Significance
Inositol phosphates (IPs) are a class of signaling molecules regulating cell physiology. The best-characterized IP, the calcium release factor IP3, is generated by phospholipase C hydrolysis of phosphoinositides lipids. For historical and technical reasons, IPs synthesis is believed to originate from the lipid-generated IP3. While this is true in yeast, our work has demonstrated that other organisms use a “soluble” (nonlipid) route to synthesize IPs. This soluble pathway depends on the metabolic status of the cells, and is under the control of the kinase ITPK1, which phosphorylates inositol monophosphate likely generated from glucose. The data shed light on the evolutionary origin of IPs, signaling and tightening the link between these small molecules and basic metabolism.
Keywords: inositol phosphate, phospholipase C, glucose, phosphate, metabolism
Abstract
Inositol phosphates (IPs) comprise a network of phosphorylated molecules that play multiple signaling roles in eukaryotes. IPs synthesis is believed to originate with IP3 generated from PIP2 by phospholipase C (PLC). Here, we report that in mammalian cells PLC-generated IPs are rapidly recycled to inositol, and uncover the enzymology behind an alternative “soluble” route to synthesis of IPs. Inositol tetrakisphosphate 1-kinase 1 (ITPK1)—found in Asgard archaea, social amoeba, plants, and animals—phosphorylates I(3)P1 originating from glucose-6-phosphate, and I(1)P1 generated from sphingolipids, to enable synthesis of IP6. We also found using PAGE mass assay that metabolic blockage by phosphate starvation surprisingly increased IP6 levels in a ITPK1-dependent manner, establishing a route to IP6 controlled by cellular metabolic status, that is not detectable by traditional [3H]-inositol labeling. The presence of ITPK1 in archaeal clades thought to define eukaryogenesis indicates that IPs had functional roles before the appearance of the eukaryote.
Inositol phosphates (IPs) are a class of small molecules displaying a large number of functional roles across the spectrum of life (1). In metazoa, IPs are essential for calcium signaling, thanks to the calcium release factor I(1,4,5)P3 (IP3) that is produced by the action of phospholipase C (PLC) on PI(4,5)P2. Many other IPs are found in eukaryotic cells, with the fully phosphorylated IP6 the most abundant. Surprisingly, even more highly phosphorylated species exist: The IP6-derived inositol pyrophosphates IP7 and IP8 (2). Among the numerous physiological functions regulated by IPs are energy metabolism (3), phosphate homeostasis (4), virus structure stability (5), insulin signaling (6, 7), and necroptosis (8).
The biosynthetic pathway of IPs synthesis was initially defined through biochemical characterization of kinase and phosphatase activities present in cell extracts that converted one IP into another (9–11). These early efforts led to the definition of biochemical fluxes between IPs, but rarely to identification of the specific enzymes. A subsequent wave of studies used the genetic power of Saccharomyces cerevisiae to discover the genes involved. A yeast genetic screen that aimed to identify genes affecting nuclear mRNA export led to the identification of PLC, and two kinases that sequentially acted on IP3 to produce IP6 (12). This resulted in the cloning of inositol polyphosphate multikinase (IPMK; in yeast known as Arg82, Ipk2) (13, 14) and inositol pentakisphosphate kinase (IPPK; in yeast called Ipk1) (12, 15). The budding yeast was also instrumental in identifying and characterizing both known classes of inositol pyrophosphates synthesizing enzymes (16, 17). The highly influential role played by S. cerevisiae research in discovering the IP kinases, and in defining a simple linear IPs biosynthetic pathway starting from PLC, generated trust in the universal nature of this pathway. However, we should always keep in mind that yeast have lost much genetic information present in other eukaryotes, and have simplified metabolic and regulatory signaling networks (18).
The potential pathways to IP6-7-8 synthesis have a profound impact on how we interpret their signaling roles. If they are synthesized from IP3, their function has evolved in relation to lipid-dependent PLC and/or calcium signaling. This constraint is relieved if IP6 synthesis occurs lipid-independently, originating directly from inositol, a “soluble” pathway. In yeast, which do not have IP3-regulated calcium signaling, IP6 synthesis strictly depends on the PLC-generated IP3 (Figs. 1A and 2D). The higher complexity of other eukaryotes might allow different pathways for IPs synthesis. For example, the social amoeba Dictyostelium discoideum (10) and plants (19) have been proposed to possess a PLC-independent route to produce IPs. However, the enzymatic machinery remains elusive.
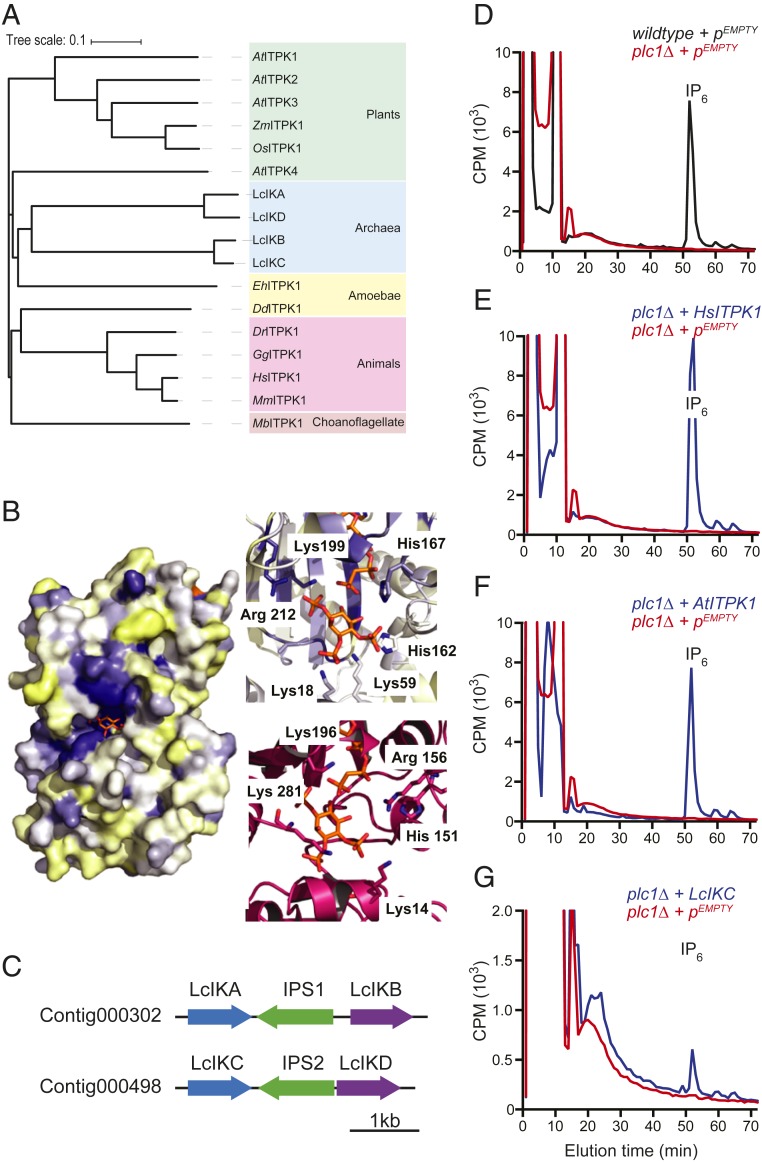
Fig. 1.
PLC-generated IP3 is converted to inositol but not to IP6. (A) Depiction of the pathway for the synthesis of phosphorylated inositol derivatives. Soluble inositol is incorporated into lipids by the action of the phosphatidylinositol synthase (PIS) to produce PI. After successive phosphorylation, PI(4,5)P2 (PIP2) is generated. Upon activation, PLC hydrolyzes PI(4,5)P2 and releases two second messengers: IP3 and DAG. IP3 can be recycled to inositol through dephosphorylation or be used to generate highly phosphorylated species. A second route, strictly lipid-independent, has also been proposed in social amoeba and plants. Here, inositol is directly phosphorylated successively to generate IP6. (B) Activation of PLC induces the recycling of IP3 but not the synthesis of IP6. SAX-HPLC profile of HEK293T cells transiently overexpressing the constitutively active mutant G protein α subunit (pGαqQL, red line) or an empty vector (pEMPTY, blue line) pulse labeled for 5 h with [3H]-inositol. Elution positions of radiolabeled standards ([3H]I(1,4,5)P3, [3H]I(1,3,4,5,6)P5, and [3H]IP6) are shown on top. (C) Quantification of results in B. The fold-change of the different IPs species over the control cells is presented, n = 6. Significant differences are shown as P < 0.01 (**), P < 0.001 (***), and P < 0.0001 (****). n.s., not significant.
Fig. 2.
ITPK1 is evolutionarily conserved and mediates the synthesis of IPs independently of Plc1. (A) Phylogenetic tree of ITPK1. At: A. thaliana, Dd: D. discoideum, Dr: Danio rerio; Eh: E. histolytica; Gg: Gallus gallus; Hs: H. sapiens; Mm: Mus musculus; Mb: Monosiga brevicollis; Os: O. sativa; Zm: Z. mays. (B) A surface representation of human ITPK1 (2qb5) docked with I(1,3,4)P3 and colored according to sequence conservation between ITPK1s from human, E. histolytica, and L. candidatus (blue is highly conserved, yellow is low conservation). The IP binding site for human ITPK1 (2QB5) (Upper) and predicted LcIKC homology model (Lower) are shown in close-up. The human structure is again colored by sequence conservation. Both structures have I(1,3,4)P3 docked into the binding sites. Key catalytic residues are marked. (C) Schematic representation of the localization of the four L. candidatus inositol kinase (LcIK) genes relative to the position of two IPS genes. The sequences of the contigs were retrieved from Spang et al. (22). (D–G) SAX-HPLC analysis of IPs from the indicated [3H]-inositol-labeled yeast strains. A plc1∆ strain (red line) does not accumulate IPs as illustrated by the complete absence of IP6 compared to a wild-type strain (blue line) (D). Expression of H. sapiens (E), A. thaliana (F), or L. candidatus (LcIKC) (G) ITPK1 (blue line) restored synthesis of IP6. Shown are representative HPLC traces of at least 3 independent experiments.
Another difficult issue is the source of inositol used to synthesize IPs. Myo-inositol, the most abundant stereoisomer (hereafter referred to as inositol), in mammalian cells and yeast, can be acquired from the extracellular milieu, or be generated endogenously by isomerization of glucose-6-phosphate (G6P) into I(3)P1. This reaction is catalyzed by inositol 3-phosphate synthase (IPS; known as ISYNA1 in mammals; Ino1 in yeast) (20). I(3)P1 can then be dephosphorylated to inositol by the inositol monophosphatase IMPA1 (21). Most studies of IPs metabolism over the past 4 decades have used [3H]-inositol labeling to monitor IP6-7-8 synthesis, an experimental set-up that precludes the analysis of G6P-derived inositol. Therefore, virtually no attention has been given to the contribution of endogenously generated inositol to the synthesis of IPs.
A recent study reported the genome of the archaea Lokiarcheum candidatus, which shed new light on eukaryogenesis (22). Archaea, including L. candidatus, usually possess a rudimental inositol metabolism (SI Appendix, Fig. S1), able to synthesize I(3)P1 and inositol. Surprisingly, genes encoding homologs of the inositol tetrakisphosphate 1-kinase 1 (ITPK1) were also found in this organism. ITPK1 has been described to phosphorylate I(1,3,4)P3 and I(1,3,4,5)P4 to generate I(1,3,4,5,6)P5 (23). Its presence in this organism suggested that ITPK1 may have additional substrate specificity.
Here, we investigated the possibility of ITPK1 as the enzyme responsible for the first steps of the soluble route to IPs synthesis. We show that ITPK1 phosphorylates IP1 [I(1)P1 and I(3)P1] to produce substrates for other IP kinases, such as IPMK and IPPK. Mammalian cells depleted for ITPK1 show a strong decrease in IP6, indicating that ITPK1 is a crucial enzyme of the pathway. More importantly, we propose that the PLC-independent route to IP6 synthesis is coupled with de novo inositol synthesis. The ITPK1-dependent increase in IP6 we observed after phosphate starvation when visualizing IP6 with PAGE is much smaller when observed by metabolic labeling using [3H]-inositol. This suggests that the two pathways (lipid-dependent and soluble) are independently regulated.
Results
PLC-Generated IP3 Is Rapidly Recycled to Inositol.
To assess the relevance of PLC-dependent IP3 production for IP6 synthesis in mammalian cells, we activated PLC by expressing the constitutively active G protein mutant GαqQL (24), and studied rapid [3H]-inositol metabolism by following its fate after 5 h of labeling. The PLC-activated cells showed a 5- to 16-fold increase in IP1-2-3-4 levels. In contrast, IP5-6 levels remained unchanged following PLC activation (Fig. 1 B and C). It is clear that, rather than being used for higher IPs synthesis, most of the PLC-generated IP3 was recycled back to inositol. This is consistent with inositol fluxes studied in rat cortical neurons after PLC activation induced by carbachol or neuronal depolarization (25). The observed IP3 recycling was greater than what has been observed after steady-state metabolic labeling (26), which highlights the importance of studying rapid IP fluxes. The increase in IP4 probably results from the activity of IP3–3-kinases that are activated by calcium (27–29). Therefore, similarly to what has been proposed for the amoeba D. discoideum and plants (10, 30), it is possible that IP6 in mammals is synthesized by an uncharacterized lipid-independent, soluble pathway.
ITPK1 Is a Conserved IP Kinase.
While inositol synthesis is a shared characteristic of two of the three kingdoms of life, archaea, and eukaryotes (31), IPs have been proposed to be a hallmark of eukaryotic cells (1, 32). However, the sequencing of Asgard archaea revealed the presence of IP kinases in a prokaryotic genome (22). We identified 4 genes homologous to ITPK1 (23) in the genome of L. candidatus, which we named LcIKA to LcIKD (Fig. 2A). Structural and homology modeling found that these proteins share the ATP-grasp domain found in both Homo sapiens HsITPK1 and Entamoeba histolytica EhITPK1 (Fig. 2B). Comparison of HsITPK1, EhITPK1 and LcIKs found that ATP and IP binding sites were the most conserved regions and, importantly, most residues involved in catalysis were conserved. The most highly conserved IPs-contact residues (H167, K199, and R212 in HsITPK1) are proximal to the ATP binding site, while less conserved residues (K18, K59, H162, and G301 in HsITPK1) sit on the opposite side of the IPs substrate from the active site. This suggests that these less-conserved distal residues contribute to variations in substrate selectivity across the ITPK1 homologs. For example, when I(1,3,4)P3 was modeled into the human structure, a steric clash with H162 was reported, which likely contributes to its greater selectivity compared to the EhITPK1 enzyme (33). In the LcIKC model, this histidine is replaced with Arg156, indicating the LcIKC enzyme will also have a more restricted substrate selectivity than EhITPK1. In addition, the LcIKA, -B, and -C have additional basic residues modeled to sit in the IP-binding pocket that are absent in the E. histolytica and human structures.
L. candidatus also possesses the typical archaeal inositol-related genes, and is therefore able to synthesize a range of inositol-containing molecules: I(3)P1, inositol, the archaea-specific di-inositol phosphate, and both archaeal- and eukaryotic-type phosphoinositides (SI Appendix, Fig. S1). Given that L. candidatus cannot synthesize the known substrates of ITPK1, IP3, or IP4, we predicted that ITPK1 is, and was originally, an inositol or an IP1 kinase. Thus, ITPK1 using these substrates might be responsible for the soluble route of IP6 synthesis. This idea is further reinforced by the observation that the two pairs of LcIK genes flank two IPS genes in the L. candidatus genome, suggesting a functional link between I(3)P1 synthesis and LcIKA-D activity (Fig. 2C). To test this hypothesis, we expressed ITPK1, which is naturally absent in yeast, in a yeast plc1∆ strain. Strains lacking PLC1 are completely devoid of IP6 and other higher IPs (Fig. 2D). The expression of human or plant (Arabidopsis thaliana and Oryza sativa) ITPK1 restored wild-type level of IP6 (Fig. 2 E and F and SI Appendix, Fig. S2A). Production of IP6 was also observed using D. discoideum and L. candidatus homologs, although to a lesser extent presumably due to suboptimal reaction conditions and/or expression (Fig. 2G and SI Appendix, Fig. S2 B and D). These data indicate that ITPK1 is able to bypass the yeast requirement for PLC in the synthesis of IPs, and that this activity is conserved through evolution.
Sphingolipid Hydrolysis Generates I(1)P1 as Substrate for ITPK1.
Recombinant ITPK1s were unable to phosphorylate [3H]-inositol; a phosphorylated inositol must therefore be required as substrate. To identify candidates that could perform this first step of the soluble pathway in generating the in vivo initial substrate of ITPK1, we looked for putative inositol kinases in the yeast genome that could potentially generate IP1. Two genes, MAK32 and RBK1, encode members of the ribokinase family and have high homology with social amoeba and plant inositol kinases. However, the simultaneous depletion of these genes did not prevent ITPK1-mediated IP6 synthesis (SI Appendix, Fig. S3A). We therefore searched for alternative sources of IP1. Besides phosphoinositides, radiolabeled [3H]-inositol can be incorporated into glycophosphatidylinositol (GPI) anchors and sphingolipids (SI Appendix, Fig. S3B). Phospholipase D (yeast Spo14) uses phosphatidylinositol (PI) as substrate, releasing I(1)P1 (34, 35). However, expression of ITPK1 in plc1∆spo14∆ still generates IP6 (SI Appendix, Fig. S3C).
Next, we tested the hypothesis that the ITPK1 substrate originates from the degradation of GPI anchors. However, IP6 is still generated in the plc1∆gpi1∆ strain harboring ITPK1 (SI Appendix, Fig. S3D). Yeast, like plants and protists, produce sphingolipids called inositol phosphorylceramides (IPC) (Fig. 3A). These lipids are degraded by the inositol phosphosphingolipid phospholipase (Isc1), an ortholog of the mammalian neutral sphingomyelinase, to generate I(1)P1 and phytoceramide (36) (Fig. 3A). IP6 is present in the isc1∆ strain, but absent in the plc1∆isc1∆ strain (SI Appendix, Fig. S4 A and B). Expression of ITPK1 in the plc1∆isc1∆ strain did not rescue IP6 synthesis, thereby demonstrating that degradation of sphingolipids generates the [3H]-I(1)P1 used by ITPK1, which leads to IP6 synthesis (Fig. 3B and SI Appendix, Fig. S4A). This result was confirmed by pharmacological inhibition of sphingolipid synthesis using Myriocin or Aureobasidin A (SI Appendix, Fig. S4 C and D). In plc1∆ expressing ITPK1, IP6 synthesis was blocked upon incubation with the drugs, while the wild-type was only slightly affected (Fig. 3C). Given the rapid synthesis of wild-type level of [3H]-IP6 in a plc1∆ strain expressing human or plant ITPK1 (SI Appendix, Fig. S3A), our data indicate that sphingolipids turnover rapidly in exponentially growing yeast.
Fig. 3.
ITPK1 uses I(1)P1 generated from sphingolipids. (A) Schematic of IPC degradation by Isc1 that releases I(1)P1. (B) SAX-HPLC analysis of labeled IPs from a strain deleted for both PLC1 and ISC1, and expressing HsITPK1 or the empty vector. (C) SAX-HPLC analysis of IPs from a wild-type strain expressing empty vector (Left) and a plc1∆ strain expressing HsITPK1 (Right) treated with Myriocin (1 µM) or Aureobasidin A (1 µM) for 1.5 h. [3H]-inositol was added to the culture medium and the cells were pulse labeled for 2 h in the presence of the drugs. (D) Vacuole morphology of the wild-type and the plc1∆ strains expressing GST or GST-HsITPK1 observed after staining with FM4-64. (Scale bar, 5 µm.) (E) PolyP was extracted from exponentially growing cells and analyzed by PAGE. PolyP45 and PolyP13 are synthetic polyP standards of average chain length of 45 and 13 phosphate units, respectively. The gel is representative of 3 experiments. (F) PolyP was quantified using a Malachite Green assay after acidic extraction and column purification, n = 3. (G) ATP levels measured by luciferase assay for the indicated strains expressing GST or GST-HsITPK1. Results are shown relative to wild-type, n = 4. Significant differences are shown as P<0.01 (**) and P<0.0001 (****).
ITPK1 Expression in Yeast Allows Uncoupling of PLC Signaling from IP-Dependent Phenotypes.
PLC generates two intracellular messengers: diacylglycerol (DAG) and IP3, which in yeast is converted to higher IPs. The rescue of IP6-7-8 level by overexpressing ITPK1 in plc1∆ allows the distinguishing of phenotypes caused by accumulation of PI(4,5)P2 or lack of DAG, or loss of IPs. The fragmented vacuoles phenotype observed in plc1∆ was not rescued by ITPK1 expression, while the synthesis of inorganic polyphosphate (polyP) was restored (Fig. 3 D–F) (4, 37). Therefore, in plc1∆ yeast the fragmented vacuoles are a consequence of both PIP2 accumulation (or lack of DAG) and lack of inositol pyrophosphates, since the latter are known to regulate vacuole physiology (38). Conversely, polyP synthesis depends only on the presence of IPs. The inositol pyrophosphates IP7 and IP8 are involved in energy homeostasis, and are required to maintain a normal cytosolic ATP concentration (3). In accordance with this, we found high levels of ATP in a plc1∆ strain. Expression of ITPK1 restored completely the levels of ATP, indicating that only highly phosphorylated IPs are required for controlling cellular energetics (Fig. 3G).
ITPK1 Can Use I(3)P1 to Generate Higher IPs.
We have demonstrated the ability of yeast to take advantage of exogenously expressed ITPK1 for IP6 synthesis from sphingolipids rather than from phosphoinositides. Mammals and L. candidatus have ITPK1 but do not contain IPC to provide I(1)P1: Their soluble pathways require an alternate source of IP1. L. candidatus, like virtually all eukaryotes, is able to convert G6P to I(3)P1 with its IPS (SI Appendix, Fig. S1). Therefore, we wondered if I(3)P1 could be used by ITPK1 to initiate IP6 synthesis in a lipid-independent fashion. To test this hypothesis, we used the Thermococcus kodakarensis inositol kinase TK2285, which selectively generates I(3)P1 (39). In vitro reactions conducted in the presence of [3H]-inositol and both TK2285 and ITPK1 showed a time-dependent synthesis of IP5 (Fig. 4A). This enzymatic plasticity of ITPK1 confirms the structure-based prediction of catalytic flexibility; this enzyme can accommodate several different IP substrates (40). We then analyzed ITPK1 activity on I(1)P1 and I(3)P1 using PAGE with Toluidine staining. This technique allows visualization of highly phosphorylated IPs only, since lower phosphorylated species stain poorly. Recombinant ITPK1 phosphorylated both isoforms I(1)P1 and I(3)P1 to IP5 (Fig. 4B). Simultaneous expression of ITPK1 with the inositol kinase TK2285 in plc∆isc1∆ yeast strain restored the IP6 level (Fig. 4C), which confirmed that ITPK1 can use I(3)P1 both in vitro and in vivo. These data implied that I(3)P1 de novo synthesis from G6P could be involved in IPs synthesis. To verify the importance of ITPK1 in IP6 synthesis and the role of the soluble route in IPs metabolism, we developed human cell lines devoid of ITPK1.
Fig. 4.
ITPK1 phosphorylates I(3)P1 and contributes to IPs synthesis in mammalian cells. (A) In vitro assay with [3H]-inositol and recombinant ITPK1 and the inositol kinase TK2285. Reactions were stopped after 30 min (blue line), 60 min (red line), or 120 min (green line) before IPs were extracted and analyzed by SAX-HPLC. (B) In vitro kinase assay using I(1)P1 and I(3)P1 as substrates. The products of the reaction were analyzed by PAGE followed by Toluidine blue staining. IP5 and IP6 standards were loaded as controls. The gel is representative of 3 experiments. (C) Expression of the archaeal inositol kinase TK2285 with HsITPK1 (blue line) or AtITPK1 (red line) rescues IPs synthesis in the plc1∆isc1∆ strain, which does not otherwise accumulate IPs (red line). (D) Verification by Western blotting of the ITPK1−/− clones generated in HCT116 by CRISPR/Cas9. Histone H3 was used as a loading control. (E and F) PAGE analysis of IPs extracted from wild-type HCT116 and 2 knockout clones, ITPK1−/− cl1 and ITPK1−/− cl2, grown in DMEM supplemented with 10% serum (E). Quantification of IP6 in the indicated lines by densitometry, n = 6 (F). (G and H) SAX-HPLC analysis of IPs from the same cells as in E labeled with [3H]-inositol and grown in inositol-free DMEM supplemented with 10% dialyzed serum. The chromatogram (G) is representative of 4 experiments. Quantification of IP6 in the indicated lines, n = 3 (H). (I and J) A region of the chromatogram presented in G is enlarged to highlight the basal level of I(1,4,5)P3 (I). Quantification of I(1,4,5)P3 in the indicated strains, n = 3 (J). The elution time of standard I(1,4,5)P3 is shown on top. Significant differences are shown as P < 0.001 (***) and P < 0.0001 (****).
ITPK1 Is Required for Mammalian IPs Metabolism and IP6 Synthesis.
The role of ITPK1 in higher IPs synthesis in mammalian cells was assessed using CRISPR/Cas9-generated knockout in the human colon carcinoma cell line HCT116. We obtained several clonal lines, and focused our study on two randomly selected lines devoid of ITPK1 protein (Fig. 4D), called ITPK1−/− cl1 and ITPK1−/− cl2. Both lines showed a 90% reduction of IP6 level when analyzed by PAGE (Fig. 4 E and F) with cells grown on normal media. Analysis by [3H]-inositol metabolic labeling, performed using inositol-free media to improve sensitivity, and strong anion exchange (SAX) high-performance liquic chromatography (HPLC), not only confirmed the dramatic reduction of IP6 observed by PAGE but revealed a similar decrease in IP5 levels (Fig. 4 G and H). To eliminate any artifact arising from using different media composition, we performed a PAGE analysis with extracts from cells grown in normal media and in inositol-free media. This analysis demonstrated that the IP6 reduction observable in ITPK1−/− cl1 and ITPK1−/− cl2 is independent of the presence of inositol in the growing media (SI Appendix, Fig. S5).
These data confirm the results from the ITPK1 knockout recently generated in a different human colon carcinoma cell line, HT29 (8). That paper reported similar levels of IP3 and IP4 in wild-type and ITPK1−/− cells but did not closely analyze lower IPs. Our full SAX-HPLC analysis showed substantial reduction in the levels of all IPs in ITPK1−/− cl1 and ITPK1−/− cl2, including even the lower phosphorylated IP1 and IP2 (Fig. 4G). Notably, the level of PLC-generated [3H]-I(1,4,5)P3 was not affected in ITPK1−/− cells, suggesting that its synthesis is not altered (Fig. 4 I and J). Conversely, a second IP3 isomer, likely I(1,3,4)P3, appears to be down-regulated in ITPK1−/− cells. These analyses demonstrate that ITPK1 plays a key role in IPs metabolism.
Metabolism Controls the Soluble Pathway.
We next assessed the physiological relevance of the soluble IPs synthesis pathway. We hypothesized that the presence of ITPK1 in Asgard archaea suggests an evolutionarily ancient origin for the soluble route. It might be regulated not by receptor activation, absent in archaea, but by core regulatory networks, such as alteration of cellular metabolic status. Therefore, we investigated whether metabolic signals could regulate IP6 synthesis through ITPK1 in mammalian cells. The regulation of IPs levels by phosphate (Pi) has been reported in several organisms (41–43). Particularly, a decrease in Pi availability has been shown to result in a decrease of the inositol pyrophosphates IP7-8. To investigate this in the context of the lipid-independent pathway, cells were cultivated in Pi-free media as previously described (44, 45). We also observed a decrease in IP7-8 during Pi starvation (Fig. 5A). This decrease occurred concomitantly with a reduction in ATP, as well as ADP and AMP levels (SI Appendix, Fig. S6). Surprisingly, we recorded a 150% increase in IP6 level after 24 h of Pi starvation (Fig. 5 A and B). This observation was not restricted to HCT116 cells, as a similar increase was observed in all mammalian cell lines analyzed (SI Appendix, Fig. S7). The PAGE analysis detects total IP6 cellular mass: The bands seen are a mixture of molecules originating from environmentally acquired inositol, or from inositol synthesized from G6P. A similar Pi starvation protocol was applied to cells metabolically labeled with [3H]-inositol for 5 d. SAX-HPLC analysis, which can only detect IPs arising from exogenous [3H]-inositol, gave only a 15% increase in [3H]-IP6 (Fig. 5 C and D). Although inositol media availability does not affect the decrease in IP6 level observable in ITPK1−/− cells (Fig. 4 E and F and SI Appendix, Fig. S5), we further validated our Pi starvation observations by repeating the experiment using cells grown in inositol-free media for matched SAX-HPLC and PAGE analysis. We also performed an additional SAX-HPLC [3H]-inositol labeling analysis in the presence of 40 μM inositol throughout. These experiments confirmed that the increase in IP6 level is only clearly observable by PAGE analysis (SI Appendix, Fig. S8).
Fig. 5.
Phosphate starvation induces an ITPK1-dependent IP6 accumulation. (A) PAGE analysis of IPs extracted from wild-type HCT116 cells during phosphate (Pi) starvation. Cells were collected at different time points (0, 6, 24, 48 h). Synthetic polyP of average length 13 phosphate units was used as ladder. (B) Quantification of IP6 after 24 h in the absence or presence of Pi (0.9 mM) in HCT116 by densitometry, n = 4. (C) SAX-HPLC analysis of IPs from wild-type HCT116 cells labeled with [3H]-inositol inositol-free DMEM supplemented with 10% dialyzed serum. Cells were labeled for 4 d, then cultivated for a further 24 h of labeling in absence or presence of 0.9 mM Pi in Pi-free DMEM supplemented with 10% dialyzed serum. (D) Quantification of IP6 in chromatogram shown in C, n = 4. (E) PAGE analysis of IPs extracted from wild-type and ITPK1−/− HCT116 cells following 24 h in absence or presence of 0.9 mM Pi in Pi-free DMEM supplemented with 10% dialyzed serum. (F) Synthesis of IP6 from glucose in HCT116 cells determined by SAX-HPLC analysis. Elution profile of extract prepared from cells labeled for 2 d with [3H]-glucose in low glucose DMEM supplemented with 10% dialyzed serum (blue trace) and after phytase treatment (red trace). The elution times of ATP and [3H]-IP6 standards are indicated on top. Significant differences are shown as P < 0.05 (*), P < 0.001 (***).
These results suggest that de novo synthesis of IP6 from G6P is induced upon Pi starvation, while exogenous inositol contributes minimally to the increase in IP6. As ITPK1 is able to act on G6P-generated I(3)P1 (Fig. 4), we tested ITPK1’s involvement in the induction of IP6 on Pi starvation. Unlike wild-type HCT116, depriving the ITPK1−/− cl1 and ITPK1−/− cl2 cells of Pi did not result in an increase in IP6 (Fig. 5E). Altogether, the data demonstrate that ITPK1 is a key enzyme in the metabolism-regulated synthesis of IPs from G6P. Next, we labeled HCT116 cell with [3H]-glucose to demonstrate a carbon flux from glucose to IP6. All organic molecules of mammalian cells are generated from the glucose carbons backbone. Therefore, a very small fraction of radioactivity may end up accumulating in IPs and ultimately in IP6. The SAX-HPLC analysis of [3H]-glucose labeled cells confirmed this prediction, with most of the radiolabeled molecules eluting in the first 20 min (Fig. 5F). The highly charged nature of IP6 results in late elution of this metabolite in a clean section of the chromatogram. We confirmed the IP6 genuine nature by comparison of the elution time with a [3H]-IP6 standard, and by treating the extract with phytase (Fig. 5F). These data clearly demonstrate that inositol first and then IP6 can be synthesized from glucose in vivo.
Discussion
IPs comprise an important family of signaling molecules, far more complex than the lipid phosphoinositides. Only seven phosphoinositides exist, while the combinatorial attachment of phosphate groups on the inositol ring allows 64 different IPs: These possibilities define the “inositol phosphate code” (1). This number is an underestimation of the actual complexity of this family, since inositol pyrophosphates are also commonly present in eukaryotic cells (2). The evolution of the elaborate IPs network underlines their fundamental roles in biology. However, IPs are less well understood than their phosphoinositide cousins. Other factors, aside from the complexity, have contributed to delaying our understanding and our full appreciation of IP signaling roles. In particular, the almost dogmatic translation of S. cerevisiae findings to other organisms may have misguided our investigation. Here we have demonstrated that ITPK1, a kinase absent in the yeast genome, is responsible for a “soluble” lipid-independent metabolic pathway leading to IP6-7-8 synthesis, a biosynthetic route that can originate directly from the conversion of G6P to I(3)P1 (Fig. 6). This brings into focus a major issue present in most IPs-related literature, which is based on exogenously added [3H]-inositol: the inability to measure IPs originating endogenously from glucose. The demonstration that phosphate starvation leads to a substantial IP6 increase when analyzed by PAGE but not by SAX-HPLC analysis demonstrates the failure of traditional [3H]-inositol labeling to account for the complete IPs metabolism and signaling. By defining the enzymology of the IPs soluble pathway, and revealing the IP6 change following metabolic alteration, our work gives a fresh perspective and impetus to future IPs research.
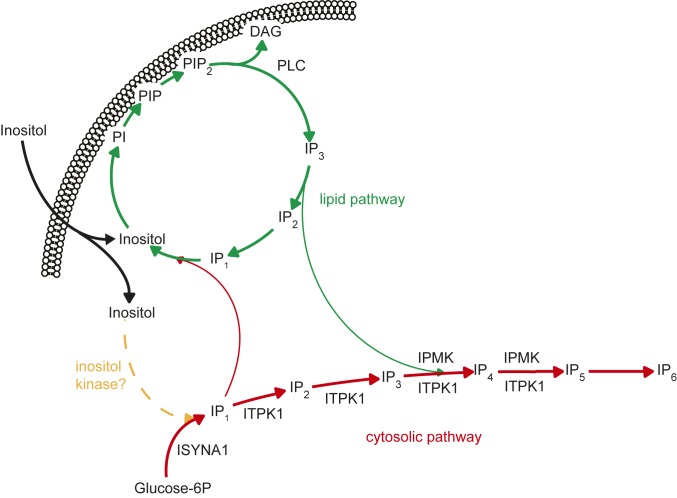
Fig. 6.
Schematic of the proposed pathway of IPs synthesis. The lipid pathway of IPs synthesis (green), foresees the synthesis of IP3 by PLC as the starting point for the generation of the whole spectrum of IPs. Our data indicate that the soluble route (red) starts from the conversion of G6P to IP1 by ISYNA1, and continues with ITPK1 converting IP1 to higher phosphorylated species. The existence of an inositol kinase, described in plants, amoebae, and archaea, remains to be demonstrated in mammals.
Mammalian ITPK1 was originally characterized as a kinase able to phosphorylate I(1,3,4)P3 at positions 5 and 6 of the inositol ring (23). Over time, its catalytic flexibility was discovered, and it has been demonstrated that this enzyme can also act as phosphatase to dephosphorylate position 1 (46, 47). Initial work mainly in vitro suggested that this activity was behind the synthesis of the plasma membrane chloride channel inhibitor I(3,4,5,6)P4, but ITPK1 was found not to be responsible in vivo (48). Still, the catalytic flexibility was further underlined by the crystal structures of ITPK1 from E. histolytica and H. sapiens (33, 40). This important work predicted that up to 18 different IP3 and IP4 substrates could occupy the ITPK1 catalytic pocket. We show that it can act on even lower IPs. The presence of ITPK1 in the Asgard archaea L. candidatus, which possesses a rudimental inositol metabolism (SI Appendix, Fig. S1), led us first to hypothesize then to demonstrate in vitro and in vivo that ITPK1 uses IP1 as a substrate to feed the soluble route of IPs synthesis. Evolution has likely taken advantage of the general substrate flexibility of ITPK1 and might have adapted this kinase to species-specific preferred substrate. Mammalian ITPK1 are prone to convert I(1,3,4)P3 to I(1,3,4,5,6)P5 (49). Conversely, plant ITPK1 might even have evolved to use IP6 as a substrate to synthesize IP7 (50).
IPC Are a Relevant Source of Substrate for the Soluble Route.
We first demonstrated that ITPK1 could use I(1)P1 as substrate. This molecule is generated in yeast from the hydrolysis of IPC (36). We showed that the human and plant ITPK1 fully rescued IP6 synthesis in plc1∆ yeast, while D. discoideum and LcIKC proteins only partially restored IP6 levels. The poor rescue might be due to the nonoptimal heterologous expression conditions, particularly related to culture temperature. It is also possible that L. candidatus, expected to have a simple IP network, does not require the same degree of catalytic flexibility for its ITPK1 as higher eukaryotes. IPC could be a relevant source of I(1)P1 substrate in plants, especially where this type of sphingolipid is particularly abundant (51). We have uncovered an unexpected rapid turnover of IPC in yeast cells, potentially representing an unexpected cross-talk between IPC and IPs signaling. In plants, IP6 is called phytic acid (from phyton, meaning part of a plant); it is the major phosphate storage molecule in seeds (52). The biotechnological objective of producing crops with low accumulation of IP6 (53, 54) in the seeds should therefore consider IPC metabolism. Interestingly, the Zea mays (maize) low-phytic acid mutant lpa2 is caused by mutation in one of its six ITPK1 enzymes (55). Additionally, an Arabidopsis thaliana mutant with a mutation in one of its four ITPK1 genes (itpk4) also shows a decrease in IP6 levels compared to wild-type plants after labeling with [32Pi] (42). These observations support a major role for the soluble route mediated by ITPK1 for synthesizing plant phytic acid.
Compartmentalization of the IPs Metabolic Routes.
The compartmentalization of the inositol acquired from the extracellular milieu, the inositol synthesized from G6P, and diverse IPs pools, is an important concept that comes with the description of the soluble route. Results obtained in Trypanosoma support the idea that exogenously acquired inositol has a different fate from G6P-derived inositol. It has been clearly demonstrated that the synthesis of phosphoinositides relies only on inositol uptake from the environment, while the synthesis of GPI anchors requires de novo synthesis of inositol from G6P (56–58). Exogenous inositol is rapidly incorporated into PI, suggesting that PI synthesis may even be coupled to cellular import (56). In addition, G6P-derived inositol must be synthesized and utilized in a different location (56). In line with these observations, IPs analysis in Trypanosoma revealed that although this organism possesses IP6 as demonstrated by PAGE analysis, radiolabeling using [3H]-inositol failed to detect significant amount of the same metabolite (59), indicating that in Trypanosoma, highly phosphorylated IPs synthesis is achieved through conversion of G6P-derived inositol and the cytosolic route.
ITPK1 Is a Tool to Uncouple Phenotypes Induced by Loss of PI(4,5)P2/DAG or IPs.
Yeast IPC regulates diverse cellular processes including cell growth and stress responses. The turnover of IPC that we observed during our short time-course experiments (SI Appendix, Fig. S3A) is probably an underestimation of the actual rate. The activity of Isc1 is known to be down-regulated by Slm1/2 proteins when bound to PI(4,5)P2 (60). Our experiments were performed in a plc1∆ strain that is unable to hydrolyze, and thus accumulates, PI(4,5)P2. Therefore, it is likely that the actual turnover rate is higher in wild-type cells. Although the focus of past literature was on the lipid side of sphingolipids, it seems natural to ask about the fate of the other degradation products of inositol-containing sphingolipids. Our data indicate that exponentially growing yeast hydrolyze IPC, and probably mannosylinositol phosphorylceramide (MIPC) and mannosyldiinositol phosphorylceramide [M(IP)2C], as fast as PI(4,5)P2. Therefore, I(1)P1, MIP, and M(IP)2, will be also released in abundance. The fate of these products, and their potential as signaling molecules, remains to be tested.
In yeast, most experiments affecting the levels of PI(4,5)P2 also affect the level of IPs, which themselves control a plethora of cellular activities. Expressing ITPK1 in plc1∆ yeast is therefore a powerful tool to properly dissect phosphoinositide signaling. We can now separate the phenotypes originating from the loss of IPs and identify those induced by altered DAG signaling or phospholipid accumulation. For example, invasive filamentous growth is required for virulence of pathogenic fungi. Both Plc1 and Kcs1 are required for filamentous growth (61, 62); however, the importance of Plc1 activity in this process could not be definitely ascertained as the knockout was also missing inositol pyrophosphates.
ITPK1 Is Essential for IPs Synthesis in Mammals.
The analysis of mammalian HCT116 or HT29 (8) ITPK1−/− cell lines demonstrates that this enzyme plays a major role in maintaining IP6 cellular levels. However, the other multikinase, IPMK, is also important: Mouse embryonic fibroblast cell lines from Ipmk−/− mice, or CRISPR-generated human cell lines, possess <20% of wild-type IP6 levels (8, 63, 64). It is likely that ITPK1 catalyzes the first steps of I(3)P1 phosphorylation, with IPMK phosphorylating a subsequent substrate to I(1,3,4,5,6)P5, which is then converted to IP6 by IPPK (yeast Ipk1). While in vitro ITPK1 synthesized IP5, we did not succeed in characterizing specifically which IP5 isomer was made. Nevertheless, the IP5 synthesized by ITPK1 is not the I(1,3,4,5,6)P5 substrate of IPPK, as expected, since expression of ITPK1 in yeast null for both PLC and IPMK (plc1∆arg82∆) failed to rescue the normal IP6 level.
One surprising observation is the strong decrease in IP1 and IP2 observed in the ITPK1−/− cells. The lifespan of the intermediates of IP6 synthesis depends on the activity of kinases and phosphatases. It is possible that in the absence of ITPK1, the IP1 and IP2 cannot be phosphorylated and trapped trough the cytosolic route, and are therefore rapidly degraded to inositol. Alternatively, we can speculate that these species are degradation products of IP6. Since the IP6 level is highly reduced in ITPK1−/− cell lines, the levels of those metabolites are necessarily lower. More work is required to infer metabolic relationships between the different metabolites.
The efficient recycling of PLC-dependent IP3 to inositol, and the PLC-independent increase in IP6 during Pi starvation in mammalian cells, are suggestive that the soluble pathway that produces highly phosphorylated IPs is compartmentalized from the lipid route. This could be a physical compartmentalization in different areas of the cell: It becomes imperative to localize the endogenous untagged IP kinases to avoid GFP overexpression artifacts. Even more important would be to localize the IPs themselves. This will certainly be challenging. However, the ability of Raman spectroscopy to identify IP6 in plant seeds (65) opens an investigative path to follow. We cannot exclude a possible cross-talk between the PLC-dependent and soluble pathways. Inactivation of one may be compensated for by an overactivation of the other, although this appears not to be the case in our HCT116 ITPK1−/− cell lines. Nor can we assume that the soluble route contributes to IPs synthesis to the same extent in all cell types or tissues.
IPs Synthesis Is Tightly Connected to the Metabolic Status of the Cell.
Until now, the cellular level of IP6 was considered to be highly stable, in contrast to IP7-8 that are known to have a very rapid turnover. The view of IPs synthesis was restricted by their quantification by metabolic labeling using [3H]-inositol as a precursor. Our work now demonstrates that during phosphate starvation most IP6 in mammalian cells is generated from G6P. One essential next step is to characterize the external or endogenous signals that regulate the soluble pathway, including deciphering the relationship between basic metabolism and IPs synthesis. A multilayered connection between IP7-8 and ATP had already been discovered. First, IP6Ks have been described as ATP sensors, their Km for ATP being close to 1.0 to 1.5 mM in vitro (14, 66). Second, the cellular ATP level is increased in IP6K-null cells (3). Third, IP7 controls metabolism by altering Akt signaling, and mice devoid of IP7 are resistant to obesity (7). Besides IPs, G6P is the starting point of three fundamental metabolic pathways: glycolysis, the pentose phosphate pathway and, in liver and muscle, glycogenesis. The diversionary metabolic route to inositol and then to IP6 is a new angle on how IP6-7-8 regulate metabolism. Interestingly, the IPS genes, INO1 in yeast and ISYNA1 in mammals, are transcriptionally regulated by IP7 (67, 68). This could indicate a feedback loop, in which reduction in IP7 levels during metabolic inhibition, for example during phosphate starvation, de-represses ISYNA1 transcription and enables the synthesis of inositol from G6P. Further supporting a close metabolic connection between glycolysis and inositol synthesis, knockout of the D. discoideum INO1 leads to the accumulation of 2,3-bisphosphoglycerate (69), derived from the glycolytic intermediate 1,3-bisphosphoglyceric acid. Our results and this model give new perspectives on how highly phosphorylated IPs, and especially inositol pyrophosphates, regulate basic metabolism. One future direction of this work will be to investigate the synthesis of IPs in cell types with different metabolic activities, such as quiescent cells.
Our data demonstrate that the IPs signaling network, which has been considered exclusive to nucleated cells, originated in the ancestors of the Asgard archaea L. candidatus, and that highly phosphorylated IPs evolved independently of, and before, PLC-dependent IP3 synthesis. Evolution has taken advantage of inositol’s metabolic stability to create a sophisticated network of signaling molecules (32). The signaling paradigm of G protein-coupled receptor activating the PLC-IP3-Ca2+ axis likely represents the evolutionary pinnacle for the lipid-/PLC-dependent IPs pathway. However, our work proposes an independent evolutionary history for higher phosphorylated IPs signaling, with a noteworthy connection to basic metabolism, whose pinnacle is yet to be discovered.
Materials and Methods
Detailed methods are provided in SI Appendix, Supplementary Information Text.
Genetic Manipulations.
CRISPR/Cas9 mutation of ITPK1 in HCT116 cells.
Low-passage HCT116 cells were originally purchased from ATCC. An ITPK1-specific crRNA oligo (5′-AltR1- GGUUGGCGAGUCCUACACCGGUUUUAGAGCUAUGCU-AltR2-3′) was obtained from IDT (ID: Hs.Cas9.ITPK1.1.AA; Integrated DNA Technologies). Cells were transfected with the crRNA oligo and the Alt-R S.p. HiFi Cas9 Nuclease V3 using Amaxa Nucleofector kit (Lonza) following the manufacturer’s instructions. Single clones were isolated by limiting dilution in 96-well plates and confirmed by visual examination. The clonal cell lines were amplified before screening by Western blotting using an anti ITPK1 antibody (ab175222, Abcam).
Transfection of mammalian cells.
For plasmid transient transfection experiments, cells were seeded into 6-well plates and transfected 24 h later. Lipofectamine 2000 was used with 1 µg DNA per well unless otherwise stated. Cells were harvested 24-h posttransfection.
Isotope Labeling and Inositol Phosphate Extraction.
Labeling of yeast strains.
Labeling and extraction of IPs were performed as described previously (70). Briefly, yeast precultures in synthetic complete media without uracil (SC-URA) were used to inoculate 5 mL of SC-URA-Inositol containing 5 µCi/mL of [3H]-inositol. Cells were grown overnight to logarithmic phase at 30 °C (or 22 °C in the case of the temperature-sensitive gpi1∆ derivatives), collected by centrifugation (2 min, 2000 × g), and washed once in water. IPs were extracted by adding 1 M perchloric acid containing 3 mM EDTA and glass beads. Cells were broken by shaking with a vortex (5 min, 4 °C) and debris were removed by centrifugation (5 min, 15, 000 × g). The supernatant, obtained as described above, was neutralized by adding potassium carbonate. Samples were kept on ice for 2 h to allow for salt precipitation. Insoluble material was removed by centrifugation (5 min, 15,000 × g) and the supernatant was kept at 4 °C until HPLC analysis.
For the labeling of the plc1∆mak32∆rbk1∆ strain, the indicated strains were grown overnight at 30 °C in SC-URA. In the morning, the cells were diluted to OD = 0.3 and incubated for 2 h at the same temperature. Next, the cells were washed twice in SC-URA–inositol and resuspended in this media supplemented with 5 µCi/mL of [3H]-inositol. The cells were incubated for 2 h at 30 °C before IPs were extracted as described below.
For the labeling in the presence of inhibitors of sphingolipid synthesis, the indicated strains were grown exponentially in SC-URA, washed 3 times in SC-URA–inositol, and resuspended at OD = 0.4 in the same medium in the presence of 1 µM Myriocin (Sigma-Aldrich) or Aureobasidin A (Clontech), or an equal volume of methanol. After 1.5 h of incubation at 30 °C, 5 µCi/mL of [3H]-inositol was added to the cell suspensions. After 2 h of labeling, IPs were extracted as described below.
Labeling of mammalian cell lines.
Cells were seeded into 6-well plates and grown for 5 d in inositol-free DMEM (MP Biomedicals) containing 10% dialyzed FBS (Sigma-Aldrich) and 5 µCi/mL of [3H]-inositol (PerkinElmer). The medium was partially renewed once during the labeling. To harvest, cells were washed once with PBS before adding 400 µL of 1 M perchloric acid directly onto the cells. After 10 min of incubation on ice, the supernatant was collected and centrifuged (16,000 × g, 5 min, 4 °C) to remove cell debris. The soluble extract was neutralized as described for yeast. For the [3H]-inositol turnover experiments, cells were seeded into 6-well plates to 70% confluence in inositol-free DMEM, and transfected after 24 h with 1 µg pcDNA3.1+ G protein α q Q209L (pGαqQL, UMR cDNA Resource Center). After a further 24 h, cells were treated with 5 µCi/mL of [3H]-inositol for 5 h before harvesting. For Pi starvation experiments, cells were seeded into 6-well plates and labeled for 4 d, as above. Cells were then washed in Pi-free DMEM (Thermo Fisher Scientific), before incubation in Pi-free DMEM containing 10% dialyzed FBS and 5 µCi/mL of [3H]-inositol for 24 h, then harvesting as described. Of note: Pi-free DMEM contains 40 μM inositol. Alternatively, to keep inositol concentration constant throughout the experiment, cells were seeded into 6-well plates and labeled in normal DMEM containing 10% dialyzed FBS for 4 d before incubation in Pi-free DMEM containing 10% dialyzed FBS. For [3H]-glucose labeling experiments, cells were grown in low glucose DMEM containing 10% dialyzed FBS in the presence of 20 µCi/mL of [3H]-glucose (PerkinElmer NET100C001MC) for 2 d before harvesting, as described for [3H]-inositol labeling experiments. Phytase treatment was performed by adding recombinant Aspergillus niger phytase PhyA (Natuphos, a gift from BASF, Ludwigshafen, Germany) to the neutralized extract and incubation for 1 h at room temperature before precipitating the added phytase with perchloric acid, followed by neutralization.
SAX Analysis of Labeled IPs.
The extraction and analysis of IPs was performed as described (70). The samples were separated onto a PartiSphere SAX (4.6 × 125 mm) column (Hichrom). The column was eluted with a gradient generated by mixing 1 mM EDTA and buffer B [1 mM EDTA/1.3 M (NH4)2HPO4, pH 3.8]; 0 to 5 min, 0% B; 5 to 10 min, 0 to 10% B; 10 to 60 min, 10 to 100% B; 60 to 80 min 100% B. To resolve [3H]-glucose labeled extracts a stepper gradient was employed 0 to 5 min, 0% B; 5 to 10 min, 0 to 30% B; 10 to 60 min, 30 to 100% B; 60 to 80 min 100% B. Fractions (1 mL) were collected and mixed with 4 mL of Ultima-FLO AP liquid scintillation mixture (PerkinElmer). The number of counts was estimated over 3 min. The different IPs species were identified using the following standards: [3H]-I(1,4,5)P3 (PerkinElmer), [3H]-I(1,3,4,5,6)P5, and [3H]-IP6. The latter two were purified from [3H]-inositol labeled ipk1∆kcs1∆ and kcs1∆ yeast, respectively, and desalted using a Sep-Pak Accell Plus QMA cartridge, as previously described (71).
Analysis of Inorganic Polyphosphate and Unlabeled IPs by PAGE.
For inorganic polyphosphate, 15 µg of RNA were loaded onto of a 25% polyacrylamide gel and run overnight at 4 °C at 5 mA and a maximum of 400V, as previously described (72). The gel was then stained with Toluidine blue and destained in 20% methanol. The migration of the samples was compared with those of standards of defined chain lengths. Images were obtained using an Epson desktop scanner.
Analysis of mammalian IPs was performed as previously described (73). Briefly, subconfluent cells in 15-cm dishes were trypsinized and extracted using 1 M perchloric acid (Sigma). Titanium dioxide beads (Titansphere TiO 5 μm; GL Sciences) were used to pull down IPs and other phosphate-rich molecules from the extracts. These extracts, normalized to protein concentration, were resolved using 35% PAGE gels and visualized by Toluidine blue. Standards used were synthetic inorganic polyphosphate of average length P13 (Sigma), or IP5 and IP6 (Sichem).
Data Availability.
The raw data are publicly available through the University College London Research Data Repository (https://doi.org/10.5522/04/10265318.v1).
Supplementary Material
Acknowledgments
The authors thank the members of the A.S. laboratory for their comments and for reading the manuscript; Haruyuki Atomi (Kyoto University) for sharing the plasmid for the expression of TK2285; and Dieter Feuerstein (BASF Animal Nutrition) for the gift of Natuphos. This work was supported by the Medical Research Council (MRC) core support to the MRC/UCL Laboratory for Molecular Cell Biology Unit, MC_UU12018/4 and MC_UU_00012/4 (to A.S.); the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Grant agreement PHEMDD 752903 (to Y.D.); and by the National Institute of General Medical Sciences of the National Institutes of Health under Award R15 GM106322-01A1 (to G.J.M.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The raw data are publicly available through the University College London Research Data Repository (https://doi.org/10.5522/04/10265318.v1).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911431116/-/DCSupplemental.
References
- 1.Irvine R. F., Schell M. J., Back in the water: The return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2, 327–338 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Wilson M. S., Livermore T. M., Saiardi A., Inositol pyrophosphates: Between signalling and metabolism. Biochem. J. 452, 369–379 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Szijgyarto Z., Garedew A., Azevedo C., Saiardi A., Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Wild R., et al. , Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Mallery D. L., et al. , IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 7, e35335 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Illies C., et al. , Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science 318, 1299–1302 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty A., et al. , Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 143, 897–910 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dovey C. M., et al. , MLKL requires the inositol phosphate code to execute necroptosis. Mol. Cell 70, 936–948.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Europe-Finner G. N., Gammon B., Wood C. A., Newell P. C., Inositol tris- and polyphosphate formation during chemotaxis of Dictyostelium. J. Cell Sci. 93, 585–592 (1989). [DOI] [PubMed] [Google Scholar]
- 10.Stephens L. R., Irvine R. F., Stepwise phosphorylation of myo-inositol leading to myo-inositol hexakisphosphate in Dictyostelium. Nature 346, 580–583 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Stephens L. R., Downes C. P., Product-precursor relationships amongst inositol polyphosphates. Incorporation of [32P]Pi into myo-inositol 1,3,4,6-tetrakisphosphate, myo-inositol 1,3,4,5-tetrakisphosphate, myo-inositol 3,4,5,6-tetrakisphosphate and myo-inositol 1,3,4,5,6-pentakisphosphate in intact avian erythrocytes. Biochem. J. 265, 435–452 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.York J. D., Odom A. R., Murphy R., Ives E. B., Wente S. R., A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285, 96–100 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Odom A. R., Stahlberg A., Wente S. R., York J. D., A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287, 2026–2029 (2000). [DOI] [PubMed] [Google Scholar]
- 14.Saiardi A., Erdjument-Bromage H., Snowman A. M., Tempst P., Snyder S. H., Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9, 1323–1326 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Ives E. B., Nichols J., Wente S. R., York J. D., Biochemical and functional characterization of inositol 1,3,4,5, 6-pentakisphosphate 2-kinases. J. Biol. Chem. 275, 36575–36583 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Mulugu S., et al. , A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Saiardi A., Caffrey J. J., Snyder S. H., Shears S. B., The inositol hexakisphosphate kinase family. Catalytic flexibility and function in yeast vacuole biogenesis. J. Biol. Chem. 275, 24686–24692 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Goffeau A., et al. , Life with 6000 genes. Science 274, 546–567 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Brearley C. A., Hanke D. E., Metabolic evidence for the order of addition of individual phosphate esters in the myo-inositol moiety of inositol hexakisphosphate in the duckweed Spirodela polyrhiza L. Biochem. J. 314, 227–233 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donahue T. F., Henry S. A., myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J. Biol. Chem. 256, 7077–7085 (1981). [PubMed] [Google Scholar]
- 21.Berridge M. J., Irvine R. F., Inositol phosphates and cell signalling. Nature 341, 197–205 (1989). [DOI] [PubMed] [Google Scholar]
- 22.Spang A., et al. , Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson M. P., Majerus P. W., Isolation of inositol 1,3,4-trisphosphate 5/6-kinase, cDNA cloning and expression of the recombinant enzyme. J. Biol. Chem. 271, 11904–11910 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Osawa S., Dhanasekaran N., Woon C. W., Johnson G. L., G alpha i-G alpha s chimeras define the function of alpha chain domains in control of G protein activation and beta gamma subunit complex interactions. Cell 63, 697–706 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Saiardi A., Mudge A. W., Lithium and fluoxetine regulate the rate of phosphoinositide synthesis in neurons: A new view of their mechanisms of action in bipolar disorder. Transl. Psychiatry 8, 175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto J. C., Kelly P., Chiou S. T., York J. D., Alterations in an inositol phosphate code through synergistic activation of a G protein and inositol phosphate kinases. Proc. Natl. Acad. Sci. U.S.A. 104, 15653–15658 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lloyd-Burton S. M., Yu J. C., Irvine R. F., Schell M. J., Regulation of inositol 1,4,5-trisphosphate 3-kinases by calcium and localization in cells. J. Biol. Chem. 282, 9526–9535 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Morris A. J., Downes C. P., Harden T. K., Michell R. H., Turkey erythrocytes possess a membrane-associated inositol 1,4,5-trisphosphate 3-kinase that is activated by Ca2+ in the presence of calmodulin. Biochem. J. 248, 489–493 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schell M. J., Inositol trisphosphate 3-kinases: Focus on immune and neuronal signaling. Cell. Mol. Life Sci. 67, 1755–1778 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J., Wang H., Hazebroek J., Ertl D. S., Harp T., The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42, 708–719 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Michell R. H., Inositol derivatives: Evolution and functions. Nat. Rev. Mol. Cell Biol. 9, 151–161 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Livermore T. M., Azevedo C., Kolozsvari B., Wilson M. S., Saiardi A., Phosphate, inositol and polyphosphates. Biochem. Soc. Trans. 44, 253–259 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Chamberlain P. P., et al. , Integration of inositol phosphate signaling pathways via human ITPK1. J. Biol. Chem. 282, 28117–28125 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang C., Wykle R. L., Daniel L. W., Cabot M. C., Identification of phosphatidylcholine-selective and phosphatidylinositol-selective phospholipases D in Madin-Darby canine kidney cells. J. Biol. Chem. 267, 16859–16865 (1992). [PubMed] [Google Scholar]
- 35.Balsinde J., Diez E., Fernandez B., Mollinedo F., Biochemical characterization of phospholipase D activity from human neutrophils. Eur. J. Biochem. 186, 717–724 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Sawai H., et al. , Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 275, 39793–39798 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Jun Y., Fratti R. A., Wickner W., Diacylglycerol and its formation by phospholipase C regulate Rab- and SNARE-dependent yeast vacuole fusion. J. Biol. Chem. 279, 53186–53195 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Saiardi A., Sciambi C., McCaffery J. M., Wendland B., Snyder S. H., Inositol pyrophosphates regulate endocytic trafficking. Proc. Natl. Acad. Sci. U.S.A. 99, 14206–14211 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagata R., Fujihashi M., Sato T., Atomi H., Miki K., Crystal structure and product analysis of an archaeal myo-inositol kinase reveal substrate recognition mode and 3-OH phosphorylation. Biochemistry 54, 3494–3503 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Miller G. J., Wilson M. P., Majerus P. W., Hurley J. H., Specificity determinants in inositol polyphosphate synthesis: Crystal structure of inositol 1,3,4-trisphosphate 5/6-kinase. Mol. Cell 18, 201–212 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Lonetti A., et al. , Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuo H. F., et al. , Arabidopsis inositol phosphate kinases IPK1 and ITPK1 constitute a metabolic pathway in maintaining phosphate homeostasis. Plant J., 10.1111/tpj.13974 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Gu C., et al. , The significance of the bifunctional kinase/phosphatase activities of diphosphoinositol pentakisphosphate kinases (PPIP5Ks) for coupling inositol pyrophosphate cell signaling to cellular phosphate homeostasis. J. Biol. Chem. 292, 4544–4555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawai M., Kinoshita S., Ozono K., Michigami T., Inorganic phosphate activates the AKT/mTORC1 pathway and shortens the life span of an α-Klotho-deficient model. J. Am. Soc. Nephrol. 27, 2810–2824 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bon N., et al. , Phosphate-dependent FGF23 secretion is modulated by PiT2/Slc20a2. Mol. Metab. 11, 197–204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho M. W., et al. , Regulation of Ins(3,4,5,6)P(4) signaling by a reversible kinase/phosphatase. Curr. Biol. 12, 477–482 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Saiardi A., Cockcroft S., Human ITPK1: A reversible inositol phosphate kinase/phosphatase that links receptor-dependent phospholipase C to Ca2+-activated chloride channels. Sci. Signal. 1, pe5 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Gokhale N. A., Zaremba A., Janoshazi A. K., Weaver J. D., Shears S. B., PPIP5K1 modulates ligand competition between diphosphoinositol polyphosphates and PtdIns(3,4,5)P3 for polyphosphoinositide-binding domains. Biochem. J. 453, 413–426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X., Shears S. B., Multitasking in signal transduction by a promiscuous human Ins(3,4,5,6)P(4) 1-kinase/Ins(1,3,4)P(3) 5/6-kinase. Biochem. J. 351, 551–555 (2000). [PMC free article] [PubMed] [Google Scholar]
- 50.Laha D., et al. , Arabidopsis ITPK1 and ITPK2 have an evolutionarily conserved phytic acid kinase activity. ACS Chem. Biol. 14, 2127–2133 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Gronnier J., Germain V., Gouguet P., Cacas J. L., Mongrand S., GIPC: Glycosyl inositol phospho ceramides, the major sphingolipids on earth. Plant Signal. Behav. 11, e1152438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raboy V., myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64, 1033–1043 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Raboy V., The ABCs of low-phytate crops. Nat. Biotechnol. 25, 874–875 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Reddy C. S., Kim S. C., Kaul T., Genetically modified phytase crops role in sustainable plant and animal nutrition and ecological development: A review. 3 Biotech 7, 195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi J., et al. , The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 131, 507–515 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin K. L., Smith T. K., The glycosylphosphatidylinositol (GPI) biosynthetic pathway of bloodstream-form Trypanosoma brucei is dependent on the de novo synthesis of inositol. Mol. Microbiol. 61, 89–105 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez-Salgado A., et al. , myo-Inositol uptake is essential for bulk inositol phospholipid but not glycosylphosphatidylinositol synthesis in Trypanosoma brucei. J. Biol. Chem. 287, 13313–13323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González-Salgado A., et al. , Trypanosoma brucei bloodstream forms depend upon uptake of myo-inositol for Golgi complex phosphatidylinositol synthesis and normal cell growth. Eukaryot. Cell 14, 616–624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordeiro C. D., Saiardi A., Docampo R., The inositol pyrophosphate synthesis pathway in Trypanosoma brucei is linked to polyphosphate synthesis in acidocalcisomes. Mol. Microbiol. 106, 319–333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tabuchi M., Audhya A., Parsons A. B., Boone C., Emr S. D., The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 26, 5861–5875 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ansari K., Martin S., Farkasovsky M., Ehbrecht I. M., Küntzel H., Phospholipase C binds to the receptor-like GPR1 protein and controls pseudohyphal differentiation in Saccharomyces cerevisiae. J. Biol. Chem. 274, 30052–30058 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Norman K. L., et al. , Inositol polyphosphates regulate and predict yeast pseudohyphal growth phenotypes. PLoS Genet. 14, e1007493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frederick J. P., et al. , An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. U.S.A. 102, 8454–8459 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Resnick A. C., et al. , Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. U.S.A. 102, 12783–12788 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kolozsvari B., Firth S., Saiardi A., Raman spectroscopy detection of phytic acid in plant seeds reveals the absence of inorganic polyphosphate. Mol. Plant 8, 826–828 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Voglmaier S. M., et al. , Purified inositol hexakisphosphate kinase is an ATP synthase: Diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc. Natl. Acad. Sci. U.S.A. 93, 4305–4310 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye C., Bandara W. M. M. S., Greenberg M. L., Regulation of inositol metabolism is fine-tuned by inositol pyrophosphates in Saccharomyces cerevisiae. J. Biol. Chem. 288, 24898–24908 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu W., Ye C., Greenberg M. L., Inositol hexakisphosphate kinase 1 (IP6K1) regulates inositol synthesis in mammalian cells. J. Biol. Chem. 291, 10437–10444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fischbach A., Adelt S., Müller A., Vogel G., Disruption of inositol biosynthesis through targeted mutagenesis in Dictyostelium discoideum: Generation and characterization of inositol-auxotrophic mutants. Biochem. J. 397, 509–518 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Azevedo C., Saiardi A., Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 1, 2416–2422 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Azevedo C., Burton A., Bennett M., Onnebo S. M., Saiardi A., Synthesis of InsP7 by the inositol hexakisphosphate kinase 1 (IP6K1). Methods Mol. Biol. 645, 73–85 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Losito O., Szijgyarto Z., Resnick A. C., Saiardi A., Inositol pyrophosphates and their unique metabolic complexity: Analysis by gel electrophoresis. PLoS One 4, e5580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson M. S., Bulley S. J., Pisani F., Irvine R. F., Saiardi A., A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open Biol. 5, 150014 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data are publicly available through the University College London Research Data Repository (https://doi.org/10.5522/04/10265318.v1).