Significance
We inferred the phylogeny and evolution of beetles using genomic data of an unprecedented scale. Moreover, we documented the diversification of plant-feeding (herbivorous) beetles, which account for nearly half of all beetle species and a similar proportion of herbivorous insects, following convergent horizontal transfers of bacterial and fungal genes enabling the digestion of lignocellulose in plant cell walls. Our findings clarify beetle phylogenetic relationships and reveal new insights into the evolution of specialized herbivory and why there are so many species of beetles. Furthermore, they underscore the intimacy and complexity of the evolutionary relationships between insects, plants, and microorganisms and show how analyses of large-scale genomic data are revealing the evolution and genomic basis of insect biodiversity.
Keywords: adaptive radiation, herbivory, horizontal gene transfer, microbes, phylogeny
Abstract
The order Coleoptera (beetles) is arguably the most speciose group of animals, but the evolutionary history of beetles, including the impacts of plant feeding (herbivory) on beetle diversification, remain poorly understood. We inferred the phylogeny of beetles using 4,818 genes for 146 species, estimated timing and rates of beetle diversification using 89 genes for 521 species representing all major lineages and traced the evolution of beetle genes enabling symbiont-independent digestion of lignocellulose using 154 genomes or transcriptomes. Phylogenomic analyses of these uniquely comprehensive datasets resolved previously controversial beetle relationships, dated the origin of Coleoptera to the Carboniferous, and supported the codiversification of beetles and angiosperms. Moreover, plant cell wall-degrading enzymes (PCWDEs) obtained from bacteria and fungi via horizontal gene transfers may have been key to the Mesozoic diversification of herbivorous beetles—remarkably, both major independent origins of specialized herbivory in beetles coincide with the first appearances of an arsenal of PCWDEs encoded in their genomes. Furthermore, corresponding (Jurassic) diversification rate increases suggest that these novel genes triggered adaptive radiations that resulted in nearly half of all living beetle species. We propose that PCWDEs enabled efficient digestion of plant tissues, including lignocellulose in cell walls, facilitating the evolution of uniquely specialized plant-feeding habits, such as leaf mining and stem and wood boring. Beetle diversity thus appears to have resulted from multiple factors, including low extinction rates over a long evolutionary history, codiversification with angiosperms, and adaptive radiations of specialized herbivorous beetles following convergent horizontal transfers of microbial genes encoding PCWDEs.
The extraordinary diversity of beetles (order Coleoptera; >400,000 species) has been attributed chiefly to the adaptive radiation of specialized herbivorous beetles feeding on flowering plants (angiosperms) (1–4). However, the evolution of herbivory and its impacts on beetle diversification remain poorly understood. We used large-scale genomic data to infer the phylogeny of beetles, reconstruct timing and patterns of beetle diversification, and trace the evolution of beetle genes enabling specialized herbivory. Our results shed light on the evolution of plant feeding and reveal endogenous plant cell wall-degrading enzymes (PCWDEs) as a key innovation in the adaptive radiation of beetles on plants.
Nearly half of all herbivorous insect species are beetles (1), and most herbivorous beetles feed on angiosperms. Nonetheless, recent studies have failed to find a strong positive relationship between herbivory—including herbivory on angiosperms—and beetle diversification (4–7). In contrast to strictly ecological explanations, recent studies of beetle genomes (8) and beetle digestive physiology (8–12) have speculated that the diversity of herbivorous beetles may have its origins in genomic innovation, specifically the evolution of endogenous PCWDEs enabling symbiont-independent digestion of lignocellulose in plant cell walls.
Beetle-encoded PCWDEs are mainly carbohydrate esterases (CE), polysaccharide lyases (PL), and glycoside hydrolases (GH) (13). They allow beetles to digest cellulose, hemicelluloses, and pectin in plant cell walls—the most abundant source of carbohydrates on Earth—liberating sugars, amino acids, and other essential nutrients (SI Appendix, Table S1) (9, 10). Apart from GH families 1 and 9, which have ancient origins in animals (14), beetle-encoded PCWDEs are thought to have originated from bacteria and fungi via horizontal gene transfer (HGT) (9, 15). Gene duplication and functional diversification post-HGT has created multigene families of PCWDEs with complementary catalytic activities (16).
PCWDEs (other than GH1 and GH9) are reported from the genomes of fewer than 50 beetle species—all specialized herbivores—representing 5 of the 190 described extant beetle families (SI Appendix, Table S2). Their presence in beetle genomes is notable because it was previously thought that beetles, like many other herbivorous animals, lack the capacity for significant endogenous lignocellulose digestion (13) and therefore require symbionts (whose genomes encode PCWDEs) for successful herbivory. While PCWDEs have been shown to play essential roles in beetle–plant interactions (8, 11), their phylogenetic distribution and impacts on beetle diversification remain virtually unknown (17).
Results and Discussion
Phylogenetic Relationships.
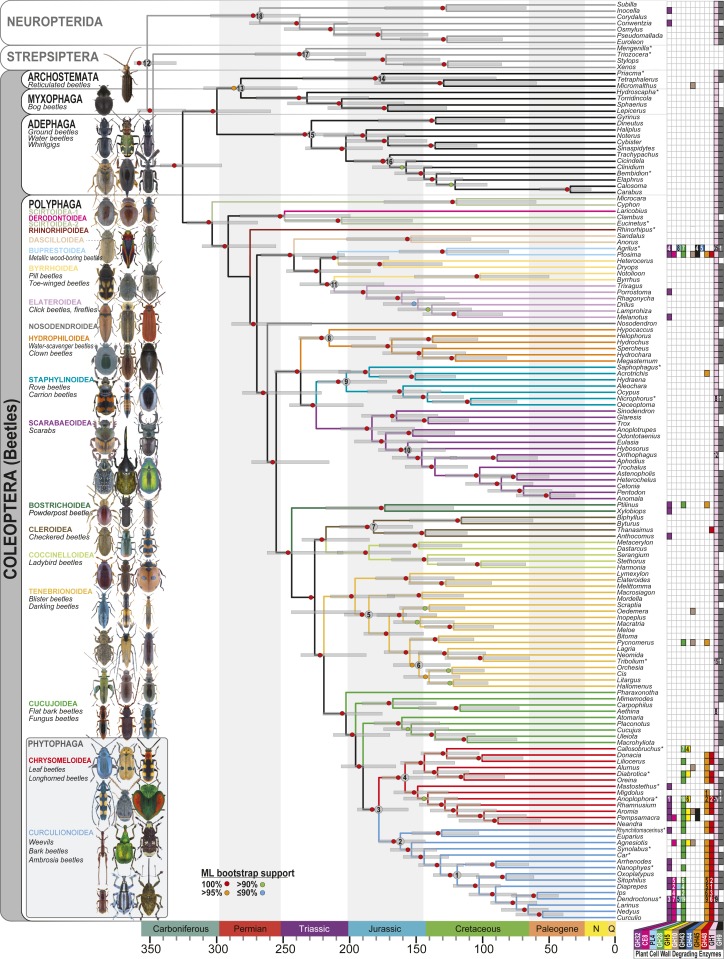
We reconstructed the phylogeny of beetles using 1,907,014 amino acid sites from 4,818 nuclear genes obtained via RNA sequencing (RNA-Seq) or genome skimming of 146 beetle species and near relatives of beetles in the orders Megaloptera, Neuroptera, Raphidioptera, and Strepsiptera (Fig. 1, Methods, SI Appendix, Tables S3 and S4, and Datasets S1–S4). Maximum likelihood (ML) phylogenetic analyses yielded consistently strong statistical support for the interrelationships among all major groups of beetles (SI Appendix, Figs. S1–S7). We recovered 2 main groupings: 1) ground, tiger, diving, whirligig, minute bog, and reticulated beetles (suborders Adephaga, Myxophaga, and Archostemata) and 2) scarab, rove, metallic wood-boring, click, firefly, ladybird, leaf, darkling, longhorn, and weevil beetles (suborder Polyphaga).
Fig. 1.
A dated phylogeny of beetles showing the distribution of putative PCWDEs, inferred from 4,818 nuclear genes (Methods and SI Appendix). Branches in suborder Polyphaga are color-coded by superfamily. Numbers indicate nodes constrained by fossil priors (SI Appendix, Table S5). Filled squares indicate the presence of putative PCWDEs and GH32 invertases (color-coded by gene family) based on analyses of whole-genome (asterisk) or RNA-Seq data. GH1 and GH9 have known ancient origins in metazoans (14, 47) and were expected to occur in most species. Asterisks denote results from the analysis of WGS (versus RNA-Seq) data. Numbers of homologs are indicated in each box when previously published (SI Appendix, Table S1). Note that Rhinorhipus was added after the initial analyses were completed based on a new ML tree search, which recovered the same topology. Bootstrapping was not conducted due to computational constraints. However, its placement was the same in the 521-taxon tree, where it had 100% ML bootstrap support. All higher taxa shown to illustrate morphological diversity (but not all species) were sampled. Cupes image courtesy of Matthew Bertone (North Carolina State University, Raleigh, NC). All other photos courtesy of Udo Schmidt (photographer).
Previously controversial beetle relationships were resolved with strong statistical support, including the paraphyly of aquatic Adephaga (diving water beetles) (18), placement of Jacobsoniidae in Staphylinoidea (19), and recovery of Nosodendron (Nosodendridae) (3) and the monotypic Rhinorhipus (Rhinorhipidae) (20) as relatively old lineages of suborder Polyphaga. Superfamilies Byrrhoidea, Cucujoidea, and Scirtoidea were paraphyletic, and Phytophaga (longhorn beetles, leaf beetles, and weevils; >125,000 described extant species)—arguably the most species-rich radiation of herbivorous insects—was recovered within Cucujoidea. Testing of alternative phylogenetic hypotheses using 4-cluster likelihood mapping supported the placement of Phytophaga within Cucujoidea and indicated lingering uncertainty in the relationships among suborders Adephaga, Archostemata, and Myxophaga (Methods and SI Appendix, Figs. S8 and S9).
Timing and Patterns of Beetle Diversification.
We recovered a Carboniferous origin of Coleoptera (327 Ma, 95% CI: 297.3 to 343.9 Ma) and subsequent episodic radiations. Evidence for these radiations included a late Carboniferous net diversification rate increase within suborder Polyphaga (∼304.7 Ma), followed by 13 post-Permian increases in diversification rate involving descendant lineages (Fig. 1, Methods, and SI Appendix, Figs. S2, S10–S14, and Tables S3–S7). More than 95% of extant beetle families originated before the end of the Cretaceous (Fig. 2). Beetles therefore appear to have radiated alongside other major infraordinal groups of holometabolous insects (21, 22) and survived 3 of the “big 5” Phanerozoic mass extinctions.
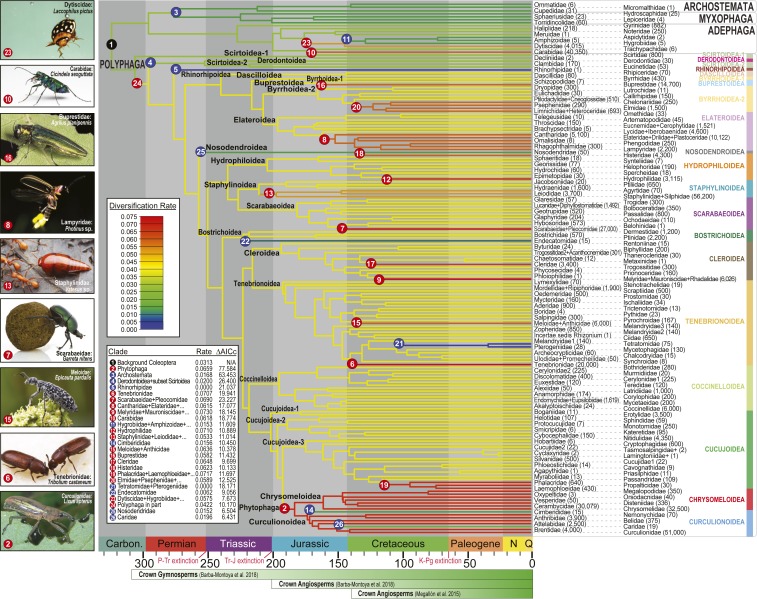
Fig. 2.
Timing and rates of beetle diversification. Family-level net diversification rates and rate shifts for 188/190 beetle families (full tree; SI Appendix, Fig. S10 and Tables S5–S7). Branch colors indicate net diversification rates. Numbered circles indicate the locations of significant net diversification rate shifts (red/increases; blue/decreases). Approximate numbers of described extant species are indicated next to family-level taxon names (76). Note that the right column entries are interdigitated between the left ones, as connected by dashed lines. The beetle suborders and polyphagan beetle superfamilies are labeled on the right side of the tree. Outgroups are not shown. The timeline indicates mass extinctions and significant events in the history of seed plants (39, 77). Photos show select beetle groups that experienced significant diversification rate increases. Lixus image courtesy of D.D.M. Garretes image courtesy of Piotr Naskrecki (Harvard University, Cambridge, MA). Agrilus image courtesy of David Cappaert (photographer). All other photos courtesy of Alex Wild (photographer).
While our analyses suggest an origin of Coleoptera near the Early Carboniferous–Late Carboniferous boundary interval (Fig. 1), this is not directly supported by the beetle fossil record, which begins in the lower Permian with †Tshecardocoleidae (Asselian or Sakmarian) and †Moravocoleidae (Artinskian) (23–25). Coleopsis archaica from the earliest Permian of Germany (Grügelborn/Saarland) was recently identified as the oldest known beetle species (26), with an elytral venation typical of the Early Permian †Tshekardocoleidae. †Adiphlebia lacoana from the Carboniferous of Mazon Creek, IL was identified as the “oldest beetle” by ref. 27. However, it was shown by ref. 23 that this species does not belong to Coleoptera (see also ref. 24). Likewise, the Carboniferous †Skleroptera does not belong to the stem group of Coleoptera (28). The earliest described holometabolous insect larva (29, 30) is 311 My old. However, the orthognathous head, clawless legs, and serially arranged leglets make close phylogenetic affinities with Coleoptera unlikely.
Permian †Tshecardocoleidae were undoubtedly associated with wood, very likely with a preference for subcortical spaces as adults, and possibly occurred in a semiaquatic environment (25). The earliest Lower Permian beetles (23, 26, 31) display a prognathous head, a characteristic elytral pattern with window punctures, a cuticular surface with tubercles (or scales), and a plesiomorphic pattern of ventral sclerites, very similar to the extant wood-associated Ommatidae and Cupedidae (32–34). Plesiomorphic features of the Lower Permian †Protocoleoptera (†Tshecardocoleidae and †Moravocoleidae) are antennae with 13 segments (31), a broad prothoracic postcoxal bridge, and the lack of a tightly sealed subelytral space, whereas a prominent pointed ovipositor is arguably a synapomorphy of the 2 families (33, 35). Upper Permian (Changhsingian Stage, ca. 254 to 252 Ma, China) wood borings were recently attributed to beetles (36, 37), indicating an early origin of wood-boring habits in Coleoptera.
Our estimates for beetle net diversification rates were not exceptional, consistent with previous studies (3–5). Furthermore, significant increases or decreases in net diversification rate were recovered for most of the same higher-level taxa of beetles as previous studies (Fig. 2) (3–5). The timing of branching events in the beetle phylogeny was in general agreement with ref. 4 and ref. 38 for deep nodes but somewhat older than most other (earlier) studies (e.g., refs. 3 and 5). However, node ages within series Cucujiformia, which contains most herbivorous beetle species, were mostly much younger than in ref. 38, similar to the results of ref. 4.
We recovered evidence for adaptive radiations of beetles into a variety of ecological niches. For example, predaceous ground beetles (Carabidae) and aquatic diving beetles (Dytiscidae and relatives) experienced near-coincident radiations following mid-Jurassic increases in diversification rate. Early splits in herbivorous Buprestoidea (metallic wood-boring beetles) and Phytophaga, and herbivorous/saprophagous scarabs (Scarabaeidae) occurred in the Jurassic and early Cretaceous, which is also when core angiosperms diversified (39, 40). These same beetle groups subsequently underwent crown diversification coeval with the rise of angiosperms to ecological dominance during the mid-Cretaceous (41) (Figs. 1 and 2 and SI Appendix, Figs. S11–S14). One-fourth of all rate increases in the beetle phylogeny were associated with herbivory (Fig. 2)—which is more than any other factor (Fig. 3).
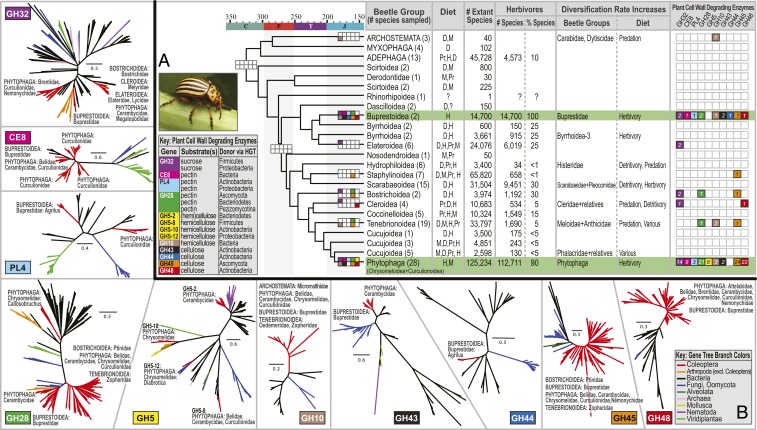
Fig. 3.
Adaptive radiation of specialized herbivorous beetles after the acquisition of PCWDEs from microbes. (A) Summary of beetle time tree showing 2 major independent origins of novel PCWDEs from bacteria and fungi and coincident net diversification rate increases among specialized herbivorous beetles (green background; summarized from Figs. 1 and 2): 1) along the stem of Buprestidae (Buprestoidea) and 2) along the stem of Phytophaga (Chrysomeloidea + Curculionoidea). Colorized boxes indicate the presence of candidate PCWDEs. The number of beetle species sampled that contain at least one PCWDE gene family member is indicated in each box. Empty/white boxes indicate a gene family was not observed. Beetle diets: detritivory (D), mycophagy (M), predation (Pr), herbivory (H), and unknown (?). Most data were obtained from ref. 6 and are ordered by decreasing prevalence. Percent herbivores >1% is shown to the nearest 5% and was estimated based on our collective knowledge of these beetle groups. (B) Unrooted phylogenetic trees for PCWDE gene families illustrating the taxonomic origins of beetle-encoded genes (SI Appendix, Figs. S15–S26 and Datasets S1–S3). The beetle groups represented in each gene tree are labeled. Leptinotarsa image courtesy of the USDA Agricultural Research Service/Scott Bauer, licensed under CC BY 3.0.
Other groups of beetles that are species-rich in the modern fauna and associated with living plants or plant litter, including detritivores, saprophages, and mycophages, also diversified during the middle to late Mesozoic (e.g., rove beetles [Staphylinidae], scarabs [Scarabaeidae], and darkling beetles [Tenebrionidae]), consistent with the view that ecologically diverse groups of modern beetles codiversified with angiosperms and in the novel habitats that angiosperms created (3). Within Phytophaga, the weevil families Caridae and Cimberididae experienced significant decreases in diversification rate during the Mesozoic (Fig. 2). These groups are thought to have ancestral associations with gymnosperms (specifically, conifers) (2), in contrast to the angiosperm associations of most extant Phytophaga.
Beetle groups that are today associated with gymnosperm pollen, including some early Cucujoidea and Phytophaga (Boganiidae: Paracucujinae, Cimberididae, most Nemonychidae, and Erotylidae: Pharaxonothinae), appeared by the late Jurassic, well before bees and butterflies (21, 42). They were likely among the first insect pollinators of gymnosperms and early angiosperms. Furthermore, our results are consistent with fossil evidence in suggesting that pollenivory was a transitional state between detritivory, mycophagy, and saprophagy (Cucujoidea) and specialized herbivory (Phytophaga) (43–45) (Fig. 1 and SI Appendix). The apparent prevalence of transitions in Coleoptera from generalized diets, such as detritivory and saprophagy, toward more specialized diets, such as mycophagy and herbivory, is consistent with the high rate of such transitions across insects (6).
Comparative Genomics of Beetle-Encoded PCWDEs.
We studied putative PCWDEs encoded in 154 transcriptomes or genomes corresponding to the 147 taxa in Fig. 1 (SI Appendix, Figs. S15–S27, and Tables S1, S2, and S8). We also studied GH32 invertases, which catalyze the conversion of sucrose—the primary form of photoassimilated carbon in plant vascular tissues—to glucose and fructose. Like PCWDEs, invertases have played a potentially important role in the evolution of specialized herbivory (e.g., see ref. 46).
GH1 and GH9 have ancient origins in animals (14, 47) and were nearly ubiquitous in our study. The other gene families we studied were found almost exclusively in Buprestoidea and Phytophaga, which encoded an expansive and remarkably similar array of PCWDEs (Figs. 1 and 3). Buprestoidea and Phytophaga are the most species-rich and most specialized radiations of herbivorous beetles (1). Their feeding habits collectively include chewing, mining, and boring of virtually all kinds of plant tissues (living or dead) and plant taxa.
Outside of Buprestoidea and Phytophaga, 10 beetle species scattered widely across the phylogeny had matches to 1 PCWDE gene family (other than GH1 and GH9), and 2 beetle species each matched 3 gene families (Figs. 1 and 3). These included Bostrichidae (Xylobiops, GH32), Cleridae (Thanasimus, GH48), Elateridae (Melanotus, GH32), Lycidae (Porrostoma, GH32), Melyridae (Anthocomus, GH32), Micromalthidae (Micromalthus, GH10), Oedemeridae (Oedemera, GH10), Ptiliidae (Acrotrichis, GH45), Ptinidae (Ptilinus, GH28, GH32, and GH45), and Zopheridae (Pycnomerus, GH10, GH28, and GH45). In contrast, Buprestoidea and Phytophaga had species with matches from up to 7 families of PCWDEs, often with multiple apparent homologs from each family. Independent losses and reacquisitions of PCWDEs are known in Phytophaga (10) and were observed in this study.
Overall, we documented putative endogenous PCWDEs (including GH32 invertases, excluding GH1 and GH9) from 22 families of beetles. Previous to this study, they were known from only 5 beetle families (Figs. 1 and 3 and SI Appendix, Tables S1 and S2, and Fig. S27). Within Buprestoidea, we report GH10, GH45, GH48, and CE8, in addition to previously reported genes. Within Phytophaga, we report GH43, in addition to previously reported genes. Also within Phytophaga, we document PCWDEs from the families Attelabidae, Belidae, Brentidae, Caridae, Megalopodidae, and Nemonychidae, in addition to the families of Phytophaga from which these genes have been previously reported. Thus, we significantly expand knowledge of the phylogenetic distribution of putative PCWDEs encoded in the genomes of Coleoptera, while at the same time establishing that they are particularly diverse in the 2 lineages of specialized herbivorous beetles—Buprestoidea and Phytophaga.
Microbial Donors of Beetle PCWDEs via HGT.
The inferred last common ancestors and potential donors of beetle PCWDEs were bacteria and fungi, including taxonomic groups that are today quintessential degraders of lignocellulose and other complex polysaccharides in plant and soil detritus (Fig. 3, SI Appendix, Figs. S15–S26, and Datasets S1–S3) (48). Some of these groups of bacteria and fungi are also found in beetle guts (49). Beetle-derived PCWDEs nonetheless formed well-supported clades distinct from microbial taxa in our phylogenies. Moreover, they were largely placed within the same clades as their homologs derived from other insect genomes and transcriptomes, including those derived from high-quality draft genomes. Within these clades, some gene families contained clusters of closely related sequences from the same beetle higher taxa or species, consistent with lineage-specific gene duplications post-HGT.
Physical incorporation of genes encoding PCWDEs into the genome of one or more beetle species has been documented for Buprestoidea-derived GH28, GH32, GH43, GH44, and PL4 and for Phytophaga-derived GH5-2, GH5-8, GH10, GH28, GH32, GH45, GH48, and CE8 (16, 50, 51) (SI Appendix, Table S1). Enzyme product functionality (metabolic integration) has been demonstrated for Buprestoidea-derived GH43, GH44, and PL4 and for Phytophaga-derived GH5-2, GH5-8, GH10, GH11, GH28, GH32, GH45, and CE8 (16, 50, 51). For GH5-2, GH28, GH45, and GH32, a similar gene has been independently horizontally transferred to a plant-feeding organism outside of Insecta (16).
Evidence from the available high-quality draft genomes of Buprestoidea and Phytophaga further indicates that these genes are encoded in beetle genomes and are not the result of contamination (8, 17). The emerald ash borer beetle (Agrilus planipennis; Buprestoidea) genome encodes GH28, GH32, GH43, GH44, and PL4 (each represented by multiple copies in the genome; Fig. 1), all of which have been PCR-amplified from adult A. planipennis elytra and legs—tissues not known to contain symbionts (50, 51). These genes are also frequently arranged in tandem arrays on scaffolds containing other beetle genes. For example, 3 of the 5 A. planipennis GH43 genes reside in the same genomic scaffold (8, 51). Four of the 5 GH43 genes have single exons (51), which is unusual, since almost all of the GH genes we studied in beetle genomes contain multiple exons. Furthermore, the 5 gene families encoding A. planipennis PCWDEs are almost exclusively expressed in the larval midgut (50, 51). Together with the presence of N-terminal secretory signal peptides in the putative proteins, this suggests that these enzymes are secreted to facilitate plant cell wall digestion (51).
Similar observations have been made using the genomes of herbivorous Phytophaga. For example, the Asian longhorned beetle (Anoplophora glabripennis; Phytophaga: Chrysomeloidea) genome contains multiple copies of GH5, GH28, GH45, and GH48 and a single copy of GH32 (8, 52), and the mountain pine beetle (Dendroctonus ponderosae; Phytophaga: Curculionoidea) genome contains GH28, GH32, GH45, GH48, CE8, and PL4, all of which are multicopy in the genome (10, 52–55). Many of the GH family genes from A. glabripennis and D. ponderosae have been PCR-amplified from beetle tissues not known to contain microbial symbionts (10, 53–55). Similarly, many of these genes have been functionally characterized in one or both species using complementary DNAs generated from RNA samples obtained from individual beetles different from the ones used for genome sequencing (8, 10, 55). Furthermore, the A. glabripennis and D. ponderosae PCWDEs (like those encoded in the A. planipennis genome) are frequently arranged in tandem arrays on genomic scaffolds containing other beetle genes (8, 54).
Convergent Evolution of Beetle PCWDEs.
The appearance of similar expansive arrays of PCWDEs in Buprestoidea and Phytophaga, separated by over 250 Ma of evolution, appears to result from convergent evolution via HGT, rather than vertical transmission from a common ancestor (Figs. 1 and 3). However, the mechanisms behind these HGT events remain obscure. Gene families in common (excluding GH1 and GH9) and thus candidates for convergence included GH10, GH28, GH32, GH43, GH45, GH48, CE8, and PL4; only GH5 and GH44 cellulases were not shared, perhaps reflecting slightly different strategies for PCW digestion. PCWDEs were absent from the near relatives of Buprestoidea and Phytophaga and most other Coleoptera. Furthermore, we recovered separate clades of genes corresponding to PCWDEs from Buprestoidea and Phytophaga.
Among the Phytophaga, the diversity of gene families encoding PCWDEs and the number of gene family members both appear to increase in the phylogeny from root to tips (Fig. 1). This suggests the stepwise evolution of symbiotic-independent mechanisms for plant cell wall degradation from symbiotic-dependent ones, although other scenarios are possible. Amplification and functional divergence of PCWDEs post-HGT (8) may have facilitated the evolution of increasingly specialized plant-feeding habits in Buprestoidea and Phytophaga, including the exploitation of woody tissues and pectin-rich young leaves and stems, seeds, and fruits (2, 19, 56). The existence of multiple copies of these genes in most beetle genomes may reduce constraints on their functional evolution, facilitating substrate diversification (activity toward additional/different plant cell wall polysaccharides) post-HGT. GH48 genes, which were found in most Buprestoidea and Phytophaga, may also help degrade fungal chitin (8, 57) and were likely relevant to the repeated evolution of specialized fungus-feeding habits in Phytophaga (2).
Adaptive Radiation of Herbivorous Beetles Post-HGT.
Remarkably, in both Buprestoidea and Phytophaga, the origins of PCWDEs are phylogenetically and temporally linked to significant increases in net diversification rate—1) along the stem of Buprestoidea and 2) along the stem of Phytophaga—indicative of adaptive radiations (58) (Figs. 2 and 3). This suggests that PCWDEs enabled Buprestoidea and Phytophaga entry into new adaptive zones, notably including penetration of the woody plant barrier, without needing to obtain and maintain symbionts for PCW degradation. Pectinases and invertases likely promoted feeding on cambium and sapwood, fruits, leaves, and seeds, while (hemi)cellulases may have promoted feeding on wood (59). Endogenous PCWDEs were therefore likely key to the evolution of specialized plant-feeding habits, such as leaf mining and seed, stem, and wood boring, which also required dealing with novel plant allelochemicals, nutritional and defensive barriers, and recalcitrant plant biopolymers (57).
Phytophaga began to diversify ∼50 Ma earlier than Buprestoidea, perhaps giving the former an evolutionary advantage and accounting for the lesser taxonomic and ecological diversity of extant Buprestoidea—typically, the first organisms to enter an adaptive zone have an advantage (60). Furthermore, while the wood-feeding habits of Buprestidae are similar to those of certain Phytophaga, the Buprestoidea and Phytophaga also exhibit significant differences in life history, behavior, and trophic habits, with potential implications for their abilities to transition between ecological adaptive zones and their resulting diversification rates. More in-depth taxon sampling is needed to further elucidate timing and patterns of gene gain, loss, and amplification, especially in Buprestoidea. Additionally, large-scale functional genomic studies are needed to characterize the roles of the candidate genes we have identified in herbivory and diet specialization across Coleoptera, that is, beyond the relatively few beetle species and genes that have been studied to date. Moreover, additional genes are known to play roles in plant cell wall degradation (and detoxification of plant alleleochemicals) but remain little studied in beetles (8, 14).
Ecological Opportunity and Evolutionary Innovation at the Beetle–Plant Interface.
Buprestoidea and Phytophaga exhibit a similar pattern of increasing host tissue specificity temporally and phylogenetically linked to the evolution of endogenous PCWDEs. We propose that the specialized but versatile trophic apparatus and evolving metabolic repertoire of Buprestoidea and Phytophaga helped them adapt to and track the increasing diversity of angiosperms during the Mesozoic, despite an escalation in the potency and variety of angiosperm chemical defenses (61) and diversification of angiosperm plant cell walls (62). Moreover, facultative symbionts that code for some of the same PCWDEs as beetle genomes (57) likely increased digestive efficiency. We speculate that ecological constraints proposed to render host specialists more susceptible to extinction, such as prolonged larval development and small population and range sizes, were ameliorated by the increasing efficiency of plant biomass assimilation afforded by endogenous PCWDEs, and by amplification and functional divergence of PCWDEs post-HGT. Moreover, this may have facilitated an even greater degree of host specificity, resulting in the evolution of increasingly more varied and specialized plant-feeding habits (63).
Early associations of Phytophaga with gymnosperms, while perhaps initially limited to cone and pollen feeding, thus expanded over time to include virtually all kinds of plant tissues and taxa. Transitions within Buprestoidea from external root feeding to specialized wood boring and leaf mining (56) may also be interpretable in this framework. Mycophagy and feeding on fungus-infested plant tissues were potentially important transitional states to specialized feeding on wood in Phytophaga and their ancestors (3, 64, 65). Phytophaga likely also transitioned from mutualistic interactions (pollenivory/pollination) to antagonistic ones (specialized herbivory on leaves and stems) with concomitant ecological impacts. The phylogenetic placement of beetle taxa that are today associated with gymnosperm cones or pollen, for example Boganiidae: Paracucujinae and Erotylidae: Pharaxonothinae (Cucujoidea) and early lineages of Phytophaga, for example Cimberididae, Nemonychidae (except Nemonyx), Belidae: Allocorynina, and Megalopodidae: Palophaginae, are consistent with this hypothesis.
Preadaptations, including hard mandibles for fragmenting plant material, expansions in gene families used in detoxifying plant allelochemicals (e.g., glutathione S-transferases and carboxylesterases) (8), the absence of external wing buds in larvae, and elytra protecting delicate body parts, may also have facilitated the evolution of uniquely specialized forms of herbivory in beetles. Moreover, these traits, in combination, may help explain the apparent lack of success of most other herbivorous insects in exploiting woody plant tissues.
Nevertheless, herbivorous beetles have taken alternate paths to taxonomic richness. The phytophagous scarabs are relative host generalists compared to Phytophaga and Buprestoidea. Most species feed externally on roots as larvae and leaves (if feeding at all) as adults, and none are known to feed internally on living plant tissues. Their genomes do not appear to encode PCWDEs other than the widespread GH9 cellulases. However, scarab larvae have an alkaline midgut for extracting plant cell wall polymers and proteins and a large hindgut fermentation chamber that houses symbiotic microbes with roles in plant cell wall degradation (66). Thus, they have a symbiont-dependent trophic apparatus and exhibit a lesser diversity of specialized plant-feeding habits compared to the Phytophaga and Buprestoidea. Some Phytophaga (notably certain Cerambycidae) also have very alkaline regions of the gut, which have been proposed to play a role in the digestion and solubilization of lignin in plant cell walls (49).
Conclusions
PCWDEs originally obtained by beetles from bacteria and fungi via HGT enabled efficient symbiont-independent digestion of plant biomass, the most abundant source of carbohydrates on Earth. We propose that this key innovation facilitated the evolution of uniquely specialized plant-feeding habits, such as leaf and seed mining and stem and wood boring, and likely also some forms of specialized fungus feeding, for example fungus farming in Platypodinae and Scolytinae (67). While this remains uncertain, the appearance and expansions of putative PCWDEs and invertases in beetle genomes are correlated with significant increases in diversification rate among specialized herbivorous beetles (Buprestoidea and Phytophaga). Our findings may help explain the disparity in the degree of feeding specialization and species richness observed among groups of herbivorous beetles possessing or lacking a diverse repertoire of PCWDEs, as well as the existence of groups of beetles that feed on plants (notably including angiosperms) but are not unusually species-rich. PCWDEs originally obtained via HGT likely played an important role in the adaptive radiation of other groups of herbivorous insects, for example certain Lepidoptera and Hemiptera, which have at least some of these gene families (8, 14, 68).
The extraordinary diversity of beetles thus appears to have resulted from multiple factors, including a low rate of lineage extinction over a long evolutionary history (2, 5), codiversification with angiosperms (2), and the adaptive radiation of specialized herbivorous beetles following convergent horizontal transfers (and “domestication”) of microbial genes encoding PCWDEs. More broadly, our findings show how large-scale genomic data can reveal new insights into the evolution and genomic basis of insect biodiversity and underscore the intimacy and complexity of the relationships between insects, plants, and microorganisms, as well as their concerted roles in the “origins of terrestrial organic diversity” (60).
Methods
For more information, see SI Appendix, Supplementary Information Text.
Phylogenetic Inference.
We reconstructed the phylogeny of Coleoptera using phylogenomic data from 135 species representing 90 families, plus 11 outgroups (SI Appendix, Fig. S2 and Table S3). We used ML inference implemented in the program RAxML v8.2.10 (69) for phylogeny reconstruction. We then conducted a Bayesian relaxed clock divergence time analysis in MCMCTree (70) with fossil constraints applied to 18 nodes in the phylogeny (Fig. 1 and SI Appendix, Table S5). Our study was designed to address concerns of taxon and locus diversity, outgroups, fossil selection, and age constraints that have been proposed to impact other molecular phylogenetic studies of beetles (e.g., refs. 3–5 and 38).
Diversification Rates.
Net diversification rates and the temporal and phylogenetic locations of diversification rate shifts were estimated using a near-comprehensive family-level time tree generated for the same 146 species as above, plus Rhinorhipus and 374 additional species from ref. 4. (521 total species in 140 beetle families; hereafter, the 89-gene tree; SI Appendix, Table S6 and Fig. S10). This tree was subsequently expanded to include 188/190 extant beetle families (SI Appendix, Supplementary Information Text), followed by analysis in MCMCTree with age constraints applied to 22 nodes (SI Appendix, Figs. S8 and S11 and Table S5). Only the beetle families Jurodidae and Crowsoniellidae, each with one extant species known only from their original type series, were missing from these analyses. Net diversification rates were estimated using MEDUSA in the Geiger package (71).
PCWDEs.
We searched the whole-genome sequencing (WGS) and transcriptome data for the presence of GH genes encoding PCWDEs (SI Appendix, Table S8) (9, 10). GH genes encode enzymes that catalyze the hydrolysis of glycosidic bonds in oligo- and disaccharide sugars and play essential roles in degrading lignocellulose in plant cell walls and other plant polysaccharides. Some also play a role in the detoxification of plant allelochemicals (72). We also searched for pectin methylesterases belonging to carbohydrate esterase family 8 (CE8), which catalyze the deesterification of pectin into pectate and methanol, and pectin lyases belonging to polysaccharide lyase family 4 (PL4), which cleave alpha-1,4 glycosidic bonds between l-rhamnose and d-galacturonic acids in pectin (SI Appendix, Table S8). Pectinolytic enzymes facilitate feeding on the pectin-rich primary cell walls of sapwood and cambium, leaves, fruits, and seeds, while (hemi)cellulolytic enzymes are beneficial for feeding on tissues rich in secondary plant cell walls, including heartwood. Finally, we searched for GH32 invertases, which catalyze the conversion of sucrose to glucose and fructose (46) (SI Appendix, Table S8).
The phylogenetic distribution of these gene families in Coleoptera was determined by identifying protein family domains in our WGS and transcriptome data (SI Appendix, Table S3) using PfamScan (73). We then used BLASTp (74) to refine these results. Subsequently, we gathered homologs for each gene of interest, and after alignment, implemented ML phylogenetic analyses using IQ-TREE (75). We explored the impact of endogenous PCWDEs on beetle diversification rates by comparing the phylogenetic location of significant increases in beetle net diversification rate with the phylogenetic distribution of gene families encoding putative PCWDEs in the 4,818-gene beetle phylogeny. Whole-genome or RNA-Seq data were available for every terminal in this tree. The 1KITE transcriptomes reported herein are permanently archived at under the Umbrella BioProject ID PRJNA 183205. Datasets S1 through S4 are archived at the Zenodo Digital Repository at 10.5281/zenodo.3522944 (78).
Data Use Statement.
Data on genetic material contained in this paper are published for noncommercial use only. Utilization by third parties for purposes other than noncommercial scientific research may infringe the conditions under which the genetic resources were originally accessed, and should not be undertaken without obtaining consent from the corresponding author of the paper and/or obtaining permission from the original provider of the genetic material.
Supplementary Material
Acknowledgments
This research was supported by funding to D.D.M. from the National Science Foundation (DEB1355169) and the University of Memphis FedEx Institute, funding to X.Z. from the China National GeneBank and BGI-Shenzhen, and funding to R.G.B. and B.M. from the German Research Foundation. We thank E. Anton, J. Beller, S. Dobler, J. Eberle, S. Fabrizi, C. Greve, J. Gutowski, S. Haddad, J. Hulcr, F. Jia, K. Kjer, M. Kubiak, S. Lapointe, K. Meusemann, C. Minteer, S. Nekum, S. Salom, G. Sabatinelli, K. Schuette, M. Thayer, X. Wang, and S. Xu for specimens studied; D. Bartel, K. Meusemann, and M. Petersen for analytical methods development; G. Meng, C. Yang, and other BGI staff for assistance with sample and data curation; and the University of Memphis and the University of Vienna for use of their high-performance computing clusters.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The 1KITE transcriptomes reported herein are permanently archived at the National Center for Biotechnology Information (NCBI), https://www.ncbi.nlm.nih.gov/bioproject/183205 under the Umbrella BioProject ID PRJNA 183205. Datasets S1 through S4, including the resulting alignments of phylogenomic data from beetles and their putative PCWDES, and gene trees and BLAST hits for candidate PCWDEs, are archived at Zenodo, DOI: 10.5281/zenodo.3522944.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909655116/-/DCSupplemental.
References
- 1.Farrell B. D., “Inordinate fondness” explained: Why are there so many beetles? Science 281, 555–559 (1998). [DOI] [PubMed] [Google Scholar]
- 2.McKenna D. D., Sequeira A. S., Marvaldi A. E., Farrell B. D., Temporal lags and overlap in the diversification of weevils and flowering plants. Proc. Natl. Acad. Sci. U.S.A. 106, 7083–7088 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenna D. D., et al. , The beetle tree of life reveals that Coleoptera survived end-Permian mass extinction to diversify during the Cretaceous terrestrial revolution. Syst. Entomol. 40, 835–880 (2015). [Google Scholar]
- 4.Zhang S. Q., et al. , Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commun. 9, 205 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunt T., et al. , A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318, 1913–1916 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Rainford J. L., Mayhew P. J., Diet evolution and clade richness in Hexapoda: A phylogenetic study of higher taxa. Am. Nat. 186, 777–791 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Wiens J. J., Lapoint R. T., Whiteman N. K., Herbivory increases diversification across insect clades. Nat. Commun. 6, 8370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKenna D. D., et al. , Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle-plant interface. Genome Biol. 17, 227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirsch R., et al. , Horizontal gene transfer and functional diversification of plant cell wall degrading polygalacturonases: Key events in the evolution of herbivory in beetles. Insect Biochem. Mol. Biol. 52, 33–50 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Pauchet Y., Wilkinson P., Chauhan R., Ffrench-Constant R. H., Diversity of beetle genes encoding novel plant cell wall degrading enzymes. PLoS One 5, e15635 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salem H., et al. , Drastic genome reduction in an herbivore's pectinolytic symbiont. Cell 171, 1520–1531.e13 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Busch A., Danchin E. G. J., Pauchet Y., Functional diversification of horizontally acquired glycoside hydrolase family 45 (GH45) proteins in Phytophaga beetles. BMC Evol. Biol. 19, 100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldeŕon-Cort́es N., Quesada M., Watanabe H., Cano-Camacho H., Oyama K., Endogenous plant cell wall digestion: A key mechanism in insect evolution. Annu. Rev. Ecol. Evol. Syst. 43, 45–71 (2012). [Google Scholar]
- 14.Chang W. H., Lai A. G., Mixed evolutionary origins of endogenous biomass-depolymerizing enzymes in animals. BMC Genomics 19, 483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderón-Cortés N., Watanabe H., Cano-Camacho H., Zavala-Páramo G., Quesada M., cDNA cloning, homology modelling and evolutionary insights into novel endogenous cellulases of the borer beetle Oncideres albomarginata chamela (Cerambycidae). Insect Mol. Biol. 19, 323–336 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Wybouw N., Pauchet Y., Heckel D. G., Van Leeuwen T., Horizontal gene transfer contributes to the evolution of arthropod herbivory. Genome Biol. Evol. 8, 1785–1801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna D. D., Beetle genomes in the 21st century: Prospects, progress and priorities. Curr. Opin. Insect Sci. 25, 76–82 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Beutel R. G., Roughley R. E., On the systematic position of the family Gyrinidae (Coleoptera: Adephaga). J. Zoological Syst. Evol. Res. 26, 380–400 (1988). [Google Scholar]
- 19.McKenna D. D., “Molecular phylogenetics and evolution of Coleoptera” in Handbook of Zoology. Arthropoda: Insecta. Coleoptera, Beetles: Morphology and Systematics (Phytophaga), Beutel R. G., Leschen R. A. B., Eds. (Walter de Gruyter, Berlin, 2014), pp 1–10. [Google Scholar]
- 20.Kusy D., et al. , Genome sequencing of Rhinorhipus Lawrence exposes an early branch of the Coleoptera. Front. Zool. 15, 21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misof B., et al. , Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Wiegmann B. M., et al. , Episodic radiations in the fly tree of life. Proc. Natl. Acad. Sci. U.S.A. 108, 5690–5695 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kukalová-Peck J., Beutel R. G., Is the Carboniferous †Adiphlebia lacoana really the “oldest beetle”? Critical reassessment and description of a new Permian beetle family. Eur. J. Entomol. 109, 633–645 (2012). [Google Scholar]
- 24.Nel A., et al. , The earliest known holometabolous insects. Nature 503, 257–261 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Ponomarenko A. G., Prokin A. A., Review of paleontological data on the evolution of aquatic beetles (Coleoptera). Paleontol. J. 49, 1383 (2015). [Google Scholar]
- 26.Kirejtshuk A. G., Poschmann M., Prokop J., Garrouste R., Nel A., Evolution of the elytral venation and structural adaptations in the oldest Palaeozoic beetles (Insecta: Coleoptera: Tshekardocoleidae). J. Syst. Palaeontology 12, 575–600 (2014). [Google Scholar]
- 27.Béthoux O., The earliest beetle identified. J. Paleontol. 83, 931–937 (2009). [Google Scholar]
- 28.Beutel R. G., Yan E. V., Kukalova-Peck J., Is †skleroptera (†Stephanastus) really an order in the stemgroup of Coleopterida (Insecta)? Insect Syst. Evol. 50, 670–678 (2018). [Google Scholar]
- 29.Haug J. T., Labandeira C. C., Santiago-Blay J. A., Haug C., Brown S., Life habits, hox genes, and affinities of a 311 million-year-old holometabolan larva. BMC Evol. Biol. 15, 208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haug J. T., Labandeira C. C., Santiago-Blay J. A., Haug C., Brown S., Erratum to: Life habits, hox genes, and affinities of a 311 million-year-old holometabolan larva. BMC Evol. Biol. 16, 169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponomarenko A. G., The historical development of archostematan beetles. (in Russian). Trudi paleont. Inst. Akad. Nauk SSSR 125, 1–238 (1969). [Google Scholar]
- 32.Lawrence J. F., The Australian Ommatidae (Coleoptera: Archostemata): New species, larva and discussion of relationships. Invertebr. Syst. 13, 369–390 (1999). [Google Scholar]
- 33.Beutel R. G., Ge S.-Q., Hörnschemeyer T., On the head morphology of Tetraphalerus, the phylogeny of Archostemata and the basal branching events in Coleoptera. Cladistics 23, 270–298 (2008). [Google Scholar]
- 34.Friedrich F., Farrell B. D., Beutel R. G., The thoracic morphology of Archostemata and the relationships of the extant suborders of Coleoptera (Hexapoda). Cladistics 25, 1–37 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Beutel R. G., Über Phylogenese und Evolution der Coleoptera (Insecta), insbesondere der Adephaga. Verhandlungen des Naturwissenschaftlichen Vereins in Hamburg NF 31, 1–164 (1997). [Google Scholar]
- 36.Feng Z., Wang J., Rößler R., Ślipiński A., Labandeira C., Late Permian wood-borings reveal an intricate network of ecological relationships. Nat. Commun. 8, 556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Z., et al. , Beetle borings in wood with host response in early Permian conifers from Germany. Paläontol. Z. 93, 409–421 (2019). [Google Scholar]
- 38.Toussaint E. F. A., et al. , The peril of dating beetles. Syst. Entomol. 42, 1–10 (2017). [Google Scholar]
- 39.Magallón S., Gómez-Acevedo S., Sánchez-Reyes L. L., Hernández-Hernández T., A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 207, 437–453 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Li H. T., et al. , Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Coiffard C., Gomez B., Daviero-Gomez V., Dilcher D. L., Rise to dominance of angiosperm pioneers in European Cretaceous environments. Proc. Natl. Acad. Sci. U.S.A. 109, 20955–20959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters R. S., et al. , Evolutionary history of the Hymenoptera. Curr. Biol. 27, 1013–1018 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Cai C., et al. , Beetle pollination of cycads in the Mesozoic. Curr. Biol. 28, 2806–2812.e1 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Peris D., et al. , False blister beetles and the expansion of Gymnosperm-insect pollination modes before Angiosperm dominance. Curr. Biol. 27, 897–904 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Liu Z., Ślipiński A., Lawrence J. F., Ren D., Pang H., Palaeoboganium gen. nov. from the Middle Jurassic of China (Coleoptera: Cucujoidea: Boganiidae): The first cycad pollinators? J. Syst. Palaeontology 16, 351–360 (2018). [Google Scholar]
- 46.Pedezzi R., et al. , A novel β-fructofuranosidase in Coleoptera: Characterization of a β-fructofuranosidase from the sugarcane weevil, Sphenophorus levis. Insect Biochem. Mol. Biol. 55, 31–38 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Davison A., Blaxter M., Ancient origin of glycosyl hydrolase family 9 cellulase genes. Mol. Biol. Evol. 22, 1273–1284 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Cragg S. M., et al. , Lignocellulose degradation mechanisms across the tree of life. Curr. Opin. Chem. Biol. 29, 108–119 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scully E. D., et al. , Metagenomic profiling reveals lignocellulose degrading system in a microbial community associated with a wood-feeding beetle. PLoS One 8, e73827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao C., Doucet D., Mittapalli O., Horizontal transfer genes in Agrilus planipennis. Insect Mol. Biol. 23, 821–832 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Zhao C., Mittapallii O., Horizontal gene transfer and gene duplication of plant cell wall degrading enzyme genes in an invasive insect pest (2014) https://kb.osu.edu/bitstream/handle/1811/60352/OARDCposter2014-2.pdf. Accessed 1 February 2019.
- 52.Evans J. D., et al. , Genome of the small hive beetle (Aethina tumida, Coleoptera: Nitidulidae), a worldwide parasite of social bee colonies, provides insights into detoxification and herbivory. Gigascience 7, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aw T., et al. , Functional genomics of mountain pine beetle (Dendroctonus ponderosae) midguts and fat bodies. BMC Genomics 11, 215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keeling C. I., et al. , Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 14, R27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Busch A., Kunert G., Heckel D. G., Pauchet Y., Evolution and functional characterization of CAZymes belonging to subfamily 10 of glycoside hydrolase family 5 (GH5_10) in two species of phytophagous beetles. PLoS One 12, e0184305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Evans A. M., McKenna D. D., Bellamy C. L., Farrell B. D., Large‐scale molecular phylogeny of metallic wood‐boring beetles (Coleoptera: Buprestoidea) provides new insights into relationships and reveals multiple evolutionary origins of the larval leaf‐mining habit. Syst. Entomol. 40, 385–400 (2015). [Google Scholar]
- 57.Scully E. D., et al. , Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genomics 15, 1096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernández-Hernández T., Evolutionary rates and adaptive radiations. T. Biol. Philos. 34, 41 (2019). [Google Scholar]
- 59.Pauchet Y., Kirsch R., Giraud S., Vogel H., Heckel D. G., Identification and characterization of plant cell wall degrading enzymes from three glycoside hydrolase families in the cerambycid beetle Apriona japonica. Insect Biochem. Mol. Biol. 49, 1–13 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Ehrlich P. R., Raven P. H., Butterflies and plants: A study in co-evolution. Evolution 18, 586–608 (1964). [Google Scholar]
- 61.Futuyma D. J., Agrawal A. A., Macroevolution and the biological diversity of plants and herbivores. Proc. Natl. Acad. Sci. U.S.A. 106, 18054–18061 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amborella Genome P.; Amborella Genome Project , The Amborella genome and the evolution of flowering plants. Science 342, 1241089 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Futuyma D. J., Moreno G., The evolution of ecological specialization. Annu. Rev. Ecol. Syst. 19, 207–233 (1988). [Google Scholar]
- 64.Leschen R. A., Buckley T. R., Multistate characters and diet shifts: Evolution of Erotylidae (Coleoptera). Syst. Biol. 56, 97–112 (2007). [DOI] [PubMed] [Google Scholar]
- 65.Haddad S., et al. , Anchored hybrid enrichment provides new insights into the phylogeny and evolution of longhorned beetles (Cerambycidae). Syst. Entomol. 43, 68–89 (2018). [Google Scholar]
- 66.Huang S. W., Zhang H. Y., Marshall S., Jackson T. A., The scarab gut: A potential bioreactor for bio-fuel production. J. Insect Sci. 17, 175–183 (2010). [Google Scholar]
- 67.Johnson A. J., et al. , Phylogenomics clarifies repeated evolutionary origins of inbreeding and fungus farming in bark beetles (Curculionidae, Scolytinae). Mol. Phylogenet. Evol. 127, 229–238 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Wheeler D., Redding A. J., Werren J. H., Characterization of an ancient lepidopteran lateral gene transfer. PLoS One 8, e59262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.dos Reis M., Donoghue P. C., Yang Z., Bayesian molecular clock dating of species divergences in the genomics era. Nat. Rev. Genet. 17, 71–80 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Harmon L. J., Rabosky D. L., FitzJohn R. G., Brown J. W., MEDUSA: Modeling Evolutionary Diversification Using Stepwise AIC. (Version: 0.12, 2011).
- 72.Hopkins R. J., van Dam N. M., van Loon J. J., Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54, 57–83 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Finn R. D., et al. , Pfam: The protein families database. Nucleic Acids Res. 42, D222–D230 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Camacho C., et al. , BLAST+: Architecture and applications. BMC Bioinformatics 10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ślipiński S. A., Leschen R. A. B., Lawrence J. F., Order Coleoptera Linnaeus, 1758. In: Zhang, Z.-Q. (Ed.) Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness Zootaxa 3148, 203–208 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Barba-Montoya J., Dos Reis M., Schneider H., Donoghue P. C. J., Yang Z., Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a Cretaceous Terrestrial Revolution. New Phytol. 218, 819–834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKenna D. D., et al. , Alignments of phylogenomic data, putative PCWDES, and gene trees, and BLAST hits for candidate PCWDEs. Zenodo. 10.5281/zenodo.3522944. Deposited 30 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.