Significance
Multiple families of DNA viruses including herpesviruses amplify their genomes in nuclear sites termed replication compartments. What benefits the viruses gain by this spatial and temporal control is unclear. We have analyzed the replication compartments induced by Epstein–Barr virus (EBV) and its DNA amplification in detail to elucidate their functions and regulation in EBV’s productive cycle. We found that EBV uses its replication compartments to coordinate the amplification of its genomes: Each compartment is seeded by single viral DNAs, each compartment supports similar levels of viral DNA synthesis, and each completes this synthesis as the replication machinery declines within it. Thus, replication compartments not only exclude cellular DNA synthesis but are hubs for the coordination of viral DNA amplification.
Keywords: Epstein–Barr virus, viral DNA replication, replication compartments
Abstract
Herpesviruses must amplify their DNA to load viral particles and they do so in replication compartments. The development and functions of replication compartments during DNA amplification are poorly understood, though. Here we examine 2 functionally distinct replicons in the same cells to dissect DNA amplification within replication compartments. Using a combination of single-cell assays, computational modeling, and population approaches, we show that compartments initially were seeded by single genomes of Epstein–Barr virus (EBV). Their amplification subsequently took 13 to 14 h in individual cells during which their compartments occupied up to 30% of the nucleus and the nuclear volume grew by 50%. The compartmental volumes increased in proportion to the amount of DNA and viral replication proteins they contained. Each compartment synthesized similar levels of DNA, indicating that the total number of compartments determined the total levels of DNA amplification. Further, the amplification, which depended on the number of origins, was regulated differently early and late during the lytic phase; early during the lytic phase, the templates limited DNA synthesis, while later the templates were in excess, coinciding with a decline in levels of the viral replication protein, BMRF1, in the replication compartments. These findings show that replication compartments are factories in which EBV DNA amplification is both clonal and coordinated.
Mammalian DNA synthesis is well orchestrated to achieve consistent outcomes. Each DNA template is synthesized once and only once during each S phase (1). Regions of DNA on the order of a megabase are synthesized with multiple internal sites of DNA initiation and completed independently of adjacent regions (2). Chromosomes are synthesized in their entirety with the possible exception of a small number of their telomeric repeats (3). DNA viruses, being obligate cellular parasites, have evolved to synthesize their genomes by capitalizing on different cellular components. Not only herpesviruses (4, 5) but also adenoviruses (6) and parvoviruses (7, 8) form discrete sites in the nuclei of infected cells in which they amplify their DNAs. These virus families have genomes spanning in length from 3 to 250 kbp and include both single- and double-stranded DNAs, illustrating the diversity of cellular parasites that have evolved to revamp nuclear structure for their replication. It is not clear, though, what advantages this compartmentalization provides them. We have examined Epstein–Barr virus (EBV) DNA amplification to elucidate the role of replication compartments in the amplification of its DNA.
In latently infected cells, EBV DNA is synthesized similarly to cellular DNA with the significant exception that only ∼85% of its genomes are duplicated each S phase (9). During the lytic phase, EBV, as with other herpesviruses, undergoes up to several hundredfold amplification of its DNA (10–12) to provide the genomes to be housed in progeny viruses. The amplification is mediated by cis and trans elements—cis elements include the origins of DNA synthesis and the trans elements include replication machinery encoded by EBV and the cell. Its DNA amplification is carried out in replication compartments that are devoid of detectable histones including H2A, H3.1, and H3.3 (4). While a single genome per se is insignificant compared to the size of the host genome, several hundredfold amplifications generate a viral DNA mass approaching that of the host genome. Replication compartments which house the amplified viral DNA therefore significantly reorganize the landscape of the host nucleus. For example, the herpes simplex virus-1(HSV-1) lytic phase correlates with an increase in the volume of host nucleus by up to 2-fold (5). This reorganization of the nucleus facilitates transport of HSV-1 capsids (13).
While replication compartments can only be visualized via microscopy (4, 13), examining DNA synthetic events often requires a population approach (10, 14). Another difficulty in understanding viral DNA amplification comes from its dependence on the cell cycle rendering it asynchronous in a population of cells (4, 15). To overcome these difficulties in understanding EBV DNA amplification, we analyzed 2 functionally distinct replicons in the same cells. Computational modeling was used to simulate features that fit their DNA amplification observed in bulk populations. The computational simulations made 3 striking predictions: 1) The lengths of the replicons could be compensated by the number of origins per template, 2) the initial number of viral DNA molecules determines their ultimate amplification, and 3) the cessation of amplification requires cessation of DNA synthesis per se. To test these predictions, single-cell assays, including live-cell imaging, fluorescence in situ hybridization (FISH), and 5‐ethynyl‐2′‐deoxyuridine (EdU) pulse labeling, in combination with population approaches were used with a cell line harboring 2 EBV DNA replicons, P3HR1 and 4012. These replicons are present in the same cells but differ in their lengths and numbers of origins. This testing confirmed the 3 predictions. P3HR1 and 4012 DNAs were synthesized to similar levels per unit of time, despite the differences in their lengths and numbers of origins, consistent with the first prediction. The replication compartments were seeded by single genomes and each compartment amplified this single genome several hundredfold. The number of compartments that were seeded by P3HR1 and 4012 were proportional to the number of molecules of P3HR1 and 4012 that were present at the start of the lytic phase. During the 13 to 14 h of DNA amplification in single cells, the replication compartments collectively eventually occupied 30% of the nucleus while the nuclei grew by 50%. Each replication compartment synthesized similar levels of DNA and therefore the number of compartments directly determined the total DNA synthesized in each cell, consistent with the second prediction derived from the simulations. Early in this amplification process, DNA templates were a limiting factor for DNA synthesis. Late in the amplification process, unused DNA templates were in excess, coinciding with a drop in the levels of viral synthetic machinery in the replication compartments, consistent with the third prediction derived from the simulations. Our results establish that replication compartments represent clonal factories for DNA amplification that coordinate DNA synthesis during the lytic phase.
Results
EBV DNA Accumulation Appears to Be Independent of the Length of the Template.
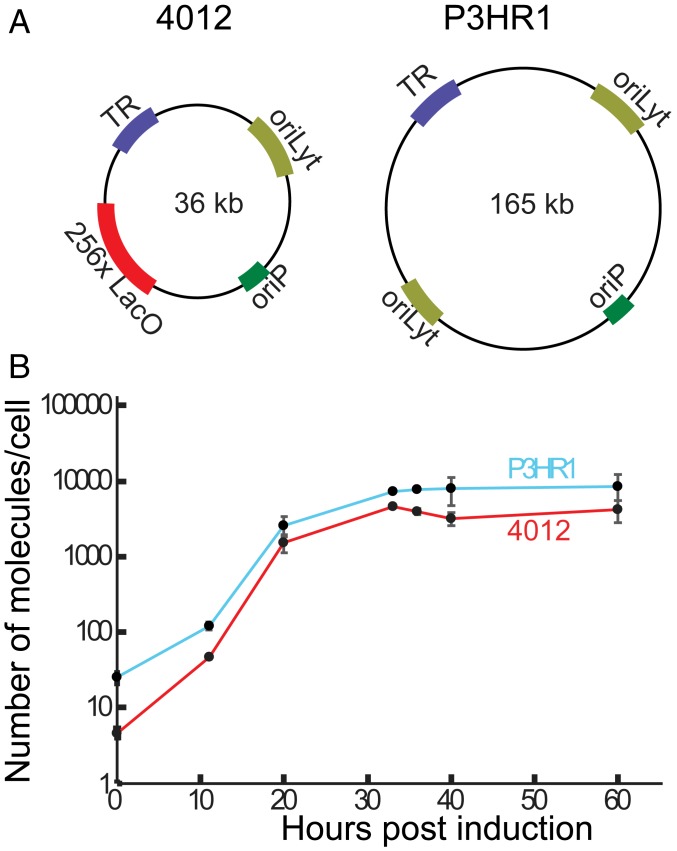
iD98HR1-4012 cells harboring P3HR1, a 165-kb strain of EBV, and 4012, which is a 36-kb amplicon derived from EBV, were used to study DNA amplification (Fig. 1A). P3HR1 and 4012 are each present in multiple copies in iD98HR1-4012 cells—an average of ∼25 copies of P3HR1 and ∼5 to 10 copies of 4012 per cell, while the copy numbers vary between clones of these cells. The 4012 contains 256 LacO repeats, encodes the LacI-td tomato gene, and has all of the elements required in cis to support lytic DNA replication (4). The needed trans-acting elements are encoded by P3HR1 and by the cell. Amplification of 4012 DNA gives increasingly intense, visible red fluorescent signals when observed by fluorescence microscopy. iD98HR1-4012 cells also express Z-ER, which is a fusion protein consisting of the immediate early gene BZLF1 fused to the ligand-binding domain of the estrogen receptor (4). Addition of tamoxifen to cells expressing Z-ER leads to translocation of the fused protein to the nucleus, where Z-ER initiates EBV’s lytic phase. The levels of both P3HR1 and 4012 DNAs at different times after induction of the lytic phase were quantified by qPCR. These bulk measurements showed that the accumulation of both replicons was rapid from 10 to 20 h postinduction (hpi), then slowed, and eventually plateaued by 40 hpi (Fig. 1B). Surprisingly, 4012, which is one-fourth the length of P3HR1, accumulated to a similar level as did P3HR1 (∼50% of P3HR1) (Fig. 1B). If the length of a replicon solely determined its rate of amplification, then 4012 would have outpaced P3HR1.
Fig. 1.
Measuring the synthesis of 2 replicons during EBV’s lytic phase. (A) The 2 replicons in iD98HR1-4012 cells are depicted. P3HR1 is a strain of EBV derived from the P3HR1 cell line and is 165 kb in length; 4012, a derivative of EBV, is 36 kb. The cis-acting elements essential for these studies are depicted and include oriP (latent origin of replication), oriLyt (the lytic origin of DNA synthesis), TR (the terminal repeats required for encapsidation of synthesized DNA), and LacO (the site to which the Lac repressor binds). (B) iD98HR1-4012 cells were induced to support the lytic phase by treatment with tamoxifen. The immediate early gene BZLF1 fused to the estrogen receptor is expressed constitutively in these cells. DNA was isolated from these cells at the indicated times and assayed for the amount of viral DNA and cellular DNA using qPCR (for time points 0, 20, 40, and 60 hpi, n [biological replicates] = 4; for 11, 33, and 36 hpi, n = 2). The error bars indicate the SD of the mean of the levels of EBV DNA/cell (with the assumption that each cell contains 2 copies of the rhodopsin gene).
Encapsidation Does Not Regulate DNA Accumulation.
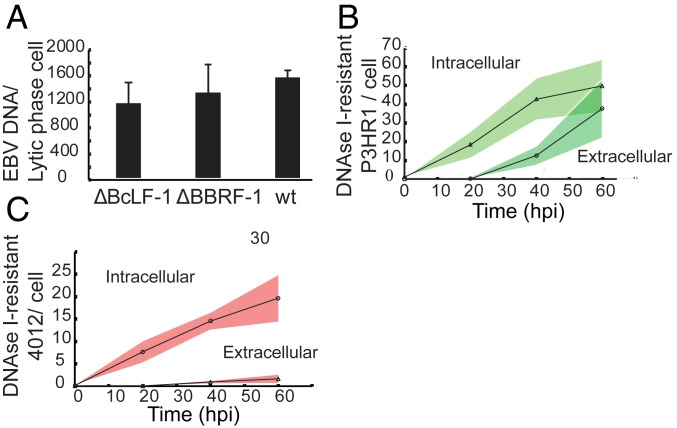
Viral DNA accumulated most rapidly during the first 20 hpi followed by a slowdown in accumulation from 20 hpi to 40 hpi (Fig. 1) and then plateaued. We tested if encapsidation could limit the late stages of viral DNA amplification by removing potential templates from DNA synthesis. This possibility could apply if EBV were to encapsidate its DNA as extensively as other herpesviruses do (16, 17). Furthermore, perhaps 4012 is preferentially encapsidated relative to P3HR1, explaining its surprisingly slow accumulation. Derivatives of EBV that could not form capsids or could not sequester DNA in capsids were assayed to learn if these defects abolished the slowing accumulation of viral DNA during the lytic phase. HEK 293 cells maintaining EBV mutants which cannot express either the major capsid protein (BcLF-1) or the portal protein (BBRF-1) were generated and analyzed. The lytic phase was induced in the cells by transfection of vectors encoding the immediate early proteins BZLF1 and BRLF1. In addition, a plasmid encoding the minor capsid protein fused to green fluorescent protein (GFP) (p18-GFP) under the control of its native promoter was transfected to quantify the fraction of cells that supported the lytic phase. Neither of these induced cell populations released detectable, extracellular virions (SI Appendix, Fig. S1), nor did they contain increased levels of EBV DNA when compared to induced cells carrying wild-type (WT) EBV (Fig. 2A). The inability of EBV to encapsidate its DNA, therefore, is not sufficient to prevent the slowing accumulation of its DNA late in EBV’s lytic phase. The number of intracellular and extracellular capsids measured from cells supporting WT EBV (Fig. 2 B and C) was similar to that found in other studies (18, 19). These measurements also showed that ∼1% of each replicon was encapsidated (Fig. 2 B and C), ruling out the possibility that preferential encapsidation could explain the failure of 4012’s synthesis to outpace that of P3HR1.
Fig. 2.
DNA encapsidation does not affect DNA accumulation. (A) The levels of viral DNA synthesized in 293 cells harboring WT EBV or 1 of 2 EBV mutants were measured by qPCR at 48 hpi. The mutants do not express either the portal protein, BBRF1, or the major capsid protein, BcLF1, and therefore cannot encapsidate viral DNA. The lytic phase was induced by transfection of plasmids encoding the immediate early genes BRLF1 and BZLF1. In addition, a plasmid encoding the late gene product p18 fused to GFP was transfected to evaluate the fraction of cells supporting the lytic phase (n [biological replicates] = 3, error bars indicate SD). An unpaired Student’s t test was carried out to determine P values (P [BcLF1 vs. WT] = 0.1; P [BBRF1 vs. WT] = 0.4). (B and C) The intracellular and extracellular encapsidated DNAs were assayed by measuring the levels of DNaseI-resistant P3HR1 and 4012 DNAs using qPCR at the indicated times from iD98HR1-4012 cells. n (biological replicates) = 3 (the shaded areas indicate the SD of the mean of the 3 biological replicates).
Modeling EBV’s DNA Amplification.
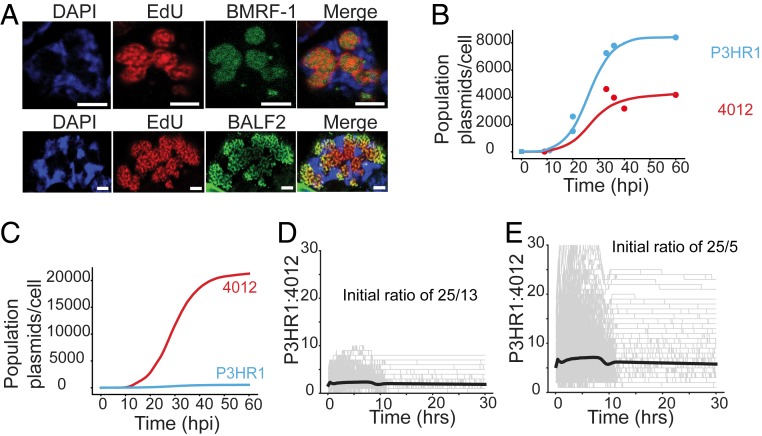
We developed simulations to predict features of the DNA replication of 4012 and P3HR1 that led to their similar accumulation during the lytic phase (Fig. 1B). The only additional data used in this modeling were measurements of the fraction of cells supporting the lytic cycle determined by the incorporation of EdU at each time point (Fig. 3A and SI Appendix, Table S1). Cells supporting EBV DNA amplification displayed distinctive EdU signals which were confirmed by staining for viral replication proteins localized to the sites of amplification (Fig. 3A). The simulations probed the following parameters: 1) replicons with short and long lengths, with 1 and 2 origins, respectively, were followed and their daughter molecules in individual cells per amplification cycle were tallied; 2) initiation was allowed to be independent at each origin; and 3) synthesis was not allowed to reinitiate at an origin until it was completed for that replicon. Three predictions of this model prompted further research. First, the model simulated the data best when the longer of the 2 replicons had 2 origins (Fig. 3B). Under these conditions, the synthesis of the shorter replicon did not outpace that of the longer having 2 origins. When a longer replicon with only 1 origin was considered, the simulation predicted that the smaller replicon would outpace the longer replicon rapidly (Fig. 3C). This prediction arises from the fact that a molecule that has 2 origins has a greater chance of being replicated due to higher chances of initiation compared to the molecule that has 1 origin. Therefore, despite the 4-fold length difference in case of 4012 and P3HR1, the 2 molecules were predicted to take a similar time to complete an amplification of 500- to 1,000-fold. Second, the overall levels of the 2 replicons synthesized were dependent on their initial numbers per cell prior to the onset of their amplification (Fig. 3 D and E). Third, the plateauing of the synthesis of each replicon late in the lytic cycle required the cessation of synthesis per se.
Fig. 3.
Computational simulations predict the properties of DNA synthesis during the lytic phase. (A) The fraction of iD98HR1-4012 cells supporting the lytic phase was measured using EdU incorporation. To establish that the detected, compartmentalized, EdU incorporation identified cells supporting the lytic phase, cells were also assayed for the early gene products BMRF1 (EBV DNA clamp protein) and BALF2 (single-stranded DNA binding protein) via immunofluorescence. They were detected only in cells exhibiting replication compartments as shown in A. (Scale bars, 2 µm.) (B) Shown is the best fit (in solid lines) of the optimal simulation of the measurements of Fig. 1B (in data points) corrected for the fraction of cells supporting the lytic phase (SI Appendix, Table S1). (C) The simulation predicts that were the P3HR1replicon to have only one origin, the 4012 replicon would amplify faster. (D and E) The predictions of the simulation in which the initial ratios of P3HR1 and 4012 were varied in cells and found to determine the ultimate levels of their synthesis in each population. In gray are shown the results of simulation of individual cells, with the solid lines reflecting the averages of all predicted cells.
P3HR1 and 4012 Are Synthesized at Similar Levels; Early and Late Lytic Replication Are Regulated Differently.
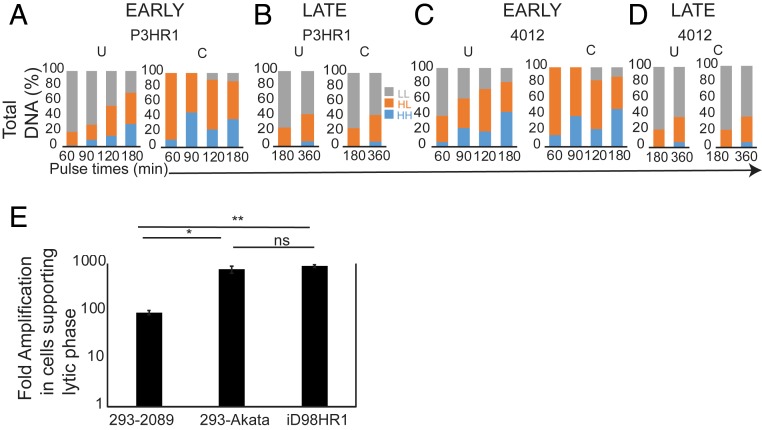
The simulations predicted that DNA synthetic activity is similar for both P3HR1 and 4012 due to the number of origins and probability of the occupancy of these origins. If this prediction were correct, then similar numbers of molecules of P3HR1 and 4012 would be synthesized per unit of time despite the differences in length. In order to test this prediction, density-shift experiments were used to follow the synthesis of P3HR1 and 4012 DNAs into daughter molecules. iD98HR1-4012 cells were pulsed with BrdU, a heavy thymidine analog, for increasing times beginning 8 hpi of EBV’s lytic phase, the time when viral DNA accumulation could first be detected in density-shift experiments. The cells were harvested and their DNA was purified, sheared, and separated by equilibrium density centrifugation in CsCl gradients. The DNA in each fraction was measured by qPCR, detecting DNAs encoding BALF5 of P3HR1, LacI-tdtomato of 4012, and rhodopsin for cellular DNA. Pairs of cell cultures were labeled for 60 and 120 or 90 and 180 min (Fig. 4 A and C and SI Appendix, Figs. S2 A and B and S3 A and B) and the experiments were performed with 2 biological replicates. In order to correct for the viral DNA present only in cells supporting EBV DNA amplification, EdU was used to identify cells undergoing viral DNA amplification. The EdU pulse-labeling experiments done in parallel to density-shift assays provided a correction factor for interpreting the density-shift assays. The density profiles of the separated DNAs demonstrated that both the fractions of the P3HR1 and of the 4012 DNAs increased in the heavy/light (HL) and the heavy/heavy (HH) portions of the gradients over time as a result of DNA replication in the presence of BrdU and decreased accordingly in the light/light (LL) portions (Fig. 4 A–D and SI Appendix, Figs. S2 and S3 and Tables S2 and S3). The LL portions also contained viral DNA from the cells that had not entered the lytic phase or entered it during the pulse time and masked the actual levels of the LL DNA remaining in only those cells supporting viral DNA amplification (Fig. 4 A–D and SI Appendix, Figs. S2 and S3 and Tables S2 and S3).
Fig. 4.
Density-shift assays reveal differences in DNA synthesis early and late during the lytic phase. (A–D) Bar graphs representing uncorrected (U) and corrected (C) data from the density-shift assays in which both P3HR1 and 4012 DNAs were measured with increasing pulse lengths at 8 hpi and 30 hpi. The corrected values at 30 to 36 hpi are close to the uncorrected values because most LL DNA is in the cells supporting the lytic phase. The density-shift assay profiles are shown in SI Appendix, Figs. S2 and S3. The area under the curve for each peak is tabulated in SI Appendix, Tables S2–S5. The density-shift experiments were repeated 2 times and the mean of the area under the curves are represented in A–D. The fraction of BrdU-labeled DNA was corrected for each sample by the fraction of cells supporting the lytic phase as determined in SI Appendix, Table S1. (E) A bar graph representing quantitative measurements of total viral DNA per cell in cells supporting the lytic phase measured using qPCR are shown. The different strains of EBV assayed are 2089 (a derivative of B95-8 which has 1 origin) in 293 cells, Akata (2 origins) in 293 cells and P3HR1 (2 origins) in iD98HR1 cells (t test; ns, not significant, P = 0.56; *P = 0.017; **P = 0.0002).
The numbers of P3HR1 and 4012 DNA molecules in the LL, HL, and HH fractions per cell supporting the lytic phase were measured (SI Appendix, Tables S2 and S3). The numbers per cell were derived by normalizing them to the cellular DNA detected with the rhodopsin probe and the fraction of the cells supporting the lytic cycle was determined by EdU pulse labeling. These measurements revealed 2 striking findings. First, the number of HL molecules at all time points early during the lytic phase for both P3HR1 and 4012 exceeded that of the LL molecules by about 10-fold. Effectively, 90% or more of all DNAs available were being used as templates. These analyses show that while 4012 DNA is one-fourth the length of P3HR1 DNA, the number of 4012 molecules per cell synthesized by the end of the labeling period (HL plus HH) was similar (60% of P3HR1) to the P3HR1 DNA synthesized (SI Appendix, Tables S2 and S3). These results indicated that at early times in the lytic phase the number of available templates and the number of origins on each DNA, but not its length, correlated with the rate of viral DNA amplification, consistent with the predictions of the simulations.
Parallel experiments were conducted between 30 and 36 hpi, when ∼60% of the cells supported EBV’s lytic phase (SI Appendix, Table S1). Even after labeling for 6 h, a time twice that of the longest labeling period used early in EBV’s lytic phase, the majority of both the P3HR1 and 4012 DNAs remained in the unreplicated LL fraction (Fig. 4 B and D and SI Appendix, Figs. S2 C and D and S3 C and D). These results indicated that at late time points EBV DNA synthesis is no longer regulated by the availability of DNA templates. This finding explains the slowing of accumulation of EBV DNA over time (Fig. 1B). Similar numbers of 4012 DNA molecules (about 50%) were synthesized during the labeling period between 30 and 36 hpi as were P3HR1 DNA molecules (SI Appendix, Tables S4 and S5), consistent again with the predictions of the simulations.
We also tested the first prediction of the simulations by measuring the levels of EBV DNA amplification for 3 full-length replicons which have 1 origin of DNA synthesis, 2089, a derivative of the B95-8 strain, or 2 origins of DNA synthesis, the EBV strains, Akata and P3HR1. Akata and P3HR1 DNAs were amplified on average 6- to 8-fold more than 2089 (Fig. 4E), independently confirming the prediction that the number of origins per replicon of a given length affects its total accumulation.
Single-Cell Assays Demonstrate the Clonality and Coordination of DNA Amplification in Replication Compartments.
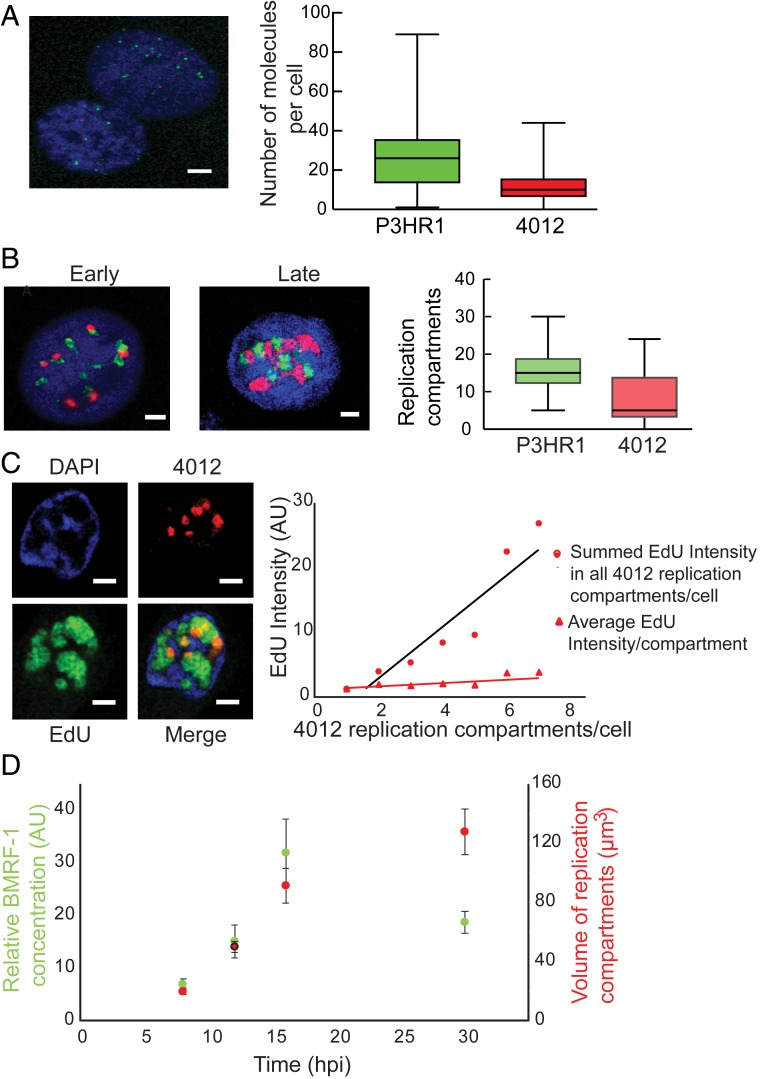
Analyses by qPCR (Figs. 1B and 4E), computational modeling, and density-shift assays (Fig. 4 A–D) established that both the rate and extent of DNA synthesis were similar for P3HR1, Akata, and 4012 DNAs despite the functional differences in these replicons while that of 2089 DNA was predictably slower. In addition to highlighting a role for the number of origins per template during DNA synthesis, the simulations also predicted that the number of molecules at the beginning of the lytic phase was critical in determining the total DNA eventually synthesized. We hypothesized that the number of molecules at the beginning of the lytic phase is proportional to the number of replication compartments formed. Single-cell assays were used therefore to address 2 questions relevant to this hypothesis: Are replication compartments seeded by individual DNA molecules, and do replication compartments coordinate their synthesis? To address the first question, FISH was used to detect P3HR1 and 4012 DNAs individually. We found that during the lytic phase P3HR1 and 4012 were in their own compartments, with only 0.2% of them being colocalized (in 1 out of 33 cells having on average 20 compartments per cell, only 1 compartment displayed colocalization [Fig. 5B; colocalization is indicated by yellow signals]). This finding indicates that single molecules serve as precursors for replication compartments. The replication compartments in some cases were adjacent to each other, as has been observed for HSV-1 (20, 21). It is in these adjacent sites that viral recombination likely occurs, as has been found for HSV-1 (21). In addition, the number of compartments formed by P3HR1 and 4012 per cell correlated with the number of viral DNA molecules per cell at the beginning of the lytic phase (Fig. 5 A and B), showing that the number of viral DNA molecules present at the onset of the lytic phase correlated with the number of compartments ultimately formed. Counting the number of compartments in Fig. 5B was restricted to cells with 30 replication compartments or fewer (90% of all cells) to maintain accuracy (in cells with >30 compartments, it was difficult to distinguish individual compartments resulting from their proximity to one another).
Fig. 5.
Single-cell assays show that the number of replication compartments determine the levels of DNA amplification. (A and B) FISH identifies P3HR1 (green) and 4012 (red) DNAs prior to the onset of the lytic phase in A and during the lytic phase in B. (Scale bars, 5 μm.) (C) Quantitative analysis of 4012 DNA synthesis using EdU pulse labeling and 3D reconstruction (n = 51 cells). On the left is a representative image of a cell containing 4012 compartments in red and undergoing lytic DNA amplification as measured with EdU in green. The chromatin is represented by DAPI in blue. The black line (R2 = 0.88) indicates the average EdU intensity per cell having the indicated number of 4012 compartments on the x axis. The red line (R2 = 0.75) indicates the average EdU intensity per replication compartment in any cell with the indicated number of 4012 compartments on the x axis. Only cells with up to 7 compartments were counted to ensure the accuracy of counting. (Scale bars, 2 µm.) (D) iD98HR1-4012 cells were induced with 200 nM tamoxifen, fixed at the indicated times and probed with anti-BMRF1 antibody. The volumes of the 4012 compartments and the intensities of anti-BMRF1 staining in green were calculated by reconstruction of 4012 compartments (n = 29 to 32 cells; error bars indicate SEM).
To address whether the number of compartments determined the overall levels of DNA synthesis and consequently the levels of DNA amplification, the cells were pulsed with EdU for 30 min at 37 °C at 13 to 14 hpi and analyzed. Replication compartments seeded by 4012 DNA were reconstructed and EdU incorporation in these compartments was measured. The analysis was restricted to cells with 7 or fewer replication compartments containing 4012 (∼80% of all cells) to ensure they were sufficiently distinct to be counted accurately (these cells had multiple, additional replication compartments housing P3HR1). The average levels of 4012 DNA synthesized were directly proportional to the number of 4012 compartments present, indicating that the number of compartments determined the total DNA accumulated during the lytic phase (Fig. 5C). Each replication compartment also supported similar levels of DNA synthesis, indicating that each compartment, in addition to housing a single genome, apparently had the resources required for comparable levels of DNA synthesis. This interpretation was further supported by single-cell assays measuring levels of BMRF1, the viral protein mediating the processivity of DNA synthesis in replication compartments. The level of BMRF1 (probed using an antibody to detect BMRF1) within the replication compartments increased early in the lytic phase in proportion to the volumes of the replication compartments and thus in proportion to the newly synthesized DNA they contained (Fig. 5D). Late in the lytic phase, the levels of BMRF1 decreased relative to the volumes of the compartments (Fig. 5D), coinciding with the majority of the DNA no longer serving as a template for DNA synthesis (Fig. 4 B and D). The observation that the distribution of BMRF1 within the replication compartments correlated with the number of viral DNA templates at times when 90% or more of the templates were supporting DNA synthesis indicates that the compartments are hubs in which DNA amplification is coordinated.
Single-Cell Assays Reveal the Evolution of EBV’s Replication Compartments over Time.
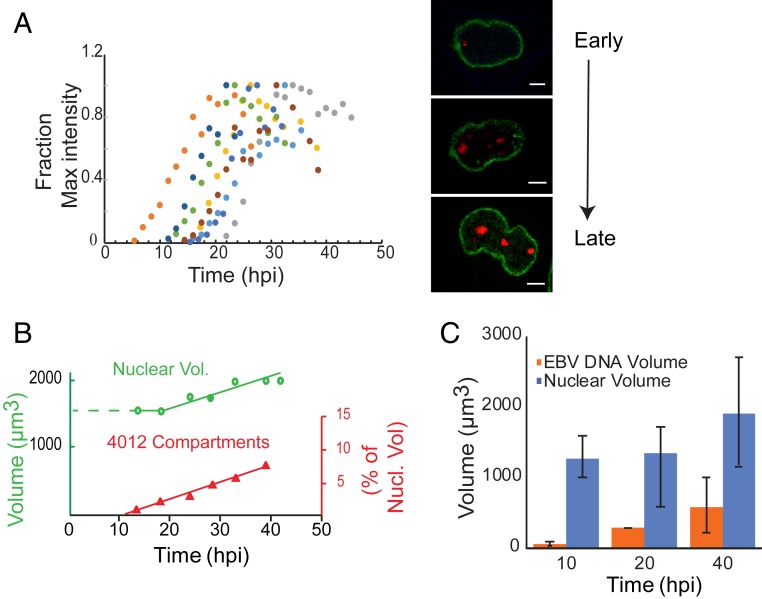
Live-cell imaging was used to follow viral DNA amplification in individual replication compartments and their DNAs were summed over time to define the time course of amplification in cells. The amplification of 4012 in replication compartments (Fig. 6A) could first be detected between 5 and 22 h after addition of tamoxifen in different cells. This broad range in time likely reflects that while EBV begins its DNA amplification at the G1/S border (4) the cells were asynchronously distributed throughout the cell cycle when tamoxifen was added. The peak accumulation of 4012 DNA took ∼13 to14 h on average in each cell (Fig. 6A). We also found that after reaching peak intensity, the 4012 signals gradually decreased by 20 to 50%. This decrease in signal was not observed in cells that were not supporting the lytic cycle, making it unlikely that the decrease resulted from photobleaching. The decrease in 4012 signals also did not result from a loss of amplified viral DNA, as evidenced by the intensities of DAPI staining within the compartments, which increased throughout the lytic cycle (SI Appendix, Fig. S4 A and B). A likely explanation for the late, observed decrease in the intensity of the tomato red signals is that its RNA is degraded by BGLF5, which is known to shut off host gene expression. For example, BGLF5 has been shown to shut off both GFP expression and that of many other genes (22). The time course of viral DNA accumulation in a population of cells entering the lytic cycle (Fig. 1B) therefore reflects the summation of the sigmoidal accrual of each replicon in single cells, each initiating asynchronously over 22 to 24 h and lasting 13 to 14 h.
Fig. 6.
Single-cell assays show spatial and temporal changes in the replication compartments during the lytic phase. (A) iD98R1-4012 cells were induced to enter the lytic phase and followed from 5 to 45 hpi. The replication compartments were then reconstructed and their total intensities were measured. Each colored dot represents the summed intensities of replication compartments in an individual cell at a given time. The fraction of maximum intensity (y) for the sum of all replication compartments for each cell was plotted as a function of time (x). The images on the right represent a single cell going through the lytic phase. Green indicates Lamin A/C-GFP, which marks the nuclear membrane; red indicates LacI-td tomato, which marks the replication compartments. (Scale bars, 5 µm.) (B) The volume of nuclei was measured by measuring the area within the Lamin A/C-GFP boundaries and summed over all of the stacks (0.36 µm per z-stack) for each time point, n = 8 cells. (C) Cells were pulsed with EdU for 30 min at the indicated times after induction and the EdU and chromatin volume (DAPI) were reconstructed for 15 to 20 cells at the indicated times and the volumes were computed.
The live-cell imaging also showed that replication compartments formed by 4012 occupied ∼7% of the nuclear volume (Fig. 6B). To extend this finding to include all compartments, we used EdU pulse labeling and 3-dimensional (3D) reconstruction of the marked replication compartments. Approximately 30 to 35% of the nucleus was eventually occupied by all of the replication compartments (Fig. 6C), while the nuclei grew by 50% in volume over the course of EBV DNA amplification (Fig. 6 B and C). This volume was consistent with the replicons together having accumulated approximately one-third the mass of the cellular DNA (Fig. 1B), while the ratio of the final volume occupied by 4012 to that occupied by P3HR1 reflected the individual levels of their DNA accumulation. Thus, single-cell assays establish that the volume of a given replication compartment is proportional to the amount of viral DNA in it (Fig. 6 B and C). Put together, the above results show that DNA amplification in individual cells proceeds for 13 to 14 h, after which it ceases, as does the growth in volume of replication compartments. This cessation of amplification is consistent with both the eventual decrease in the fraction of DNA templates used for DNA synthesis (Fig. 4 B and E) and in the levels of the viral replication protein BMRF1 in compartments (Fig. 5D). These findings, therefore, confirm the third prediction made by the simulations, that is, the plateau observed in DNA amplification in bulk measurements (Fig. 1B) reflects the cessation of DNA synthesis per se. These analyses together reveal that replication compartments are hubs in which DNA amplification occurs clonally and coordinately.
Discussion
Previous studies have shown that herpesviruses reorganize the nucleus during the lytic phase (4, 13, 23, 24) and induce specialized regions in the nucleus termed replication compartments in which viral DNA is amplified. Multiple additional DNA viruses also form replication compartments (25). While it is has been shown that inhibition of DNA synthesis leads to a collapse of replication compartments (26), it is not clear what advantage these compartments provide these viruses, including herpesviruses.
We examined the amplification of EBV DNA during the viral lytic phase to characterize it spatially and temporally. We used 2 fundamentally distinct viral replicons within the same cells to determine how their lengths and number of origins of DNA synthesis affected their amplification. Population assays of their synthesis showed surprisingly their ultimate levels of amplification to be within 2-fold of each other. We therefore asked how viral DNA synthesis within cells and between cells was coordinated to yield this unexpected, similar amplification of different replicons in the population of induced cells.
We used computational modeling to simulate the DNA synthesis measured in bulk populations with variables including the length of templates and number of origins per template. The simulation that modeled the bulk synthesis best made 3 predictions: 1) The longer replicon would keep pace with the shorter so long as the longer had 2 origins; 2) the number of DNA molecules initially present per cell determined the final level of molecules synthesized per cell; and 3) DNA synthesis per se must cease to allow the observed bulk synthesis. These 3 predictions were independently confirmed with multiple experiments. Density-shift assays proved that both replicons were synthesized to similar levels, although both templates were limiting early during the lytic phase consistent with the first prediction made by the simulations. Measurement of the amplification of different full-length replicons with either 1 or 2 origins confirmed that 2 origins per replicon increase the total amplification of the replicon. In addition, the density-shift assays proved that only a small fraction of templates was used for DNA synthesis late in the lytic phase coincident with a drop in the levels of the viral replication protein BMRF1 in the replication compartments. Additional studies have shown that there is a decline in multiple viral replication proteins late in the lytic phase (27, 28). These observations confirm the third prediction that DNA synthesis must cease to yield the eventual plateau in DNA amplification. Live-cell imaging, FISH, and EdU pulse labeling documented that each replication compartment was seeded by single DNA molecules, that the number of replication compartments per cell was proportional to the number of DNA molecules at the onset of the lytic phase, and that the levels of DNA synthesis were similar in each replication compartment. These findings demonstrate that the amplification of EBV DNA in a population of cells is coordinated through the seeding and subsequent DNA synthesis supported in each replication compartment. While our study has focused on the DNA templates and the machinery that synthesizes them, much else in the cell will contribute to the success of EBV’s lytic phase. For example, HSV-1 inhibits expression of arginosuccinate synthetase, fostering an increase in nucleotides (29), and human cytomegalovirus induces a dramatic accumulation of fatty acid elongase 7 needed to produce infectious progeny (30). EBV likely acts similarly to commandeer cellular metabolic pathways to optimize its viral replication.
The findings embodied in the model (Fig. 7) illuminate similarities and differences between the replication of EBV and alpha-herpesviruses such as HSV-1. EBV enters its lytic phase from cells already supporting the latent phase, while alpha-herpesviruses enter their lytic phase in most studies in cell culture soon after infecting cells and inducing and commandeering a DNA damage response (31, 32). In both cases replication compartments are seeded by single DNA molecules (Fig. 5 and refs. 20 and 33). In both cases viral DNAs are amplified 100- to 1,000-fold. How this amplification is achieved for different herpesviruses is not clear, though. While HSV-1 circularizes its DNA shortly after infection (34), it is not known whether the circularized DNA replicates extensively via a theta intermediate or uses rolling circle replication or some other highly branched intermediate (35). EBV uses a theta intermediate (36) for its replication at least until the synthesized DNAs are largely free of nucleosomes. If EBV were to use its theta mechanism more extensively than HSV-1, then this usage could underlie EBV’s low levels of encapsidation relative to HSV-1, given that linear polymers are a required precursor for encapsidation. While HSV-1 usually enters its lytic phase shortly after infection of most cells in culture, in vivo it establishes latency in trigeminal neurons. The transition to its lytic phase in these neurons results in low levels of viral particles (37), as opposed to the 30 to 50% encapsidation observed in cells in culture (16). One insight this conjecture supports is that latent herpesviral infections may yield inefficient encapsidation relative to those initiated by primary infections.
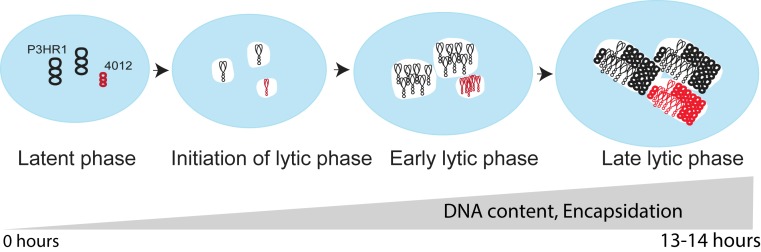
Fig. 7.
A model for DNA amplification during the lytic phase. During the latent phase each molecule of EBV is replicated no more than once per cell cycle. The mature viral DNA (both P3HR1 in black and 4012 in red) is associated with chromosomal DNA represented in blue. On entering the lytic phase, each DNA molecule (shown as a replicative intermediate) has seeded a replication compartment (in white) which is devoid of cellular chromatin. During the early lytic phase, 90% of all viral DNAs are used as templates and compartments become juxtaposed but do not intermingle. Late in the lytic phase, the nuclear volume has increased by 50% and the compartments have occupied up to 30% of the nuclear volume. The compartments still have not intermingled, only ∼30% of the viral DNA serving as templates for DNA synthesis. The cellular chromatin is now restricted to the periphery of the nucleus. Late in the lytic phase, the ratio of each viral DNA (P3HR1:4012) remains similar to that in the beginning in the lytic phase, illustrating the coordination of DNA amplification by replication compartments. The amplified DNA is associated with and becomes encapsidated by viral particles late in the lytic phase.
Materials and Methods
qPCR.
Probes were labeled with 5′-FAMRA and 3′-TAMRA (Integrated DNA Technologies). For detection of EBV DNA, 4 μL of purified total DNA per reaction was used as template, with primers and probe specific for the BALF5 sequence (as an internal control, cellular DNA was also detected using primers and probe specific to rhodopsin). The reactions were incubated at 50 °C for 2 min, then at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Data were collected on a 7900HT real-time instrument (Applied Biosystems) and analyzed using SDS Version 2.2.2 or 2.2.4 software. Fold changes were based on the assumption that amplicon levels increase by 1.8 times every cycle. Linearized plasmids were used for standard controls (38).
Live-Cell Imaging.
A Leica TCS-SP8 laser scanning confocal microscope was used to follow live cells maintained in a Tokei-Hit Bio chamber at 37 °C, 5% CO2 for ∼40 h. Cells were imaged every 90 min and 60 to 70 z-slices were captured with a distance of 0.35 µm between each successive z-plane.
EdU Pulse Labeling.
Cells were pulsed with EdU at a final concentration of 10 μM for various amounts of time depending on the experiment. The cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (PFA) for 10 min, washed with PBS, and permeablized with 0.2 to 0.5% Triton X-100 for 15 min. Cells were washed after permeablization and incubated with a labeling mixture as described in ref. 39 for 30 min. Cells were washed with PBS and mounted onto coverslips with media containing DAPI.
EdU Pulse Labeling and Immunofluorescence.
Cells pulsed with 10 μM final concentration of EdU were washed and fixed with 4% PFA for 10 min, washed with PBS, and permeablized with 0.2 to 0.5% Triton X-100 for 15 min. EdU was labeled as described above. The cells were incubated with PBS+2% bovine serum albumin (BSA) for 2 h and labeled with a primary antibody in 2% BSA (anti-BMRF1 at 1:3,000 or anti- hemagglutinin at 1:2,000) for 2 h. The cells were washed 3 times with PBS+2% BSA and incubated with a secondary antibody (fluorescent dye-conjugated) for 2 h. The cells were washed 3 times with 2% BSA and mounted with mounting media containing 2 μg/mL DAPI.
Density-Shift Experiment.
Approximately 5 × 106 to 1 × 107 cells were plated on 15-cm dishes and allowed to adhere to the surface (approximately 16 to 18 h). The cells were induced with 200 nM tamoxifen for 8 to 10 h; 100 μM of BrdU and 200 μM deoxycytidine were added for either 30, 60, 90, 120, or 180 min. The cells were washed with ice-cold PBS, trypsinized, pelleted, resuspended in 1 mL PBS, and sonicated 3 times with a 10-s-on, 30-s-off cycle at an amplitude of 2 (model CV18 ultrasonic processor). The DNA was purified by phenol–chloroform extraction followed by ethanol precipitation and resuspended in 500 μL of 10 mM Tris and 1 mM EDTA. The DNA was sheared by passing through a 26-gauge (1/2-inch) needle 6 to 9 times; 10 to 20 μg of DNA from each sample were mixed with 9.4 mL of CsCl chloride solution, reaching a final refractive index between 1.4002 and 1.402 in a Beckmann Coulter 5/8- × 3-inch centrifuge tube. The tubes were spun at 40,000 rpm in an 80Ti rotor for ∼63 h and fractionated into 250-μL fractions; 100 μL from each fraction was dialyzed using drop dialysis (2 times) on a membrane (0.025-μm VSWP filter type from Millipore) for 1 h each time. Four microliters from each DNA fraction was assayed for the amounts of viral DNA (probe against BALF5) and cellular DNA (probe against rhodopsin) using qPCR (ABI7900). Input DNA was measured for viral and cellular DNAs using qPCR to monitor their recovery.
Analysis of Extracellular Viral Particles.
Cell supernatants (1.2 mL) were collected and clarified by centrifugation at 12,000 rpm for 5 min to remove the cellular debris; 1 mL was carefully collected and treated with 10 μg of DNase in the presence of 2 mM MgCl2 at 37 °C for 30 min. A final concentration of 4 mM of EDTA was added to quench the reaction. The sample was treated with proteinase K at a final concentration of 100 μg/mL for 2 h at 56 °C. The DNA was purified by phenol–chloroform extraction followed by ethanol precipitation and quantified using qPCR.
Analysis of Intracellular Capsids.
Cells (1 to 2 × 106) were harvested and lysed by sonication (model CV18 ultrasonic processor; 10 s on, 30 s off at an amplitude of 2) in PBS. The supernatant was clarified by spinning at 2,000 rpm for 10 min in an Allegra X-15R centrifuge in a swinging bucket rotor and layered onto a 20% sucrose cushion and spun at 24,000 rpm in an SW50.1 rotor for 1 h (40). The supernatant was carefully removed and the pellet was resuspended in TE (Tris 10 mM and EDTA 1 mM) for 2 h and processed just as was done for analysis of extracellular capsids above.
Generation of HEK 293 Cells Containing EBV Mutants.
EBV bacmids MI-197 (Δ BcLF-1) and MI 371 (Δ BBRF-1) were gifts from Ya-Fang Chiu, Chang-Gung University, Taoyuan, Taiwan (41). BM2710 bacteria were transformed with the above bacmids. BM2710 carrying EBV bacmids (ΔBcLF-1and ΔBBRF-1) were grown overnight for 16 h at 32 °C. HEK 293 cells in a 6-well dish at 70% confluency were infected with 500 μL of the overnight-grown culture for 2 to 3 h at 37 °C. The medium was aspirated and replaced with fresh medium. Gentamycin at 50 µg/mL was added. Two days later, the cells were split into 15-cm dishes and 2 d later 300 μg/mL of hygromycin was added. Three weeks later, 6 colonies for each mutant were picked and expanded.
Induction of Lytic Cycle of HEK 293 Cells Carrying EBV Bacmids.
HEK 293 cells carrying EBV bacmids on 10-cm dishes at 25% confluency were transfected with plasmids encoding immediate early genes BZLF1 and BRLF1 and a late gene, p18, fused to GFP expressed from its native promoter (42) at a concentration of 1 μg of DNA per 106 cells. Lipofectamine 2000 was used for the transfections at a ratio of 2 μL of lipofectamine per μg of DNA.
Three-Dimensional Reconstruction of Cells.
Amira from Thermo Fisher Scientific and Imaris from Bitplane were used for reconstruction. The signals from various channels were manually thresholded and the signal intensities and volumes enclosed within the region thresholded were calculated. As a control, beads (Abberior Instruments) ranging from 1 μm to 6 μm were used for reconstruction and the inaccuracies ranged from 100 to 2%, respectively.
FISH.
FISH analysis was performed as described (9). In brief, cells were treated with 0.075 M KCl for 20 min at 37 °C, fixed in methanol:acetic acid at a 3:1 ratio for 30 min at room temperature, and spread on cold slides. Slides were dried and prehybridized in a buffer with 4× standard saline citrate (SSC) and 0.5% (vol/vol) Nonidet P-40 (Sigma-Aldrich) for 30 min at 37 °C, dehydrated in a cold ethanol series (70, 80, and 90%) for 2 min each, air-dried at 50 °C, denatured in a buffer with 70% formamide and 2× SSC, pH 5.3, for 2 min at 72 °C followed by dehydration with a cold ethanol series, and air-dried. A plasmid encoding LacO (2348) and one encoding LacI-tdtomato (4001.6) were labeled with digoxigenin-11-2′-deoxy-uridine-5′-triphosphate (Roche), and a plasmid encoding the HindIII C fragment of EBV (43) was labeled with biotin-16-dUTP (Roche) by nick translation as hybridization probes for detection of 4012 and P3HR1 in cells, respectively. Twenty microliters of the labeled probe was precipitated along with 6 µg salmon sperm DNA and 4 µg human Cot-1 DNA (Invitrogen) and resuspended in 15 µL of CEP hybridization buffer (55% formamide, 1× SSC, pH 7.0, and 10% dextran sulfate) followed by denaturation at 70 °C for 10 min. Three to four microliters of the probe was hybridized with each sample at 37 °C overnight in a moist chamber. The slides were washed twice in 2× SSC containing 50% formamide for 30 min at 50 °C and twice in 2× SSC for 30 min at 50 °C. The hybridized probe was detected by incubation with 10 µL of a detection solution containing a mouse monoclonal anti-digoxin–FITC conjugate (Sigma-Aldrich) and a streptavidin–Cy3 conjugate (Sigma-Aldrich) for 20 min at 37 °C. The slides were washed twice in 4× SSC containing 0.05% Triton X-100 for 5 min at room temperature and mounted in Vectashield antifading media with DAPI (Vector Laboratories) for staining chromosomes. FISH slides were imaged on Leica confocal stimulated emission depletion microscope.
Simulation of Plasmid Replication.
We developed a simulation of viral DNA replication in a population of cells that contained 2 different types of plasmids, one of which had 1 origin of replication and the other had 2 origins of replication and was 4 times longer. Our simulations contained one variable to describe the rate of elongation of DNA synthesis (varied to fit only the P3HR1 data and then fixed for these simulations at 1.37 kb/min), one for the fraction of origins that are active, and one for the mean number of plasmids per cell. The fraction of origins that are active reflects, in part, the turnover of the replication machinery over time and has therefore been fitted as a sigmoid function. This sigmoid function captures both the limits of the fraction of origins that can be active early and late in the lytic phase. The sigmoid function required 3 variables: a maximal probability of plasmid replication (eventually held constant at 0.9), the slope at which replication decreased (varied to fit the data and then held at 0.0005), and the half-maximal value of the sigmoid, which roughly describes the maximal number of origins before which replication ceases (varied to fit the data and then held at 15,500). The initial populations were drawn from a normal distribution with a SD of one-fifth of the mean (varied as described in the main text, for example set at means of 5 ± 1 and 25 ± 5 for 4012 and P3HR1, respectively). Given these conditions, we simulated time steps (6 to 48 per replication of 4012) during which origins of replication could fire, replication could extend, and origins could be released back to the pool after a plasmid completed replication. We determined that the number of time steps did not substantially affect the results above 12 steps per replication, or approximately one step every 2.5 min. Each cell was assigned a time at which it would enter the lytic cycle, determined from experimental data. Each cell was assigned a random number of plasmids of each type, as described above. Then, we stepped through time. At each step, we determined which cells had entered into the lytic phase. Any origins of just-completed plasmids were returned to the open pool and 2 plasmids were returned to the pool for each one completed. We determined the total number of open origins of replication per cell, treating each origin independently. Because we were stepping through time, we had to adjust the probability of origin firing by the size of the time step (otherwise every origin would fire essentially immediately). We scaled down the maximal probability of plasmid replication per step by the number of time steps such that the sigmoid above describes the probability of firing across the replication time of 4012. We used random numbers derived from the binomial distribution and the sigmoid described above to determine the number of origins of replication that would fire during the time step. We then drew from a random multinomial distribution that represented the origins of each plasmid to assign which origins would fire. Finally, we calculated the time remaining until the plasmid was replicated with the assumption that the rate of replication was fixed and initiation from the second origin of P3HR1 would halve the time remaining until the plasmid completed replication.
Data Availability.
The computer program developed here can be found at the following link: https://bitbucket.org/asugden/rocsim/src.
Supplementary Material
Acknowledgments
We thank Donata Oertel and Andrea Bilger for all the helpful discussions and critical inputs for the manuscript, Makoto Ohashi for 293 cells carrying the Akata strain, and Ya-Fang Chiu for bacmids encoding EBV mutants. We thank Beth Weaver, Aussie Suzuki, Dave Vereide, Eric Johannsen, Nate Sherer, and our laboratory colleagues for all their help with the manuscript. We thank the University of Wisconsin–Madison Optical Core for giving us access to the microscopy facility. This work was supported by the NIH Grant P01 CA022443. B.S. is an American Cancer Society Research Professor.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The computer program developed here can be found at the following link: https://bitbucket.org/asugden/rocsim/src.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913992116/-/DCSupplemental.
References
- 1.Takeda D. Y., Dutta A., DNA replication and progression through S phase. Oncogene 24, 2827–2843 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Pope B. D., et al. , Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Y., Shay J. W., Wright W. E., Telomere terminal G/C strand synthesis: Measuring telomerase action and C-rich fill-in. Methods Mol. Biol. 735, 63–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu Y.-F., Sugden A. U., Sugden B., Epstein-Barr viral productive amplification reprograms nuclear architecture, DNA replication, and histone deposition. Cell Host Microbe 14, 607–618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monier K., Armas J. C. G., Etteldorf S., Ghazal P., Sullivan K. F., Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2, 661–665 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Pombo A., Ferreira J., Bridge E., Carmo-Fonseca M., Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13, 5075–5085 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weitzman M. D., Fisher K. J., Wilson J. M., Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70, 1845–1854 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cziepluch C., et al. , H-1 parvovirus-associated replication bodies: A distinct virus-induced nuclear structure. J. Virol. 74, 4807–4815 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nanbo A., Sugden A., Sugden B., The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J. 26, 4252–4262 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dembowski J. A., Dremel S. E., DeLuca N. A., Replication-coupled recruitment of viral and cellular factors to herpes simplex virus type 1 replication forks for the maintenance and expression of viral genomes. PLoS Pathog. 13, e1006166 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prichard M. N., Duke G. M., Mocarski E. S., Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J. Virol. 70, 3018–3025 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammerschmidt W., Sugden B., Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55, 427–433 (1988). [DOI] [PubMed] [Google Scholar]
- 13.Bosse J. B., et al. , Remodeling nuclear architecture allows efficient transport of herpesvirus capsids by diffusion. Proc. Natl. Acad. Sci. U.S.A. 112, E5725–E5733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y., Cei S. A., Rodriguez Huete A., Colletti K. S., Pari G. S., Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J. Virol. 78, 11664–11677 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobbs W. E. 2nd, DeLuca N. A., Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73, 8245–8255 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Church G. A., Dasgupta A., Wilson D. W., Herpes simplex virus DNA packaging without measurable DNA synthesis. J. Virol. 72, 2745–2751 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson L. K., et al. , Murine cytomegalovirus capsid assembly is dependent on US22 family gene M140 in infected macrophages. J. Virol. 83, 7449–7456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granato M., et al. , Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82, 4042–4051 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlova S., et al. , An Epstein-Barr virus mutant produces immunogenic defective particles devoid of viral DNA. J. Virol. 87, 2011–2022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobiler O., Brodersen P., Taylor M. P., Ludmir E. B., Enquist L. W., Herpesvirus replication compartments originate with single incoming viral genomes. MBio 2, e00278–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomer E., et al. , Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses. FASEB J. 33, 9388–9403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowe M., et al. , Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U.S.A. 104, 3366–3371 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu Y.-F., Sugden B., Epstein-Barr virus: The path from latent to productive infection. Annu. Rev. Virol. 3, 359–372 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Chang L., et al. , Herpesviral replication compartments move and coalesce at nuclear speckles to enhance export of viral late mRNA. Proc. Natl. Acad. Sci. U.S.A. 108, E136–E144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid M., Speiseder T., Dobner T., Gonzalez R. A., DNA virus replication compartments. J. Virol. 88, 1404–1420 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strang B. L., et al. , Human cytomegalovirus UL44 concentrates at the periphery of replication compartments, the site of viral DNA synthesis. J. Virol. 86, 2089–2095 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ersing I., et al. , A temporal proteomic map of Epstein-Barr virus lytic replication in B cells. Cell Rep. 19, 1479–1493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu C.-C., et al. , Genome-wide transcription program and expression of the Rta responsive gene of Epstein-Barr virus. Virology 345, 358–372 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Grady S. L., Purdy J. G., Rabinowitz J. D., Shenk T., Argininosuccinate synthetase 1 depletion produces a metabolic state conducive to herpes simplex virus 1 infection. Proc. Natl. Acad. Sci. U.S.A. 110, E5006–E5015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purdy J. G., Shenk T., Rabinowitz J. D., Fatty acid elongase 7 catalyzes lipidome remodeling essential for human cytomegalovirus replication. Cell Rep. 10, 1375–1385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohni K. N., Smith S., Dee A. R., Schumacher A. J., Weller S. K., Herpes simplex virus type 1 single strand DNA binding protein and helicase/primase complex disable cellular ATR signaling. PLoS Pathog. 9, e1003652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dembowski J. A., DeLuca N. A., Temporal viral genome-protein interactions define distinct stages of productive herpesviral infection. MBio 9, e01182–e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekine E., Schmidt N., Gaboriau D., O’Hare P., Spatiotemporal dynamics of HSV genome nuclear entry and compaction state transitions using bioorthogonal chemistry and super-resolution microscopy. PLoS Pathog. 13, e1006721 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strang B. L., Stow N. D., Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79, 12487–12494 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weller S. K., Coen D. M., Herpes simplex viruses: Mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 4, a013011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfüller R., Hammerschmidt W., Plasmid-like replicative intermediates of the Epstein-Barr virus lytic origin of DNA replication. J. Virol. 70, 3423–3431 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sawtell N. M., Quantitative analysis of herpes simplex virus reactivation in vivo demonstrates that reactivation in the nervous system is not inhibited at early times postinoculation. J. Virol. 77, 4127–4138 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vereide D. T., Sugden B., Lymphomas differ in their dependence on Epstein-Barr virus. Blood 117, 1977–1985 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salic A., Mitchison T. J., A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 2415–2420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grady L. M., et al. , The exonuclease activity of HSV-1 UL12 is required for the production of viral DNA that can be packaged to produce infectious virus. J. Virol. 91, e01380–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu Y.-F., et al. , A comprehensive library of mutations of Epstein Barr virus. J. Gen. Virol. 88, 2463–2472 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Djavadian R., Chiu Y.-F., Johannsen E., An Epstein-Barr virus-encoded protein complex requires an origin of lytic replication in cis to mediate late gene transcription. PLoS Pathog. 12, e1005718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammerschmidt W., Sugden B., Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340, 393–397 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The computer program developed here can be found at the following link: https://bitbucket.org/asugden/rocsim/src.