Significance
Interactions between crowded cytosols and membrane surfaces are unavoidable inside cells. This all-atom simulation study suggests that nonspecific protein–membrane interactions create a water-rich protein depletion zone between the membrane and the crowded environment, leading to an increased propensity of proteins to aggregate in bulk, but also allow for accelerated diffusion on the surface of the membrane when proteins come closer to the surface occasionally. The simulation results furthermore provide evidence of a nonspecific mechanism for protein-induced membrane curvature formation as a result of crowding near the membrane.
Keywords: molecular dynamics, diffusion, crowding, membrane curvature, phospholipids
Abstract
Atomistic molecular dynamics simulations of concentrated protein solutions in the presence of a phospholipid bilayer are presented to gain insights into the dynamics and interactions at the cytosol–membrane interface. The main finding is that proteins that are not known to specifically interact with membranes are preferentially excluded from the membrane, leaving a depletion zone near the membrane surface. As a consequence, effective protein concentrations increase, leading to increased protein contacts and clustering, whereas protein diffusion becomes faster near the membrane for proteins that do occasionally enter the depletion zone. Since protein–membrane contacts are infrequent and short-lived in this study, the structure of the lipid bilayer remains largely unaffected by the crowded protein solution, but when proteins do contact lipid head groups, small but statistically significant local membrane curvature is induced, on average.
Proteins and nucleic acids have to function under highly crowded conditions inside cells (1). An unresolved question is how such environments impact biomolecular structure and dynamics compared with the in vitro noncrowded conditions in most experimental and computational studies (2, 3). Earlier work has described the volume exclusion effect of crowding (4), but more recent studies emphasize the role of weak, nonspecific interactions between biomolecules in the cell (5–11). There is increasing evidence that protein–protein interactions in highly concentrated environments can potentially destabilize native folds, contrary to what the volume-exclusion effect predicts (12–16). Transient molecular cluster formation between biomolecules has emerged as the primary determinant of reduced diffusion in crowded cellular environments (10, 11, 17–19).
In addition to studies of biomolecular crowding in cytoplasmic environments, crowding inside or near membranes has also been examined (20–24). Membrane surfaces are ubiquitous not just at the boundaries of cells but also as part of lipid vesicles and cellular organelles such as the endoplasmic reticulum (ER). Protein crowding within the membrane slows down dynamics as in the cytoplasm (23), but the effects are more complicated as a result of domain formation and confinement by cytoskeletal elements (25). Moreover, as predicted by Saffman-Delbrück theory (26), diffusion of proteins inside the membrane is only weakly dependent on particle size R (23). In contrast, the Stokes-Einstein model for isotropic solvent describes an 1/R dependence. As a consequence, molecular association and clustering are not expected to strongly impact diffusion within the membrane.
Less is known about the interface between membrane surfaces and crowded cytoplasmic environments, especially when nonmembrane binding proteins are involved. Previous studies have found a role of protein crowding in inducing membrane curvature (27). This effect was attributed to the anisotropic pressure that is generated by proteins moving laterally on a membrane surface. More recent work argues that this effect is much less significant compared with membrane curvature induced by hydrophobic insertion of peripherally associated membrane proteins (28, 29). Moreover, it seems that a high fraction of the membrane surface needs to be covered by proteins, and an asymmetric distribution of crowding between membrane leaflets is needed to realize significant overall curvature (28). It remains unclear how high concentrations of proteins may modulate other membrane properties. There is also little insight into how the properties of cytoplasmic proteins may be affected by the presence of a membrane surface under crowded conditions. The structural and dynamic properties of proteins in concentrated solutions could be altered in the presence of a membrane. It may be expected, for example, that proteins forced to interact with a membrane as a result of a crowding experience reduced diffusion and are subject to destabilization when surrounded partially by a nonaqueous environment.
To examine these questions in molecular detail, we present 10 μs-scale atomistic molecular dynamics (MD) simulations of mixtures of proteins in the presence and absence of a membrane. The simulations suggest that the presence of the membrane increases protein clustering and allows proteins to diffuse faster on the membrane surface than in the crowded milieu. However, the proteins affect the membrane properties only to a small extent, and the membrane has little effect on protein stability.
Results and Discussion
We carried out simulations of concentrated mixtures of villin, protein G, and ubiquitin in the absence and presence of a lipid bilayer composed of 1-palmitoyl-2-oleyol-phosphatidylcholine (POPC), sphingomyelin, and cholesterol (Fig. 1), initially with NAMD on submicrosecond time scales, followed by 10 μs on Anton2, for which results are reported here (SI Appendix, Table S1). The simulations follow well-tested protocols for describing protein–membrane interactions that have resulted in excellent agreement between simulation and experiment in many previous studies (30–33). To avoid the overestimation of protein–protein interactions, we have applied a recently introduced force field modification (10).
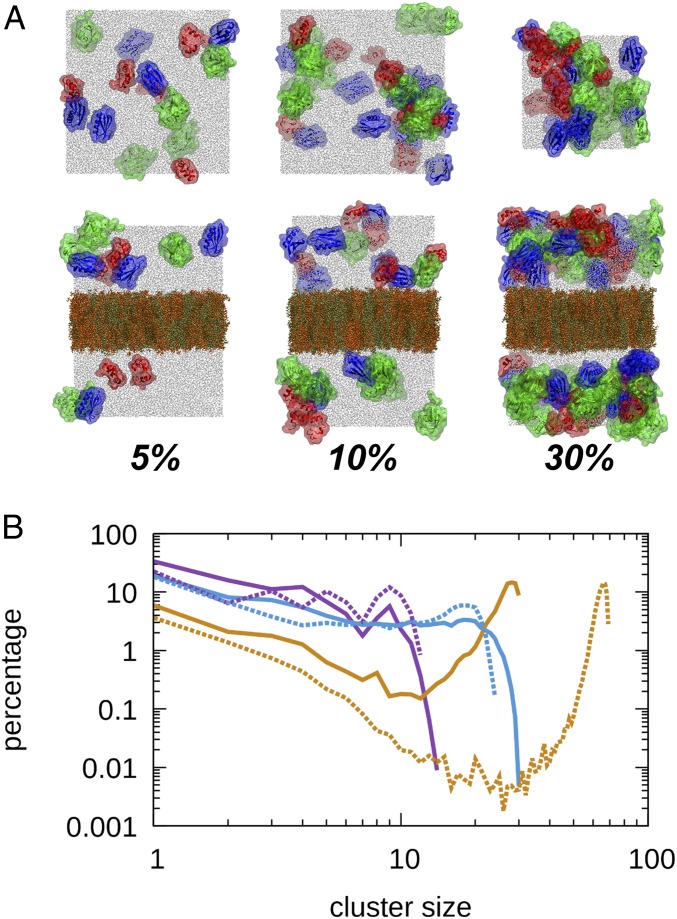
Fig. 1.
Systems and protein clustering. (A) Overview of simulated systems without (Top) and with (Bottom) phospholipid bilayers at protein volume fractions of 5%, 10%, and 30% with villin (red), protein G (blue), ubiquitin (green), POPC (tan), sphingomyelin (orange), cholesterol (dark brown), and water (gray). (B) Cluster size distributions between all proteins at 5% (purple), 10% (light blue), and 30% (tan) in the absence (solid lines) and presence (dashed lines) of the membrane based on protein contacts with minimum Cα–Cα distances <7 Å. A cluster size of 1 corresponds to monomers.
Stability of Proteins.
Most of the proteins remained stable near their native structure, but at least 1 villin in each of the simulations and 2 copies of protein G in the simulation at 10% without the membrane deviated significantly from the experimental reference structures after several microseconds (SI Appendix, Figs. S1–S3). Increased RMSD was correlated with increases in the radius of gyration (SI Appendix, Figs. S4–S6) and indicates unfolding. In previous simulations of similar protein solutions, partial unfolding was also observed, especially for villin (10, 13). Villin has marginal stability around 4 kcal/mol to full unfolding, and 2 to 3 kcal/mol to partially unfolded states with the force field used here (10). Experimental stability is estimated to be 2 to 3 kcal/mol at 298 K, with unfolding rates around 100 μs or less (34). Some unfolding during 10 μs may thus be expected. However, the main conclusions are not significantly affected when only the first 2 μs are analyzed to avoid partially unfolded structures (SI Appendix, Figs. S15, S32, and S34).
The volume exclusion effect of crowding is expected to increase the stability of compact native states (35), but a destabilization of the native state because of protein–protein interactions upon crowding has also been proposed (13). To analyze whether protein structures may be altered at higher concentrations and whether the presence of the membrane affects stability, average RMSD and radii of gyration for each type of protein were calculated as a function of concentration (SI Appendix, Table S2). Small differences may be consistent with native state destabilization (such as a slight increase in average RMSD for folded villin with an RMSD below 2.5 Å) or with native state compaction due to volume exclusion (such as a slight decrease in the radius of gyration for ubiquitin) when comparing the most (30%) and least (5%) concentrated systems. However, P values of 0.2 or larger suggest only weak significance of these observations. The presence of the membrane also does not significantly affect protein stability (SI Appendix, Table S3).
Similar conclusions are found when comparing root mean square fluctuations (RMSF) (SI Appendix, Figs. S7–S9). For most residues, different concentrations and the presence of the membrane have only a small effect, but the loop in ubiquitin around residue 35 fluctuates significantly less at higher protein concentrations. A decreased RMSF is consistent with the smaller radius of gyration in ubiquitin on crowding.
Protein Contacts and Clustering.
At the high concentrations considered here, interactions between proteins are unavoidable. Indeed, we find extensive contacts between proteins at all concentrations (SI Appendix, Fig. S10). In the absence of the membrane, about 5% to 10% of the theoretical maximum contacts are formed at any time, largely independent of concentration. When contacts are analyzed between the same type of protein, we find that contacts between ubiquitin are more likely than between villin or protein G (SI Appendix, Table S4). This finding generally holds even after normalizing the number of contacts by the surface area of spheres with volumes equivalent to the proteins (SI Appendix, Table S4), and can be understood based on the differences in net charges. Ubiquitin is neutral, and villin and protein G are positively and negatively charged, respectively. Overall, interactions between different proteins are nonspecific without a strong bias toward specific protein–protein interfaces. However, there are some preferences for involving certain residues in protein–protein contacts (Fig. 2 and SI Appendix, Figs. S11–S13) that vary only slightly, depending on what the other protein partner is. Interestingly, the RMSF is elevated for many of the residues involved in forming contacts (SI Appendix, Figs. S7–S9).
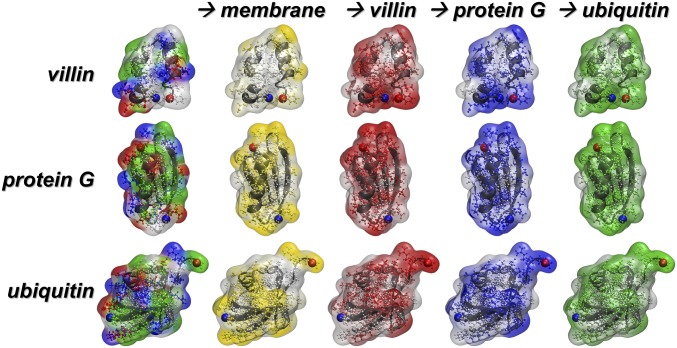
Fig. 2.
Protein interactions. Normalized preferences for residue–residue interactions projected onto the molecular structures of villin (+2e), protein G (−4e), and ubiquitin (neutral) from simulation at 30% volume fraction in the presence of the membrane. The coloring in the leftmost column reflects residue types (polar, green; basic, blue; acidic, red; hydrophobic, white). Subsequent coloring reflects preferences for interactions with the membrane (yellow), villin (red), protein G (blue), or ubiquitin (green). More saturated colors indicate stronger preferences. The N and C termini are shown as blue and red spheres, respectively.
When the membrane is introduced, the number of contacts per protein increases, with the most significant change at 5% and 30% (based on P values of 0.03 [5%], 0.55 [10%], and 0.07 [30%]; SI Appendix, Fig. S10 and Table S4). The trend is less clear for contacts between proteins of the same type (SI Appendix, Table S4). This contrasts with a geometrically expected decrease in contacts resulting from the membrane, as contacts along z between proteins above and below the membrane are prevented.
We further analyzed the formation of clusters based on the protein contacts. Cluster size distributions based on Cα–Cα contacts are shown in Fig. 1B and are very similar results to those of contacts based on heavy atom distances (SI Appendix, Fig. S14). A simple hard-sphere model without attraction results in significantly smaller cluster sizes (SI Appendix, Fig. S16).
At 5%, we find a decaying cluster size distribution indicative of transient cluster formation similar to what we described previously for concentrated villin solutions below the solubility limit (10). At 30%, most proteins are found in a single large cluster consistent with a phase change to an aggregated form (36). The presence of the membrane generally shifts the cluster size distribution to larger cluster sizes. This can be seen most clearly from a reduction in monomers and other small clusters. This effect is greater at 5% and 30%, consistent with a greater increase in contacts (SI Appendix, Table S4). We note that differences at the largest cluster sizes reflect, in part, different numbers of protein copies in nonmembrane and membrane systems (SI Appendix, Table S1).
The increased clustering in the presence of the membrane indicates a decrease in solubility that could be observable macroscopically. Here, the protein solutions are sandwiched between 2 membrane surfaces (when considering the periodic images along z), with a gap of about 150 Å. Such conditions may be found in the ER or Golgi apparatus. Although our systems are only a simple approximation of these complex biological environments, our results are consistent with previous observations of condensation of enzymes in the rough ER (37) and the aggregation and sorting of secretory proteins in the ER and Golgi apparatus (38, 39). Although specific mechanisms involving membrane-bound receptors likely play a role as well (40), this study proposes a generic mechanism for selective protein aggregation in the ER based on close membrane surfaces.
Protein–Membrane Interactions.
None of the proteins studied here is expected to bind the membrane peripherally or via insertion. However, crowding may be expected to force proteins to make close contacts with the membrane surface. Density profiles of proteins relative to the membrane show that this is not the case (Fig. 3 and SI Appendix, Fig. S17). Instead, the proteins are preferentially excluded from the membrane surface, even at 30%. Next to the membrane surface, a water and ion layer of about 10 to 15 Å is formed where protein concentrations are low. This finding is independent of the scaling of protein–water interactions (SI Appendix, Fig. S18). For comparison, we simulated hard spheres with radii equivalent to the proteins in the presence of a hard surface (SI Appendix, Fig. S18). The hard spheres pack more closely to the surface compared with the proteins, where density increases slowly up to 70 Å away from the membrane center. Our results also do not agree with the increased membrane interactions reported from a simple hydrodynamic model (41). This indicates clearly that the proteins are thermodynamically excluded from the membrane. This means that protein–membrane binding affinities are low, and that solvation of the membrane lipid head groups with water and ions is preferred over membrane–protein interactions. This finding is consistent with previous analyses showing weak protein–membrane binding unless anionic lipids are present (42–44). Moreover, examples of strongly binding peripheral membrane proteins typically involve hydrophobic anchors that are partially inserted into the lipid bilayer to interact with the lipid acyl chains (45) and/or π–cation interactions between aromatic residues and choline headgroups (30, 44). The proteins studied here do not have a large fraction of aromatic residues, surface-exposed hydrophobic elements, or a lipidation modification such as myristoylation suitable for partial membrane insertion.
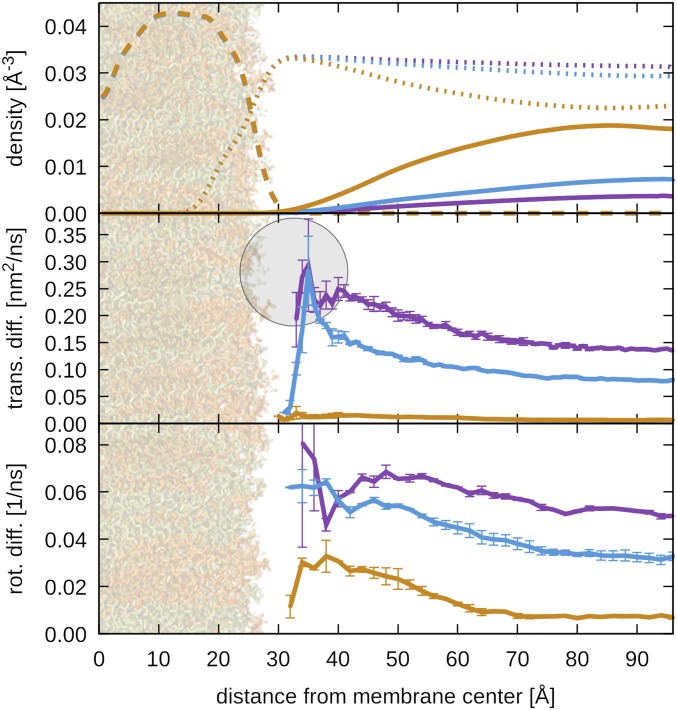
Fig. 3.
Density and diffusion near the membrane. Heavy atom density distributions of molecular components (Top), translational diffusion constants parallel to the membrane (Center), and rotational diffusion constants (Bottom) as a function of the distance from the membrane center along the membrane normal for systems with proteins at 5% (purple), 10% (light blue), and 30% (tan). Densities are shown for proteins (solid lines), lipids (long dashes), and water molecules (short dashes). Translational and rotational diffusion constants were assigned to the center of mass of a given protein at the beginning of the intervals for which mean-square displacements and rotational correlation functions were obtained. Diffusion on shorter time scales, along the membrane normal, and based on mid- or endpoints of diffusion time intervals is shown in SI Appendix, Figs. S26–S28. Translational diffusion was estimated from mean-square displacement vs. time during 10 to 100 ns. Statistical errors for density distributions are less than 1%. The lipid bilayer projected at scale and a gray sphere at the size of protein G at the point of closest membrane contact are shown for perspective.
Protein exclusion from the membrane surface results in an increased protein concentration away from the membrane. This is consistent with increased protein contacts and clustering in the presence of the membrane. However, all types of proteins occasionally come into contact with the lipid headgroups (SI Appendix, Fig. S19). The interactions are slightly more frequent for villin and protein G than for ubiquitin (SI Appendix, Fig. S19). This is expected from stronger electrostatic interactions between the charged proteins and the zwitterionic lipid head group. Protein residues that are preferred in protein–membrane interactions vary by protein (Fig. 2) and only partially overlap with the residues involved in protein–protein contacts (SI Appendix, Figs. S11–S13). The residues that are most likely to be involved in protein–membrane contacts are tyrosine, which is known to interact strongly with choline head groups (30, 44); hydrophobic alkanes, which interact favorably with the lipid acyl chains; and polar residues, which can form hydrogen bonds with the lipid head groups (SI Appendix, Table S5).
An analysis of contact residence times (based on protein–lipid heavy atom distances within 5 Å) revealed a typical contact time of around 2 ns for protein G and ubiquitin; villin remained bound slightly longer (i.e., 4 to 6 ns; SI Appendix, Table S6). A much longer time component of around 1 μs is attributed to unbinding, reinsertion into the main protein cluster, and later rebinding.
The predictions from the simulations could be tested experimentally by employing X-ray or neutron scattering to study the density variations. Protein–membrane interactions could be probed via site-directed spin labels in combination with paramagnetic resonance or other fluorescence techniques. We are not aware of such experiments for comparable systems.
Protein Diffusion.
Retarded diffusion of proteins on crowding is well known. Recent work (10, 17, 19, 46) has suggested that this is largely a result of transient cluster formation. Here, we analyzed whether the presence of the membrane affects diffusive properties. Translational and rotational diffusion coefficients in bulk solutions without the membrane bilayer match previously reported values for villin (10). SI Appendix provides averages in SI Appendix, Tables S7–S8, mean-squared displacement in SI Appendix, Figs. S20–S22, and correlation functions in SI Appendix, Figs. S23–S25. The TIP3P water model used here underestimates solvent viscosity about 3-fold, and reported diffusion rates are thus 3 times faster than in experiment. Diffusion coefficients for the ubiquitin are retarded more than for villin or protein G, as expected based on size. There is only a moderate decrease in translational diffusion between short (<1 ns) and longer (>10 ns) time scales for 5% and 10%, consistent with extensive cluster formation that results in a lack of transiently varying diffusion rates (10). At 30%, diffusion on longer time scales is retarded more significantly compared with shorter times, reflecting cage effects (SI Appendix, Table S7).
In the presence of the membrane, diffusion rates vary depending on the location of a protein with respect to the membrane (Fig. 3). The discussion here primarily focuses on motion parallel to the membrane. Diffusion perpendicular to the membrane (SI Appendix, Fig. S27) shows similar trends. Diffusion is slowest at the farthest point from the membrane and increases toward the membrane surface up until 35 Å from the membrane center. When proteins come into direct contact with the lipids, sharply reduced diffusion is observed in most cases. Translational and rotational diffusion are affected similarly, but translational diffusion is accelerated more strongly near the membrane than rotational diffusion at 5% and 10%. The opposite trend is found at 30%. At 5%, rotational diffusion appears to slow down significantly near 40 Å, but the translational diffusion is affected less. This may indicate that proteins near the membrane are subject to orientational restraints as a result of preferential involvement of certain protein residues in membrane interactions (Fig. 2), but as this observation is only made at 5%, an artifact resulting from limited sampling is also possible. Increased diffusion near the membrane is most pronounced for long-time diffusion (>10 ns; SI Appendix, Figs. S26 and S27). This finding is largely independent of whether the initial, mid-, or endpoint of a diffusion interval is used to assign the distance from the membrane (SI Appendix, Fig. S28).
The translational diffusion of proteins parallel to the membrane in a confined membrane system can be estimated from bulk 3D diffusion in a nonmembrane system (47). The long-time x-y diffusion far away from the membrane is lower than or equal to the bulk-based estimate (using Eq. 4 in SI Appendix), but diffusion near the membrane surface is significantly faster (SI Appendix, Fig. S29). Faster diffusion near the membrane can be understood from the protein concentration gradient in the presence of the membrane, where there are fewer obstacles in the depletion zone near the membrane surface, but the reduced protein concentration also means that proteins are less likely to experience the faster diffusion near the membrane surface. However, there is still a net effect of accelerated diffusion parallel to the membrane, when the probability of finding a protein close to the membrane is considered (SI Appendix, Fig. S30). Villin and protein G benefit most when they move from the bulk to about 50 to 60 Å from the membrane center, and the acceleration is greater at 30% than at 5% and 10%, with an increase of up to almost 90% for protein G. The increase in diffusion described here is in disagreement with the retarded diffusion found near the cell wall, based on a simplified model that emphasizes hydrodynamic effects but neglects the details of protein–protein and protein–lipid interactions (41).
Rotational diffusion is not expected to be affected strongly in the presence of confinement when the membrane surfaces are separated by more than several times the size of the proteins. Therefore, we compared the rotational diffusion rates directly between the nonmembrane and membrane systems (SI Appendix, Fig. S31). In general, we find a similar conclusion of rotational diffusion far away from the membrane being slower than or equal to bulk diffusion, whereas diffusion near the membrane surface exceeds bulk diffusion values at 10% and 30% volume fractions. At 5% volume fraction, only villin surpasses bulk diffusion rates, at around 47 Å from the membrane center, whereas protein G and ubiquitin remain below bulk diffusion rates for all distances from the membrane center. This can be understood from a greater sensitivity of rotational diffusion to contact formation (10) and the greater increase in the number of protein contacts at 5% in the presence of the membrane relative to the nonmembrane systems (SI Appendix, Fig. S10), as well as apparently longer-lasting protein–membrane contacts.
The overall picture that is emerging from the above analysis is that proteins near a membrane surface may diffuse faster than in the crowded bulk solution because of the protein depletion zone that is formed by nonmembrane interacting proteins in the vicinity of a membrane bilayer. The faster diffusion is most evident in translational diffusion over longer time scales and could suggest a mechanism for circumventing the challenge of slow transport of biomolecules within the crowded cellular milieu and to reach membrane-embedded receptors and transporters. One could test this idea experimentally by comparing diffusion via NMR or fluorescence recovery after photobleaching in a very membrane-rich environment of a cell, such as the ER; with other parts of the cell; or by studying comparable in vitro systems.
Membrane Properties.
Lipid order parameters as a function of protein concentration (SI Appendix, Fig. S33) were found to be very similar values to previous results for pure POPC–sphingomyelin–cholesterol mixtures with the same force field that was used here (48). Protein concentration had virtually no effect, indicating that the internal structure and dynamics of the lipid bilayer and the liquid-ordered state expected for the lipid composition in this study are not affected by the presence of the proteins.
Previous studies have suggested that crowding may introduce membrane bending (27). Membrane deformations as a result of protein–membrane contacts were examined by averaging the distance of lipid phosphate atoms from the membrane center for phosphates near the closest protein–membrane contact point as a function of the protein–membrane distance (Fig. 4). The average phosphate distance from the membrane center is decreased by as much as 0.2 Å as proteins approach the membrane to within 3 to 10 Å of heavy atom distances. This suggests membrane indentation on either direct contact or via indirect interactions mediated, for example, by water or longer-range electrostatics. This effect is most pronounced for villin, which also has the longest protein–membrane contact residence times. The weakest effect is observed for ubiquitin, for which the membrane is only slightly indented on contact at 5% and 10%, while the membrane is slightly wider when ubiquitin touches the membrane at 30% (Fig. 4). For very short protein–membrane contacts (<3 Å), the membrane appears to become distorted more strongly, with indentations and extrusions varying by protein and concentration, but there are high statistical uncertainties.
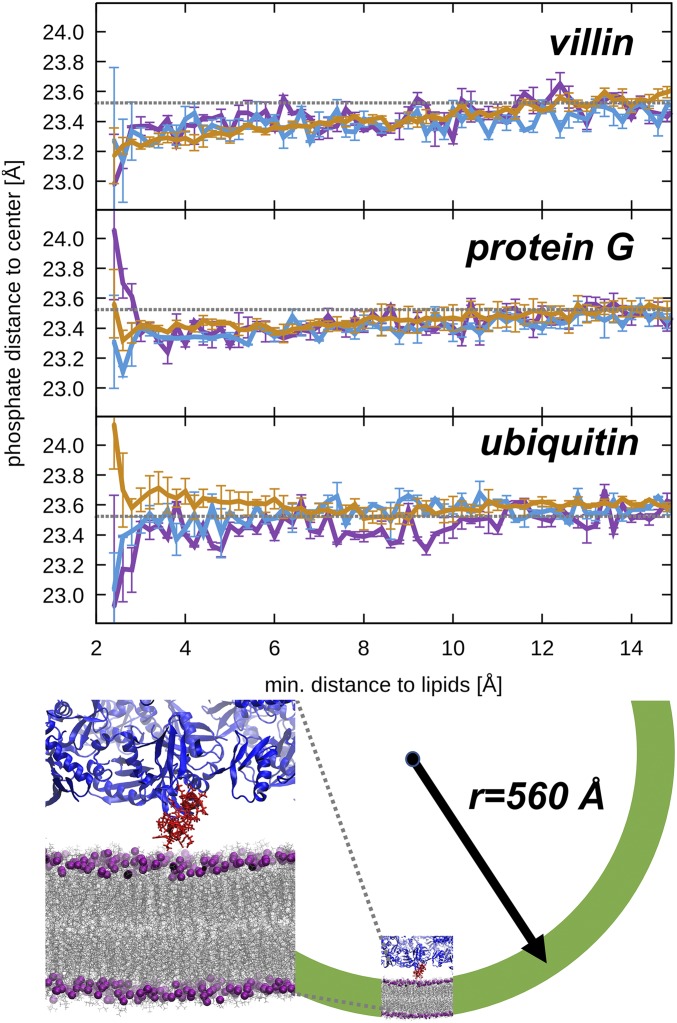
Fig. 4.
Crowding-induced membrane deformations.(Top) Membrane distortion as a function of protein interactions at 5% (purple), 10% (light blue), and 30% (tan). Protein–membrane distances are defined based on minimum heavy atom distances between proteins and lipids. Membrane distortions are characterized by average phosphate distances from the membrane center for phosphate atoms within a 15-Å radius from the lipid atom in closest contact with the protein. The average phosphate distance to the center irrespective of any protein contact is indicated as a gray line. (Bottom, Left): Snapshot of a curved membrane (gray with purple phosphates) when in contact with villin (red) from the simulation at 30%. Other proteins are shown in blue. (Bottom, Right) Projection of a sphere with a radius compatible with the induced curvature (green).
In the absence of any specific interactions, it is expected that proteins are most likely to bump into membrane surfaces that extrude furthest into the aqueous solvent. In principle, lipid head groups could also be pulled away from the membrane as a result of electrostatic attraction. However, the opposite finding of an indented membrane on protein contact suggests a specific mechanism for inducing membrane curvature. We did not observe any overall net membrane bending, presumably because of periodic boundary conditions and an equal distribution of proteins on either side of the membrane. However, our results predict that net bending would arise if proteins are unequally distributed on either side of the membrane, in terms of concentration and/or composition. An indentation of 0.2 Å over a 30-Å-diameter disk (the diameter of the spherical region within which phosphates around the protein–membrane contact point were analyzed) is equivalent to the curvature on the surface of a sphere with a 560-Å radius (Fig. 4). Such spheres are in the range of lipid vesicle sizes, suggesting that crowding could stabilize such vesicles. Although the effect may appear small, we find that at about 25% of the time there is either a villin or protein G within 10 Å of a lipid (the maximum distance at which we see an effect on membrane curvature). This translates into 1 protein per 700 nm2, or about 56 proteins bound to the membrane of an entire vesicle. This could provide significant overall stabilization when the contributions of all proteins are summed up. The stabilization of vesicles or formation of curved membranes could be tested experimentally in the presence of proteins at high concentration.
Conclusions
In this study, we are reporting atomistic simulations of concentrated protein solutions near a neutral phospholipid bilayer. The model proteins considered here, villin, protein G, and ubiquitin, are not known to interact specifically with phospholipid membranes, and we found that even at the highest concentrations of 30% volume fraction, the proteins are preferentially excluded from the membrane surface. This finding has 2 major consequences: the proteins effectively experience a higher concentration as they occupy a smaller volume in the presence of the membrane, which leads to increased contacts and increased clustering, and when proteins enter the depletion zone of 10 to 15 Å near the membrane surface, they can diffuse significantly faster than in the crowded environment, especially over longer (>10 ns) time scales. Additional insights from this work are that the membrane structure remains largely unperturbed in the presence of the crowding proteins, which may be expected because the proteins largely avoid the membrane. However, when proteins do contact the membrane, they appear to be able to induce local curvature that could support lipid vesicles. The conclusions are experimentally testable hypotheses that we hope will stimulate new studies of the interaction between crowded cellular environments and membrane surfaces either in vitro or in vivo.
A major limitation of this work is the relatively small size of the systems dictated by the available computer resources; future work will aim at extending the spatial and temporal scales via coarse-grained modeling. Another limitation is that proteins that are expected to interact with the membrane were not included and that no integral membrane proteins were present in the phospholipid bilayer. In both cases, we would expect that cytoplasmic proteins may interact more extensively with the membrane surface. This could mitigate the membrane-induced increase in clustering described here, and alter the diffusive characteristics of proteins near the membrane. Extending our current work to such more complicated systems will be another aim of future work.
Methods
Systems.
Concentrated solutions of proteins with and without a lipid bilayer were constructed (Fig. 1 and SI Appendix, Table S1). All systems contained equal numbers of 3 types of proteins: the chicken villin head piece (HP-36; “villin”), the B1 domain of streptococcal protein G (“protein G”), and human ubiquitin (“ubiquitin”). The proteins were chosen because of their small size, variation in charge (villin: +2e, protein G: −4e, ubiquitin: 0), and variation in secondary structures. Phospholipid bilayers consisted of equal numbers of POPC, sphingomyelin, and cholesterol to mimic the typical composition of animal cell membranes. Crowded systems were prepared at 3 protein concentrations (5%, 10%, and 30%) based on the total volume of the proteins relative to the aqueous solvent. In terms of weight, the concentrations were about 40, 80, and 250 g/L. All systems were solvated in explicit water, and K+/Cl− were added to neutralize the systems and achieve excess KCl concentrations of about 150 mM (SI Appendix, Table S1) to reflect typical physiological conditions (49).
MD Simulations.
The initial systems were equilibrated before commencing production MD simulations. The equilibration of the systems was performed using NAMD (version 2.10) (50) and the CHARMM-GUI protocol (51, 52). After initial setup and equilibration (SI Appendix), production simulations of the 10% and 30% systems were then carried out without any restraints, using NAMD (version 2.10) (50) for periods of 300 to 800 ns (SI Appendix, Table S1). Subsequently, simulations were extended for 10 μs, using the special-purpose Anton2 hardware (53). Additional simulations at 5%, with and without a membrane, were also carried out on Anton2 over 10 μs each. All systems were simulated under periodic boundaries in the NPT ensemble. In the production simulations, temperature was set to 310 K, and the pressure to 1.01325 bar. See SI Appendix for further details.
Data Availability.
All data discussed in the paper will be made available to readers.
Supplementary Material
Acknowledgments
Funding was provided by the National Science Foundation Grants MCB 1330560, MCB 1817307, and MCB 1810695; by the National Institutes of Health (National Institute of General Medical Sciences) Grant R35 GM126948; by RIKEN pioneering research projects on “dynamic structural biology” and “glycolipidologue”; and by a grant for the RIKEN Center for Biosystems Dynamics Research. Computer time was used on XSEDE resources (TG-MCB090003) and on the Anton2 special-purpose supercomputer at the Pittsburgh Supercomputing Center (PSCA18053P).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The simulations were run on the Anton2 supercomputer at Pittsburgh Supercomputing Center which requires that all generated data are automatically archived and made available to the public. Otherwise, any and all data described in this manuscript will be shared upon request.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910771116/-/DCSupplemental.
References
- 1.Ellis R. J., Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 11, 114–119 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Danielsson J., Oliveberg M., Comparing protein behaviour in vitro and in vivo, what does the data really tell us? Curr. Opin. Struct. Biol. 42, 129–135 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Rivas G., Minton A. P., Macromolecular crowding in vitro, in vivo, and in between. Trends Biochem. Sci. 41, 970–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman S. B., Minton A. P., Macromolecular crowding: Biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 22, 27–65 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Feig M., Yu I., Wang P. H., Nawrocki G., Sugita Y., Crowding in cellular environments at an atomistic level from computer simulations. J. Phys. Chem. B 121, 8009–8025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu I., et al. , Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm. eLife 5, e19274 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monteith W. B., Cohen R. D., Smith A. E., Guzman-Cisneros E., Pielak G. J., Quinary structure modulates protein stability in cells. Proc. Natl. Acad. Sci. U.S.A. 112, 1739–1742 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majumder S., et al. , Probing protein quinary interactions by in-cell nuclear magnetic resonance spectroscopy. Biochemistry 54, 2727–2738 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roos M., et al. , Coupling and decoupling of rotational and translational diffusion of proteins under crowding conditions. J. Am. Chem. Soc. 138, 10365–10372 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Nawrocki G., Wang P. H., Yu I., Sugita Y., Feig M., Slow-down in diffusion in crowded protein solutions correlates with transient cluster formation. J. Phys. Chem. B 121, 11072–11084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etoc F., et al. , Non-specific interactions govern cytosolic diffusion of nanosized objects in mammalian cells. Nat. Mater. 17, 740–746 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Inomata K., et al. , High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature 458, 106–109 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Harada R., Tochio N., Kigawa T., Sugita Y., Feig M., Reduced native state stability in crowded cellular environment due to protein-protein interactions. J. Am. Chem. Soc. 135, 3696–3701 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feig M., Sugita Y., Variable interactions between protein crowders and biomolecular solutes are important in understanding cellular crowding. J. Phys. Chem. B 116, 599–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miklos A. C., Sarkar M., Wang Y., Pielak G. J., Protein crowding tunes protein stability. J. Am. Chem. Soc. 133, 7116–7120 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Wang Q., Zhuravleva A., Gierasch L. M., Exploring weak, transient protein–protein interactions in crowded in vivo environments by in-cell nuclear magnetic resonance spectroscopy. Biochemistry 50, 9225–9236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nawrocki G., Karaboga A., Sugita Y., Feig M., Effect of protein-protein interactions and solvent viscosity on the rotational diffusion of proteins in crowded environments. Phys. Chem. Chem. Phys. 21, 876–883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck C., et al. , Nanosecond tracer diffusion as a probe of the solution structure and molecular mobility of protein assemblies: The case of ovalbumin. J. Phys. Chem. B 122, 8343–8350 (2018). [DOI] [PubMed] [Google Scholar]
- 19.von Bülow S., Siggel M., Linke M., Hummer G., Dynamic cluster formation determines viscosity and diffusion in dense protein solutions. Proc. Natl. Acad. Sci. U.S.A. 116, 9843–9852 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchhoff H., Haferkamp S., Allen J. F., Epstein D. B., Mullineaux C. W., Protein diffusion and macromolecular crowding in thylakoid membranes. Plant Physiol. 146, 1571–1578 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H.-X., Crowding effects of membrane proteins. J. Phys. Chem. B 113, 7995–8005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindén M., Sens P., Phillips R., Entropic tension in crowded membranes. PLoS Comput. Biol. 8, e1002431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramadurai S., et al. , Lateral diffusion of membrane proteins. J. Am. Chem. Soc. 131, 12650–12656 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Javanainen M., Martinez-Seara H., Metzler R., Vattulainen I., Diffusion of integral membrane proteins in protein-rich membranes. J. Phys. Chem. Lett. 8, 4308–4313 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Dix J. A., Verkman A. S., Crowding effects on diffusion in solutions and cells. Annu. Rev. Biophys. 37, 247–263 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Saffman P. G., Delbrück M., Brownian motion in biological membranes. Proc. Natl. Acad. Sci. U.S.A. 72, 3111–3113 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stachowiak J. C., et al. , Membrane bending by protein-protein crowding. Nat. Cell Biol. 14, 944–949 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Kozlov M. M., et al. , Mechanisms shaping cell membranes. Curr. Opin. Cell Biol. 29, 53–60 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z., Atefi E., Baumgart T., Membrane shape instability induced by protein crowding. Biophys. J. 111, 1823–1826 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schillinger A.-S., Grauffel C., Khan H. M., Halskau O., Reuter N., Two homologous neutrophil serine proteases bind to POPC vesicles with different affinities: When aromatic amino acids matter. Biochim. Biophys. Acta 1838, 3191–3202 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Muller M. P., et al. , Characterization of lipid-protein interactions and lipid-mediated modulation of membrane protein function through molecular simulation. Chem. Rev. 119, 6086–6161 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monje-Galvan V., Klauda J. B., Peripheral membrane proteins: Tying the knot between experiment and computation. Biochim. Biophys. Acta 1858, 1584–1593 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Schwierz N., Krysiak S., Hugel T., Zacharias M., Mechanism of reversible peptide-bilayer attachment: Combined simulation and experimental single-molecule study. Langmuir 32, 810–821 (2016). [DOI] [PubMed] [Google Scholar]
- 34.Buscaglia M., Kubelka J., Eaton W. A., Hofrichter J., Determination of ultrafast protein folding rates from loop formation dynamics. J. Mol. Biol. 347, 657–664 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Zhou H.-X., Rivas G., Minton A. P., Macromolecular crowding and confinement: Biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 37, 375–397 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karandur D., Wong K.-Y., Pettitt B. M., Solubility and aggregation of Gly(5) in water. J. Phys. Chem. B 118, 9565–9572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tooze J., Kern H. F., Fuller S. D., Howell K. E., Condensation-sorting events in the rough endoplasmic reticulum of exocrine pancreatic cells. J. Cell Biol. 109, 35–50 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chanat E., Huttner W. B., Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J. Cell Biol. 115, 1505–1519 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizuno M., Singer S. J., A soluble secretory protein is first concentrated in the endoplasmic reticulum before transfer to the Golgi apparatus. Proc. Natl. Acad. Sci. U.S.A. 90, 5732–5736 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dancourt J., Barlowe C., Protein sorting receptors in the early secretory pathway. Annu. Rev. Biochem. 79, 777–802 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Chow E., Skolnick J., Effects of confinement on models of intracellular macromolecular dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 14846–14851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogaski B., Klauda J. B., Membrane-binding mechanism of a peripheral membrane protein through microsecond molecular dynamics simulations. J. Mol. Biol. 423, 847–861 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Heimburg T., Marsh D., “Thermodynamics of the Interaction of Proteins with Lipid Membranes” in Biological Membranes: A Molecular Perspective from Computation and Experiment, Merz K. M., Roux B., Eds. (Birkhäuser Boston, Boston, MA, 1996), pp. 405–462. [Google Scholar]
- 44.Khan H. M., et al. , A role for weak electrostatic interactions in peripheral membrane protein binding. Biophys. J. 110, 1367–1378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sankaram M. B., Marsh D., “Protein-lipid interactions with peripheral membrane proteins” in New Comprehensive Biochemistry, Watts A., Ed. (Elsevier, 1993), vol. 25, chap. 6, pp. 127–162. [Google Scholar]
- 46.Rothe M., et al. , Transient binding accounts for apparent violation of the generalized Stokes-Einstein relation in crowded protein solutions. Phys. Chem. Chem. Phys. 18, 18006–18014 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Simonnin P., Noetinger B. T., Nieto-Draghi C., Marry V., Rotenberg B., Diffusion under confinement: Hydrodynamic finite-size effects in simulation. J. Chem. Theory Comput. 13, 2881–2889 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Bera I., Klauda J. B., Molecular simulations of mixed lipid bilayers with sphingomyelin, glycerophospholipids, and cholesterol. J. Phys. Chem. B 121, 5197–5208 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Martin R. B., “Bioinorganic chemistry” in Encyclopedia of Molecular Cell Biology and Molecular Medicine, Meyers R. A., Ed. (Wiley-VCH, 2004). [Google Scholar]
- 50.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Jo S., Lim J. B., Klauda J. B., Im W., CHARMM-GUI Membrane Builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaw D. E., et al. , “Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer” in Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (IEEE Press, New Orleans, LA, 2014), pp 41-53. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper will be made available to readers.