Significance
Cell adhesion is indispensable for multicellular organisms to form cohesive sheets and to serve as tissues and organs. In addition to the adhesive activity, it exhibits the signaling ability to organize a variety of cellular processes. However, the mechanism by which the cell adhesion signaling influences the cellular events, especially on regulation of gene expression, remains obscure. We determine that the cell adhesion signaling originated from the tight-junction protein claudin-6, which targets members of the nuclear receptor superfamily, such as retinoic acid receptor γ and estrogen receptor α, thereby regulating expression of the respective target genes. This signaling provides insight into regulation of nuclear receptor activity by cell adhesion.
Keywords: tight junction, claudin, signal transduction, retinoic acid receptor, estrogen receptor
Abstract
Cell adhesion is essential for proper tissue architecture and function in multicellular organisms. Cell adhesion molecules not only maintain tissue integrity but also possess signaling properties that contribute to diverse cellular events such as cell growth, survival, differentiation, polarity, and migration; however, the underlying molecular basis remains poorly defined. Here we identify that the cell adhesion signal initiated by the tight-junction protein claudin-6 (CLDN6) regulates nuclear receptor activity. We show that CLDN6 recruits and activates Src-family kinases (SFKs) in second extracellular domain-dependent and Y196/200-dependent manners, and SFKs in turn phosphorylate CLDN6 at Y196/200. We demonstrate that the CLDN6/SFK/PI3K/AKT axis targets the AKT phosphorylation sites in the retinoic acid receptor γ (RARγ) and the estrogen receptor α (ERα) and stimulates their activities. Interestingly, these phosphorylation motifs are conserved in 14 of 48 members of human nuclear receptors. We propose that a similar link between diverse cell adhesion and nuclear receptor signalings coordinates a wide variety of physiological and pathological processes.
Cell adhesion molecules connect cells to each other and to the extracellular matrix, thereby participating in tissue formation and homeostasis. In addition to their adhesive properties, they propagate the intracellular signaling that organizes a broad range of cell behaviors, including cell growth, survival, differentiation, polarity, and migration, possibly via activation or repression of transcription factors that regulate the expression of target genes (1–3). However, it is still fragmentary how cell adhesion signaling is transduced to the nucleus and regulates gene expression.
Claudins (CLDNs) are fundamental transmembrane proteins of tight junctions, the apical-most constituents of apical junctional complexes (4–10). The CLDN family consists of 27 members in mammals and exhibits distinct expression patterns in tissue- and cell-specific manners. They are tetraspan proteins with a short cytoplasmic N terminus, 2 extracellular domains (EC1 and EC2), and a C-terminal cytoplasmic domain. The CLDN-EC1, in which there is a large variation in the position and number of charged amino acids depending on each CLDN subtype, creates paracellular barriers or pores for selective ions and solutes between neighboring cells. Conversely, the CLDN-EC2 contributes not only to the binding for Clostridium perfringens enterotoxin (CPE) (11–15) but also to trans-interaction between the plasma membranes of opposing cells (16, 17). Moreover, several signaling proteins are known to be localized at tight junctions; nevertheless, it has not yet been established whether and how CLDNs may propagate intracellular signals (18).
The retinoic acid receptors (RARs), members of the nuclear receptor superfamily, transcriptionally regulate the expression of numerous target genes via forming heterodimers with the retinoid X receptors (RXRs) (19–22). The multiple RAR (RARα, β, and γ) and RXR (RXRα, β, and γ) isotypes and isoforms are conserved among vertebrates and exert pleiotropic effects on development, cellular differentiation, proliferation, apoptosis, and homeostasis. Using F9 stem cells (23), we previously demonstrated that both RXRα/RARγ heterodimers and another member of nuclear receptors, hepatocyte nuclear factor α (HNF4α), triggered epithelial differentiation (24–27). These effects were very similar to the CLDN6-initiated cellular processes (28), implying a possible link between them.
Here we report that CLDN6-mediated cell–cell adhesion activates the Src-family kinase (SFK)/PI3K/AKT signaling pathway and stimulates the RARγ and estrogen receptor α (ERα) activities. The identification of this machinery highlights regulation of the nuclear receptor activity by cell adhesion to control diverse cellular events.
Results
The EC2 and Y196/200 of CLDN6 Are Required for the Signaling Activity.
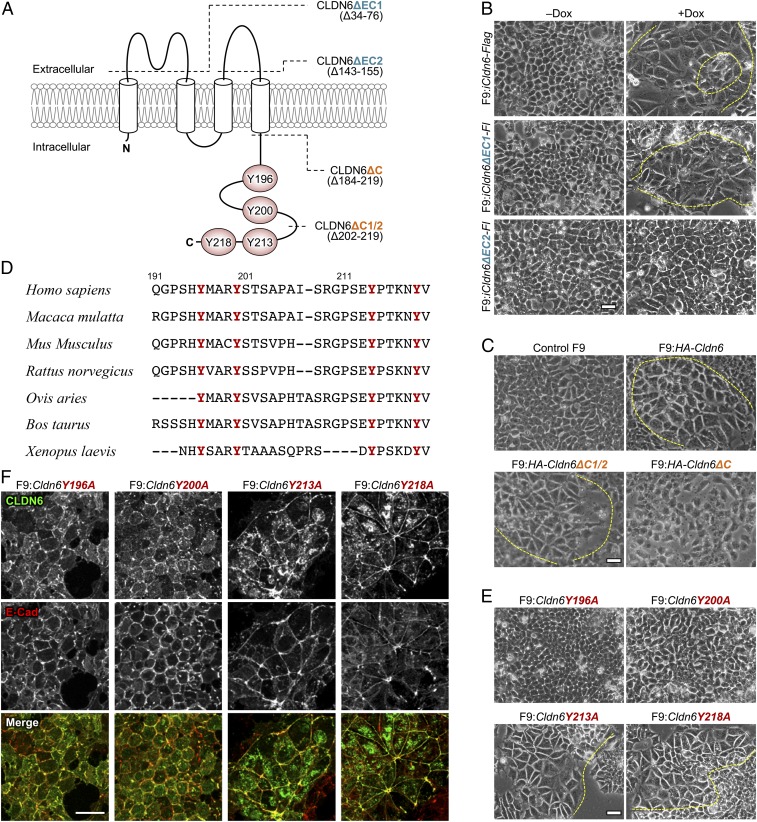
To verify whether the extracellular and C-terminal cytoplasmic domains of CLDN6 are involved in the CLDN6-initiated signaling, we generated F9 cells that constitutively or inducibly expressed the corresponding CLDN6 deletion mutants (Fig. 1A). Morphological differentiation and mature cell–cell junctions appeared in F9:iCldn6-Flag (hereafter, i means doxycycline [Dox]-inducible expression of a given gene), F9:iCldn6ΔEC1-Flag, F9:HA-Cldn6, and F9:HA-Cldn6ΔC1/2 cells, but not in F9:iCldn6ΔEC2-Flag or F9:HA-Cldn6ΔC cells (Fig. 1 B and C and SI Appendix, Figs. S1 A–C and S2 A and B). It should be noted that epithelial differentiation is not triggered in the entire cells because F9 cells, similar to other stem cells, symmetrically or asymmetrically divide. In addition, the expression of the tight junction markers (CLDN6, CLDN7, occludin [OCLN], ZO-1α+ variant) and the microvilli marker (ezrin/radixin/moesin-binding phosphoprotein 50 [EBP50]) was not enhanced in F9:HA-Cldn6ΔC cells (SI Appendix, Fig. S2C). Taken collectively, these results revealed that the CLDN6-adhesion signal was transduced through the EC2 and C-terminal domains.
Fig. 1.
The CLDN6-triggered signaling is mediated via the EC2 and Y196/200. (A) Predicted topology of mouse CLDN6. The conserved tyrosine residues are highlighted as red circles. Dashed lines indicate the location of truncated domains. (B, C, and E) Morphological appearance of the indicated F9 cell lines. F9:iCldn6-Flag, F9:iCldn6ΔEC1-Flag, and F9:iCldn6ΔEC2-Flag cells were treated for 72 h with either the vehicle or 1.0 μg/mL doxycycline (Dox). The boundaries between undifferentiated and epithelial cells are shown in the dashed yellow lines. (D) Amino acid sequences of the C-terminal cytoplasmic domains of vertebrate CLDN6 homologs with conserved tyrosine residues in red. (F) Confocal images of the revealed cells stained for CLDN6 together with E-Cad. (Scale bars, 20 μm.)
Because the 4 tyrosine residues in the C-terminal cytoplasmic domain of CLDN6 are completely conserved among vertebrates (Fig. 1D), we next established F9 cell lines expressing CLDN6 mutants with a substitution of each tyrosine for an alanine residue (Y196A, Y200A, Y213A, and Y218A; Fig. 1A). Interestingly, epithelial differentiation was induced in F9:Cldn6Y213A and F9:Cldn6Y218A cells, but neither in F9:Cldn6Y196A nor F9:Cldn6Y200A cells (Fig. 1E and SI Appendix, Figs. S1D and S2D). Furthermore, both Cldn6Y196A and Cldn6Y200A mutants were accumulated to primordial cell junctions, together with E-cadherin (E-Cad; Fig. 1F).
The CLDN6-Adhesion Signaling Activates the SFK/PI3K/AKT Pathway.
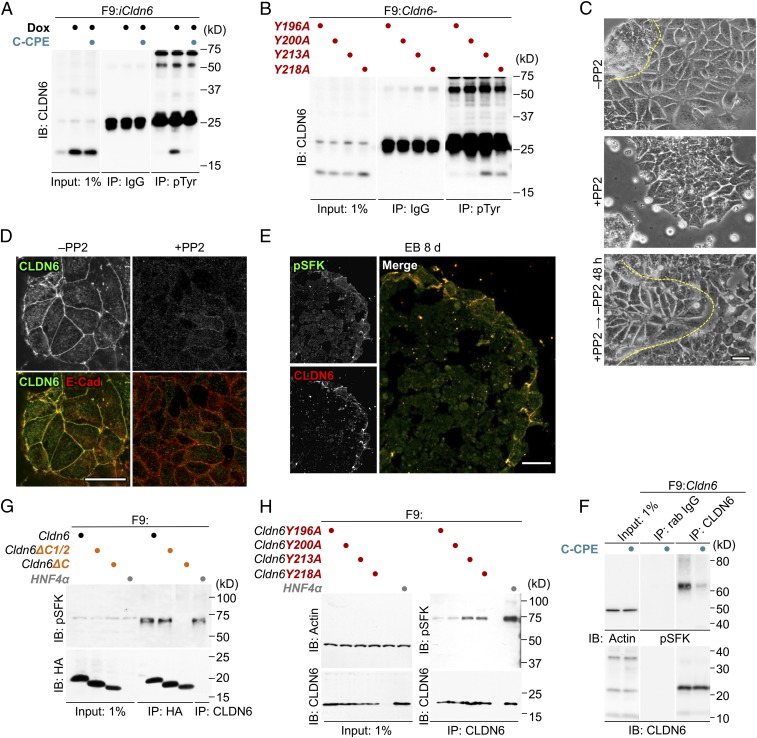
Since Y196 and Y200 of CLDN6 were required for its signaling activity, we subsequently determined whether CLDN6 was tyrosine-phosphorylated during epithelial differentiation of F9 stem cells. As shown in Fig. 2A, CLDN6 was heavily tyrosine-phosphorylated in F9:iCldn6 cells, implying the involvement of tyrosine kinases in CLDN6-induced signaling. In addition, the phospho-tyrosine levels of CLDN6 were suppressed by the treatment with the C-terminal half of CPE (C-CPE), which binds to the EC2 of CLDN6 with high affinity and eliminates CLDN6 from cell–cell junctions without any changes in its total mRNA or protein levels in F9 and mouse ES cells (28). Moreover, the phospho-tyrosine amounts were decreased in F9:Cldn6Y196A and F9:Cldn6Y200A cells (Fig. 2B).
Fig. 2.
SFKs contribute to the CLDN6-adhesion signaling. (A and B) Tyrosine-phosphorylation of CLDN6 in F9:iCldn6 (A) and the indicated F9 mutant cells (B). F9:iCldn6 cells were exposed for 48 h to the vehicle, 1.0 μg/mL doxycycline (Dox) alone, or together with 1.0 μg/mL C-CPE. (C) Effect of PP2 (10 μM) on morphological appearance of F9:Cldn6 cells. The borders between undifferentiated and epithelial cells are shown in the dashed yellow lines. (D) Confocal images of vehicle- and PP2-treated F9:Cldn6 cells stained for the indicated markers. (E) Confocal images of the embryoid body (EB) stained for the indicated markers. (F–H) Association between CLDN6 and pSFK in the indicated F9 cell lines. F9:Cldn6 cells were exposed for 72 h to the vehicle or 1.0 μg/mL C-CPE (F). (Scale bars, 20 μm.)
Among the various tyrosine kinases, we focused on the SFKs, because they are activated via the engagement of cell–cell and cell–matrix adhesion proteins lacking intrinsic kinase activity, such as cadherins and integrins (29, 30). When F9:Cldn6 and Dox-treated F9:iCldn6 cells were exposed to the SFK inhibitor PP2, epithelial differentiation was markedly inhibited (Fig. 2 C and D and SI Appendix, Figs. S1E and S3A). The inhibitory effect on morphological appearance was recovered 48 h after PP2 removal (Fig. 2C and SI Appendix, Fig. S1E). Similar results were obtained in F9:Cldn6 cells exposed to other SFK inhibitors (i.e., PP1, SU6656, and aminogenistein) without influence on cell viability at the concentrations used (SI Appendix, Fig. S3 B and C). Collectively, these results indicated that SFKs contributed to CLDN6-provoked signaling.
The levels of pSFK were decreased in F9 cells by CLDN6 knockdown, as well as in F9:HA-Cldn6ΔC cells and in F9:Cldn6Y196A and F9:Cldn6Y200A cells (SI Appendix, Fig. S3 D–F). The pSFK amounts were also reduced in F9:iCldn6 on C-CPE treatment (SI Appendix, Fig. S3G). These results revealed that the EC2 and Y196/200 of CLDN6 were required to activate SFKs at cell-cell junctions.
Double immunofluorescence staining showed that pSFK was colocalized with CLDN6 along cell borders in F9:Cldn6 cells and both signals had almost disappeared upon PP2 treatment (SI Appendix, Fig. S3H). The pSFK signal was also observed with CLDN6 and OCLN along the cell boundaries in F9:Cldn6Y218A cells (SI Appendix, Fig. S3I). In contrast, both pSFK and CLDN6, but not OCLN, were concentrated to nascent cell–cell junctions of F9:Cldn6Y196A cells. The pSFK immunoreactivity was also detected with CLDN6 at cell–cell junctions of epithelia in embryoid bodies (EBs; Fig. 2E). Immunoprecipitation assay revealed that CLDN6 was associated with pSFK in F9:Cldn6 cells, and their interaction was prominently decreased by C-CPE treatment (Fig. 2F). In addition, a complex between HA-CLDN6 and pSFK was observed in F9:HA-Cldn6 and F9:HA-Cldn6ΔC1/2 cells, but not in F9:HA-Cldn6ΔC cells (Fig. 2G). Furthermore, the CLDN6-pSFK complex was formed in F9:Cldn6Y213A or F9:Cldn6Y218A cells, and their association was reduced in F9:Cldn6Y196A and F9:Cldn6Y200A cells (Fig. 2H).
We next determined whether the PI3K/AKT axis, the well-established downstream signaling pathway of SFKs, participated in the CLDN6/SFK signal, using the respective protein kinase inhibitors LY294002 and AKT inhibitor VIII. Exposure to these inhibitors strikingly prevented epithelial differentiation in F9:Cldn6 cells and reduced the pAKT levels in F9:iCldn6 cells (SI Appendix, Fig. S4 A–D). These inhibitors and PP2 also significantly blocked CLDN6-induced increases in the proportion of EBs delineated by epithelia (solid form with clear zone, and cystic form; SI Appendix, Fig. S4E). Thus, the SFK/PI3K/AKT cascade was involved in the CLDN6-adhesion signaling.
The CLDN6/SFK/PI3K/AKT Axis Targets RARγ.
To verify the potential link between either retinoid receptors or HNF4α and the CLDN6 signalings, we then suppressed the low levels of HNF4α expression in F9:Cldn6 cells and generated F9:Rxra−/−:Rarg−/− cells expressing CLDN6. HNF4α knockdown did not affect the morphological appearance, mature cell junction formation, or expression of CLDN7, OCLN, ZO-1α+, and EZRIN in F9:Cldn6 cells (SI Appendix, Fig. S5 A–C). However, HNF4α suppression markedly blocked the CLDN6-triggered EBP50 expression and the enrichment of EZRIN on apical cell surfaces, in agreement with our previous work (27). In contrast, the CLDN6-induced epithelial differentiation was hindered in F9:Rxra−/−:Rarg−/−:Cldn6 cells, except for the CLDN7 expression (SI Appendix, Fig. S6 A and B). Induction of CLDN7 expression was also observed at mRNA levels in 3 F9:Rxra−/−:Rarg−/−:Cldn6 clones (SI Appendix, Fig. S6C), suggesting that the CLDN6 signal activates CLDN7 expression via RXR(β+γ)/RARα or RXR(β+γ)/RARβ pairs due to the redundant function of RXR/RAR heterodimers, as reported previously (31, 32). It is noteworthy that epithelial differentiation was never observed in F9:Rxra−/−:Rarg−/−:Cldn6 cells, even though SFK was activated in the cells (SI Appendix, Fig. S6D). In addition, no HNF4α expression was observed in F9:Rxra−/−:Rarg−/− and F9:Rxra−/−:Rarg−/−:Cldn6 cells. Hence, these results strongly suggested that the CLDN6-adhesion signaling links to RXRα/RARγ, as well as HNF4α, via these retinoid receptors.
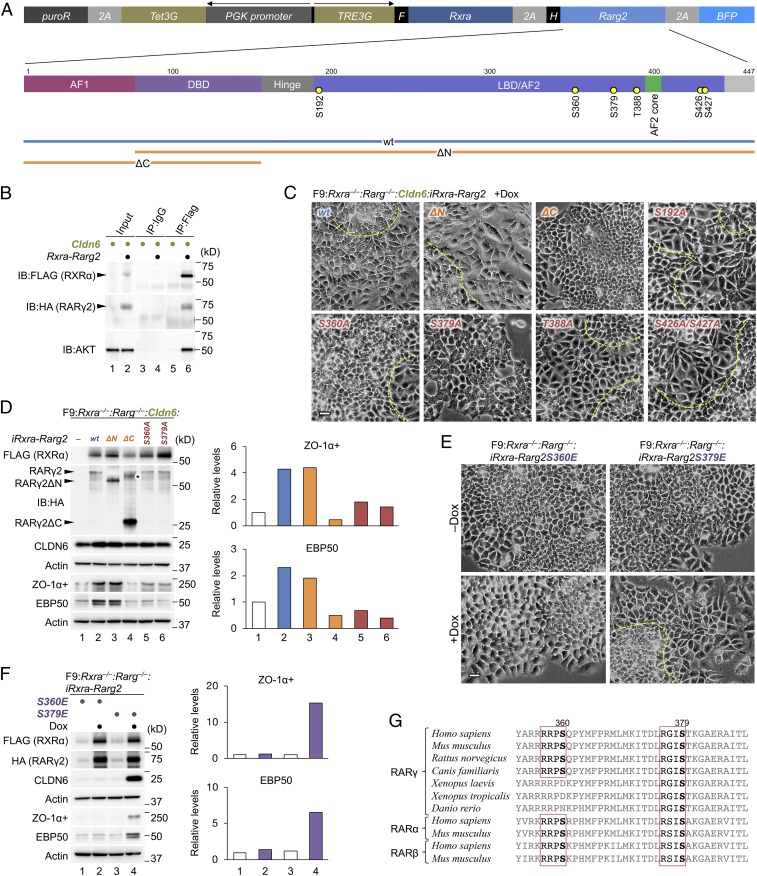
We subsequently demonstrated that AKT was associated with RXRα and RARγ2 in HEK293T cells transiently transfected with the RXRα-RARγ2 and the Cldn6 expression vectors (Fig. 3 A and B). We then evaluated the phenotype of F9:Rxra−/−:Rarg−/−:Cldn6 cells exhibiting the Dox-induced expression of RXRα-RARγ2, RXRα-RARγ2ΔN, or RXRα-RARγ2ΔC (Fig. 3A). Epithelial differentiation was only prevented in F9:Rxra−/−:Rarg−/−:Cldn6:iRxra-Rarg2ΔC cells (Fig. 3 C and D and SI Appendix, Fig. S1F), indicating that the CLDN6 signaling targets the Hinge, LBD/AF2, or F region of RARγ2. Because 5 putative phosphorylation sites for AKT were identified in these RARγ2 regions (Fig. 3A) by the GPS3.0 program (33), we next established F9:Rxra−/−:Rarg−/−:Cldn6 cells expressing the Dox-inducible RXRα-RARγ2 mutants with a substitution of each serine/threonine for an alanine residue. Among them, epithelial differentiation was inhibited in F9:Rxra−/−:Rarg−/−:Cldn6:iRxra-Rarg2S360A and F9:Rxra−/−:Rarg−/−:Cldn6:iRxra-Rarg2S379A cells (Fig. 3 C and D and SI Appendix, Fig. S1F). Importantly, however, epithelialization was evident in F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S379E cells, in which phosphorylation of RARγ2S379 was mimicked, but not in F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S360E cells (Fig. 3 E and F and SI Appendix, Fig. S1G). Moreover, CLDN6 phosphorylated RARγ2 in the AKT-dependent manner, and the phosphorylation level was decreased in RARγ2S379A mutant (SI Appendix, Fig. S7A). This AKT-consensus phosphorylation motif (RXXS, aa 376 to 379) was conserved not only among the 3 RARs but also among RARs of various vertebrates (Fig. 3G). Taken together, these data disclosed the importance of phosphorylation of mouse RARγ at S379, but not at S360, which corresponds to S371 in human RARγ and is shown to be phosphorylated by protein kinase A (34, 35), in CLDN6-trigggered signaling.
Fig. 3.
S379 of RARγ is indispensable for the CLDN6-triggered cellular events. (A) The construct of Dox-inducible wild-type and mutant RXRα/RARγ2 expression vectors. AF1, activation function-1; DBD, DNA-binding domain; LBD, ligand-binding domain; AF2, activation function-2. The putative AKT-phosphorylation sites in the Hinge, LBD/AF2, and F regions of RARγ2 are indicated. (B) Association between AKT and RXRα-RARγ2 in HEK293T cells that were transiently transfected with the RXRα-RARγ2 expression vector illustrated in A, along with the Cldn6 expression vector, and exposed for 72 h to either the vehicle or 1.0 μg/mL doxycycline (Dox). In the input lanes, 1% for HA and FLAG, or 0.1% for AKT of the input protein samples were loaded. (C and E) Morphological appearance of the indicated F9 cell lines. The cells were treated for 72 h with 1.0 μg/mL Dox. The borders between undifferentiated and epithelial cells are shown in the dashed yellow lines. (Scale bars, 20 μm.) (D and F) Western blot for the indicated proteins in various F9 cells. The revealed cell lines are exposed for 72 h with 1.0 μg/mL Dox. The asterisk shows the uncut form of RXRα-P2A-RARγ2ΔC. Quantification of the protein levels is shown in the histograms. (G) Conservation of the AKT-consensus phosphorylation motif (RXXS, aa 376 to 379 for mouse RARγ) in the LBD/AF2 domain of RARs among vertebrates. The conserved serine residues and AKT-consensus phosphorylation motif are indicated in bold and red rectangle lines, respectively.
Since the transcriptional corepressors interact with unliganded nuclear receptors and are released upon ligand binding (19, 21, 36–39), we next validated by chromatin immunoprecipitation assay the effects of the CLDN6 signal and RARγS379 phosphorylation on the recruitment of the nuclear receptor corepressor (NCoR) to putative and established RA response elements (RAREs) in the promoters of Rarb, Hnf4a, and Cldn6 genes (40). The binding of NCoR to 5, 2, and 1 RAREs of Rarb, Hnf4a, and Cldn6 genes, respectively, was significantly decreased in Dox-treated F9:iCldn6 cells compared with vehicle-exposed cells (SI Appendix, Fig. S8 A and B). Similar dissociations of NCoR from these sites were observed in Dox-treated F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S379E cells. In contrast, the recruitment of NCoR to these sites was significantly increased in F9:Rxra−/−:Rarg−/−:Cldn6:iRxra-Rarg2S379A cells compared with F9:Rxra−/−:Rarg−/−:Cldn6:iRxra-Rarg2 cells. Thus, CLDN6-triggered RARγS379 phosphorylation resulted in releasing NCoR from several RAREs of 3 distinct RA target genes. In addition, neither the CLDN6 signal, RARγS379A, nor RARγ S379E affected the binding of RXRα/RARγ to these RAREs as expected (SI Appendix, Fig. S8C).
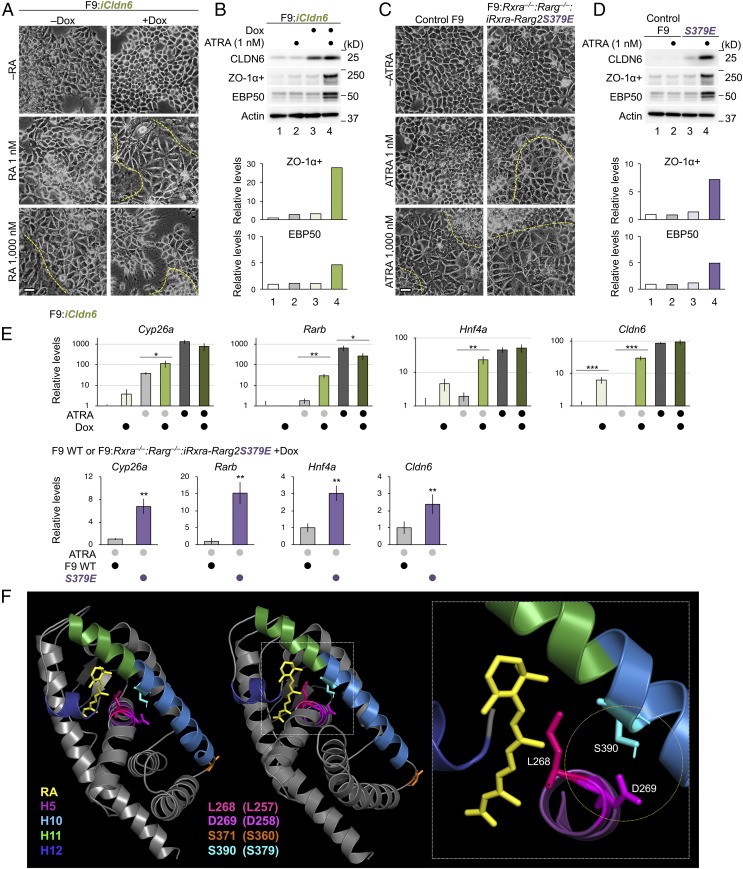
We subsequently determined whether suboptimal concentrations of all-trans retinoic acid (ATRA) influenced these cellular events. To this end, various F9 cells were grown in a culture condition, using charcoal-treated FBS to eliminate fat-soluble ligands. Under this culture condition, morphological differentiation and expression of ZO-1α+ and EBP50 protein were induced in F9:iCldn6 and F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S379E cells exposed to both Dox and 1 nM ATRA, but not in those treated with either one (Fig. 4 A–D and SI Appendix, Fig. S1 H and I). The amounts of CLDN6 protein were also synergistically increased in F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S379E cells upon Dox and 1 nM ATRA treatment. Dox-induced CLDN6 expression significantly increased the expression of endogenous Cldn6 mRNA in F9:iCldn6 cells (Fig. 4E). In addition, Dox and 1 nM ATRA synergistically induced the expression of Cyp26a, Rarb, Hnf4a, and Cldn6 genes in F9:iCldn6. Moreover, 1 nM ATRA treatment significantly increased expression levels of these RA target genes in Dox-exposed F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S379E cells compared with WT F9 cells. We also revealed that knockdown of endogenous CLDN6 expression repressed amounts of ZO-1 and pSFK/pPI3K/pAKT proteins, as well as the expression of Cyp26a and Cldn6 genes in F9:iHNF4α (SI Appendix, Fig. S9 A–D).
Fig. 4.
The CLDN6-adhesion signaling stimulates the RAR activity. (A and C) Morphological appearance of the indicated F9 cell lines. The cells were grown for 72 h in the presence or absence of 1.0 μg/mL Dox, 1 nM and 1 μM of ATRA, or the combination. The borders between undifferentiated and epithelial cells are shown in the dashed yellow lines. (Scale bars, 20 μm.) (B and D) Western blot for the indicated proteins in the revealed cell lines. Cells were cultured as in A and C. Quantification of the protein levels is shown in the histograms. (E) Quantitative RT-PCR for the indicated molecules in F9:iCldn6 and F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S379E cells. The cells were treated for 24 h with 1.0 µg/mL Dox, ATRA (1 nM or 1,000 nM) or both together. The relative expression levels are shown in the histograms (mean ± SD; n = 3 to 4). *P < 0.05; **P < 0.01; ***P < 0.001. (F) Structural representation of the holo-LBD of human RARγ (49) was visualized by PyMOL. The corresponding amino acids in mouse RARγ are indicated in parentheses.
The CLDN6-Adhesion Signaling Stimulates ERα Function.
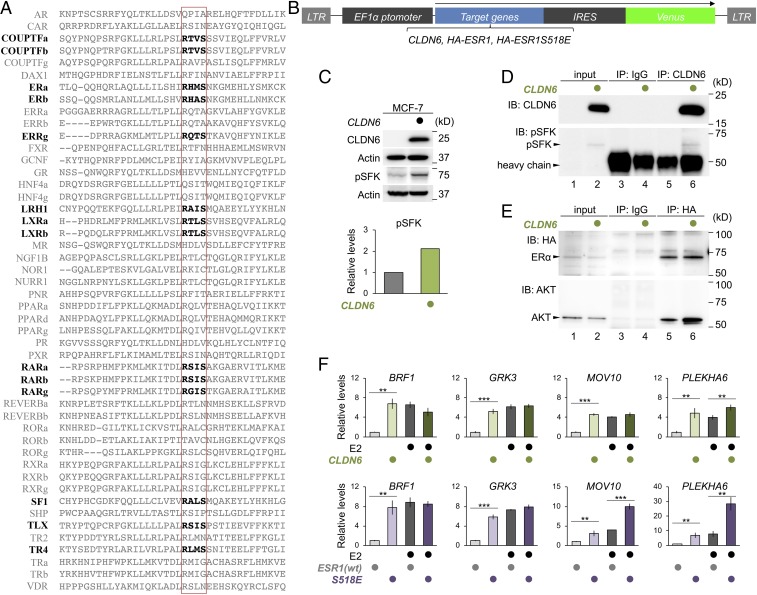
The AKT-consensus phosphorylation motif in RARs (RXXS, aa 376 to 379 in mouse RARγ) was conserved among closely related nuclear receptors in invertebrates (SI Appendix, Fig. S10A). Interestingly, this phosphorylation motif was present in 14 of 48 members of human nuclear receptors (Fig. 5A), and was fully conserved among ERα and ERβ in vertebrates (SI Appendix, Fig. S10B). Therefore, to determine whether the CLDN6-initiated signal also targets ERα, the ERα-positive human breast cancer cell line MCF-7 was transiently transfected with CLDN6 (Fig. 5B). As expected, the pSFK levels were higher in MCF-7:CLDN6 cells than in the parental MCF-7 cells (Fig. 5C). In addition, a complex between CLDN6 and pSFK was observed in MCF-7:CLDN6 cells (Fig. 5D). Furthermore, AKT interacted with the transiently introduced HA-ERα in MCF-7 cells, and their association was increased in MCF-7:CLDN6 cells (Fig. 5E). Interestingly, the transcript levels of 4 ER target genes (BRF1, GRK3, MOV10 and PLAKHA6) were higher in MCF-7:CLDN6 cells than in MCF-7 cells when grown in the absence of estradiol (E2) (Fig. 5F). The levels of these transcripts were similar to those in MCF-7 cells cultured in the presence of 1 μM E2. Moreover, the expression of these target genes was induced in MCF-7:ESR1S518E cells compared with that in MCF-7:ESR1 cells, and the mRNA levels were comparable to those in MCF-7:ESR1 cells exposed to 1 μM E2. In addition, the amounts of MOV10 and PLAKHA6 transcripts, but not BRF1 or GRK3, additively and synergistically increased in MCF-7:ESR1S518E cells upon addition of 1 μM E2, respectively. Also, CLDN6 phosphorylated ERα via the AKT, and the phosphorylation intensity was reduced in ERαS518A mutant (SI Appendix, Fig. S7B). Thus, the CLDN6-adhesion signaling targets not only to RARγ but also to ERα. We also showed that IGF1, another upstream signal of AKT, activated RAR and ER target genes in F9 and MCF-7 cells, respectively (SI Appendix, Fig. S11 A–C).
Fig. 5.
The CLDN6-adhesion signaling regulates the ERα activity. (A) Conservation of the AKT phosphorylation motif in human nuclear receptor proteins. The AKT-consensus phosphorylation motif and the corresponding sites in nuclear receptors are indicated in bold and the red rectangle lines, respectively. (B) The construct of CLDN6, HA-ESR1, and HA-ESR1S518E expression vectors. (C) Western blot for the indicated proteins in MCF-7 and MCF-7:CLDN6 cells. Quantification of the protein levels is shown in the histogram. (D) Association between CLDN6 and pSFK in MCF-7:CLDN6 cells. In the input lanes, 2% of the input protein samples were loaded. (E) Association between AKT and HA-ERα in MCF-7:HA-ESR1 and MCF-7:CLDN6:ESR1 cells. In the input lanes, 1% for HA or 0.1% for AKT of the input protein samples were loaded. (F) Quantitative RT-PCR for the indicated molecules in the MCF-7 cells. The cells were treated for 24 h with vehicle (ethanol) or 1.0 µM Estradiol (E2). The relative expression levels are shown in the histograms (mean ± SD; n = 4). **P < 0.01; ***P < 0.001.
Discussion
In the present study, we showed that the CLDN6-adhesion signaling activated SFKs to initiate the cellular processes. This conclusion was drawn from the following results: the pSFK levels were increased in F9:iCldn6 and MCF-7:CLDN6 cells compared with the respective parental cells, and were reversed by CLDN6 knockdown in F9:Cldn6 cells; various SFK inhibitors strikingly inhibited morphological differentiation and/or formation of mature cell–cell junctions in F9:Cldn6 cells; PP2 treatment of F9:Cldn6 and Dox-treated F9:iHNF4α cells failed to induce the expression of the tight-junction markers or EBP50; CLDN6 was colocalized with pSFK along cell boundaries in F9:Cldn6 cells and epithelial cells of EBs; and 5) an CLDN6-pSFK complex was formed in F9:Cldn6 and MCF-7:CLDN6 cells.
We previously reported that C-CPE exposure to F9:iCldn6 cells and mouse EBs markedly eliminated CLDN6 from cell junctions, resulting in the prevention of epithelial differentiation (28). Although C-CPE binds to EC2 of CLDN3, CLDN4, CLDN6, and CLDN9 with high affinity (11–14), the effects of C-CPE in these cells were attributed to its binding to CLDN6 (28), suggesting that the CLDN6-driving cellular events were mediated via the EC2-dependent CLDN6 engagement at a cell–cell junctions. We demonstrated in the present work that epithelial differentiation was evident in F9:iCldn6ΔEC1-FLAG cells, but not in F9: iCldn6ΔEC2-FLAG cells. Our results also revealed that C-CPE treatment of F9:iCldn6 and F9:Cldn6 cells led to a decrease in both pSFK levels and CLDN6-pSFK complex formation. Taken collectively, we conclude that the EC2 domain contributes to the CLDN6-adhesion signaling.
We also demonstrated that CLDN6-triggered signal was transmitted through the first half C-terminal cytoplasmic domain, in which both Y196 and Y200 were absolutely required to activate SFKs. In addition, CLDN6 formed a complex with pSFK in both EC2- and Y196/200-dependent manners. Furthermore, CLDN6 was tyrosine-phosphorylated at both Y196 and Y200, and tyrosine-phosphorylation of CLDN6 was governed by the EC2 domain. Taken together with the results of previous reports showing that the SH2 domain binds to the pY-containing peptide ligands 1,000 times greater than to the nonphosphorylated ones (41, 42), we speculated the mechanism behind the CLDN6/SFK signaling to be as follows: the EC2-oriented trans-interaction of CLDN6 recruits SFKs probably via the SH2 domain to Y196/200 and activates them, the weakly activated SFKs sequentially phosphorylate CLDN6 at Y196/200, and strong binding of SFKs-SH2 to CLDN6-pY196/200 fully activates SFKs, leading to stimulation of the downstream PI3K/AKT pathway (Fig. 6). It is unknown how the EC2-dependent engagement of CLDN6 drives outside-in signaling. However, since the ligand binding of integrins causes conformational changes in the cytoplasmic domain (43–45), similar conversion of the CLDN6 structure may disclose the Y196/200-containing amino acid sequence in the C-terminal cytoplasmic domain. In this regard, it should be noted that CAR (Coxsackie and Adenovirus receptor)–JAML (junctional adhesion molecule-like) ectodomain interaction engages and stimulates PI3K (46). Note also that, upon search for amino acid sequences of mouse CLDN members, CLDN6 is the only CLDN subtype in which both Y196 and Y200 are conserved (SI Appendix, Fig. S12A), strongly suggesting the functional specificity of CLDN6. In fact, overexpression of CLDN4 in ES cells was unable to provoke epithelial differentiation (SI Appendix, Fig. S12B). On the other hand, Li et al. (47) have recently shown that CLDN11 is phosphorylated at 2 adjacent tyrosine residues and activates SRC, which is required for facilitating collective migration of head and neck squamous cell carcinoma.
Fig. 6.
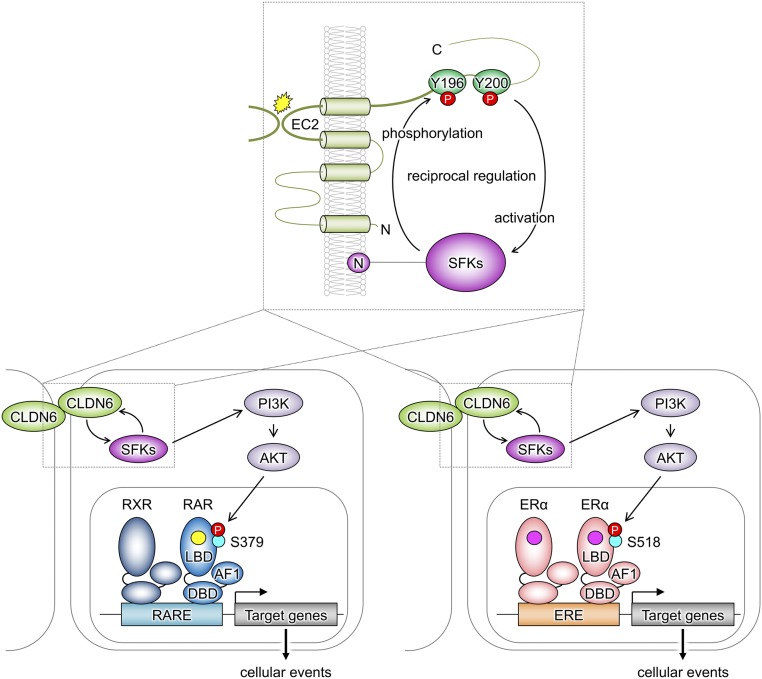
Schematic model for regulation of the nuclear receptor activity by the CLDN6-adhesion signaling. The EC2-dependent trans-interaction of CLDN6 recruits and activates SFKs at tyrosine residues 196 and 200. SFKs in turn phosphorylate CLDN6 at Y196/200, and strong binding of SFKs to CLDN6-pY196/200 fully activates SFKs, resulting in stimulating the downstream PI3K/AKT pathway. This CLDN6 adhesion signal phosphorylates mouse RARγ and human ERα at S379 and S518, respectively, leading to stimulation of the RARγ and ERα activities (RA; yellow circle, estrogen; pink circle).
The most important finding of the present study is that the CLDN6/SFK/PI3K/AKT signaling controls the RARγ and ERα activities (Fig. 6). This conclusion was apparent because the CLDN6-induced cellular processes were abolished in 3 distinct F9:Rxra−/−:Rarg−/−:Cldn6 cell lines, even though SFKs were activated. In addition, AKT formed a complex with either RXRα/RARγ2 or ERα, further strengthening the conclusion. Moreover, characterization of F9:Rxra−/−:Rarg−/−:Cldn6:iRxra-Rarg2S379A and F9:Rxra−/−:Rarg−/−:iRxra-Rarg2S379E cells, as well as that of MCF-7:ESR1S518E cells, indicated that the CLDN6 signaling directs S379 and S518 in mouse RARγ and human ERα, respectively. Based on the 3-dimensional structure of human RARγ-LBD (ligand-binding domain) bound to ATRA (19, 48, 49), S390, which corresponds to S379 in mouse RARγ, was located in helix 10 (H10; Fig. 4F). The hydrophobic RA-binding pocket in human RARγ is constituted by 24 amino acids, and L268 is one of such residues. Therefore, it is worth noting that S390 is opposed to D269 in H5 and that the repulsion force between them may be weakened by serine-phosphorylation, thereby promoting the ability of L268 to hold ligands in the pocket and releasing NCoR. This would be how the phosphorylation of mouse RARγS379 by the CLDN6/SFK/PI3K/AKT axis greatly increases sensitivity to ATRA, leading to synergistic induction of the expression of RA target genes. Interestingly, the CLDN6-adhesion signal ligand-independently activated the expression of endogenous Cldn6 and 4 distinct estrogen target genes in F9 and MCF-7 cells, respectively. In addition, the expression of these estrogen target genes was induced in MCF-7:ESR1S518E cells at levels similar to those in MCF-7:ESR1 cells treated with the pharmacological concentration of E2. The crystal structure of human ERα holo-LBD (50, 51) reveled that the positional relationship between S518 (in H11) and L384/E385 (in H6) in human ERα was similar to that between S390 and L268/D269 in human RARγ (SI Appendix, Fig. S10C). This may cause additive or synergistic induction of some estrogen target genes (e.g., MOV10 and PLAKHA6) in MCF-7:ESR1S518E cells upon addition of 1 μM E2. It is also noteworthy that the AKT-consensus phosphorylation motif (RXXS, aa 376 to 379 in mouse RARγ) is conserved not only in RARs and ERs across species but also in an additional 9 members of human nuclear receptors (Fig. 5A), further pointing to the biological relevance of this phosphorylation site. Thus, not only growth factors (52–55) but also cell-adhesion signaling modulate the activity of nuclear receptors.
In conclusion, we suppose that the EC2-dependent trans-interaction of CLDN6 recruits and activates SFKs, which phosphorylate CLDN6 at Y196/200 and propagate the PI3K/AKT cascade, and this signaling module extensively stimulates the RARγ and ERα activities (Fig. 6). The link between transcription factors and the signals originating from cell–cell adhesion and possibly cell–matrix interaction, which are critical for the appropriate tissue organization, will contribute not only to a range of physiological cellular events but also to various diseases.
Materials and Methods
The materials and methods used in this study are described in detail in SI Appendix, Supplementary Materials and Methods. Antibodies are listed in SI Appendix, Table S1; the sequences of siRNAs are shown in SI Appendix, Table S2; and PCR primers are listed in SI Appendix, Table S3. The data are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Dr. Shigeaki Kato (Iryo Sosei University) and Dr. Daniel Metzger (Institut de Génétique et de Biologie Moléculaire et Cellulaire) for discussion, A. Hozumi and K. Watari for their technical assistance, and the Scientific English Editing Section of the Medical Research Promotion Office, Fukushima Medical University, for their help with editing and proofreading the manuscript. This work was supported by JSPS KAKENHI Grant Numbers 17K08699 and 17K17978 and by the Uehara Memorial Foundation and the Takeda Science Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913346116/-/DCSupplemental.
References
- 1.Gumbiner B. M., Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell 84, 345–357 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Hynes R. O., Integrins: Bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Perez-Moreno M., Jamora C., Fuchs E., Sticky business: Orchestrating cellular signals at adherens junctions. Cell 112, 535–548 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Van Itallie C. M., Anderson J. M., Claudins and epithelial paracellular transport. Annu. Rev. Physiol. 68, 403–429 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Chiba H., Osanai M., Murata M., Kojima T., Sawada N., Transmembrane proteins of tight junctions. Biochim. Biophys. Acta 1778, 588–600 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Zihni C., Mills C., Matter K., Balda M. S., Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 17, 564–580 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Tsukita S., Tanaka H., Tamura A., The claudins: From tight junctions to biological systems. Trends Biochem. Sci. 44, 141–152 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Furuse M., Sasaki H., Fujimoto K., Tsukita S., A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 143, 391–401 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuse M., Fujita K., Hiiragi T., Fujimoto K., Tsukita S., Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 141, 1539–1550 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuse M., Tsukita S., Claudins in occluding junctions of humans and flies. Trends Cell Biol. 16, 181–188 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Fujita K., et al. , Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 476, 258–261 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Kimura J., et al. , Clostridium perfringens enterotoxin interacts with claudins via electrostatic attraction. J. Biol. Chem. 285, 401–408 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winkler L., et al. , Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J. Biol. Chem. 284, 18863–18872 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veshnyakova A., et al. , Determinants contributing to claudin ion channel formation. Ann. N. Y. Acad. Sci. 1257, 45–53 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Nakamura S., et al. , Morphologic determinant of tight junctions revealed by claudin-3 structures. Nat. Commun. 10, 816 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause G., et al. , Structure and function of claudins. Biochim. Biophys. Acta 1778, 631–645 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Piontek J., et al. , Formation of tight junction: Determinants of homophilic interaction between classic claudins. FASEB J. 22, 146–158 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Cavallaro U., Dejana E., Adhesion molecule signalling: Not always a sticky business. Nat. Rev. Mol. Cell Biol. 12, 189–197 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Chambon P., A decade of molecular biology of retinoic acid receptors. FASEB J. 10, 940–954 (1996). [PubMed] [Google Scholar]
- 20.Germain P., et al. , International union of pharmacology. LXIII. Retinoid X receptors. Pharmacol. Rev. 58, 760–772 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Cunningham T. J., Duester G., Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 16, 110–123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans R. M., Mangelsdorf D. J., Nuclear receptors, RXR, and the big bang. Cell 157, 255–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan B. L., Taylor A., Adamson E., Cell interactions modulate embryonal carcinoma cell differentiation into parietal or visceral endoderm. Nature 291, 235–237 (1981). [DOI] [PubMed] [Google Scholar]
- 24.Kubota H., et al. , Retinoid X receptor alpha and retinoic acid receptor gamma mediate expression of genes encoding tight-junction proteins and barrier function in F9 cells during visceral endodermal differentiation. Exp. Cell Res. 263, 163–172 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Chiba H., et al. , Hepatocyte nuclear factor (HNF)-4alpha triggers formation of functional tight junctions and establishment of polarized epithelial morphology in F9 embryonal carcinoma cells. Exp. Cell Res. 286, 288–297 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Satohisa S., et al. , Behavior of tight-junction, adherens-junction and cell polarity proteins during HNF-4α-induced epithelial polarization. Exp. Cell Res. 310, 66–78 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Chiba H., et al. , The nuclear receptor hepatocyte nuclear factor 4alpha acts as a morphogen to induce the formation of microvilli. J. Cell Biol. 175, 971–980 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugimoto K., et al. , The tight-junction protein claudin-6 induces epithelial differentiation from mouse F9 and embryonic stem cells. PLoS One 8, e75106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLachlan R. W., Kraemer A., Helwani F. M., Kovacs E. M., Yap A. S., E-cadherin adhesion activates c-Src signaling at cell-cell contacts. Mol. Biol. Cell 18, 3214–3223 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canel M., Serrels A., Frame M. C., Brunton V. G., E-cadherin-integrin crosstalk in cancer invasion and metastasis. J. Cell Sci. 126, 393–401 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Chiba H., Clifford J., Metzger D., Chambon P., Specific and redundant functions of retinoid X Receptor/Retinoic acid receptor heterodimers in differentiation, proliferation, and apoptosis of F9 embryonal carcinoma cells. J. Cell Biol. 139, 735–747 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiba H., Clifford J., Metzger D., Chambon P., Distinct retinoid X receptor-retinoic acid receptor heterodimers are differentially involved in the control of expression of retinoid target genes in F9 embryonal carcinoma cells. Mol. Cell. Biol. 17, 3013–3020 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue Y., et al. , GPS: A comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 33, W184–W187 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rochette-Egly C., et al. , Phosphorylation of the retinoic acid receptor-alpha by protein kinase A. Mol. Endocrinol. 9, 860–871 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Chebaro Y., et al. , Phosphorylation of the retinoic acid receptor alpha induces a mechanical allosteric regulation and changes in internal dynamics. PLoS Comput. Biol. 9, e1003012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J. D., Evans R. M., A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377, 454–457 (1995). [DOI] [PubMed] [Google Scholar]
- 37.Hörlein A. J., et al. , Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377, 397–404 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Glass C. K., Rosenfeld M. G., The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14, 121–141 (2000). [PubMed] [Google Scholar]
- 39.Mottis A., Mouchiroud L., Auwerx J., Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 27, 819–835 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delacroix L., et al. , Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol. Cell. Biol. 30, 231–244 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olsen J. V., et al. , Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Huttlin E. L., et al. , A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leisner T. M., Wencel-Drake J. D., Wang W., Lam S. C., Bidirectional transmembrane modulation of integrin alphaIIbbeta3 conformations. J. Biol. Chem. 274, 12945–12949 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Legate K. R., Wickström S. A., Fässler R., Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 23, 397–418 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Durrant T. N., van den Bosch M. T., Hers I.. Integrin αIIbβ3 outside-in signaling. Blood 130, 1607–1619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witherden D. A., et al. , The junctional adhesion molecule JAML is a costimulatory receptor for epithelial gammadelta T cell activation. Science 329, 1205–1210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C.-F., et al. , Snail-induced claudin-11 prompts collective migration for tumour progression. Nat. Cell Biol. 21, 251–262 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Renaud J. P., et al. , Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature 378, 681–689 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Klaholz B. P., et al. , Conformational adaptation of agonists to the human nuclear receptor RAR gamma. Nat. Struct. Biol. 5, 199–202 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Pike A. C., et al. , Structure of the ligand-binding domain of oestrogen receptor beta in the presence of a partial agonist and a full antagonist. EMBO J. 18, 4608–4618 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delfosse V., et al. , Structural and mechanistic insights into bisphenols action provide guidelines for risk assessment and discovery of bisphenol A substitutes. Proc. Natl. Acad. Sci. U.S.A. 109, 14930–14935 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kato S., et al. , Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science 270, 1491–1494 (1995). [DOI] [PubMed] [Google Scholar]
- 53.Bunone G., Briand P. A., Miksicek R. J., Picard D., Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 15, 2174–2183 (1996). [PMC free article] [PubMed] [Google Scholar]
- 54.Hu E., Kim J. B., Sarraf P., Spiegelman B. M., Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science 274, 2100–2103 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Shupnik M. A., Crosstalk between steroid receptors and the c-Src-receptor tyrosine kinase pathways: Implications for cell proliferation. Oncogene 23, 7979–7989 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.