Significance
How blood stem cells within the bone marrow respond to infection is still unclear. Here we study if mitochondrial transfer from the stromal cells to blood stem cells is required for the rapid generation of leukocytes which are required for the immune system to respond to bacterial infection. We find that mitochondria are transferred to blood stem cells within 2 h of sensing infection and show that direct contact between stromal cells and blood stem cells is required for mitochondrial transfer to occur. The metabolic changes that follow mitochondrial transfer into hematopoietic stem cells underpin the rapid mammalian response to infection. Finally, we find that, if mitochondrial transfer is blocked, an increase in bacterial colonization in the mammalian system occurs.
Keywords: ROS, infection, mitochondria, hematopoietic stem cell, PI3K
Abstract
Hematopoietic stem cells (HSCs) undergo rapid expansion in response to stress stimuli. Here we investigate the bioenergetic processes which facilitate the HSC expansion in response to infection. We find that infection by Gram-negative bacteria drives an increase in mitochondrial mass in mammalian HSCs, which results in a metabolic transition from glycolysis toward oxidative phosphorylation. The initial increase in mitochondrial mass occurs as a result of mitochondrial transfer from the bone marrow stromal cells (BMSCs) to HSCs through a reactive oxygen species (ROS)-dependent mechanism. Mechanistically, ROS-induced oxidative stress regulates the opening of connexin channels in a system mediated by phosphoinositide 3-kinase (PI3K) activation, which allows the mitochondria to transfer from BMSCs into HSCs. Moreover, mitochondria transfer from BMSCs into HSCs, in the response to bacterial infection, occurs before the HSCs activate their own transcriptional program for mitochondrial biogenesis. Our discovery demonstrates that mitochondrial transfer from the bone marrow microenvironment to HSCs is an early physiologic event in the mammalian response to acute bacterial infection and results in bioenergetic changes which underpin emergency granulopoiesis.
The lifelong maintenance of hematopoiesis is dependent on the ability of hematopoietic stem cells (HSCs) to self-renew and differentiate into all subtypes of mature blood cells (1, 2). Hematopoiesis, however, is dynamic, and the HSCs, while predominantly quiescent, rapidly enter the cell cycle upon exposure to specific stimuli. Rapid expansion of leukocytes in response to pathogens underpins the mammalian response to infection. Presently, the mechanisms by which HSC metabolism is regulated in response to the challenges of pathogenic stress are not fully understood.
HSCs reside in a specialized niche within the bone marrow (BM) microenvironment (3). The BM microenvironment regulates the production of both hematopoietic and nonhematopoietic cells for the maintenance of blood production under normal and stressed conditions (3, 4). HSCs reside in a hypoxic niche, and studies suggest that quiescent HSC favor anaerobic glycolysis to generate the energy required for cell maintenance under baseline conditions (5, 6). Furthermore, quiescent HSCs have also been reported to possess low mitochondrial mass (5, 7, 8), sustained by active elimination of mitochondria through mitophagy, which has been suggested to be critical for HSC maintenance (8, 9). However, a more recent study shows that mitochondrial mass in HSCs is stable throughout their life span, while mitochondrial turnover capacity is in fact lower in HSCs than in progenitors (10). In addition to regulation of mitochondrial mass, the maintenance of HSC quiescence requires the limiting of mitochondrial respiration (11). Reactive oxygen species (ROS) levels, related to mitochondrial activity, are much lower in the HSCs compared to the committed progenitors (12). Taken together, the baseline maintenance of HSC quiescence depends on a set of specific metabolic conditions regulated in the HSC niche.
Pathogens have been a significant selective pressure in the history of mammalian evolution and remain a leading cause of global mortality (13). In response to infection HSCs require a swift metabolic switch from glycolysis to mitochondrial oxidative phosphorylation (OXPHOS) (14). This switch allows for some of the HSCs to rapidly cycle and differentiate into the required progenitor cells while other HSCs are maintained in the multipotent state. Increasing ROS levels, as in response to stress infection, promote the differentiation of a subset of HSCs whereas low levels of ROS maintain quiescence (15). HSC expansion favors the utilization of OXPHOS as the principal energy source in contrast to quiescent cells which favor glycolysis; accordingly, the changes in cellular metabolism from dormant to cycling is clearly of fundamental importance for determining HSC fate (16). This implies that differentiation of HSCs requires metabolic conditions that include oxidative stress as well as a rapid burst of energy supplied via mitochondrial OXPHOS.
Previously, we and others have shown that mitochondria can transfer from bone marrow stromal cells (BMSCs) to acute myeloid leukemia (AML) blasts to enhance their proliferation in vitro and in vivo through a mechanism which increases OXPHOS in the leukemic blasts (17, 18). Moreover, as part of that study we observed that human CD34+ hematopoietic progenitor cells (HPCs) could also acquire mitochondria from BMSCs under oxidative stress (17), leading us to hypothesize that the mitochondrial transfer-dependent metabolic switch in AML has its “origins” in the physiology of the HSC response to infection.
Most cases of invasive nontyphoidal Salmonella infection are caused by Salmonella typhimurium (S. typhimurium), and invasive strains of nontyphoidal salmonellae have emerged as a prominent cause of bloodstream infection in African adults and children, with an associated case fatality of 20 to 25% (19). In the context of the challenge of acute bacterial infection, the hematopoietic system needs to drive rapid HSC and leukocyte expansion, necessary for host survival (20). Therefore, through studies using S. typhimurium and its outer membrane lipopolysaccharide (LPS) to model acute bacterial infection, we aim to understand if and how the mammalian response to infection involves the acquisition of mitochondria by HSC from cells within the BM microenvironment and the mechanisms and processes by which this facilitates the bio-energetic and oxidative changes required for rapid leukocyte generation.
Results
Mitochondria Are Transferred from the BM Microenvironment to the HSC Populations In Vivo in Response to LPS.
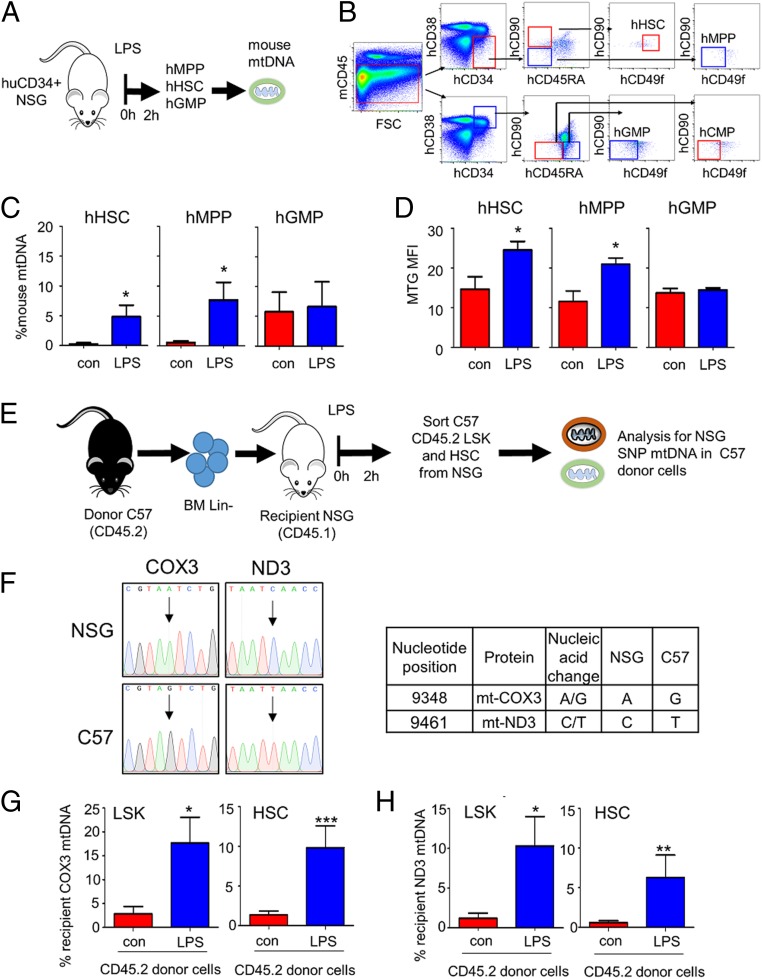
To determine if mitochondria are transferred to HSCs in conditions of stressed hematopoiesis, we used a humanized nonobese diabetic (NOD) severe combined immunodeficiency (SCID) Il2rg knockout NOD.Cg.PrkdscidIL2rgtm1Wji/SzJ (NSG) mouse model to assess mitochondrial transfer from mouse BM to human CD34+ HSCs, employing species-specific mitochondrial DNA (mtDNA) detection as a surrogate tracker as previously shown (21). Humanized (Hu)-NSG mice were created (Fig. 1 A and B) and were then treated with LPS for 2 h. Animals were then killed and human multipotent progenitors (MPPs), HSCs, and granulocyte macrophage progenitors (GMPs) were isolated and analyzed for the presence of mouse mtDNA (Fig. 1 C and D). SI Appendix, Fig. S1 A and B, confirms engraftment of human CD34+ HSCs in recipient NSG mice. Fig. 1C shows that mouse mtDNA is significantly increased in MPPs and HSCs but not in GMPs from LPS-treated C57BL/6 hu-NSG mice. Fig. 1D confirms mitochondrial mass increase in MPPs and HSCs from LPS-treated C57BL/6 mice.
Fig. 1.
Mitochondria are transferred from the BM microenvironment to the HSC populations in vivo in response to LPS. (A) Schematic diagram of experimental design. Post engraftment hu-NSG mice were injected with LPS for 2 h. (B) The BM was extracted, and the cells were analyzed by flow cytometry or sorted for hHSC (mCD45−, hCD34+, hCD38−, hCD45RA−, hCD90+, hCD49f+), hMPP (mCD45−, hCD34+, hCD38−, hCD45RA−, hCD90−, hCD49f−), and hGMP (mCD45−, hCD34+, hCD38+, hCD45RA+, hCD90−, hCD49f−) cell populations using FACS. (C) mtDNA transfer was quantified using Taqman qPCR with species-specific probes. Bars represent the percentage of mouse mtDNA in the hHSC, hMPP, or hGMP cells after 2 h treatment with LPS versus the control (untreated cells). Human genomic DNA was used to standardize mtDNA. Data shown are means ± SD of n = 5 mice. *P < 0.05. (D) Flow cytometry analysis of MTG fluorescence was assessed in the progenitor cells to quantify mitochondrial content. (E) Schematic diagram of experimental design. Recipient CD45.1 NSG mice were treated with busulfan prior to tail-vein injections of lineage-negative CD54.2 C57BL/6 donor cells. One month post engraftment mice were treated with LPS or vehicle PBS for 2 h. BM was isolated and sorted for donor LSK (Lin−, Sca-1+, c-Kit+) and donor HSC (Lin−, Sca-1+, c-Kit+, CD48−, CD150+) cell populations expressing CD45.2 using FACS. The sorted donor cells were analyzed for specific donor or host mtDNA by TaqMan PCR using ND3 or COX3 probes designed to detect strain-specific SNPs in the mitochondrial genome. (F) SNP differences in mtDNA between NSG mice and C57BL/6 mice. (G) The percentage of recipient mtDNA in donor cells after a 2-h treatment with LPS versus control (untreated) cells using the COX3 TaqMan probe. (H) The percentage of recipient mtDNA in donor cells after a 2-h treatment with LPS versus control (untreated) cells using the ND3 TaqMan probe. Donor genomic DNA was used to standardize mtDNA copy number. Data shown are means ± SD of n = 5 mice. *P < 0.05; **P < 0.01; ***P < 0.001.
As a second model to confirm transfer of mtDNA, we used NSG (CD45.1: recipient) animals transplanted with C57BL/6 lineage-negative cells (CD45.2: donor) (Fig. 1E). Sanger sequencing of the mtDNA from NSG and C57BL/6 mice identified 2 single-nucleotide polymorphisms (SNP) that exist in the mitochondrial genomes and which are capable of distinguishing the 2 murine strains (Fig. 1F and SI Appendix, Fig. S2 A and B). SI Appendix, Fig. S1 C and D, confirmed engraftment of the donor (CD45.2) in the recipient (CD45.1). SNP analysis of sorted Lineage−Sca-1+c-Kit+ (LSK) cells and HSCs (lin−, Sca-1+, c-Kit+, CD48−, CD150+) showed that transfer of mitochondrial DNA from recipient to donor cells had occurred in vivo in LPS-treated animals (Fig. 1 G and H). Together, these 2 animal models confirm that mitochondrial DNA is transferred from the BM microenvironment to HSC populations.
S. typhimurium Infection Increases Mitochondrial Potential and Expansion of HSCs.
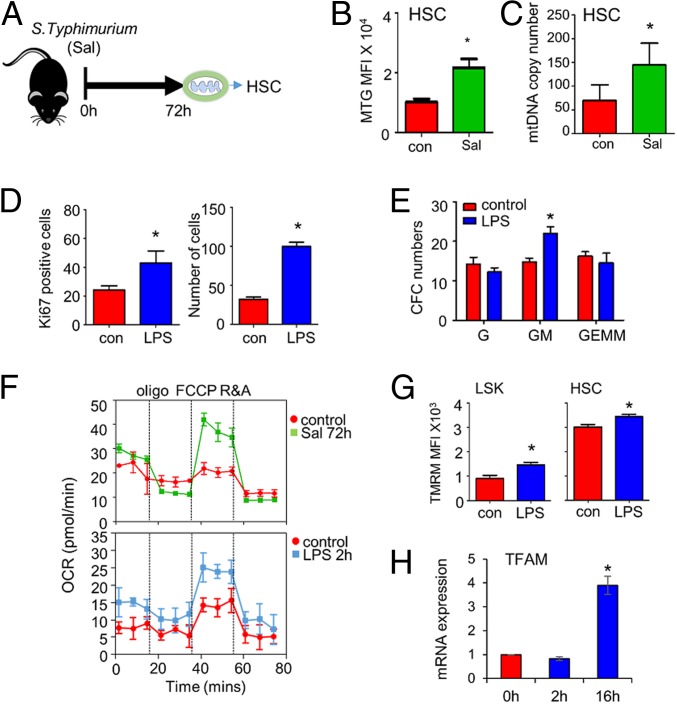
To confirm that mitochondrial mass increases in HSC populations in response to S. typhimurium MitoTracker Green (MTG) and mtDNA mass measured by RT-PCR, were used (Fig. 2A). C57BL/6 mice at 8 wk of age were infected with S. typhimurium for 72 h, and HSCs showed an increase in MTG fluorescence and mtDNA (Fig. 2 B and C). Fig. 2D confirms the HSC expansion at 2 h post LPS treatment, and Fig. 2E shows expansion of the GMP. Seahorse metabolic flux analysis measuring oxygen consumption rates (OCR) confirmed increased oxidative phosphorylation levels in LSK from LPS (2 h) and S. typhimurium-treated (72 h) C57BL/6 mice (Fig. 2F). Extracellular acidification rate, which is a method to monitor glycolysis, was not significantly up-regulated (SI Appendix, Fig. S3). Fig. 2G shows increased tetramethylrhodamine, methyl ester (TMRM) staining in LPS-treated LSK cells and HSCs in response to LPS, indicating increased mitochondrial activity in these cells. The messenger RNA (mRNA) expression of mitochondrial transcription factor A (TFAM), which is a regulator of mitochondrial biogenesis, was not up-regulated by 2 h post LPS stimulation (Fig. 2H). Together, these data confirm that infection with Gram-negative bacteria increases mitochondrial content and function in the HSC population; however, this was not associated with increased mitochondrial biogenesis in HSCs.
Fig. 2.
S. typhimurium infection increases mitochondrial potential and expansion of HSC. (A) Schematic of in vivo experiment in which C57/Bl6 mice were infected with S. typhimurium (Sal) for 72 h and analyzed for HSC by flow cytometry and PCR. (B) MTG fluorescence was assessed in the HSC cells by flow cytometry. (C) mtDNA analysis of the HSC population sorted (Lin−, Sca-1+, c-Kit+, CD48−, CD150+) from control and S. typhimurium treated mice. (D) Percentage of cycling HSC as measured by Ki67-positive cells after 16 h of LPS treatment. Number of HSC after 16 h of LPS treatment. (E) BM was isolated from control and LPS-treated animals at 2 h. BM cells (1 × 104) were seeded in a semisolid Methocult for 7 d. Granulocytic progenitor (G), granulocyte monocyte progenitor (GM), and granulocyte monocyte megakaryocyte progenitor (GEMM) colonies were then counted. (F) OXPHOS levels were measured by OCR in freshly isolated LSK from control, LPS, and S. typhimurium treated mice. (G) C57BL/6 mice were subjected to LPS or control PBS i.p. injections. After 2 h the mice were killed, and the BM was extracted. The populations were analyzed by flow cytometry for mean TMRM fluorescence intensity within each population. (H) LSK were FACS-sorted from control and LPS-treated animals, RNA was analyzed for TFAM gene expression. Data shown are means ± SD of n = 5 mice. *P < 0.05.
Superoxide Drives Mitochondrial Transfer to HSCs.
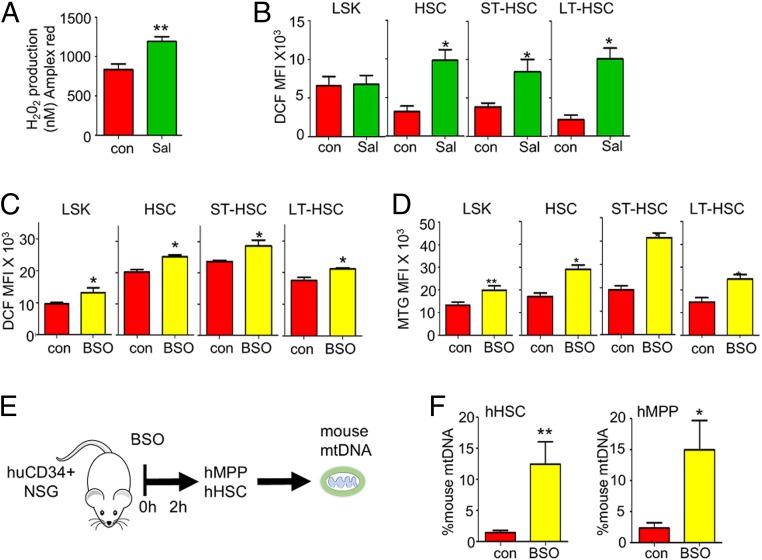
In previous work we have reported that transfer of functional mitochondria from the BM to AML is driven by AML-derived NADPH oxidase 2 (NOX2)-dependent ROS (17). We used the Amplex Red assay to determine if superoxide is elevated in C57BL/6 BM of S. typhimurium and LPS-treated animals. Amplex Red reacts with H2O2 to produce a fluorescent signal, and, following inoculation of animals with either S. typhimurium (72 h) or LPS (2 h), we observed an increase in H2O2 in the BM of the C57BL/6 mice (Fig. 3A and SI Appendix, Fig. S4A). Moreover, ROS (measured by H2DCFDA fluorescence) was elevated in the LSK and HSC populations from S. typhimurium (72 h) and LPS (2 h) treated C57BL/6 mice compared to control animals (Fig. 3B and SI Appendix, Fig. S4B).
Fig. 3.
Superoxide drives mitochondrial increase to the HSC. (A) C57/Bl6 mice were infected with S. typhimurium for 72 h, and then 1 × 106 BM cells were analyzed by Amplex Red assay. (B) C57/Bl6 mice were infected with S. typhimurium for 72 h and analyzed by flow cytometry of H2DCFDA, and fluorescence was assessed in the HSC populations to quantify ROS levels. (C) C57/Bl6 mice were treated with BSO for 16 h and analyzed by flow cytometry, and H2DCFDA fluorescence was assessed. (D) C57/Bl6 mice were treated with BSO for 16 h and analyzed by flow cytometry, and MTG fluorescence was assessed. (E) Schematic diagram of experimental design. Post engraftment hu-NSG mice were injected with BSO for 2 h. The BM was extracted, and the cells were analyzed by flow cytometry or sorted for hHSC (mCD45−, hCD34+, hCD38−, hCD45RA−, hCD90+, hCD49f+), hMPP (mCD45−, hCD34+, hCD38−, hCD45RA−, hCD90−, hCD49f−), and hGMP (mCD45−, hCD34+, hCD38+, hCD45RA+, hCD90−, hCD49f−) cell populations using FACS. (F) mtDNA transfer was quantified using Taqman qPCR with species-specific probes. The graph represents the percentage of mouse mtDNA in the hHSC and hMPP cells after 2 h treatment with LPS versus control (untreated) cells. Human genomic DNA was used to standardize mtDNA. Data shown are means ± SD of n = 5 mice. *P < 0.05; **P < 0.01.
Next, to confirm ROS as the stimulus for mitochondrial transfer, we treated mice with l-buthionine-sulfoximine (BSO) as previously described, to mimic a rise in intracellular ROS concentrations in vivo but in the absence of infection (20). The dose of BSO was selected to induce an increase in intracellular ROS in LSK cells and HSCs at levels comparable to those that we observed in LPS and S. typhimurium-induced ROS elevation (Fig. 3C). We found that mitochondrial content was increased in LSK and HSC populations in BSO-treated mice compared to controls (Fig. 3D). Next, Hu-NSG animals were treated with BSO for 2 h, and then human MPPs and HSCs were isolated and analyzed for mouse mtDNA (Fig. 3E). Fig. 3F shows that mouse mtDNA is significantly increased in the human MPPs and HSCs from BSO-treated Hu-NSG mice. For further assurance that ROS mediates LPS-driven mitochondrial content increase in HSCs, we repeated the experiments in Fig. 3 E and F, this time using the ROS scavenger N-acetyl-cysteine (NAC) to reduce ROS levels. Treatment with NAC suppressed LPS-induced mitochondrial content increase in the LSK and HSC populations and led to a reduced level of ROS in the LSK and HSC populations (SI Appendix, Fig. S5 A and B). To determine if the ROS was derived from NOX2, we investigated LPS-induced mitochondrial transfer in the chronic granulomatous disease (CGD) mouse model, in which the gp91 subunit of NOX2 has been deleted (22). We found that LPS did not increase mitochondrial content in HSCs in the gp91 knockout CGD mice (SI Appendix, Fig. S6 A and B). Together, these data show that ROS is a signal which mediates mitochondrial transfer from the BM microenvironment to HSC populations.
BMSCs but Not Osteoblasts or Macrophages Donate Their Mitochondria to HSCs in Response to Infection.
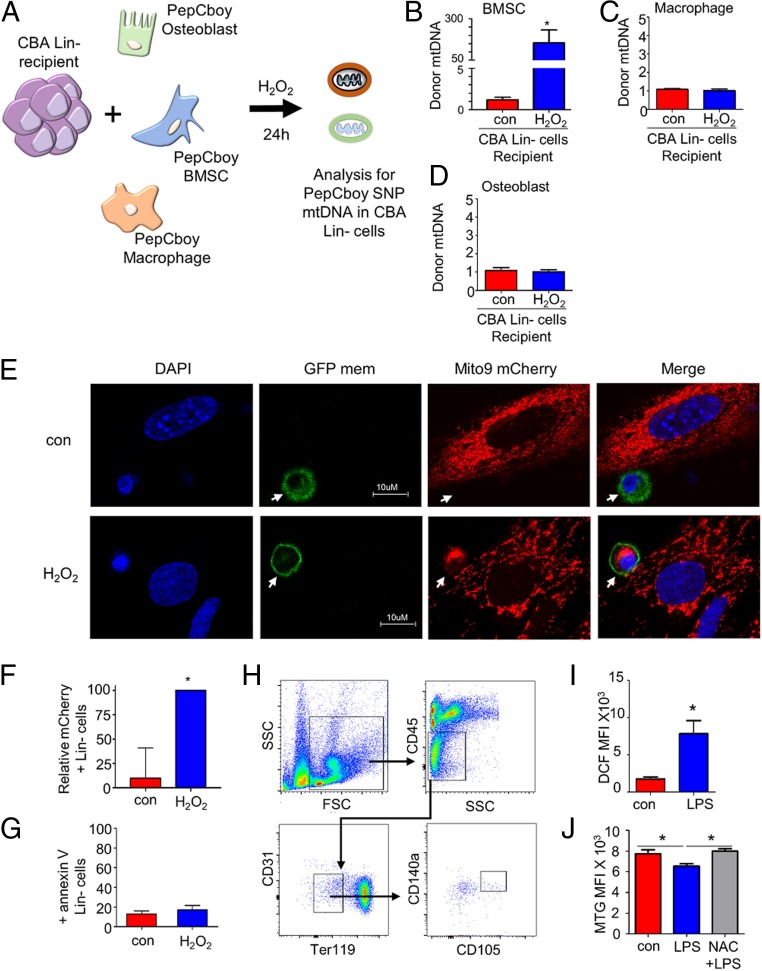
Multiple cell types make up the BM microenvironment (17); moreover, mitochondrial transfer from BMSCs to pulmonary alveoli has been shown to protect against acute lung injury (23). Therefore, we investigated which cells of the BM transfer their mitochondria to HSCs during stressed hematopoiesis (Fig. 4A and SI Appendix, Fig. S7). Fig. 4B shows that BMSCs transfer mitochondria to HSCs in vitro when cultured in the presence of H2O2. There was no increase in mitochondrial transfer from the macrophage or osteoblast when cultured with H2O2 (Fig. 4 C and D). We infected primary mouse BMSCs (mBMSCs) with an rLV.EF1.mCherry lentivirus for stable production of mitochondria-incorporated mCherry-tagged protein, and then lineage-negative cells transduced with rLV.EF1.AcGFP-Mem9 lentivirus (enabling stable labeling of the cell membrane) were cocultured with the rLV.EF1.mCherry mBMSCs for 24 h in the presence or absence of H2O2. Fig. 4E shows that, upon the coculture, the lineage-negative cells acquire the mitochondria-incorporated mCherry fluorescence in the presence of H2O2. Quantification of lineage-negative cells positive for rLV.EF1.mCherry is shown in Fig. 4F. H2O2 treatment of lineage-negative BMSC cocultures did not induce significant apoptosis (Fig. 4G).
Fig. 4.
BMSCs supply mitochondria to HSCs in response to infection. (A) Schematic diagram of experimental design. CBA lineage-negative cells (recipient) were cocultured with C57BL/6 bone marrow-derived macrophage cells, BM osteoblasts, and BMSCs (donor) for 24 h with and without H2O2. The lineage-negative cells were removed and analyzed for PepCboy SNP mtDNA in CBA lineage-negative cells by TaqMan PCR using ND3 probes. (B–D) The percentage of PepCboy mtDNA in CBA lineage-negative cells after 24 h treatment with H2O2 versus control (untreated) cells using the ND3 TaqMan probe. CBA gDNA was used to standardize mtDNA copy number. (E) Representative fluorescent microscopy images of lineage-negative cells (white arrow) lentivirally transduced with the rLV.EF1.AcGFP-Mem9 virus cultured with mBMSCs transduced with rLV.EF1.mCherry-Mito-9 in the absence or presence of 10 µM H2O2 for 24 h. (F) Quantification of rLV.EF1.mCherry-Mito-9 in lineage-negative cells from images presented is taken from 3 independent experiments and 20 lineage cells from each experiment (E). (G) Lineage-negative cells cultured with mBMSCs transduced in the absence or presence of 10 µM H2O2 for 24 h and then stained for Annexin V and analyzed by flow cytometry. (H) Flow cytometry-based analysis of BMSC. (I) Relative ROS in BMSC as measured by H2DCFDA fluorescence. (J) Mitochondrial content in BMSC as measured by MTG after treatment with NAC and LPS or LPS alone. Data shown are means ± SD of n = 5 mice. *P < 0.05.
To determine if infection induced ROS in BMSCs in vivo, we analyzed ROS levels in BMSCs using H2DCFDA (Fig. 4H). Elevated ROS levels were detected in BMSCs from LPS-treated C57BL/6 compared to control animals (Fig. 4I). Moreover, mitochondrial levels were reduced in the BMSCs of LPS-treated C57BL/6 compared to control animals, and this reduction was inhibited by the addition of NAC (Fig. 4J). Using both in vitro and in vivo modeling, these results show that BMSCs are the source of the majority of mitochondria transferred to LSK cells and HSCs in stress hematopoiesis.
PI3K Activates Connexin 43 Gap Junctions to Enable Mitochondrial Movement by Phosphorylation of AKT.
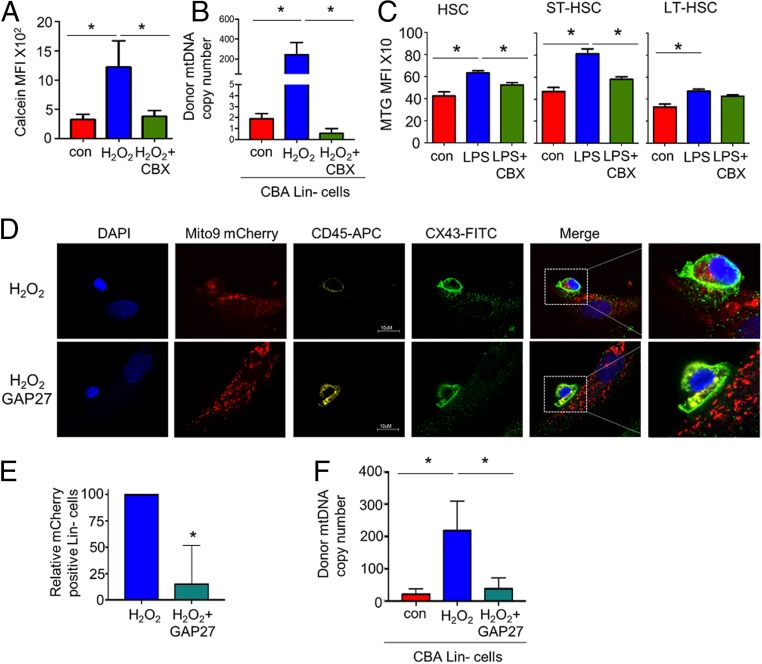
It has been reported that mitochondria can be transferred intracellularly through gap junctions (24). We therefore investigated whether gap junctions regulated the transfer of mitochondria into HSCs in response to LPS. To determine if gap junctions are involved in intercellular communication between the HSCs and the BMSCs, we used the mobile dye calcein (25, 26). Fig. 5A shows that calcein flows from the BMSC into the HSC in the presence of H2O2 versus the control. This transfer of calcein dye was reduced when the BMSCs were cultured in the presence of the gap junction inhibitor carbenoxolone before the H2O2 treatment. Fig. 5B shows that there is also a significant decrease in mtDNA transfer from the BMSCs to the HSCs in vitro when cultured in the presence of carbenoxolone and H2O2 compared to H2O2 alone. To establish if the increase in mitochondrial content in the HSCs after treatment with LPS is regulated by CX43 gap junctions in vivo, we injected C57BL/6 mice with the gap junction inhibitor carbenoxolone before LPS administration and compared them to C57BL/6 mice treated with LPS alone. We observed a decrease in the mitochondrial content within the HSC population when pretreated with carbenoxolone before LPS compared to the animals treated with LPS alone (Fig. 5C). To further evaluate the role of CX43 gap junctions in mitochondrial transfer from BMSCs to HSCs, we used the peptide inhibitor of CX43, GAP27. Immunofluorescent images show that CX43 is expressed in both BMSCs and HSCs and is colocated in the area of mitochondrial transfer, which is inhibited further in the presence of GAP27 (Fig. 5 D and E). Movies S1 and S2 show z-stacks which confirm that the rLV.EF1.mCherry is inside the lineage-negative cells and not on the cell membrane in the H2O2-treated cells but also not in the GAP27 and the H2O2-treated cells. Fig. 5F shows that there is also a significant decrease in mtDNA transfer from the BMSCs to the HSCs in vitro when cultured in the presence of GAP27 and H2O2 compared to H2O2 alone. Taken together, including methods to inhibit gap junctions, these data show that CX43 gap junctions mediate transfer of mitochondria from the BM microenvironment to HSC populations in models of stress hematopoiesis.
Fig. 5.
Connexin channels regulate mitochondrial movement from BMSCs to HSCs. (A) Flow cytometry-based analysis of lineage-negative cells cocultured with mBMSC stained with calcein. The cocultures were treated with 10 µM H2O2 or carbenoxolone and 10 µM H2O2 for 24 h, and the lineage-negative cells were removed and analyzed for calcein mean fluorescence intensity (MFI). (B) CBA-derived lineage-negative cells and PepCboy-derived BMSCs were cocultured with H2O2 or carbenoxolone and 10 µM H2O2 for 24 h. The lineage-negative cells were removed and analyzed for C57BL/6 SNP mtDNA in CBA lineage-negative cells by TaqMan PCR using ND3 probes. (C) C57BL/6 mice were subjected to carbenoxolone (CBX) pretreatment before LPS or control PBS i.p. injections. After 2 h the mice were killed, and the BM was extracted. The populations were analyzed by flow cytometry for mean MTG fluorescence intensity within each population. (D) Immunofluorescent staining of CX43 of lineage-negative cells and BMSCs cocultured with 10 µM H2O2 with and without pretreatment with GAP27 (100 µM). (Magnification: D, 63×.) (E) Quantification of images shown in E numbers of lineage-negative cells positive for mito-9 mCherry. (F) CBA lineage-negative cells cocultured with C57 mBMSC. The cocultures were treated with 10 µM H2O2 or GAP27 (100 µM) for 24 h, and the lineage-negative cells were removed and analyzed for SNP mtDNA in CBA lineage-negative cells by TaqMan PCR using ND3 probes. *P < 0.05.
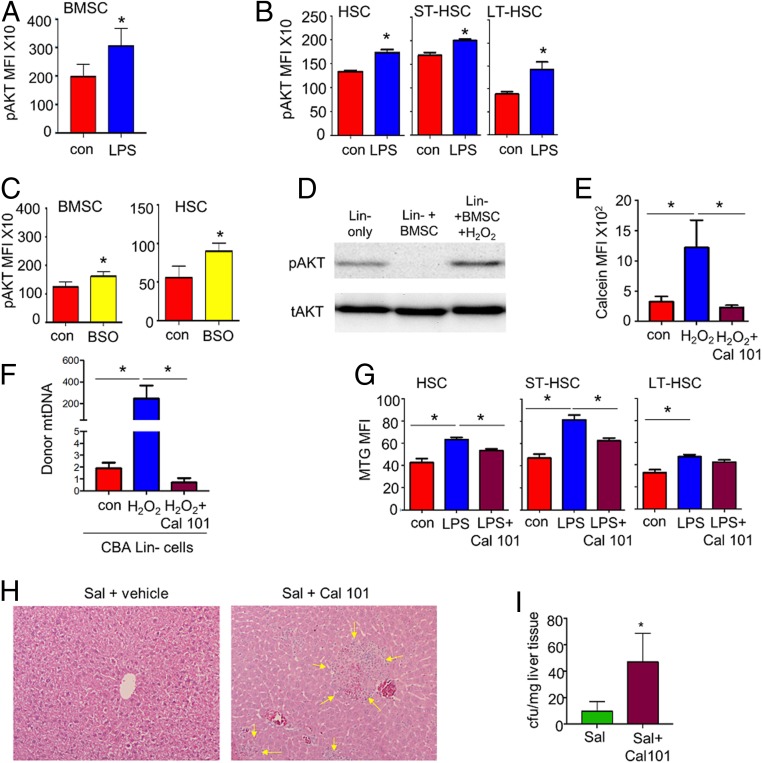
It has previously been shown that increased ROS levels are associated with an increase in the signaling of phosphoinositide-3,4,5-trisphosphate (PIP3) via oxidation of PTEN and subsequent activation of PI3K (27–29). PIP3 activates the downstream signaling component protein kinase AKT, which is required for cell growth and survival (29). Therefore, we looked at AKT phosphorylation (S473) in the BMSCs and HSCs after LPS treatment. Fig. 6 A and B showed that AKT phosphorylation was elevated in both the BMSCs and the HSCs of LPS-treated animals when compared to the untreated controls. BSO treatment also induced AKT phosphorylation in both HSCs and BMSCs (Fig. 6C). Fig. 6D shows increased phosphorylation of AKT (pAKT-S473) in the lineage-negative cells cocultured with BMSCs in the presence of H2O2. Fig. 6E shows that H2O2-induced transfer of calcein dye to lineage-negative cells was inhibited when the BMSCs were cultured in the presence of CAL-101 [inhibitor of PI3 kinase delta (PI3Kδ)]. Moreover, CAL-101 decreased mitochondrial transfer from the BMSCs to HSCs in vitro (Fig. 6F). C57BL/6 mice pretreated with CAL-101 resulted in a decrease in mitochondrial content within the HSC population in response to LPS (Fig. 6G). Finally, to determine the impact of PI3Kδ inhibition on S. typhimurium infection, mice were infected with S. typhimurium for 5 d and treated daily with CAL-101. At day 5, surviving animals were killed and histopathological analysis of liver sections showed increased infection associated with liver injury in CAL101-treated mice as evidenced by areas of hepatocellular death and immune cell infiltration (Fig. 6H). Accordingly, further analysis showed increased bacterial colony-forming units (CFUs) in CAL101-treated livers after Salmonella infection (Fig. 6I). Results show that inhibition of PI3Kδ renders the immune system less capable of fighting S. typhimurium infection. Taken together, these data indicate that PI3K signaling regulates mitochondrial transfer from BMSCs to HSCs through a pAKT- and CX43-dependent mechanism without which the immune response to acute bacterial infection is compromised.
Fig. 6.
ROS-derived pAKT drives mitochondrial transfer. (A) C57BL/6 mice were subjected to LPS or control PBS i.p. injections. After 2 h the mice were killed, and the BM extracted. BMSCs (A) and HSCs (B) were analyzed by flow cytometry for pAKT. (C) C57BL/6 mice were subjected to BSO or control PBS i.p. injections. After 2 h the mice were killed, and the BM was extracted. BMSCs and HSCs were analyzed by flow cytometry for pAKT. (D) Western bot analysis of lineage-negative cells cocultured with mBMSCs. The cocultures were treated with 10 µM H2O2 for 30 min, and the lineage-negative cells were removed and analyzed for pAKT expression. (E and F) Flow cytometry-based analysis of lineage-negative cells cocultured with mBMSCs stained with calcein. The cocultures were treated with 10 µM H2O2 or CAL-101 and H2O2 for 24 h, and the lineage-negative cells were removed and analyzed for calcein MFI (E) or (F) for SNP mtDNA in CBA lineage-negative cells by TaqMan PCR using ND3 probes. (G) C57BL/6 mice were subjected to LPS with or without pretreatment with CAL-101 or control PBS i.p. injections. After 2 h the mice were killed, and the BM was extracted. HSCs were analyzed for mitochondrial content using MTG. (H and I) C57/Bl6 mice were infected with S. typhimurium and then split into 2 groups. The first group received vehicle control and the test group received a daily dose of CAL-101 (30 mg/kg) by oral gavage. Animals were killed on day 5, and livers were isolated and analyzed for CFUs/mg of tissue and sectioned and stained with hematoxylin and eosin. (Magnification: H, 63×.) *P < 0.05.
Conclusions
Here we report that HSCs acquire mitochondria from BMSCs within the protective BM microenvironment in response to acute bacterial infection. This process facilitates a rapid shift within the HSCs from the baseline, quiescent state—glycolytic metabolism—toward OXPHOS, which is subsequently followed by leukocyte expansion. Mitochondria transfer occurs early in the mammalian response to infection, preceding the onset of the transcriptional changes necessary for increased mitochondrial biogenesis within HSCs and thus facilitating rapid leukocyte expansion. Furthermore, we identified that increased ROS level mediates PI3 kinase activation which drives mitochondrial transfer from the microenvironment to the HSCs. Overall, these results provide insights into the bioenergetic changes within the hematopoietic system which underpin the mammalian response to bacterial infection.
The challenge of infection has been a significant selective pressure on mammalian evolution and remains a leading global cause of mortality. Of the top 10 global causes of death in 2016, 3 are infectious or communicable diseases: lower respiratory tract infection, diarrheal illness, and tuberculosis (13). Faced with the challenge of infection, the hematopoietic response must be robust and rapid and result in a massive amplification of myeloid cells over a short period of time. Neutrophils are the most abundant leukocyte in mammals and contribute to the resolution of infection through a series of mechanisms including phagocytosis, enzyme-mediated lysis, capture within neutrophil extracellular traps, and activation of lymphocytes (30). Neutrophils are used up in large quantities, and the hematopoietic system rapidly adapts to the increased demand by switching from steady-state to emergency granulopoiesis (31). The transition from steady-state to emergency hematopoiesis is established to involve a complex remodeling and interplay between hematopoietic and nonhematopoietic cells of the BM microenvironment mediated by cytokines and growth factors (32–34) and places a significant metabolic demand on the hematopoietic system. Moreover, BMSCs have been shown to enhance hematopoiesis during inflammatory conditions to support the replenishment of innate effector cells and to prevent the exhaustion of the HSC pool (35). Here, we show how mitochondrial transfer facilitates the rapid onset of emergency hematopoiesis by allowing an increase in mitochondrial mass and subsequent metabolic changes in HSCs without the need to “wait” for intracellular increase in mitochondrial biogenesis.
Mitochondrial trafficking between cells is now increasingly recognized as a fundamental interaction in both malignant and nonmalignant tissue. Others have reported mitochondrial transfer from stromal cells to cancer cell lines including to breast (36), lung (37, 38), and melanoma (39) cells. Moreover, a recent paper by Bajzikova et al. (40) has shown that cancer cells acquire mitochondria from host stroma to drive OXPHOS-derived dihydroorotate dehydrogenase pyrimidine synthesis and that this process does not require mitochondrial ATP. In addition, we have previously shown how AML-derived NADPH oxidase-2–derived superoxide drives mitochondrial transfer from BMSCs to leukemic blasts in a paracrine protumoral mechanism. This transfer of mitochondria promoted OXPHOS in the tumor cells, and inhibition of NADPH oxidase-2 was able to prevent mitochondrial transfer, increase AML apoptosis, and improve NSG AML mouse survival (17). However, an inhibitor of dihydroorotate dehydrogenase, BAY2402234, has been shown to induce differentiation of AML blasts, which suggests that multiple cellular functions require import of mitochondria, other than just OXPHOS-driven ATP production (41).
In the nonmalignant setting, mitochondrial transfer from BM-derived stromal cells to pulmonary alveoli protects against acute lung injury (23), and transient focal cerebral ischemia in mice induces entry of astrocytic mitochondria into adjacent neurons, which amplifies cell survival signals (42). At a similar time, others reported that BM microenvironment myeloid cell-derived NADPH oxidase-2–derived ROS plays a critical role in mediating emergency granulopoiesis during acute infection (20). Therefore, in context, our present study identifies ROS-driven protumoral mitochondrial transfer in AML as a process which appears to have been hijacked from the highly evolved mammalian HSC response to infection. In doing so, we provide a paradigm to be investigated in other malignancies which are established to rely on mitochondrial transfer for tumor growth; such cancers evolve from tissue-specific stem and progenitor cells which utilize mitochondrial transfer as a response to injury or cellular stress.
In conclusion, we report that mitochondria are transferred from the BMSCs into HSCs under the regulation of superoxide in response to bacterial infection. Importantly, we have discovered that mitochondrial transfer occurs before mitochondrial biogenesis in a system which has evolved in mammals to support the required rapid granulocytic response to acute bacterial infection.
Materials and Methods
Animals.
NOD SCID Il2rg knockout NSG (CD45.1) and B6.SJL-PtprcaPep3b/BoyJ (CD45.1) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). C57BL/6J mice (CD45.2), B6.129S-Cybbtm1Din/J mice, and CBA mice were purchased from Charles River Laboratories. Animals were housed in a specific pathogen-free facility. All animal work in this study was carried out in accordance with regulations set by the United Kingdom Home Office and the Animal Scientific Procedures Act of 1986. Mice were 8 to 12 wk of age (with the exception of mice that were used for transplantation in which case 3- to 4-wk-old mice were used), and both genders were used for experiments.
Human Cord Blood.
HPCs were isolated from cord blood at the Norfolk and Norwich University Hospital following informed consent and under approval of the UK NHS Health Research Authority (ref07/H0310/146). The samples collected were de-identified before use in this study.
Cells.
Hematopoietic populations were isolated from mouse BM. Mouse lineage-negative cells were depleted from mouse BM using a direct cell lineage depletion kit. (Miltenyi Biotec). The lineage-negative cells were expanded in Dulbecco’s Modified Eagle Media (DMEM) containing 10% fetal bovine serum (FBS) plus 1% penicillin-streptomycin supplemented with mSCF (100 ng/mL), mIL3 (10 ng/mL), and hIL6 (100 ng/mL) (Peptrotech). mBMSCs were isolated from the mouse BM by adherence to tissue culture plastic and then expanded in minimum essential media (MEM) containing 20% FBS plus 1% penicillin-streptomycin. mBMSC markers were confirmed by flow cytometry for expression of CD105+, CD140a+ CD31−, Ter119−, and CD45−. mBM macrophages were isolated by adherence when cultured in MEM containing 20% FBS plus 1% penicillin-streptomycin supplemented with macrophage colony-stimulating factor (CSF) (20 ng/mL). The media were refreshed daily until day 3 when the media were replaced with MEM containing 20% FBS plus 1% penicillin-streptomycin supplemented with macrophage CSF (10 ng/mL) (Peptrotech). Osteoblasts were isolated from mouse BM by adherence when cultured in MEM containing 20% FBS plus 1% penicillin-streptomycin supplemented with macrophage CSF (10 ng/mL) (Peptrotech) and Prostaglandin E2 (PGE2) (10−7M) (Peptrotech) for 3 d. On day 3 the media were changed to MEM containing 20% FBS plus 1% penicillin-streptomycin supplemented with macrophage CSF (10 ng/mL), PGE2 (10−7M), and RANKL (10 ng/mL) (Peptrotech). Human CD34+ cells were enriched from whole cord blood using CD34+ Magnetic Bead separation (Miltenyi Biotec). Culture of all hematopoietic populations was carried out in 5% CO2 at 37 °C.
Method Details.
Cell isolation and preparation.
BM isolation was prepared by isolating the tibia, femur, and pelvis of each mouse. The bone was cut in the middle and placed in a 0.5-mL Eppendorf tube in which a hole was made to allow the removal of the BM, placed in an intact 1.5-mL Eppendorf tube, and centrifuged on full power for 6 s to collect the BM cells. The BM pellet from each mouse was pooled and washed in phosphate buffer solution (PBS), and, where needed, the red blood cells were lysed using 1× red blood cell lysis buffer (ThermoFisher, Waltham, MA) and centrifuged at 1,200 × g for 5 min, and the pellet was resuspended in antibody mixtures in PBS.
Flow cytometry and cell sorting.
Antibody mixtures were prepared in 1× PBS and incubated with BM cells for at least 30 min at 4 °C. In experiments using MTG, the cells were incubated with MTG (30 nM, Invitrogen) with verapamil (50 μM, Sigma-Aldrich, St. Louis) at room temperature for 30 min, washed twice in 1× PBS, and centrifuged at 1,200 × g for 5 min before staining with antibody mixture. For experiments using H2DCFDA (1 nM, Invitrogen), the cells were incubated with H2DCFDA at room temperature for 30 min, washed twice in 1× PBS, and centrifuged at 1,200 × g for 5 min before staining with antibody mixture. For flow cytometric cell sorting of BM cell populations, pellet was resuspended in antibody mix, and cells were sorted directly into lysis buffer. Flow cytometry was carried out using a FACSCanto II flow cytometer (BD Bioscience), and cell sorting was performed on a BD FACSMelody (BD Bioscience). Data were analyzed using FlowJo (TreeStar, Ashland, OR). See Fig. 1B for specific gating strategies.
BM transplantation.
For the C57 NSG allograft model (C57NSG), the NSG mouse model expressing the CD45.1 allele antigen was used in transplant experiments in which isolated lineage-negative BM cells from C57 Bl/6 mice expressing the CD45.2 allele were transplanted. The 2 × 105 isolated C57BL/6 lineage-negative BM cells were then injected into the tail vein of nonirradiated 3- to 4-wk-old NSG mice. Engraftment of the C57BL/6 BM was analyzed by flow cytometry through analysis of CD45.2 cells in the peripheral blood. These animals were then treated with 1 mg/kg LPS or control PBS for 2 h and killed by exposure to CO2. BM was isolated and sorted for donor LSK (Lin−, Sca-1+, c-Kit) and donor HSC (Lin− Sca-1+ c-Kit CD48−CD150+).
For the CD34+ hematopoietic progenitor cell xenograft model (hu-NSG), 2 × 105 CD34+ cord blood cells were injected into the tail vein of nonirradiated 3- to 4-wk-old female NSG mice. CD34+ cell engraftment was monitored by peripheral blood analysis for human CD45 cells. At 12 wk post injection, mice were treated with 1 mg/kg LPS for 2 h and killed by exposure to CO2. BM was harvested, and cell sorting was performed for human HSCs (hHSCs) (mCD45−, hCD34+, hCD38−, hCD45RA−, hCD90+, hCD49f+), hMPPs (mCD45-, hCD34+, hCD38−, hCD45RA−, hCD90−, hCD49f−), and hGMPs (mCD45−, hCD34+, hCD38+, hCD45RA+, hCD90−, hCD49f−). BM was also analyzed for human cell engraftment by human CD45+ cells. If more than 1% of human CD45 cells was detected in the BM, the CD34+ cells were determined to be engrafted.
mtDNA quantification.
DNA was extracted from sorted or cultured hematopoietic populations using the GenElute Mammalian Genomic DNA Miniprep Kit (Sigma Aldrich, St. Louis) according to the manufacturer’s instructions.
For the C57NSG model and coculture experiments, relative quantitative real-time PCR using the Taqman probes ND3 and COX3 (ThermoFisher) was performed on the purified DNA. ND3 and COX3 Taqman assays were run in simplex reactions, and master mixes were generated. One microliter of Taqman assay (containing primers and probe) was mixed with 2.5 μL of TaqPath ProAmp enzyme (ThermoFisher) and 1.5 μL of water. After preamplification (60 °C for 30 s and 95 °C for 5 min), the PCRs were amplified for 50 cycles (95 °C for 15 s and 60 °C for 1 min) and cooled (40 °C for 30 s) on a 384-well LightCycler 480 (Roche, Burgess Hill, UK). Each mtDNA level was normalized against the genomic DNA (gDNA). The relative mtDNA:gDNA ratio was calculated using the ΔΔCt method.
In the hu-NSG model, predesigned Taqman assays were obtained from (ThermoFisher), encompassing both human and mouse mtDNA and gDNA. The human and mouse mtDNA Taqman assays were run in duplex reactions. The human and mouse gDNA Taqman assays were ran in simplex reactions, and simplex and duplex master mixes were generated. For the simplex master mix, 1 μL of Taqman assay (containing primers and probe) was mixed with 2.5 μL of TaqPath ProAmp enzyme (ThermoFisher) and 1.5 μL water. For the duplex master mix, 0.5 μL of each Taqman assay (total 1 μL) was mixed with 2.5 μL of TaqPath ProAmp enzyme and 1.5 μL water. This master mix (5 μL) was then added to the DNA (1 μL) on the PCR plate. The PCR plate was sealed, centrifuged at 1,000 × g for 1 min and loaded into the Lightcycler using the program previously described. mtDNA copy numbers were determined for both human and mouse mitochondria applying the ∆∆Ct method, using human genomic telomerase reverse transcriptase to normalize results. These values were used to generate the percentage of mouse mitochondria in the human cells to quantify mitochondrial transfer.
Amplex Red superoxide detection assay.
Amplex Red superoxide detection assay (ThermoFisher) was used to specifically measure the superoxide levels generated. This reaction was carried out per the manufacturer’s specifications. BM was plated on a black 96-well plate with a transparent base in a volume of 50 μL FluoroBrite DMEM supplemented with 10% FCS. A master mix was created by mixing 0.5 μL Amplex Red reagent, 1 μL horseradish peroxidase, and 48.5 μL 1× reaction buffer per sample. Fifty microliters of this master mix was added to the cell suspension, and fluorescence was measured using the FLUOstar Omega microplate reader (BMG LABTECH, Ortenberg, Germany). A hydrogen peroxide standard curve was performed on each 96-well plate measured to determine an accurate superoxide concentration in the samples tested.
Coculture experiments.
mBMSCs, mBM osteoblasts, or mBM macrophages were seeded in a 24-well plate in normal growth media. Once 70% confluent 0.25 × 106 lineage-negative cells were then cocultured with the mBM cells in the absence or presence of H2O2 for 24 h and the CX43 peptide inhibitor GAP27. Lineage-negative cells were removed and sorted for CD45-specific markers into cell lysis solution. DNA was extracted, and mtDNA quantification was performed using qPCR.
Immunofluorescence.
mBMSCs were isolated from the bone morrow as previously described and seeded at a density of 5 × 105 BMSCs in 500 μL of penicillin-streptomycin–free DMEM supplemented with 10% FCS. A total of 0.5 μL of the rLV.EF1.mCherry-Mito-9 lentivirus (Clontech Takara Bio Europe, Saint-Germain-en-Laye, France) (0.5 × 106 virus particles) was added to the medium. One milliliter of DMEM was added after 24 h, and mBMSCs were cultured for a further week to ensure that no residual lentivirus remained. Successful transduction was assessed by the detection of mCherry fluorescence in the mBMSCs by fluorescent microscopy. In addition, the rLV.EF1.AcGFP-Mem9 lentivirus was purchased from Clontech Takara Bio Europe, enabling the stable tagging of plasma membranes with a green fluorescent protein (GFP) fluorophore. Mouse lineage-negative cells were depleted from mouse BM using a direct cell lineage depletion kit (Miltenyi Biotec), and cells were transduced with this lentivirus and cultured for 72 h prior to use. Successful transduction was assessed by the detection of GFP fluorescence in the lineage-negative cells by fluorescent microscopy. If successful transduction was observed, mBMSCs were seeded at 5 × 104 cells per well of a 24-well plate on tissue culture coverslips coated with 1 μg/mL poly-d-lysine (Sigma-Aldrich). Transduced lineage-negative cells (0.25 × 106) were then cocultured with the mBMSCs for 24 h in the absence or presence of H2O2. Cells were fixed in 4% paraformaldehyde and stained for 15 min in DAPI (ThermoFisher). After washing 3 times with 1× PBS, cells were mounted with mounting media (ThermoFisher). Immunofluorescent images were acquired with a Zeiss Axioplan 2ie microscope. Confocal images were acquired on Zeiss LSM 800 Axio Observer.Z1 confocal microscope with a 63× water objective (Carl Zeiss).
Seahorse.
XFp flux cartridges were hydrated in XF Calibrant overnight at 37 °C. LSK cells were isolated from the bone marrow of 2 mice, and 2 mice were treated either with LPS or PBS by fluorescence-activated cell sorting (FACS) into conventional Seahorse base media. A total of 50,000 cells were plated onto one well of a Seahorse XFp culture plate coated with poly-d-lysine and centrifuged briefly to achieve a uniform monolayer of cells. Cells were equilibrated in a humidified non-CO2 incubator until the start of the assay. Flux cartridges were loaded with Oligomycin (2 μM), carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (1 μM), and Rotenone (0.5 μM) according to the manufacturer’s instructions. Oxygen consumption rate and basal extracellular acidification rate values were obtained using the XFp Mito Stress Kit. Metabolic parameters were derived from calculations based on the manufacturer’s instructions. All results were normalized to input cell number. Due to the feasibility of cell number isolation, experiments are represented as one replicate per cell type, per condition.
S. typhimurium and LPS elicited stressed hematopoiesis.
Glycerol stock of S. typhimurium (SL1344-JH3009) was a kind gift from Isabelle Hautefort, Quadram Institute Bioscience, Norwich, United Kingdom. The stock was plated on Luria Broth agar plates, and the colonies were inoculated and grown overnight in 5 mL of Luria Broth with 0.3 M NaCl (LBS). The overnight culture was then diluted 1:100 in LBS and grown until the culture optical density (ΔOD600nm) of 1.2 to 1.4 (late exponential phase). This is the time point at which SPI1 invasion genes are turned on in S. typhimurium. The bacterial culture was then centrifuged at 8,000 × g for 7 min before washing bacterial cells twice in 25 mL of sterile Dulbecco’s phosphate-buffered saline (DPBS) at room temperature. Finally, we resuspended the bacterial cells in sterile DPBS at concentration of 1 to 5 × 108 CFUs per 100 μL of DPBS (knowing that ΔOD600nm 1.26 corresponds to 7.53 × 108 CFUs/mL).
C57BL/6 mice were treated with streptomycin (20 mg/mL) 24 h prior to S. typhimurium infection. Mice were then left untreated or infected with 100 μL of 1 × 108 CFUs of S. typhimurium (SL1344-JH3009) by oral gavage for 72 h. C57BL/6 mice, CGD mice, or Hu-NSG mice were treated with LPS or control PBS for 2 or 16 h. The mice were killed by exposure to CO2, and the BM was analyzed by flow cytometry and cell sorting.
Analysis of ROS in vivo.
C57BL/6 mice were treated with 100 mg/kg BSO for 2 h and killed by exposure to CO2. The BM was extracted for flow cytometry-based lineage analysis. C57BL/6 mice were also pretreated with 500 mg/kg NAC for 1 h, followed by 1 mg/kg LPS, and after 2 h the mice were killed, and the BM was extracted for flow cytometry-based lineage analysis. The BM was extracted for flow cytometry-based lineage analysis of the LSK and HSC populations for mean MTG intensity as a measure for mitochondrial content and H2DCFDA as a measure of ROS.
Real-time PCR for mitochondrial biogenesis genes.
The ReliaPrep RNA cell miniprep system (Promega, Southampton, UK) was used to extract whole-cell RNA. Nugen PicoSL WTA (Redwood City, CA) was used to generate first-strand, followed by double-strand complementary DNA (cDNA), followed by cDNA amplification. A qRT-PCR assay was performed with the SYBR-green technology (PCR Biosystems). PCRs were amplified for 45 cycles (95 °C/15 s, 60 °C/10 s, 72 °C/10 s), after preamplification (95 °C/60 s), on a Roche 384-well/96-well LightCycler480. mRNA expression was normalized against glyceraldehyde 3-phosphate dehydrogenase using the comparative cycle threshold method.
Granulocyte and monocyte CFUs.
BM cells (1 × 104) from control and LPS-treated mice were seeded in a semisolid Methocult (Stemcell Technologies, Cambridge, UK) containing penicillin-streptomycin for detection of granulocytes, granulocyte monocytes, granulocyte monocyte megakaryocytes. After 1 wk of culture colony types were counted per well.
Bacterial CFUs and liver histology.
Livers were homogenized in sterile PBS solution and plated in LB agar plates for 24 h, after which colonies were counted. Data are shown in CFUs relative to milligrams of liver tissue. Liver tissues were fixed in neutral formalin for 24 h and embedded afterward in paraffin blocks that were further sectioned, dewaxed, and hydrated. Slides were stained with hematoxylin and eosin for histopathological analysis.
Quantification and Statistical Analysis.
For statistical comparison of 2 groups, unpaired two-tailed Student’s t test was used. When more than 2 groups were compared, one-way ANOVA followed by Kruskal–Wallis test was performed using Prism version 7.00 for Windows (GraphPad, La Jolla, CA). Due to variability in the data, statistical comparison of in vivo work was performed without assumption of normal distribution using the Mann–Whitney U test. Differences among group means were considered significant when the probability value, P, was less than 0.05*, 0.01**, or 0.001***. Sample size (n) represents the number of biological replicates. No statistical methods were used to predetermine sample size.
Data Availability Statement.
All relevant data are within the manuscript and its supporting information files. The sequences for the mitochondrial SNP analysis are in ref. 43.
Supplementary Material
Acknowledgments
We thank the Norwich Research Park (NRP), The Rosetrees Trust, The Big C, and The National Health Service; Dr. Allyson Tyler, Dr. Ian Thirkettle, and Dr. Karen Ashurst (Norfolk and Norwich University Hospital) for technical assistance; Professor Richard Ball, Professor Mark Wilkinson, and Dr. Rachel Stanley at the NRP Biorepository (UK) for supporting primary tissue collection; the team at the Disease Modelling Unit of the University of East Anglia for assistance with the in vivo studies; and Robert Kingsley, Quadram Institute, for providing S. typhimurium for the in vivo studies.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913278116/-/DCSupplemental.
References
- 1.Orkin S. H., Zon L. I., Hematopoiesis: An evolving paradigm for stem cell biology. Cell 132, 631–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCracken M. N., et al. , Normal and neoplastic stem cells. Cold Spring Harb. Symp. Quant. Biol. 81, 1–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mendelson A., Frenette P. S., Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 20, 833–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anthony B. A., Link D. C., Regulation of hematopoietic stem cells by bone marrow stromal cells. Trends Immunol. 35, 32–37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simsek T., et al. , The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nombela-Arrieta C., et al. , Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat. Cell Biol. 15, 533–543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takubo K., et al. , Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell 12, 49–61 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vannini N., et al. , Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat. Commun. 7, 13125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito K., et al. , Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 354, 1156–1160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Almeida M. J., Luchsinger L. L., Corrigan D. J., Williams L. J., Snoeck H. W., Dye-independent methods reveal elevated mitochondrial mass in hematopoietic stem cells. Cell Stem Cell 21, 725–729.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warr M. R., Passegué E., Metabolic makeover for HSCs. Cell Stem Cell 12, 1–3 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Inoue S., et al. , Mitochondrial respiration defects modulate differentiation but not proliferation of hematopoietic stem and progenitor cells. FEBS Lett. 584, 3402–3409 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Anonymous ; World Health Organization , Global health estimates 2016: Deaths by cause, age, sex, by country and by region, 2000-2016. https://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death. 2018. Accessed 18 December 2018.
- 14.Ho T. T., et al. , Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludin A., et al. , Reactive oxygen species regulate hematopoietic stem cell self-renewal, migration and development, as well as their bone marrow microenvironment. Antioxid. Redox Signal. 21, 1605–1619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGuire V. A., Arthur J. S., Stress-induced haematopoietic stem cell proliferation: New roles for p38α and purine metabolism. Stem Cell Investig. 3, 64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marlein C. R., et al. , NADPH oxidase-2 derived superoxide drives mitochondrial transfer from bone marrow stromal cells to leukemic blasts. Blood 130, 1649–1660 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Moschoi R., et al. , Protective mitochondrial transfer from bone marrow stromal cells to acute myeloid leukemic cells during chemotherapy. Blood 128, 253–264 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Feasey N. A., Dougan G., Kingsley R. A., Heyderman R. S., Gordon M. A., Invasive non-typhoidal salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 379, 2489–2499 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak H.-J., et al. , Myeloid cell-derived reactive oxygen species externally regulate the proliferation of myeloid progenitors in emergency granulopoiesis. Immunity 42, 159–171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong L. F., et al. , Horizontal transfer of whole mitochondria restores tumorigenic potential in mitochondrial DNA-deficient cancer cells. eLife 6, e22187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollock J. D., et al. , Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat. Genet. 9, 202–209 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Islam M. N., et al. , Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat. Med. 18, 759–765 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osswald M., et al. , Brain tumour cells interconnect to a functional and resistant network. Nature 528, 93–98 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Juul M. H., Rivedal E., Stokke T., Sanner T., Quantitative determination of gap junction intercellular communication using flow cytometric measurement of fluorescent dye transfer. Cell Adhes. Commun. 7, 501–512 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Tomasetto C., Neveu M. J., Daley J., Horan P. K., Sager R., Specificity of gap junction communication among human mammary cells and connexin transfectants in culture. J. Cell Biol. 122, 157–167 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor K. M., et al. , Mitochondrial H2O2 regulates the angiogenic phenotype via PTEN oxidation. J. Biol. Chem. 280, 16916–16924 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Covey T. M., Edes K., Fitzpatrick F. A., Akt activation by arachidonic acid metabolism occurs via oxidation and inactivation of PTEN tumor suppressor. Oncogene 26, 5784–5792 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kwon J., et al. , Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc. Natl. Acad. Sci. U.S.A. 101, 16419–16424 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brinkmann V., et al. , Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Manz M. G., Boettcher S., Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302–314 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Hérault A., et al. , Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature 544, 53–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boettcher S., Manz M. G., Regulation of inflammation- and infection-driven hematopoiesis. Trends Immunol. 38, 345–357 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Silberstein L., et al. , Proximity-based differential single-cell analysis of the niche to identify stem/progenitor cell regulators. Cell Stem Cell 19, 530–543 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ziegler P., Boettcher S., Takizawa H., Manz M. G., Brümmendorf T. H., LPS-stimulated human bone marrow stroma cells support myeloid cell development and progenitor cell maintenance. Ann. Hematol. 95, 173–178 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Pasquier J., et al. , Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J. Transl. Med. 11, 94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spees J. L., Olson S. D., Whitney M. J., Prockop D. J., Mitochondrial transfer between cells can rescue aerobic respiration. Proc. Natl. Acad. Sci. U.S.A. 103, 1283–1288 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmad T., et al. , Miro1 regulates intercellular mitochondrial transport & enhances mesenchymal stem cell rescue efficacy. EMBO J. 33, 994–1010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan A. S., et al. , Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab. 21, 81–94 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Bajzikova M., et al. , Reactivation of dihydroorotate dehydrogenase-driven pyrimidine biosynthesis restores tumor growth of respiration-deficient cancer cells. Cell Metab. 29, 399–416.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christian S., et al. , The novel dihydroorotate dehydrogenase (DHODH) inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia 33, 2403–2415 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Hayakawa K., et al. , Transfer of mitochondria from astrocytes to neurons after stroke. Nature 535, 551–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X., et al. , Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res. 19, 159–165 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and its supporting information files. The sequences for the mitochondrial SNP analysis are in ref. 43.