Abstract
Sirtuins (SIRTs) are nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases with diverse physiological functions. A variety of small molecules have been developed to interrogate the physiological function of SIRTs. Therefore, it is desirable to establish efficient and convenient assays to screen SIRTs modulators. In this study, we designed a series of fluorescent nonapeptide probes derived from substrates of SIRT1–SIRT3. Fluorescence increment of these probes is based on SIRT-mediated removal of the acyl side chain with fluorophore, which makes this system free of lysine-recognizing protease. Comparing the reaction of these fluorescent nonapeptide substrates with SIRT1–SIRT3 and SIRT6, it was confirmed that this assessment system was the most suitable for SIRT2 activity detection. Thus, SIRT2 was used to modify substrates by truncating the amino acids or lysine side chain of nonapeptide. Finally, two specific and efficient fluorescent probes for SIRT2, ne-D9 and ne-K4a, were developed. Evaluation of the results revealed that ne-K4a based assay was more suitable for modulators screening in vitro, while the other specific substrate ne-D9 was stable in cell lysate and could detect the activity of SIRT2 in the same. In summary, this study presents a novel strategy for detecting SIRT2 activity in vitro and in cell lysate.
Key words: Deacetylate, Deacylate, Fluorescent probe, One-step assay, Sirtuins, SIRT2
Abbreviations: DLAT, dihydrolipoyl transacetylase; FRET, fluorescence resonance energy transfer; NAD+, nicotinamide adenine dinucleotide; SIRT, sirtuin; TNF-α, tumor necrosis factor α
Graphical abstract
Sirtuins (SIRTs) are NAD+-dependent histone deacetylases with extensive physiological functions. We established a novel strategy for detecting the activity of SIRT2 in vitro and in cell lysate. Fluorescence increment of these probes is based on SIRT-mediated removal of the acyl side chain with fluorophore.
1. Introduction
Sirtuins (SIRTs), NAD+-dependent histone deacetylases, are mammalian orthologues of yeast silent information regular 2. There are seven isoforms (SIRT1–SIRT7) of the SIRTs protein family and the subcellular locations of these isoforms were different1, 2. The SIRT1, SIRT6 and SIRT7 are largely reported to reside in the nucleus. SIRT2 primarily resides in the cytoplasm while SIRT3–SIRT5 were found in the mitochondria. Besides their different subcellular localization, SIRTs also greatly differ in catalytic activity. SIRTs were characterized as histone deacetylase from the very beginning. Research and development revealed that SIRTs were discovered to have the ability to deacylate the lysine residues from a plenty of non-histone protein. Among the seven isoforms, only SIRT1–SIRT3 are verified as the robust deacetylase. Yet they are not restricted to deacetylation as SIRT1–SIRT3 are also defatty-acylases, especially exhibited potent activity to remove long chain acyl groups (e.g., C8–C16)3. The other isotype enzymes, they perform weak deacetylase activity but are able to remove relatively acylated lysine side chain. For instance, SIRT5 exhibits the better capacity to demalonylate, desuccinylate and deglutarylate lysine residues than deacetylate lysine4, 5. SIRT4 prefers delipoyl and debiotinyl lysine side chains6. Recently, it was found that SIRT4 could remove methylglutaryl group, hydroxymethylglutaryl (HMG) group and 3-methylglutaconyl group from the modificated lysine residues7. Further, Bao et al.8 found that SIRT5 could regulate HMGylation in cells. SIRT6 can remove long acyl chains9 while SIRT7 was reported to be a desuccinylase10.
Through regulating the acylation condition of widespread proteins in vivo, SIRT enzymes possess extensive physiological functions, including stress responses, metabolic control, stabilization of genomic DNA and aging11. Therefore, SIRTs are considered as attractive therapeutic targets for plenty of diseases, such as obesity, cardiovascular diseases, diabetes, cancer and neurodegenerative diseases12. SIRT1 and SIRT3 were reported as therapeutic targets for breast and oral cancer13. SIRT2 inhibitors impair hepatic glucose uptake, which enables SIRT2 to be a new therapeutic target for type 2 diabetes14. SIRT6 activator is considered as a potential approach for the treatment of hepatocellular carcinoma15. Therefore, variety of SIRT modulators were developed and explored to further interrogate the physiological function of SIRTs and drug discovery. At the same time, diverse methodologies for activity assessment of SIRT enzymes were established for discovery and characterization of these modulators. In vitro methods for assessing SIRT modulators include the use of radioisotope-labeled histone16, high-performance liquid chromatography (HPLC)17 and fluorometric assay18. Both radioisotope-based method and HPLC assay require complex sample handling, which constrains their utilization and makes them inconvenient in high-throughput screening. Due to the convenience and efficiency, fluorescence probe has been broadly applied in detecting the activity of SIRTs and high-throughput screening of SIRTs modulators19. However, most of these fluorometric assays work based on a two-step principle, which needs trypsin to digest the C-terminal lysine residual. During the first step, SIRTs deacetylate the fluorescent peptide. While in the second step the C-terminal lysine residual of deacetylated peptide is detected and digested by trypsin. There are two strategies based on the two-step principle: one uses a fluorescent group 7-amino-4-methylcoumarin (AMC), which fluoresces in a free state; the other uses fluorescence resonance energy transfer (FRET) principle, which connects a donor dye and an accepter dye to the ends of peptide19. The lysine side chain of the fluorescent probe is not restricted to acetyl group. For instance, the fluorogenic succinyl and myristoyl peptides were used to detect the activity of SIRT5 and SIRT6, respectively20, 21. The introduction of trypsin may increase false-positive or false-negative signals when the detected molecules affect the activity of trypsin, limiting the measurement range of the assay22. This two-step assay is usually discontinuous, which impedes the enzymatic kinetics research.
Based on the discovery that SIRT6 is an efficient long-chain defatty acylase of tumor necrosis factor α (TNF-α)9. Schuster et al.23 designed a one-step SIRT assay. They synthesized a nonapeptide substrate from TNF-α and joined the fluorophore to lysine side chains directly, which was different from the former two-step design where the fluorophore and quenching groups replaced the peptide terminal parts. The principle of one-step assay is shown in Fig. 1, whereby the initial fluorescence of the substrate is suppressed by the quenching residue. After incubating with SIRTs which possess large hydrophobic pockets to establish the fatty acyl groups17 and NAD+, fluorescently labeled acyl residues could be recognized and transferred from lysine side chain to ADP-ribose leading to a fluorescence increase. In this study, the one-step assay was further explored with a target on SIRT1–SIRT3 for their similar protein structure and excellent ability to recognize fatty acyl groups. Variant fluorescent probes were designed to establish the influence of the different peptides, the length of lysine side chain and the amount of amino acid residues on this technique. By comparing these fluorescent peptides with four reported long-chain defatty acylases SIRT1–SIRT3 and SIRT6, this work revealed two convenient and specific SIRT2 fluorescent peptides, ne-K4a and ne-D9, which could continuously detect SIRT2 activity without using trypsin. Also, by comparing the two specific probes, it was confirmed that ne-K4a was more suitable for high throughput screening of SIRT2 modulators in vitro, while the other specific fluorescent peptide ne-D9 was verified to be stable in cell lysis and could be hydrolyzed by overexpressed SIRT2 in cell lysis, extending its application in cellular systems. In conclusion, the one-step assay for SIRT2 activity we established provides a novel strategy for screening SIRT2 modulators in vitro and detecting the activity of SIRT2 in cell lysis.
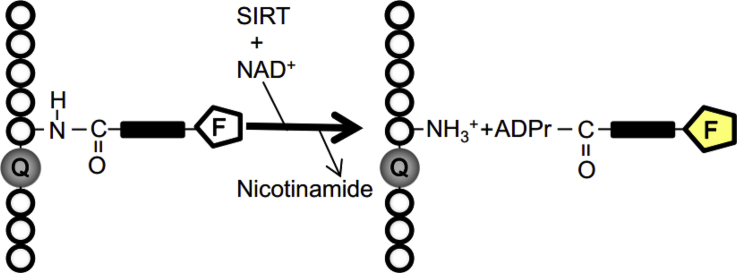
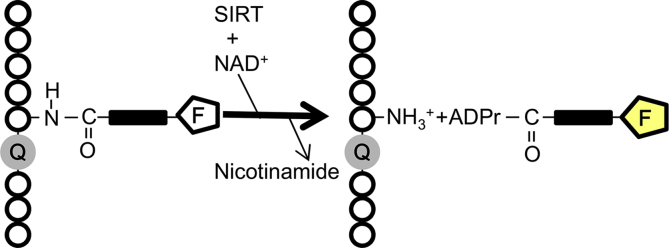
Figure 1.
Principle of the one-step-based assay. The initial fluorescence of the substrate would be suppressed by the quenching residue. After reacting with SIRTs and NAD+, fluorescently labeled acyl residues could be recognized and transferred from lysine side chain to ADP-ribose leading to a fluorescence increase. (Q: Quencher, F: Fluorophore, ADPr: ADP ribose).
2. Materials and methods
2.1. Design and synthesis of fluorescent peptides
All fluorescent substrates were designed in our laboratory, and synthesized by a commissioned company GL Biochem (Shanghai, China). The synthesized peptides were analyzed by mass spectra and HPLC with the purity of above 90%.
2.2. Expression and purification of recombinant SIRT1, SIRT2, SIRT3, and SIRT6
The expression and purification of recombinant human SIRT1 (full-length, aa 1–556), SIRT2 (full-length, aa 1–389), SIRT2 (aa 34–356), SIRT3 (aa 101–399)24 and SIRT6 (aa 13–308)25 were determined based on the previous studies and further analyzed in this study.
The fragments of human SIRT1, SIRT2, SIRT3, and SIRT6 were cloned from HEK293A cDNA libraries and inserted into the bacterial expression vectors pET28a(+) (EMD Biosciences, Darmstadt, Germany) by PCR-amplified and confirmed by sequencing. His-tagged SIRT1 and SIRT3 were overexpressed in E. coli BL21 (DE3) (Weidi Biotechnology, Shanghai, China). His-tagged SIRT2 and SIRT6 were transformed into E. coli BL21 (DE3) Condon Plus and E. coli Rosseta (DE3) (Weidi Biotechnology), respectively. E. coli expressing pET28a(+)/SIRT1–SIRT3 and pET28a(+)/SIRT6 were cultured with shaking at 37 °C until OD600≈0.6, 0.5 mmol/L of isopropyl-d-thiogalactopyranoside was added and cells were cultured at 16 °C for 20 h. For recombinant proteins purification, nickel magnetic beads for His-tag proteins (Bimake, Houston, TX, USA) were used to bind these extracellular-expressed proteins in binding buffer (20 mmol/L sodium phosphate, 500 mmol/L NaCl, 5 mmol/L imidazole, pH 8.0). The unspecific proteins were eliminated using a washing buffer (20 mmol/L sodium phosphate, 500 mmol/L NaCl, 40–80 mmol/L imidazole, pH 8.0). Finally, the proteins of interest were eluted by eluting buffer (20 mmol/L sodium phosphate, 500 mmol/L NaCl, 500 mmol/L imidazole, pH 8.0). After concentrating and dialyzing, the required proteins were obtained and stored in 20% glycerol at −80 °C. The purity and molecular weight of these proteins were analyzed by SDS-PAGE. The amount of target proteins for assay development were calculated based on the purity. Protein concentration was analyzed using the BCA protein assay kit (Thermo Fisher, Waltham, MA, USA).
2.3. Enzymatic reaction and fluorescence detection on 384-well microplate
The continuous fluorescent reaction proceeded in white 384-well assay plate (CORNING, NY, USA) and was performed on the FlexStation3 (Molecular Devices, Silicon Valley, CA, USA) at excitation of 305 nm and emission of 416 nm. After adding assay buffer 1 (50 mmol/L HEPES, 100 mmol/L KCl, 0.001% Tween-20, 0.05 mg/mL BSA, 200 mmol/L TCEP, pH 7.4 for SIRT2) or assay buffer 2 (50 mmol/L Tris-HCl, 1 mmol/L MgCl2, 137 mmol/L NaCl, 2.7 mmol/L KCl, pH 8.0 for SIRT1, SIRT3 and SIRT6), NAD+, peptides and SIRTs successively (total volume 50 μL), the white 384-well assay plate was preincubated in a shaker for 5 min at 37 °C and 1200×g. The fluorescent intensity was recorded at 37 °C for 30 min.
2.4. Calibration lines
1 mmol/L of NAD+, peptides with gradient concentration (0–20 μmol/L) and 2 μmol/L of SIRT2 were successively added in assay buffer 1, followed by preincubation for 5 min at 37 °C, 1200×g. The 384-well assay plate was put in the FlexStation 3 microplate reader to monitor the incremental fluorescent signals until all the fluorescent signals was no longer increasing (about 2 h), which means peptide substrates completely turnover. The linear regression of the final fluorescent signals against peptide concentration was drawn by GraphPad Prism 6 (GraphPad Software, Inc., La Jolla, CA 92037 USA).
2.5. Determination of kinetic parameters
0.3 μmol/L of SIRT2 or 0.4 μmol/L of SIRT3 was mixed with fluorescent probes (0.375–24 μmol/L) and 1 mmol/L of NAD+ fafter cobnce in assay buffer 1 for SIRT2 or assay buffer 2 for SIRT3. After preincubation for 5 min at 37 °C, 1200×g, the fluorescent intensity was recorded at 37 °C for 5–15 min. Calibration lines were using to convert the fluorescent signal to product concentration. The initial velocity (μmol/L∙s) of enzymatic reaction was plotted against peptides concentration. Km and Kcat were acquired by GraphPad Prism 6.
2.6. Fluorescence spectroscopy of SIRT2 special fluorescent probes
The assay of lacking NAD+ or SIRT2 for the whole system (including NAD+, SIRT2 and fluorescent) was performed at 37 °C for 2 h. The fluorescence spectra were measured at emission wavelength from 380 nm to 580 nm with excitation wavelength of 305 nm using the FlexStation 3 (Molecular Devices) microplate reader.
2.7. Evaluation of SIRT2 inhibitors by ne-D9 and ne-K4a
The activities of SIRT2 against ne-D9 or ne-K4a were evaluated in buffer 1 under different concentration of AGK2 (MCE, Monmouth, NJ, USA) or SirReal2 (MCE). 0.3 μmol/L of SIRT2 was incubated with 20 μmol/L of ne-D9 or 20 μmol/L of ne-K4a, 1 mmol/L of NAD+ and inhibitor (AGK2 or SirReal2: 0, 0.2, 1, 6, 30, 100 and 500 μmol/L) in assay buffer 1. Reaction condition and detection methods were the same as enzymatic reaction and fluorescence detection on 384-well microplate in Section 2.3. The IC50 value of SIRT2 inhibitors were calculated by Graphpad Prism 6.
2.8. Cell culture and transfection
HeLa and HEK293A were cultured in DMEM (Thermo Fisher) supplemented with 10% (v/v) fetal bovine serum (Thermo Fisher) and 10 U/mL of penicillin–streptomycin (Thermo Fisher) at 37 °C in a humidified 5% CO2 incubator. The plasmids pcDNA3.1(+)-myc-SIRT1, pcDNA3.1(+)-flag-SIRT3, pcDNA3.1(+)-SIRT5,and pcDNA3.1(+)-flag-SIRT6 were purchased from Addgene. p3×FLAG-CMV-10-SIRT7 was provided by Dr. Baohua Liu (Department of Biochemistry and Molecular Biology, Shenzhen University Health Science Center, China)26. The plasmid pcDNA3.1(+)-his-SIRT2 and pEGFP-N3-SIRT4 were constructed by our laboratory. For transient protein expression, cells were split in 12-well plates and transfected with each plasmid for 24 h using Lipofetamine3000 (Thermo Fisher).
2.9. Western blotting
Cell lysis was obtained through RIPA lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitor (Thermo Fisher). After protein denaturation, 20 μg of cell protein was separated by SDS-PAGE and transferred to PVDF membranes (Millipore, Billerica, MA, USA). The following primary antibodies were obtained for immunoblotting: anti-SIRT1 (Santa Cruz Biotechnology, Dallas, TX, USA), anti-SIRT2 (ABclonal, Wuhan, China), anti-SIRT3 (Cell Signaling Technology, Danvers, MA, USA), anti-GFP (Santa Cruz Biotechnology, Dallas, Texas, USA), anti-SIRT5 (Abcam, Cambridge, UK), anti-SIRT6 (Cell Signaling Technology), anti-FLAG (Cell Signaling Technology), anti-GAPDH (Proteintech, Rosemont, IL, USA). After using HRP-conjugated secondary antibodies (Thermo Fisher), immunoblot images were obtained by ImageQuat LAS 4000 mini (GE, Fairfield, Sweden).
2.10. Detection of the activity of SIRT2 in cell lysates
After transfecting plasmid, HEK293A and HeLa cells in 12-well plates were rinsed with PBS for three times, then RIPA lysis buffer (Beyotime) supplemented with protease inhibitor (Thermo Fisher) was added to lyse cells. After scraped off the 12-well plates, the lysed cells were centrifuged by Heraeus Fresco 17 (Thermo Fisher) at 11,200×g for 15 min at 4 °C. The supernatant was collected and the protein concentration of supernatant was determined by BCA protein assay kit (Thermo Fisher). After collecting proteins of lysed cells, 45 μg of supernatant was added to the solutions which included 20 μmol/L of fluorescent substrates and 1 mmol/L of NAD+ in assay buffer 2. Reaction condition and detection method were the same as enzymatic reaction and fluorescent detection on 384-well microplate in Section 2.3.
2.11. Z factor, signal-to-noise ratio (S/N), coefficient of variation (CV) analysis
It was determined through the measurement of fluorescence increment velocity at 0% and 100% activity (n=48). The calculation of Z factor, S/N, CV was according to Eqs. (1), (2), (3):
| Z′ = 1–[(3·SD100%+3·SD0%)/|mean100%–mean0%|] | (1) |
| S/N = (Mean100%–Mean0%)/SD0% | (2) |
| CV = SD100%/Mean100%×100% | (3) |
2.12. Data analysis and Statistics
All results are expressed as means±standard deviation (SD) of at least three independent experiments. The data analysis and statistics were obtained using Graphpad Prism 6. Comparison among various was identified by one-way analysis of variance (ANOVA) test. Values of P<0.05 were considered to be statistically significant.
3. Results and Discussion
3.1. Initial fluorescent nonapeptide probes design and evaluation
Based on previous reports and the accurate conditions, expression and purification protocols of SIRT1–SIRT3 and SIRT6 were established, which can be used to stably obtain these enzymes with higher purity for use in subsequent experiments (Supporting Information Fig. S1A). The amount of target proteins were calculated based on the purity. The fragmented SIRT2 (aa 34–356) has been reported to have the same activity as full-length SIRT224. In addition, the protein yield of full-length SIRT2 is far lower than that of the fragmented SIRT2 (aa 34–356). Thus, fragmented SIRT2 is used as SIRT2 in most assays if not stated.
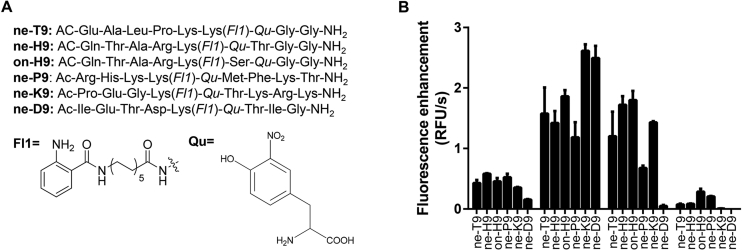
Early studies revealed that SIRTs were insensitive to the substrate amino acid sequence27. However, based on the discovery that SIRTs could interact with not only acetyl lysine but also acetyl lysine, it was found that the peptide sequence had a greater influence on removal of acyl groups of lysine28. Prompted by recent work by Schuster et al.23, it was envisioned that changing the peptide context could provide the desired specific and more sensitive one-step-based fluorescent probe for detecting SIRT activity and screening their inhibitors or agonists. Based on the common substrates of SIRT1–SIRT3, we designed five fluorescent nonapeptide probes including ne-H9 (Histone H36–14), on-H9 (Histone H36–14), ne-P9 (P53379–397), ne-K9 (Ku70558–566) and ne-D9 (dihydrolipoyl transacetylase, DLAT255–263). Ne-T9 (TNFα6–14) was also synthesized according to the method described by Schuster et al. as a positive control (Fig. 2A). The fluorescent assay is based on FRET-principle in which a small fluorophore 2-aminobenzoylamides is attached to the terminal of lysine side chain and the quencher moiety 3-nitrotyrosine replaces the amino acid in +1 or +2 position of lysine29. Initial evaluation of the interaction of these new designed fluorescent substrates interacting with SIRT1, full-length SIRT2, SIRT3, and SIRT6 was carried out through examining the of fluorescence increments in 30 min (Fig. 2B). It was obviously shown that at the same concentration, the fluorescent increment velocities of these fluorescent peptides were much higher after incubating with SIRT2 and SIRT3 than incubating with SIRT1 and SIRT6. These results signified that the one-step-based method was more adapted to SIRT2 and SIRT3 regardless of the alteration of fluorescent peptide context. Interestingly, it was found that the fluorescent probe ne-D9 was specific to SIRT2. The peptide sequence of ne-D9 was from DLAT, which is a SIRT4-mediated substrate in vivo6. Moreover, DLAT256–259 is found to be specific for SIRT2 in a two-step principle assay in vitro3. The results of the current study indicated that a segment of DLAT was also specific for SIRT2 in one-step method.
Figure 2.
The initial assessment of newly designed fluorescent nonapeptide substrates. (A) The structures of reference nonapeptide substrate ne-T9 and five new designed nonapetide substrates, ne-H9, on-H9, ne-P9, ne-K9, and ne-D9. (Qu: Quencher, Fl: Fluorophore) (B) Fluorescence enhancement velocity due to the fluorescence increment of variant fluorescent nonapeptide substrates reacting with SIRT1, full-length SIRT2, SIRT3, and SIRT6 for 30 min. Enzymatic reactions were performed with 40 μmol/L of fluorescent nonapeptide substrate (ne-T9, ne-H9, on-H9, ne-P9, ne-K9, or ne-D9), 1 mmol/L of NAD+, and 0.5 μmol/L enzyme (SIRT1, full-length SIRT2, SIRT3, or SIRT6) in assay buffer at 37 °C. Data are presented as mean±SD, n=3.
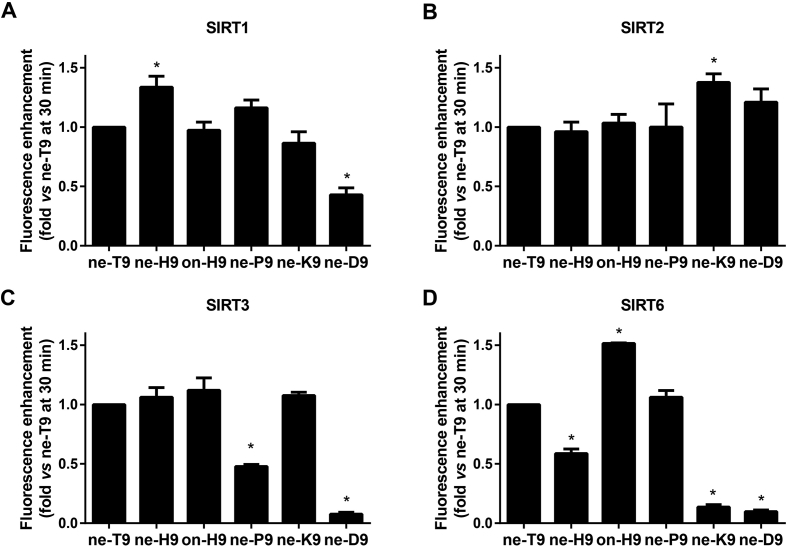
3.2. Enzymatic reactions and kinetics evaluation
For exact evaluation of the five nonapeptide substrates with SIRT1–SIRT3 and SIRT6, the fluorescent variation of the five peptides was detected under at appropriate enzyme concentrations of SIRT1–SIRT3 and SIRT6. Through the initial evaluation, it was obvious that SIRT2 and SIRT3 performed satisfactorily in cleaving most nonapeptide fluorescent substrates. Considering the linear increase in fluorescence in 30 min and the observation of obvious fluorescence increment, we used 20 μmol/L of control substrate ne-T9 reacted with gradient concentration of SIRT1–SIRT3 and SIRT6 to establish the appropriate enzyme concentration (Fig. S1B). Finally, the concentrations of SIRT1, SIRT2 and SIRT3 which could react with 20 μmol/L of peptides were found to be 1, 0.3 and 0.4 μmol/L, respectively. However, when the concentration of SIRT6 was up to 4 μmol/L, the fluorescent increment velocity was still not saturated. Considering the loading volume, 2 μmol/L of SIRT6 was used in the one-step system. Under the suitable concentration, it was possible to estimate these different sequence probes using the four SIRT isoforms (Fig. 3). Ne-H9 caused the highest conversion of SIRT1 and it was evidently better than the control substrate ne-T9. Due to the excellent ability to cleave fluorescent labeled acyl residue, SIRT2 seemed to convert each substrate well and only ne-K9 showed a statistically significant difference in comparison with ne-T9. Although SIRT3 performed well in this assay method using ne-T9, the fluorescence enhancement of newly designed substrates were not higher than that of ne-T9. For SIRT6, there was a big difference in most of newly designed substrates as compared with ne-T9. The substrate on-H9 was the best nonapeptide for SIRT6 while ne-H9, ne-K9, and ne-D9 could not be well hydrolyzed by SIRT6. Overall comparison showed that SIRT2 was insensitive to the variant peptide sequence contexts whereas SIRT6 was significantly affected by the sequence of amino acid substrates. This phenomenon was consistent with the hypothesis that the deacylation would be greatly dependent on peptide sequence contexts in vitro if the acyl group of lysine has a weak affinity with SIRT28. It was found that the fluorescence increment of ne-D9 was not observed with SIRT1, SIRT3 and SIRT6 even under the appropriate concentration, which reconfirmed that ne-D9 sequence had a specific efficient effect on SIRT2.
Figure 3.
Evaluation of fluorescent nonapeptide substrates at appropriate enzyme concentration. Fluorescence enhancement velocity of 20 μmol/L fluorescent nonapeptide substrate (ne-T9, ne-H9, on-H9, ne-P9, ne-K9, or ne-D9) incubating with different SIRTs [1 μmol/L SIRT1(A), 0.3 μmol/L SIRT2(B), 0.4 μmol/L SIRT3(C), or 2 μmol/L SIRT6(D)], and 1 mmol/L NAD+ in assay buffer at 37 °C for 30 min. Data are presented as mean±SD, n=3; ∗P<0.05 vs. ne-T9.
As a major step in development of continuous assay, the kinetic parameters in enzymatic reactions of SIRT2 and SIRT3 were then determined. The applicability of Michaelis–Menten equation requires that the enzyme concentration is much lower than substrate concentration, which means that SIRT1 and SIRT6 were not appropriate for kinetic analysis. The calibration curves of the fluorescent intensity against different concentrations of these fluorescent probes were established through complete conversion assays, which were performed by incubating excessive SIRT2 with fluorescent probes at gradient concentrations (Supporting Information Fig. S2). The Kcat/Km values of each fluorescent nonapeptide probes were basically consistent with the results of fluorescence increment velocity (Table 1, Supporting Information Figs. S3 and S4). From the above, we considered this one-step-based assay as the most adaptive application on SIRT2 and a specific fluorescent probe ne-D9 for SIRT2 was found. Comparing the fluorescent increment velocity and Kcat/Km values, ne-K9 was considered the optimum substrate of SIRT2 among the five fluorescent nonapeptide probes and the structure of ne-K9 was modified for further research.
Table 1.
The kinetic parameters of different peptides towards SIRT2 and SIRT3.
| Peptide | SIRT2 (0.3 μmol/L) |
SIRT3 (0.4 μmol/L) |
||||
|---|---|---|---|---|---|---|
| Km (μmol/L) | 10−3×Kcat (1/s) | Kcat/Km (L/mol∙s) | Km (μmol/L) | 10−3×Kcat (1/s) | Kcat/Km (L/mol∙s) | |
| ne-T9 | 6.58±1.78 | 9.68±1.04 | 1471.12 | 3.49±0.47 | 3.98±0.18 | 1140.40 |
| ne-H9 | 1.81±0.21 | 4.85±0.15 | 2679.56 | 2.92±0.47 | 4.91±0.25 | 1681.51 |
| on-H9 | 2.40±0.46 | 7.56±0.44 | 3150.00 | 1.03±0.18 | 3.74±0.16 | 3631.17 |
| ne-P9 | 14.77±3.20 | 1.03±1.16 | 69.74 | 0.74±0.01 | 0.74±0.01 | 317.60 |
| ne-K9 | 1.05±0.17 | 5.00±0.20 | 4347.83 | 4.75±0.79 | 4.75±0.28 | 1000.00 |
| ne-D9 | 1.48±0.24 | 6.63±0.28 | 4479.73 | – | – | – |
| ne-K4a | 1.98±0.26 | 5.85±0.22 | 2954.55 | – | – | – |
–Not detected.
3.3. Development of the optimum substrate for SIRT2
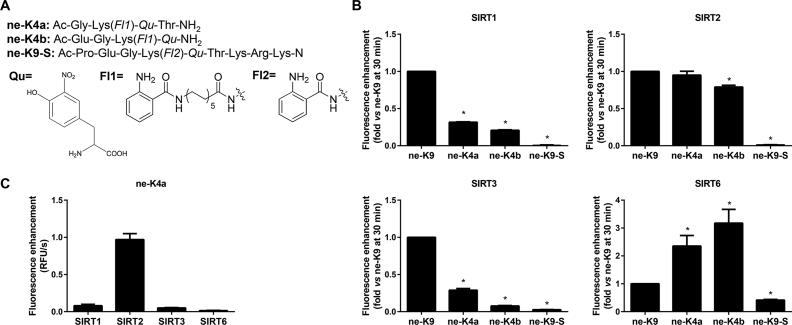
Ne-K9 was modified from the length of lysine side chain and the number of amino acid in the peptide. Considering that SIRT2 is a robust deacetylase, ne-K9-S was designed, of which the lysine side chain was short. Referring to most of tetrapeptide substrates in commercial SIRTs assay kits, ne-K9 substrate was truncated nonapeptide to two tetrapeptide substrates, ne-K4a and ne-K4b (Fig. 4A). However, no fluorescence increments of ne-K9-S were observed with any SIRTs, which was probably because SIRTs could not hydrolyze aromatic carboxylates30. For the truncated tetrapeptide substrates, ne-K4a and ne-K4b, it was unpredictable that SIRT1 and SIRT3 obviously declined in ability to deacylate ne-K4a and ne-K4b, whereas the opposite results were obtained for SIRT6. The fluorescence enhancement velocity of ne-K4a with SIRT2 did not have significant variation compared with ne-K9 but the fluorescence enhancement velocity of ne-K4b was obviously lower than that of ne-K9. All SIRT1–SIRT3 had better fluorescent increments with ne-K4a than ne-K4b. However, ne-K4b was more adapted to SIRT6 compared with ne-K4a and only SIRT6 showed an improved hydrolysis ability to ne-K4a and ne-K4b contrasting to ne-K9 (Fig. 4B). Given the results of substrates selectivity for SIRT1–SIRT3 and SIRT6, it was inferred that: (1) the new truncating substrate ne-K4a might be a specific and efficient fluorescent probe for SIRT2. The specificity of ne-K4a was confirmed by comparing the fluorescent increment velocity of ne-K4a after reacting with SIRT1–SIRT3 and SIRT6 at the same concentration, respectively (Fig. 4C). (2) the number of amino acids of fluorescent peptide is likely to affect the deacylation of SIRTs in vitro.
Figure 4.
Evaluation of newly developed fluorescent substrates, ne-K4a, ne-K4b, and ne-K9-S. (A) The structures of three developed ne-K9 fluorescent substrates, ne-K4a, ne-K4b and ne-K9-S. (Qu: Quencher, Fl: Fluorophore) (B) Fluorescence enhancement velocity of 20 μmol/L fluorescent substrate (ne-K9, ne-K4a, ne-K4b, or ne-K9-S) incubating with different SIRTs (1 μmol/L SIRT1, 0.3 μmol/L SIRT2, 0.4 μmol/L SIRT3 or 2 μmol/L SIRT6), and 1 mmol/L NAD+ in assay buffer at 37 °C for 30 min. Data are presented as mean±SD, n=3; ∗P<0.05 vs. ne-K9. (C) Fluorescence enhancement velocity of 20 μmol/L ne-K4a incubating with 0.3 μmol/L of different SIRTs (SIRT1-SIRT3 or SIRT6) and 1 mmol/L NAD+ in assay buffer at 37 °C for 30 min. Data are presented as mean±SD, n=3.
3.4. Comparison and further study of the two SIRT2 specific substrates
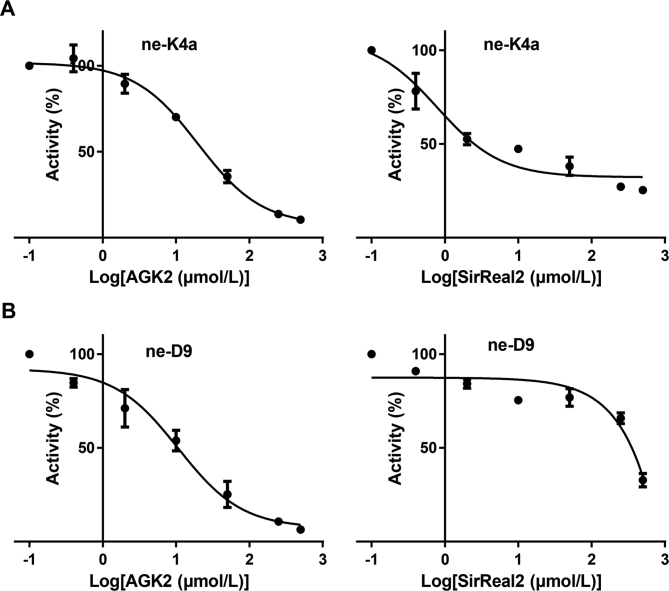
In this study, two SIRT2 specific substrates, ne-D9 and ne-K4a, were discovered. Both ne-D9 and ne-K4a had negligible fluorescence at excitation wavelength of 305 nm without enzyme or NAD+. An intense emission peak was observed when SIRT2, NAD+ and peptides were added simultaneously (Supporting Information Fig. S5). In terms of kinetic parameters, the Kcat/Km values reflected a slightly lower affinity between ne-K4a and SIRT2 than that between ne-D9 and SIRT2 (Table 1 and Supporting Information Fig. S3). The effects of two SIRT2 inhibitors, AGK2 and SirReal2, on SIRT2-mediated hydrolysis of ne-K4a and ne-D9 were then measured. AGK2 is a selective SIRT2 inhibitor and has slightly inhibiting effect on SIRT1 and SIRT331. SirReal2 is a specific and potent SIRT2 inhibitor, which has very little influence on SIRT3–SIRT5 and affects SIRT1 and SIRT6 at high concentrations32. In ne-K4a assay system, AGK2 and SirReal2 inhibited SIRT2 with the IC50 of 20.02 and 0.96 μmol/L, respectively (Fig. 5A). For ne-D9 assessment system, AGK2 showed a strong inhibition of SIRT2 with the IC50 of 10.71 μmol/L but SirReal2 had a significantly lower effect on SIRT2 (Fig. 5B). Since the principle of detection and different substrates, it was not surprising that the IC50 values of AGK2 and SirReal2 measured through ne-K4a or ne-D9 assessment system were different from those reported by other researchers (IC50 of 3.5 μmol/L33 and 140 nmol/L32, respectively). Moreover, the reported inhibitory effect of both AGK2 and SirReal2 was in terms of deacetylase activity. Comparing the two specific probes in different aspects, it was thought that ne-K4a may be more suitable for screening SIRT2 modulators in vitro. Although the kinetic parameters of ne-K4a are slightly inferior to those of ne-D9, the existing inhibitors could be well recognized in ne-K4a-based assay system. Therefore, a Z factor of 0.66, S/N of 124.13 and CV of 12.22% were detected for the ne-K4a assay system after optimizing the assay conditions (Supporting Information Fig. S6). The result illustrates that ne-K4a system is feasible for high-throughput screening34.
Figure 5.
Dose–response curves and IC50 value for AGK2 and SirReal2 examined by ne-D9-based assay and ne-K4a-based assay. Dose–response inhibition experiments for AGK2 and SirReal2 in (A) ne-K4a-based assay and (B) ne-D9-based assay. The IC50 values of AGK2 and SirReal2 were determined to be 20.02±2.47 μmol/L and 0.96±0.56 μmol/L, respectively, in ne-K4a-based assay. The IC50 value of AGK2 was determined to be 10.71±3.88 μmol/L. The reactions were performed with 20 μmol/L ne-K4a or ne-D9, 1 mmol/L NAD+, 0.3 μmol/L SIRT2 and inhibitor (AGK2 or SiReal2) at gradient concentrations (0, 0.2, 1, 6, 30, 100, and 500 μmol/L) in assay buffer at 37 °C for 30 min. Data are presented as mean±SD, n=3.
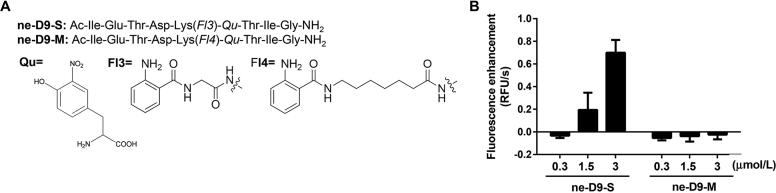
It was reported that the inhibition effect of NAM on SIRTs was varied in substrates with different lysine acyl chain length35. Analogously, the selective inhibitor SirReal2 of SIRT2 was reported to show lower inhibiting effect against demyristoylation activity, but no distinct effects on inhibiting acetyl and decanoyl deacylation were noted3. On the basis of these findings, the fluorescently labeled lysine acyl side chain of ne-D9 was truncated into two new substrates, ne-D9-S and ne-D9-M (Fig. 6A). The aim was to demonstrate whether altering the acyl chain length of substrates would influence the assessment of the inhibitory abilities of AGK2 or SirReal2 to SIRT2. However, it was found that neither ne-D9-S nor ne-D9-M could be hydrolyzed by SIRT2 at 0.3 μmol/L. An obvious fluorescent increment would appear for ne-D9-S while the concentration of SIRT2 was up to 3 μmol/L, and ne-D9-M was hard to be hydrolyzed by SIRT2 even at a high concentration (Fig. 6B).
Figure 6.
Evaluation of newly developed fluorescent substrates, ne-D9-S and ne-d9-M. (A) The structures of two developed ne-D9 fluorescent substrates, ne-D9-S and ne-D9-M. (B) Fluorescence enhancement velocity of 20 μmol/L of substrates (ne-D9-S and ne-D9-M) incubating with SIRT2 at gradient concentrations (0.3, 1.5 and 3 μmol/L), 1 mmol/L NAD+ in assay buffer at 37 °C for 30 min. Data are presented as mean±SD, n=3.
Considering all the above results, we infer that ne-K4a may be a specific fluorescent probe more suitable for screening SIRT2 modulators in vitro. In addition, all the three substrates which were modified on the acyl side chain length, ne-K9-S, ne-D9-S, ne-D9-M, showed a sharp decrease in fluorescence enhancement velocity under the action of SIRT2, implying that SIRT2 was highly sensitive to the fluorescent-labeled acyl side chain length of the substrates.
3.5. The application of continuous SIRT2 assay in cell lysis
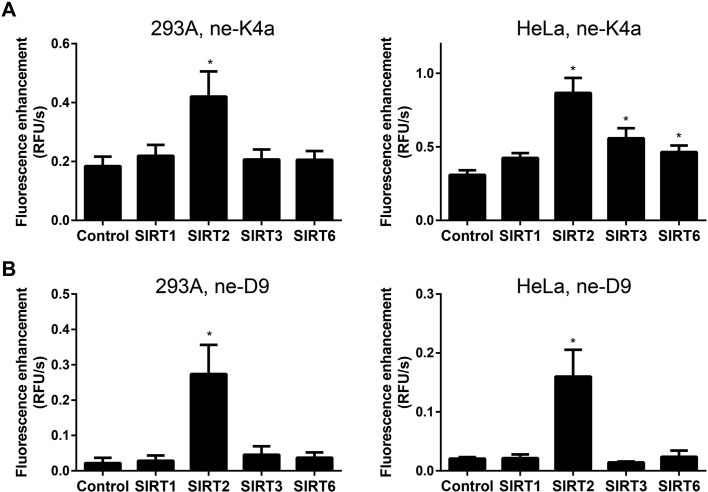
In view of the specificity of ne-D9 and ne-K4a for SIRT2, we postulated that the two fluorescent probes may be involved in cell lysis. Therefore, cell lysates of HeLa and HEK293A with or without overexpressing SIRT1, SIRT2, SIRT3 and SIRT6 were prepared (Supporting Information Fig. S7). They were added to the SIRT2 activity evaluating system using substrates, ne-D9 or ne-K4a, to confirm two aspects: (1) the stability of ne-D9 and ne-K4a in cell lysates; (2) the specificity of ne-D9 and ne-K4a for SIRT2 in cell lysates. It was apparent that fluorescence intensity increased for ne-K4a even in wide-type cell lysis, reflecting that ne-K4a was unstable in cell lysis. However, the increase in fluorescence velocity of SIRT2 group was higher compared to other groups obviously, confirming the specificity of ne-K4a for SIRT2 (Fig. 7A). A weak fluorescence intensity was observed when ne-D9 reacted with the cell lysis sample with only basal level of SIRT1, SIRT3, and SIRT6. In contrast, when SIRT2 was overexpressed, the fluorescence intensity of substrate ne-D9 appearred increased significantly (Fig. 7B). This indicated that ne-D9 was stable in the cell lysis and thus may be applied in cell lysis to assess the activity of cell SIRT2 enzyme. In contrast, when the cell lysate overexpressing SIRT4, SIRT5 and SIRT7 was reacted with ne-K4a and ne-D9, unlike SIRT2 group, the fluorescence velocity was not increased, which eliminated the possibility that the remaining members of SIRTs could react with ne-K4a and ne-D9 (Supporting Information Fig. S8). Based on these findings, we speculate that ne-D9 is suitable to detect the agonist of SIRT2 in cells.
Figure 7.
Application of two specific fluorescent probes in cell lysis. Fluorescence enhancement velocity of 20 μmol/L of substrates, (A) ne-K4a and (B) ne-D9, incubating with 45 μg of supernatant protein derived from cell lysis (gathered from 293A or HeLa cells transfected with or without SIRT1–SIRT3 or SIRT6 plasmids), and 1 mmol/L NAD+ in assay buffer at 37 °C for 30 min. Data are presented as mean±SD, n=3; ∗P<0.05 vs. control.
4. Conclusions
We developed a continuous, specific fluorescent probe, ne-K4a, for screening SIRT2 modulators in vitro. Compared with the current assay methods for SIRT2, including two-step method, radioisotope-labeled method and HPLC method, the novel ne-K4a assay system has three major advantages. First, the ne-K4a assay system is free from interference with trypsin modulators. Second, the detection duration of the ne-K4a assay system is less than 30 min, which is different from the detection duration of some commercial kits based on two-step method, being nearly 2 h, because both SIRT and trypsin reactions requires longer time to reach completion. Third, the whole ne-K4a system only requires addition of ne-K4a, NAD+ and SIRT2 in the buffer, which greatly simplifies the sample handling and experimental operation. In addition to in vitro application, another specific probe, ne-D9, displayed good stability in cell lysates, indicating that it can be applied in the detection of SIRT2 activity in cell lysates. However, ne-D9 cannot be applied in live cell imaging of SIRT2 activity because its excitation/emission wavelength of the fluorophore is unsuitable. This can be addressed by improving the one-step method through changing the fluorophore. Finally, this study examined the influence of fluorescent peptide contexts, number of amino acids in fluorescent substrates and fluorescent-labeled acyl side chain length on the one-step based assay, thereby providing reference for the design of SIRTs fluorescent substrates. Particularly, the sequence of substrate ne-D9 is highly specific to SIRT2 in vitro because it was verified to be recognized only by SIRT2 in the traditional two-step based assay and in the same condition in our one-step based assay. We envision that our probes can be useful tools for screening SIRT2 modulators, and will promote the development of new one-step-based SIRT assay in the future.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (31671437), the Natural Science Foundation of Guangdong Province, China (2016A030313335), and the Guangdong Provincial Key Laboratory of Construction Foundation, China (2017B030314030), and Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program, China (2017BT01Y093). We thank Dr. Baohua Liu (Department of Biochemistry and Molecular Biology, Shenzhen University, Health Science Center, China) for the gift of plasmid of SIRT7.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2019.05.007.
Contributor Information
Peiqing Liu, Email: liupq@mail.sysu.edu.cn.
Min Li, Email: limin65@mail.sysu.edu.cn.
Appendix A. Supporting information
The following are the Supporting data to this article:
References
- 1.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 3.Galleano I., Schiedel M., Jung M., Madsen A.S., Olsen C.A. A continuous, fluorogenic sirtuin 2 deacylase assay: substrate screening and inhibitor evaluation. J Med Chem. 2016;59:1021–1031. doi: 10.1021/acs.jmedchem.5b01532. [DOI] [PubMed] [Google Scholar]
- 4.Du J., Zhou Y., Su X., Yu J.J., Khan S., Jiang H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan M., Peng C., Anderson K.A., Chhoy P., Xie Z., Dai L. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metabol. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathias R.A., Greco T.M., Oberstein A., Budayeva H.G., Chakrabarti R., Rowland E.A. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159:1615–1625. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson K.A., Huynh F.K., Fisher-Wellman K., Stuart J.D., Peterson B.S., Douros J.D. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metabol. 2017;25:838–855. doi: 10.1016/j.cmet.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao X., Xiong Y., Li X., Li X.D. A chemical reporter facilitates the detection and identification of lysine HMGylation on histones. Chem Sci. 2018;9:7797–7801. doi: 10.1039/c8sc02483a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H., Khan S., Wang Y., Charron G., He B., Sebastian C. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L., Shi L., Yang S., Yan R., Zhang D., Yang J. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun. 2016;7:12235. doi: 10.1038/ncomms12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haigis M.C., Guarente L.P. Mammalian sirtuins—emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 12.Hershberger K.A., Martin A.S., Hirschey M.D. Role of NAD+ and mitochondrial sirtuins in cardiac and renal diseases. Nat Rev Nephrol. 2017;13:213–225. doi: 10.1038/nrneph.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Fidalgo S., Villegas I., Sanchez-Hidalgo M., de la Lastra C.A. Sirtuin modulators: mechanisms and potential clinical implications. Curr Med Chem. 2012;19:2414–2441. doi: 10.2174/092986712800269272. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe H., Inaba Y., Kimura K., Matsumoto M., Kaneko S., Kasuga M. Sirt2 facilitates hepatic glucose uptake by deacetylating glucokinase regulatory protein. Nat Commun. 2018;9:30. doi: 10.1038/s41467-017-02537-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Z., Zhao J., Deng W., Chen Y., Shang J., Song K. Identification of a cellularly active SIRT6 allosteric activator. Nat Chem Biol. 2018;14:1118–1126. doi: 10.1038/s41589-018-0150-0. [DOI] [PubMed] [Google Scholar]
- 16.Borra M.T., Denu J.M. Quantitative assays for characterization of the Sir2 family of NAD+-dependent deacetylases. Methods Enzymol. 2004;376:171–187. doi: 10.1016/S0076-6879(03)76011-X. [DOI] [PubMed] [Google Scholar]
- 17.Dancy B.C., Ming S.A., Papazyan R., Jelinek C.A., Majumdar A., Sun Y. Azalysine analogues as probes for protein lysine deacetylation and demethylation. J Am Chem Soc. 2012;134:5138–5148. doi: 10.1021/ja209574z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borra M.T., Smith B.C., Denu J.M. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Liu T., Liao S., Li Y., Lan Y., Wang A. A mini-review on Sirtuin activity assays. Biochem Biophys Res Commun. 2015;467:459–466. doi: 10.1016/j.bbrc.2015.09.172. [DOI] [PubMed] [Google Scholar]
- 20.Li Y., You L., Huang W., Liu J., Zhu H., He B. A FRET-based assay for screening SIRT6 modulators. Eur J Med Chem. 2015;96:245–249. doi: 10.1016/j.ejmech.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Li Y., Huang W., You L., Xie T., He B. A FRET-based assay for screening SIRT5 specific modulators. Bioorg Med Chem Lett. 2015;25:1671–1674. doi: 10.1016/j.bmcl.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Kawaguchi M., Ikegawa S., Ieda N., Nakagawa H. A fluorescent probe for imaging Sirtuin activity in living cells, based on one-step cleavage of the dabcyl quencher. Chembiochem. 2016;17:1961–1967. doi: 10.1002/cbic.201600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuster S., Roessler C., Meleshin M., Zimmermann P., Simic Z., Kambach C. A continuous sirtuin activity assay without any coupling to enzymatic or chemical reactions. Sci Rep. 2016;6:22643. doi: 10.1038/srep22643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang Y.L., Lin H. An improved fluorogenic assay for SIRT1, SIRT2, and SIRT3. Org Biomol Chem. 2016;14:2186–2190. doi: 10.1039/c5ob02609a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlicker C., Boanca G., Lakshminarasimhan M., Steegborn C. Structure-based development of novel sirtuin inhibitors. Aging (Albany NY) 2011;3:852–872. doi: 10.18632/aging.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X., Shi L., Xie N., Liu Z., Qian M., Meng F. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat Commun. 2017;8:318. doi: 10.1038/s41467-017-00396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avalos J.L., Celic I., Muhammad S., Cosgrove M.S., Boeke J.D., Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 28.Bheda P., Jing H., Wolberger C., Lin H. The substrate specificity of Sirtuins. Annu Rev Biochem. 2016;85:405–429. doi: 10.1146/annurev-biochem-060815-014537. [DOI] [PubMed] [Google Scholar]
- 29.Roessler C., Tuting C., Meleshin M., Steegborn C., Schutkowski M. A novel continuous assay for the deacylase Sirtuin 5 and other deacetylases. J Med Chem. 2015;58:7217–7223. doi: 10.1021/acs.jmedchem.5b00293. [DOI] [PubMed] [Google Scholar]
- 30.Feldman J.L., Baeza J., Denu J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Outeiro T.F., Kontopoulos E., Altmann S.M., Kufareva I., Strathearn K.E., Amore A.M. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science. 2007;317:516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 32.Rumpf T., Schiedel M., Karaman B., Roessler C., North B.J., Lehotzky A. Selective Sirt2 inhibition by ligand-induced rearrangement of the active site. Nat Commun. 2015;6:6263. doi: 10.1038/ncomms7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatum P.R., Sawada H., Ota Y., Itoh Y., Zhan P., Ieda N. Identification of novel SIRT2-selective inhibitors using a click chemistry approach. Bioorg Med Chem Lett. 2014;24:1871–1874. doi: 10.1016/j.bmcl.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 35.Feldman J.L., Dittenhafer-Reed K.E., Kudo N., Thelen J.N., Ito A., Yoshida M. Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry. 2015;54:3037–3050. doi: 10.1021/acs.biochem.5b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.