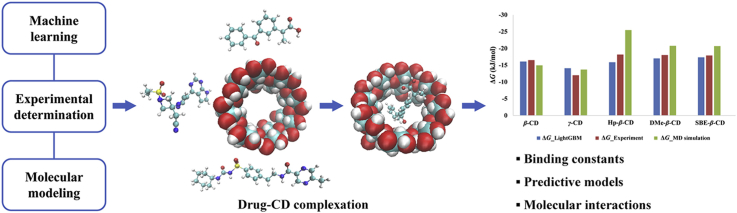
Abstract
Most pharmaceutical formulation developments are complex and ideal formulations are generally obtained after extensive experimentation. Machine learning is increasingly advancing many aspects in modern society and has achieved significant success in multiple subjects. Current research demonstrated that machine learning can be adopted to build up high-accurate predictive models in drugs/cyclodextrins (CDs) systems. Molecular descriptors of compounds and experimental conditions were employed as inputs, while complexation free energy as outputs. Results showed that the light gradient boosting machine provided significantly improved predictive performance over random forest and deep learning. The mean absolute error was 1.38 kJ/mol and squared correlation coefficient was 0.86. The evaluation of relative importance of molecular descriptors further demonstrated the key factors affecting molecular interactions in drugs/CD systems. In the specific ketoprofen–CD systems, machine learning model showed better predictive performance than molecular modeling calculation, while molecular simulation could provide structural, dynamic and energetic information. The integration of machine learning and molecular simulation could produce synergistic effect for interpreting and predicting pharmaceutical formulations. In conclusion, the developed predictive models were able to quickly and accurately predict the solubilizing capacity of CD systems. Current research has taken an important step toward the application of machine learning in pharmaceutical formulation design.
Key words: Machine learning, Deep learning, LightGBM, Random forest, Cyclodextrin, Binding free energy, Molecular modeling, Ketoprofen
Graphical abstract
Current research presented an alternative to traditionally trial-and-error experimentation for drug–CD formulation development. It revealed that the integration of experimental determinations, molecular modeling calculation and data-driven machine learning techniques could provide a new solution for highly efficient and accurate formulation development in the future.
1. Introduction
In pharmaceutical industry, the formulation developments intend to optimize the excipient composition and process parameters to acquire optimal formulations with satisfactory physicochemical and biological characteristics1. However, the process of current formulation development highly depends on the trial-and-error experimentation by the experiences of individual formulation scientists. Thus, the multivariable and complex nature of formulation design seriously hinders the efficiency and success rate of product development2, 3. Moreover, the successful application of high-throughput experimentation in the field of drug discovery in the past decade has contributed to significant increase of active compounds, which put forward constant pressure for the productivity of formulation development4, 5. Although the statistical experimental designs and optimization techniques, such as orthogonal design or response surface methodology, to some extent simplify the formulation screening process, the extensive experimentation is still needed due to the unpredictable formulation performance6. It is necessary to seek for a high efficiency and accuracy methodology for the prediction and virtual screening of pharmaceutical formulations.
Currently molecular simulation-based calculations in the formulation development provide an alternative technique to the traditional trial-and-error experiments. Numerous molecular modeling methods, like molecular dynamics (MD), docking, Monte Carlo and even quantum mechanics, have been extensively applied3, 7, 8. The molecular modeling approaches are highly capable of the explanation for the molecular mechanism for specific formulations and delivery systems, but have the limited ability to distinguish key index in some specific formulations. Another existed significant limitation is that the modeling systems with limited molecule number may not represent the real experiments due to the limited computing capability. These methods have difficulties in making balance between time consumption and accuracy8. In any case, from the wide studies performed to date, it is highly necessary and urgent to build up predictive models which allow a high-accuracy prediction in the pharmaceutical formulations and contributing factors analysis.
Machine learning is able to learn from input data, automate analytical model building and even update outputs as new data becomes available by computer algorithms9, 10. The latest machine learning algorithm—deep learning garnered significant attention, which is representation-learning methods with multiple levels of representation11, 12. These algorithms are able to make high-accuracy predictions for the classification and regression tasks, which lead to extensive application in the bioinformatics and computational biology. However, the application of machine learning in pharmaceutical research especially formulations development, is still very limited13. Given commercial confidentiality and long research periods, the pharmaceutical datasets start out too small to guarantee implementation of these algorithms. Despite many challenges, two recent studies have advanced the application of machine learning in the pharmaceutical field14, 15, 16, 17. The remote liposome study has used machine learning approaches to distinguish the drug loading efficiency and to predict the drug loading on the 366 formulation dataset14. Another attempt was the combination of machine learning and nondestructive ultrasonic to develop a predictive model for the tablet breaking force and disintegration time for advancing the quality-by-design paradigm18. Because of the complex nature of formulation design process and multidimensionality of chemical structures, it is really necessary to explore whether machine learning could be further expanded into formulation prediction based on available data.
Current research selected the cyclodextrin (CD) complexation systems as the model system for the predictive model development because of its extensive application in pharmaceutical field, relatively controllable variables and specific evaluation index. CDs can accommodate varieties of non-polar molecules into the inner cavity to form non-covalent host-guest inclusion complexes, which has been successfully used in pharmaceutical formulations. The guest/CDs binding process is not fixed or permanent, but dynamic and reversible19. Complexation free energy or equilibrium constant is a key index to evaluate the strength of host–guest complexation and disassociation/association stability of the complex, which can decide the usage of a specific drug–CDs inclusion complex in the formulation design20, 21. Many experimental techniques have been developed to measure the complexation free energy including phase solubility measurements, calorimetric titration, nuclear magnetic resonance chemical shift, freezing point depression, pH-metric methods and so on, among which phase solubility method described by Higuchi and Connors is the most widely used22, 23, 24, 25. However, these experimental measurements often bring serious challenges for the pharmaceutical scientists because of the limited synthetic amount of new compounds and the limited API's solubility at the early stage of drug discovery20.
This study was proposed to develop high-accurate predictive models of complexation free energy between CDs and guest molecules based on a dataset of 3000 date points by three machine learning techniques (e.g., light gradient boosting machine [LightGBM], random forest [RF] and deep learning [DL]). The relative contributions of molecular descriptors were analyzed to infer underlying molecular interactions in the drug–CD complexation systems. Furthermore, ketoprofen (KTP) was used as a model drug and the KTP–CDs inclusion complexes were studied by the experimental methods and the MD simulations. The comparison of complexation free energy by experimental determination, simulation-based calculation and machine learning model was further conducted.
2. Materials and methods
2.1. Dataset preparation and distribution
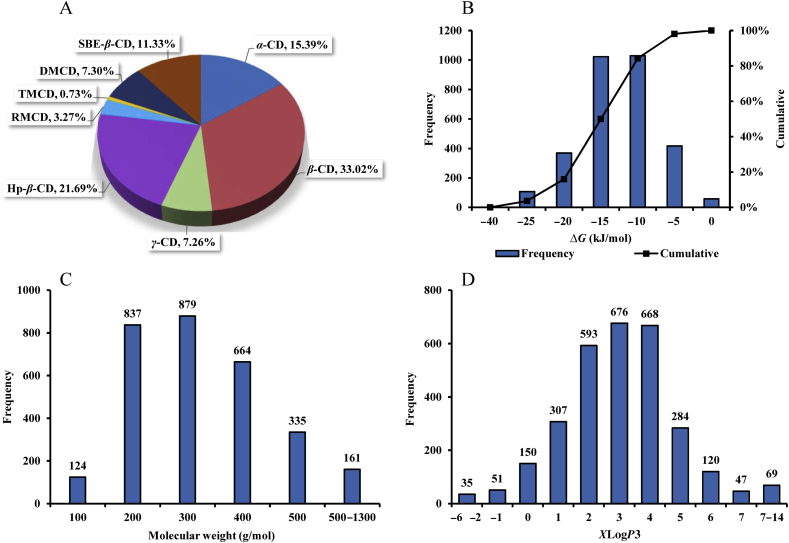
The data for the complexation free energy between CDs and a diverse set of organic molecules were collected from published literatures from the year 1990 to 2018. The literature data were considered acceptable if they belonged to binary CD complexation system with the complexation free energy or the equilibrium constant by phase solubility study. When the experimental pH values were not clear or reported as aqueous solution, they were considered to be neural (pH = 7). The temperatures were assumed to be room temperature (T = 25 °C) if the specific temperature was not noted or reported. The available dataset was further processed to eliminate duplicates, to eliminate entries for the same compounds with different complexation free energies in the same experimental conditions, as well as to remove experiments under uncommonly used conditions (e.g., partially or completely organic solvent). The final dataset with 3000 formulations for the predictive model building contained 1320 guest molecules and 8 CDs, of which the proportions of each CD were shown in Fig. 1A. The complexation free energies between guest molecules and CDs were distributed over the range of 0 to −40 kJ/mol with the mean of −15.12 kJ/mol, as shown in Fig. 1B. The guest molecule set contained a large amount of structurally diverse organic compounds, such as phenols, ketones, alcohols, steroids, nitriles, barbitals, and hydrochloride.
Figure 1.
Relative distribution of data points in the full dataset: (A) The percentage of experimental data for each cyclodextrin. (B) The distribution of complexation free energy between cyclodextrins and guest molecules. (C) The molecular weight distribution of guest molecules. (D) The XlogP3 distribution of guest molecules. The full dataset was comprised of 3000 data points with 1320 different structures. Hp-β-CD, 2-hydroxypropyl-β-cyclodextrin; RMCD, randomly methylated β-cyclodextrin; TMCD, (2,3,6-tri-O-methyl)-β-cyclodextrin; DMCD, (2,6-di-O-methyl)-β-cyclodextrin; SBE-β-CD, sulfobutylether-β-cyclodextrin.
2.2. Molecular descriptors and dataset slitting
Both the guest molecules and CDs were characterized with an appropriate set of molecular descriptors. The molecular descriptors used in this study were computed using ALOGPS 2.1 program (VCCLAB, Virtual Computational Chemistry Laboratory, http://www.vcclab.org) and ChemAxon program (https://chemaxon.com/company)26. The total of 17 and 22 molecular descriptors were used to characterize the guest molecules and CDs, respectively. These calculated molecular descriptors were then combined with the corresponding experimental conditions (described as above, pH value and temperature) to assemble a hybrid set of molecular descriptors for each entry in the dataset.
Relative distributions of data points used in the dataset were shown in Fig. 1, which shows that the percentages of data points for each CD in the dataset vary greatly. Thus, the dataset was split by the method of stratified random sampling, which is a method of sampling that random samples extracted from each CD group was proportional to the size of the original group. The dataset was randomly split into training set (80%), test set (10%) and validation set (10%), among which the data points of each CD kept the same percentage as that in the original dataset. It guaranteed that the composition of subsets was representative and avoided the unbalance in these three datasets. The test set is completely independent from the training set and validation set. Running a training set and validation set through an algorithm teaches the model how to weigh different features, adjust their parameters according to their likelihood of minimizing errors in the results, while the test set serves to evaluate the generalization ability of the predictive model.
2.3. Machine learning methods
Currently there have been developed many predicting methods in machine learning. This research compared the predictive models from three widely used algorithms—LightGBM, RF and DL. LightGBM was the first time to be used in the pharmaceutical system.
LightGBM uses an open source gradient boosting decision tree (GBDT) algorithm by Microsoft (http://lightgbm.apachecn.org/cn/latest/). LightGBM is a fast, distributed and high-performance gradient boosting framework with tree-based learning algorithm, which has been extensively used for both classification and regression tasks. It uses histogram-based algorithms, which buckets continuous feature values into discrete bins27. Compared with other decision tree algorithms, it can reduce the calculation cost of the gain for each split and speed up training. Furthermore, LightGBM grows tree leaf-wise and vertically, while most decision tree learning algorithms grow trees horizontally by level-wise. In fact, LightGBM tends to obtain lower loss than level-wise algorithms by choosing the leaf with max delta loss to grow when holding leaf fixed28. Leaf-wise may cause over-fitting when the dataset is small. The max_depth in LightGBM is an important parameter in handling with over-fitting. In current research, the max_depth for tree model was limited by set max_depth to 4. The complexity of the tree model was controlled by the parameter of num_leaves, which was set to 9. N_estimators was used to control the number of boosted trees to fit, it was set to 800. The number of boosting iterations was set to 1000. When the LightGBM was used on the sparse datasets, each parameter had a small adjustment.
Random forest uses sklearn.ensemble RandomForestRegressor (http://scikitlearn.org/stable/modules/generated/sklearn.ensemble.RandomForestRegressor.html). RF is an ensemble method for classification or regression tasks by using bootstrap samples of the training data and random feature selection in tree construction29. Each bootstrap samples grow a tree and the best split at each node is defined among a randomly selected subset of mtry descriptors30. When the data is limited, random forest estimates the ensemble prediction performance by performing a type of cross-validation in parallel with the bootstrap procedure with the out-of-bag data22. The RF prediction of new data is made by aggregating the prediction of the ensemble. In addition to built-in estimation of prediction accuracy, RF can also calculate the importance of each molecular descriptor based on the out-of-bag predictions with each descriptor permuted22, 30. During the data training, n_estimators controlled the number of trees in the forest was set to 300, while max_depth controlled the maximum depth of the tree was set to 14. When looking for the best split, the number of features (max_features) to consider was set to 32. Min_sample_split, represented the minimum number of samples required to split an internal node, was set to 2. Min_samples_leaf, controlled the minimum number of samples required to be at a leaf node, was set to 3.
Deep Learning using Keras deep learning library in python (http://keras.io/). DL is a form of machine learning method, which comprises of multiple processing layers to learn representations of data with multiple levels of abstraction12. Unlike simple artificial neural networks, DL has multiple hidden layers with different weights, and then passes the signals successively deeper in the network until the output layer. During training procedure, the deep network trains the first layer as an auto-encoder with the training sets as the input until the average of the objective function stops decreasing, and then trains the second layer as an auto-encoder taking the first layer's output as the input. The process is iterated for the desired number of layers and finally the output of the last layer as the input for the prediction layer31. In this research, the neural network included three hidden layers—the first layer containing 512 neurons, the second layer containing 256 neurons, and the third layer containing 64 neurons. The first three hidden layers used Rectified Linear Units (ReLU) as the activate function, while the last layer used a linear function. The number of samples that going to be propagated through the network was controlled by the batch_size, which was set to 16. The parameter of epoch indicated the times of the entire dataset passed through the neural network, which was set to 3600. During the training process, it would shuffle data for each epoch to have different data for each batch.
2.4. Model selection and comparison
The predictive performance had been characterized by three statistics, which were the mean absolute error (MAE), the root means square error (RMSE) and the squared correlation coefficient (R2). They were defined as:
| (1) |
| (2) |
| (3) |
where and were the experimentally observed and theoretically predicted complexation free energy between molecule i and corresponding CD, respectively. yobs,mean was the mean of experimental value and n was the number of data points.
2.5. Experimental study of KTP–CDs inclusion complexes
2.5.1. Materials and reagents
α-, β-, and γ-CD were purchased from J&K Scientific Co., Ltd. (Beijing, China). 2-Hydroxypropyl-β-cyclodextrin (Hp-β-CD; FW = 1541.54 g/mol, purity > 99%), dimethyl-β-cyclodextrin (DMe-β-CD, FW = 1331 g/mol, purity > 99%), and sulfobutylether-β-cyclodextrin (SBE-β-CD; FW = 2242.01 g/mol, purity > 99%) were purchased from Shanghai Aladdin Bio-Chem Technology Co., Ltd. (Shanghai, China). Ketoprofen (KTP, purity > 99%) was purchased from Dongkangyuan Technology Co., Ltd. (Wuhan, China). All other chemicals in the study were of analytical grade.
Phase solubility study was performed to determine the binding constant (K) according to the methods described by Higuchi and Connors24. Excess amounts of KTP were added to 3 mL of various concentrations of aqueous CDs solutions. The concentration of three parent CDs were between the ranges of 0 to 15 mmol/L, while the concentration of the other three CD derivatives were 0 to 9 mmol/L. The mixtures were shaken at 37 ± 0.5 °C for two days until equilibrium. Afterward, the samples were filtered through a 0.45 μm Millipore filter and then assayed by UV spectrophotometer at 260 nm. The binding constant (K, L/mol) of the KTP–CD complex was calculated from the phase solubility diagrams using Eq. (4) and the complexation free energy (ΔG, kJ/mol) could be further calculated according to Eq. (5).
| (4) |
| (5) |
where S0 was the intrinsic solubility of KTP in distilled water, and the slope was obtained from the phase solubility curve by linear regression between KTP and CD concentration. R is the gas constant, and T is Kelvin temperature.
2.6. Molecular dynamic simulation of KTP–CDs inclusion complexes
The molecular dynamic (MD) simulations of KTP–CDs inclusion complexes were carried out by using AMBER14 software package and Generalized Amber Force Field (GAFF)32. The initial structures were generated by docking KTP with various CDs by AutoDock Tools package and then these structures were loaded into LEAP model to construct the solvated systems by using the TIP3P water box with 20 Å radius33. All systems were equilibrated in the NPT ensembles at T = 310 K and P = 1 bar, and then ran MD simulation for 100 ns with a time step of 2 fs. The molecular mechanics with a Possion–Boltzmann/surface area solvent (MM-PBSA) method was used to calculate the binding affinity “ΔGbinding”. The binding entropy (ΔS) was calculated by normal mode analysis using the ptraj program in AMBER Tools. The binding enthalpy (ΔE) comprising of electrostatic energy (ELE), van der Waals (VDW), non-polar and polar contributions to solvation (PBSOL), were calculated for the complexes according to Eq. (6). The binding free energy (ΔG) was further calculated according to Eq. (7).
| (6) |
| (7) |
3. Results and discussion
3.1. Predictive performance of three machine learning methods
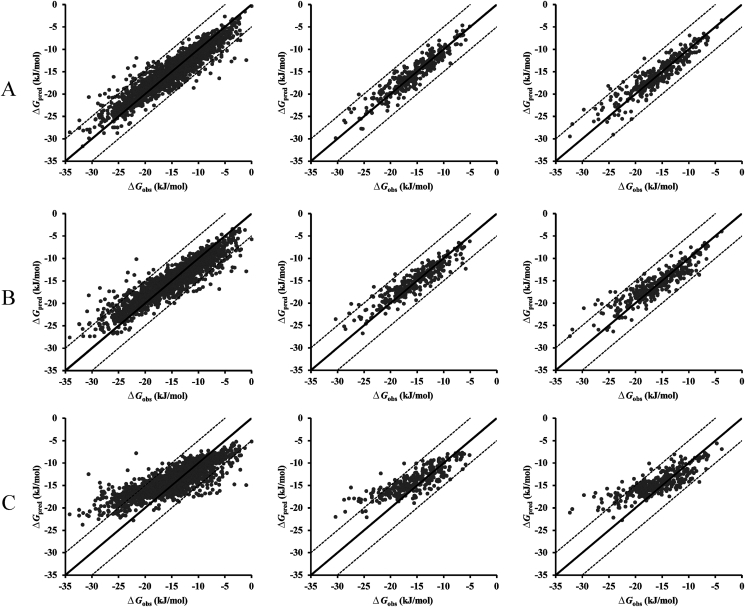
With these 3000 data points available in hand, the predictive accuracies of three machine learning model were evaluated by using 80% of the data as training set and 10% of the data as validation set to predict the remaining 10% dataset (as test set). Fig. 2 showed the scatter plots of the predicted vs. observed complexation free energy on full dataset by three machine learning methods and the detailed statistical performance of the predictive models for CDs complexation free energy were presented in Table 1. For the LightGBM model, the MSE and RMSE of the training set was 1.41 and 1.94 kJ/mol, respectively, with a coefficient (R2) value of 0.86. Generally, the predictive accuracy of the built model highly relied on the similarity degrees of the molecules to be predicted and the molecules in the training set. In order to measure the built model's generalizability, an independent and new test set was performed to calculate. While applying the LightGBM model for calculations on the test set, MAE, RMSE and R2 were 1.38 kJ/mol, 1.83 kJ/mol and 0.86, respectively. Thus, the built LightGBM model showed reasonable statistical criteria for new dataset. Turning to random forest model, it did not show overfitting and also had achieved good predictive performance on both the training set and test set. However, the predictive performance of the random forest model did not exceed that of the LightGBM model. The statistical performance of the deep learning predictive models showed the MAE for the training set was 3.34 kJ/mol with a R2 value of 0.76, and the MAE for the testing set was 3.36 kJ/mol with a R2 value of 0.62. Compared with LightGBM model and random forest model, deep learning model showed a relatively poor predictive performance.
Figure 2.
Scatter plot of predicted vs. observed complexation free energy on full dataset by three machine learning methods. The predicted results of (A) LightGBM model, (B) random forest model, and (C) deep learning model. For all the models, an 80/10/10 split of training, validation and test set was used to measure the predictive performance.
Table 1.
Statistical performance of the predictive models for CDs complexation free energy.
| Method | Statistical parameter | Training set | Validation set | Test set |
|---|---|---|---|---|
| LightGBM | R2 | 0.86 | 0.87 | 0.86 |
| MAE (kJ/mol) | 1.41 | 1.28 | 1.38 | |
| RMSE (kJ/mol) | 1.94 | 1.63 | 1.83 | |
| RF | R2 | 0.83 | 0.83 | 0.81 |
| MAE (kJ/mol) | 1.54 | 1.39 | 1.54 | |
| RMSE (kJ/mol) | 2.17 | 1.89 | 2.11 | |
| DL | R2 | 0.76 | 0.63 | 0.62 |
| MAE (kJ/mol) | 2.53 | 2.41 | 2.56 | |
| RMSE (kJ/mol) | 3.34 | 3.12 | 3.36 |
RF, random forest; DL, deep learning.
Although three predictive models shown high prediction accuracy both in the training and test set, there still were large deviations for some data points between the observed values and predicted values in the test set, especially when the value of complexation free energy was less than −20 kJ/mol, as shown in Fig. 2. Significant proportion of the deviations may be caused by the value distribution of complexation free energy in the original dataset, as show in Fig. 1B. The value of complexation free energy in the original data set between −5 and −20 kJ/mol accounted for more than 90% data points in the full dataset. Therefore, the imbalance distribution of dataset had a serious influence on the model construction and the predictive accuracy. In addition, the size of the dataset also had a great impact on the model construction and its predictive accuracy, which had been verified by the following research on the sparse data.
By comparing with the statistical performance of the predictive models on the training and test set, the LightGBM model can deliver high predictive accuracy without succumbing to overfitting. Moreover, LightGBM has the unique advantages with a faster training speed, higher efficiency and better accuracy than any other boosting algorithm because its histogram algorithm can produce much more complex trees by leaf-wise split approach rather than a level-wise approach. But the biggest challenge for the LightGBM algorithm is that it is sensitive to overfitting, especially on the small dataset. Therefore, the parameters of the LightGBM model should be carefully tested, such as the maximum depth of tree, the minimum number of the records for a leaf, and the fraction of data to be used for each iteration, etc.
Random forest has become one of the most widely used machine learning algorithms in the pharmaceutical researches because it can be used for both classification and regression tasks. In addition, random forest also has another two advantages, which attract the pharmaceutical scientists to bring this algorithm in their research. One advantage is that random forest can randomly sample the data and construct decision trees and aggregate many decision tress to limit overfitting, which is one of the biggest problem in machine learning30. The other advantage is that random forest can rank the relative importance of each feature on the prediction. Recently, Ahneman et al.10 used various machine learning techniques to predict the reaction performance in C–N cross-coupling. The results showed that the random forest algorithm provided better predictive performance than the other five techniques. The random forest algorithm was also used to rank the important descriptors and the relative experiments verified its effectiveness.
Deep learning uses multi-layered artificial neural network to characterize the input features at different levels and then optimizes the model performance through various training skills. In fact, deep learning algorithm can not only build the predictive models, but also have the potential to automatically learn representations from data without introducing hand-coded rules or human domain knowledge. Therefore, compared with simple artificial neural networks and other machine learning techniques, deep learning can learn directly from raw data and be more useful for more complex problems. However, many parameters in deep learning algorithms have to be tuned and large amount of data is needed to come up with somewhat generalizable models. Generally speaking, the predictive accuracy of the models increases with increased data. In the pharmaceutical formulation research, however, the available datasets are quite small, which greatly limit further application of deep learning. Our group compared the predictive performance of single-layer neural network model and deep learning model for predicting the disintegration time of oral disintegrating tablets. The raw data comprised of 145 formulations. Due to small dataset, the accuracy of the prediction was only 80% on the test set15. Although deep learning has strong predictive potential, the challenges for pharmaceutical researchers is the limited data in the formulation prediction. Meanwhile, the black-box of deep learning models cause many criticisms because it is difficult to provide the explanation between input variables and the built model. Thus, there is still lots of work to successfully introduce deep learning into the pharmaceutical formulation research.
As the results shown in Table 1, all three models of LightGBM, random forest and deep learning showed high predictive accuracy for the complexation free energy between CDs and guest molecules. Based on the comparison of the MAE and R2, the predictive performance of LightGBM model was the best, followed by the random forest model, and then deep learning. Meanwhile, the LightGBM algorithm had the highest efficiency as well as the fastest training speed than another two algorithms. Furthermore, LightGBM algorithm can also directly calculate the feature importance, which helped to guide theoretical analysis. Thus, the LightGBM model was the optimal model for the following research.
3.2. Predictive performance of LightGBM model on sparse data
Machine learning is a sub-field of artificial intelligence and data science for new algorithms and the predictions. It is generally accepted that gathering as much and high-quality data as possible is the key to ensure high predictability of machine learning algorithms. However, it is really difficult to obtain a large quantity of formulation data in pharmaceutics because of long research cycle and high cost in formulation development. Therefore, it is significantly important to explore the exact relationship between the size of dataset and the predictive accuracy of a machine learning algorithm in the formulation prediction.
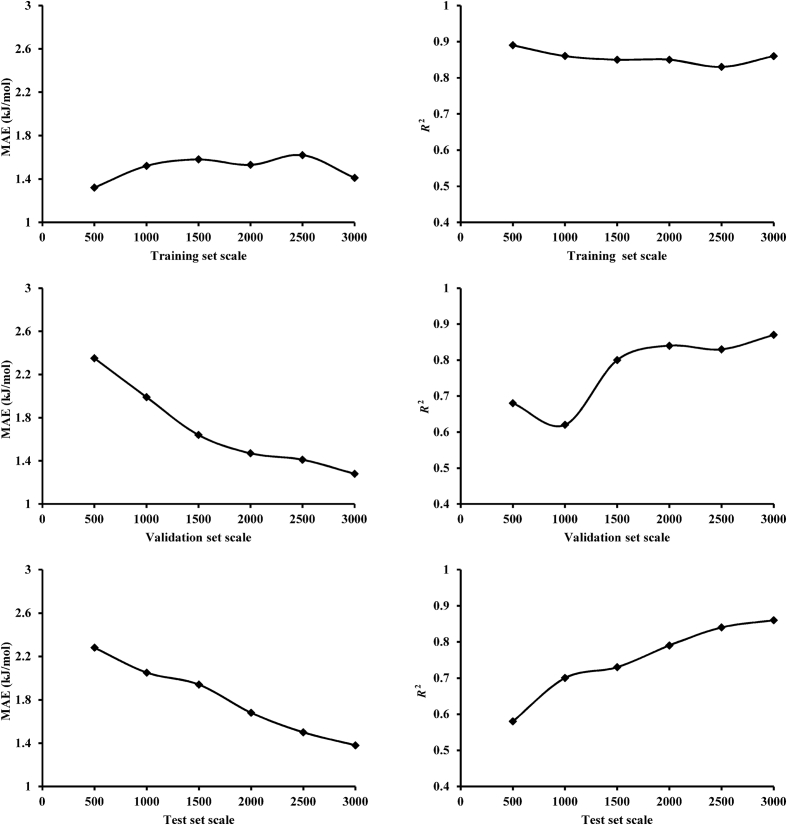
Current research has investigated the predictive performance of the LightGBM model with sparse data, varying from 3000, 2500, 2000, 1500, 1000 to 500. Each dataset was split by the method of stratified random sampling as described above. Predictive performance on sparse data by the method of LightGBM for the training and test set was shown in Fig. 3. In the training set, the value of MAE did not change significantly with the decrease of the dataset, fluctuating between 1.3 and 1.65 kJ/mol, and the R2 value had small fluctuation between 0.8 and 0.9. For the test set, however, it was clearly demonstrated that a gradual erosion in the predictive accuracy occurred from 3000 to 500. The value of MAE increased dramatically with the decrease of dataset, from 1.38 to 2.28 kJ/mol, while the value of R2 also decreased significantly with the decrease of dataset, from 0.86 to 0.58. Results showed that the LightGBM predictive model still had good performance on the training set even when the data points in dataset decreased to 500, which demonstrated that the molecular descriptors used in the current research could exactly captured the properties of input variables. But the predictive performance on the test set became significantly worse with the smaller dataset size, which showed that the LightGBM model had a poor generalization ability on small dataset.
Figure 3.
Training and test set performance of the LightGBM model with sparse data. The data points were gradually reduced from 3000 to 500 and the smaller datasets were randomly extracted from the full datasets. A gradual increase of MAE and a significant drop of R2 occurred with the decrease of dataset size.
In fact, machine learning tends to encounter predictive limitation when substantially different guest molecules are used in the testing set. Even though some predictive models for complexation free energy between CDs and guest molecules had been built in previously published papers, these models were limited to the dataset with no more than 300 data point20, 34. In current research, the full dataset with 3000 data points had about 1320 different chemical structures, which highly contributed to the high-accuracy predictive ability. Therefore, one method for maximizing the extrapolative ability of a model with limited formulation data maybe spread the training data across its interesting chemical space.
On the other hand, higher quality data contributed to higher predictive accuracy. In pharmaceutical industry, formulation data can be gathered from the published literature and patents, pharmaceutical companies or self-measured data. However, very little data can be shared from pharmaceutical companies because of confidentiality. Moreover, very few applications of high-throughput screening methods in formulation development leads to very limited self-measured data. The CD formulation data in current research was collected from the published literature, but almost all datasets from the published literature are flawed, such as missed, repeated, experimental or analytical errors. Furthermore, the reproductivity crisis of scientific literature leads to worse situation of the reliability of data. Thus, the preparation of high-quality dataset is the key step in machine learning process.
3.3. Molecular descriptor contributions for LightGBM model
In order to reveal the molecular mechanism and facilitate the future formulation design of drug–CD complexes, the relative importance of molecular descriptors in the model construction were calculated. Fig. 4 listed the ranking of relative importance of the molecular descriptors in the LightGBM model for the prediction of complexation free energy, which based on the used times of the feature in the model.
Figure 4.
The relative importance of the molecular descriptors in the LightGBM model. Molecular descriptor with the suffix of x represented the descriptor of guest molecules, while with the suffix of y meant the descriptor of cyclodextrins. pH and T (K) represented the experimental conditions in the phase solubility study. The relative importance of molecular descriptors was determined by the measuring the number of times of the features used in the LightGBM model.
It can be clearly remarked that the major contribution to the complexation free energy stemmed from the guest molecules, followed by experimental conditions and CDs. The top five important molecular descriptors with the contribution value above 700 in the LightGBM model were minimum projection radius_x, solvent accessible surface area_x, complexity_x, XLogP3_x and maximum projection radius_x. The minimum projection radius_x and maximum projection radius_x both characterized the size of guest molecules, where the minimum projection radius was more important. The geometrical or steric parameters of guest molecules are decisive rather than chemical factors because the guest compounds with proper size are accommodated into the inner cavity of CDs. During the process of the molecular dynamic simulation of drug–CD complexes, the preferred binding orientation of guest molecules are that the smaller ends have priority to insert into the inner cavity of CDs. Therefore, the minimum projection radius played the most important role in the complexation process.
Solvent accessible surface area_x is the surface area of the guest molecules accessible to a solvent. In the CD complexation process, the solvent accessible surface area of guest molecules is closely related to the transfer free energy from the aqueous environment to the non-polar CD inner cavity. The process that guest molecules displace the polar water molecules from the apolar CD cavity is an energetically favorable interactions, which can help to shift the equilibrium to form the inclusion complexation. In general, one of the main driving force of CD complexation is the release of enthalpy-rich water molecules from the inner cavity, and to gain an apolar–apolar association between guest molecules and inner cavity35.
XLogP3_x is the partition coefficient, which represents the ratio of concentrations of the guest molecules in a mixture of water and oil at equilibrium. The value of XLogP3 is proportional to the hydrophobicity of a molecule, which is related to the hydrophobic interaction during the drug–CD complexation process. Guest molecules with strong hydrophobicity can contribute to the stability of the CD complexation system because of the favored hydrophobic effect. The complexity_x of the guest molecules in current research is defined as the topological complexity, which can be characterized as a steric descriptor of guest molecules by the number of atoms or bonds, branching and cyclicity, etc. To some extent, it represents the flexibility or the rigidity of guest molecules contributing to the CD complexation36. In a word, three of the five most important molecular descriptors were the steric descriptors of guest molecules, while another two were hydrophobic descriptors.
Apart from the five most important descriptors, molecular weight_x, topological polar surface area_x and van der Waals surface area_x of the guest molecules also correlated with the complexation free energy. pH value of the experimental conditions with the contribution value over 600 also showed the importance because most of the guest molecules were either weak acids or weak bases. The ionization of drug molecules has a great influence on the complexation process. For example, the acidic molecule has weak complexation capability in the alkaline solution than neutral environment. Therefore, the value of complexation free energy was strongly affected by pH or the temperature of the experimental conditions.
3.4. Experimental determination of KTP–CDs complexation free energy
Phase solubility study has been a widely applied technique to determine the complexation free energy for the drug–CD inclusion complexes. Phase solubility curves of KTP with different CDs in distilled water at 37 ± 0.5 °C clearly dedicated that the aqueous solubility of KTP linearly increased with the increasing concentrations of CDs except for α-CD with a low value of R2 (shown in Table 2), which could be identified as AL type24. Concerning that the value of all the slopes in KTP–CD solubility curves was less than unity, the stoichiometry may be supposed to be 1:1 (except for α-CD). In order to avoid the deviations of intercepts, the stability constants were calculated based on the intrinsic solubility of KTP. The complexation free energy was calculated to −18.178 kJ/mol for Hp-β-CD, −18.021 kJ/mol for DMe-β-CD and −17.876 kJ/mol for SBE, which showed no big difference between KTP and these three β-CD derivatives. But the calculated binding free energy between KTP and three parent CDs showed an order of β-CD (−16.489 kJ/mol) < γ-CD (−11.986 kJ/mol) < α-CD. Therefore, the solubilization effect of CD derivatives on KTP was significantly stronger than that of native CDs.
Table 2.
Phase solubility study of ketoprofen with different CDs in distilled water at 37 ± 0.5 °C.
| System | Equation | Kca (L/mol) | R2b | ΔGc (kJ/mol) |
|---|---|---|---|---|
| KTP-α-CD | y = 0.007x + 1.157 | 6.408 | 0.578 | N/A |
| KTP-β-CD | y = 0.397x + 1.038 | 598.523 | 0.993 | −16.489 |
| KTP-γ-CD | y = 0.103x + 0.999 | 104.388 | 0.983 | −11.986 |
| KTP-Hp-β-CD | y = 0.559x + 0.744 | 1152.340 | 0.985 | −18.178 |
| KTP-DMe-β-CD | y = 0.544x + 1.046 | 1084.530 | 0.999 | −18.021 |
| KTP-SBE-β-CD | y = 0.530x + 1.423 | 1025.145 | 0.993 | −17.876 |
Stability constant.
Correlation coefficient.
Gibbs free energy at the temperature of 37 ± 0.5 °C.
3.5. MD simulation calculation of KTP–CDs complexation free energy
The MM-PBSA is a widely used method to calculate the complexation free energies for the CD complexation in the solvent by MD simulation. The calculated complexation free energy and energy components of KTP–CD inclusion complexes by MD simulations were shown in Table 3. The binding free energy by the MM-PBSA showed the order of Hp-β-CD < Me-β-CD < SBE-β-CD < β-CD < γ-CD < α-CD, which was well in agreement with the order determined by the phase solubility study. However, the absolute values by the MD simulation were relatively larger than those of the experimental determinations because MD simulations were performed in the ideal state without any consideration for the aggregation of CD molecules. In addition, KTP molecule could not be entrapped by the inner cavity of α-CD and the KTP-α-CD system did not obtain equilibrium. Thus, the binding free energy of the KTP-α-CD system could not be calculated.
Table 3.
Calculated complexation free energy and energy components of KTP–CD inclusion complexes by MD simulation.
| System | ELE | VDW | PBSOL | PBTOT (kcal/mol) | TΔS (kcal/mol) | ΔG (kcal/mol) | ΔG (kJ/mol) |
|---|---|---|---|---|---|---|---|
| α-CD | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| β-CD | −7.82 | −25.91 | 16.95 | −16.78 | −13.21 | −3.57 | −14.94 |
| γ-CD | −9.41 | −26.38 | 18.71 | −17.08 | −13.81 | −3.27 | −13.68 |
| Hp-β-CD | −10.33 | −33.21 | 23.41 | −20.14 | −14.06 | −6.08 | −25.44 |
| DMe-β-CD | −6.89 | −29.06 | 16.7 | −19.24 | −14.28 | −4.96 | −20.75 |
| SBE-β-CD | −10.25 | −27.45 | 19.02 | −18.68 | −13.44 | −4.94 | −20.67 |
ELE, electrostatic energy as calculated by the MM force field; VDW, van der Waals contribution from MM; PBSOL, non-polar and polar contributions to solvation; PBTOT, binding enthalpy; TΔS, total Entropy. Temperature has been multiplied in as 310 K.
The final binding free energy was made up of four energy components: non-bonded ELE, VDW, PBSOL and total entropy. The VDW showed the highest contribution to the stability of the complexes, followed by TΔS and the ELE. In fact, the inclusion of a guest molecules in the CD cavity is the displacement of entrapped polar water molecules from the hydrophobic inner cavity by the polar guest molecules. Thus, the complexation equilibrium was driven by the favorable electrostatic energy and van der Waals, which was closely related to the geometric properties, ionized conditions, hydrogen bond donors or acceptors of the guest molecules. The dis-favorable solvation free energy stemmed from two factors: (1) the polar/apoplar interaction between entrapped water molecules and the CD inner cavity, and (2) the polar/apoplar interaction between free water molecules and guest molecules, which heavily depended on the hydrophobic properties of guest molecules. According to the analysis of relative importance of the molecular descriptors (shown in Fig. 4), six of top ten important descriptors in predicting the complexation free energy were the geometric properties, while three reflected the hydrophobic properties and one related to the ionized conditions of guest molecules. Thus, the analytical results of machine learning model further elucidated the factors and specific properties governing the binding free energy of the drug–CD complexation, which provided deep insight into the molecular interaction between the guest molecules and CDs.
3.6. Comparison of the experimental, theoretical and predicting results of KTP–CDs complexes
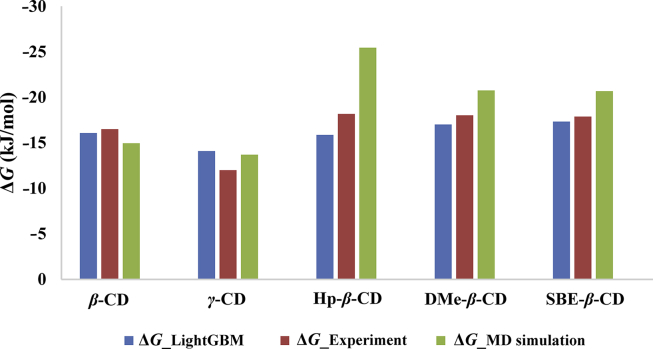
To demonstrate the predictive performance of built LightGBM model relative to typical MD simulation calculation, the comparison of complexation free energy between KTP and CDs provided by the experimental determination, MD simulation calculation and LightGBM prediction were performed as shown in Fig. 5. Taking the experimental results as a reference, LightGBM model showed high accuracy predictability with the MAE value of 1.282 kJ/mol, while the MD simulation calculation presented a large deviation with the MAE value of 3.206 kJ/mol. Traditional development of CD formulations primarily relies on the experimental trial-and-error and limited understanding of the physicochemical properties of guest molecules, which lead to high cost and long research process. Simulation-based calculation and data-driven predictive model provide alternatives to experimental determination as implementation of pre-formulation screens. Due to the limitation of computing capacity and empirical force field, molecular simulations encounter the difficulty to mimic the real experimental process and thus generate larger deviation. However, molecular modeling is able to provide the structural, dynamic and energetic information of the CDs systems. Thus, among these three major strategies of formulation design, machine learning predictive model demonstrated significant advantages, not only with high predicting accuracy and efficiency, but also increasing understanding about the molecular interaction of the drug–CD complexation process. On the other hand, the main difficulty of data-driven machine learning model is large amount of reliable data. The integration of these three approaches (experiment, molecular modeling and data-driven machine learning) will be the main challenge in the next step.
Figure 5.
Complexation free energy between ketoprofen and various cyclodextrins by the experimental determination (ΔG_Experiment), MD simulation calculation (ΔG_MD simulation) and LightGBM prediction (ΔG_LightGBM).
Linear regression is the traditional tool to establish a predicting model in CD systems20. Katritzky and co-workers built QSAR multiple regression models with a dataset of 218 compounds to predict the complexation free properties between diverse guest molecules and β-CDs37. Many other published reports also applied multiple linear regression analysis to build predicting models between guests and α-CDs/γ-CDs36, 37, 38, 39. Even though some of these models showed good fit between input variables and complexation free energy, their application range were severely limited because their predictive ability was generally applicable to only guest molecules with similarly chemical structures10, 40, 41.
In addition, there were several prediction models by the machine learning methods, most of the predicting results for the test set were significantly poorer than those for the training set because of very limited dataset of no more than 300 data points, which made susceptible to overfitting and chance correlation42. Meanwhile, previous efforts primarily focused on the native CDs, while there were very few applications of machine learning to predict the complexation free energy of CD derivatives. Merzlikine and co-workers22 applied cubist and random forest to build up the β-CDs and SBE-β-CDs complexation models, which was the first study to establish a predictive model for β-CDs derivatives. Furthermore, these predictive models were only applicable for one specific CDs, not general to all CDs. The dataset of current research comprised of 3000 data points with 1320 different structures of chemicals. Thus, the prediction models presented high accuracy both on the training and test set, which demonstrated the generalization ability of prediction models was significantly better than those of the past models.
Until now, great efforts have been made to elucidate the possible intermolecular interaction and molecular forces which may contribute to the stability of the inclusion complexes43, 44, 45. The binding forces for the formation and affinity of CD inclusion complexes include hydrophobic interaction, van der Waals interaction, hydrogen bonding, relief of high-energy water, and relief of conformational strain energy8, 46, 47. However, the relative contributions and even the nature of the different forces in the complexation process are still uncertain48, 49. Thus, although complexation free energy had been determined for thousands of drug–CD complexes, it is still unable to fast and accurately predict the actual complexation free energy of a complexation between a given CD and a given drug molecule in a given condition, and then to identify whether the drug–CD complexation could be feasible or successful in the formulations. In current research, the relative importance of each molecular descriptor in the LightGBM model for the prediction of complexation free energy not only help us to understand the molecular mechanism of CD complexation, but also to make a pre-formulation screening for the complexation free energy between CDs and guest molecules.
4. Conclusions
Vast resources and time are currently spending on the determination or calculation of complexation free energy between CDs and molecules. The research successfully established predictive models for the complexation free energy between CDs and guest molecules by three machine learning methods. The LightGBM model showed the highest prediction accuracy. The full dataset comprised of 3000 formulation with 1320 different structures. Therefore, the prediction models showed high predictive accuracy and good generalization ability without succumbing to overfitting. Furthermore, input variables included the molecular descriptors of compounds and experimental conditions, which indicated that the prediction models were not only applicable to specific CDs but to all CDs. Introducing experimental conditions into input variables to construct predictive models was also the first attempt in CD systems, which has significantly practical guidance on the CD formulation preparation. In addition, the predictive accuracy of LightGBM model on the sparse data decreased with the decreased size of dataset. The calculation of molecular descriptor contributions provided a clear framework about the factors that affect the complexation free energy between the CDs and guest molecules. Comparison with molecular modeling, the predictive models showed better agreement with experimental determination. However, currently research was only applicable to the drug–CD binary systems. Our future research will build the predictive models for the ternary or multiple-component CD systems and improved understanding of the molecular mechanism of these systems by integrated machine learning and molecular modeling techniques.
In a word, the inspiration from current research is that machine learning method can help to rule out useless avenues of formulation design and increase the efficiency of formulation researches, which will be especially important to early drug discovery with very scarce or expensive compounds and to the complicated formulation composition. Moreover, the prediction models can also help us to identify the key factors of formulation components and process parameters. The integrated methodology of machine learning and molecular modeling approaches will also help us to simplify formulation design and improve the productivity of other pharmaceutical formulation development.
Acknowledgments
We thank Mr. Weifeng Peng for the technical support in the research about the machine learning. The research was supported by the University of Macau Research Grants (MYRG2016-00038-ICMS-QRCM and MYRG2016-00040-ICMS-QRCM, Macau, China). This work was performed in part at the High-Performance Computing Cluster (HPCC) which is supported by Information and Communication Technology Office (ICTO) of the University of Macau, China.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Campisi B., Chicco D., Vojnovic D., Phan-Tan-Luu R. Experimental design for a pharmaceutical formulation: optimisation and robustness. J Pharm Biomed Anal. 1998;18:57–65. doi: 10.1016/s0731-7085(98)00175-7. [DOI] [PubMed] [Google Scholar]
- 2.Hussain A.S., Shivanand P., Johnson R.D. Application of neural computing in pharmaceutical product development: computer aided formulation design. Drug Dev Ind Pharm. 1994;20:1739–1752. [Google Scholar]
- 3.Ouyang D., Smith S.C. 1st ed. Wiley; Chichester: 2015. Computational pharmaceutics: application of molecular modeling in drug delivery. [Google Scholar]
- 4.Light D.W., Lexchin J.R. Pharmaceutical research and development: what do we get for all that money? BMJ. 2012;345:1–5. doi: 10.1136/bmj.e4348. [DOI] [PubMed] [Google Scholar]
- 5.Santanilla A.B., Regalado E.L., Pereira T., Shevlin M., Bateman K., Campeau L.-C. Nanomole-scale high-throughput chemistry for the synthesis of complex molecules. Science. 2015;347:49–53. doi: 10.1126/science.1259203. [DOI] [PubMed] [Google Scholar]
- 6.Lewis G.A., Mathieu D., Phan-Tan-Luu R. 1st ed. Taylor; Boca Raton: 1998. Pharmaceutical experimental design. [Google Scholar]
- 7.Zhao Q., Zhang W., Wang R., Wang Y., Ouyang D. Research advances in molecular modeling in cyclodextrins. Curr Pharm Des. 2017;23:522–531. doi: 10.2174/1381612822666161208142617. [DOI] [PubMed] [Google Scholar]
- 8.Lipkowitz K.B. Applications of computational chemistry to the study of cyclodextrins. Chem Rev. 1998;98:1829–1874. doi: 10.1021/cr9700179. [DOI] [PubMed] [Google Scholar]
- 9.Jordan M.I., Mitchell T.M. Machine learning: trends, perspectives, and prospects. Science. 2015;349:255–260. doi: 10.1126/science.aaa8415. [DOI] [PubMed] [Google Scholar]
- 10.Ahneman D.T., Estrada J.G., Lin S., Dreher S.D., Doyle A.G. Predicting reaction performance in C–N cross-coupling using machine learning. Science. 2018;360:186–190. doi: 10.1126/science.aar5169. [DOI] [PubMed] [Google Scholar]
- 11.Ekins S. The next era: deep learning in pharmaceutical research. Pharm Res. 2016;33:2594–2603. doi: 10.1007/s11095-016-2029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 13.Akbal-Delibas B., Farhoodi R., Pomplun M., Haspel N. Accurate refinement of docked protein complexes using evolutionary information and deep learning. J Bioinform Comput Biol. 2016;14:1642002. doi: 10.1142/S0219720016420026. [DOI] [PubMed] [Google Scholar]
- 14.Cern A., Golbraikh A., Sedykh A., Tropsha A., Barenholz Y., Goldblum A. Quantitative structure–property relationship modeling of remote liposome loading of drugs. J Control Release. 2012;160:147–157. doi: 10.1016/j.jconrel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han R., Yang Y., Li X., Ouyang D. Predicting oral disintegrating tablet formulations by neural network techniques. Asian J Pharm Sci. 2018;13:336–342. doi: 10.1016/j.ajps.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aliper A., Plis S., Artemov A., Ulloa A., Mamoshina P., Zhavoronkov A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol Pharm. 2016;13:2524–2530. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Ye Z., Su Y., Zhao Q., Li X., Ouyang D. Deep learning for in vitro prediction of pharmaceutical formulations. Acta Pharm Sin B. 2019;9:177–185. doi: 10.1016/j.apsb.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akseli I., Xie J., Schultz L., Ladyzhynsky N., Bramante T., He X. A practical framework toward prediction of breaking force and disintegration of tablet formulations using machine learning tools. J Pharm Sci. 2017;106:234–247. doi: 10.1016/j.xphs.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Loftsson T., Brewster M.E. Cyclodextrins as functional excipients: methods to enhance complexation efficiency. J Pharm Sci. 2012;101:3019–3032. doi: 10.1002/jps.23077. [DOI] [PubMed] [Google Scholar]
- 20.Xu Q., Wei C., Liu R., Gu S., Xu J. Quantitative structure–property relationship study of β-cyclodextrin complexation free energies of organic compounds. Chemometr Intell Lab Syst. 2015;146:313–321. [Google Scholar]
- 21.Zhu Q., Guo T., Xia D., Li X., Zhu C., Li H. Pluronic F127-modified liposome-containing tacrolimus–cyclodextrin inclusion complexes: improved solubility, cellular uptake and intestinal penetration. J Pharm Pharmacol. 2013;65:1107–1117. doi: 10.1111/jphp.12074. [DOI] [PubMed] [Google Scholar]
- 22.Merzlikine A., Abramov Y.A., Kowsz S.J., Thomas V.H., Mano T. Development of machine learning models of β-cyclodextrin and sulfobutylether-β-cyclodextrin complexation free energies. Int J Pharm. 2011;418:207–216. doi: 10.1016/j.ijpharm.2011.03.065. [DOI] [PubMed] [Google Scholar]
- 23.Dodziuk H. 1st ed. Wiley; Weinheim: 2006. Cyclodextrins and their complexes: chemistry, analytical methods, applications. [Google Scholar]
- 24.Higuchi T. A phase solubility technique. Adv Anal Chem Instrum. 1965;4:117–211. [Google Scholar]
- 25.Li S., Yuan L., Chen Y., Zhou W., Wang X. Studies on the inclusion complexes of daidzein with β-cyclodextrin and derivatives. Molecules. 2017;22:1–18. doi: 10.3390/molecules22122183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tetko I.V., Gasteiger J., Todeschini R., Mauri A., Livingstone D., Ertl P. Virtual computational chemistry laboratory–design and description. J Comput Aided Mol Des. 2005;19:453–463. doi: 10.1007/s10822-005-8694-y. [DOI] [PubMed] [Google Scholar]
- 27.Meng Q., Ke G., Wang T., Chen W., Ye Q., Ma Z.-M. NIPS; Barcelona: 2016. A communication–efficient parallel algorithm for decision tree; pp. 1279–1287. [Google Scholar]
- 28.Wang D., Zhang Y., Zhao Y. ICCBB; Newark: 2017. LightGBM: an effective miRNA classification method in breast cancer patients; pp. 7–11. [Google Scholar]
- 29.Svetnik V., Liaw A., Tong C., Culberson J.C., Sheridan R.P., Feuston B.P. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 30.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 31.Lv Y., Duan Y., Kang W., Li Z., Wang F.-Y. Traffic flow prediction with big data: a deep learning approach. IEEE Trans Intell Transp Syst. 2015;16:865–873. [Google Scholar]
- 32.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 33.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Solovev A., Solov'ev V. 3D molecular fragment descriptors for structure–property modeling: predicting the free energies for the complexation between antipodal guests and β-cyclodextrins. J Incl Phenom Macrocycl Chem. 2017;89:167–175. [Google Scholar]
- 35.Szejtli J. Introduction and general overview of cyclodextrin chemistry. Chem Rev. 1998;98:1743–1754. doi: 10.1021/cr970022c. [DOI] [PubMed] [Google Scholar]
- 36.Klein C.T., Polheim D., Viernstein H., Wolschann P. A method for predicting the free energies of complexation between β-cyclodextrin and guest molecules. J Incl Phenom Macrocycl Chem. 2000;36:409–423. doi: 10.1023/a:1007565409407. [DOI] [PubMed] [Google Scholar]
- 37.Katritzky A.R., Fara D.C., Yang H., Karelson M., Suzuki T., Solov'ev V.P. Quantitative structure–property relationship modeling of β-cyclodextrin complexation free energies. J Chem Inf Comput Sci. 2004;44:529–541. doi: 10.1021/ci034190j. [DOI] [PubMed] [Google Scholar]
- 38.Klein C.T., Polheim D., Viernstein H., Wolschann P. Predicting the free energies of complexation between cyclodextrins and guest molecules: linear versus nonlinear models. Pharm Res. 2000;17:358–365. doi: 10.1023/a:1007565409407. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki T., Ishida M., Fabian W.M. Classical QSAR and comparative molecular field analyses of the host–guest interaction of organic molecules with cyclodextrins. J Comput Aided Mol Des. 2000;14:669–678. doi: 10.1023/a:1008103122313. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Garrido A., Helguera A.M., Cordeiro M.N.D., Escudero A.G. QSPR modelling with the topological substructural molecular design approach: β-cyclodextrin complexation. J Pharm Sci. 2009;98:4557–4576. doi: 10.1002/jps.21747. [DOI] [PubMed] [Google Scholar]
- 41.Faucci M.T., Melani F., Mura P. Computer-aided molecular modeling techniques for predicting the stability of drug–cyclodextrin inclusion complexes in aqueous solutions. Chem Phys Lett. 2002;358:383–390. [Google Scholar]
- 42.Hawkins D.M. The problem of overfitting. J Chem Inf Comput Sci. 2004;44:1–12. doi: 10.1021/ci0342472. [DOI] [PubMed] [Google Scholar]
- 43.Devasari N., Dora C.P., Singh C., Paidi S.R., Kumar V., Sobhia M.E. Inclusion complex of erlotinib with sulfobutyl ether-β-cyclodextrin: preparation, characterization, in silico, in vitro and in vivo evaluation. Carbohydr Polym. 2015;134:547–556. doi: 10.1016/j.carbpol.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 44.Zhao Q., Miriyala N., Su Y., Chen W., Gao X., Shao L. Computer-aided formulation design for a highly soluble lutein–cyclodextrin multiple-component delivery system. Mol Pharm. 2018;15:1664–1673. doi: 10.1021/acs.molpharmaceut.8b00056. [DOI] [PubMed] [Google Scholar]
- 45.Sherje A.P., Kulkarni V., Murahari M., Nayak U.Y., Bhat P., Suvarna V. Inclusion complexation of etodolac with hydroxypropyl-beta-cyclodextrin and auxiliary agents: formulation characterization and molecular modeling studies. Mol Pharm. 2017;14:1231–1242. doi: 10.1021/acs.molpharmaceut.6b01115. [DOI] [PubMed] [Google Scholar]
- 46.Connors K.A. The stability of cyclodextrin complexes in solution. Chem Rev. 1997;97:1325–1358. doi: 10.1021/cr960371r. [DOI] [PubMed] [Google Scholar]
- 47.Hilschmann J., Kali G., Wenz G. Rotaxanation of polyisoprene to render it soluble in water. Macromolecules. 2017;50:1312–1318. [Google Scholar]
- 48.López C.A., de Vries A.H., Marrink S.J. Molecular mechanism of cyclodextrin mediated cholesterol extraction. PLoS Comput Biol. 2011;7:e1002020. doi: 10.1371/journal.pcbi.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brewster M.E., Loftsson T. Cyclodextrins as pharmaceutical solubilizers. Adv Drug Deliv Rev. 2007;59:645–666. doi: 10.1016/j.addr.2007.05.012. [DOI] [PubMed] [Google Scholar]