Significance
Many bacteria capable of causing disease utilize a nanomachine to inject bacterial proteins into mammalian cells. Those proteins have the capacity to modulate cellular functions for the pathogen’s benefit. In this study, we visualized components of this injection machine at very high resolution, which has allowed us to obtain very important structural and functional information critical for the understanding of its function.
Keywords: molecular machines, protein secretion, bacterial pathogenesis, cryoelectron microscopy, cryoelectron tomography
Abstract
Type III protein secretion systems are essential virulence factors for many important pathogenic bacteria. The entire protein secretion machine is composed of several substructures that organize into a holostructure or injectisome. The core component of the injectisome is the needle complex, which houses the export apparatus that serves as a gate for the passage of the secreted proteins through the bacterial inner membrane. Here, we describe a high-resolution structure of the export apparatus of the Salmonella type III secretion system in association with the needle complex and the underlying bacterial membrane, both in isolation and in situ. We show the precise location of the core export apparatus components within the injectisome and bacterial envelope and demonstrate that their deployment results in major membrane remodeling and thinning, which may be central for the protein translocation process. We also show that InvA, a critical export apparatus component, forms a multiring cytoplasmic conduit that provides a pathway for the type III secretion substrates to reach the entrance of the export gate. Combined with structure-guided mutagenesis, our studies provide major insight into potential mechanisms of protein translocation and injectisome assembly.
Type III protein secretion systems (T3SSs) have the remarkable capacity to deliver multiple bacterially encoded effector proteins into target eukaryotic cells (1–3). They are central for the virulence of many important bacterial pathogens and therefore have been long considered prime targets for the development of novel antiinfective strategies (4–6). T3SSs are evolutionary related to flagella and, consequently, some of its components share amino acid sequence or structural similarities with components of the flagellar apparatus (7).
The central element of the protein secretion machine is the injectisome, a multiprotein structure composed of the needle complex and the cytoplasmic sorting platform (Fig. 1A) (8, 9). The needle complex is made up of a base structure, embedded in the bacterial envelope, and a filamentous extension or needle that projects several nanometers from the bacterial surface. The base is composed of a double-ring structure, inner ring 1 (IR1) and inner ring 2 (IR2), which are made up of PrgH and PrgK in the Salmonella pathogenicity island 1 (SPI-1) T3SS, and the outer rings (OR1 and OR2) and neck, which are made up of InvG (10, 11). Buried within the needle complex lies the export apparatus, a complex of several membrane proteins that facilitate the passage of type III secreted substrates through the bacterial inner membrane (12). The entire needle complex is traversed by a narrow (∼3 nm) channel, which serves as a conduit for the proteins transiting the type III secretion pathway. On the cytoplasmic side of the needle complex lies the sorting platform, a multiprotein complex that engages, sorts, and initiates substrates into the secretion pathway (9).
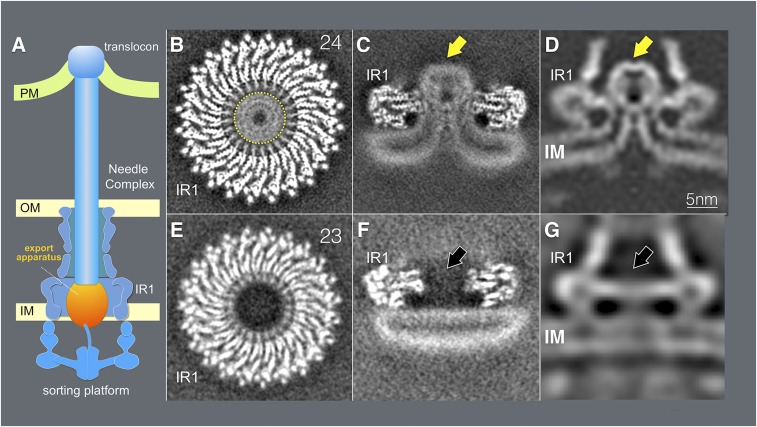
Fig. 1.
Cryo-EM and cryo-ET analyses of the core components the T3SS export apparatus. (A) A schematic representation of the T3SS injectisome. (B and C) Horizontal (B) and vertical (C) sections of a 3D reconstruction of a cryo-EM structure of the inner rings of the needle complex containing the export apparatus (denoted with a yellow arrow). (D) A vertical section from an in situ cryo-ET structure of the injectisome from a S. Typhimurium ∆invA mutant strain. The export apparatus is denoted with a yellow arrow. (E and F) Horizontal (E) and vertical (F) sections of a 3D reconstruction of a cryo-EM structure of the inner rings of the needle complex obtained from a S. Typhimurium mutant strain (∆spaP ∆spaQ ∆spaR ∆spaS) lacking the core components of the export apparatus. (G) A vertical section from in situ cryo-ET structure of the injectisome from a ∆spaP ∆spaQ ∆spaR ∆spaS S. Typhimurium strain. The position where the export apparatus would be located is denoted with a black arrow. IM, inner membrane; IR1, inner ring 1; OM, bacterial outer membrane; PM, target host cell plasma membrane.
The complexity of the injectisome dictates that its assembly must occur in a highly coordinated, step-wise manner that is initiated by the formation of a complex of the core membrane protein components of the export apparatus, which are thought to nucleate the assembly of the inner rings (12, 13). Following the recruitment of the independently assembled outer rings, the assembly of the injectisome is completed by the recruitment of the cytoplasmic sorting platform and the polymerization of the needle filament (14–16).
Previous studies have provided structural information on the different substructures that make up the injectisome (10, 11, 17–25). However, there are still significant knowledge gaps, and importantly, it remains unclear how some of the structural information generated with isolated subcomplexes may ultimately relate to the injectisome structure in situ. This is particularly the case for the export apparatus, which is composed of 5 predicted membrane proteins (SpaP, SpaQ, SpaR, SpaS, and InvA) in the Salmonella (S.) Typhimurium SPI-1 T3SS (also known as SctR, SctS, SctT, SctU, and SctV in the proposed unifying nomenclature) that are highly conserved in both the virulence-associated type III secretion and flagellar assembly systems (12, 26). A recent single-particle cryoelectron microscopy (cryo-EM) structure of a FliP/FliQ/FliR complex of the flagellar export apparatus (homologs of SpaP/SpaQ/SpaR) surprisingly showed that, in isolation, these proteins do not adopt typical integral membrane protein topologies but rather, they organize in a helical assembly that was proposed to be located largely within the periplasmic portion of the secretion machine (27). Therefore, it remains unclear how this complex facilitates the passage of type III secreted proteins through the inner membrane. Even less structural information is available about InvA (28), which is thought to play a central role in energizing the secretion process (29). Although it has been proposed that the cytoplasmic domain of this protein family forms a nonameric ring interfacing with components of the sorting platform and export apparatus (25, 30), the localization of its critical transmembrane domain region within the secretion machine has remained elusive.
In this study, we have utilized single-particle cryo-EM and cryoelectron tomography (cryo-ET) to obtain a high resolution view of the export apparatus of the S. Typhimurium T3SS encoded within its pathogenicity island 1 (SPI-1), both in situ and in the context of needle complexes obtained with an isolation protocol optimized to maintain its native structural organization. These approaches have allowed us to determine the precise topological organization of the export apparatus relative to other components of the type III secretion injectisome, to characterize its interface with the base substructure, and to reveal major membrane remodeling concomitant with the assembly of the export apparatus. Furthermore, we have defined the structural organization of InvA, which outlines a multiring conduit for the type III secretion substrates to reach the entrance of the export apparatus gate. These studies provide major insight into the structure and assembly of the type III secretion injectisomes and suggest a pathway for the type III secreted substrates to cross the bacterial membrane.
Results
Cryo-EM and Cryo-ET Analysis of the Core Components of the T3SS Export Apparatus and Associated Structures Reveals Major Remodeling of the Bacterial Inner Membrane.
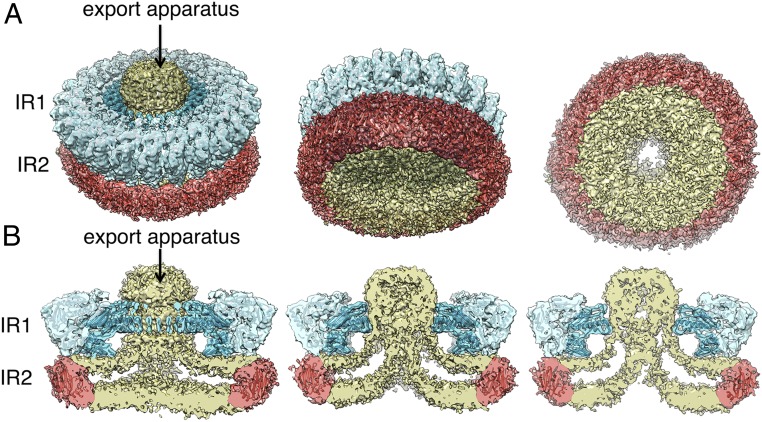
Previous structural studies of isolated T3SS injectisomes have relied on isolation procedures that result in the loss of some of its components or the disruption of its interaction with surrounding structures (10, 11, 24). To address this limitation, we developed a needle complex isolation protocol under mild conditions that results in minimal losses of export apparatus components. Extraction of the needle complex from the bacterial envelope under these mild conditions was facilitated by the genetic removal of the outer rings and neck components, which anchor this structure to the bacterial outer membrane. Liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis of the isolated structures detected the presence of the lower ring components PrgH and PrgK, the inner rod and filament proteins PrgJ and PrgI, as well as the export apparatus components SpaP, SpaQ, and SpaS (SI Appendix, Table S1). The apparent absence of the core component of the export apparatus SpaR was likely due to its lower stoichiometry in the complex (27, 31) and difficulties in its detection by LC-MS/MS under the conditions used in our analysis. Although InvA was also present in the sample, only few peptides were detected despite its large size and high stoichiometry. This indicates that, most likely, InvA was absent from most of the particles, which is consistent with previous observations suggesting its loose association to the core components of the export apparatus (12). Cryo-EM grids with the isolated structures (SI Appendix, Fig. S1) were examined by single-particle analysis as described in Materials and Methods. We found that the most abundant population of substructures consisted of inner rings with the associated export apparatus but lacking the needle filaments (Fig. 1 and SI Appendix, Fig. S1). However, inner rings with associated needle structures were also observed in a proportion (∼5%) of the total number of particles compiled from the electron micrographs (SI Appendix, Fig. S1). Reference-free two-dimensional (2D) class average images of the inner rings with the associated export apparatus revealed elements of secondary structure in the IR1 (SI Appendix, Fig. S1). However, the IR2 was poorly defined in all 2D class averages (Fig. 1 and SI Appendix, Fig. S2), which is expected for this substructure that undergoes a significant repositioning upon its isolation leading to varying locations relative to the rest of the needle complex structure (25), thus hampering its visualization at high resolution. After subtraction of the less-resolved IR2 signal and imposing 24-fold symmetry, the resolution of the IR1 could be further improved (SI Appendix, Fig. S1). Residues 171–364 of PrgH and 20–203 of PrgK could therefore be docked with an excellent fit into the densities of this ring (Fig. 2).
Fig. 2.
Three-dimensional reconstructions of the needle complex IR1 and IR2 containing the core components of the export apparatus and associated membranes. (A and B) Three-dimensional density maps of the inner membrane rings containing the export apparatus with the crystal structures of PrgH (residues 171–364, light blue) and PrgK (residues 20–203, dark blue) docked into the density corresponding to the IR1. Although the resolution is not sufficient for confident assignment, a modeled structure of residues 11–120 of PrgH (red) docked into the density corresponding to the IR2 is shown to mark the expected position of this structural element. Tilted (A) and cut-away (B) views are shown. Densities corresponding to the export apparatus and associated membranes are shown in yellow. IR1, inner ring 1; IR2, inner ring 2.
Further analysis of the “side views” in the 2D class averages of structures lacking the needle filament revealed a cloud of density spanning the space between IR1 and IR2 and extending above IR1 (Figs. 1C and 2 A and B and SI Appendix, Figs. S1–S3). We assigned this cloud of density to the core protein components of the export apparatus since this density was absent in structures isolated from an isogenic ∆spaP ∆spaQ ∆spaR ∆spaS S. Typhimurium mutant strain (Fig. 1 E and F and SI Appendix, Fig. S4). The 2D class averages of tilted views of the rings and the 3-dimensional (3D) reconstruction of structures isolated from this S. Typhimurium mutant strain showed an empty space at the center of the double ring structure (Fig. 1 E and F and SI Appendix, Fig. S4).
Comparison of this inner ring substructure with previously reported needle complex structures showed a notable difference, characterized by the presence of a large double-layered density surrounded by the IR2 that occupies the entire cytoplasmic side of the double ring structure (Figs. 1 and 2 and SI Appendix, Fig. S5). This density, which cannot be accounted for by the cytoplasmic domain of PrgH (Fig. 2), forms a funnel-like structure with its center leading to the entrance of the export apparatus (Figs. 1 and 2 and SI Appendix, Fig. S5). The overall thickness (∼4 nm) and double layer appearance of this density indicates that this structure corresponds to the bacterial inner membrane, which under our mild isolation conditions has been retained in the isolated structures. Consistent with this hypothesis, the 3D reconstruction of structures isolated from the strain lacking the export apparatus showed that in this mutant, the double layer density no longer organizes in a funnel shape but, rather, appears flat, completely sealing the space surrounded by the IR2 (Fig. 1F and SI Appendix, Fig. S4). These results indicate that the deployment of the core components of the export apparatus results in a rather significant remodeling of the surrounding bacterial membrane.
To ascertain whether the membrane remodeling observed in isolated structures is also present in in situ structures embedded within the bacterial envelope, we examined by cryo-ET, injectisomes in bacterial minicells obtained from an S. Typhimurium strain lacking invA (which was absent in our samples thus better resembling the isolated complex and thus facilitating direct comparison). Subtomogram average from 5,929 injectisome reconstructions shows a rather pronounced rearrangement of the inner membrane in the vicinity of the density corresponding to the export apparatus (Fig. 1D). Like in the isolated complex, in the in situ structure the membrane is seen adopting a funnel-like shape with its narrow end in close proximity to the density that corresponds to the core components of the export apparatus. The remodeled membrane was not observed in in situ structures obtained from a mutant lacking all of the core components of the export apparatus (subtomogram average from 2,368 injectisomes reconstructions) (Fig. 1G), indicating that membrane remodeling is a consequence of the deployment of the core components of the export apparatus. The isolated cryo-EM injectisome structures and associated membranes could be overlaid onto the cryo-ET structures with an excellent fit (SI Appendix, Fig. S6), indicating that the isolation procedure did not disrupt the topology of the core components of the export apparatus relative to the surrounding membranes and injectisome substructures. Importantly, the membrane remodeling was also observed in cryo-ET structures from WT injectisomes (SI Appendix, Fig. S7), although the presence of InvA resulted in differences in the densities surrounding the core components of the export apparatus (see below). Furthermore, the core-components of the export apparatus in injectisome structures obtained from WT and ∆invA mutant strains completely overlap, indicating that the observed membrane remodeling and the position of the core components of the export apparatus are not significantly influenced by the presence or absence of InvA (SI Appendix, Fig. S7).
Taken together, these results indicate that the mild isolation protocol resulted in needle complex substructures that have retained the export apparatus and associated membranes, thus maintaining the in situ topology. More importantly, these observations indicate that the bacterial inner membrane undergoes a significant remodeling upon deployment of the core components of the export apparatus.
Needle Complex Inner Rings Assembled in the Absence of the Export Apparatus Exhibit a Different Symmetry.
The 2D class averages and the 3D reconstruction of structures obtained from the S. Typhimurium mutant strain lacking the export apparatus showed that the vast majority (∼80%) of the inner membrane rings exhibited 23-fold symmetry (Fig. 1E and SI Appendix, Fig. S4). This observation is significant since it is in marked contrast to the inner membrane rings assembled in the presence of the export apparatus, which invariably exhibited 24-fold symmetry (Fig. 1B and SI Appendix, Figs. S1 and S2). We have previously shown that the efficiency of assembly of the needle complex is markedly reduced in S. Typhimurium mutants lacking the core components of the export apparatus, which in combination with additional biochemical data led us to propose that these components of the export apparatus templates the assembly of the inner rings (12). The observation that inner rings assembled in the absence of the export apparatus exhibit symmetry never observed in the rings assembled in its presence is consistent with this model and strongly supports the notion that during assembly of a functional injectisome the export apparatus must be deployed prior to the assembly of the inner rings.
Overall Architecture of the Core Components of the Export Apparatus in Association with the Inner Rings and the Bacterial Membrane.
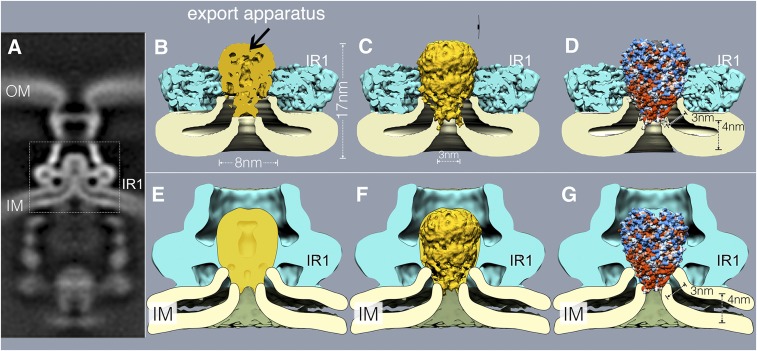
Overall, the density associated with the core export apparatus in the context of needle complex rings is ∼11 nm long from top to bottom and ∼8 nm at its widest (SI Appendix, Fig. S8). The cryo-EM structure of an isolated subcomplex of the flagellar-associated export apparatus made of FliP, FliQ, and FliR (homologs of SpaP, SpaQ, and SpaR, respectively) (27) revealed that none of its subunits adopt a typical membrane protein topology, but rather they participate in the formation of a helical structure that was postulated to be largely located in the periplasm (27). This observation raised the question how this complex may insert in the membrane, an essential requirement to mediate the passage of secreted proteins through the membrane barrier. Analysis of the surface hydrophobicity of the FliP/FliQ/FliR complex revealed only a small hydrophobic strip at the base of the structure, which was hypothesized to be buried in the membrane (27) (SI Appendix, Fig. S9). The length of this segment (∼30 Å) would predict that such hydrophobic region would not span a typical membrane, thus raising intriguing questions about the mechanism by which this complex may mediate protein translocation across the membrane. Although attempts were made to place the isolated structure onto a low-resolution cryo-ET map of the export apparatus (27), it has remained unclear what the precise location of the core component of the export apparatus relative to the bacterial inner membrane is. We therefore docked a cryo-EM structure of the isolated FliP/FliQ/FliR complex of the flagellar-associated export apparatus into our structures, which allowed us to precisely place this export apparatus subcomplex in its appropriate topological context. Overall, there is good agreement between the FliP/FliQ/FliR isolated complex structure and the cryo-EM and cryo-ET structures of the S. Typhimurium export apparatus embedded within the needle complex, particularly in the densities that correspond to the periplasmic domain of the export apparatus (Fig. 3 B–G and SI Appendix, Fig. S9). Close examination of the overlay of the structures reveals that the hydrophobic vertex of the FliP/FliQ/FliR complex is predicted to be located at the expected plane of the membrane (Fig. 3 B–G and SI Appendix, Fig. S9). Importantly, both our cryo-EM and cryo-ET structures suggest that the substantial remodeling of the inner membrane associated with the deployment of the export apparatus results in a significant membrane thinning in the immediate vicinity of the export apparatus (Fig. 3 B–G and SI Appendix, Fig. S9). The thinned membrane allows the hydrophobic vertex of the export apparatus core complex to span the membrane in its entire width (Fig. 3 B–G, SI Appendix, Fig. S9, and Movie S1). This architecture places the cavity observed at the vertex of the cryo-EM structure of the FliP/FliQ/FliR complex in direct communication with the cytoplasm of the bacterial cell (Fig. 3 B–G, SI Appendix, Fig. S9, and Movie S1), an arrangement that may facilitate the passage of the substrates through the membrane. These observations have important implication for the understanding of the protein translocation mechanism across the bacterial inner membrane during type III secretion.
Fig. 3.
Assembly of the export apparatus results in pronounced membrane remodeling. (A) A vertical section from in situ cryo-ET structure of the injectisome from a ∆invA S. Typhimurium strain (the dotted box denotes area of detail shown in B–G). (B–G) Vertical sections of cryo-EM (B–D) and cryo-ET (E–G) structures of the inner rings of the needle complex T3SS complex overlaid with a section (B and E), surface rendering (C and F), or hydrophobicity surface representation (D and G) of the FliP/FliQ/FliR (homologs of SpaP/SpaQ/SpaR from S. Typhimurium) isolated complex (PDB ID code 6F2E). Red and blue colors (D and G) on the surface representation of the FliP/FliQ/FliR isolated complex represent hydrophobic and charge residues, respectively. IM, inner membrane; IR1, inner ring 1; OM, bacterial outer membrane.
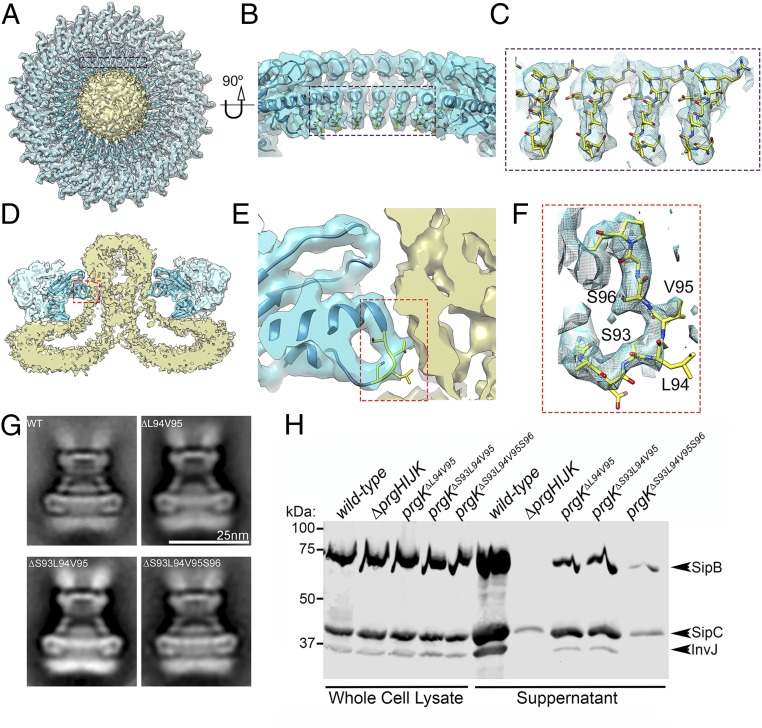
Interactions of a Domain of the Periplasmic PrgK Ring with the Core Components of the Export Apparatus Are Required for Type III Secretion.
Close examination of the interface between the needle complex IR1 and the export apparatus revealed a segment of PrgK that makes the most intimate contact with the periplasmic region of the export apparatus (Fig. 4 A–F). This region consists of a 7 amino acid loop (residues 92–98), which is anchored by 2 helixes (Fig. 4F). To examine the functional significance of this intimate association between IR1 and the export apparatus, we introduced discrete mutations in this loop region of PrgK and examined the structure and function of the needle complex. We found that structures isolated from S. Typhimurium strains expressing PrgK∆94–95, PrgK∆93–95, or PrgK∆93–96 were indistinguishable from WT except for the reduced number of needle filaments associated to these structures, which was particularly apparent in the structures obtained from the strain expressing the PrgK∆93–96 mutant (Fig. 4G and SI Appendix, Figs. S10 and S11). The export apparatus was readily detectable in the needle complexes isolated from all mutants, which at this level of resolution appeared indistinguishable from WT (Fig. 4G and SI Appendix, Fig. S10). Despite the apparently normal appearance of the injectisomes and the equivalent yield during their isolation, all of the mutants showed a marked decrease in type III secretion function as measured by the ability of these S. Typhimurium prgK mutant strains to secret various substrates of the SPI-1 T3SS (Fig. 4H). The mutant phenotype was particularly apparent for the prgK∆S93-S96 mutant strain, which showed almost complete loss of function (Fig. 4H). These observations indicate that this intimate interaction between PrgK and the export apparatus is central for type III secretion but not for the assembly of the needle complex and the deployment of the export apparatus. These observations suggest an active role for the PrgK ring substructure in T3SS function beyond the scaffolding of the export apparatus in the bacterial membrane.
Fig. 4.
Interaction of the periplasmic PrgK ring with the export apparatus is required for type III secretion function but not for injectisome assembly. (A–F) Top view (A) and cross section (D) of the 3D reconstruction of the IR1 and IR2 rings of the needle complex containing the core components of the export apparatus. The interface between PrgK and the export apparatus is highlighted (in blue and red rectangles). The specific densities corresponding to PrgK residues 93–96 targeted for mutagenesis and functional analysis are shown in more detailed in the zoom-in views depicted in B–F. (G) Representative class averages of needle complexes isolated from S. Typhimurium expressing WT PrgK or the indicated deletion mutants. (H) Type III protein secretion analysis of S. Typhimurium strains expressing the PrgK mutants with the indicated deletions in the region that interacts with the core components of the export apparatus. Whole-cell lysates and cultured supernatants of the indicated strains were analyzed by Western immunoblot for the presence of SipB, SipC, and InvJ, which are substrates of the S. Typhimurium type III secretion system. This experiment was repeated 3 times with equivalent results.
In Situ Structure of InvA, a Highly Conserved Component of the Export Apparatus.
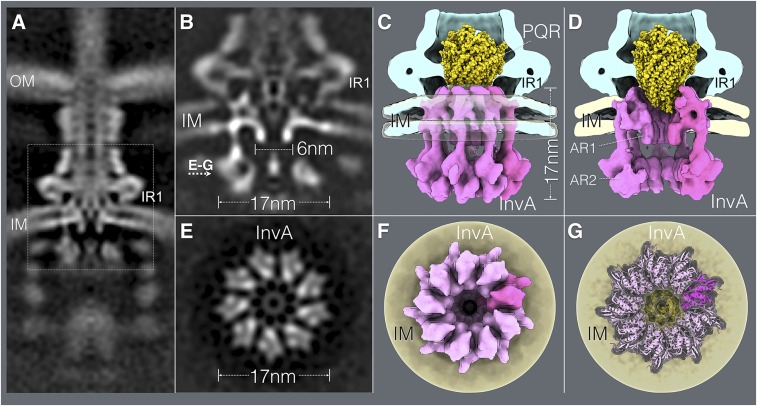
InvA (FlhA in the flagellar system and SctV in the proposed unifying nomenclature) is 1 of the most highly conserved components of the export apparatus (28, 32). However, its role in the protein secretion process is poorly understood. Secondary structure predictions and molecular modeling indicate that the amino terminal half of this protein family contains several transmembrane segments, and its large carboxyl-terminal domain is located entirely within the bacterial cytoplasm (32, 33). It has been proposed that this protein family, alone or in association with other components of the export apparatus, may function as a proton or Na+ channel that contributes to energizing the secretion process (29, 34, 35). However, how this protein may contribute to this activity is still uncertain, and it is unclear whether it forms a physical complex with other components of the export apparatus. Of critical importance for the understanding of the function of this crucial component of the T3SS is to identify the location of the predicted transmembrane region within the context of the injectisome structure. Efforts to isolate InvA (or any of its homologs) in association with the injectisome have failed. The crystal structure of a region of the carboxy-terminal cytoplasmic domain of a member of this protein family suggests that it can form a nonameric ring (30), which previous cryo-ET imaging has located in the bacterial cytoplasm, in close apposition to components of the sorting platform (25). However, there is no structural information on the critical amino terminal half of this protein, which is responsible for positioning it within the structure of the injectisomes. We therefore applied a high-throughput cryo-ET pipeline to image minicells obtained from a S. Typhimurium strain expressing a WT SPI-1 T3SS and applied subtomogram averaging to image InvA. We found that InvA forms a sea horse-like structure, with the head embedded in the membrane and the body and tail located entirely within the cytoplasm (Fig. 5 A–D and Movie S2). Consistent with the crystal structure (30), the cytoplasmic domain of InvA forms a nonameric ring 17 nm in diameter and 5.5 nm in height (Fig. 5). This ring is enclosed within the sorting platform immediately above a density we have previously assigned to the ATPase InvC (25). Importantly, immediately above this ring and closer to the membrane but still within the bacterial cytoplasm, we detected an additional ring 6 nm in diameter and 8 nm in height (Fig. 5 and Movie S2). This smaller ring aligns very well with the larger ring and marks the entrance to a broad conduit that narrows as it gets closer to the export apparatus gate. This conduit is bounded by densities that we postulate correspond to the transmembrane domains of InvA, whose arrangement tracks the contour of the drastically remodeled bacterial membrane. Consistent with this notion, subtomogram averages of injectisome from an S. Typhimurium strain expressing an InvA mutant lacking its last 329 amino acids, which removes its entire cytoplasmic domain, lack the larger cytoplasmic ring but retained the smaller ring and membrane proximal structures (SI Appendix, Fig. S12). None of these structures were present in a strain lacking InvA (Fig. 1D) although they were readily detected in a strain lacking the export apparatus component SpaS (SI Appendix, Figs. S12 and S13). All combined, the 2 rings and membrane-associated densities build a contiguous substructure that we hypothesize initiates the substrates of the type III secretion machine into the secretion pathway (Movie S3). Of note, sagittal sections through the injectisome cutting through the PrgH cytoplasmic IR2 domain showed an intimate association between this structure of the needle complex and a surface region (dubbed SD2 in previous studies; ref. 30) within the larger InvA cytoplasmic ring (Fig. 6). Previous studies in Shigella flexneri have shown that mutations in SD2 domain of MxiA, a homolog of InvA, impaired type III secretion function but did not affect its oligomerization (30). These observations suggest a potential mechanism by which conformational changes in the needle complex could modulate the activity of InvA, which plays a central role in the initiation of substrates into the secretion pathway.
Fig. 5.
In situ structure of InvA, an essential component of the type III secretion export apparatus. (A) A vertical section from the in situ cryo-ET structure of the injectisome from WT S. Typhimurium (the dotted box denotes area of detail shown in B–G). (B–G) Vertical (B–D) and horizontal (through the plane indicated in B) (E–G) sections highlighting the structure of InvA. Side (C) and cut-away (D) views of the InvA structure (in pink) are shown and the location of the small (AR1) and large (AR2) rings are denoted. The overlaid of the structure of the FliP/FliQ/FliR isolated complex is shown in yellow. (F and G) Bottom views of the InvA nonameric ring, with the overlaid atomic structure of the cytoplasmic domain of an InvA homolog (PDB ID code 4A5P) (G). No symmetry was applied for A and B. However, to visualize the InvA ring, symmetry was applied in C–G. IM, inner membrane; IR1, inner ring 1; OM, bacterial outer membrane.
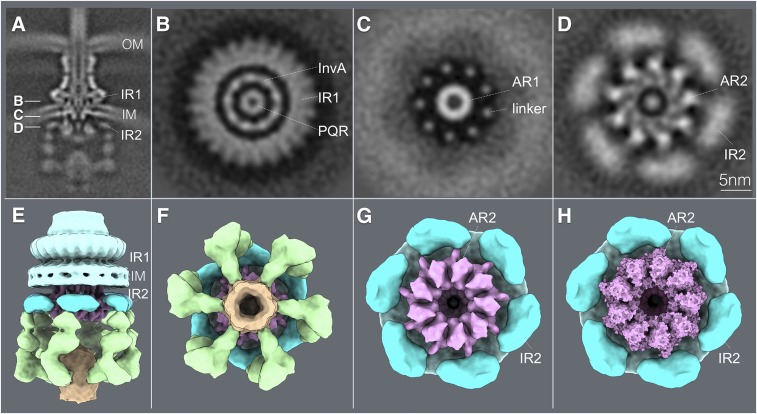
Fig. 6.
Close association of a cytoplasmic ring of InvA with the sorting platform and cytoplasmic domain of the needle complex component PrgH. (A) A vertical section from the in situ cryo-ET structure of the injectisome from WT S. Typhimurium indicating the section planes of the structures shown in B–D. (B–D) Horizontal sections through the planes indicated in B depicting the InvA nonameric ring within the inner ring 1 (IR1) of the needle complex structure (B), the linker region that joins the membrane and cytoplasmic domains of InvA (C), and its tight association with the cytoplasmic domain of the inner ring 2 (IR2) of the needle complex (D). The position of the core component of the export apparatus (labeled PQR) is also shown. (E and F) Side (E) and bottom (F) views of the surface rendering of the cytoplasmic elements of the T3SS injectisome. (G) Bottom view of the T3SS injectisome after removal of the sorting platform elements to highlight the close association between the larger cytoplasmic ring of InvA (AR2) with the cytoplasmic ring of PrgH (IR2). (H) Bottom view of the InvA nonameric ring (AR2) with the overlaid atomic structure of the cytoplasmic domain of an InvA holog (PDB-4A5P). No symmetry was applied in A–D but to visualize the InvA ring, symmetry was applied in E–H. AR1, InvA-associated ring 1; AR2, InvA-associated ring 2. IM, bacterial inner membrane; OM, bacterial outer membrane.
Discussion
The export apparatus is a central core component of T3SS that is thought to mediate the passage of type III secreted substrates through the bacterial membrane, a critical step during the protein translocation process. All of the core components of the export apparatus (SpaP, SpaQ, SpaR, SpaS, and InvA in the S. Typhimurium SPI-1 T3SS) are predicted to be integral membrane proteins (33). However, a recent cryo-EM structure of an isolated subcomplex of the flagellar-associated export apparatus made up of FliP, FliQ, and FliR (homologs of SpaP, SpaQ, and SpaR) revealed that none of these proteins adopt a typical membrane protein topology, and, instead, they assemble into a helical structure that through fitting into a low resolution in situ structure of the injectisomes was proposed to be largely located within the bacterial periplasm (27). The absence of typical transmembrane regions in this subcomplex is intriguing in light of their proposed role in mediating the passage of type III secreted proteins through the bacterial membrane and raised the question whether other components of the export apparatus could fulfill this role. To address these issues, we have utilized single-particle cryo-EM and cryo-ET to determine the precise architecture of the export apparatus in situ and, in particular, to determine the topological organization of its components relative to the bacterial membrane.
To this aim, we developed an isolation protocol that allowed us to obtain needle complex substructures containing the core components of the export apparatus in association with the bacterial membrane. Analysis of this structure by single-particle cryo-EM allowed us to determine the precise topological relationship between the core components of the export apparatus, the needle complex, and the associated bacterial membrane. Notably, we found that the bacterial membrane undergoes a profound remodeling in the immediate vicinity of the export apparatus. This remodeling, which is also observed in the cryo-ET in situ structure, is characterized by a significant outward “pinching” of the membrane, which organizes in a “funnel-like” fashion with its narrower end in immediate contact with the core components of the export apparatus. Notably, this reorganization was not observed in injectisome subcomplexes isolated from a ∆spaP∆spaQ∆spaR∆spaS S. Typhimurium mutant strain, indicating that membrane remodeling is strictly dependent on the deployment of the core components of export apparatus. The membrane remodeling was also accompanied by a significant thinning of the bacterial membrane, particularly in the immediate vicinity of its interface with the core components of the export apparatus. The thinning of the membrane may allow the relatively short (∼30 Å) hydrophobic vertex of the core complex of the export apparatus to span the entire membrane, thus facilitating the movement of the type III secreted proteins through the bacterial inner membrane. Furthermore, this topological organization would allow a central cavity present within this vertex to be in direct communication with the bacterial cytoplasm, thus facilitating substrate translocation across the membrane. These observations have important implications for the understanding of the mechanisms involved in the protein translocation function of the export apparatus as it implies that this complex would have all of the necessary structural elements to form the channel that mediate the passage of the secreted substrates through the bacterial inner membrane. It is noteworthy that the thinning of the membrane and a similar topological organization have also been observed in the context of other protein conducting channels such as the Hrd1, twin arginine translocator (Tat), and YidC protein translocation machines (36–38), suggesting that this type of membrane remodeling may be a more general feature associated with some protein translocation machines.
We have identified in our structure a region of PrgK, an inner ring component of the needle complex, which makes intimate contact with the periplasmic region of the core complex of the export apparatus. Introduction of discrete mutations in this domain resulted in loss of function, although these mutations did not detectably affect the assembly of the PrgK rings or the deployment of the export apparatus. It is therefore possible that PrgK may trigger conformational changes in the export apparatus that may be necessary to “prime” its secretion function. These observations point to an unexpected active role of the PrgK rings in the type III secretion process, presumably involving more than just serving as a scaffold for the export apparatus.
We have previously shown that in the absence of the export apparatus the efficiency of assembly of the needle complex is significantly diminished, which in combination with other observations led us to propose an injectisome assembly pathway that is initiated by the deployment of the core components of the export apparatus (12). However, alternative models of assembly have also been proposed (16). We have shown here that needle complexes assembled in the absence of the export apparatus exhibit a fold symmetry that is never observed in fully assembled needle complexes. Therefore, this observation demonstrates that the assembly of the core components of the export apparatus must be the first step in the injectisome assembly pathway.
Despite their central role in protein secretion, the mechanisms by which the InvA protein family contributes to type III secretion has remained elusive. Although it has been proposed to exert its function as a proton channel that energizes the secretion process (29), direct demonstration of its proton transport activity is lacking. Furthermore, it is unclear how a putative proton channel activity contributes to the secretion process. A major limitation for the understanding of InvA function has been the lack of information on its precise localization within the secretion apparatus. The crystal structure of the carboxy-terminal domain of InvA forms a nonameric ring, and this structural feature has been proposed to correlate with a toroidal density observed within the cytoplasmic sorting platform of the injectisome (25, 30). However, there has been no information on the localization of the amino-terminal half of InvA, which contains multiple predicted transmembrane domains. Here, using a high-throughput cryo-ET pipeline, we have precisely determined the topological organization of the entire InvA molecule. Our in situ structure indicates that, as predicted by the crystal structure (30), the carboxy-terminal domain of InvA (amino acids 357–685) forms a nonameric ring (17 nm in diameter) located in the apical side of the sorting platform cavity. In addition, our in situ structure revealed that InvA forms a second cytoplasmic ring 6 nm in diameter that is located in close apposition to the bacterial membrane and in alignment with the larger ring. In keeping with a previous proposal (35), we hypothesize that this ring is formed by a large cytoplasmic loop (amino acids 131–196) predicted by secondary structure and molecular modeling within the amino terminal half of InvA (33). Consistent with this hypothesis, removal of the large cytoplasmic domain of InvA did not affect the localization and organization of its membrane proximal ring. Our structure also detected densities that most likely represent the membrane-embedded domain of InvA, which are located immediately adjacent (although apparently not in intimate contact) to the core components of the export apparatus, and within the remodeled funnel-shaped inner membrane. These observations have significant implications for the understanding of InvA function. First, the observed topological organization does not support a direct role for the transmembrane domains of InvA in the building of the putative protein translocation channel. This is significant since the lack of an obvious transmembrane regions in the SpaPQR complex that would be able to span the entire membrane had raised the possibility that this complex may work in conjunction with InvA to form such a channel (27). These observations coupled to the observed thinning of the membrane around the core components of the export apparatus strongly suggest that the SpaPQR complex (perhaps with SpaS) may be sufficient to form such channel as its entire vertex would be able to span the thinned membrane. Rather, the topology of InvA raises the exciting possibility that through its series of cytoplasmic rings, InvA “guides” the substrates to the entrance of the export apparatus channel (Fig. 7 and Movie S3), a function that could be aided by its putative proton channel activities as previously suggested (35).
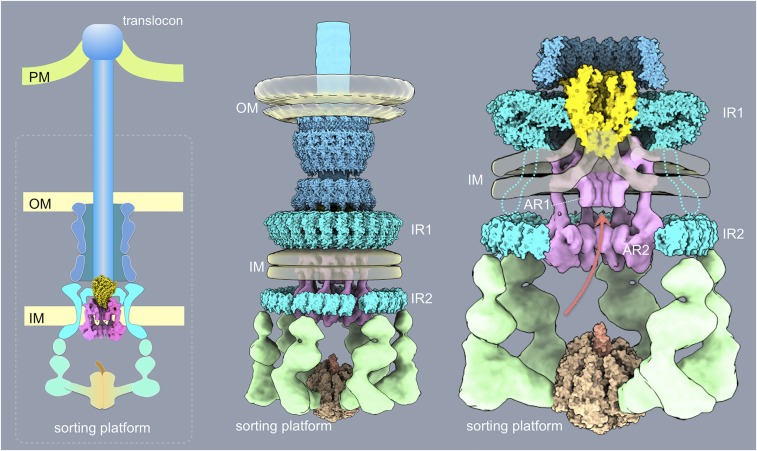
Fig. 7.
Schematic model of the export apparatus in the context of a fully assembled injectisome structure and the different bacterial envelope elements. AR1 and AR2, membrane proximal (AR1) and distal (AR2) cytoplasmic rings of InvA; IM, bacterial inner membrane; IR1 and IR2, inner rings 1 and 2 of the T3SS needle complex; OM, bacterial outer membrane; PM, target host cell plasma membrane.
A central feature of virulence-associated T3SS is that their protein-secretion activity is triggered only upon contact with eukaryotic cells (39, 40). Consequently, it is expected that machines visualized in isolation or in situ after growth in culture medium would be in the “close” or inactive conformation. Since the activation of the machine is triggered by contact with target eukaryotic cells, the activating signal is likely to emerge from the tip structure of the needle filament and eventually transmitted to the export apparatus through the needle and inner rod substructures (i.e., “top down”) (41). How this activating signal is relayed to the export apparatus is unclear. However, our observation that a discrete domain of the large cytoplasmic ring of InvA shown to be essential for type III secretion function (30) makes intimate contact with the IR2 cytoplasmic domain of the needle complex protein PrgH suggests a potential functional link between these 2 structures that may be important for transducing the activation signal. In this context, the close proximity of the core components of the export apparatus to both the transmembrane domains of InvA as well as a defined periplasmic domain of the needle complex protein PrgK (see above) may define potential sites of interactions that may serve as relay points for signals emanating from the needle tip complex. These signals would be then transduced through the needle filament and PrgJ ring to the export apparatus and subsequent relay points to coordinate substrate engagement with the opening of the secretion channel. Furthermore, these signals may also lead to the activation/coordination of the putative proton motive force of InvA that may be required to advance the substrates through its multiring conduit that lead to the entrance of the export channel.
In summary, these studies have provided a close-up view of the export apparatus in its in situ topological context and in association with other injectisomes components and the bacterial membrane. The topological relationship of these components to each other and to the bacterial membrane combined with the multiring organization of InvA suggest a potential pathway for the type III-secreted substrates during their translocation through the bacterial envelope.
Materials and Methods
Bacterial Strains and Plasmids.
All bacterial strains used in this study are listed in SI Appendix, Table S2. All strains were constructed by allelic exchange (42), and all plasmids were constructed by Gibson assembly (43).
Analysis of Type III Protein Secretion Function.
The functionality of the SPI-1 T3SS in the S. Typhimurium strains was assessed by examining their ability to secrete type III-secreted proteins to the culture supernatant as previously described (44).
Isolation of Needle Complexes Substructures.
Needle complexes from the different S. Typhimurium strains were isolated by an affinity purification protocol described in SI Appendix.
Single-Particle Cryo-EM.
Vitrified specimens were imaged at the NIH or at the Yale West Campus microscopy facility on a Titan Krios microscope (Thermo Fisher Scientific) operated at 300-kV cryoelectron microscope equipped with an energy filter (Gatan GIF Quantum), and a post-GIF Gatan K2 Summit direct electron detector.
Single-Particle Cryo-EM Image.
The frames from each exposure were aligned to compensate for drift and beam-induced motion using MotionCor2 (45) in Relion 2.1 (46, 47). Single particles were manually picked using Relion 2.1, and defocus and astigmatism were estimated with CTFFIND4 (48). Local resolution variations were estimated from 2 half data maps using ResMap (49). Additional details on EM image processing can be found in SI Appendix. Model fitting of cryo-EM structures was carried out using UCSF Chimera (50) and refined using PHENIX real-space refinement (51). The outliers were corrected in Coot (52). Overall model quality and geometry outliers for final models were reported using MolProbity (53). More detail can be found in SI Appendix, Table S3.
EM and Negative Staining.
Samples containing needle complex preparations were applied to glow-discharged, carbon-coated copper grids, stained with 2% (wt/vol) solution of uranyl acetate, and examined under a FEI Technai T12 (120 KV) microscope equipped with a Gatan 4k-by-4k CCD camera. Particle picking and several cycles of classification and averaging were performed with Relion 2.1 (46, 47).
Cryo-ET Sample Preparation, Data Collection, and Subtomogram Analysis.
Cryo-ET samples obtained from WT S. Typhimurium and several export apparatus mutants (SI Appendix) were prepared as previously described (25). The frozen-hydrated specimens were imaged with a 300-kV electron microscope (Titan Krios, Thermo Fisher Scientific) equipped with an energy filter (Gatan) with VPP. Raw images were processed using MotionCor2 (47). The tilt series were aligned automatically using IMOD (54). Tomograms were generated by using TOMO3D (55). Subtomogram analysis was accomplished as described previously (25). More details are provided in SI Appendix, Table S4.
Three-Dimensional Visualization and Modeling.
IMOD (54) was used to take snapshots of 2D slices from 3D tomograms, and UCSF Chimera (50) was used for surface rendering of 3D averaged structures. Animations were generated using UCSF ChimeraX and edited with iMovie.
Data Availability.
Bacterial strains generated in this study are available upon request. The coordinates of the EM structures shown in this manuscript can be found at the Electron Microscopy Data Bank with accession nos. 20830–20833 and 20838 and Protein Data Bank (PDB) ID codes 6UOT and 6UOV.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants AI030492 (to J.E.G.), AI126158 (to M.L.-T.), and GM107629 and AI087946 (to J.L.).
Footnotes
The authors declare no competing interest.
Data deposition: Coordinates have been deposited in Electron Microscopy Data Bank (accession nos. 20830–20833 and 20838); and Protein Data Bank (ID codes 6UOT and 6UOV).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1916331116/-/DCSupplemental.
References
- 1.Galán J. E., Lara-Tejero M., Marlovits T. C., Wagner S., Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 68, 415–438 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornelis G. R., The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biol. Chem. 391, 745–751 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Deng W., et al. , Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Gu L., Zhou S., Zhu L., Liang C., Chen X., Small-molecule inhibitors of the type III secretion system. Molecules 20, 17659–17674 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charro N., Mota L. J., Approaches targeting the type III secretion system to treat or prevent bacterial infections. Expert Opin. Drug Discov. 10, 373–387 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Tsou L. K., et al. , Antibacterial flavonoids from medicinal plants covalently inactivate type III protein secretion substrates. J. Am. Chem. Soc. 138, 2209–2218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diepold A., Armitage J. P., Type III secretion systems: The bacterial flagellum and the injectisome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubori T., et al. , Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Lara-Tejero M., Kato J., Wagner S., Liu X., Galán J. E., A sorting platform determines the order of protein secretion in bacterial type III systems. Science 331, 1188–1191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marlovits T. C., et al. , Structural insights into the assembly of the type III secretion needle complex. Science 306, 1040–1042 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schraidt O., Marlovits T. C., Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331, 1192–1195 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Wagner S., et al. , Organization and coordinated assembly of the type III secretion export apparatus. Proc. Natl. Acad. Sci. U.S.A. 107, 17745–17750 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukhan A., Kubori T., Wilson J., Galán J. E., Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J. Bacteriol. 183, 1159–1167 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diepold A., Wagner S., Assembly of the bacterial type III secretion machinery. FEMS Microbiol. Rev. 38, 802–822 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Kubori T., Sukhan A., Aizawa S. I., Galán J. E., Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc. Natl. Acad. Sci. U.S.A. 97, 10225–10230 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diepold A., et al. , Deciphering the assembly of the Yersinia type III secretion injectisome. EMBO J. 29, 1928–1940 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkinshaw B. J., Strynadka N. C., Assembly and structure of the T3SS. Biochim. Biophys. Acta 1843, 1649–1663 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Kosarewicz A., Königsmaier L., Marlovits T. C., The blueprint of the type-3 injectisome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1140–1154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee S., Chaudhury S., McShan A. C., Kaur K., De Guzman R. N., Structure and biophysics of type III secretion in bacteria. Biochemistry 52, 2508–2517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erhardt M., Namba K., Hughes K. T., Bacterial nanomachines: The flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2, a000299 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marlovits T. C., et al. , Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Schraidt O., et al. , Topology and organization of the Salmonella typhimurium type III secretion needle complex components. PLoS Pathog. 6, e1000824 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B., et al. , Visualization of the type III secretion sorting platform of Shigella flexneri. Proc. Natl. Acad. Sci. U.S.A. 112, 1047–1052 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu J., et al. , Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat. Commun. 9, 3840–3855 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu B., Lara-Tejero M., Kong Q., Galán J. E., Liu J., In situ molecular architecture of the Salmonella type III secretion machine. Cell 168, 1065–1074.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minamino T., Protein export through the bacterial flagellar type III export pathway. Biochim. Biophys. Acta 1843, 1642–1648 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Kuhlen L., et al. , Structure of the core of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 25, 583–590 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galán J. E., Ginocchio C., Costeas P., Molecular and functional characterization of the Salmonella invasion gene invA: Homology of InvA to members of a new protein family. J. Bacteriol. 174, 4338–4349 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minamino T., Morimoto Y. V., Hara N., Aldridge P. D., Namba K., The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog. 12, e1005495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abrusci P., et al. , Architecture of the major component of the type III secretion system export apparatus. Nat. Struct. Mol. Biol. 20, 99–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zilkenat S., et al. , Determination of the stoichiometry of the complete bacterial type III secretion needle complex using a combined quantitative proteomic approach. Mol. Cell. Proteomics 15, 1598–1609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginocchio C. C., Galán J. E., Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect. Immun. 63, 729–732 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor W. R., Matthews-Palmer T. R., Beeby M., Molecular models for the core components of the flagellar type-III secretion complex. PLoS One 11, e0164047 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minamino T., Morimoto Y. V., Hara N., Namba K., An energy transduction mechanism used in bacterial flagellar type III protein export. Nat. Commun. 2, 475–483 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erhardt M., et al. , Mechanism of type-III protein secretion: Regulation of FlhA conformation by a functionally critical charged-residue cluster. Mol. Microbiol. 104, 234–249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Sa’adedin F., Deme B., Rao P., Bradshaw J., Insertion of TAT peptide and perturbation of negatively charged model phospholipid bilayer revealed by neutron diffraction. Biochim. Biophys. Acta 1828, 1982–1988 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., et al. , YidC insertase of Escherichia coli: Water accessibility and membrane shaping. Structure 25, 1403–1414.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoebel S., et al. , Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 548, 352–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ménard R., Sansonetti P., Parsot C., The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 13, 5293–5302 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zierler M. K., Galán J. E., Contact with cultured epithelial cells stimulates secretion of Salmonella typhimurium invasion protein InvJ. Infect. Immun. 63, 4024–4028 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blocker A. J., et al. , What’s the point of the type III secretion system needle? Proc. Natl. Acad. Sci. U.S.A. 105, 6507–6513 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaniga K., Bossio J. C., Galán J. E., The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13, 555–568 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Lara-Tejero M., et al. , Role of SpaO in the assembly of the sorting platform of a Salmonella type III secretion system. PLoS Pathog. 15, e1007565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheres S. H., A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415, 406–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scheres S. H., RELION: Implementation of a bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastronarde D. N., Held S. R., Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agulleiro J. I., Fernandez J. J., Tomo3D 2.0–Exploitation of advanced vector extensions (AVX) for 3D reconstruction. J. Struct. Biol. 189, 147–152 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bacterial strains generated in this study are available upon request. The coordinates of the EM structures shown in this manuscript can be found at the Electron Microscopy Data Bank with accession nos. 20830–20833 and 20838 and Protein Data Bank (PDB) ID codes 6UOT and 6UOV.