Abstract
Two and half million red blood cells (RBC) are generated every second in a healthy adult. The process of RBC production known as erythropoiesis requires a meticulous synchrony between signaling processes and the activity of many transcription factor complexes. FOXO3 is a transcription factor that is responsive to signaling processes and essential for the erythroid proliferation and maturation, RBC formation, and lifespan. Here, we discuss how using an integrated computational and experimental systems biology approach new and unanticipated FOXO3 functions in terminal erythropoiesis were uncovered. These combinatory approaches identified FOXO3 as a key regulator of terminal erythropoiesis. As a result, a new mode of FOXO3 participation in erythroid transcription complex formation has been proposed.
Keywords: Erythropoiesis, Red blood cell (RBC), Enucleation, Mitochondria, Mitophagy, FOXO, FOXO3, Transcriptome, RNA sequencing
1. Introduction
Erythropoiesis is the complex process of RBC generation through stepwise differentiation and lineage restriction of hematopoietic stem cells (HSC). This process begins with HSC differentiation to multipotent progenitor cells, which in turn generate erythroid lineage-committed progenitors. Primitive erythroid progenitors called burst-forming unit erythroid cells (BFU-Es) then further differentiate into colony-forming unit erythroid cells (CFU-Es). Finally, the last stages of RBC maturation involved four progressively differentiated erythroblast stages culminating in reticulocyte formation and maturation into an RBC. Terminal erythroblast maturation is characterized by reduced cell size, chromatin condensation, and synthesis and accumulation of hemoglobin. To completely mature, erythroblasts also need to expulse their nuclei through a process called enucleation. The ensuing reticulocytes remodel their membrane and clear first mitochondria—through autophagy—and then the remaining organelles before transitioning into fully mature erythrocytes. These processes are coordinated by signals generated by the erythropoietin receptor (EpoR) and other growth factor/cytokine receptors in crosstalk with erythroid transcription factors including GATA-1, KLF-1, TAL-1, and their cofactors as well as FOXO3 [1, 2]. While the function of these transcription factors in early stages of erythroblast differentiation has been known, their participation in terminal erythroblast maturation has just begun to be elucidated. Impaired maturation of erythroblasts and subsequent alterations in kinetics of RBC production (decreased or increased) are associated with different blood disorders ranging from anemia to leukemia [3, 4].
FOXO3 belongs to the FOXO (four-related mammalian FOXO1, 3, 4, 6) family of transcription factors [5, 6]. FOXO proteins are involved in various cellular processes ranging from DNA damage response to metabolism. FOXO3 is regulated by erythropoietin receptor (EpoR) signaling [7–10] and is the main active FOXO in primary erythroid cells [11]. During erythropoiesis the expression and activity of FOXO3 increase with erythroblast maturation in mice and humans supporting FOXO3 involvement in terminal erythropoiesis. In agreement with this, FOXO3 has crucial functions in homeostatic and stress-induced erythropoiesis [11–15]. In primary erythroid cells FOXO3 regulates oxidative stress and metabolic processes and is in cross talk with mTOR signaling [11, 16].
To elucidate the exact function FOXO3 carries out during terminal erythroid maturation, the transcriptome of adult bone marrow WT andFoxo3−/− mouse erythroblasts at three distinct and increasing stages of maturation that precede erythroid enucleation were compared using deep sequencing of isolated RNA. Hierarchical clustering of gene expression patterns identified a cluster of genes that are normally upregulated as erythroblasts mature, but fail to become induced in Foxo3−/− erythroblasts. Pathway enrichment analysis of these clusters revealed multiple cellular processes linked to enucleation and autophagy. In parallel, functional assays involving imaging flow cytometry, confocal immunofluorescence imaging, and ex vivo maturation also revealed defects in autophagic flux. Similar approaches exposed defective enucleation in Foxo3−/− erythroblasts. Subsequently, many genes involved in enucleation and autophagy pathways were validated to be FOXO3 direct targets by qRT-PCR and chromatin immunoprecipitation. In addition, restoration of Foxo3−/− through viral transduction rescued aberrant expression of autophagy and enucleation genes and rescued the maturation defects of Foxo3−/− erythroblasts. Together, these analyses enabled the generation of a testable transcription complex model that suggests FOXO3 interacts with other erythroid transcription factors (i.e., GATA-1, TAL1, and LDB1) to activate erythroid-specific processes during the terminal stages of erythroid maturation.
In this protocol, we describe the methods for RNA-seq processing and subsequent in silico analyses that uncovered new and unanticipated functions of FOXO3 that are critical for terminal erythroid maturation.
2. Materials
2.1. Bone Marrow Isolation and FACS Sorting
Wild type and Foxo3−/− mice aged 9–12 weeks.
Sorting Media—IMDM + 2% FBS.
Staining Media—PBS + 2% FBS.
Cell Counting Media–Water + 3% Acetic Acid.
TER119, CD44 antibodies.
DAPI.
FACS machine.
5 mL FACS tubes, 15 mL Falcons, microcentrifuge tubes, 1 mL syringe, 26 ½ gauge needles.
2.2. RNA Extraction and RNA-seq Processing
RNAeasy Micro Plus Kit (Qiagen).
Illumina True Seq RNA prep Kit.
Hi Seq 2000 platform (Ilumina).
2.3. Chromatin Immunoprecipitation
Tris–HCl pH 8.0.
0.4% formaldehyde in PBS.
NaCl.
NP-40.
SDS.
EDTA.
FOXO3a antibody.
Magna ChIPTM Protein A + G magnetic beads.
Lysis Buffer (10 mM Tris–HCl pH 8.0, 10 mM NaCl, 0.2% NP40).
ChIP dilution buffer (20 mM Tris–HCl, pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton, 0.01% SDS).
Wash Buffer (20 mM Tris–HCl, pH 8.0, 2 mM EDTA, 50 mM NaCl, 1% Triton, 0.1% SDS).
DNA isolation kit.
3. Methods
3.1. Bone Marrow Isolation
Sacrifice mouse and separate both hind legs from skin of mouse.
Clear off muscle from tibia and femur and proceed to flushing.
Fill syringe with sorting media and insert into ends of tibia or femur and dispense media to flush out bone marrow cells into 15 mL falcon tube containing 3 mL of sorting media. Repeat until bones appear clear of all cells.
Pool bone marrow cells from up to 3 mice in one tube.
3.2. FACS Sorting (see Note 1)
Filter isolated bone marrow cells through 35 μm nylon mesh cap FACS tube.
Pellet cells by centrifuging at 200 × g and resuspend in 1 mL of cell staining media.
Dilute 10 μL of BM cells into 990 μL of cell counting media to lyse RBCs and count on hemocytometer.
Pellet cells again and resuspend to 150 × 106 cells/mL— subsequent step will consider antibody staining in 1 mL of staining buffer with 150 × 106 nucleated cells.
Add 40 μL of TER119 and CD44 fluorescence-conjugated antibodies and mix thoroughly by pipetting. (Additionally, prepare unstained and single stained controls using 0.5 × 106 cells in 100 μL of staining buffer).
Incubate at 4 °C in the dark for 15 min.
Wash off excess antibody by adding an additional 1 mL of staining buffer and pellet cells at 200 g for 5 min.
Aspirate the supernatant and resuspend in 1 mL of sorting media containing 1 μg/mL of DAPI to distinguish viable cells (DAPI negative).
Further dilute cells to a concentration of 15 × 106 cells/mL (nucleated) and filter into the required number of 35 μm nylon mesh cap FACS tubes.
Prepare 3 FACS tubes containing 1 mL of sorting media each as collection tubes for the three erythroblast populations to be sorted.
Bring cells to FACS machine and run control samples to determine proper gating of subsequent positive and negative populations.
Gate out debris, non-singlet, and dead cells based on forward scatter, side scatter, and DAPI fluorescence.
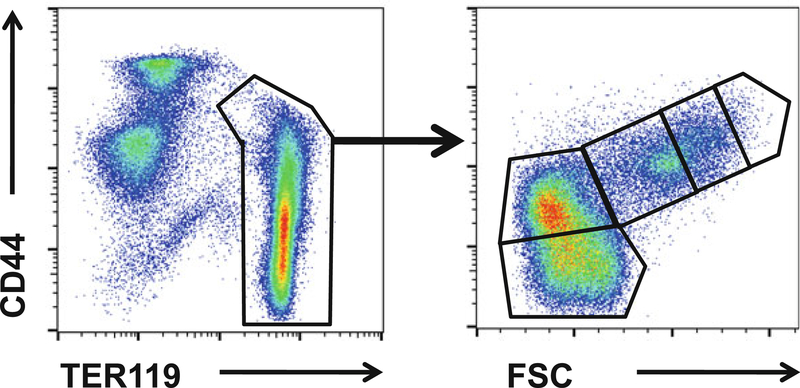
Mouse erythroblasts are specifically detected by the TER119 antibody. The progressive stages of erythroblast maturation can be resolved by CD44 expression and FSC parameters that decrease with maturation (Fig. 1).
Sort proerythroblasts (Gate 1), basophilic erythroblasts, polychromatic erythroblasts (Gate 2), and orthochromatic erythroblasts (Gate 3) into collection tubes until all BM cells are sorted (see Note 2).
Fig. 1.
Gating strategy for flow cytometry: pro-, basophilic, polychromatophilic and orthochromatic erythroblasts from WT and Foxo3−/− BM were FACS-sorted according to their forward scatter and cell surface expression of CD44 and TER119
3.3. RNA Extraction
Transfer cells to 5 mL conical tubes or multiple microcentrifuge tubes and pellet at 200 × g for 5 min (see Note 3).
Wash cells once with 1 mL of staining buffer (consolidate cells into a single tube if multiple tubes were used), pellet cells again at 200 g for 5 min, and carefully aspirate all supernatant.
Add 300 μL RLT buffer from the RNAeasy kit with 2-mercaptoethanol added at the concentration noted to the cell pellet.
Fully lyse cells by pipetting and mixing further by vortexing for up to a minute.
Lysed cells in RLT buffer can be stored at −80 °C for extended periods of time (our lab has tested samples stored up to 6 months with no RNA degradation).
From here, follow the manufacturer’s instructions to isolate RNA.
Analyze RNA quality and quantity using bioanalyzer to determine if RNA quality and quantity is sufficient for cDNA synthesis, library prep, and subsequent sequencing, dependent on type of next-gen sequencer being used.
Following RNA extraction, cDNA synthesis is carried out using the SuperScript IITM Reverse Transcriptase from Invitrogen. qRT-PCR is carried out using SYBR Premix Ex TaqTm II (Tli RNase H Plus).
For mRNAseq libraries, RNA is extracted as mentioned above and mRNAseq libraries are prepared using the Illumina True Seq RNA prep kit. A Hi Seq™ 2000 platform (Illumina) is used to sequence the samples in parallel lanes in order to obtain around107 single end 100 bp reads per sample.
3.4. RNA-Seq Processing
Map RNA-seq reads to mouse genome with TopHat or preferred method.
Process mapped reads to calculate fragments per kilo-base permillion reads (FPKM) or other method of normalization to allow comparison between transcriptome profiles of WT and Foxo3−/− cells.
Test for differentially expressed genes between WT and Foxo3−/− populations using Cufflinks or preferred method.
For downstream routes of analysis only genes with a FPKM >1 and at least 1.8 fold difference between WT and Foxo3−/− samples are good starting points.
The total number of reads would be in the range of 30–50 million for the WT and around 10–50 million for the Foxo3−/− samples.
3.5. Identifying FOXO3 Regulated Cell Processes with RNAseq Data
If distinct populations of WT and Foxo3−/− cells are used as with erythroblast populations, the gene expression data can be clustered (Otherwise genes will either cluster as upregulated or downregulated when comparing just one population of WT and Foxo3−/− cells). Using programs such as Cluster 3.0 or GENE-E, perform k-means clustering by gene to reveal how gene expression patterns change from population to population and how it differs in Foxo3−/− populations.
Perform pathway enrichment analysis of differentially expressed genes or clusters of genes with programs such as FuncAssociate, ENRICHR, or others. Such analysis should reveal multiple statistically significant pathways.
Additionally, chromatin immunoprecipitation enrichment analysis (ChEA) can reveal other transcription factors that regulate similar sets of genes based on published ChIP-seq datasets [17]. The known function of these transcription factors can be correlated with pathways identified from the previous step as a starting point for functional analysis.
3.6. Chromatin Immunoprecipitation
Chromatin immunoprecipitation or ChIP is carried out in TER119+ cells derived from WT total bone marrow cells. Following FACS, the cells are fixed in 0.4% formaldehyde / 1XPBS and subsequently lysed (see Note 4).
The lysate is then sonicated using a diagenode bioruptor sonicator for 30 cycles of 30 s on/30 s off at 4 °C (to be optimized according to the sonicator available). This is then further diluted in ChIP dilution buffer. The diluted cell lysate is incubated overnight at 4 °C in anti-FOXO3a antibody and Magna ChIPTM Protein A + G magnetic beads.
The beads are washed in and the bound complexes are reverse cross-linked after which DNA isolation and qPCR analysis is carried out.
As a negative control, TER119+ cells derived from Foxo3−/− bone marrow are used.
The putative binding sites are identified using MatInspector from Genomatix.
3.7. Validating FOXO3 Regulated Cell Processes with Functional Analysis
Validate differential expression of key genes within pathway by qRT-PCR comparing WT and Foxo3−/− cells of interest.
Dependent on the pathway being deregulated in the absence of FOXO3, specific functional assays should be performed to validate defects in the identified pathways (e.g., we identified a failure to upregulate autophagy genes in Foxo3−/− erythroblasts and determined defective autophagy with multiple methods of measuring autophagic flux).
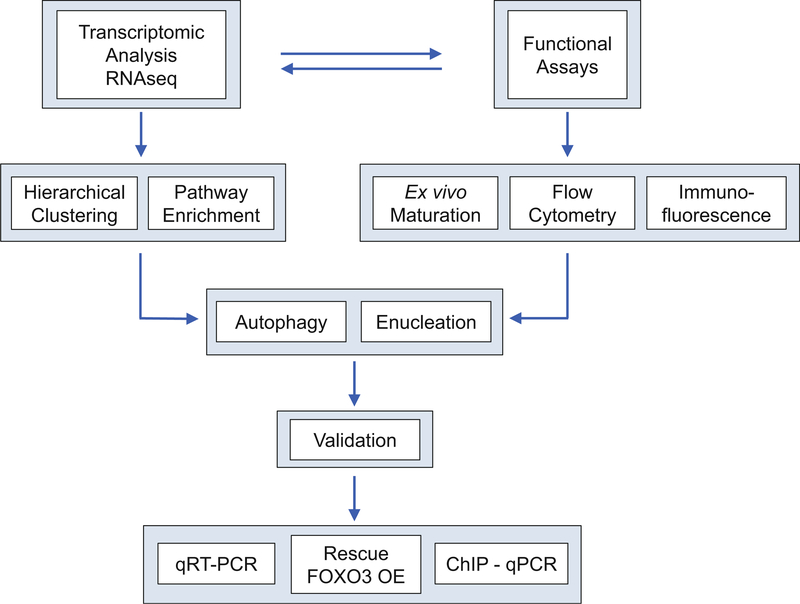
Ectopic expression of FOXO3 in Foxo3−/− cell type should restore expression of deregulated genes to WT levels and rescue functional defects of Foxo3−/− cells. To confirm if various pathways are also directly contributing to defect seen in Foxo3−/− cells, aberrant expression of key genes in the defective pathway should also be ectopically expressed and analyzed for rescue. Figure 2 illustrates the strategy for identifying FOXO3 functions in terminal erythropoiesis (see Note 5).
Fig. 2.
Strategy for identifying FOXO3 functions in terminal erythropoiesis
4. Notes
This protocol can apply to other cell populations of blood and would require different combinations of cell surface markers to sort for the populations of interest.
Due to the nature of sorted erythroblasts being sequential in maturation subsequent analysis did not require biological replicates. However for comparisons between WT and Foxo3−/− cells of other populations at least three replicates are most likely required to obtain statistical significance in finding differential expression.
At least 300,000 cells per population are required to comfortably produce enough RNA for subsequent steps. If cells will be used for other types of sequencing (e.g., single-cell RNAseq) or validation by qRT-PCR much fewer amounts of cells are required to produce usable amounts of cDNA. Our lab has been able to robustly perform qPCR with 5000–10,000 cells.
To confirm direct regulation of specific target genes, chromatin immunoprecipitation (ChIP) should be performed with FOXO3-specific antibody (see protocol).
As each individual study can be highly specific and different from each other, this protocol provides just a guideline to how to analyze and identify FOXO3 regulated pathways in blood cells. Different types of studies most likely will involve specialized toolsets and would require additional types of analysis to adapt to the specific study.
Acknowledgments
The work in the Ghaffari lab is supported by grants from National Institutes of Health (NCI and NHLBI). Raymond Liang is supported by a pre-doctoral fellowship from the American Heart Association.
References
- 1.Kerenyi MA, Orkin SH (2010) Networking erythropoiesis. J Exp Med 207:2537–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira R, Ohneda K, Yamamoto M, Philipsen S (2005) GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol Cell Biol 25:1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kassebaum NJ, Jasrasaria R, Naghavi M, Wulf SK, Johns N, Lozano R, Regan M, Weatherall D, Chou DP, Eisele TP, Flaxman SR, Pullan RL, Brooker SJ, Murray CJ (2014) A systematic analysis of global anemia burden from 1990 to 2010. Blood 123:615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman SB, Block MH (1967) Increased red blood cell production in chronic myelocytic leukemia. JAMA 200:621–624 [PubMed] [Google Scholar]
- 5.Salih DA, Brunet A (2008) FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 20:126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eijkelenboom A, Burgering BM (2013) FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 14:83–97 [DOI] [PubMed] [Google Scholar]
- 7.Bakker WJ, Blazquez-Domingo M, Kolbus A, Besooyen J, Steinlein P, Beug H, Coffer PJ, Lowenberg B, von Lindern M, van Dijk TB (2004) FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J Cell Biol 164:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakker WJ, van Dijk TB, Parren-van Amelsvoort M, Kolbus A, Yamamoto K, Steinlein P, Verhaak RG, Mak TW, Beug H, Lowenberg B, von Lindern M (2007) Differential regulation of Foxo3a target genes in erythropoiesis. Mol Cell Biol 27:3839–3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghaffari S, Jagani Z, Kitidis C, Lodish HF, Khosravi-Far R (2003) Cytokines and BCR-ABL mediate suppression of TRAIL induced apoptosis through inhibition of forkhead FOXO3a transcription factor. Proc Natl Acad Sci U S A 100:6523–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kashii Y, Uchida M, Kirito K, Tanaka M, Nishijima K, Toshima M, Ando T, Koizumi K, Endoh T, Sawada K, Momoi M, Miura Y, Ozawa K, Komatsu N (2000) A member of Forkhead family transcription factor, FKHRL1, is one of the downstream molecules of phosphatidylinositol 3-kinase-Akt activation pathway in erythropoietin signal transduction. Blood 96:941–949 [PubMed] [Google Scholar]
- 11.Marinkovic D, Zhang X, Yalcin S, Luciano JP, Brugnara C, Huber T, Ghaffari S (2007) Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J Clin Invest 117:2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang R, Camprecios G, Kou Y, McGrath K, Nowak R, Catherman S, Bigarella CL, Rimmele P, Zhang X, Gnanapragasam MN, Bieker JJ, Papatsenko D, Ma’ayan A, Bresnick E, Fowler V, Palis J, Ghaffari S (2015) A systems approach identifies essential FOXO3 functions at key steps of terminal erythropoiesis. PLoS Genet 11:e1005526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franco SS, De Falco L, Ghaffari S, Brugnara C, Sinclair DA, Matte A, Iolascon A, Mohandas N, Bertoldi M, An X, Siciliano A, Rimmele P, Cappellini MD, Michan S, Zoratti E, Anne J, De Franceschi L (2014) Resveratrol accelerates erythroid maturation by activation of FoxO3 and ameliorates anemia in beta-thalassemic mice. Haematologica 99:267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, Dore LC, Yao Y, D’Souza J, Zhang Z, Ghaffari S, Choi J, Friend S, Tong W, Orange JS, Paw BH, Weiss MJ (2010) miR-451 protects against erythroid oxidant stress by repressing 14–3-3zeta. Genes Dev 24:1620–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McIver SC, Kang YA, DeVilbiss AW, O’Driscoll CA, Ouellette JN, Pope NJ, Camprecios G, Chang CJ, Yang D, Bouhassira EE, Ghaffari S, Bresnick EH (2014) The exosome complex establishes a barricade to erythroid maturation. Blood 124:2285–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Camprecios G, Rimmele P, Liang R,Yalcin S, Mungamuri SK, Barminko J, D’Escamard V, Baron MH, Brugnara C, Papatsenko D, Rivella S, Ghaffari S (2014) FOXO3-mTOR metabolic cooperation in the regulation of erythroid cell maturation and homeostasis. Am J Hematol 89:954–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lachmann A, Xu H, Krishnan J, Berger SI,Mazloom AR, Ma’ayan A (2010) ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26:2438–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]