Significance
Multisubunit motor nucleases recognize, unwind, and resect DSB ends during homologous recombination in bacteria. RecBCD, AddAB, and AdnAB exemplify three distinct clades of end-resection machines in different bacterial taxa. AdnAB is thought to represent an ancestral state of the machine. We present here cryo-EM structures of mycobacterial AdnAB, in the absence of DNA and bound to a forked duplex DNA, that illuminate its domain architecture, the structural basis for 5′ strand cleavage by the AdnA nuclease, and DNA interactions of the AdnB motor that ensure unidirectional translocation. We employ DNA curtain technology to study DSB resection dynamics on a single-molecule level. Our findings add to an emerging appreciation of the evolutionary path and functional diversity of bacterial DSB resection systems.
Keywords: homologous recombination, DNA end resection, cryoelectron microscopy, DNA curtain
Abstract
Mycobacterial AdnAB is a heterodimeric helicase–nuclease that initiates homologous recombination by resecting DNA double-strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N-terminal motor domain and a C-terminal nuclease domain. Here we report cryoelectron microscopy (cryo-EM) structures of AdnAB in three functional states: in the absence of DNA and in complex with forked duplex DNAs before and after cleavage of the 5′ single-strand DNA (ssDNA) tail by the AdnA nuclease. The structures reveal the path of the 5′ ssDNA through the AdnA nuclease domain and the mechanism of 5′ strand cleavage; the path of the 3′ tracking strand through the AdnB motor and the DNA contacts that couple ATP hydrolysis to mechanical work; the position of the AdnA iron–sulfur cluster subdomain at the Y junction and its likely role in maintaining the split trajectories of the unwound 5′ and 3′ strands. Single-molecule DNA curtain analysis of DSB resection reveals that AdnAB is highly processive but prone to spontaneous pausing at random sites on duplex DNA. A striking property of AdnAB is that the velocity of DSB resection slows after the enzyme experiences a spontaneous pause. Our results highlight shared as well as distinctive properties of AdnAB vis-à-vis the RecBCD and AddAB clades of bacterial DSB-resecting motor nucleases.
The repair of DNA double-strand breaks (DSBs) in bacterial chromosomes is essential for bacteria to thrive. Mycobacteria, including the human TB pathogen Mycobacterium tuberculosis and its avirulent relative Mycobacterium smegmatis, deploy three distinct DSB repair mechanisms: RecA-dependent homologous recombination (HR), RecA-independent single-strand annealing, and Ku/LigD-dependent nonhomologous end joining (1–3). Among these repair pathway options, only HR ensures faithful repair of the damaged chromosome without mutation at the DSB site. Bacterial HR initiates when a multisubunit helicase–nuclease machine unwinds and resects the DSB ends to generate 3′ tailed single-strand DNA (ssDNA) that is initially bound by single-strand DNA binding protein (SSB). SSB is subsequently replaced by RecA, the DNA strand exchange protein that performs the homology search and strand invasion steps of the HR pathway (4).
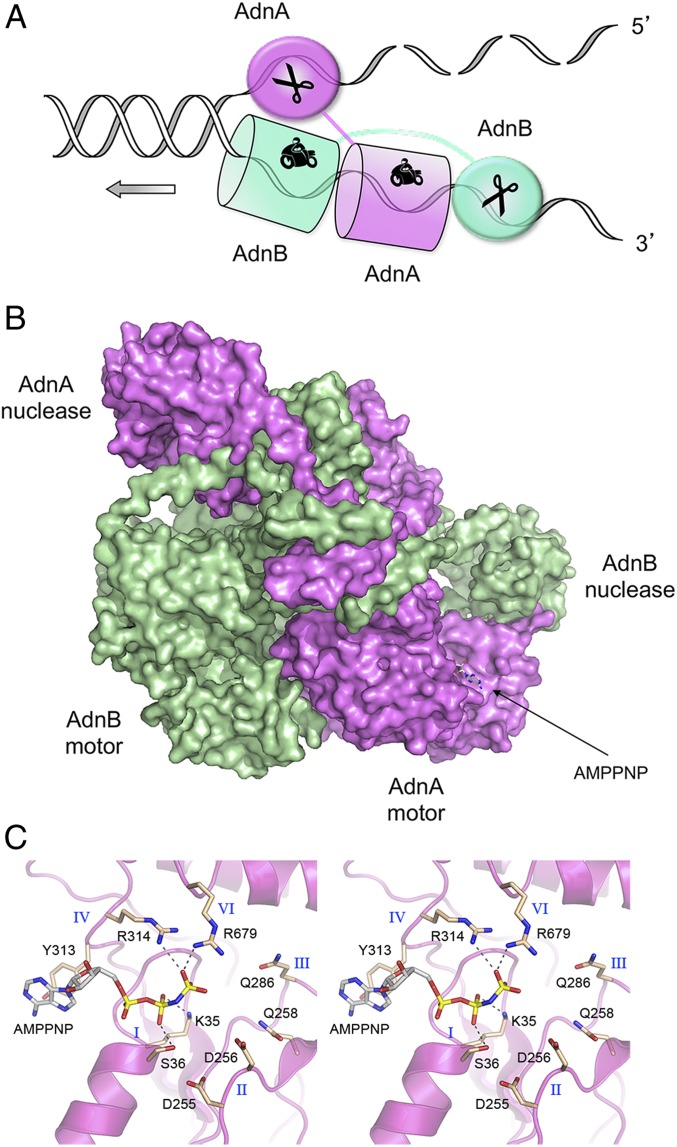
Bacterial DSB-resecting helicase–nucleases come in three flavors. In Escherichia coli, resection is initiated by the heterotrimeric RecBCD enzyme (5), whereas in Bacillus and Mycobacterium, this function is performed by the heterodimeric enzymes AddAB (6) and AdnAB (7), respectively. The mycobacterial AdnA and AdnB subunits, encoded in a two-gene operon, are each composed of an N-terminal UvrD-like superfamily I helicase motor domain and a C-terminal RecB-like nuclease module (7) (SI Appendix, Fig. S1A). Purified AdnAB displays DNA-dependent ATPase, double-strand DNA (dsDNA) helicase, 3′-to-5′ ssDNA translocase, ssDNA nuclease, DSB end-binding, and DSB end-resection activities in vitro (7–10). Analyses of the effects of inactivating mutations in the nuclease and ATPase active sites on these activities revealed a division of labor between the two subunits, and a working model for DSB resection. In brief, ATP-independent binding of AdnAB to a DSB end melts a short segment of the duplex and allows access of the 5′ terminal dinucleotide to the AdnA nuclease-active site. ATP hydrolysis catalyzed by the AdnB motor then drives translocation of the two tandem motor domains in the 3′-to-5′ direction along the same DNA strand, with the “leading” AdnB motor domain in the vanguard (Fig. 1A). Unwinding of the DNA duplex by the advancing tandem motor “pumps” the 3′ ssDNA strand into the AdnB nuclease domain and simultaneously threads the 5′ ssDNA strand through the AdnA nuclease domain (7–10). Bacillus AddAB displays an analogous division of labor whereby its two C-terminal nuclease modules each digest one of the single strands displaced by the 3′-to-5′ motor (11, 12). AdnAB and AddAB differ in that the “lagging” motor domain in AdnAB has a seemingly intact ATPase active site, whereas that of AddAB lacks several functional groups needed for ATP hydrolysis.
Fig. 1.
AdnAB helicase–nuclease. (A) Model for division of labor during AdnAB unwinding and resection at a DNA double-strand break. (B) Surface model of the AdnA(D934A)-AdnB(D1014A) heterodimer. AdnA is colored magenta and AdnB is colored green. AMPPNP bound to AdnA is depicted as a stick model. (C) Stereoview of the AMPPNP-binding site in the AdnA subunit. Amino acids from conserved superfamily I ATPase motifs I, II, III, IV, and VI are shown as stick models with beige carbons.
RecBCD is composed of a single nuclease module (in RecB) that catalyzes digestion of either displaced strand, a 3′-to-5′ helicase (in RecB, analogous to the lead motors in AddAB and AdnAB), a vestigial inactive motor-like domain (in RecC, analogous to the lagging motor domains of AddAB and AdnAB), a vestigial inactive nuclease module (in RecC, analogous to the 5′ processing nuclease in AddAB and AdnAB), and a distinctive 5′-to-3′ RecD helicase subunit not found in AddAB or AdnAB (13–16).
An evolutionary scheme has been suggested whereby: 1) AdnAB exemplifies an ancestral state of the bacterial end resection machine, composed of two motors and two nucleases; 2) AddAB arose from an AdnAB-like precursor via loss of function of its lagging motor (while repurposing the inactive motor to recognize a χ sequence in the tracking strand that controls nuclease activity); and 3) RecBCD arose from an AddAB-like enzyme by degeneration of one of the nuclease modules, further erosion of the vestigial RecC motor leading to complete loss of the canonical ATPase motifs, and gain of a helicase subunit (RecD) that translocates on the displaced 5′ strand (8, 17). This scheme is supported by structural analyses, via X-ray crystallography or cryoelectron microscopy (cryo-EM), of E. coli RecBCD and Bacillus AddAB in complexes with DNA that exemplify several functional states along the DSB recognition and processing pathways (13–15, 18, 19).
Here we report cryo-EM structures of M. smegmatis AdnAB, in the absence of DNA and bound to a duplex DNA with 5′ and 3′ tails. These structures reveal: 1) the path of the displaced 5′ strand and its interactions with the AdnA nuclease; 2) the path of the 3′ strand within the AdnB motor and the nature of the protein ratchet that ensures unidirectional translocation; and 3) presence of AMPPNP in an apparently complete ATPase active site in the AdnA subunit. We also employ single-molecule DNA curtain methods (20) to gauge the kinetic behavior of AdnAB and the role of the AdnA motor in DSB end resection in vitro.
Results
Cryo-EM Structure of AdnAB.
An initial experiment aimed at capturing AdnAB in complex with DNA was performed by preincubating a helicase-active, nuclease-inactive mutant heterodimer AdnA(D934A)-AdnB(D1014A) (7) with a forked DNA substrate (comprising a 19-bp duplex with a 5-nucleotide hairpin on one end and a 6-nucleotide 5′ tail and a 21-nucleotide 3′ tail on the other end) in the presence of 1 mM AMPPNP (adenylyl-imidodiphosphate) and 3 mM magnesium. The mixture was subjected to gel filtration and the peak AdnAB fraction (which contained DNA, as gauged by the A260/280 profile) was applied to grids for cryo-EM analysis. Sequential 2D and 3D classification (SI Appendix, Fig. S2) highlighted distinct DNA-free and DNA-bound particle populations. An ensemble of 80,396 particles (28% of total 3D class) was used to generate a model of the DNA-free AdnAB structure at an overall resolution of 3.5 Å (SI Appendix, Table S1). The model is shown in surface view in Fig. 1B with AdnA colored magenta and AdnB colored green. The N-terminal motor domains and C-terminal nuclease domains are arranged in the quaternary structure as hypothesized (Fig. 1A). AMPPNP was bound in the putative ATPase active site of the AdnA motor domain (Fig. 1B) but was absent from the AdnB subunit.
The AdnA Motor Domain Has an Intact Site for ATP Binding and Hydrolysis.
A detailed view of the complex of AMPPNP with the AdnA motor domain is shown in Fig. 1C. The AdnA nucleotide-binding pocket is formed by constituents of ATPase motifs I, II, III, IV, and VI that are conserved in E. coli UvrD and other superfamily I helicases (21) (SI Appendix, Fig. S1A). The β-phosphate is engaged by Lys35 and Ser36 of motif I (the P loop). The γ-phosphate is coordinated by Arg314 (motif IV) and Arg679 (motif VI). The adenine base makes a π-stack on Tyr313 (motif IV) (Fig. 1C). Although we did not see a divalent cation in the AdnA•AMPPNP complex, Asp255, Asp256, and Gln258 of the conserved metal-binding motif II were positioned near the β- and γ-phosphates (Fig. 1C), consistent with the disposition of motif II in the structure of UvrD in complex with ADP•MgF3 (a mimetic of the penta-coordinate phosphorane transition state) and the magnesium cofactor for ATP hydrolysis (21) (SI Appendix, Fig. S1B). Indeed, the composition of the AMPPNP site in the AdnA motor domain closely resembles that seen in the UvrD structure (compare Fig. 1C and SI Appendix, Fig. S1B). The corresponding domain of Bacillus AddAB binds AMPPNP•Mg2+ via intact motifs I and IV and the first aspartate of motif II, but it lacks the motif VI arginine and the second carboxylate side chain of motif II that are essential for ATP hydrolysis (19). The second carboxylate of motif II and the glutamine of motif III, both of which are conserved in AdnA (Fig. 1C, Asp256 and Gln286), promote ATP hydrolysis by orienting and activating the water nucleophile for its attack on the γ-phosphate (SI Appendix, Fig. S1B).
Structure of AdnAB in Complex with a Forked Duplex DNA.
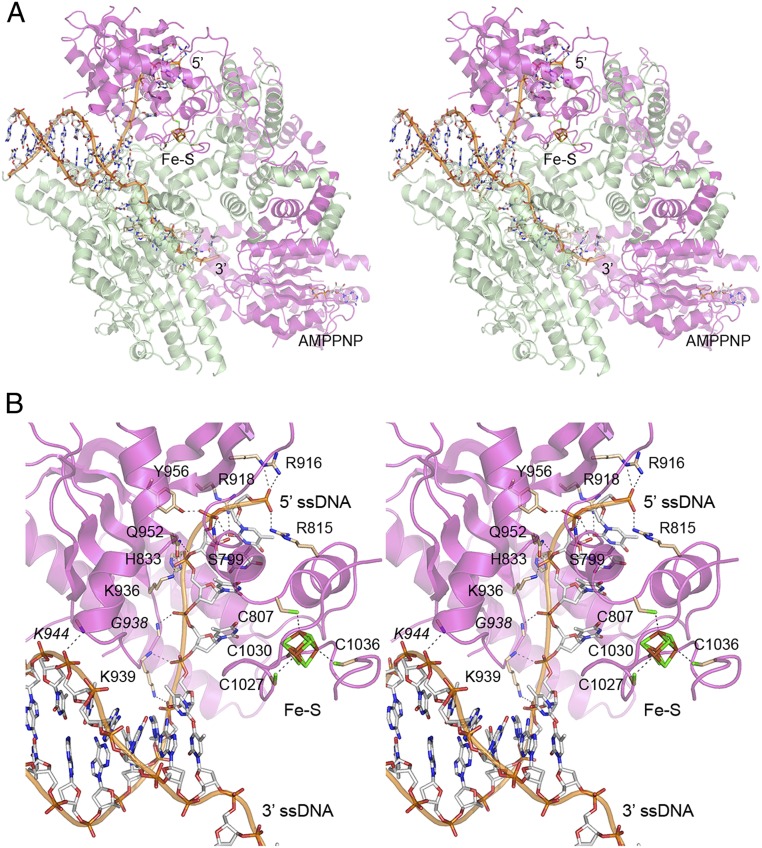
A set of 61,579 particles (21% of total 3D class) (SI Appendix, Fig. S2) was used to generate a model of the DNA-bound AdnA(D934A)-AdnB(D1014A) heterodimer at an overall resolution of 3.5 Å (SI Appendix, Table S1). The structure is shown in Fig. 2A with the protein subunits depicted as semitransparent cartoon models and the DNA as a stick model with a cartoon trace through the phosphodiester backbone. AMPPNP was present in the AdnA motor domain, but not in the AdnB motor domain. (The AdnB nuclease domain was not visible in the DNA-bound complex.) A portion of the input forked DNA was modeled, comprising the 10-bp duplex segment at the junction from which a 4-nucleotide 5′ tail and an 8-nucleotide 3′ tail emanate. The terminal 2 nucleotides of the 5′ single strand and the terminal 13 nucleotides of the 3′ single strand were not visible. The 3′ single strand tail completely traverses the AdnB motor domain such that the last visible 3′ nucleotide is oriented toward the AdnA motor (Fig. 2A). The trajectory of the 5′ single strand tail is nearly orthogonal to the duplex axis and the path of the 3′ tail. The 5′ tail runs through the AdnA nuclease domain. A Fe4S4 iron–sulfur cluster in the AdnA subunit organizes the C-terminal segment into a subdomain that insinuates into the fork junction (Fig. 2A, shown in detail in Fig. 2B), where it can act as a wedge to pry the strands apart as the AdnB motor tracks along the 3′ tail.
Fig. 2.
AdnAB complex with forked duplex DNA. (A) Stereoview of the AdnA(D934A)-AdnB(D1014A) heterodimer (semitransparent cartoon model; magenta AdnA and green AdnB) in complex with forked duplex DNA (stick model with cartoon trace through the phosphodiester backbone). The 5′ and 3′ ssDNA ends are indicated. The iron–sulfur cluster (Fe–S) in the AdnA nuclease domain and AMPPNP in the AdnA motor-like domain are shown as stick models. (B) Detailed stereoview of the AdnA nuclease and its interface with the fork junction and the 5′ ssDNA. Amino acids are rendered as stick models with beige carbons; DNA contacts are indicated by dashed lines. The Fe4S4 cluster and four cysteine ligands in AdnA are shown as stick models with green sulfur atoms.
Interface of the AdnA Nuclease Domain with the Duplex Junction and Displaced 5′ ssDNA.
A stereoview of the AdnA–DNA interface in Fig. 2B highlights atomic contacts to the first six phosphates of the 5′ strand (5′-pT1pT2pT3pT4pT5pC6) that includes the four-nucleotide ssDNA tail and the two phosphates of the duplex segment of the 5′ strand at the dsDNA–ssDNA junction. The terminal T1 phosphate is coordinated by the Arg815 and Arg916 side chains. The penultimate T2 phosphate receives hydrogen bonds from the Ser799 and Tyr956 side chains. The adjacent T3 phosphate is engaged by the His833, Lys936, and Gln952 side chains. The Gly938 main chain amide donates a hydrogen bond to the T4 phosphate. The T5 and C6 phosphates of the junction duplex are engaged by the Lys939 main chain amide and Lys939 side chain Nζ, respectively. The Arg918 side chain makes a hydrogen bond to the O2 atom of the T1 nucleobase. AdnA’s interaction with the 3′ strand is limited to a single hydrogen bond from the Lys944 main chain amide to a phosphate in the dsDNA segment situated four nucleotides upstream of the ds–ss junction. These AdnA–DNA contacts are summarized in cartoon form in Fig. 3B. It is noteworthy that the Ser799, Arg815, His833, Arg916, Arg918, Lys936, Gly938, Gln952, and Tyr956 residues of AdnAB that contact the 5′ ssDNA strand are all conserved in the nuclease domain of AdnB (SI Appendix, Fig. S1A), suggesting that the AdnB nuclease interface with the 3′ ssDNA tracking strand after it passes through the AdnA motor is likely to resemble that of AdnA with the 5′ ssDNA strand. Previous mutational analyses showed that AdnA Lys936 is essential for the AdnA nuclease activity of the AdnAB heterodimer and that the corresponding AdnB Lys side chain is essential for the AdnB nuclease activity (7). By contrast, the AdnA nuclease Lys939 side chain that interacts with the duplex junction is conspicuously not conserved in AdnB (which has an aspartate at the equivalent position); this is consistent with the fact that the AdnB nuclease has no call to recognize a duplex junction or interact with duplex DNA.
Fig. 3.
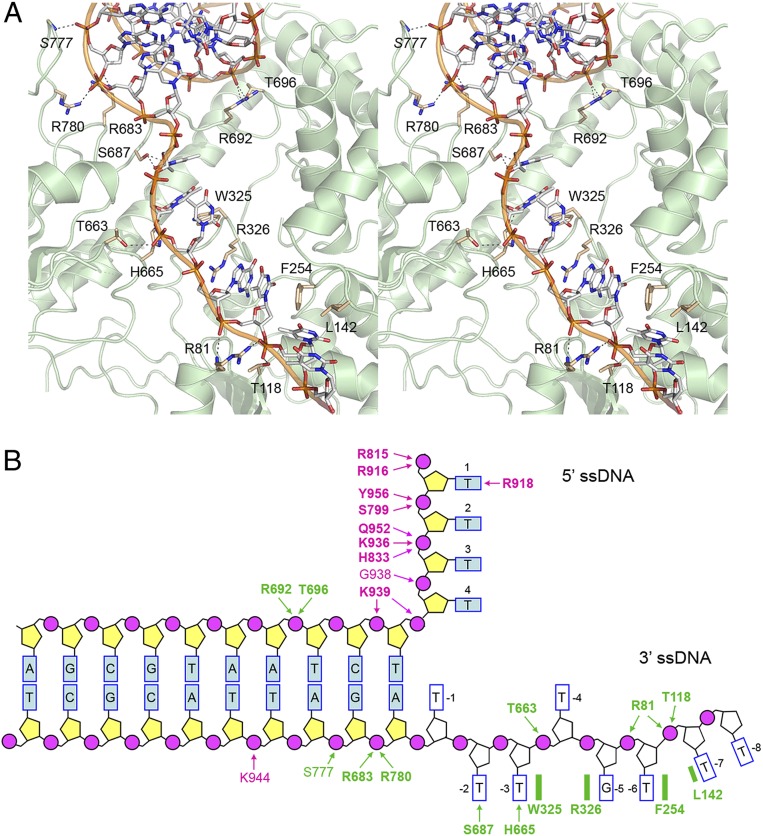
AdnAB motor domain interface with the fork junction and 3′ ssDNA tracking strand. (A) Detailed stereoview of the AdnB–DNA interaction. DNA is depicted as a stick model with gray carbons. Amino acids are rendered as stick models with beige carbons. DNA contacts are indicated by dashed lines. (B) Cartoon summary of DNA contacts in the complex with the AdnA(D934A)-AdnB(D1014A) heterodimer. Amino acids in the AdnA subunit are colored magenta; amino acids in the AdnB subunit are colored green. Protein main chain contacts are shown in plain font. Protein side chain contacts are in bold font.
Interface of the AdnB Motor Domain with the Duplex Junction and 3′ ssDNA Tracking Strand.
A stereoview of the AdnB–DNA interface in Fig. 3A shows an extensive network of atomic contacts to the phosphates and nucleobases of the eight-nucleotide 3′ ssDNA tail, to the two phosphates of the duplex segment of the 3′ strand at the ssDNA–dsDNA junction, and to a single phosphate of the duplex portion of the 5′ strand three nucleotides upstream of the dsDNA–ssDNA junction. The contacts are depicted in cartoon form in Fig. 3B. The 3′ tail of the tracking DNA strand (5′-pT−1pT−2pT−3pT−4pG−5pT−6pT−7pT−8-3′) is partitioned by the AdnB motor into three distinct base-stacked segments by virtue of insertion of amino acids that stack on the nucleobases and splay apart the nucleosides (Fig. 3 A and B). In particular, the Trp325 indole ring forms a π-stack on the T−3 base and the Arg326 guanidinium forms a π-cation stack on the G−5 base and, in doing so, they flip out the T−4 nucleoside. Further downstream, the Phe254 aromatic ring makes a π-stack on the T−6 base and the Leu142 side chain stacks on the T−7, thereby forming a wedge between the −6 and −7 nucleotides. (As discussed in a section below, we hypothesize that these base-stacking interactions provide a ratchet to ensure unidirectional translocation of the AdnB motor.) T−1, the first unpaired base of the 3′ strand flanking the duplex junction, remains stacked on adenine of the T:A base pair at the junction (Fig. 3B). The Ser687 side chain makes a bifurcated hydrogen bond to O2 and O4 of the T−2 base and the His665 side chain donates a hydrogen bond to O2 of the T−3 base. The T−6 and T−7 phosphates are engaged by the Arg81 main chain amide and side chain guanidinium, respectively. The T−7 phosphate also receives a hydrogen bond from the Thr118 side chain. The phosphate of the flipped out T−4 nucleotide makes a hydrogen bond with the Thr663 side chain.
Seven of the 3′ tail-binding amino acids of the AdnB motor are constituents of superfamily I helicase motifs. All 7 are conserved in UvrD (21) and 5/7 are conserved in the AdnA motor-like domain (SI Appendix, Fig. S1A). They include: Thr118 in motif Ib (corresponding to Thr78 in AdnA); Trp325 and Arg326 in motif III (Tyr291 and Arg292 in AdnA); Thr663 and His665 in motif V (Ser607 and His609 in AdnA); Phe254 in motif Id (replaced by alanine in AdnA); and Ser687 in motif Va (replaced by asparagine in AdnA). AdnB Leu142 is conserved as isoleucine in UvrD and leucine in AdnA but is not part of a heretofore identified helicase motif (SI Appendix, Fig. S1A). AdnB 3′ tail-binding residue Arg81 (nominally part of motif Ia) is not conserved in either UvrD or AdnA (SI Appendix, Fig. S1A).
Five amino acids of the AdnB motor domain make contacts with the duplex DNA junction (Fig. 3 A and B). The Arg683 and Arg780 side chains engage the first 3′ strand phosphate of the junction duplex while the S777 main chain amide makes a hydrogen bond to the second phosphate of the duplex. The Arg692 and Thr696 side chains coordinate a 5′ strand phosphate of the duplex located three nucleotides upstream of the dsDNA–ssDNA junction. Arg692 and Thr696 reside in a segment of the AdnB motor that has no counterpart in UvrD (SI Appendix, Fig. S1B). Arg692 is conserved in AdnA but Thr696 is not. Arg683 and Arg780 are not conserved in UvrD or AdnA (SI Appendix, Fig. S1B). We surmise that AdnB relies on unique structural elements to engage the duplex junction that are not present in UvrD or AdnA. The lack of conservation of the junction-binding residues in AdnA is sensible, given that AdnA is poised to receive the ssDNA tracking strand that is pumped through the AdnB motor and therefore has no cause to recognize dsDNA.
AdnAB in Complex with Forked DNA Subsequent to 5′ Cleavage by AdnA Nuclease.
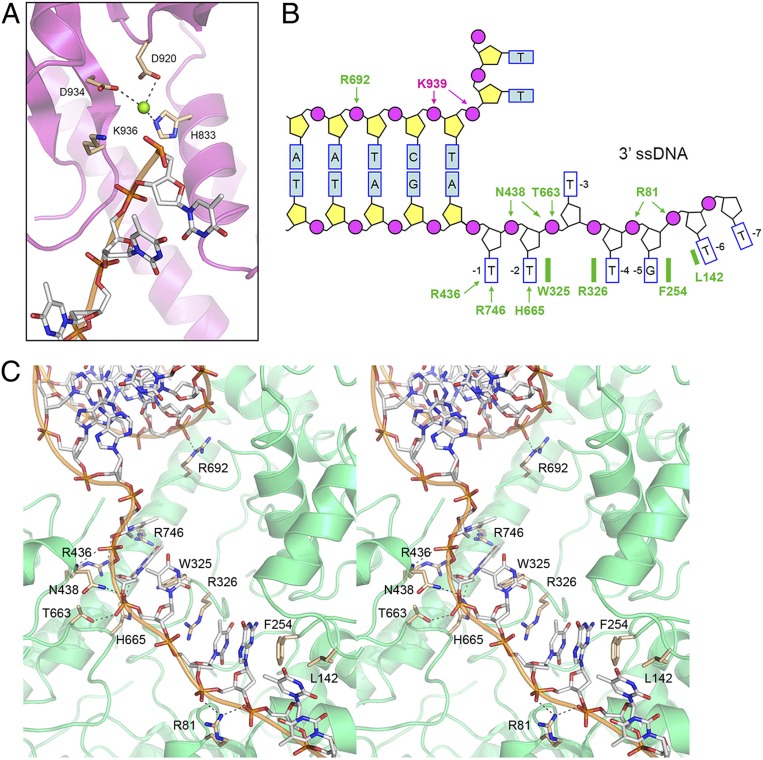
In a second series of cryo-EM experiments, we focused on a complex of wild-type (WT) AdnAB with the forked duplex DNA ligand prepared in the presence of AMPPNP and magnesium. A set of 60,108 particles was used to generate a model of the DNA-bound wild-type AdnAB heterodimer at an overall resolution of 3.56 Å (SI Appendix, Table S1). As in the previous DNA complex, we did not visualize the AdnB nuclease domain. The N terminus of the AdnA motor domain was not visible, though AMPPNP was present in the AdnA ATPase site (albeit not in the AdnB ATPase site). The forked DNA comprised the same 10-bp duplex segment seen in the nuclease-inactive AdnAB–DNA complex, but the 5′ ssDNA tail within the AdnA nuclease domain was two nucleotides shorter in the wild-type complex (Fig. 4A) than in the nuclease-inactive AdnAB structure. Moreover, we saw that the wild-type AdnA subunit now had a magnesium ion in the active site of the AdnA nuclease domain, situated adjacent to the terminal 5′ phosphate of the ssDNA tail (Fig. 4A). The magnesium is coordinated by three amino acid side chains: His833, Asp920, and Asp934 (Fig. 4A). We surmise that the 5′ ssDNA tail of the input forked DNA was incised by the AdnA nuclease module of the wild-type AdnAB in the presence of magnesium and that the structure observed with respect to the AdnA nuclease-active site reflects that of a postcleavage product complex. In the nuclease-inactive AdnAB–DNA complex, the 5′ strand was uncleaved and magnesium was not visible in the active site because one of the metal ligands, Asp934, had been mutated to alanine.
Fig. 4.
AdnAB in complex with forked DNA after 5′ cleavage by AdnA nuclease and with an alternative conformation of the 3′ tracking strand. (A) View of the AdnA nuclease domain in the complex of wild-type AdnAB with forked duplex DNA. A Mg2+ ion (green sphere) is present in the nuclease active site and the 5′ ssDNA is shortened by two nucleotides compared to the nuclease-inactive AdnAB–DNA complex, indicating that the 5′ strand was cleaved. Amino acids that bind the metal cofactor adjacent to the scissile phosphodiester (the terminal 5′ phosphate in the postcleavage complex) are shown as stick models. Lys936 is imputed to interact with the scissile phosphodiester and stabilize the transition state. (B) Cartoon summary of DNA contacts in the complex with the wild-type AdnAB heterodimer. Amino acids in the AdnA subunit are colored magenta; amino acids in the AdnB subunit are colored green. Protein side chain contacts are in bold font. Note that the position and base-stacking patterns of the nucleosides of the 3′ ssDNA strand in the wild-type AdnAB complex shown here differ from those in the nuclease-inactive AdnAB complex shown in Fig. 3B. (C) Detailed stereoview of the AdnB–DNA interaction. DNA is depicted as a stick model with gray carbons and a cartoon trace through the phosphodiester backbone. Amino acids are rendered as stick models with beige carbons. DNA contacts are indicated by dashed lines.
Comparison of precleavage (nuclease-inactive) and post-5′ cleavage (wild-type) structures pinpoints the scissile phosphodiester in the 5′-pT1pT2pT3pT4pT5pC6 strand (highlighted in bold font) and indicates that AdnA incises the 5′ tail two nucleotides away from the duplex junction. AdnA Lys939 retains its contacts to the phosphates at the duplex junction (Fig. 4B). The structures suggest a mechanism of phosphodiester hydrolysis by AdnA nuclease whereby the metal ion and the essential Lys936 side chain (situated next to the 5′ terminal phosphate in the product complex; Fig. 4A) interact with the scissile phosphate and stabilize a penta-coordinate phosphorane transition state of the nuclease reaction.
An Alternative Conformation of the 3′ ssDNA Tail in the AdnB Motor.
A stereoview of the 3′ ssDNA interface with the AdnB motor in the wild-type AdnAB–DNA complex is shown in Fig. 4C and summarized in cartoon form in Fig. 4B. The disposition of the 3′ ssDNA within the AdnB motor domain was different vis-à-vis the nuclease-inactive AdnAB–DNA complex (affirmed by the density maps shown in SI Appendix, Fig. S3). Specifically, the ssDNA nucleotides are shifted forward within the AdnB motor by a 1-nucleotide step, such that the T−1 base is no longer stacked on the duplex junction (as in the nuclease-inactive AdnAB complex) but rather now occupies the position previously taken by the T−2 base. T−1 has acquired new hydrogen bond contacts to O2 and O5 from Arg436 (a conserved motif IVa side chain; SI Appendix, Fig. S1A) and Arg746, respectively. Consequently, Trp325 now forms a π-stack on the T−2 base, Arg326 forms a π-cation stack on the T−4 base, and, the T−3 nucleoside is flipped out. Phe254 makes a π-stack on the G−5 base and Leu142 stacks on the T−6 base (Fig. 4 B and C). In effect, the protein side chains that splay apart the bases between base-stacked segments of the tracking strand stay in place, while the tracking strand itself can toggle between 1-nucleotide changes in register. Although we cannot correlate this shift with the ATPase cycle of AdnAB motor (i.e., because we do not see AMPPNP in the AdnB active site), we presume that such changes do indeed occur during translocation of the AdnB motor on the tracking strand.
AdnB Trp325-Arg326 Couples ATP Hydrolysis to DNA Translocation.
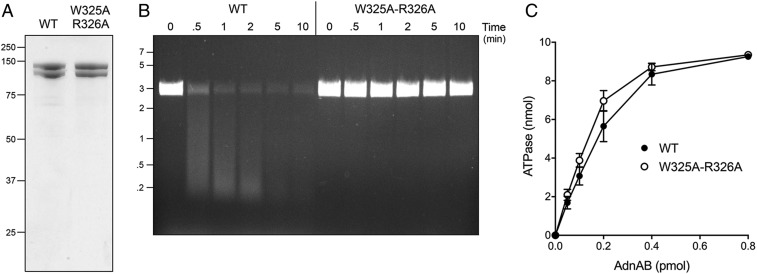
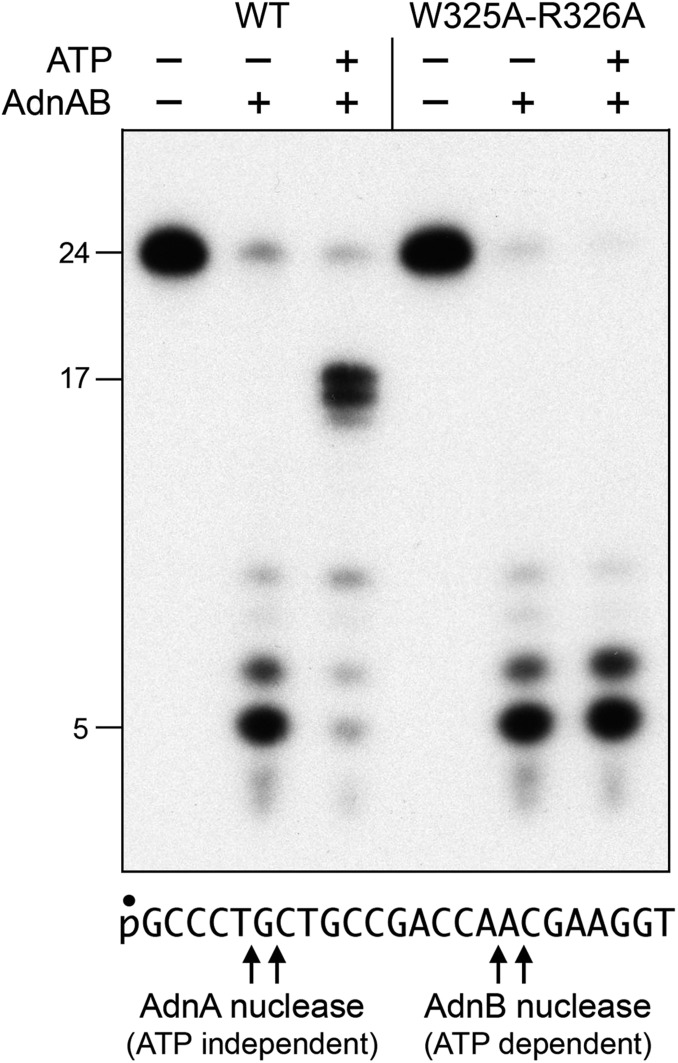
Chemomechanical coupling by ssDNA translocating helicases is driven by iterative protein domain motions synchronized to the steps of the ATP hydrolytic cycle. ATP hydrolysis is mechanically productive when the protein domain motions ratchet the DNA tracking strand forward in one direction, typically by a single-nucleotide step. For AdnAB, we envision (based on the cryo-EM structures) that the step motion of the tracking strand through the AdnB motor entails the flipping of nucleobases between amino acid “bookends” that demarcate the base-stacked segments of the tracking strand. To test this hypothesis, we mutated AdnB motor domain residues Trp325 and Arg326 to alanine and purified recombinant AdnA-AdnB(W325A-R326A) heterodimer in parallel with wild-type AdnAB (Fig. 5A). We found that whereas the specific activity of the W325A-R326A mutant in ssDNA-dependent ATP hydrolysis was unchanged vis-à-vis wild-type AdnAB (Fig. 5C), the mutant enzyme was unable to catalyze ATP-dependent resection of linear duplex plasmid DNA (Fig. 5B). As a gauge of ssDNA translocation, we assayed the wild-type and mutant AdnAB enzymes for nuclease activity on a 5′ 32P-labeled 24-mer ssDNA substrate (Fig. 6). As shown previously (7, 8), the 5′ end of the ssDNA can access the AdnA nuclease from solution and thread into the active site without need for ATP hydrolysis, resulting in cleavage by the AdnA nuclease at two principal sites to yield radiolabeled 5-mer and 6-mer products (Fig. 6). The wild-type AdnAB and AdnA-AdnB(W325A-R326A) mutant heterodimers were equally adept at ATP-independent cleavage by the AdnA nuclease (Fig. 6). In the presence of ATP, wild-type AdnAB couples ATP hydrolysis to unidirectional translocation (3′ to 5′) on the 24-mer ssDNA and thereby pumps the 3′ end of the ssDNA through the tandem AdnB and AdnA motor domains and into the AdnB nuclease domain. AdnB nuclease cleaves the ssDNA as soon as the 5′ end clears the AdnB motor (at which point pumping stops). The 16- to 17-nucleotide products of ATP-dependent cleavage of the 24-mer by the AdnB nuclease reflect the distance along the DNA path from the end of the AdnB motor to the AdnB nuclease-active site (7, 8). The salient finding was that the W325A-R326A mutant AdnB motor cannot translocate 3′ to 5′ on ssDNA when it hydrolyzes ATP and thus cannot pump the ssDNA into the AdnB nuclease (Fig. 6). These results implicate the Trp-Arg motif as a pawl in the molecular ratchet required for unidirectional movement of the AdnB motor in response to ATP hydrolysis.
Fig. 5.
AdnB Trp325-Arg326 mutation uncouples ATP hydrolysis from DSB resection. (A) Aliquots (5 µg) of recombinant wild-type AdnAB and AdnA-AdnB(W325A-R326A) mutant heterodimers were analyzed by SDS/PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (in kilodaltons) of marker polypeptides are indicated on the Left. (B) DSB resection. Reaction mixtures (50 μL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM ATP, 200 ng linear pUC19 DNA (cut with SmaI), and 2.5 pmol of wild-type or mutant AdnAB were incubated at 37 °C. Aliquots (10 µL) were withdrawn at the times specified and the reactions were quenched immediately by adding 2.5 µL of 500 mM EDTA (pH 8.0). The reactions were supplemented with 5 μL of a solution containing 10 mM Tris⋅HCl (pH 7.6), 60% glycerol, 60 mM EDTA, and 0.15% Orange G and then analyzed by electrophoresis through a horizontal 0.8% agarose gel containing 45 mM Tris-borate, 1.2 mM EDTA, and 0.5 μg/mL ethidium bromide. DNA was visualized under shortwave UV illumination. (C) DNA-dependent ATPase activity. Reaction mixtures (10 µL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, 1 mM (10 nmol) [α32P]ATP, 50 μM 85-mer ssDNA oligonucleotide, and AdnAB as specified were incubated for 10 min at 37 °C. The reactions were quenched by adding 2 μL of 5 M formic acid. Aliquots (2 μL) of the mixtures were applied to PEI-cellulose TLC plates, which were developed with 0.45 M ammonium sulfate. [α-32P]ATP and [α-32P]ADP were quantified by scanning the plates with a Fujix BAS2500 imager. The extent of ATP hydrolysis is plotted as function of input AdnAB. Each datum in the graph is the average of three independent titration experiments ± SEM.
Fig. 6.
AdnB Trp325-Arg326 mutation uncouples ATP hydrolysis from ssDNA translocation. Reaction mixtures (10 μL) containing 20 mM Tris⋅HCl (pH 8.0), 2 mM MgCl2, 1 mM DTT, 0.1 μM 5′ 32P-labeled ssDNA substrate (shown at Bottom), 1 mM ATP, and 0.1 pmol wild-type or mutant AdnAB (where indicated by +) were incubated for 20 min at 37 °C. The reactions were quenched by adjustment of the mixtures to 45% formamide and 25 mM EDTA. The products were analyzed by electrophoresis through a 15-cm 18% polyacrylamide gel containing 7 M urea in 45 mM Tris-borate and 1.2 mM EDTA and then visualized by autoradiography. The sites of cleavage of the 24-mer ssDNA by the AdnA and AdnB nuclease domains are indicated by arrows at Bottom.
Single-Molecule DNA Curtain Analysis of DSB Resection by AdnAB.
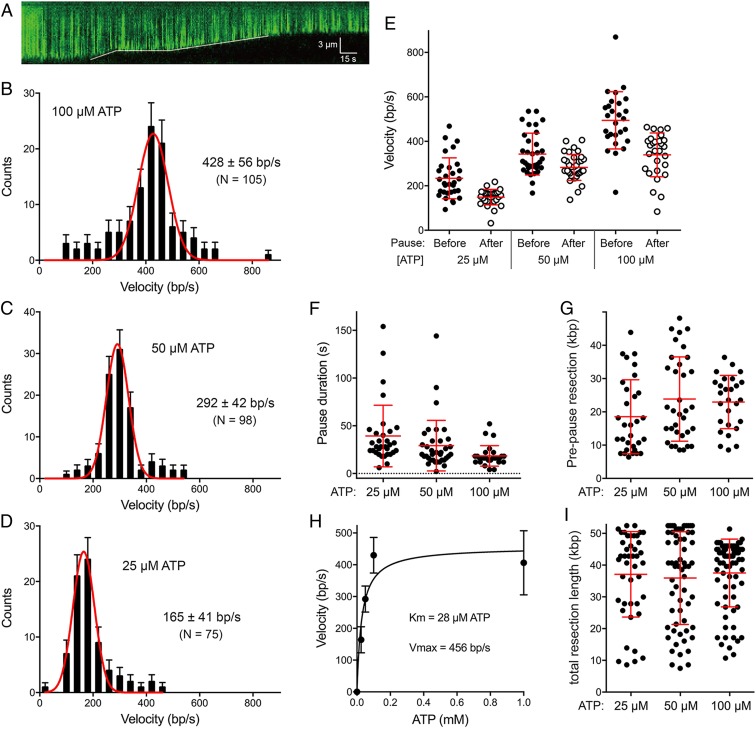
Single-molecule studies of DSB resection by RecBCD and AddAB have yielded valuable mechanistic insights not attainable via ensemble biochemistry (22–30). Here we employed single-molecule DNA curtain methods (20, 26) to study the dynamics of DSB resection by AdnAB. In brief, the curtain consists of a parallel array of bacteriophage lambda DNAs tethered at one end to a lipid bilayer surface along a metallic barrier oriented perpendicular to the direction of liquid flow. The DNAs are stained with fluorescent dye YOYO-1 and visualized by total internal reflection fluorescence (TIRF) microscopy. Engagement of the free DNA ends by AdnAB in the presence of ATP and magnesium results in a time-dependent shortening of the fluorescent linear DNAs. DNA shortening is taken to be synonymous with helicase unwinding and concomitant DSB resection. The kymograph in Fig. 7A exemplifies an event in which AdnAB resects the DNA at an apparently uniform rate, pauses transiently, then resumes resection at a different rate. The reaction endpoint at which no further shortening is observed is presumed to reflect dissociation of AdnAB from the DNA substrate. Analysis of 103 DSB resections in the presence of 1 mM ATP revealed that 67 events (65%) entailed no pausing, 32 events (31%) were punctuated by one pause, and four events (4%) entailed two pauses (SI Appendix, Fig. S4C). The percentage of DSB resections with one or more pauses was the same at 100 µM ATP (35%) but increased as the ATP concentration was reduced to 50 µM (to 55%) and 25 µM (72%).
Fig. 7.
Single-molecule DNA curtain analysis of DSB resection by AdnAB. (A) Kymograph showing the resection of YOYO1-stained lambda DNA (green) upon the injection of 5 nM AdnAB within a 1-min injection window. The solid white line indicates the dsDNA end position. (B–D) Velocity distributions of all individual AdnAB resection tracts (including no-pause, before-pause, and after-pause resection tracts) at 100 µM ATP (n = 105) (B), at 50 µM ATP (n = 98) (C), and at 25 µM ATP (n = 75) (D). Black error bars represent 95% confidence intervals calculated from bootstrap analysis. The solid red lines represent Gaussian fits to the data. The mean velocities (±SD) derived from the Gaussian fit are shown in each panel. (E, G, and I) Scatter plots are shown of resection velocities prior to and after AdnAB pausing events (E); AdnAB pause durations (F); resection tract lengths prior to pausing (G); and resection tract lengths at termination (I; a gauge of processivity). Data were collected from DSB resection reactions containing 25 µM ATP (n = 31), 50 µM ATP (n = 34), and 100 µM ATP (n = 27). Red lines indicate the mean and 68% confidence intervals. (H) Plot of resection velocities (with error bars obtained from Gaussian fits) as a function of ATP concentration. The data were fit to the Michaelis–Menten equation, with Km and Vmax values as indicated.
The distributions of velocities of DNA shortening during individual resection tracts are shown for ATP concentrations of 100 µM (Fig. 7B), 50 µM (Fig. 7C), and 25 µM (Fig. 7D). (Velocities at 1 mM ATP are shown in SI Appendix, Fig. S4A.) The data were fit to a Gaussian distribution (red curves) and the mean DSB resection velocities were plotted as a function of ATP concentration (Fig. 7H), showing that resection velocity increased with [ATP] from 25 to 100 µM and plateaued at 1 mM. The apparent Km for ATP was 28 µM (±14 µM SE) and the Vmax was 456 bp/s (±58 bp/s SE) (Fig. 7H). Because the Vmax of DSB resection is similar to the apparent kcat for ssDNA-dependent ATP hydrolysis by AdnAB (415 s−1) (8), we surmise that 1 bp is unwound by AdnAB per ATP hydrolyzed.
For those AdnAB resection events involving pausing and resumption of resection, we plotted the distribution of resection velocities before and after the pause. As shown in Fig. 7E, the rates of AdnAB resection were lower after the pause than before the pause; this was the case at every concentration of ATP tested. For example, at 100 µM ATP, the mean prepause rate was 661 ± 42 bp/s and the postpause rate was 427 ± 31 bp/s (P < 0.0001). Of the 27 molecules that underwent a pause at 100 µM ATP, 26 displayed a slower velocity after the pause; in only 1 out of 27 molecules did the velocity increase after the pause. At 50 µM ATP, the mean pre- and postpause rates were 343 ± 16 bp/s and 283 ± 10 bp/s, respectively (P < 0.002). At 25 µM ATP, the mean pre- and postpause rates were 234 ± 17 bp/s and 149 ± 16 bp/s, respectively (P < 0.0001). These results suggest that spontaneous pausing by AdnAB can elicit a persistent conformational change that slows the motor upon resumption of end resection.
The durations of the pause events were quite variable (ranging from 2 s to 154 s) with mean pause times of 39 s, 29 s, and 18 s at 25 µM, 50 µM, and 100 μM ATP, respectively (Fig. 7F). There was no correlation between the duration of the pause and velocity of end resection after the pause. The sites of pausing were scattered along the linear duplex DNA in a seemingly random fashion (Fig. 7G). The mean lengths of the prepause resection tracts were 18.5 kbp, 23.8 kbp, and 22.9 kbp at 25 µM, 50 µM, and 100 µM ATP, respectively. The sites at which resection terminated were distributed broadly but were concentrated in the distal half of the DNA molecule (Fig. 7I). The mean lengths of DNA resected at the termination point, which are indicative of AdnAB’s processivity, were 37.1 kbp, 35.9 kbp, and 37.5 kbp at 25 µM, 50 µM, and 100 µM ATP, respectively (Fig. 7I).
Gauging the Effect of Mutating AdnA’s ATPase Site on DSB Resection Dynamics.
The AdnAB cryo-EM structures indicate that the AdnA motor-like domain has a seemingly intact ATPase active site. Previous ensemble biochemical studies showed that alanine mutation of AdnA Asp255—the motif II side chain that coordinates the metal cofactor for ATP hydrolysis—did not exert a significant effect on the rate of plasmid DNA unwinding (9). Here, we performed single-molecule DNA curtain assays of DSB resection by wild-type AdnAB and the AdnA(D255A)-AdnB mutant in the presence of 1 mM ATP. The two enzymes behaved similarly with respect to the distribution of resection velocities (SI Appendix, Fig. S4 A and B), the frequency of pausing (SI Appendix, Fig. S4C), and the reduction in resection velocity after recovery from a pause event (SI Appendix, Fig. S4D). Thus, it appears that ATP hydrolysis by the lagging AdnA motor-like domain is not contributing in a major way to the overall motor activity of the AdnAB complex.
Discussion
The present study provides insights into the structure of the mycobacterial motor-nuclease AdnAB and its mechanism of DSB end resection. Cryo-EM structures of M. smegmatis AdnAB in three functional states (in the absence of DNA and in complex with forked DNAs before and after cleavage of the 5′ ssDNA tail by the AdnA nuclease domain) affirm the domain organization and division of labor deduced from biochemical studies and mutagenesis, whereby unidirectional translocation of the AdnB motor on the unwound 3′ ssDNA strand drives the displaced 5′ ssDNA into the AdnA nuclease domain. The structures are especially instructive regarding: the path of the 5′ ssDNA through the AdnA nuclease and the mechanism of 5′ strand cleavage; the path of the 3′ tracking strand through the AdnB motor and the DNA contacts that couple ATP hydrolysis to mechanical work; and the position of the AdnA iron–sulfur cluster subdomain at the fork junction and its likely role in maintaining the split paths of the unwound 5′ and 3′ strands.
The AdnA nuclease-active site is shown to consist of an ensemble of amino acids that bind the metal cofactor, coordinate the scissile phosphate, and interact with five of the backbone phosphates of the 5′ strand flanking the scissile phosphate (two on the 5′ side and three on the 3′ side, including one phosphate at the dsDNA junction). In the previously reported structures of Bacillus AddAB in complex with DNA, no 5′ ssDNA strand was modeled (18, 19), even though the input DNA ligand was similar to that used in the present study: a forked hairpin duplex that has both 5′ and 3′ single strand tails. The structure of an E. coli RecBCD–DNA complex (13) did not have DNA in the RecB nuclease domain, though it did reveal a calcium ion (analog of the Mg cofactor) coordinated by two aspartates and a histidine (the counterparts of the AdnAB magnesium-binding residues).
The nuclease domains of AdnA, AdnB, AddA, AddB, and RecB are structurally homologous and share several motifs at their active sites. The key distinctions are the directionality of ingress of the displaced ssDNA substrates into the nuclease-active site. In the case of AdnA nuclease, our structures show that the displaced 5′ end coming off the duplex junction is fed into the active site and that cleavage of the 5′ strand occurs two nucleotides upstream of the fork junction. Although we do not visualize the 3′ tracking strand beyond the AdnB motor, we presume that during translocation of the AdnB motor from a DSB end the unwound 3′ end is threaded through the AdnA motor-like domain and then enters the AdnB nuclease-active site (i.e., in the oppose polarity used by the AdnA nuclease). This situation is captured in the structure of AddAB bound to a forked DNA with a χ site, in which the 3′ ssDNA tracking strand traverses the lagging motor-like domain and terminates at the entrance to the 3′ processing nuclease domain (19). Superimposing the mycobacterial 5′ processing nuclease domain on the Bacillus 3′ processing nuclease domain in their respective DNA complexes (SI Appendix, Fig. S5) highlights both conserved and distinctive features. The scissile phosphodiesters (depicted with yellow phosphorus atoms) are intact because one of the equivalent metal binding residues is mutated to alanine in both structures (SI Appendix, Fig. S5, denoted by the asterisks). Thus, whereas the metal is not present in either structure, the other Asp and His metal ligands are similarly disposed next to the cleavage site. A conserved Ser-Lys-Gln triad coordinates the scissile phosphate and a conserved tyrosine coordinates the neighboring phosphate on the 5′ side. Whereas the mycobacterial nuclease contacts the phosphate and nucleobase located two nucleotides 5′ of the cleavage site (via two arginines; SI Appendix, Fig. S5A), the corresponding amino acids of the Bacillus enzyme (glutamine and isoleucine) make stacking and hydrogen-bond interactions with the nucleobases two and three nucleotides 5′ of the scissile phosphate (SI Appendix, Fig. S5B).
The path of the 3′ tracking strand through the AdnB motor is punctuated by a series of amino acid “baffles” (i.e., Trp325, Arg326, Phe254, and Leu142) that stack on and between the nucleobases to partition the 3′ ssDNA into discrete base-stacked segments. The partitioning of the tracking strand into such segments is seen in two different cryo-EM structures in which the register of the nucleobases is shifted by a 1-nucleotide step (Figs. 3B and 4B and SI Appendix, Fig. S3). We envision that this register shift mimics (in part) the changes occurring during the mechanical power stroke, in which protein subdomain movements within the AdnB motor, synchronized to the ATP hydrolytic cycle, drive passage of the nucleobases over the amino acid baffles in the 3′-to-5′ direction, and the baffles prevent backward sliding in a pawl-and-ratchet–like fashion. Consistent with this model, we report that alanine mutation of the Trp325-Arg326 motif uncouples DNA-dependent ATP hydrolysis (which is unperturbed by the mutation) from mechanical work by the AdnB motor (which is abolished by the mutation). An analogous ensemble of four amino acids in the lead motor domain of Bacillus AddAB (Phe, Arg, Phe, and Ile) makes similar nucleobase stacking interactions with the 3′ ssDNA tracking strand (19).
The requirement for AdnB Trp-Arg to achieve translocation raises interesting questions about how AdnAB initiates unwinding at a DSB end. The structure of the WT AdnAB–DNA complex shows that Trp325 and Arg326 stack on the second and fourth single-stranded nucleobases of the 3′ strand flanking the duplex junction (Fig. 4B). Thus, in order for the AdnB motor to be “in gear” it would require that a four-nucleotide 3′ single-strand tail be available at a DSB end. We showed previously that ATP-independent binding of AdnAB to a blunt DSB end melts out a short segment of the duplex and allows access of the 5′ strand to the AdnA nuclease-active site, which cleaves the melted 5′ strand two and three nucleotides from the 5′ end to liberate dinucleotide and trinucleotide products (10). With the present insight that the AdnA nuclease cleaves the displaced 5′ strand two nucleotides from the duplex junction, we can deduce that the ATP-independent end-melting step unpairs four or five nucleotides at the blunt DSB end, which would suffice for the 3′ strand to be engaged by the AdnB motor.
Our structures show that AdnAB contacts the phosphates of both strands of the DNA duplex at sites close to the fork junction (Fig. 3B). Whereas we see no direct protein contacts to the distal end of the duplex, three α-helices of the AdnB motor domain are situated below the duplex in the view shown in Fig. 2A; the connecting loop between two of the duplexes (210GPYQRDR216) is disordered. It is conceivable that interaction of AdnAB with the distal duplex is tuned to the ATPase cycle of the motor, as suggested for AddAB (19). Alternatively, the junction duplex interactions might suffice for AdnAB’s processive DSB resection activity. Our structures show that AdnAB does not have a counterpart of the “arm domain” of the AddAB lead motor that engages the DNA duplex one helical turn upstream of the dsDNA–ssDNA junction (18, 19).
Our single-molecule DNA curtain analysis affords valuable insights into the dynamics of DSB resection by AdnAB. To wit: 1) the enzyme is highly processive, but prone to spontaneous pausing; 2) the velocity of the AdnAB motor speeds up, while the frequency and duration of pausing declines, with increasing ATP concentration in the range of 25 to 100 µM; and 3) overall AdnAB processivity is not sensitive to ATP concentration in this range. The striking property of AdnAB is that the velocity of DSB resection slows after the enzyme experiences a spontaneous pause. This contrasts with the ergodic behavior of RecBCD when DSB resection is arrested by instantaneous depletion of ATP and magnesium and then allowed to resume resection when ATP-Mg is replenished (27). In this nonspontaneous pause regime, individual RecBCD molecules that had been resecting at a defined velocity prior to the pause resumed resection at variable velocities reflecting the full spectrum of prepause velocities in the ensemble, i.e., the postpause rates could be higher or lower than the prepause rates (27). The postpause slowing by AdnAB is more akin to the behavior of RecBCD and AddAB after they pause transiently at their respective χ sequences, whereby the velocity of the helicase motor slows after the encounter with χ (24–26, 29). In the case of AdnAB, we see no evidence for specific sites of pausing during single-molecule DSB resection. We favor the idea that there is no specific χ sequence for AdnAB. (Note, the amino acids in the lagging motor-like domain of Bacillus AddAB that recognize the Bacillus χ sequence are not conserved in mycobacterial AdnA.) Rather, we envision that stochastic conformational switches in AdnAB during translocation elicit a clamping down on the 3′ ssDNA tracking as it traverses the lagging AdnA motor that basically mimics the analogous switches that χ recognition is thought to trigger in RecBCD and AddAB (30). One attractive model was that the conformational switch might be dictated by the ATPase activity of AdnA. However, we find here that the AdnA-D255A mutation did not affect the dynamics of DSB resection by the AdnAB complex in the single-molecule DNA curtain assay. Thus, if the AdnA motor-like domain does modulate AdnAB activity, it may do so via ATP binding (without a requirement for ATP hydrolysis) or via an ATP-dependent effect on the nuclease activity rather than the helicase. The AdnA ATPase site is functionally relevant, insofar as a M. smegmatis adnA-D255A strain displayed a 12-fold decrement in the frequency of DSB repair by homologous recombination vis-à-vis a wild-type control (10).
Methods
The procedures for cryo-EM data acquisition and structure modeling, ensemble assays of AdnAB activity, and single-molecule DNA curtain analysis of DSB resection are described in detail in SI Appendix. Atomic structure models of AdnAB have been deposited in the PBD/EMDB databases under ID codes 6PPJ/EMD-20440 (DNA-free), 6PPR/EMD-20446 (precleavage DNA complex), and 6PPU/EMD-20447 (postcleavage DNA complex).
Supplementary Material
Acknowledgments
This work was supported by NIH grants R35-GM126945 and R01-AI64693 (to S.S.) and R35-GM118026 (to E.C.G.). D.J.P. is supported by a grant from the Maoris Foundation. The Memorial Sloan Kettering Cancer Center structural biology core facility is supported by National Cancer Institute grant P30-CA008748.
Footnotes
The authors declare no competing interest.
Data deposition: Atomic structure models of AdnAB have been deposited in the Protein Data Bank, http://www.rcsb.org (PDB ID codes 6PPJ, 6PPR, and 6PPU) and the Electron Microscopy Data Bank (EMDB ID codes EMD-20440, EMD-20446, and EMD-20447).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913546116/-/DCSupplemental.
References
- 1.Gupta R., Barkan D., Redelman-Sidi G., Shuman S., Glickman M. S., Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 79, 316–330 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aniukwu J., Glickman M. S., Shuman S., The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 22, 512–527 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta R., Shuman S., Glickman M. S., RecF and RecR play critical roles in the homologous recombination and single-strand annealing pathways of mycobacteria. J. Bacteriol. 197, 3121–3132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell J. C., Kowalczykowski S. C., RecA: Regulation and mechanism of a molecular search engine. Trends Biochem. Sci. 41, 491–507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dillingham M. S., Kowalczykowski S. C., RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72, 642–671 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chédin F., Kowalczykowski S. C., A novel family of regulated helicases/nucleases from gram-positive bacteria: Insights into the initiation of DNA recombination. Mol. Microbiol. 43, 823–834 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Sinha K. M., Unciuleac M. C., Glickman M. S., Shuman S., AdnAB: A new DSB-resecting motor-nuclease from mycobacteria. Genes Dev. 23, 1423–1437 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Unciuleac M. C., Shuman S., Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor-nuclease machines. J. Biol. Chem. 285, 2632–2641 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Unciuleac M. C., Shuman S., Double strand break unwinding and resection by the mycobacterial helicase-nuclease AdnAB in the presence of single strand DNA-binding protein (SSB).J. Biol. Chem. 285, 34319–34329 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta R., Unciuleac M. C., Shuman S., Glickman M. S., Homologous recombination mediated by the mycobacterial AdnAB helicase without end resection by the AdnAB nucleases. Nucleic Acids Res. 45, 762–774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeeles J. T., Dillingham M. S., A dual-nuclease mechanism for DNA break processing by AddAB-type helicase-nucleases. J. Mol. Biol. 371, 66–78 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Yeeles J. T., Gwynn E. J., Webb M. R., Dillingham M. S., The AddAB helicase-nuclease catalyses rapid and processive DNA unwinding using a single Superfamily 1A motor domain. Nucleic Acids Res. 39, 2271–2285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B., Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432, 187–193 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Saikrishnan K., Griffiths S. P., Cook N., Court R., Wigley D. B., DNA binding to RecD: Role of the 1B domain in SF1B helicase activity. EMBO J. 27, 2222–2229 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson M., Chaban Y., Wigley D. B., Mechanism for nuclease regulation in RecBCD. eLife 5, e18277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigden D. J., An inactivated nuclease-like domain in RecC with novel function: Implications for evolution. BMC Struct. Biol. 5, 9 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wigley D. B., Bacterial DNA repair: Recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11, 9–13 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Saikrishnan K., et al. , Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 31, 1568–1578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krajewski W. W., et al. , Structural basis for translocation by AddAB helicase-nuclease and its arrest at χ sites. Nature 508, 416–419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins B. E., Ye L. F., Duzdevich D., Greene E. C., DNA curtains: Novel tools for imaging protein-nucleic acid interactions at the single-molecule level. Methods Cell Biol. 123, 217–234 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Lee J. Y., Yang W., UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bianco P. R., et al. , Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature 409, 374–378 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Dohoney K. M., Gelles J., Chi-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature 409, 370–374 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Spies M., et al. , A molecular throttle: The recombination hotspot χ controls DNA translocation by the RecBCD helicase. Cell 114, 647–654 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Spies M., Amitani I., Baskin R. J., Kowalczykowski S. C., RecBCD enzyme switches lead motor subunits in response to χ recognition. Cell 131, 694–705 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finkelstein I. J., Visnapuu M. L., Greene E. C., Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature 468, 983–987 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu B., Baskin R. J., Kowalczykowski S. C., DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate. Nature 500, 482–485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reuter M., Parry F., Dryden D. T., Blakely G. W., Single-molecule imaging of Bacteroides fragilis AddAB reveals the highly processive translocation of a single motor helicase. Nucleic Acids Res. 38, 3721–3731 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrasco C., Gilhooly N. S., Dillingham M. S., Moreno-Herrero F., On the mechanism of recombination hotspot scanning during double-stranded DNA break resection. Proc. Natl. Acad. Sci. U.S.A. 110, E2562–E2571 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrasco C., Dillingham M. S., Moreno-Herrero F., Single molecule approaches to monitor the recognition and resection of double-stranded DNA breaks during homologous recombination. DNA Repair (Amst.) 20, 119–129 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.