Significance
In organisms composed of a single cell, RNAs of viral origin may be transmitted to daughter cells at cell division without passing through an extracellular virion stage. These RNAs usually encode an RNA-dependent RNA polymerase that enables their replication. For some of these agents, such as Narnaviruses, no capsid protein is expressed, and thus, they are called capsidless viruses. Here, we identify putative capsidless viral RNAs in animals, in nematodes closely related to the model organism Caenorhabditis elegans. We show that these RNAs are transmitted vertically through the host germline. Our work provides evidence that animal cells harbor capsidless viruses.
Keywords: Caenorhabditis, vertical transmission, viral RNA-dependent RNA polymerase, bunyavirus, narnavirus
Abstract
Here, we report on the discovery in Caenorhabditis nematodes of multiple vertically transmitted RNAs coding for putative RNA-dependent RNA polymerases. Their sequences share similarity to distinct RNA viruses, including bunyaviruses, narnaviruses, and sobemoviruses. The sequences are present exclusively as RNA and are not found in DNA form. The RNAs persist in progeny after bleach treatment of adult animals, indicating vertical transmission of the RNAs. We tested one of the infected strains for transmission to an uninfected strain and found that mating of infected animals with uninfected animals resulted in infected progeny. By in situ hybridization, we detected several of these RNAs in the cytoplasm of the male and female germline of the nematode host. The Caenorhabditis hosts were found defective in degrading exogenous double-stranded RNAs, which may explain retention of viral-like RNAs. Strikingly, one strain, QG551, harbored three distinct virus-like RNA elements. Specific patterns of small RNAs complementary to the different viral-like RNAs were observed, suggesting that the different RNAs are differentially recognized by the RNA interference (RNAi) machinery. While vertical transmission of viruses in the family Narnaviridae, which are known as capsidless viruses, has been described in fungi, these observations provide evidence that multicellular animal cells harbor similar viruses.
Symbionts are transmitted through host generations in different manners (we use here the word “symbiont” in the sense of “living together”). Schematically, the smaller symbiotic partner may have intracellular or extracellular forms and be transmitted horizontally or vertically to progeny. An example of a vertically transmitted symbiont is the intracellular bacteria Wolbachia, which resides directly inside the host oocyte cytoplasm and is thus passed to the next generation (1, 2).
Eukaryotic viruses are often horizontally transmitted, but some are vertically transmitted: for example, fungal totiviruses (3) or the Drosophila melanogaster sigma virus (4). An extreme form of vertical transmission is the integration of retroviruses or transposons in the host genome, for which the distinction between host and symbiont becomes unclear (e.g., ref. 5 in Caenorhabditis). In unicellular organisms, vertically transmitted viral nucleic acids may be replicated and passed to the mitotic or meiotic progeny cells without going through a capsid-covered virion stage. Such entities are called capsidless viruses, and their nucleic acid usually encodes the polymerase that enables their replication (6).
The best-studied capsidless viruses are the narnaviruses of fungi coding for an RNA-dependent RNA polymerase (RdRP) only (7–9). The narnaviral RNA of the yeast Saccharomyces cerevisiae (7–9) encodes an RdRP and is passed through cell divisions. Some Narnaviridae family members of the Mitovirus genus replicate in mitochondria (10) and possibly evolved from bacteriophages (6). Narnaviridae are also found in other protists, such as oomycetes (11). Related RNA sequences were detected by sequencing animals (12), although it remains unclear whether the animals or associated fungi or protists are the actual hosts. A further reduction is known with plant viroids: the viroid RNA does not encode any protein and depends solely on host proteins for replication (13, 14). To our knowledge, capsidless viruses have been described in plants, oomycetes, and fungi but not in animals.
Bunyaviruses are negative-strand RNA viruses, meaning that the noncoding strand is packaged in the virion. Note that the notion of negative- or positive-strand viruses depends on the presence of a virion stage and may thus be inapplicable if no such stage takes place in a cellular vertical transmission mode. Bunyaviruses usually include 3 segments, encoding an RdRP, a nucleoprotein, and an accessory glycoprotein. The RNAs are 5′ capped by a cap-snatching mechanism (15) and enveloped in Golgi-derived membranes (16–18). So far, no form of bunyaviruses without an enveloped stage has been demonstrated, but in some cases, only an RdRP has been reported by sequencing (compare with in Drosophila flies) (table S2 of ref. 19).
Sobemoviruses are positive-stranded RNA viruses. They infect plants but are also found associated with insects, such as the Ixodes scapularis (tick) Associated Virus 2 (20). The RdRP protein is expressed via a ribosomal frameshifting mechanism facilitated by a canonical slippery sequence UUUAAAC (21).
The nematode Caenorhabditis elegans has become a prime model to study interactions with other organisms, such as bacteria, fungi, microsporidia, oomycetes, or viruses (22). Yet so far, no vertically transmitted symbiont has been found other than transposons and retroelements in the genome (5, 23) or an artificially introduced virus in a mutant background (24). Intracellular Wolbachia bacteria have been found to infect other nematodes but not Caenorhabditis (25). We previously found the first viruses to infect Caenorhabditis by noticing intestinal cell degeneration (26). However, all viruses isolated in this manner belonged to a single family related to nodaviruses; they infect intestinal cells only and are horizontally transmitted through the fecal–oral route (26–28).
High-throughput sequencing has revolutionized virus discovery by the ability to detect viral-like DNA or RNA sequences in diverse sample types (12, 19, 29–31). Sequencing alone does not suffice to determine in which hosts these nucleic acids reside: animal or plant samples may contain other eukaryotes, such as fungi. We here report on sequencing of RNA preparations of cultures of wild-caught Caenorhabditis nematodes and the discovery of vertically transmitted viral-like RNA sequences coding for putative RdRPs. These sequences are not present as DNA and must thus replicate exclusively as RNAs. By in situ hybridization, we localize these RNAs to the nematode germline as well as soma. We further show that the infected host strains are defective in degrading exogenous double-stranded RNAs (dsRNAs) and detected specific patterns of small RNAs (sRNAs) complementary to the different viral-like RNAs. Caenorhabditis nematodes are thus animal hosts harboring narnavirus sequences. Furthermore, the detection of bunya-like sequences without evidence of an accompanying capsid as well as sobemo-like viral RNAs suggests that a wide range of such viral RNA elements can persist in animal hosts.
Results
Vertically Transmitted RNA Molecules Coding for Viral RdRPs in Wild Caenorhabditis.
Horizontally transmitted intestinal viruses related to nodaviruses (26–28) had previously been identified in wild nematodes displaying intestinal symptoms. Here, we screened for viral sequences in an unbiased manner by sequencing RNA preparations of Caenorhabditis natural strains without selection for a disease phenotype. Multiple samples yielded noda-like viruses as described elsewhere (28).
Other than noda-like viruses, we found in the RNA preparations of a small number of Caenorhabditis strains (of ca. 200 processed strains) one or more RNA molecules encoding putative RdRPs related to those of RNA viruses (Table 1). In Caenorhabditis nematodes, bleach treatment kills all developmental stages except the embryos (SI Appendix) and eliminates microbial associates from the culture. For each RNA, we thus tested for their direct association with the nematode and their transmission mode using a bleach treatment of the nematode culture followed by RT-PCR and/or in situ hybridization.
Table 1.
List of detected RdRP-encoding RNA molecules in Caenorhabditis natural isolates confirmed by RT-PCR and/or FISH
| Nematode strain | Nematode species | Geographic origin | Year of isolation | Sequence length, nt | Relatedness of RdRP | Accession no. | Name |
| JU1396 | C. brenneri | Near Medellin, Colombia | 2008 | 7,968 | Bunyavirus | KM580531 | C. brenneri Colombia bunya-like virus |
| QG551 | C. remanei | Okinawa, Japan | 2011 | 3,841 | Bunyavirus | MN272066 | C. remanei Okinawa bunya-like virus |
| QG551 | C. remanei | Okinawa, Japan | 2011 | 3,189 | Narnavirus | MN272067 | C. remanei Okinawa narnavirus |
| QG551 | C. remanei | Okinawa, Japan | 2011 | 1,844 | Sobemovirus | MN272068 | C. remanei Okinawa sobemo-like virus |
| JU2557 | C. remanei | Illkirch, France | 2013 | 2,440 | Bunyavirus | MN272069 | C. remanei Illkirch bunya-like virus |
| JU3236 | C. zanzibari | M'Tsangamouji, Mayotte | 2017 | 1,538 | Bunyavirus | MN272070 | C. zanzibari Mayotte bunya-like virus |
Identification of bunya-like viral RdRPs.
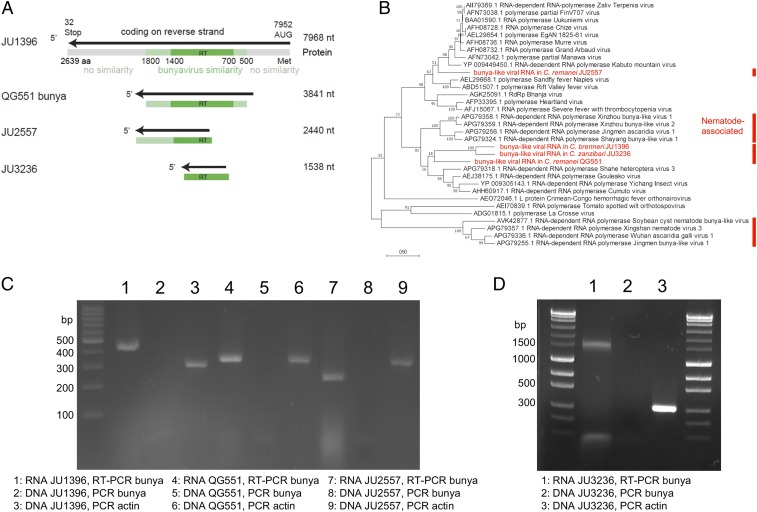
The first viral RdRP detected by sequencing was found in the Caenorhabditis brenneri strain JU1396 from Colombia. Combining next generation sequencing (NGS) and 3′ rapid amplification of complementary DNA ends-PCR, we obtained a contig of 7,968 nucleotides (nt) encoding an open reading frame (ORF) of 2,639 amino acids (aa) (Fig. 1A). Its sequence is highly divergent from any known sequence. By basic local alignment search tool (BLAST) analysis, its central region (aa 500 to 1,800) displays greatest similarity to RdRP sequences of viral family Phenuiviridae in the order Bunyavirales (32); the N-terminal and C-terminal regions do not match anything in the databases. Specifically, the protein shares 29 and 28% amino acid identity with Gouleako (17) and Cumuto (18) goukoviruses, respectively. Phylogenetic analysis (Fig. 1B) demonstrated that this sequence is also related to bunya viral-like sequences recently found associated with insect or vertebrate parasitic nematodes (12).
Fig. 1.
Several RNAs detected in Caenorhabditis wild isolates code for viral RdRPs related to bunyaviruses. (A) Schematic representation of the bunyavirus-like contigs. The black line represents the noncoding strand (as is usual for bunyaviruses), with the arrow indicating the coding direction. The protein is represented below in gray, with the RdRP similarity in green and the reverse transcriptase (RT) domain in dark green. (B) Phylogenetic analysis of the 4 Caenorhabditis RdRPs aligning to bunyaviruses using our sequences (in red), their best BLAST counterparts, a representative subset of bunyavirus sequences from ref. 18, and those associated with vertebrate or plant nematodes (12, 66) (marked with red lines). The relationships were inferred using maximum likelihood based on 333 alignable positions and tested using 200 bootstraps. Branch lengths are scaled to the number of amino acid substitutions per site. The percentage of trees in which the associated taxa clustered together in the bootstrap analysis is shown next to the branches. (C and D) The bunyavirus-like sequences are found under an RNA form and not under a DNA form. The electrophoresis gels show the products of RT-PCR on RNA preparations and of direct PCR on RNase-treated DNA preparations, with a positive PCR control using actin primers. The PCRs within each panel were performed in parallel. Sequencing of PCR products yielded the same sequences as found earlier. Bp, base pair.
The genomes of canonical bunyaviruses, including Gouleako and Cumuto goukoviruses, are composed of 3 negative–single-strand RNA [(−)ssRNA] segments: large, medium, and small. In the JU1396 strain, however, no other viral-like RNA molecules could be detected by read alignment. The 2 other segments may thus be absent in JU1396; alternatively, they may have diverged beyond our ability to recognize them using alignment tools, such as BLAST.
Through further sampling and sequencing, we found 3 additional bunya-like viral RNAs encoding RdRPs: 2 in Caenorhabditis remanei (a 3,841-nt contig in strain QG551 from Japan; a 2,440-nt contig in JU2557 from France) and 1 in Caenorhabditis zanzibari (a 1,538-nt contig in JU3236 from Mayotte). As with JU1396, no medium or small segment-like RNAs were detected (Table 1). Their relationships to the closest RdRP protein sequences in the databases are shown in Fig. 1B. Three of them appear in a clade including insect-borne goukoviruses and nematode-associated sequences (Fig. 1B), while the RdRP sequence in C. remanei JU2557 was more closely related to insect-transmitted phleboviruses, such as Kabuto mountain virus (42% identity) and Sandfly and Rift Valley fever viruses (Fig. 1B).
All 4 tested bunya-like RdRP RNAs could still be detected after several rounds of bleaching and culture from the bleached embryos (Fig. 1 C and D), demonstrating that the RNA was directly associated with the nematode and likely vertically transmitted through the germline.
We tested whether these viral-like RNA molecules could be transcribed from DNA. In all 3 tested strains, PCR amplification occurred after the RNA preparation was reverse transcribed but not when a DNA preparation was used (Fig. 1 C and D), demonstrating that these sequences were not present in DNA form.
Narnaviral RdRP.
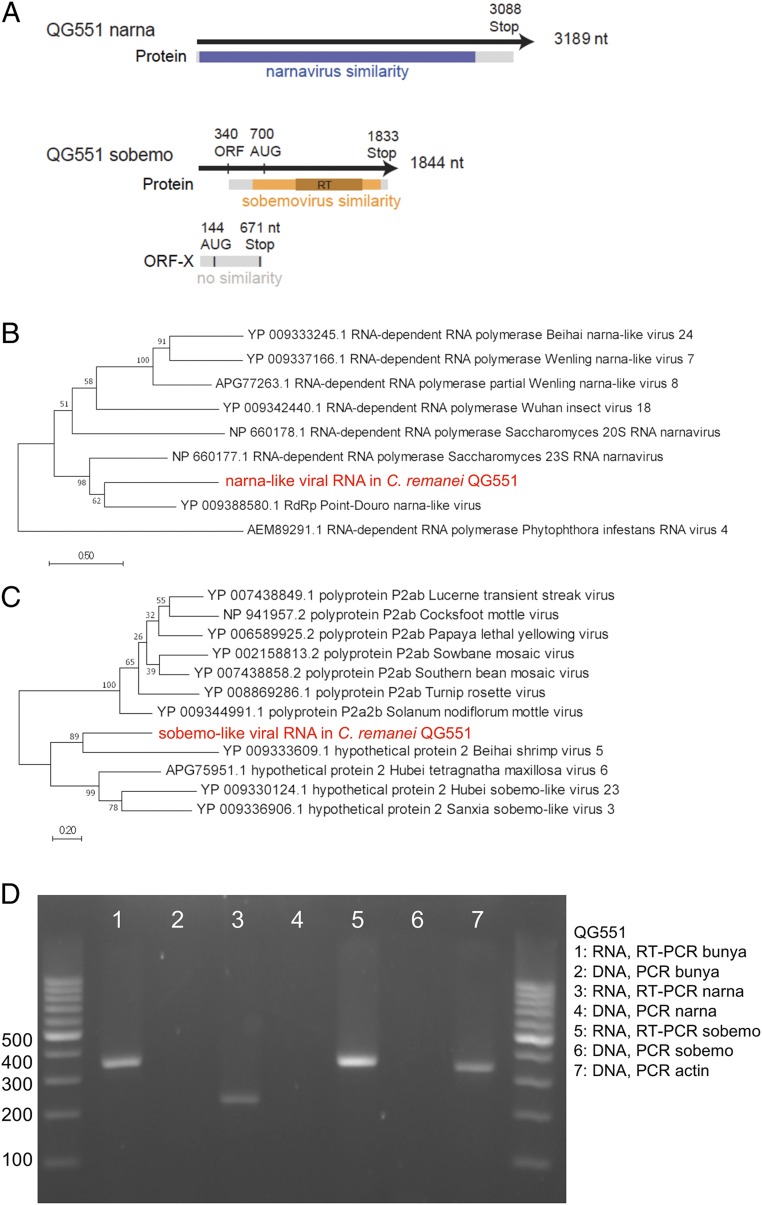
Next, we detected RNA sequences coding for an RdRP with similarity to narnaviruses (3,189-nt contig in C. remanei QG551) (Fig. 2A). Narnaviruses are capsidless viruses of yeasts with a single RNA segment encoding an RdRP (6, 8, 9, 33). The QG551 narnavirus-like RdRP protein sequence was most closely related to those of the 23S narnavirus of S. cerevisiae (33) (954 aa, 34% amino acid similarity) and Point Douro narnavirus (841 aa, 32% similarity) found by sequencing mosquitoes (34) (Fig. 2B). The current QG551 narnavirus contig is slightly longer than the complete 23S narnavirus sequence and may be near full length. At the 3′ end, the sequence includes a stop codon and a 99-nt untranslated region (Fig. 2A) containing 3 potential hairpin structures as reported for 2 S. cerevisiae narnaviruses (33). At the 5′ end, the 7th amino acid of the ORF aligns with the 11th of the 23S narnavirus, suggesting that, while the ORF lacks an initiation codon, very few amino acids may be missing.
Fig. 2.
RNAs detected in C. remanei QG551 code for various viral RdRPs. (A) Schematic representation of the narnavirus- and sobemovirus-like contigs. The black line represents the contig. The protein is represented in gray, with the regions similar to narnaviruses in blue and to sobemoviruses in orange. A putative upstream ORF is indicated (ORF-X). RT, reverse transcriptase domain. (B) Maximum likelihood phylogenetic analysis of the QG551 narnaviral-like RdRP (in red) using best BLAST counterparts and the Phytophthora infestans virus 4 RdRP chosen as a distant relative based on ref. 8. The relationships were inferred based on 562 positions and tested using 100 bootstraps. (C) Maximum likelihood phylogenetic analysis of the QG551 sobemo-like RdRP (in red) using best BLAST counterparts and sobemovirus sequences chosen using ref. 21. The relationships were inferred based on 323 positions and tested using 100 bootstraps. (D) RT-PCR on RNA preparations and direct PCR on DNA preparations for sequences from C. remanei QG551 aligning to various viruses. The PCRs were performed in parallel with those in Fig. 1C. Sequencing of PCR products yielded the same sequences as found earlier.
After bleaching and RT-PCR, we found that the QG551 narnavirus RNA is transmitted vertically (Fig. 2D). Again, PCR amplification only occurred after the nucleic acid preparation was reverse transcribed (Fig. 2D), showing that the sequence was not present as DNA.
Sobemo-like viral RdRP.
Additionally, we detected in C. remanei QG551 a 1,844-nt RNA sequence encoding a putative RdRP (Fig. 2A) with a best BLAST to Behai shrimp virus 5 and Hubei sobemo-like virus 23 found in an Odanata mix in China (12). These sequences are related to the (+)ssRNA genus Sobemovirus, which infects plants, arthropods being putative vectors (Fig. 2C). The QG551 sobemo-like sequence codes a predicted protein of 382 aa from AUG to Stop. The reading frame is open for 263 nt (121 aa) 5′ to the first AUG at nucleotide 702; thus, it is possible that non-AUG translation initiation leads to expression of a larger protein (Fig. 2A). The RNA upstream region in addition contains a putative ORF possibly extending on 223 aa, with no similarity in databases (called ORF-X in Fig. 2A). Sobemoviruses characteristically encode a slippery sequence AAAUUUC for ribosomal frameshifting, but this sequence was not detected in this contig; however, we cannot rule out a frameshift at the end of ORF-X. After bleaching and RT-PCR, we found that the sobemo-like viral RNA was also transmitted vertically in C. remanei QG551 and could only be amplified after reverse transcription (Fig. 2D).
To further verify that the viral-like RdRP sequences were only found under an RNA form and not a DNA form, we sequenced DNA from each of the 4 isolates at a coverage sufficient to find hundreds of reads against single-copy genes in the nematode genomes (Table 2). Except for 1 read (likely a contamination from experiments or a rare reverse transcription event in the host or during sample preparation), we did not detect any reads mapping to viral RdRPs. We conclude from the PCR and high-throughput sequencing experiments that the viral-like RdRPs are not present under a DNA form in Caenorhabditis hosts.
Table 2.
Summary of alignments to viral contigs, single-copy genes, and reference genomes
| Host strain | Nematode species | No. of reads | No. of reads that map to viral-like contigs | Single-copy gene | Single-copy gene, bp | No. of reads that map to unspliced gene | No. of BLASTn alignments of single-copy gene to reference |
| JU1396 | C. brenneri | 76,901,650 | 0 | Cbn-hum-6 | 7,952 | 2,928 | 1 |
| Cbn-tbc-19 | 7,969 | 2,552 | 1 | ||||
| JU2557 | C. remanei | 61,931,642 | 0 | Cre-fmo-4 | 2,207 | 1,096 | 1 |
| Cre-snx-27 | 2,452 | 927 | 1 | ||||
| QG551 | C. remanei | 76,828,314 | 1 | Cre-ears-1 | 3,805 | 2,406 | 1 |
| Cre-mog-1 | 3,726 | 2,105 | 1 | ||||
| Cre-eyg-1 | 1,798 | 396 | 1 | ||||
| Cre-srx-44 | 1,765 | 1,295 | 1 | ||||
| Cre-grl-25 | 3,213 | 1,472 | 1 | ||||
| Cre-cpt-6 | 3,123 | 1,719 | 1 | ||||
| JU3236 | C. zanzibari | 99,643,102 | 0 | g22162.t1 | 1,485 | 564 | 1 |
| g22153.t1 | 1,473 | 336 | 1 |
JU1396, JU2557, and JU3236 produced no alignments to detected viral-like contigs. QG551 produced one alignment to the narna-like contig. Single-copy nematode genes produced several hundreds or thousands of reads.
Collectively, these results demonstrate that Caenorhabditis nematodes harbor multiple symbionts that are vertically transmitted in their germline under the form of RNA molecules coding for viral-like RdRPs.
Germline and Soma Localization by Single-Molecule Fluorescent In Situ Hybridization.
We tested the localization of the viral-like RNA molecules using single-molecule fluorescent in situ hybridization (smFISH) (35). For these, we designed sets of 30 to 50 oligonucleotide probes (SI Appendix, Table S2), which when hybridized as a pool, can detect a single RNA molecule appearing as a dot slightly above the background at long exposure times. The viral-like RNA molecules proved far less abundant than the RNAs of the intestinal noda-like viruses (36).
As could be expected from elements that are vertically transmitted, the bunyavirus-like and narna-like RNAs were detected in the germline, in C. brenneri JU1396 and C. remanei QG551 in both sexes (Figs. 3 and 4). Probe specificity was tested using negative control strains C. brenneri PB2801 and C. remanei PB4641 (SI Appendix, Figs. S1–S3) and a positive control probe for the elt-2 homolog (a gut-expressed gene) in the same animal. No dot staining was detected with the viral-like RNA probes, whereas an elt-2 probe showed the characteristic dot staining of single messenger RNAs, showing that this particular animal was sufficiently permeabilized.
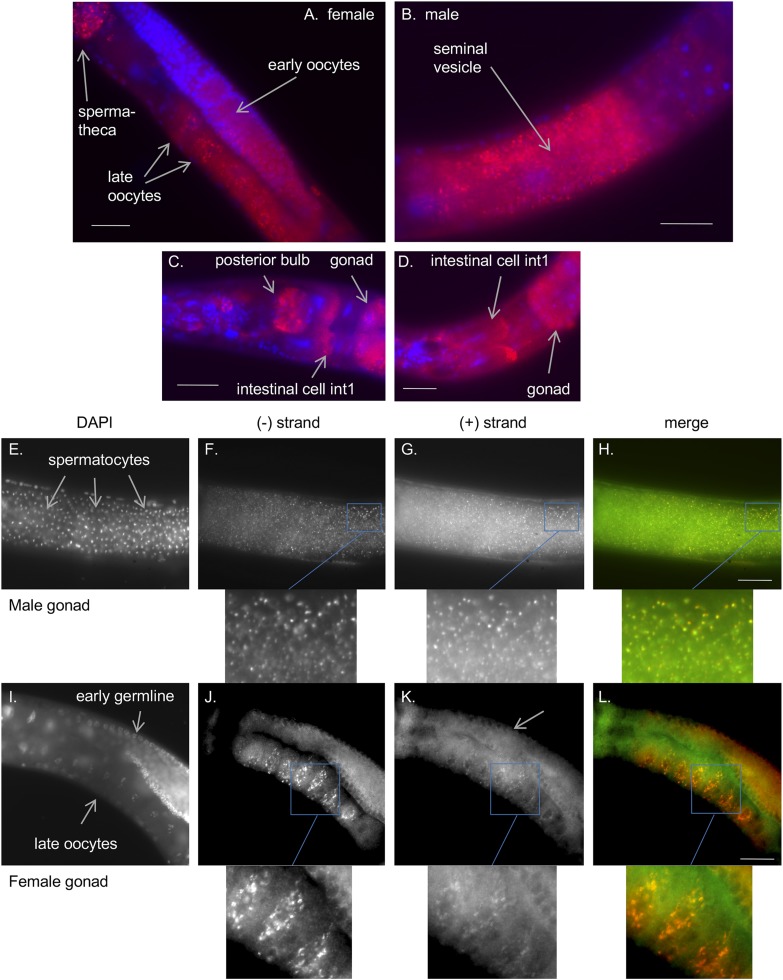
Fig. 3.
Localization of the bunyaviral-like RNA in C. brenneri JU1396 animals. (A–D) FISH against the minus strand of the JU1396 bunya-like viral sequence (using a plus-strand probe labeled with Quasar670; shown in red) and DAPI staining of nuclei (blue). The bunya-like RNA is found in germline (A and B) and somatic tissues (C and D). SI Appendix, Fig. S1 shows negative controls. (E–L) Colocalization of both RNA strands in male (E–H) and female (I–L) gonads. The nuclei are labeled with DAPI (E and I). The minus and plus strands of the bunya-like RNA are labeled with probes bound to Quasar670 (F and J) and CALFluorRed 610 (G and K), respectively. The merge of the Quasar670 and CALFluorRed 610 channels is shown in H and L, and a zoomed-in view is provided below each FISH panel. The CALFluorRed610 channel (green in the merge) yields more background than the Quasar670 channel, rendering the signal more difficult to detect. (Scale bars: 50 μm.)
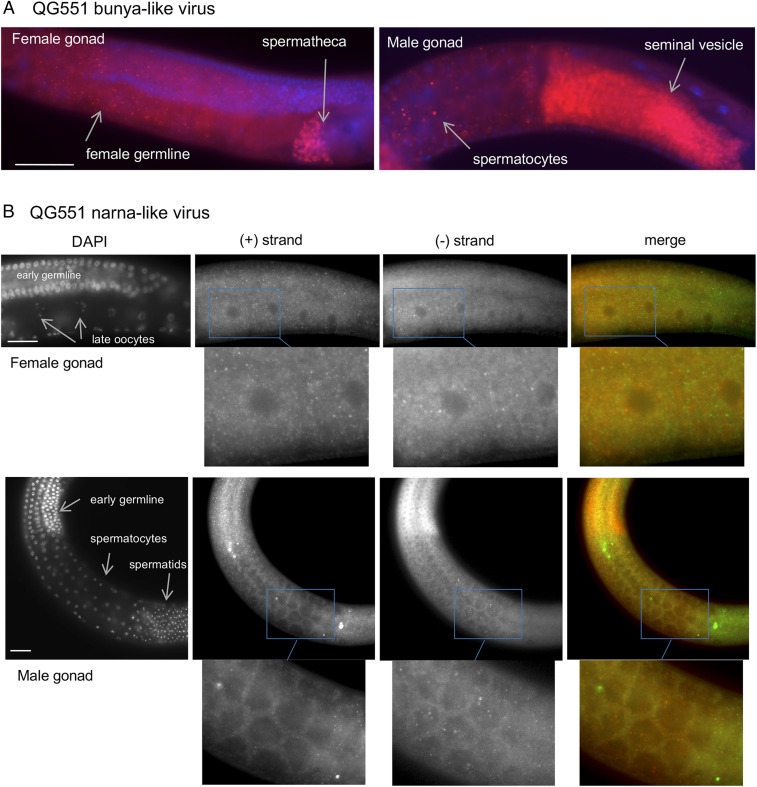
Fig. 4.
Localization of viral-like RNAs in C. remanei QG551. (A) FISH against the plus strand of the QG551 bunya-like viral sequences (probe labeled with CALFluorRed610; displayed in red) and DAPI staining (blue). (B) FISH against the minus (labeled with CALFluorRed610; red in the merge) and plus (labeled with Quasar670; green in the merge) strands of the QG551 narna-like viral sequences. The RNA molecules are found in the female and male germlines. The 2 strands do not colocalize. (Scale bars: 50 μm.)
In C. brenneri JU1396 stained for the bunya-like negative-strand RNA (using a plus-strand probe), fluorescent dots were detected in sperm cells in the male seminal vesicle or after transfer to the female spermatheca (Fig. 3 A, B, and F). In the female germline, perinuclear aggregates were observed in early meiotic oocytes before the gonadal arm turn, while the staining spread in the cytoplasm of later oocytes (Fig. 3 A and J). Staining was also detected in somatic tissues, such as the pharynx, head, and anterior gut cells called int1 (37) (Fig. 3 C and D), and in embryos. The 2 strands at least partially colocalized (Fig. 3 F–H and J–L), suggesting the possible presence of double-stranded molecules or at least colocalization at the subcellular level. From the NGS data, the relative abundance of negative strand to positive strand was ∼1.9:1 (1,896 vs. 1,000 reads).
In C. remanei QG551, the probe against the bunyaviral-like positive strand also stained the male and female germlines; sperm cells appeared particularly bright (Fig. 4A). Some staining could be seen in the soma. We found 49 reads on the negative strand and 26 reads on the positive strand (from a total of 3,056,999 reads). For the QG551 narnavirus, probes for either strand stained the male and female germlines, but in this case, the 2 strands did not colocalize (Fig. 4B).
In C. remanei JU2557, the fluorescent in situ hybridization (FISH) signal against the bunyaviral-like negative strand was weak (which may be in part due to the shorter sequences, allowing for the synthesis of only 30 probes) (SI Appendix, Table S2), with signal in the female germline and the head.
We further screened by FISH wild strains of C. brenneri and C. remanei from the same or other geographical locations but could not detect any signal, except what we took as a perhaps faint signal in C. brenneri CB5161 with the JU1396 probe (SI Appendix, Table S3). We performed RT-PCR and sequenced RNAs from the CB5161 strain, but we could not detect any bunyaviral-like molecules. Thus, if present in CB5161, the viral-like RNA molecules must be at a very low level.
Using the probe against the JU1396 bunya-like RNA, we tested whether both C. brenneri females and males could transmit the RNA molecule by performing crosses of JU1396 with JU1397 in both directions, with a single female and a single male per cross. For each combination, the transmission was clear, while animals arising from a control negative cross never show staining (SI Appendix, Table S4). The negative result in one cross with JU1396 could be a failure of detection, a genetic polymorphism in resistance segregating in one parental strain, or an incomplete transmission rate. Overall, this shows that both sexes could transmit the RNA molecule as expected from the localization of the bunyaviral RNA in both sperm and oocyte.
The Infected Caenorhabditis Strains Are Incompetent for Exogenous RNA Interference.
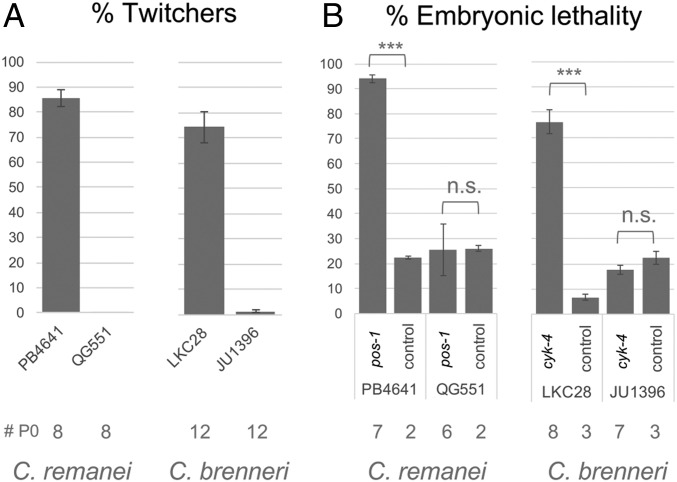
Replication by an RdRP creates at least transiently a dsRNA intermediate, which may be recognized by the host RNA interference (RNAi) machinery. As variation in RNAi pathway efficiency has been observed within species of Caenorhabditis (38–43), we wondered whether RNAi was active in the strains carrying viral-like RNAs. Because many Caenorhabditis wild strains do not respond to dsRNAs in their environment (39, 40), we injected in vitro synthesized dsRNAs into the syncytial female germline. We targeted a somatic-acting gene unc-22, in which inactivation causes the animals to twitch, and germline-acting embryonic-lethal genes cyk-4 and pos-1 (Fig. 5). We used the endogenous gene sequences of the strain and took a reference strain of the same species as a positive control for RNAi.
Fig. 5.
C. brenneri JU1396 and C. remanei QG551 are not competent for exogenous RNAi in somatic and germline cells. (A) Injection of dsRNAs against the unc-22 homolog in C. remanei and C. brenneri and scoring for twitching progeny. (B) Injection of dsRNAs against Cre-pos-1 in C. remanei and Cbn-cyk-4 in C. brenneri and scoring for embryonic lethality. The control corresponds to dsRNAs targeting GFP. The number of injected parents is indicated below. Both strains show embryonic lethality in the control, likely due to inbreeding depression. Generalized linear model testing for the effect of RNAi on the proportion of dead embryos. n.s. indicates P > 0.05. ***P < 10−15.
As expected, RNAi worked well in both soma and germline of the reference C. brenneri LKC28 strain (Fig. 5). However, C. brenneri JU1396 was incompetent for RNAi on unc-22 and cyk-4 dsRNA injections (Fig. 5). Similarly, while the C. remanei PB4641 strain was competent for RNAi, QG551 was incompetent in both soma (unc-22) and germline (pos-1). Thus, exogenous RNAi is defective in the 2 tested strains where we found vertically transmitted viral RdRPs.
sRNAs Mapping to the Viral RNAs.
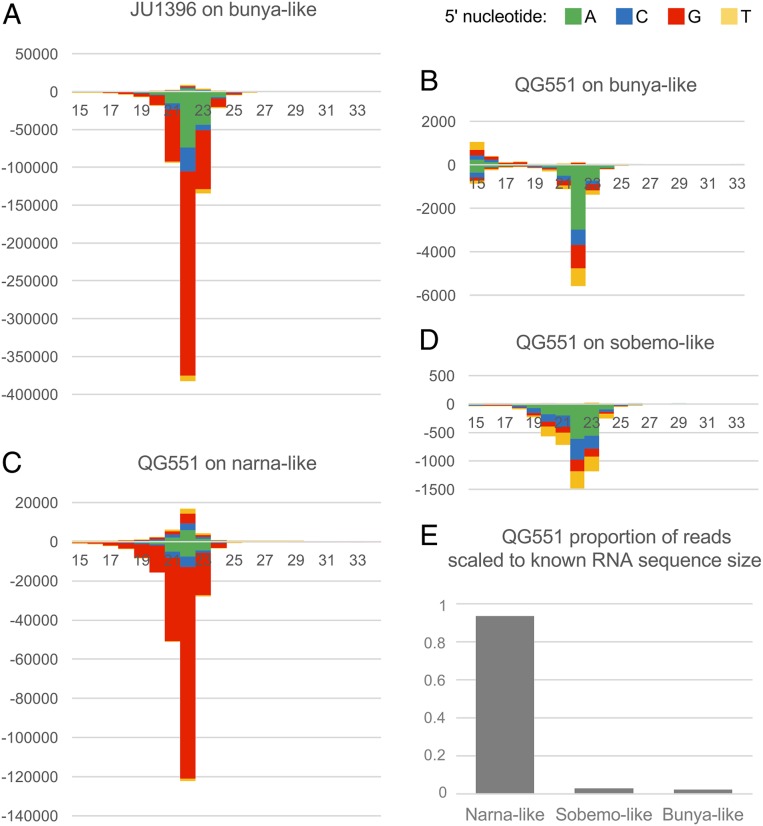
Partial defects in RNAi pathways do not prevent the formation of sRNAs, at least as degradation products. We, therefore, sought to detect sRNAs matching the viral-like RNAs. We prepared sRNA libraries of C. brenneri JU1396 and C. remanei QG551 after phosphatase treatment so that the sRNAs with 5′ mono-, di-, and triphosphates would all be included. We mapped the sRNAs to the viral-like genomes of each strain (and of the other strain as a control); 2.6% of sRNAs in C. brenneri JU1396 mapped to the bunyaviral-like RNA of that strain (and as expected, 0.00% mapped to that of C. remanei QG551). In C. remanei QG551, 0.03% mapped to the QG551 bunyavirus-like RNA (0.00% to the JU1396 bunyaviral sequences), while 1.0% mapped to the narnavirus-like RNA (Fig. 6E shows a graph of the proportion scaled to RNA genome size). The sRNAs were thus more abundant for the bunyaviral-like RNAs in JU1396 than in QG551.
Fig. 6.
Differential pattern of sRNAs mapping to the different viral-like RNA genomes. (A–D) The stack bar charts display the distribution in length and 5′ nucleotide of sRNAs mapping onto each viral-like genome. The plus strand is the coding strand. (A) JU1396 sRNAs mapping onto the bunya-like RNA. (B–D) QG551 sRNAs mapping onto the bunya-like (B), narna-like (C), and sobemo-like (D) RNAs. (E) Proportion of reads mapping to each RNA in QG551 normalized to contig length.
We analyzed the sRNA distribution according to 3 criteria (Fig. 6 A–D): 1) mapping to the positive (coding) or negative (noncoding) strand of the viral-like RNA, 2) sRNA length, and 3) first nucleotide at the 5′ end. For example, “22G” means an sRNA of 22 nt with a 5′ guanine.
The 2 bunyavirus-like RNAs elicited different responses in their respective hosts. The C. brenneri JU1396 bunya-like RNA elicited a strong sRNA response (2.6% of all sRNA reads vs. 0.03% for the QG551 bunya-like RNA) that was dominated by negative-strand 22G sRNAs (Fig. 6A), while for the C. remanei QG551 bunya-like RNA, an enrichment for negative-strand 22A sRNAs was observed (Fig. 6B). Similarly, the sobemo-like viral RdRP segment elicited a weak sRNA response with a negative-strand 22A pattern like the QG551 bunya virus (Fig. 6D). C. remanei QG551 was, however, able to mount a response against the narnavirus of predominantly 22G sRNAs (Fig. 6C). Interestingly, the sRNA patterns correlated with their abundance (i.e., the viral-like RNAs with higher relative abundance favored negative-strand 22G sRNAs).
Discussion
Vertically Transmitted RdRP-Coding RNA Molecules in Caenorhabditis.
We describe endogenous vertically transmitted RNAs of viral origin in Caenorhabditis nematodes. Vertical transmission through the germline is demonstrated by persistence of the RNAs after repeated bleach treatments and their in situ localization in the male and female germline. Whether horizontal transmission may also occur remains unknown.
Viral-like RNAs encoding an RdRP are able to replicate in the host cytoplasm and propagate through the germline without leaving the cellular environment, as narnaviruses and totiviruses do in fungi (3). The identification of narnavirus sequences in Caenorhabditis nematodes is an instance of a capsidless virus in animals. The bunyavirus-like sequences may also represent capsidless viral elements, as we were unable to identify sequences corresponding to a viral capsid-like protein in these strains. We tried to detect viral-like structures by electron microscopy in oocytes and the smaller sperm cells of C. brenneri JU1396 and failed to detect any (A.R. and M.-A.F.). Because the animal germline is a continuous cellular lineage that gives rise to both germline and soma, viral RNAs can replicate and be passed to further generations without ever exiting or entering cells. The Caenorhabditis germline is syncytial, achieving perhaps more readily the distribution of molecules into gametes. Self-replicating RNAs may be expected to find their way in the syncytial germline of nematodes, if the hosts are able to counter them by specific mechanisms.
In their extensive invertebrate virus dataset, Shi et al. (12) find nucleoproteins and glycoproteins of bunyaviruses associated with arthropods and gastropods. However, from sequencing of nematode samples, a single RdRP-encoding fragment was observed, and no nucleoproteins or glycoproteins were observed, further strengthening the possibility that nematodes may harbor RdRP-encoding RNA in the absence of other proteins typical of that viral family. We cannot, however, rule out the possibility that the strains harboring bunya-like RdRps also encode highly divergent capsid or glycoprotein sequences that are not recognizable by alignment. Viral sequences evolve rapidly, and the capsid may be unrecognizable as such, exchanged among viruses (44), or replaced by a host protein (45). Even though an infective virion is in principle not needed for vertical transmission, it may aid movement between tissues and possibly allow horizontal transmission to the same or another host species (6).
Diversity of Phylogenetic Relationships of the RdRP RNAs.
We found a diverse set of viral-like RNAs in various Caenorhabditis species. We found one example of an RdRP sequence related to those of the family Narnaviridae, known to be capsidless RNA viruses (6, 8). Here, we show that narnaviral sequences propagate vertically through the germline of a multicellular organism.
Most frequently detected were RNAs coding for RdRPs belonging to the order Bunyavirales, with at least 2 evolutionary origins (Fig. 1B). These occurrences were widespread in terms of geography (4 continents) and host species (3 Caenorhabditis species). The Bunyavirales include viruses infecting vertebrates and nonvertebrates; many are transmitted to vertebrates by arthropods (16, 46).
The diversity of phylogenetic placement of the viral-like RNA molecules found in Caenorhabditis suggests that entry and replication inside these nematodes occurred multiple times independently. Because these molecules were found in only few C. brenneri, C. remanei, and C. zanzibari strains, it is possible that they were gained independently within each species, originally by horizontal transmission.
Specific Hosts and Their sRNA Response.
Whereas other strains of C. brenneri and C. remanei are able to mount an RNAi response, the hosts C. brenneri JU1396 and C. remanei QG551 are both defective (Fig. 5) (40, 47). Remarkably, QG551 harbors at least 3 different viral-like RNAs in its germline. This is likely a result of its deficient RNAi response: a permissive state for self-replicating RNAs of this C. remanei strain may have favored the accumulation of replicating RNAs in its germline.
Despite their incompetence for RNAi, we observe in C. brenneri JU1396 and C. remanei QG551 sRNAs against the viral sequences in a proportion of a few percent of total sRNAs. The viral RNAs are thus kept at a balance between degradation and replication as observed for a Leishmania virus (48) or the Orsay noda-like virus in its original C. elegans host strain JU1580 (49). Parts of the many RNAi pathways (49) may still be active.
The sRNA patterns differ widely between the viral sequences both in the same C. remanei QG551 host and between the 2 bunya-like viruses in C. brenneri vs. C. remanei strains. Differences in sRNA distributions were previously observed for viruses infecting different wild Drosophila strains (19). The differential response in the same strain suggests that the viral RNAs are differentially recognized by the RNAi machinery. Reasons may be found in their respective biology: 1) a differential localization in soma vs. germline (the narnavirus may be the most exclusively associated with the germline, where 22G sRNAs are abundant), 2) a differential localization in males and females, 3) a different subcellular localization, and 4) different sequences (including different RdRPs), capping, and folding. We cannot rule out that the difference is due to incomplete sequences to which the sRNAs could be mapped, as some locations along the sequence trigger a sizeable proportion of the reads (SI Appendix, Fig. S4).
The JU1396 bunya-like RNA was found in large clusters around the early oocyte nucleus and in the late oocyte cytoplasm, reminiscent of ribonucleoprotein granules, such as P granules, mutator foci, etc., involved in sRNA pathways and RNA stability (50–56). Both strands of JU1396 bunyavirus-like RNA appear to colocalize for most spots, which is expected from a replication site but may not be expected to occur in such great proportion. The oocyte perinuclear localization was not observed for the QG551 bunyavirus-like RNA (note that only our probe targeted to the plus strand yielded a signal). This implies that the JU1396 and QG551 bunyavirus-like RNAs have a different biology as also suggested by the sRNA patterns.
Host Fitness and Reproductive Mode.
The impact on host fitness of harboring these viral-like RNAs is currently unknown. The infected animals appear superficially normal, but currently, isogenic lines with and without the viral-like RNAs do not exist; therefore, it is not possible to assess potential effects of these RNA elements on fitness.
Remarkably, in our surveys of Caenorhabditis nematodes, vertically transmitted viral-like sequences were found exclusively in gonochoristic species, namely C. remanei, C. brenneri, and C. zanzibari, while horizontally transmitted noda-like viruses were detected exclusively in 2 selfing species Caenorhabditis briggsae and C. elegans (26–28). How could this pattern be explained?
From the parasite viewpoint, vertical transmission is favorable if highly efficient, as it ensures transmission. Especially favorable for the parasite in the short term are selfing lineages where the progeny has the same genotype as the parent, rendering the host incapable of new immune defenses (57). For the host, it may become favorable to outcross (58). However, the very low outcrossing level of C. elegans natural populations (59–63) raises the problem for vertical parasites to persist and spread in a selfing vs. an outcrossing species. In this context, a given selfing lineage may die out either due to the vertical parasite or for any other reason; by contrast, in outcrossing species, the vertical parasites may be kept in check by sexual recombination yet be efficiently dispersed and maintained if they have a low impact on fitness.
In conclusion, we here show the presence in an animal of capsidless viruses transmitted through the germline. The detection of vertically transmitted viral-like RNA elements in multiple species of Caenorhabditis nematodes raises the possibility that similar elements may be more broadly distributed in multicellular eukaryotes. Additional efforts to identify such elements and define their potential impacts on host physiology are needed.
Methods Summary
Additional methods are in SI Appendix.
Screening for Viral Sequences.
RNAs from mixed-stage populations of Caenorhabditis wild strains were sequenced. Contigs with similarity to viruses were identified using VirusSeeker (64). Sequence reads are available at the Sequence Read Archive under accessions numbers SRR8869242 to SRR8869245 (BioProject PRJNA531652).
RT-PCR.
RNAs of the bleached strains were extracted and used to amplify the different viral-like sequences by 1-step RT-PCR with the oligonucleotides listed in SI Appendix, Table S2.
smFISH.
smFISH was performed as described (35, 65), except that the hybridization solution contained 20% formamide (probe sequences are in SI Appendix, Table S2).
sRNA Analysis.
RNAs from mixed-stage populations were treated with 5′-polyphosphatase prior to library construction.
Supplementary Material
Acknowledgments
We thank Amir Yassin for sampling JU3236 and Amhed Vargas Velazquez for help with sRNA analysis. We also thank Nimit Jain for reading of the manuscript. We are grateful to Martin Sachse for help with electron microscopy. This work was supported by NIH Grants R21 AI119917 (to D.W.) and R01 AI134967 (to D.W.), Agence Nationale pour la Recherche Grant ANR-11-BSV3-013 (to M.-A.F.), and Fondation pour la Recherche Médicale Grant DEQ20150331704 (to M.-A.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (accession nos. SRR8869242–SRR8869245 [BioProject PRJNA531652] for initial RNA sequencing, SRR9206839–SRR9206841 [BioProject PRJNA531652] for DNA sequencing, and SRX6374058–SRX6374060 [BioProject PRJNA551618] for small RNA sequencing). The viral-like RNA contigs reported in this paper have been deposited in the NCBI Nucleotide (accession no. KM580531 for the Caenorhabditis brenneri JU1396 bunya-like RNA and MN272066–MN272070 for the other viral-like RNAs; Table 1).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903903116/-/DCSupplemental.
References
- 1.Kose H., Karr T. L., Organization of Wolbachia pipientis in the Drosophila fertilized egg and embryo revealed by an anti-Wolbachia monoclonal antibody. Mech. Dev. 51, 275–288 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Landmann F., Foster J. M., Michalski M. L., Slatko B. E., Sullivan W., Co-evolution between an endosymbiont and its nematode host: Wolbachia asymmetric posterior localization and AP polarity establishment. PLoS Negl. Trop. Dis. 8, e3096 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowley P. A., The frenemies within: Viruses, retrotransposons and plasmids that naturally infect Saccharomyces yeasts. Yeast 34, 279–292 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Wilfert L., Jiggins F. M., Flies on the move: An inherited virus mirrors Drosophila melanogaster’s elusive ecology and demography. Mol. Ecol. 23, 2093–2104 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Dennis S., Sheth U., Feldman J. L., English K. A., Priess J. R., C. elegans germ cells show temperature and age-dependent expression of Cer1, a Gypsy/Ty3-related retrotransposon. PLoS Pathog. 8, e1002591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin E. V., Dolja V. V., Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 78, 278–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteban R., Vega L., Fujimura T., Launching of the yeast 20 s RNA narnavirus by expressing the genomic or antigenomic viral RNA in vivo. J. Biol. Chem. 280, 33725–33734 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Hillman B. I., Cai G., The family Narnaviridae: Simplest of RNA viruses. Adv. Virus Res. 86, 149–176 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Vega L., Sevillano L., Esteban R., Fujimura T., Resting complexes of the persistent yeast 20S RNA Narnavirus consist solely of the 20S RNA viral genome and its RNA polymerase p91. Mol. Microbiol. 93, 1119–1129 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Cole T. E., Hong Y., Brasier C. M., Buck K. W., Detection of an RNA-dependent RNA polymerase in mitochondria from a mitovirus-infected isolate of the Dutch Elm disease fungus, Ophiostoma novo-ulmi. Virology 268, 239–243 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Cai G., Krychiw J. F., Myers K., Fry W. E., Hillman B. I., A new virus from the plant pathogenic oomycete Phytophthora infestans with an 8 kb dsRNA genome: The sixth member of a proposed new virus genus. Virology 435, 341–349 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Shi M., et al. , Redefining the invertebrate RNA virosphere. Nature 540, 539–543 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Ding B., Itaya A., Viroid: A useful model for studying the basic principles of infection and RNA biology. Mol. Plant Microbe Interact. 20, 7–20 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Flores R., Gago-Zachert S., Serra P., Sanjuán R., Elena S. F., Viroids: Survivors from the RNA world? Annu. Rev. Microbiol. 68, 395–414 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Hopkins K. C., et al. , A genome-wide RNAi screen reveals that mRNA decapping restricts bunyaviral replication by limiting the pools of Dcp2-accessible targets for cap-snatching. Genes Dev. 27, 1511–1525 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott R. M., Orthobunyaviruses: Recent genetic and structural insights. Nat. Rev. Microbiol. 12, 673–685 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Marklewitz M., et al. , Gouleako virus isolated from West African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J. Virol. 85, 9227–9234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auguste A. J., et al. , Characterization of a novel Negevirus and a novel Bunyavirus isolated from Culex (Culex) declarator mosquitoes in Trinidad. J. Gen. Virol. 95, 481–485 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webster C. L., et al. , The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 13, e1002210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokarz R., et al. , Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J. Virol. 88, 11480–11492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sõmera M., Sarmiento C., Truve E., Overview on sobemoviruses and a proposal for the creation of the family Sobemoviridae. Viruses 7, 3076–3115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulenburg H., Félix M.-A., The natural biotic environment of Caenorhabditis elegans. Genetics 206, 55–86 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bessereau J.-L., “Transposons in C. elegans” in WormBook, The C. elegans Research Community, Ed. (2006). http://www.wormbook.org/chapters/www_transposons/transposons.html. Accessed 4 November 2019.
- 24.Gammon D. B., et al. , The antiviral RNA interference response provides resistance to lethal arbovirus infection and vertical transmission in Caenorhabditis elegans. Curr. Biol. 27, 795–806 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koutsovoulos G., Makepeace B., Tanya V. N., Blaxter M., Palaeosymbiosis revealed by genomic fossils of Wolbachia in a strongyloidean nematode. PLoS Genet. 10, e1004397 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Félix M.-A., et al. , Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9, e1000586 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franz C. J., Zhao G., Félix M.-A., Wang D., Complete genome sequence of Le Blanc virus, a third Caenorhabditis nematode-infecting virus. J. Virol. 86, 11940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frézal L., Jung H., Tahan S., Wang D., Félix M.-A., Noda-like RNA viruses infecting Caenorhabditis nematodes: Sympatry, diversity and reassortment. J. Virol. 93, e01170-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mokili J. L., Rohwer F., Dutilh B. E., Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2, 63–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi M., Zhang Y. Z., Holmes E. C., Meta-transcriptomics and the evolutionary biology of RNA viruses. Virus Res. 243, 83–90 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmonds P., et al. , Consensus statement: Virus taxonomy in the age of metagenomics. Nat. Rev. Microbiol. 15, 161–168 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Adams M. J., et al. , Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch. Virol. 162, 2505–2538 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Rodríguez-Cousiño N., Solórzano A., Fujimura T., Esteban R., Yeast positive-stranded virus-like RNA replicons. 20 S and 23 S RNA terminal nucleotide sequences and 3′ end secondary structures resemble those of RNA coliphages. J. Biol. Chem. 273, 20363–20371 (1998). [DOI] [PubMed] [Google Scholar]
- 34.Shi M., et al. , High-resolution metatranscriptomics reveals the ecological dynamics of mosquito-associated RNA viruses in Western Australia. J. Virol. 91, e00680-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., Tyagi S., Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franz C. J., et al. , Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology 448, 255–264 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Altun Z. F., et al. , 2002–2018 WormAtlas. https://www.wormatlas.org/. Accessed 26 December 2018.
- 38.Tijsterman M., Okihara K. L., Thijssen K., Plasterk R. H. A., PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr. Biol. 12, 1535–1540 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Winston W. M., Sutherlin M., Wright A. J., Feinberg E. H., Hunter C. P., Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl. Acad. Sci. U.S.A. 104, 10565–10570 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nuez I., Félix M.-A., Evolution of susceptibility to ingested double-stranded RNAs in Caenorhabditis nematodes. PLoS One 7, e29811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollard D. A., Rockman M. V., Resistance to germline RNA interference in a Caenorhabditis elegans wild isolate exhibits complexity and nonadditivity. G3 (Bethesda) 3, 941–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashe A., et al. , A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. eLife 2, e00994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paaby A. B., et al. , Wild worm embryogenesis harbors ubiquitous polygenic modifier variation. eLife 4, e09178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang R., et al. , A capsidless ssRNA virus hosted by an unrelated dsRNA virus. Nat. Microbiol. 1, 15001 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Krupovic M., Koonin E. V., Multiple origins of viral capsid proteins from cellular ancestors. Proc. Natl. Acad. Sci. U.S.A. 114, E2401–E2410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marklewitz M., Zirkel F., Kurth A., Drosten C., Junglen S., Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl. Acad. Sci. U.S.A. 112, 7536–7541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winston W. M., Molodowitch C., Hunter C. P., Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Brettmann E. A., et al. , Tilting the balance between RNA interference and replication eradicates Leishmania RNA virus 1 and mitigates the inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 113, 11998–12005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Billi A. C., Fischer S. E. J., Kim J. K., “Endogenous RNAi pathways in C. elegans” in WormBook, The C. elegans Research Community, Ed. (2014). http://www.wormbook.org/chapters/www_endoRNAipathwys/endoRNAipathwys.html. Accessed 4 November 2019.
- 50.Pitt J. N., Schisa J. A., Priess J. R., P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 219, 315–333 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Jud M., Razelun J., Bickel J., Czerwinski M., Schisa J. A., Conservation of large foci formation in arrested oocytes of Caenorhabditis nematodes. Dev. Genes Evol. 217, 221–226 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Updike D. L., Hachey S. J., Kreher J., Strome S., P granules extend the nuclear pore complex environment in the C. elegans germ line. J. Cell Biol. 192, 939–948 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Phillips C. M., Montgomery T. A., Breen P. C., Ruvkun G., MUT-16 promotes formation of perinuclear mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 26, 1433–1444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hubstenberger A., Noble S. L., Cameron C., Evans T. C., Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev. Cell 27, 161–173 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wan G., et al. , Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 557, 679–683 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ishidate T., et al. , ZNFX-1 functions within perinuclear nuage to balance epigenetic signals. Mol. Cell 70, 639–649.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agrawal A. F., Similarity selection and the evolution of sex: Revisiting the red queen. PLoS Biol. 4, e265 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebert D., The epidemiology and evolution of symbionts with mixed-mode transmission. Annu. Rev. Ecol. Evol. Syst. 44, 623–643 (2013). [Google Scholar]
- 59.Barrière A., Félix M.-A., High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15, 1176–1184 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Cutter A. D., Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 172, 171–184 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barrière A., Félix M.-A., Temporal dynamics and linkage disequilibrium in natural Caenorhabditis elegans populations. Genetics 176, 999–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andersen E. C., et al. , Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44, 285–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richaud A., Zhang G., Lee D., Lee J., Félix M.-A., The local co-existence pattern of selfing genotypes in Caenorhabditis elegans natural metapopulations. Genetics 208, 807–821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao G., et al. , VirusSeeker, a computational pipeline for virus discovery and virome composition analysis. Virology 503, 21–30 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barkoulas M., van Zon J. S., Milloz J., van Oudenaarden A., Félix M. A., Robustness and epistasis in the C. elegans vulval signaling network revealed by pathway dosage modulation. Dev. Cell 24, 64–75 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Ruark C. L., Gardner M., Mitchum M. G., Davis E. L., Sit T. L., Novel RNA viruses within plant parasitic cyst nematodes. PLoS One 13, e0193881 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.