Significance
DNA sequence variants in the Tbk1 gene associate with both sporadic and familial amyotrophic lateral sclerosis (ALS). We report a functional study of 25 missense TBK1 mutations scattered across all structural domains. These mutations negatively impact TBK1 functions ranging from kinase activity, substrate specificity, dimerization, and protein stability to interactions with critical autophagy receptors. Contrary to previous studies, we find that TBK1 dimerization increases protein stability and improves catalytic efficiency, but it is not required for its activation. Our results show that different TBK1 ALS mutations, which are associated with the same pathological outcome, affect distinct signaling pathways in which TBK1 is critically involved. TBK1-associated ALS provides a remarkable example of the genetic and functional complexity of ALS disease mechanisms.
Keywords: ALS, TBK1, mutations, kinase, dimerization
Abstract
Exonic DNA sequence variants in the Tbk1 gene associate with both sporadic and familial amyotrophic lateral sclerosis (ALS). Here, we examine functional defects in 25 missense TBK1 mutations, focusing on kinase activity and protein–protein interactions. We identified kinase domain (KD) mutations that abolish kinase activity or display substrate-specific defects in specific pathways, such as innate immunity and autophagy. By contrast, mutations in the scaffold dimerization domain (SDD) of TBK1 can cause the loss of kinase activity due to structural disruption, despite an intact KD. Familial ALS mutations in ubiquitin-like domain (ULD) or SDD display defects in dimerization; however, a subset retains kinase activity. These observations indicate that TBK1 dimerization is not required for kinase activation. Rather, dimerization seems to increase protein stability and enables efficient kinase–substrate interactions. Our study revealed many aspects of TBK1 activities affected by ALS mutations, highlighting the complexity of disease pathogenicity and providing insights into TBK1 activation mechanism.
TANK binding kinase 1 (TBK1) is a serine/threonine protein kinase that plays central roles in innate immunity, inflammation, selective autophagy, oncogenesis, and cell death (1–4). TBK1 was initially identified through its interaction with the adaptor proteins TANK and TRAF2 and was shown to be involved in the regulation of nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) signaling (5, 6). Subsequent studies revealed that TBK1 plays an essential role in the cellular innate immune response against viral and bacterial pathogens (7–9). Upon pathogen challenge, distinct sensors/adaptors are activated (for example, DNA and RNA virus infections activate cGAS/STING and RIG-I/MAVS pathways, respectively), which leads to the activation of TBK1 (10, 11). Phosphorylation of the transcription factors IRF3/7 by TBK1, which is required for the activation of type I interferon (IFN) genes, is a key step in the establishment of the cellular antiviral state (10, 12).
In addition to playing a crucial role in the immune response to exogenous pathogen infections, TBK1 also plays a key role in the maintenance of cellular homeostasis through its role in selective autophagy (13, 14). Specifically, TBK1 phosphorylates autophagy receptor proteins, such as SQSTM1 (p62) and Optineurin (OPTN) (13), which bind to ubiquitinated protein aggregates, damaged organelles, and other toxic cellular components (15, 16). These receptors mediate the incorporation of autophagy cargos into the autophagosome, a membranous structure that fuses with lysosomes, leading to protein degradation. ATG8 family proteins, such as microtubule-associated protein 1 light chain 3 beta (LC3), are essential autophagosome membrane proteins that directly interact with autophagy receptors (13, 14). TBK1 phosphorylation of autophagy receptors enhances interactions between receptors and cargos and/or ATG8 family members and receptors and thus, facilitates the autophagic process.
Given the importance of TBK1 in multiple signaling pathways, it is not surprising that mutations in the Tbk1 gene can lead to a number of human diseases (1). Point mutations of TBK1 were first identified in patients susceptible to childhood herpes simplex virus-1 (HSV-1) encephalitis infection (17). These mutations resulted in the loss of TBK1 function and impaired the antiviral response to HSV. In addition, Tbk1 gene duplication was shown to be associated with normal-tension glaucoma (18, 19). More recently, more than 90 TBK1 mutations were shown to associate with amyotrophic lateral sclerosis (ALS), a fatal neurodegenerative disease resulting from the loss of motor neurons (20–22). Specifically, DNA mutations in the coding region of the Tbk1 gene were found to associate with both sporadic and familial ALS cases in large-scale exome sequencing studies (20, 21). TBK1 mutations account for ∼1% of all ALS patients examined. Among these DNA sequence variants, missense mutations of TBK1 are the most frequent followed by internal deletion/insertion, splice site, frameshift, and nonsense mutations. TBK1 mutations occur along the entire coding region of TBK1 rather than clustered within a particular structural or functional domain. While frameshift and nonsense mutations can lead to the loss of protein expression, missense mutations and insertion/deletion mutations could result in the loss of TBK1 function and/or gain of detrimental function. Understanding the impact of these mutations on the structure and function of TBK1 through biochemical and biophysical studies should provide insights into ALS disease mechanisms.
Misregulation of neuroinflammation, autophagy, and stress-induced cell death are among many contributing factors of ALS (23). TBK1 plays important roles in the regulation of all of these pathways (24). Despite intensive investigation, mechanisms of TBK1 activation under physiological conditions are still poorly understood. However, recent structural studies have provided valuable insights (25–28). The N terminus of TBK1 contains a kinase domain (KD) followed by ubiquitin-like domain (ULD), scaffold dimerization domain (SDD), and a C-terminal domain (CTD) (Fig. 1A). TBK1 normally exists as a dimer, and dimerization occurs primarily through intermolecular interactions in the SDD. Additional intermolecular interactions in the KD and ULD in TBK1 also contribute to dimerization (25–27). This differs from the closely related IκBα kinase β (IKKβ), which functions in NFκB activation. In this case, SDDs comprise the entire dimerization interface (29). Structural studies suggest that TBK1 dimerization is required for activation, and a key step in TBK1 activation is the phosphorylation of a critical serine residue (S172) within the activation loop of the kinase (30). S172 phosphorylation triggers intramolecular interactions between amino acids within the KD that lock the kinase into an active conformation (25). Structural studies showed that active sites within TBK1 dimers point in opposite directions and are positioned relatively distant from each other. These observations suggest the possibility that TBK1 is activated through transautophosphorylation of dimeric TBK1 (25, 28). It has been proposed that recruitment of TBK1 by various adaptor proteins (such as MAVS and STING in innate immunity, OPTN in autophagy) leads to local high concentrations of the kinase, which triggers TBK1 autophosphorylation (25).
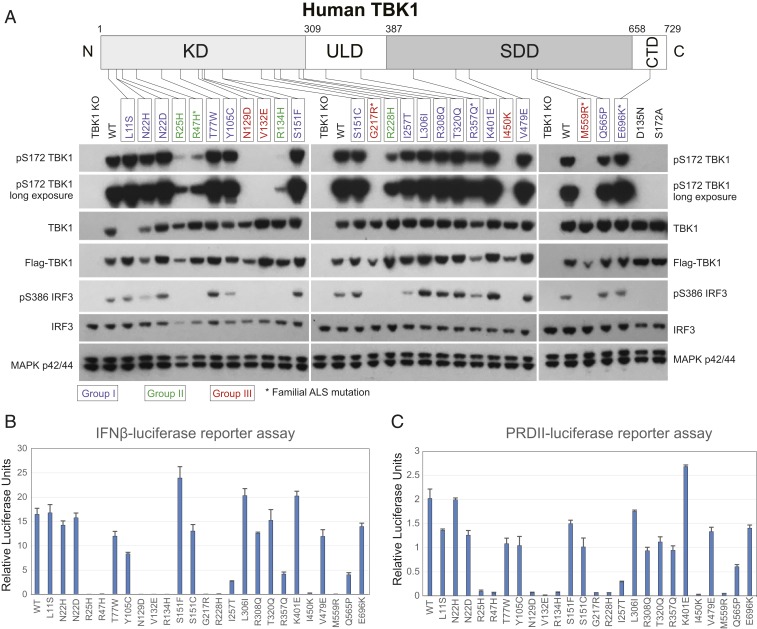
Fig. 1.
Effects of TBK1 ALS mutations on its kinase activity. (A) TBK1 autophosphorylation and IRF3 activation in overexpression experiments. Various expression constructs for TBK1 ALS mutations were transfected into TBK1 knockout 293T cells; 24 h later, total cell lysates were prepared, and the levels of pS172 TBK1, total TBK1, pS386 IRF3, total IRF3, and MAP kinase p42/44 were determined by western blots with specific antibodies. Positions of TBK1 ALS mutations in this study are indicated in the human TBK1 protein diagram. Note that TBK1 L11S was not detected by the total TBK1 antibody because of epitope disruption in the mutant protein. Groups I to III mutants are color coded, and familial mutations are labeled with asterisks. (B) Effects of mutations on the IFNβ-luciferase reporter assays. Expression constructs for various TBK1 mutations were cotransfected with an IFNβ promoter-driven firefly luciferase construct and a renilla luciferase reference construct into TBK1 knockout 293T cells; luciferase activities were measured 24 h after transfection. (C) Effects of TBK1 ALS mutations on NFκB reporter activity. Similar to experiments in B but a 2-copy PRDII element of the IFNβ enhancer-driven firefly luciferase construct was used instead of the IFNβ reporter.
While the current model of TBK1 activation can explain many observations of TBK1 activity in vitro and in studies with overexpressed TBK1, other findings are inconsistent with the model. For example, a number of studies have shown that treating cells with specific TBK1 inhibitors failed to block TBK1 S172 phosphorylation induced by a wide range of stimulators (31, 32). Thus, TBK1 can be activated by upstream kinases.
Here, we provide insights into TBK1 activation by studying the impact of TBK1 ALS mutations on function. Specifically, we provide biochemical data to show that a heterodimer of TBK1, in which 1 functional subunit is phosphorylated at S172, is active. This observation suggests that TBK1 can be activated by any kinase(s) that targets S172 under physiological conditions. We also found that TBK1 ALS mutations display a variety of defects, including loss of kinase activity, decreased protein stability, loss of dimerization, and loss of interactions with adaptor proteins. Perhaps most interesting is the finding that certain KD mutations display substrate-specific kinase defects, implicating distinct mutant deficiencies in different cellular signaling pathways. We also find that heterodimers of TBK1 formed between the wild type (WT) and dysfunctional mutants are active, albeit at about 50% of WT homodimer activity, thus supporting the proposal that TBK1 ALS mutations may elicit pathogenicity through haploinsufficiency. Finally, we identified TBK1 ALS mutations that affect dimerization without losing kinase activity, and these mutant TBK1 proteins seem less stable. Our data suggest that TBK1 dimerization enables optimal protein–substrate interaction and provides greater stability, but it is not a prerequisite for kinase activation.
Results
The Effect of TBK1 ALS Mutations on Kinase Activity.
To determine the effects of TBK1 ALS mutations on kinase activity, we expressed a number of TBK1 mutants in human 293T cells in which both Tbk1 alleles were deleted by CRISPR-Cas9 (TBK1 knockout cells). We assayed for both the level of TBK1 autophosphorylation at S172 and phosphorylation of the endogenous TBK1 substrate IRF3. A total of 25 missense mutations, with mutation sites distributed across all of the domains of TBK1 (Fig. 1A, 15 mutations within KD, 4 within ULD, 5 within SDD, and 1 in the CTD), were studied. These mutations associate with either sporadic (20 cases) or familial ALS cases (5 cases) (20, 21).
Most missense mutations showed detectable levels of expression comparable with WT TBK1, but a few were expressed at slightly lower levels (N22H, N129D, G217R, R357Q, I450K, and M559R, Fig. 1A). Sixteen mutants displayed nearly normal (or slightly reduced) autophosphorylation, with nearly equal detection of pS172 compared with WT TBK1 (Fig. 1A). We define these mutations as group I (color coded in Fig. 1A), and they are scattered across all functional domains of TBK1 and display relatively mild effects on TBK1 kinase activity. For example, the S386 amino acid residue of endogenous IRF3 is phosphorylated by these mutant TBK1 proteins at a level similar to that by WT TBK1 (S386 of IRF3 is a well-established TBK1 phosphorylation site [27]) (Fig. 1A). Four KD mutations (R25H, R47H, R134H, and R228H [group II]) showed no detectable IRF3 phosphorylation but retained a low level of autophosphorylation (Fig. 1A). Five remaining mutations (N129D, V132E, G217R, I450K, and M559R [group III]) showed neither IRF3 phosphorylation nor autophosphorylation (Fig. 1A). The complete loss of S172 phosphorylation was observed with the kinase-dead negative control, TBK1 D135N (Fig. 1A), confirming that this phosphorylation assay specifically measures TBK1 autophosphorylation. Specificity of the phospho-TBK1 antibody was validated by expression of the TBK1 S172A negative control (Fig. 1A). The observation that group II mutations abolish IRF3 phosphorylation while retaining at least residual levels of TBK1 autophosphorylation revealed that the effect of certain ALS mutations is substrate specific. Consistent with this possibility, TBK1 I257T (group I mutation) also resulted in a lower level of IRF3 phosphorylation, despite a strong level of S172 TBK1 phosphorylation (Fig. 1A).
We also conducted IFNβ luciferase reporter assays to quantitatively determine the effect of the TBK1 mutations on kinase activity. Individual expression constructs for TBK1 mutations were cotransfected with an IFNβ promoter-driven firefly luciferase construct together with a renilla luciferase reference construct, and luciferase activity was measured 24 h after transfection. Accounting for variations in expression levels seen in Fig. 1A, many group I mutations showed reporter activities in an expected range (Fig. 1B). The reduced reporter activity of R357Q (∼30% of WT TBK1) is consistent with its lower expression. Reduced reporter activity of I257T may be related to a substrate-specific effect, as this mutant does not significantly lower activation or expression levels. However, it is surprising that Q565P showed lower reporter activity considering that IRF3 S386 phosphorylation was unaffected by this mutation. It is possible that TBK1 Q565P might be defective in the phosphorylation of other sites on IRF3, such as S396, which is required for the maximal transcriptional activity of IRF3 (33); alternatively, TBK1 Q565P might have impaired ability to phosphorylate NFκB, which is also required for IFNβ reporter assays (34). Consistent with Fig. 1A, group II and group III mutations, which showed undetectable IRF3 activation, showed almost no detectable reporter activity. Similar results (Fig. 1C) were obtained using an NFκB-luciferase reporter (2 copies of the PRDII element from the IFNβ enhancer, PRDIIx2-luciferase [7]), suggesting that overexpressed TBK1 activates both IFNβ and NFκB pathways similarly, and the weak kinase activities of the group II TBK1 mutations were not sufficient to activate either pathway in overexpression assays.
Taken together, these data show that different TBK1 ALS point mutations display distinct levels of kinase activity (Table 1). Of the mutations studied, 64% retain near-intact (or slightly reduced) kinase activity, 16% had significantly reduced activity, and 20% completely lost kinase activity. Among the 5 familial mutations (R47H, G217R, R357Q, M559R, and E696K), 2 resulted in a complete loss of the kinase activity (G217R and M559R), 1 dramatically reduced activity (R47H), 1 mildly reduced activity (R357Q; largely due to lower expression level), and 1 had no detectable effect on kinase activity (E696K). TBK1 functions in multiple signaling pathways by phosphorylating distinct protein substrates. It seems that ALS-associated TBK1 mutations can either directly impact kinase activity or indirectly affect the phosphorylation of specific substrates in different signaling pathways. However, the ultimate pathological consequence of these mutations, motor neuron loss, is the same. Identification of the key target(s) that leads to ALS is a fundamentally important objective.
Table 1.
List of key findings of TBK1 activities affected by TBK1 ALS missense mutations in this study
| Domain and mutations | Kinase activity | Protein dimerization | OPTN interaction | Stability | |||
| TBK1 | IRF3 | OPTN | p62 | ||||
| KD | |||||||
| L11S* | +++ | +++ | ++ | +++ | +++ | ++ | |
| N22H* | +++ | ++ | +++ | + | +++ | +++ | |
| N22D* | +++ | +++ | +++ | +++ | +++ | +++ | |
| R25H† | + | − | + | − | +++ | +++ | |
| R47H†‡ | + | − | − | − | +++ | +++ | |
| T77W* | +++ | +++ | +++ | + | +++ | +++ | |
| Y105C* | +++ | ++ | ++ | + | +++ | + | |
| N129D§ | − | − | − | − | +++ | ++ | |
| V132E§ | − | − | − | − | +++ | +++ | |
| R134H† | ± | − | − | − | +++ | +++ | |
| S151F* | +++ | +++ | +++ | +++ | + | ++ | |
| S151C* | +++ | +++ | +++ | ± | + | ++ | |
| G217R‡§ | − | − | ± | − | + | ++ | Less stable |
| R228H† | ++ | − | ± | ± | +++ | +++ | |
| I257T* | +++ | ++ | ++ | ± | ++ | +++ | |
| ULD | |||||||
| L306I* | +++ | +++ | +++ | +++ | +++ | +++ | |
| R308Q* | +++ | +++ | ++ | +++ | +++ | + | |
| T320Q* | +++ | +++ | +++ | +++ | +++ | +++ | |
| R357Q*‡ | +++ | +++ | +++ | ± | − | ++ | Less stable |
| SDD | |||||||
| K401E* | +++ | +++ | +++ | +++ | +++ | +++ | |
| I450K§ | − | − | ± | − | − | ++ | Less stable |
| V479E* | +++ | +++ | +++ | + | ++ | +++ | |
| M559R‡§ | − | − | ± | − | − | ++ | Less stable |
| Q565P* | +++ | +++ | +++ | − | − | ++ | |
| CTD | |||||||
| E696K*‡ | +++ | +++ | +++ | +++ | +++ | − | |
Group I.
Group II.
Group III.
Familial ALS mutation.
+++, Normal activity.
++, Slightly reduced activity.
+, Significantly reduced activity.
±, Very weak activity.
−, No activity.
Effects of TBK1 Mutations on Protein Dimerization.
The 3-dimensional structure of the nearly full-length TBK1 protein was independently solved by 3 different groups (25–27), and all concluded that WT TBK1 exists as a dimer. In contrast to the dimer formed by IKKβ, where intermolecular associations within the SDD are sufficient for dimerization (29), the TBK1 dimer interface includes residues within the KD and ULD as well (25). Dimerization was proposed to be required for TBK1 activation, as mutations designed to disrupt the dimer interface showed reduced activity in reporter and kinase assays (25, 26). We, therefore, investigated whether TBK1 ALS mutations, particularly those that showed greatly reduced kinase activity, also affect dimerization.
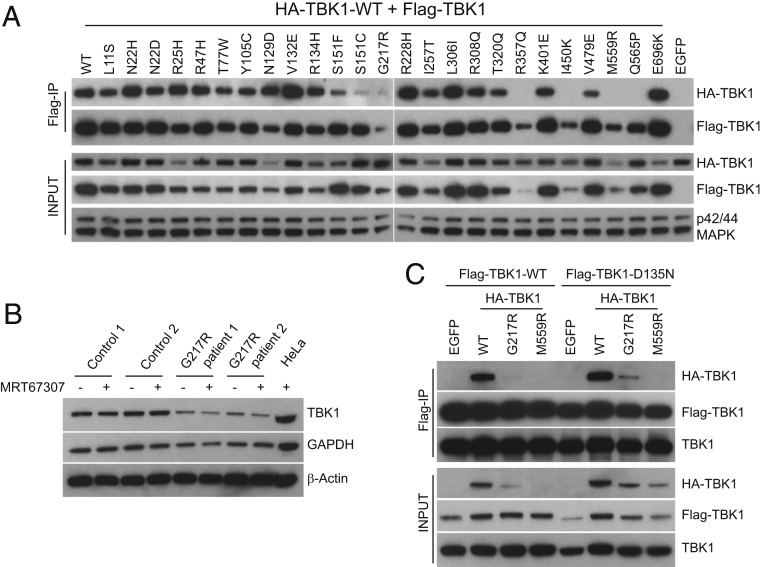
Coimmunoprecipitation (Co-IP) experiments were conducted with differentially tagged WT and mutant TBK1 to more closely mimic formation of heterozygous mutant dimers presumed in disease state (TBK1 mutations in ALS patients are heterozygous) (22). Specifically, we coexpressed HA-tagged WT and Flag-tagged mutant TBK1 and assayed for their association. As shown in Fig. 2A, we can readily detect interaction between most Flag-tagged TBK1 mutants and the HA-tagged WT partner. In general, KD mutations do not significantly interfere with dimer formation, with the exception of S151F/C mutations, which showed some reduction of association with the WT partner. S151 maps to a location near KD–SDD interacting surfaces of the dimer interface in the known TBK1 structure (25, 26). Strikingly, R357Q, I450K, M559R, and Q565P mutations in the ULD and SDD completely abolished WT TBK1 association (Fig. 2A). Although we had observed lower overexpression levels of R357Q, I450K, and M559R mutants compared with WT TBK1 (Fig. 1A), unexpectedly coexpression of these mutants with WT TBK1 further reduced expression (Fig. 2A). Similarly, the dramatically reduced G217R protein level also seems to be related to WT TBK1 coexpression, and its association with WT TBK1 was difficult to estimate from this experiment.
Fig. 2.
Effects of TBK1 ALS mutations on protein–protein interactions. (A) Impact of TBK1 ALS mutations on its dimerization. Expression constructs of Flag-tagged various TBK1 mutations from ALS patients were cotransfected with HA-tagged WT TBK1 into TBK1 knockout 293T cells, and cell lysates were prepared 24 h later and precipitated with anti-Flag antibodies (M2). The interaction between 2 differentially tagged TBK1 was assayed by western blot with anti-Flag and HA antibodies of the IP samples. (B) Reduction of TBK1 protein in cells derived from patients carrying G217R mutation. Control and patient fibroblasts were either untreated or treated with TBK1 inhibitor MRT67307 for 3 h, and total protein lysates were prepared for western blot analysis of TBK1, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-Actin expression with specific antibodies. Lysates prepared from HeLa cells were also included as a control. (C) Coexpression of TBK1 mutants with a kinase-dead TBK1 improves the stability of mutant TBK1. HA-tagged G217R, M559R TBK1, and WT control were cotransfected with Flag-tagged WT TBK1 or D135N TBK1 into 293T cells; 24 h later, cell lysates were prepared for IP with anti-Flag beads. The expression and interaction of TBK1 mutants were analyzed by western blots with antibodies against HA, Flag, and TBK1.
To provide further insights into reduced TBK1 mutant expression levels, we determined the TBK1 protein level in ALS patient-derived cells. We observed ∼50% reduction of total TBK1 in heterozygous Tbk1 G217R fibroblasts (Fig. 2B). Considering that there is no difference in the messenger RNA level between G217R patient cells and control cells, the G217R mutation seems to cause instability at protein level, and this could be a common feature of ALS mutations that affect the overall structure of the protein. Coexpression of G217R or M559R with kinase-dead TBK1 (D135N) resulted in higher expression of the mutants than with WT TBK1 (Fig. 2C). Thus, it seems that instability of certain TBK1 ALS mutations is exacerbated by the presence of functional TBK1. Using TBK1 D135N coexpression to improve expression, we show more reliably that the G217R mutation can associate with another TBK1 partner, but the efficiency is greatly reduced (Fig. 2C) compared with controls. We also confirmed that the M559R mutation abolishes association (Fig. 2C). Similarly, we performed Co-IP experiments using TBK1 D135N to provide additional evidence that I450K and Q565P mutations have diminished dimerization (SI Appendix, Fig. S1A). To exclude the possibility that homodimer formation might be different from heterodimerization, we also conducted Co-IP studies with HA and Flag-tagged R357Q, I450K, M559R, and Q565P mutations. Again, we failed to detect homodimer formation between these differentially tagged mutant TBK1 proteins in cells (SI Appendix, Fig. S1B). The steady-state level of these mutant TBK1 proteins appears lower compared with that of WT TBK1, suggesting that dimer conformation increases TBK1 stability.
Taken together, these results show that several TBK1 ALS mutations, including the familial mutations R357Q and M559R, affect protein dimerization. In addition, the dimerization defect seems to have a direct impact on TBK1 protein stability rather than kinase activity (Figs. 1A and 2A and Table 1).
Monomeric TBK1 R357Q Is a Functional Kinase.
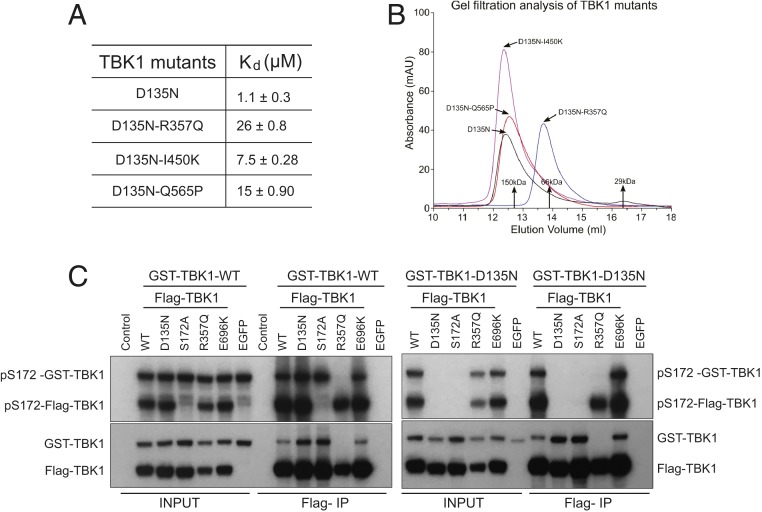
Biophysical studies were conducted on recombinant TBK1 mutants to further characterize the effect of the mutations on self-association. Previous structural studies showed that recombinant TBK1 (residues 1 to 657) behaves the same as full-length TBK1 in dimerization assays (25). Since we detected dimerization defects of TBK1 R357Q, I450K, and Q565P, we carried out sedimentation equilibrium analytical ultracentrifugation (AUC) experiments to determine the dissociation constants (Kd) of these TBK1 mutants. Control TBK1 D135N (1 to 657) had a Kd of 1.1 ± 0.3 μM, and the Kd values for TBK1 D135N-R357Q (1 to 657), D135N-I450K (1 to 657), and D135N-Q565P (1 to 657) were measured as 26 ± 0.8, 7.5 ± 0.28, and 15 ± 0.90 μM (Fig. 3A). The reduced self-association of these mutants is consistent with our Co-IP results (Fig. 2A). A dimerization defect of TBK1 R357Q was also detected by gel filtration chromatography: when the TBK1 D135N control and TBK1 D135N-R357Q, D135N-I450K, and D135N-Q565P mutants (residues 1 to 657) were injected at a concentration of 13 μM, TBK1 D135N-R357Q was the only one that eluted as a monomer (Fig. 3B). We also conducted gel filtration analysis for TBK1 D135N-M559R, and this mutant displayed nonmonodisperse solution behavior. Recombinant protein expression of the D135N-M559R mutant was poor, and protein eluted as a broad peak across a wide volume range corresponding to molecular mass sizes ranging from monomeric protein to large nonspecific aggregates. When assayed at a lower concentration (∼2 μM) by the more sensitive size exclusion chromatography–multiple angle light scattering technique, both TBK1 D135N-R357Q and D135N-Q565P showed monomeric behavior, with estimated molecular masses of 76 and 97 kDa, respectively, in comparison with 138 kDa for a dimer of TBK1 (1 to 657).
Fig. 3.
TBK1 R357Q is a monomeric and functional kinase. (A) Dissociation constant (Kd) of selected TBK1 mutants (residues 1 to 657) determined by AUC experiments. (B) Gel filtration chromatograms of the indicated recombinant TBK1 mutants showing milli-absorbance units (mAU) over elution volume. Positions of molecular mass standards were also marked. TBK1 (1 to 657) mutants were expressed in insect cells and purified. These ALS-related mutants were all in the kinase-dead D135N background. Recombinant proteins were injected onto a Superdex 200 Increase 10/300GL column (GE Healthcare) at a flow rate of 0.5 mL/min in a buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine (TCEP), and the ultraviolet absorbance (mAU) at 280 nm was recorded. (C) TBK1 R357Q can phosphorylate a different TBK1 molecule when coexpressed. GST-tagged WT TBK1 (Left) or D135N TBK1 (Right) was coexpressed with Flag-tagged WT, D135N, S172A, R357Q, E696K TBK1, or EGFP as a control in TBK1 knockout 293T cells; 24 h after transfection, cell lysates were prepared, and anti-Flag beads were used to IP Flag-tagged proteins. The profiles of pS172 TBK1 and total TBK1 from different samples were determined by western blots with respective antibodies.
In the structure of the TBK1 biological dimer, R357 is directly involved in self-association, forming the R357 to D452′ intermolecular salt bridge at the ULD–SDD interface (25, 26). This interaction seems to be critical for dimerization and is apparently abolished by the R357Q mutation. Supporting the importance of this salt bridge, a D452R mutation also significantly reduced dimerization (SI Appendix, Fig. S1C).
Biochemical and biophysical data clearly showed that TBK1 R357Q is monomeric (Figs. 2A and 3B), yet this mutant protein retains relatively high kinase activity, as overexpressed TBK1 R357Q autophosphorylates and also, phosphorylates IRF3 (Fig. 1A) in vivo. Subsequent in vitro experiments using recombinant proteins further confirmed the activity of TBK1 R357Q toward OPTN (SI Appendix, Fig. S2A). We also performed Co-IP experiments with Flag-tagged R357Q and glutathione S-transferase (GST)-tagged kinase-dead TBK1 D135N. Coexpression of these 2 mutants in the absence of endogenous TBK1 showed S172 phosphorylation of both proteins in the total lysates, while no association was detected between R357Q and D135N TBK1 subunits in the immunoprecipitated samples (Fig. 3C). Together, these results show that activation of the TBK1 R357Q kinase does not require stable dimer formation.
Finally, we compared the activity of TBK1 R357Q with a known engineered monomeric TBK1 mutant (H459E/I466E/F470E [HIF]) (25). Expression of either TBK1 R357Q or HIF mutation in TBK1 knockout 293T cells leads to a slightly decreased total level of TBK1 accompanied by a similarly lower level of phospho-IRF3 (SI Appendix, Fig. S2B). The similar behaviors of these 2 mutants confirm the monomeric nature of the TBK1 R357Q mutant protein and again, support the notion that TBK1 activation does not require dimerization.
TBK1 WT:mutant Heterodimers Are Functional.
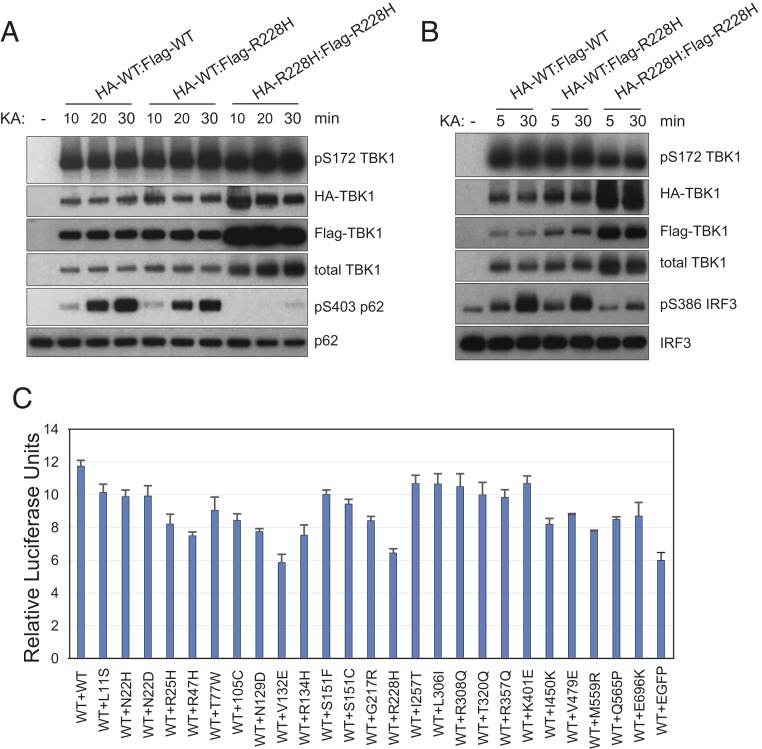
In the TBK1 structures in the phosphorylated state solved to date, all molecules in the crystal lattice are uniformly phosphorylated at S172. This is in contrast to a solved structure of an asymmetrical human IKKβ dimer containing only 1 phosphorylated subunit at S181 (an equivalent site of S172 of TBK1) (35). Asymmetry in TBK1 seems directly relevant to ALS, as TBK1 mutations identified in ALS patients are heterozygous, and a number of TBK1 mutants do not display dimerization defects (Fig. 2A). TBK1 WT:mutant heterodimers are expected to form in the disease state. To investigate whether formation of disease-related TBK1 WT:mutant heterodimers specifically affects kinase activity of the WT TBK1, we expressed and purified WT:mutant heterodimers (HA- and Flag-tagged, respectively) from transfected TBK1 knockout cells through sequential IPs and assayed for in vitro kinase activity using either p62 or IRF3 as substrates. As an example, we purified WT homodimers, WT:R228H heterodimers, and R228H homodimers and carried out kinase assays. As shown in Fig. 4 A and B, homodimers of R228H are defective in the phosphorylation of p62 (S403, a well-established TBK1 target site [36]) and IRF3 (S386), despite a relatively high level of pS172 TBK1 autophosphorylation observed with the more abundant protein. By contrast, WT homodimers showed robust activity toward both p62 and IRF3. Interestingly, WT:R228H heterodimers are active in these kinase assays; however, the total activity of the heterodimers is about half that of the WT homodimers (Fig. 4 A and B). We conclude that, although TBK1 R228H is itself enzymatically compromised, it does not interfere with the activity of the other subunit as part of a dimer complex.
Fig. 4.
WT:mutant heterodimer TBK1 is functional. (A and B) WT:R228H heterodimer retains kinase activity toward p62 and IRF3. HA-TBK1-WT or HA-TBK1-R228H constructs were coexpressed with Flag-TBK1-WT or Flag-TBK1-R228H in TBK1 knockout 293T cells. Cell lysates were prepared 24 h after transfection and subjected to anti-HA beads immunoprecipitation for 2 h at 4 °C. After extensive washes, immunoprecipitated samples were eluted with HA peptide (0.5 μg/μL in wash buffer) and subjected to anti-Flag beads IP overnight. Final immunoprecipitated samples were assayed for kinase activity with recombinant GST-p62 (A) or Flag-IRF3 (B) on beads for increasing time (10, 20, and 30 min in A and 5 and 30 min in B). The phosphorylation status of various proteins was determined by western blots probed with specific antibodies. KA, kinase assay. (C) TBK1 ALS mutations do not inhibit WT TBK1 activity in IFNβ-luciferase reporter assays. Expression constructs for various TBK1 mutations were cotransfected with the same amount of WT expression construct plus IFNβ promoter-driven firefly luciferase construct and a renilla luciferase reference construct into TBK1 knockout 293T cells; luciferase activities were measured 24 h after transfection.
To extend the study of heterodimer functionality to additional ALS mutations, we conducted IFNβ-luciferase reporter assays by cotransfecting WT TBK1 expression construct with the same amount of various ALS mutation expression constructs and assayed for reporter activity. As shown in Fig. 4C, the total luciferase reporter activities from these coexpression experiments varied between the detected activities of WT + EGFP and WT + WT samples; none of the coexpressed TBK1 ALS mutations inhibited the activity of WT TBK1. These results convincingly show that ALS missense mutations in this study do not function in a dominant negative manner. Rather, different degrees of loss of function of these mutations are most likely responsible for disease pathogenesis in patients carrying TBK1 mutations.
Complementation Between TBK1 Subunits within a Dimer.
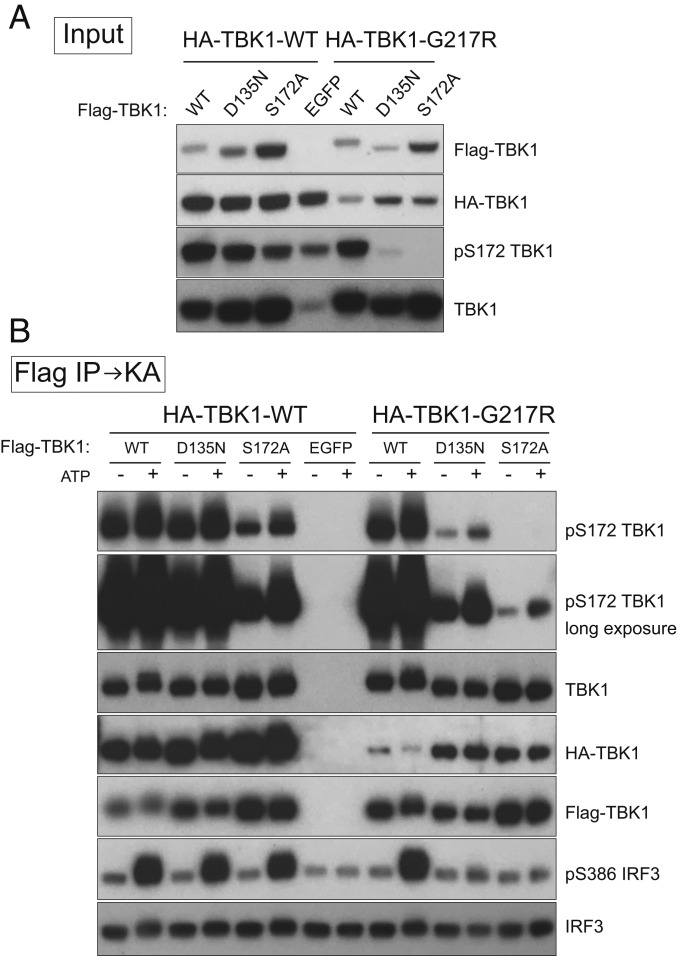
TBK1 dimerization seems to increase protein stability, as dimerization mutants are, in general, expressed at a lower level compared with WT TBK1 (Fig. 2A and SI Appendix, Fig. S1B). To explore the possibility that TBK1 dimerization also enhances enzymatic activity, we conducted experiments to test whether individual subunits that are enzymatically defective can complement each other to regain kinase activity in a heterodimer. We coexpressed HA-tagged WT or TBK1 G217R together with Flag-tagged WT, TBK1 D135N, or TBK1 S172A (Fig. 5A) and purified TBK1 complexes by anti-Flag beads followed by kinase assays with recombinant IRF3 as a substrate. Consistent with previous results, purified TBK1 heterodimers containing WT subunit (in WT:D135N, WT:S172A, and WT:G217R complexes) showed robust kinase activity with TBK1 and IRF3 substrates (Fig. 5B). The WT:S172A complexes are worth mentioning, as only S172 from the WT subunit is phosphorylated, and this single S172 phosphorylated TBK1 heterodimer is active. This finding reveals that, during TBK1 activation, S172 phosphorylation on a single functional subunit of the TBK1 dimer is sufficient to activate the kinase.
Fig. 5.
TBK1 subunits within the dimer complement each other to exert kinase activity. HA-tagged WT TBK1 or G217R TBK1 was coexpressed with Flag-tagged WT, D135N, S172A TBK1, or EGFP as a control in TBK1 knockout 293T cells (A). Cell lysates were prepared 24 h after transfection; immunoprecipitations were conducted with anti-Flag beads followed by kinase assays (KA) with recombinant IRF3 as the substrate at 30 °C for 40 min (B).
We note that TBK1 heterodimers formed between D135N and G217R gained kinase activity as indicated by S172 phosphorylation and weak activation of IRF3 (Fig. 5B), while D135N and G217R alone lack normal kinase activity (Fig. 1A and SI Appendix, Fig. S3). A similar observation was made with a G217R:S172A heterodimer, despite the lower level of phospho-S172 due to the lack of the site in TBK1 S172A mutant (Fig. 5B). Taken together, these results demonstrate that subunits within the TBK1 dimer are indeed able to complement each other to exert optimal kinase activity, a function clearly lacking in monomeric TBK1 mutants.
TBK1 Mutations Affect the Phosphorylation of Autophagy Receptors.
We also investigated the effects of TBK1 mutations on interaction and phosphorylation of autophagy receptors. Initially, we focused on the effects of TBK1 phosphorylation of OPTN, a well-studied autophagy receptor that is mutated in a small fraction of ALS patients (13, 37). We conducted coexpression experiments with Flag-tagged TBK1 mutations and OPTN-GFP and probed the interactions between TBK1 mutants with OPTN by Co-IPs. With notable exceptions, most Flag-tagged mutant TBK1 proteins coimmunoprecipitated OPTN-GFP (Fig. 6A). We confirmed loss of interaction between TBK1 E696K with OPTN (Fig. 6A) as previously reported (21), and we detected reduced interactions between TBK1 Y105C and TBK1 R308Q with OPTN (Fig. 6A). Unlike E696, Y105 and R308 are not located within the TBK1 CTD, which forms a stable interaction with the N terminus of OPTN (38). These residues are located relatively distant from the CTD, and therefore, mutations in these regions of the protein would not be expected to perturb the CTD. We conclude that stable TBK1–OPTN interaction in vivo involves regions of TBK1 more than the CTD.
Fig. 6.
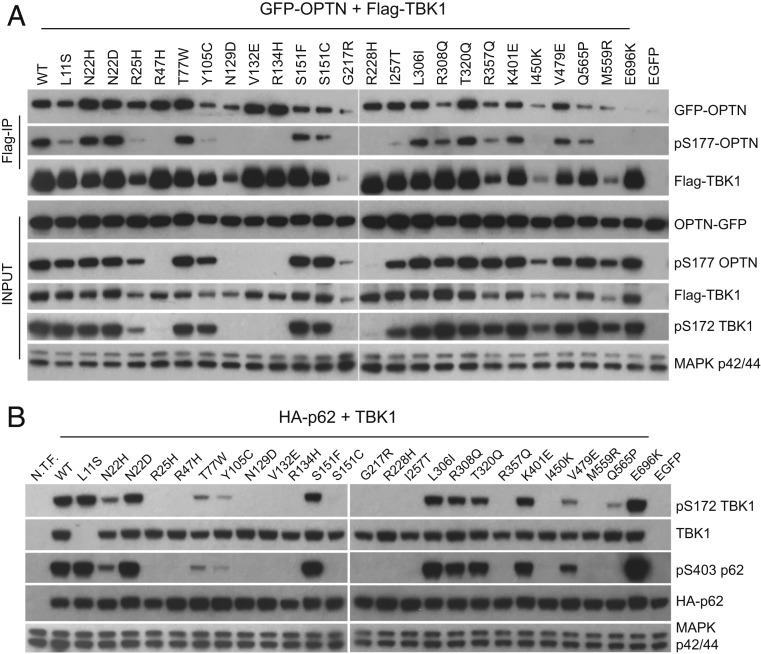
TBK1 mutations display different defects toward OPTN and p62. (A) Effects of TBK1 ALS mutations on its interaction and phosphorylation of OPTN. A GFP-tagged OPTN construct was coexpressed with various Flag-tagged TBK1 mutations in TBK1 knockout 293T cells; cell lysates were prepared 48 h later and precipitated with anti-Flag antibodies (M2). The interaction between TBK1 and GFP-OPTN was assayed by western blots with anti-Flag and GFP antibodies of the IP samples. The levels of pS177 OPTN in total lysates and IP samples and the levels of pS172 TBK1 in total lysates were determined by the anti-pS177 OPTN and anti-pS172 TBK1 antibodies, respectively. (B) Effects of TBK1 ALS mutations on the phosphorylation of p62. HA-p62 expression construct was cotransfected with various Flag-tagged TBK1 mutations into TBK1 knockout 293T cells. Total cell lysates were prepared 24 h after transfection and analyzed by western blots for the levels of phospho-S172 TBK1, phospho-S403 p62, total TBK1, and p62 by specific antibodies. TBK1 L11S mutant protein was not detected by the total TBK1 antibody due to epitope disruption. N.T.F., not transfected.
OPTN S177 has been shown to be specifically phosphorylated by TBK1, and this enhances interactions between OPTN and LC3 during selective autophagy (16). In general, OPTN S177 phosphorylation levels from whole-cell lysates in our coexpression experiments correlated with previously determined mutant TBK1 kinase activities (Fig. 1A), with the exception of TBK1 G217R, I450K, and M559R, which gained some kinase activity upon coexpression with OPTN (Fig. 6A). The E696K mutant, which disrupts OPTN binding, showed relatively high levels of total OPTN phosphorylation (Fig. 6A and SI Appendix, Fig. S2A), indicating that TBK1 binding is not required for OPTN phosphorylation. In a more detailed analysis in which phosphorylated OPTN was assayed exclusively within the Co-IP fraction, TBK1 mutants L11S, R25H, Y105C, and I257T precipitated a significantly lower fraction of phosphorylated OPTN, while OPTN coprecipitated with TBK1 I450K or M559R was not phosphorylated at S177. Thus, OPTN in these mutant TBK1–OPTN complexes serves as a poor substrate for the corresponding mutant TBK1. Since OPTN S177 phosphorylation enhances its interaction with ATG8 family proteins, such as LC3 (16), it is possible that these mutant TBK1–OPTN complexes have reduced affinity for ATG8 family protein on autophagosomes.
We also coexpressed TBK1 with p62/SQSTM1, another selective autophagy receptor implicated in ALS (39). We observed differences in p62 S403 phosphorylation, another established TBK1 target site with phosphorylation that enhances p62 binding to ubiquitinated proteins (36, 40). A significant decrease in p62 S403 phosphorylation was observed with the TBK1 N22H and S151C ALS mutant proteins compared with the same residue with different TBK1 ALS mutations (N22D and S151F) (Fig. 6B). A few TBK1 ALS mutations, including T77W, Y105C, R357Q, V479E, and Q565P, also showed selectively reduced phosphorylation of p62 (Fig. 6B). Unexpectedly, coexpression with p62 selectively inhibited S172 phosphorylation of these mutant TBK1 proteins as well (Fig. 6B). In vitro kinase assays confirmed that a majority of these mutant TBK1 proteins display reduced kinase activity toward p62 (SI Appendix, Fig. S4).
While the consequences of hypophosphorylated p62 or OPTN on autophagy are well documented (16, 40, 41), the fact that TBK1 mutations display phosphorylation defects toward specific substrates required for autophagy and innate immunity reveals the complexity of the role(s) of TBK1 in multiple cellular signaling pathways.
Discussion
TBK1 ALS Mutations Impact Multiple TBK1 Activities.
The analysis of TBK1 ALS mutations revealed a variety of defects in TBK1 activities (Table 1) ranging from kinase activity and substrate specificity to protein dimerization, protein stability, and interactions with autophagy adaptor proteins. A striking feature of these ALS mutations is that they can affect TBK1 at different levels. In the case of the most pathogenic familial mutations (R47H, G217R, R357Q, M559R, and E696K), 3 display nearly complete loss of kinase activity (R47H, G217R, and M559R), 2 fail to form TBK1 dimers (R357Q and M559R), and 1 (E696K) affects interactions with autophagy receptor proteins, such as OPTN. Since the kinase activity of TBK1 is essential for its function in multiple signaling pathways, loss of kinase activity will obviously have profound effects on cellular activities involving TBK1. In the case of monomeric TBK1 mutations, R357Q is active, but its steady-state level is lower than WT TBK1, and therefore, its activity toward certain substrates (such as IRF3, OPTN, and p62) is likely to be less efficient compared with a normal dimer. Mutations affecting the interaction and phosphorylation of autophagy receptors, such as OPTN and p62, also contribute to less efficient targeting of protein aggregates or damaged mitochondria during ALS disease progression. These observations reveal the complexity of ALS disease mechanisms, as multiple defects can be caused by mutations from a single disease-associated gene but can lead to the common pathogenic outcome. We note that not all mutations (sporadic mutations in particular) showed defects in the TBK1 activities that we studied. Additional assays, particularly those related to the nervous system, are required to better understand the mechanisms by which TBK1 mutations cause or contribute to ALS pathogenesis.
Our results support the hemizygous loss of function hypothesis of TBK1 mutations in ALS pathogenesis (21). Assays of heterodimers revealed that mutant TBK1 does not seem to inhibit the activity of WT TBK1, as none of the 25 missense mutations studied showed a dominant negative effect (Fig. 4 A–C). These results are significant, since all TBK1 mutations from ALS patients are heterozygous. TBK1 is known to play important roles in several critical signaling pathways, and partial reduction of TBK1 activity caused by ALS mutations can affect specific pathways during disease progression (ALS disease is typically late onset, ∼50 y). Consistent with this hypothesis, the loss of function of a single allele of TBK1 is sufficient to impact the ALS disease progression in a mouse model of ALS (42).
We found that TBK1 ALS mutations can differentially affect distinct downstream substrates/signaling pathways. This is particularly true for mutations in the KD. Using TBK1, IRF3, p62, and OPTN as substrates, we showed that R25H, R47H, R134H, and R228H were able to weakly phosphorylate TBK1 but not IRF3; T77W and S151C mutations also selectively affect phosphorylation of p62 but not OPTN. This selectivity is likely based on different sequences surrounding the site of phosphorylation, the accessibility of the target site within the native protein complex, and the differential soluble/insoluble cellular distribution of kinase and substrate. Our results identified sites within the KD that play roles in the access of distinct protein substrates. It will be interesting to determine whether different TBK1 ALS mutations can differentially affect disease progression in ALS animal models.
Mechanism of TBK1 Activation.
The functional studies of TBK1 ALS mutations described here also provide insights into the mechanism of TBK1 activation. For example, monomeric TBK1 can be active, as R357Q TBK1 phosphorylates TBK1, IRF3, and OPTN without forming a stable dimer with itself or an allelic TBK1 subunit. We also provide direct evidence that a TBK1 heterodimer (either formed between WT and mutant TBK1 subunits or an asymmetrical dimer with only 1 functional subunit phosphorylated at S172) can be active. These data suggest that dimer conformation is not required for TBK1 activation. Rather, TBK1 dimerization increases protein stability and improves the efficiency of kinase–substrate interaction, as heterodimers formed by 2 subunits bearing different mutations that only give rise to defective TBK1 homodimers can retain a low level of catalytic activity.
On the basis of these insights, we propose that the initial activation of TBK1 (S172 phosphorylation) under physiological conditions may require an upstream kinase (this is consistent with the finding that TBK1 small molecule inhibitors do not block S172 phosphorylation under many stimulated conditions [31, 32]), and phosphorylation of S172 on 1 subunit of the TBK1 dimer is sufficient to activate kinase activity. The asymmetric TBK1 dimer phosphorylated at 1 S172 can directly act on downstream signal-specific substrates, amplify the activation signal by phosphorylation of additional TBK1 dimers in the vicinity, or gain full activation (S172 phosphorylation on the other subunit) through this process. The nature of distinct signaling pathways, the strength of stimulation, the location of signaling complex assembly, and the involvement of other adaptor proteins all play a role in determining the outcome of TBK1 activity. In light of the important role of adaptor proteins in TBK1 signaling in innate immunity, Chen and colleagues (43) recently showed that STING, MAVS, and TRIF proteins all interact and also serve as substrates for TBK1 in their respective signaling pathways. Phosphorylated STING MAVS, and TRIF subsequently function as docking sites for TBK1 and its downstream substrates (43). It remains to be determined whether other signaling pathways involving TBK1 also use a similar mechanism to achieve specific and robust responses.
A common feature of ALS and other neurodegenerative diseases is disruption of proteostasis, the process by which the folding, trafficking, and degradation of proteins are controlled (44, 45). In most cases, the disruption of proteostasis is caused by mutations that lead to protein aggregation (e.g., SOD1, TDP-43, FUS) or in proteins required for protein turnover (e.g., TBK1, p62, OPTN). TBK1 functions at the heart of proteostasis as a critical component of autophagy, innate immunity, and the stress response. It, therefore, is not surprising that loss of function point mutations in TBK1 can cause familial ALS and associate with sporadic ALS. Here, we have taken a biochemical approach to study the effects of ALS-causing or -associated TBK1 missense mutations on the kinase function of TBK1. We provide mechanistic insights into TBK1 activation and substrate specificity. We provide evidence that dimerization is not required for TBK1 activation, consistent with the hypothesis that TBK1 can be activated by a yet to be identified protein kinase. We also provide additional evidence that the mechanism of TBK1 contribution to ALS is hemizygous loss of function rather than dominant gain of function. Finally, we show that ALS TBK1 KD mutations can selectively affect the phosphorylation of specific TBK1 target proteins. Considering that TBK1 target proteins function in distinct cellular pathways (autophagy, innate immunity, and stress response), it is not yet possible to identify the critical target(s) involved in ALS disease progression.
Numerous studies have shown that ALS is a multicellular disease, involving not only motor neurons but also, other cells in the spinal cord, including microglia, astrocytes, and oligodendrocytes (23, 46). Thus, TBK1 mutations not only affect multiple signaling pathways, they also likely differentially affect distinct spinal cord cell types. Studies of TBK1 provide an excellent model for understanding complex ALS disease mechanisms.
Materials and Methods
Missense ALS point mutations of TBK1 were generated from the pcDNA3-Flag-TBK1 expression construct by standard mutagenesis procedures. Mutations were identified from studies by Cirulli et al. (20) and Freischmidt et al. (21). A total of 25 mutations (5 from familial cases) were studied in this work. HA-tagged and GST-tagged WT or mutant TBK1 was generated by standard cloning. pOPTN-EGFP was a gift from Beatrice Yue (University of Illinois at Chicago, Chicago IL; Addgene plasmid 27052). HA-p62 plasmid was a gift from Qing Zhong (University of California, Berkeley, CA; Addgene plasmid 28027). Recombinant IRF3 protein was purified from 293T cells transfected with pcDNA3-Flag-IRF3 (human) by anti-Flag beads and eluted by Flag peptide competition. Recombinant OPTN was similarly purified by transfecting cells with the pLV-EF1a-OPTN-HA construct followed by anti-HA immunoprecipitation and elution with HA peptide. Recombinant GST-p62 was purchased from Enzo Life Sciences. Conditions of cell culture, the generation of TBK1 knockout cells, transfection and luciferase assays, Co-IP and western blots, and other experimental procedures can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Javier Coloma and Sarah Dugger for technical assistance. This work was supported by grants from Takeda Pharmaceutical Company (Project 1000344702) and Project ALS (PRALS 2017-09).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915732116/-/DCSupplemental.
References
- 1.Ahmad L., Zhang S. Y., Casanova J. L., Sancho-Shimizu V., Human TBK1: A Gatekeeper of neuroinflammation. Trends Mol. Med. 22, 511–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helgason E., Phung Q. T., Dueber E. C., Recent insights into the complexity of Tank-binding kinase 1 signaling networks: The emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett. 587, 1230–1237 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Herhaus L., Dikic I., Expanding the ubiquitin code through post-translational modification. EMBO Rep. 16, 1071–1083 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu D., et al. , TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnard M., et al. , Deficiency of T2K leads to apoptotic liver degeneration and impaired NF-kappaB-dependent gene transcription. EMBO J. 19, 4976–4985 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pomerantz J. L., Baltimore D., NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 18, 6694–6704 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald K. A., et al. , IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496 (2003). [DOI] [PubMed] [Google Scholar]
- 8.McWhirter S. M., et al. , IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 101, 233–238 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S., et al. , Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Akira S., Uematsu S., Takeuchi O., Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Wu J. X., Chen Z. J., “Innate immune sensing and signaling of cytosolic nucleic acids” in Annual Review of Immunology, Littman D. R., Yokoyama W. M., Eds. (Annual Reviews, Palo Alto, CA, 2014), vol. 32, pp. 461–488. [DOI] [PubMed] [Google Scholar]
- 12.Honda K., Takaoka A., Taniguchi T., Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Dikic I., “Proteasomal and autophagic degradation systems” in Annual Review of Biochemistry, Kornberg R. D., Ed. (Annual Reviews, Palo Alto, CA, 2017), vol. 86, pp. 193–224. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N., Komatsu M., Autophagy: Renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Richter B., et al. , Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. U.S.A. 113, 4039–4044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wild P., et al. , Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herman M., et al. , Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 209, 1567–1582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Awadalla M. S., et al. , Copy number variations of TBK1 in Australian patients with primary open-angle glaucoma. Am. J. Ophthalmol. 159, 124–30.e1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritch R., et al. , TBK1 gene duplication and normal-tension glaucoma. JAMA Ophthalmol. 132, 544–548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirulli E. T., et al. ; FALS Sequencing Consortium , Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freischmidt A., et al. , Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat. Neurosci. 18, 631–636 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Freischmidt A., Müller K., Ludolph A. C., Weishaupt J. H., Andersen P. M.; Association of Mutations in TBK1 With Sporadic and Familial Amyotrophic Lateral Sclerosis and Frontotemporal Dementia , Association of mutations in TBK1 with sporadic and familial amyotrophic lateral sclerosis and frontotemporal dementia. JAMA Neurol. 74, 110–113 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Taylor J. P., Brown R. H. Jr, Cleveland D. W., Decoding A. L. S., Decoding ALS: From genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oakes J. A., Davies M. C., Collins M. O., TBK1: A new player in ALS linking autophagy and neuroinflammation. Mol. Brain 10, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larabi A., et al. , Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 3, 734–746 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Tu D., et al. , Structure and ubiquitination-dependent activation of TANK-binding kinase 1. Cell Rep. 3, 747–758 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shu C., et al. , Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure 21, 1137–1148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma X., et al. , Molecular basis of Tank-binding kinase 1 activation by transautophosphorylation. Proc. Natl. Acad. Sci. U.S.A. 109, 9378–9383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu G., et al. , Crystal structure of inhibitor of κB kinase β. Nature 472, 325–330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishore N., et al. , IKK-i and TBK-1 are enzymatically distinct from the homologous enzyme IKK-2: Comparative analysis of recombinant human IKK-i, TBK-1, and IKK-2. J. Biol. Chem. 277, 13840–13847 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Clark K., et al. , Novel cross-talk within the IKK family controls innate immunity. Biochem. J. 434, 93–104 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Heo J.-M., Ordureau A., Paulo J. A., Rinehart J., Harper J. W., The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell 60, 7–20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Servant M. J., et al. , Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278, 9441–9447 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Thanos D., Maniatis T., Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Liu S., et al. , Crystal structure of a human IκB kinase β asymmetric dimer. J. Biol. Chem. 288, 22758–22767 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilli M., et al. , TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37, 223–234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markovinovic A., et al. , Optineurin in amyotrophic lateral sclerosis: Multifunctional adaptor protein at the crossroads of different neuroprotective mechanisms. Prog. Neurobiol. 154, 1–20 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Li F., et al. , Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins. Nat. Commun. 7, 12708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iguchi Y., Katsuno M., Ikenaka K., Ishigaki S., Sobue G., Amyotrophic lateral sclerosis: An update on recent genetic insights. J. Neurol. 260, 2917–2927 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto G., Wada K., Okuno M., Kurosawa M., Nukina N., Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto G., Shimogori T., Hattori N., Nukina N., TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum. Mol. Genet. 24, 4429–4442 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Brenner D., et al. , Heterozygous Tbk1 loss has opposing effects in early and late stages of ALS in mice. J. Exp. Med. 216, 267–278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S., et al. , Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Labbadia J., Morimoto R. I., The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84, 435–464 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hipp M. S., Park S. H., Hartl F. U., Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Burda J. E., Sofroniew M. V., Reactive gliosis and the multicellular response to CNS damage and disease. Neuron 81, 229–248 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.