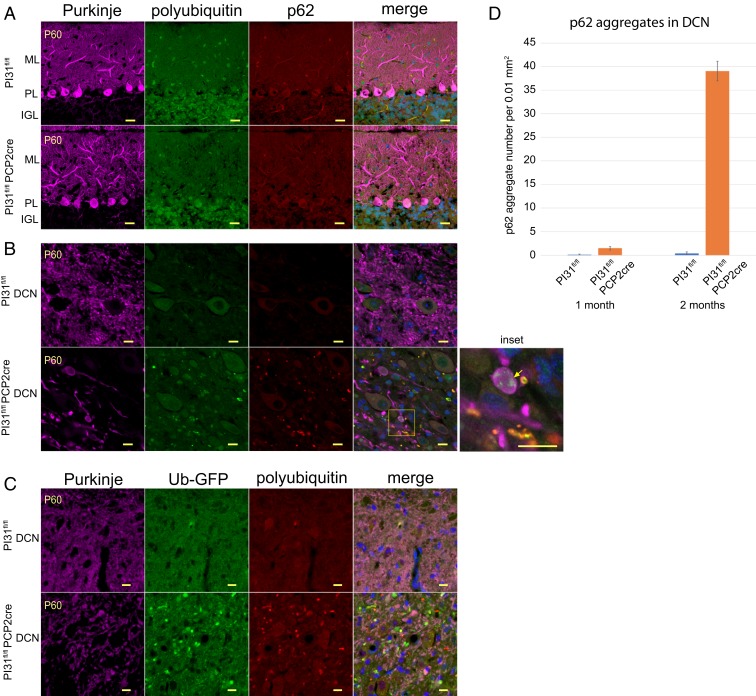
Next, we looked for evidence of changes in protein degradation and cellular structure of PCs. At P18 and P22, PCs did not appear to have overt morphological abnormalities in PI31fl/fl Pcp2-Cre mice, with well-developed dendritic arbors and axonal morphology (Fig. 4 E and F). However, we observed increased staining for ubiquitin-G76V-green fluorescent protein (Ub-GFP) expressed from the transgene (Tg(CAG-Ub*G76V/GFP)1Dant/J), a marker for proteotoxic stress (Fig. 4 F and F′ and SI Appendix, Fig. S6). By P30, PC primary and secondary dendrites appeared swollen in PI31fl/fl Pcp2-Cre mice (Fig. 4G). Significantly, PC axon terminals in the DCN had swellings with increased Ub-GFP, indicating impaired protein breakdown (Fig. 4H). We also found that loss of PI31 caused swellings along the length of the axons in the white matter. These swellings were positive for Ub-GFP, consistent with impaired protein breakdown in mutant PC axons (Fig. 4I). Interestingly, it appeared that the compartment most affected by the loss of PI31 at this stage was the PC axon. By P60, the DCN contained large aggregates throughout which were positive for both poly-Ub conjugates and p62 (Fig. 5 and SI Appendix, Fig. S7). Quantification revealed a striking increase in p62-positive aggregates between 1- and 2-mo-old mutant mice (Fig. 5D). These data demonstrate that the loss of PI31 function exposes PCs to proteotoxic stress that progressively increases over time.
Fig. 5.
Loss of PI31 in cerebellar PCs leads to axon degeneration and the appearance of aggregates positive for proteotoxic stress markers (p62, polyubiquitin and Ub-GFP). Immunostaining of cerebellum from 2-mo-old PI31fl/fl Ub-GFP (PI31fl/fl) and PI31fl/fl Pcp2-Cre Ub-GFP (PI31fl/fl Pcp2-Cre) mice. (A and B) Calbindin-staining (magenta) for PCs, and polyubiquitin (FK2, green), p62 (red), and Hoechst 33342 (blue) for nuclei. (A) Loss of PI31 did not cause noticeable accumulation of either p62, FK2, or Ub-GFP aggregates in the cell bodies or dendrites of P60 PCs. (B) Swellings of PC axon terminals in the DCN and accumulation of both poly-Ub and p62-positive aggregates was observed in the DCN of P60 mutant mice (see also SI Appendix, Fig. S7). Yellow arrow in Inset points to an example of poly-Ub aggregates (green) in axon swellings. (C) Ub-GFP−positive aggregates (green) in the DCN of P60 mutants. Images in A–C are single confocal slices. (Scale bars: A, 20 µm; B and C, 10 µm.) (D) Quantification of p62 aggregates in the DCN reveals a dramatic increase from 1- to 2-mo-old PI31fl/fl Pcp2-Cre mice. The p62-positive aggregates were counted in a 0.01-mm2 area in the DCN of P30 PI31fl/fl (n = 3) and PI31fl/fl Pcp2-Cre (n = 3) and P60 PI31fl/fl (n = 4) and PI31fl/fl Pcp2-Cre (n = 4) mice. Statistical significance between loss of PI31 mice and control mice at 2 mo is P = 6.49 × 10−5 (2-tailed t test). Error bars indicate SD.
Many neurodegenerative diseases are associated with gliosis (glial cells become reactive and proliferate), which often precedes the formation of aggregates, tangles, and plaques (80–84). Microglia, the macrophages of the brain, are very sensitive to neuronal damage. The presence of reactive microglia is a prominent marker of gliosis, and it can be assessed by Iba1 expression and cellular morphology (80, 85). We did not detect an increase in reactive microglia in the cerebellum of PI31fl/fl Pcp2-Cre mice at P18, despite the elevated expression of Ub-GFP (SI Appendix, Fig. S8). However, reactive microglia became detectable in the DCN of PI31fl/fl Pcp2-Cre mice at P22, and gliosis became more severe at P30 (SI Appendix, Fig. S8).
An increase in glial fibrillary acidic protein (GFAP) and hypertrophy of astrocytes are markers of astrogliosis (81, 86). In the mouse cerebellum, GFAP mainly stains the Bergmann glia in the Purkinje and molecular layers, and astrocytes which are predominantly found in the white matter. For PI31fl/fl Pcp2-Cre mice, the GFAP staining at P22 was very similar to their control littermates (SI Appendix, Fig. S9). However, by P30, there was a dramatic increase in reactive astrocytes in the DCN, but not the Purkinje or molecular layers (SI Appendix, Fig. S9). At P60, we observed some reactive astrocytes in the Purkinje layer, coinciding with the onset of PC loss. Collectively, these observations indicate that inactivation of PI31 causes progressive damage to PC axon terminals, which induces gliosis.
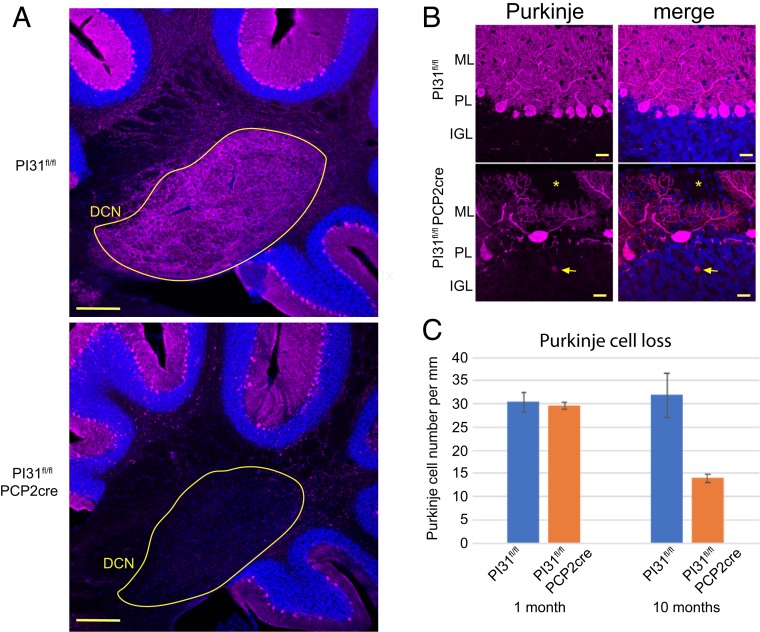
An extension of this analysis to older mice revealed that mutant PCs had lost almost all of their axon terminals in the DCN by the age of 10 mo (Fig. 6B). We also observed axonal torpedoes, focal swellings on PC axons that have long been associated with cerebellar disorders (87–90). In addition, at this time, there was a major reduction in PC number (Fig. 6 A and C). This striking loss of PCs upon inactivation of PI31 is consistent with the motor defects revealed by gait analysis in these mice. Collectively, our results indicate that loss of PI31 initially impairs local protein homeostasis in axons and dendrites of PCs and disrupts their structure, and that, over time, this compromises cell survival.
Fig. 6.
Loss of PI31 in cerebellar PCs leads to PC loss. (A) Overview showing loss of PC axons in the DCN of the PI31fl/fl Pcp2-Cre cerebellum. Immunostaining of cerebellum from 10-mo-old PI31fl/fl and PI31fl/fl Pcp2-Cre mice. PCs were marked by Calbindin staining (magenta) and nuclei with Hoechst 33342 (blue). DCN circled in yellow. Note near complete loss of Calbindin staining in the DCN of the PI31fl/fl Pcp2-Cre cerebellum. (Scale bar, 200 µm.) (B) Aberrant morphology and loss of PCs. The yellow asterisk marks a gap of PC dendrites in the molecular layer, and the arrow points to an axonal torpedo. PC dendritic arbors were also severely reduced. PCs were marked by Calbindin (magenta) and nuclei with Hoechst 33342 (blue). Maximum intensity projection (6 µm). (Scale bar, 20 µm.) (C) Graph showing number of PCs per millimeter of Purkinje layer. At 1 mo of age, the number of PCs was similar to control, but, by 10 mo, PC number had decreased to about 44% of control mice (P = 0.0175 [2-tailed t test]; P30 PI31fl/fl [n = 3] and PI31fl/fl Pcp2-Cre [n = 3]; 10 mo PI31fl/fl [n = 3] and PI31fl/fl Pcp2-Cre [n = 3] mice).
Discussion
In the present study, we addressed the physiological role of PI31 in mammals by generating complete and conditional PI31 mouse mutants. In contrast to the function implied by its original name, we provide compelling evidence for a role of PI31 in stimulating protein breakdown that is critical for maintaining synaptic structures and long-term survival of neurons. Interestingly, the first morphological defects caused by conditional inactivation of PI31, in both MNs and PCs of the cerebellum, were observed in the distal part of neurons, namely axons and synapses. These structural defects were accompanied by molecular proteotoxic stress markers (accumulation of Ub-GFP, poly-Ub proteins, and p62-positive granules) at the NMJ and in DCN. Importantly, symptoms became progressively more severe with age, culminating in the degeneration of axons and dendrites, and, eventually, cell loss; these morphological abnormalities were also associated with motoric problems, modeling ALS and cerebellar disorders, respectively. Overall, these observations suggest that PI31 activity is required for protein homeostasis in distal compartments of neurons.
A plausible mechanistic explanation for these defects comes from our recent finding that PI31 serves as an adapter protein to mediate fast axonal transport of proteasomes (43). PI31 can directly bind to dynein light chain proteins and thereby couples proteasomes to cellular motors. Significantly, inactivation of PI31 in Drosophila blocks proteasome motility and disrupts synaptic protein homeostasis and structure. This function of PI31 is conserved from insects to mammals, since inactivation of PI31 in mouse hippocampal neurons impairs proteasome movement in axons as well (43). Although we have not yet been able to perform live-cell imaging analyses to directly demonstrate a requirement of PI31 for proteasome movement in mouse MNs or PCs, impaired transport of proteasomes in axons and dendrites provides an attractive explanation for the observed defects in protein homeostasis at nerve endings. Synapses are highly dynamic sites of active local protein synthesis, which has to be balanced by local protein degradation (91). According to our model, inactivation of PI31 leads to a shortage of proteasomes at synapses, which, in turn, causes the accumulation of poly-Ub proteins and p62-positive aggregates. It appears that backup clearance mechanisms, such as autophagy and possibly also shedding of microvesicles/exosomes, can partially compensate for the loss of PI31-mediated proteasome activity in PI31 mutants (6, 92–96). The latter would also explain the gliosis that we observed already at P30 in PI31fl/fl Pcp2-Cre mice. However, even if backup clearance pathways are induced, neurons eventually succumb to defects in the absence of PI31 function.
Interestingly, conditional inactivation of PI31 in MNs resulted in kyphosis and ALS-like behavioral phenotypes comparable to inactivation of Rpt3, an essential component of the proteasome 19S regulatory particle (54). This is somewhat surprising, since Rpt3, unlike PI31, is an essential component of the proteasome regulatory particle. In the mouse embryo, complete inactivation of Rpt3 causes a much more severe phenotype (Rpt3-deficient mice die before implantation) than complete inactivation of PI31 (69). Unlike PI31, inactivation of Rpt3 primarily disrupted protein homeostasis in the MN cell body (54). We interpret this to indicate the importance of localized proteasome activity in axons and dendrites of neurons: Whereas Rpt3 function is required in every cellular compartment, including the nucleus and cell body, PI31-mediated proteasome transport appears to be particularly important for protein homeostasis in axons and dendrites. Since neuronal activity critically relies on proper synaptic function, it may be particularly sensitive to perturbation of synaptic protein homeostasis mediated by PI31. Moreover, it is also possible that proteasomes can be active independent of the main regulatory particle, at least to a certain degree and in some subcellular compartments. Together, the critical and conserved role of PI31 in proteasome transport, the accumulation of nondegraded poly-Ub conjugates and induction of proteotoxic stress in PI31 mutants, and comparison with Rpt3 inactivation in MNs all support a function of PI31 as a positive regulator of proteasome activity.
The activity of PI31 critically depends on Fbxo7, also termed PARK15 in humans, which encodes the substrate recognition component of an SCF E3 ligase complex and binds directly to PI31 (26, 30–32). Conditional inactivation of PI31 in mice causes neuronal degeneration, and human mutations in Fbxo7/PARK15 are associated with neurodegenerative diseases (33, 34, 42, 97, 98). Strikingly, inactivation of Fbxo7 leads to severe reduction of PI31 protein and reduced proteasome activity in both Drosophila and mice (26, 32, 33). This suggests the possibility of a direct mechanistic link between PI31, reduced proteasome function, and human disease. However, a role for PI31 in mediating the proteasome impairment and neuronal degeneration observed in Fbxo7/PARK15 mutants was dismissed based on results from cell culture experiments (33, 99). The current study, together with the work of Liu et al. (43), suggests that this topic deserves more careful examination. MEFs deficient for PI31 survived for extended periods of time, and we were unable to detect a major reduction of total proteasome activity in these cells. However, local defects in proteasome activity would not have been revealed by these approaches. When combined with proteasome inhibitors, inactivation of PI31 resulted in a small but significant reduction of cell survival. These findings are consistent with the reported role of the yeast PI31 ortholog to alleviate proteotoxic stress (27). We also saw that PI31-null MEFs had slight defects in proteasome assembly, as indicated by reduced numbers of single-capped 26S particles. Therefore, PI31 may regulate proteasomes by multiple mechanisms: through the assembly of functional 26S proteasomes from 19S and 20S particles, and via transport (29, 43). In any event, it appears that neurons are particularly sensitive to the loss of PI31 function, and a neuroprotective role of PI31 should not be ruled out based on results with nonneuronal immortalized cells.
A neuroprotective role of PI31 was particularly obvious in MNs and PCs, 2 major types of neurons with long axons. Therefore, it is possible that large neurons are especially sensitive to the impairment of PI31-mediated proteasome transport, since they face a particularly challenging problem in allocating proteasomes to compartments where protein breakdown has to occur. Moreover, PCs are very sensitive to proteotoxic stress, and PC pathology is a common finding in cerebellar diseases (47, 71, 79). Consistent with this, inactivation of PI31 in these cells caused proteotoxic stress as soon as these cells matured (P22), and it led to behavioral and anatomical defects that are characteristic for cerebellar diseases. These include progressively more severe loss of balance and defects in gait, which coincided with axonal swelling, torpedoes, gliosis, and, eventually, loss of PC bodies.
It is well known that protein clearance mechanisms decline during the aging process, and impairment of the UPS in neurodegenerative diseases has been widely reported (35, 36, 39, 40, 96, 100–103). Most of the attention in previous studies has been focused on the ability of aggregates to inhibit proteasome activity (37, 41). However, aggregate-mediated proteasome inhibition cannot explain the failure to degrade poly-Ub proteins and accumulation of aggregates in the first place. We suggest that impaired activity of PI31, caused either by mutations affecting PI31 function (such as Fbxo7/PARK15) or through epigenetic mechanisms during the normal aging process, contributes to age-related neuronal degeneration. The mouse mutant strains described here provide powerful models to further investigate this possibility.
Materials and Methods
Mice.
In order to generate PI31 mutant using CRISPR\Cas9 technology, we have designed a single-guide targeting RNA that will target the mouse PSMF1 gene in the first exon right after the first Methionine (Psmf1a-1 CACCGAACGGCTACTATGCCTT, Psmf1a-2 AAACAAGGCATAGTAGCCGTTC). Targeting sequence was ordered as oligos and cloned into plasmid px330 (a gift from F. Zhang at Massachusetts Institute of Technology) that contains both Cas9 and the tracer RNA sequence. ES cells were transfected with this plasmid, and Surveyor assay was performed to assess targeting efficiency. Gene targeting resulted in 5 mutant lines, all of which were heterozygous for an indel in the PSMF1 gene. After sequencing the relevant genomic loci, we have decided to continue our research with one clone, which has a 16-bp deletion in the first exon (nt103 to nt118 of open reading frame) that results in a frame shift and a premature stop codon in the second exon (nt239). Homozygous mutants are not viable and die perinatal; hence, we used tissue from E13 homozygous mutant embryos (which are morphologically similar to wild types [WTs]) to test the presence of PI31 protein.
For the generation of conditional PI31 knockout (KO) mice, we have purchased commercially available ES cells (Psmf1tm1a(EUCOMM)Hmgu). We sequenced these ES cells to validate that they harbor PI31 KO first allele with conditional potential. Chimeras were generated by injection of targeted ES cells into albino blastocysts purchased from The Jackson Laboratory (B6(Cg)-Tyr<c-2J>/J). Next, chimeras were crossed with B6 albino mice (B6(Cg)-Tyrc-2J/J) from The Jackson Laboratory. Heterozygous transgene mice were then bred to mouse expressing FLP-1 recombinase gene under the direction of the human ACTB promoter (The Jackson Laboratory #005703), to create a mouse line where the third exon PI31 is flanked by 2 LoxP sites (PI31fl/fl). Conditional PI31fl/fl were generated using CDX2-Cre, Hb9-Cre, and Ubc-CreERt, purchased from The Jackson Laboratory.
C57B1/6J PI31fl/fl mice were crossed with C57B1/6J Tg(Pcp2-Cre)GN135Gsat generated by and obtained from GENSAT (Rockefeller University). PI31fl/flPcp2-Cre mice were compared to PI31fl/fl control littermates. Genotyping of PI31fl/f and Pcp2-Cre alleles was carried out by PCR. All animal work was performed as required by the United States Animal Welfare Act and NIH's policy to ensure proper care and use of laboratory animals for research, and under established guidelines and supervision by the Institutional Animal Care and Use Committee of The Rockefeller University. Mice were housed in accredited facilities of the Association for Assessment of Laboratory Animal Care in accordance with NIH guidelines.
Behavioral Tests.
Righting reflex.
To test for defects in the righting reflex, P7, P10, P14, and P22 pups were placed in a supine position, and the time taken to right onto all 4 paws was measured. Following rest periods, this was repeated for a total of 3 trials which were then averaged (104).
Ledge test.
The mouse was placed on the ledge of the cage and scored for coordination and balance (105). A mouse which walked the ledge with few slips and descended into the cage gracefully, landing on its paws, was given a score of 0. A mouse which lost its footing received a score of 1. A mouse that did not use its hind paws well and landed on its head instead of its paws when descending into the cage received a score of 2. A mouse that refused to move, even with gentle encouragement, or fell or nearly fell while walking the ledge or lowering itself into the cage received a score of 3.
Gait analysis.
The forepaws of each mouse were painted with nontoxic red paint, and the hind paws were painted with nontoxic blue paint, after which the mice were allowed to walk on paper along a 10-cm-wide by 76-cm-long corridor with 20-cm-high opaque walls. This was done 3 times for each mouse, and each parameter was measured for the 3 longest strides in the middle of a run. The footprint patterns were analyzed for 3 step parameters (all measured in centimeters). As per Carter et al. (106), 4 parameters were measured: 1) Stride length was measured as the average distance of forward movement between each stride. 2) Hind-base width, or sway, was measured as the average distance between left and right hind footprints. These values were determined by measuring the perpendicular distance of a given step to a line connecting its opposite preceding and proceeding steps. 3) Distance from left hind footprint to right hind was measured to determine stance length.
Native Gel Electrophoresis.
Protein extracts of tissue were prepared in lysis buffer (50 mM Tris‐HCl [pH 8.0], 5 mM MgCl2, 0.5 mM ethylenediaminetetraacetic acid [EDTA], 2 mM adenosine 5′-triphosphate [ATP], 0.2% Nonidet P-40, protease inhibitor, phosphatase inhibitor) using liquid nitrogen freezing and thawing technique. Extracts were cleared by centrifugation at 14,000 rpm for 30 min to remove nuclei and cell debris, and protein concentrations were measured by Bradford assay. To resolve proteasomes, 26-well 3 to 8% Triacetate gels were used. Samples were mixed with 2× native loading buffer just before loading. Electrophoresis was carried out at room temperature (1 h at 50 V) and then at 4 °C for an additional 5 h at 120 V, in running buffer (0.45 M Tris, 0.45 M Boric Acid, 5 mM EDTA, 12.5 mM MgCl2, supplied with 0.5 mM dithiothreitol, 0.5 mM ATP). For immunoblotting, proteins in native gels were transferred to 0.45-mm poly(vinylidene difluoride) (PVDF) membranes. The 26S and 20S were detected with anti-Alpha7 antibodies (1:5,000; Enzo). Purified bovine 20S, and 26S proteasomes, were used as standards.
Antibodies and Western Blot.
Protein extracts were prepared as for native gel electrophoresis. For Western blotting, proteins were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a 0.45 mm PVDF membrane. Membranes were blocked overnight at 4 °C with 5% milk in phosphate-buffered saline (PBS) and 0.5% Tween-20 (PBST). Membranes were incubated for 60 min with primary antibodies (antibody details in SI Appendix) at room temperature. Primary antibodies were detected by secondary species-specific horseradish peroxidase-conjugated antibodies (1:5,000; Jackson ImmunoResearch). Detection was performed with Amersham ECL Western Blotting Detection Reagent.
Cell Survival Assay.
Low-cycle (4 to 6) primary MEFs from WT or PI31 KO embryos were seeded in a 96-well plate at a concentration of 15,000 cells per well and incubated overnight in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and penicillin/streptomycin at 37 °C under 5% CO2 (vol/vol). Then, cells were incubated for an additional 18 to 22 h with different concentrations of the proteasome inhibitors Bortezomib or MG132, 3 wells per concentration. Cell viability was measured using PrestoBlue viability reagent as described in the manufacture protocol. The experiment was repeated 3 independent times.
Immunofluorescence Muscle Innervation.
Whole-mount staining of triangularis sterni muscle was done as described in Brill et al. (107). Briefly, the thoracic wall containing the triangularis sterni muscle was dissected and fixed in 4% PFA for 1 h on ice. The fixed tissue was then washed for 10 min in 0.1 M glycine, and the triangularis sterni muscle was dissected out. The dissected muscle was transferred into blocking solution (10% goat serum, 0.5% TX-100 in PBS) for 1 h at room temperature. After blocking, tissue was incubated with Alexa488 conjugated primary antibodies for Tuj1 and Synapsin (antibody details in SI Appendix) overnight at 4 °C. To label postsynaptic acetylcholine receptors, samples were incubated in Alexa594-α-bungarotoxin (reagent details in SI Appendix). Tissue was washed 3× in PBS and mounted on a slide with Vectashield mounting media.
Quantification of Axonal Swellings.
For the quantification of axonal swellings, z-stack images of age-matched PI31f/f controls and PI31f/f Hb9:Cre KO animals were taken at low magnification (10×). Maximal intensity Z-projects images were generated using Fiji software, and axonal swellings were counted manually. Average number of axonal swellings was calculated based on 3 images per animal. Image size was 850 μm2; for 1-mo-old (1MO), control n = 3 and KO n = 3; for 3MO, control n = 2 and KO n = 3; for 5MO, control n = 2 and KO n = 4. P value was calculated using the Student t test.
Quantification of Axonal Sprouting.
Quantification of axonal sprouting was performed on the same images used for the quantification of axonal swelling with Fiji software tools. Briefly, for each image, three 200 × 200 pixel regions of interests (ROI) were chosen. Images were then skeletonized, and total axon length was measured for each ROI (SI Appendix, Fig. S3). For 1MO, control n = 3 and KO n = 3; for 5MO, control n = 2 and KO n = 4. P value was calculated using the Student t test.
Immunofluorescence of Spinal Cord Sections.
Mice were perfused with 4% paraformaldehyde in phosphate buffered saline (PFA/PBS), and the brains were removed and then postfixed in 4% PFA/PBS overnight and transferred to 70% ethanol. Vertebrae were removed, and cervical spinal cord (C3 to -C4) was embedded in paraffin blocks. Four μm coronal sections were subjected to immunofluorescence analysis. Sections were deparaffinized and rehydrated. Heat-induced antigen retrieval was performed in sodium citrate pH 6.00 buffer, in a pressure cooker, 125 °C for 20 min. Sections were blocked in CAS-Block for 1 h at room temperature, followed by an overnight incubation at 4 °C with primary antibodies. Primary antibodies (antibody details in SI Appendix) were diluted in CAS-Block. Subsequently, sections were washed 3 times in Tris buffered saline with 0.5% Tween 20 (TBST) 0.5% tween20. Sections were then incubated with secondary antibodies that were diluted in TBS. After incubation with the species-appropriate secondary antibody (Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647 [1/500; Invitrogen]), sections were washed 3 times in TBST and mounted with Vectashield mounting medium.
Images were captured on a Zeiss confocal LSM780 with Zen software. Images shown are single slices unless otherwise noted.
Immunofluorescence of Cerebellar Sections.
Mice were perfused with 4% PFA/PBS, and the brains were removed and then postfixed in 4% PFA/PBS overnight at 4 °C. They were then washed in PBS, soaked in 15% sucrose/PBS for several hours until they sank, and then soaked in 30% sucrose/PBS overnight at 4 °C. The brains were then embedded in Richard-Allan Scientific Neg-50 Frozen Section Medium, and 50-µm sagittal sections were made using a Leica CM 3050S cryostat.
The floating cryostat sections were permeabilized and blocked overnight in 1× PBS/0.3% Triton X-100/5% Normal Donkey Serum (NDS) at 4 °C. The sections were then incubated overnight with primary antibodies (detailed in SI Appendix) diluted in 1× PBS/0.3% Triton X-100/5% NDS. Subsequently, the sections were washed 3 times for 20 min each in 1× PBS/0.3% Triton X-100/5% NDS and incubated overnight with Alexa Fluor conjugated secondary antibodies diluted in 1× PBS/0.3% Triton X-100/5% NDS at 4 °C. The sections were then washed 4 times for 30 min in 1× PBS/0.3% Triton X-100/5% NDS and then mounted with Molecular Probes ProLong Diamond antifade mounting medium. Images were captured on a Zeiss confocal LSM780 with Zen software. Images shown are single slices unless otherwise noted.
Quantification of Ub-GFP Fluorescence in Purkinje Axons.
Confocal images of cerebellar sections stained with anti-CALB1 to mark the PCs, and anti-GFP were taken with identical confocal settings (Zeiss confocal LSM780 with Zen software). These images were used for quantification of Ub-GFP in Purkinje axons (illustrated in SI Appendix, Fig. S6). In brief, the calbindin image was used to create an ROI with ImageJ software, which was then superimposed on the Ub-GFP image to measure the integrated density and area of the ROI. For each mouse, at least 3 images per DCN were quantified and averaged, and then the average integrated density/area of PI31fl/fl Pcp2-Cre mice (n3) and the average integrated density/area of PI31fl/fl control sibling mice (n3) were determined. Excel was used to determine SD and statistical significance by 2-tailed t test.
P62 Aggregates.
Confocal images of cerebellar sections stained with anti-CALB1 to mark the PCs and anti-p62. Aggregates within the DCN were manually counted and converted to aggregates/area for P60 PI31fl/fl Pcp2-Cre mice (n3) and their control littermates (n3). Excel was used to determine SD and statistical significance by 2-tailed t test.
PC Counting.
PC bodies were manually counted from confocal images of cerebellar sections immunostained with anti-CALB1 to mark the PCs of P30 and 10-mo-old PI31fl/fl Pcp2-Cre mice (n3, n3) and their control littermates (n3, n3). This count was converted to cells per square millimeter. Excel was used to determine SD and statistical significance by 2-tailed t test.
Data Availability.
Details and availability of antibodies, mice, genotyping primers, genotyping PCR program, and reagents used in this work are included in SI Appendix, Tables S1–S5, respectively.
Supplementary Material
Acknowledgments
We thank Drs. David Ng and Thomas Jessell for advice, for invaluable support, and for providing the Hb9-Cre strain. We are also grateful to Drs. Thomas Misgeld and Monika Leischner for their helpful advice regarding the analysis of muscle innervation. We thank Dr. Ainhoa Perez for the schematic illustrating cerebellar anatomy. We thank members of the H.S. and M.E.H. laboratories for thoughtful comments and helpful advice throughout. This work was supported by NIH Grant RO1GM60124, a gift from the Loewenberg Foundation, and a grant from the Cure Alzheimer’s Foundation to H.S.; The Rockefeller University Center for Clinical and Translational Science to A.L.; and support from the Eugene W. Chinery 2012 Trust to E.-E.G. and M.E.H.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911921116/-/DCSupplemental.
References
- 1.Hershko A., Ciechanover A., The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Glickman M. H., Ciechanover A., The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82, 373–428 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Goldberg A. L., On prions, proteasomes, and mad cows. N. Engl. J. Med. 357, 1150–1152 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Dikic I., Elazar Z., Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 19, 349–364 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Labbadia J., Morimoto R. I., The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff S., Weissman J. S., Dillin A., Differential scales of protein quality control. Cell 157, 52–64 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Varshavsky A., The ubiquitin system, an immense realm. Annu. Rev. Biochem. 81, 167–176 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Levine B., Kroemer G., Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumeister W., Walz J., Zühl F., Seemüller E., The proteasome: Paradigm of a self-compartmentalizing protease. Cell 92, 367–380 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Collins G. A., Goldberg A. L., The logic of the 26S proteasome. Cell 169, 792–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hafner A. S., Donlin-Asp P. G., Leitch B., Herzog E., Schuman E. M., Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 364, eaau3644 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Biever A., Donlin-Asp P. G., Schuman E. M., Local translation in neuronal processes. Curr. Opin. Neurobiol. 57, 141–148 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Bingol B., Sheng M., Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron 69, 22–32 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Campbell D. S., Holt C. E., Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32, 1013–1026 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Hegde A. N., Haynes K. A., Bach S. V., Beckelman B. C., Local ubiquitin-proteasome-mediated proteolysis and long-term synaptic plasticity. Front. Mol. Neurosci. 7, 96 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo C. T., Jan L. Y., Jan Y. N., Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc. Natl. Acad. Sci. U.S.A. 102, 15230–15235 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watts R. J., Hoopfer E. D., Luo L., Axon pruning during Drosophila metamorphosis: Evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38, 871–885 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Yi J. J., Ehlers M. D., Ubiquitin and protein turnover in synapse function. Neuron 47, 629–632 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Tai H. C., Schuman E. M., Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Kuo C. T., Zhu S., Younger S., Jan L. Y., Jan Y. N., Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron 51, 283–290 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Schmidt M., Hanna J., Elsasser S., Finley D., Proteasome-associated proteins: Regulation of a proteolytic machine. Biol. Chem. 386, 725–737 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Besche H. C., Peth A., Goldberg A. L., Getting to first base in proteasome assembly. Cell 138, 25–28 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rousseau A., Bertolotti A., Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 19, 697–712 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Chu-Ping M., Slaughter C. A., DeMartino G. N., Purification and characterization of a protein inhibitor of the 20S proteasome (macropain). Biochim. Biophys. Acta 1119, 303–311 (1992). [DOI] [PubMed] [Google Scholar]
- 25.McCutchen-Maloney S. L., et al. , cDNA cloning, expression, and functional characterization of PI31, a proline-rich inhibitor of the proteasome. J. Biol. Chem. 275, 18557–18565 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Bader M., et al. , A conserved F box regulatory complex controls proteasome activity in Drosophila. Cell 145, 371–382 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yashiroda H., et al. , N-terminal α7 deletion of the proteasome 20S core particle substitutes for yeast PI31 function. Mol. Cell. Biol. 35, 141–152 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang B. J., et al. , Arabidopsis PROTEASOME REGULATOR1 is required for auxin-mediated suppression of proteasome activity and regulates auxin signalling. Nat. Commun. 7, 11388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho-Park P. F., Steller H., Proteasome regulation by ADP-ribosylation. Cell 153, 614–627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirk R., et al. , Structure of a conserved dimerization domain within the F-box protein Fbxo7 and the PI31 proteasome inhibitor. J. Biol. Chem. 283, 22325–22335 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Nelson D. E., Randle S. J., Laman H., Beyond ubiquitination: The atypical functions of Fbxo7 and other F-box proteins. Open Biol. 3, 130131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bader M., Arama E., Steller H., A novel F-box protein is required for caspase activation during cellular remodeling in Drosophila. Development 137, 1679–1688 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vingill S., et al. , Loss of FBXO7 (PARK15) results in reduced proteasome activity and models a parkinsonism-like phenotype in mice. EMBO J. 35, 2008–2025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stott S. R. W., et al. , Loss of FBXO7 results in a Parkinson’s-like dopaminergic degeneration via an RPL23-MDM2-TP53 pathway. J. Pathol. 249, 241–254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciechanover A., Brundin P., The ubiquitin proteasome system in neurodegenerative diseases: Sometimes the chicken, sometimes the egg. Neuron 40, 427–446 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Dantuma N. P., Bott L. C., The ubiquitin-proteasome system in neurodegenerative diseases: Precipitating factor, yet part of the solution. Front. Mol. Neurosci. 7, 70 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myeku N., et al. , Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 22, 46–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keller J. N., Hanni K. B., Markesbery W. R., Impaired proteasome function in Alzheimer’s disease. J. Neurochem. 75, 436–439 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Rubinsztein D. C., The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Schmidt M., Finley D., Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta 1843, 13–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thibaudeau T. A., Anderson R. T., Smith D. M., A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers. Nat. Commun. 9, 1097 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conedera S., et al. , FBXO7 mutations in Parkinson’s disease and multiple system atrophy. Neurobiol. Aging 40, 192.e1–192.e5 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Liu K., et al. , PI31 is an adaptor protein for proteasome transport in axons and required for synaptic development. Dev. Cell 50, 509–524.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanning K. C., Kaplan A., Henderson C. E., Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 33, 409–440 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Hatten M. E., Heintz N., Mechanisms of neural patterning and specification in the developing cerebellum. Annu. Rev. Neurosci. 18, 385–408 (1995). [DOI] [PubMed] [Google Scholar]
- 46.Palay S. L., Chan-Palay V., Cerebellar Cortex: Cytology and Organization (Springer, Berlin, Germany, 1974). [Google Scholar]
- 47.Huang M., Verbeek D. S., Why do so many genetic insults lead to Purkinje Cell degeneration and spinocerebellar ataxia? Neurosci. Lett. 688, 49–57 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Rothblum-Oviatt C., et al. , Ataxia telangiectasia: A review. Orphanet J. Rare Dis. 11, 159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Louis E. D., Faust P. L., Vonsattel J. P., Purkinje cell loss is a characteristic of essential tremor. Parkinsonism Relat. Disord. 17, 406–409 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fatemi S. H., et al. , Consensus paper: Pathological role of the cerebellum in autism. Cerebellum 11, 777–807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nijssen J., Comley L. H., Hedlund E., Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 133, 863–885 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferraiuolo L., Kirby J., Grierson A. J., Sendtner M., Shaw P. J., Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat. Rev. Neurol. 7, 616–630 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Taylor J. P., Brown R. H. Jr, Cleveland D. W., Decoding A. L. S., Decoding ALS: From genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tashiro Y., et al. , Motor neuron-specific disruption of proteasomes, but not autophagy, replicates amyotrophic lateral sclerosis. J. Biol. Chem. 287, 42984–42994 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding M., Weng C., Fan S., Cao Q., Lu Z., Purkinje cell degeneration and motor coordination deficits in a new mouse model of autosomal recessive spastic ataxia of Charlevoix-Saguenay. Front. Mol. Neurosci. 10, 121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unno T., et al. , Development of Purkinje cell degeneration in a knockin mouse model reveals lysosomal involvement in the pathogenesis of SCA6. Proc. Natl. Acad. Sci. U.S.A. 109, 17693–17698 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cendelin J., From mice to men: Lessons from mutant ataxic mice. Cerebellum Ataxias 1, 4 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ljungberg L., et al. , Transient developmental Purkinje cell axonal torpedoes in healthy and ataxic mouse cerebellum. Front. Cell. Neurosci. 10, 248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Testa G., et al. , A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38, 151–158 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Skarnes W. C., et al. , A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fujimuro M., Sawada H., Yokosawa H., Dynamics of ubiquitin conjugation during heat-shock response revealed by using a monoclonal antibody specific to multi-ubiquitin chains. Eur. J. Biochem. 249, 427–433 (1997). [DOI] [PubMed] [Google Scholar]
- 62.Katsuragi Y., Ichimura Y., Komatsu M., p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 282, 4672–4678 (2015). [DOI] [PubMed] [Google Scholar]
- 63.Moscat J., Diaz-Meco M. T., p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137, 1001–1004 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldberg A. L., Development of proteasome inhibitors as research tools and cancer drugs. J. Cell Biol. 199, 583–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arber S., et al. , Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 23, 659–674 (1999). [DOI] [PubMed] [Google Scholar]
- 66.Yang X., et al. , Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron 30, 399–410 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Anderson D. M., et al. , Severe muscle wasting and denervation in mice lacking the RNA-binding protein ZFP106. Proc. Natl. Acad. Sci. U.S.A. 113, E4494–E4503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McGoldrick P., Joyce P. I., Fisher E. M., Greensmith L., Rodent models of amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1832, 1421–1436 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Sakao Y., et al. , Mouse proteasomal ATPases Psmc3 and Psmc4: Genomic organization and gene targeting. Genomics 67, 1–7 (2000). [DOI] [PubMed] [Google Scholar]
- 70.Cummings C. J., et al. , Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 10, 1511–1518 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Ronnebaum S. M., Patterson C., Schisler J. C., Emerging evidence of coding mutations in the ubiquitin-proteasome system associated with cerebellar ataxias. Hum. Genome Var. 1, 14018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sasaki K., et al. , PAC1 gene knockout reveals an essential role of chaperone-mediated 20S proteasome biogenesis and latent 20S proteasomes in cellular homeostasis. Mol. Cell. Biol. 30, 3864–3874 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Komatsu M., et al. , Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. U.S.A. 104, 14489–14494 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishiyama J., Miura E., Mizushima N., Watanabe M., Yuzaki M., Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 3, 591–596 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Wang C., Liang C. C., Bian Z. C., Zhu Y., Guan J. L., FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat. Neurosci. 16, 532–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Redmann V., et al. , Clec16a is critical for autolysosome function and Purkinje cell survival. Sci. Rep. 6, 23326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim M., et al. , Mutation in ATG5 reduces autophagy and leads to ataxia with developmental delay. eLife 5, e12245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Margolin D. H., et al. , Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N. Engl. J. Med. 368, 1992–2003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vriend J., Ghavami S., Marzban H., The role of the ubiquitin proteasome system in cerebellar development and medulloblastoma. Mol. Brain 8, 64 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burda J. E., Sofroniew M. V., Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81, 229–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sofroniew M. V., Astrogliosis. Cold Spring Harb. Perspect. Biol. 7, a020420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Burda J. E., Sofroniew M. V., Seducing astrocytes to the dark side. Cell Res. 27, 726–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu C. Y., Yang Y., Ju W. N., Wang X., Zhang H. L., Emerging roles of astrocytes in neuro-vascular unit and the tripartite synapse with emphasis on reactive gliosis in the context of Alzheimer’s disease. Front. Cell. Neurosci. 12, 193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carter S. F., et al. , Astrocyte biomarkers in Alzheimer’s disease. Trends Mol. Med. 25, 77–95 (2019). [DOI] [PubMed] [Google Scholar]
- 85.Wolf S. A., Boddeke H. W., Kettenmann H., Microglia in physiology and disease. Annu. Rev. Physiol. 79, 619–643 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Yang Z., Wang K. K., Glial fibrillary acidic protein: From intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 38, 364–374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Louis E. D., et al. , Neuropathologic findings in essential tremor. Neurology 66, 1756–1759 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Yang Q., et al. , Morphological Purkinje cell changes in spinocerebellar ataxia type 6. Acta Neuropathol. 100, 371–376 (2000). [DOI] [PubMed] [Google Scholar]
- 89.Louis E. D., Kuo S. H., Vonsattel J. P., Faust P. L., Torpedo formation and Purkinje cell loss: Modeling their relationship in cerebellar disease. Cerebellum 13, 433–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hirano A., Dembitzer H. M., Ghatak N. R., Fan K. J., Zimmerman H. M., On the relationship between human and experimental granule cell type cerebellar degeneration. J. Neuropathol. Exp. Neurol. 32, 493–502 (1973). [DOI] [PubMed] [Google Scholar]
- 91.Alvarez-Castelao B., Schuman E. M., The regulation of synaptic protein turnover. J. Biol. Chem. 290, 28623–28630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frühbeis C., Fröhlich D., Krämer-Albers E. M., Emerging roles of exosomes in neuron-glia communication. Front. Physiol. 3, 119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Saman S., et al. , Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 287, 3842–3849 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yuyama K., Sun H., Mitsutake S., Igarashi Y., Sphingolipid-modulated exosome secretion promotes clearance of amyloid-β by microglia. J. Biol. Chem. 287, 10977–10989 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iguchi Y., et al. , Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 139, 3187–3201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Morimoto R. I., Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Di Fonzo A., et al. , FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology 72, 240–245 (2009). [DOI] [PubMed] [Google Scholar]
- 98.Paisán-Ruiz C., et al. , Early-onset L-dopa-responsive parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Mov. Disord. 25, 1791–1800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li X., Thompson D., Kumar B., DeMartino G. N., Molecular and cellular roles of PI31 (PSMF1) protein in regulation of proteasome function. J. Biol. Chem. 289, 17392–17405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deger J. M., Gerson J. E., Kayed R., The interrelationship of proteasome impairment and oligomeric intermediates in neurodegeneration. Aging Cell 14, 715–724 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tanaka K., Matsuda N., Proteostasis and neurodegeneration: The roles of proteasomal degradation and autophagy. Biochim. Biophys. Acta 1843, 197–204 (2014). [DOI] [PubMed] [Google Scholar]
- 102.Vilchez D., Saez I., Dillin A., The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat. Commun. 5, 5659 (2014). [DOI] [PubMed] [Google Scholar]
- 103.Dennissen F. J., Kholod N., van Leeuwen F. W., The ubiquitin proteasome system in neurodegenerative diseases: Culprit, accomplice or victim? Prog. Neurobiol. 96, 190–207 (2012). [DOI] [PubMed] [Google Scholar]
- 104.Crawley J. N., Behavioral phenotyping of transgenic and knockout mice: Experimental design and evaluation of general health, sensory functions, motor abilities, and specific behavioral tests. Brain Res. 835, 18–26 (1999). [DOI] [PubMed] [Google Scholar]
- 105.Guyenet S. J., et al. , A simple composite phenotype scoring system for evaluating mouse models of cerebellar ataxia. J. Vis. Exp., 1787 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Carter R. J., et al. , Characterization of progressive motor deficits in mice transgenic for the human Huntington’s disease mutation. J. Neurosci. 19, 3248–3257 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brill M. S., et al. , Branch-specific microtubule destabilization mediates axon branch loss during neuromuscular synapse elimination. Neuron 92, 845–856 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Details and availability of antibodies, mice, genotyping primers, genotyping PCR program, and reagents used in this work are included in SI Appendix, Tables S1–S5, respectively.