Abstract
Swine diarrhea can be caused by multiple agents, including porcine epidemic diarrhea virus (PEDV), porcine sapelovirus (PSV), and porcine sapovirus (SaV). We designed a one-step triplex reverse-transcription PCR (RT-PCR) detection method including 3 pairs of primers that focused on the S1 gene of PEDV, a conserved gene of PSV, and the VP1 gene of SaV. The optimal concentrations of upstream and downstream primers in the triplex RT-PCR were 0.24 μM for PEDV, 0.15 μM for PSV, and 0.2 μM for SaV, and the optimal annealing temperature was 55.5°C. Triplex RT-PCR assessment of 402 piglet diarrhea samples was compared with conventional individual RT-PCR. Concordance rates in both tests for individual viruses were 100%, 97.6%, and 94.4% for PEDV, PSV, and SaV, respectively. PEDV, PSV, and SaV were detected in 57.2%, 10.4%, and 9.0% of the samples, respectively. The high sensitivity and specificity of this triplex RT-PCR–based detection method for PEDV, PSV, and SaV could allow rapid detection and analysis of mixed infections by these 3 viruses.
Keywords: porcine epidemic diarrhea virus, porcine sapelovirus, porcine sapovirus, triplex RT-PCR
Porcine epidemic diarrhea virus (PEDV; family Coronaviridae, genus Alphacoronavirus) is the cause of PED, which is characterized by acute enteritis and watery diarrhea. The morbidity in piglets (mainly within 7 d of birth) can be close to 100%, and the mortality rate is often 80–100%.18 Porcine sapelovirus (PSV; family Picornaviridae, genus Sapelovirus, species Sapelovirus A) is a single-stranded, positive-sense RNA virus that can cause not only neurologic signs, diarrhea, reproductive disorders, and pneumonia, but also villus atrophy and crypt hyperplasia. PSV is excreted in feces, and can be found in large amounts in the environment. The infected pig continues to excrete the virus after recovery; thus, the infection rate is high in the source herd.17 The PSV-csh strain was first isolated from a case of severe gastroenteritis, respiratory distress, and polioencephalomyelitis in China.10 Among the viruses included in family Caliciviridae are noroviruses and sapoviruses, such as porcine norovirus (NoV), which causes asymptomatic infection in adult pigs, and porcine sapovirus (SaV), which infects pigs of all ages, especially weaned animals, causing intestinal disease and small bowel lesions in specific pathogen–free piglets,4 leading to diarrhea. The first outbreak of gastroenteritis caused by porcine SaV in piglets in China was reported in 2008.20 PSV or SaV may coinfect with PEDV or other porcine viruses, such as group A and C rotaviruses.11 Gastroenteritis and diarrhea caused by PSV and SaV cannot be distinguished easily from PED, that is, clinical signs and pathologic manifestations are not specific. PSV and SaV are not included in routine detection programs for porcine pathogens, indicating that insufficient attention may have been paid to these viruses in the etiologic analysis of diarrhea. Therefore, we established a triplex reverse-transcription PCR (RT-PCR) detection method for PEDV, PSV, and SaV. The technique was applied to laboratory testing of diarrhea samples to achieve rapid detection, screening, and differential detection of the 3 pathogens, providing a basis for the diagnosis and prevention of swine diarrhea.
The following genomes were selected from GenBank for homology analysis (DNASTAR; Madison, WI): 54 PEDV complete genomes including standard strain CV777 and endemic PEDV strains; 8 PSV complete genomes including YC2011, V13, csh-2011, and JD2011 strains; and 10 SaV GIII (which infects porcine species)20 complete genomes including AF182760, AY425671, FJ387164, JX678943, and KF204570. Primers were designed (Primer Premier; Premier Biosoft, Palo Alto, CA) based on: the highly conserved region of the PSV genome sequence; PEDV M, N, S genes; and SaV VP1 gene. Using the software evaluation score to avoid interference between multiple primers in the same reaction, the BLAST specificity comparison, and the highest detection rate on positive clinical samples, 3 pairs of primers were selected (Table 1) and synthesized (GeneWiz Biotec, Suzhou, Jiangsu, China). PEDV-TGEV-PoRV triple attenuated vaccine (Zhengye Biological, Jilin, Jilin, China; PEDV, transmissible gastroenteritis virus, TGEV; porcine rotavirus, PoRV), classical swine fever virus (CSFV), porcine circovirus 2 (PCV-2), porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus (PRV), PSV, SaV, and porcine parvovirus (PPV) were obtained from the Laboratory of Animal Disease Prevention and Control Center of Jinhua City, Zhejiang province, China.
Table 1.
RT-PCR primers designed for triplex detection of porcine enteric viruses.
| Primer | Sequence (5′–3′) | Location | GenBank accession |
|---|---|---|---|
| PEDV-F | CTGCCAATGTATTTGCCAC | 21341-21359 | AF353511 (CV777) |
| PEDV-R | GGAAGTTCCTTGAACCTCG | 21999-21981 | AF353511 (CV777) |
| PSV-F | TGCTTGAGGAGTCGGAGAG | 5102-5120 | JX286666 (YC2011) |
| PSV-R | GCCCTGCACAACTGCTTTC | 5529-5510 | JX286666 (YC2011) |
| SaV-F | TACGGGGGAATAGGTTT | 5855-5871 | AF182760 |
| SaV-R | CAGCCACATCTGGGTAGT | 6100-6083 | AF182760 |
PEDV = porcine epidemic diarrhea virus; PSV = porcine sapelovirus; RT-PCR = reverse-transcription PCR; SaV = sapovirus.
Because watery diarrhea is usually caused by viral infection, stool samples from 402 piglets (1–28 d old, preweaning, mainly newborn piglets 1–7 d old) with watery diarrhea were collected from 42 pig farms in 4 regions of eastern China (Zhejiang, Jiangsu, Fujian Provinces, as well as Shanghai) from 2013 to 2017. We did not include typical cases of bacterial diarrhea caused by enterotoxigenic Escherichia coli, nor did we test samples for rotaviruses. Sows on 23 of the 42 pig farms were immunized with various PEDV vaccines. PSV or SAV vaccines were not used on any of the 42 farms.
Approximately 1 g of stool sample was diluted 10-fold with 10 mmol/L of phosphate-buffered saline, oscillated and mixed for 5 min by vortexing, and centrifuged at 13,400 × g for 5 min. The supernatant was collected for further assessment. Each liquid sample (200 μL) was combined with 1 mL of total RNA extraction reagent (RNAiso Plus; Takara Bio, Dalian branch, Liaoning, China). Chloroform, isopropanol, and ethanol (Shanghai Chemical Reagent, Shanghai, China) were used in the extraction process. RNA was extracted according to the manufacturer’s instructions. All materials used for RNA extraction, including deionized water, were treated with 0.1% diethyl pyrocarbonate to avoid RNase contamination.
The optimal annealing temperature and primer concentrations for individual PCR were determined as follows. The one-step RT-PCR amplification reaction system (PrimeScript one-step RT-PCR kit v.2; Takara Bio) composition including 2 μL of PrimeScript 1-step enzyme mix (including PrimeScript reverse transcriptase, TaKaRa Ex Taq hot start and RNase inhibitor), 25 μL of 2× 1-step buffer (including buffer and dNTP mixture [final concentration 400 μM]), 0.2–1.5 μL of forward primer (10 μM), 0.2–1.5 μL of reverse primer (10 μM), 5 μL of RNA template, and RNase-free H2O added to make up 50 μL. The reaction parameters were: 50°C for 30 min, 94°C for 2 min; 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 35 cycles; 72°C for 10 min. Annealing temperature was assessed at 8 levels, including 51.0, 51.5, 52.4, 53.8, 55.5, 56.8, 57.6, and 58.0°C; upstream and downstream primers were diluted to a concentration of 10 μM, and 0.2, 0.35, 0.5, 0.75, 1.0, 1.2, 1.35, and 1.5 μL, respectively, were added to achieve final concentrations of 0.04, 0.07, 0.1, 0.15, 0.2, 0.24, 0.27, and 0.3 μM. The RT-PCR reaction was carried out in an S1000 thermal cycler (Bio-Rad, Hercules, CA). The PCR products were electrophoresed in 1.5% agarose gels (Sangon Biotech, Shanghai, China), then imaged and analyzed (Gel Doc XR+ UV gel imaging analysis system and nucleic acid electrophoresis instrument; Bio-Rad).
In specificity tests, positive samples of PEDV, PSV, SaV, PEDV-TGEV-PoRV triple attenuated vaccine, PRRSV, CSFV, PRV, PCV-2, and PPV, were used as reaction templates for our triplex RT-PCR–specific reaction (RNase-free H2O was used as a negative control). Positive samples were obtained from the Laboratory of Animal Disease Prevention and Control Center of Jinhua. The samples were determined to be positive by fluorescence quantitative PCR or RT-PCR, or conventional PCR or RT-PCR, and then confirmed by sequencing. In sensitivity tests, to prepare standard plasmids, the target gene of PEDV (659 bp) from CV777, PSV (428 bp) from YC2011, and SaV (246 bp) from AF182760 were amplified (PrimeScript one-step RT-PCR kit; Takara Bio) and cloned into a pUC57-Amp vector by GeneWiz Biotec. All selected colonies were confirmed by sequencing. Plasmids were purified (Plasmid DNA purification kit; Corning Life Sciences, Suzhou, Jiangsu, China), and quantified (NanoDrop spectrophotometer; Thermo Scientific, Waltham, MA). The copy number of the extracted plasmids was calculated.5 The highest concentrations of standard plasmid were 108, 108, and 106 viral copies/μL for PEDV, PSV, and SaV, respectively. The concentrations were mixed to make 10-fold serial dilutions (101–107) to determine the limit of detection (LOD). A triplex RT-PCR sensitivity test was carried out.
Triplex RT-PCR and conventional individual RT-PCR were used to assess 402 porcine diarrheal stool samples for comparison. Selected positive PCR products of mixed infection with 2 or 3 viruses of PEDV, PSV, and SaV were sequenced (GeneWiz Biotec), and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for sequence alignment and confirmation.
Electrophoresis results for PEDV, PSV, and SaV in individual RT-PCR at different annealing temperatures were similar, and the integrated annealing temperature was determined to be in the range of 53.8–56.8°C. Electrophoresis results of different primer concentrations were 0.2–0.27 μM, 0.04–0.3 μM, and 0.2–0.3 μM for SaV, PSV, and PEDV, respectively, for a comprehensive primer range of 0.2–0.3 μM. When the upstream and downstream primers of the 3 pairs were at 0.2, 0.24, 0.27, and 0.3 μM, primer concentrations were gradually optimized, and the optimal concentrations in triplex PCR primers were eventually determined to be 0.24 μM for PEDV, 0.15 μM for PSV, and 0.2 μM for SaV. Of the annealing temperatures tested, the optimal temperature was 55.5°C.
Next, specificity was evaluated. Under the optimal conditions determined above, triplex RT-PCR amplification was performed with PEDV+PSV+SaV RNA template; PEDV-TGEV-PoRV triple attenuated vaccine RNA; PSV, SaV, CSFV, and PRRSV RNA templates; PRV, PCV-2, and PPV DNA templates; and RNase-free H2O (Fig. 1). Only the PEDV+PSV+SaV RNA templates (Fig. 1, lane 1) and PEDV, PSV, and SaV (lanes 2–4) had bands of the same sizes as respective target fragments; the remaining templates were strip-free. In the sensitivity assay, using optimized triplex RT-PCR reaction conditions, bands could still be observed after 104-fold dilution of PEDV and PSV, and 102-fold dilution of SaV. Therefore, the detection sensitivity for PEDV, PSV, and SaV was 104, 104, and 104 viral copies/μL, respectively (Fig. 2).
Figure 1.
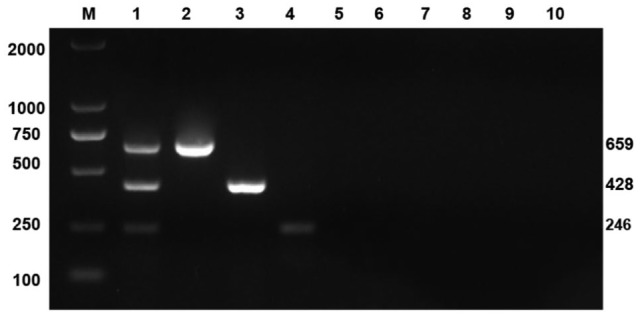
Specificity of triplex reverse-transcription PCR. M = marker DL2000; lane 1 = PEDV+PSV+SaV; lane 2 = attenuated vaccine of PEDV-TGEV-PoRV; lanes 3–6 = PSV, SaV, CSFV, and PRRSV; lanes 7–9 = PCV-2, PRV, and PPV; lane 10 = RNase-free H2O.
Figure 2.
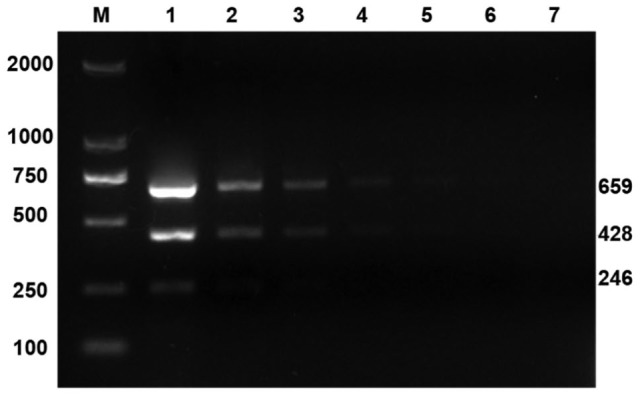
Sensitivity of triplex reverse-transcription PCR. M = marker DL2000; lanes 1–7 = copy numbers of re-cloned plasmids of PEDV and PSV at 107, 106, 105, 104, 103, 102, and 101/μL; SaV at 105, 104, 103, 102, 101, 10-1, and 10-2/μL respectively.
From 402 samples from piglets with diarrhea, RNA was extracted and divided into 2 groups for PEDV, PSV, and SaV triplex RT-PCR, and conventional individual RT-PCR detections, with the same reaction conditions and reagents. The overall concordance rate for the 2 techniques was 99.0% (305 of 308); concordance rates for detection of individual viruses were 100% (230 of 230) for PEDV, 97.6% (41 of 42) for PSV, and 94.4% (34 of 36) for SaV. Specificity was 100%, indicating maximum sensitivity and strong specificity for PEDV, PSV, and SaV using the triplex RT-PCR method. The positive rates were 57.2% (230 of 402) for PEDV, 10.4% (42 of 402) for PSV, and 9.0% (36 of 402) for SaV (Table 2). The positive rate for PSV was 7.9% (5 of 63) in a similar study19 that used conventional RT-PCR. The positive rate of SaV in piglet diarrhea samples in other studies ranged from 8.8% to 30.1% by RT-PCR or nested RT-PCR.1,8,12,15
Table 2.
Comparison of triplex and conventional individual RT-PCR methods for clinical samples.
| Year | No. of samples | Triplex RT-PCR positive number | Conventional individual RT-PCR positive number | ||||
|---|---|---|---|---|---|---|---|
| PEDV | PSV | SaV | PEDV | PSV | SaV | ||
| 2013 | 12 | 5 | 1 | 0 | 5 | 1 | 0 |
| 2014 | 66 | 39 | 5 | 4 | 39 | 5 | 5 |
| 2015 | 86 | 54 | 9 | 5 | 54 | 9 | 5 |
| 2016 | 136 | 70 | 15 | 10 | 70 | 16 | 11 |
| 2017 | 102 | 62 | 11 | 15 | 62 | 11 | 15 |
| Total | 402 | 230 | 41 | 34 | 230 | 42 | 36 |
We detected only PEDV in 187 samples, only PSV in 20 samples, and only SAV in 10 samples. We detected PEDV with PSV and SaV in 5 of 230 (2.2%) samples; PEDV-PSV were co-detected in 17 of 230 (7.4%) samples, and PEDV-SaV were co-detected in 21 of 230 (9.1%) samples, indicating that PEDV, PSV, and SaV may coexist in piglet diarrhea cases. Coinfection of PEDV with other viruses, such as TGEV,9 GARV (porcine group A rotavirus),16 and PCV-2, has become more common.7 A previous report6 co-detected SaV with PEDV, porcine rotaviruses A–C, Salmonella, and/or Brachyspira in 40 of 237 (16.9%) samples. Whether multiple infections would aggravate clinical signs or prolong the course of porcine diarrhea should be further investigated. The specificity and sensitivity of our triple RT-PCR method are similar to those of the conventional RT-PCR methods reported in the literature,6,14,16 and the LOD of the triple RT-PCR methods all ranged from 102–103 viral copies/μL. The LOD of our triple RT-PCR method is 10–100-fold less sensitive than that of real-time RT-PCR or other more advanced detection methods,2,3,13,19 in which LOD values were 100–101 viral copies/μL. However, the instruments used for conventional RT-PCR are more common and less expensive, and the cost of primer synthesis is lower. Therefore, our new triplex RT-PCR method could be used widely in the detection and screening of routine veterinary disease agents in local laboratories.
Acknowledgments
We thank the local veterinarians and farmers for their assistance in sample collection and information support.
Footnotes
Declaration of conflicting interests: The authors declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This project was supported in part by grants from the Jinhua Science and Technology Research Project (2016-2-013 to Y Wu, 2018-2-004 to H He, and 2019-2-012 to C Jiang).
ORCID iD: Chunyan Jiang  https://orcid.org/0000-0001-9258-7571
https://orcid.org/0000-0001-9258-7571
References
- 1. Barry AF, et al. High genetic diversity in RdRp gene of Brazilian porcine sapovirus strains. Vet Microbiol 2008;131:185–191. [DOI] [PubMed] [Google Scholar]
- 2. Chen J, et al. Development of a minor groove binder assay for real-time PCR detection of porcine Sapelovirus. J Virol Methods 2014;198:69–74. [DOI] [PubMed] [Google Scholar]
- 3. Gunson RN, et al. The real-time detection of sapovirus. J Clin Virol 2006;35:321–322. [DOI] [PubMed] [Google Scholar]
- 4. Guo M, et al. Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J Virol 2001;75:9239–9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang Y, et al. Development of a reverse transcription multiplex real-time PCR for the detection and genotyping of classical swine fever virus. J Virol Methods 2009;160:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jeong C, et al. Genetic diversity of porcine sapoviruses. Vet Microbiol 2007;122:246–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jung K, et al. The effects of transplacental porcine circovirus type 2 infection on porcine epidemic diarrhoea virus-induced enteritis in preweaning piglets. Vet J 2006;171:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim HJ, et al. Detection and molecular characterization of porcine enteric calicivirus in Korea, genetically related to sapoviruses. J Vet Med B Infect Dis Vet Public Health 2006;53:155–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim SH, et al. Multiplex real-time RT-PCR for the simultaneous detection and quantification of transmissible gastroenteritis virus and porcine epidemic diarrhea virus. J Virol Methods 2007;146:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lan D, et al. Isolation and characterization of the first Chinese porcine sapelovirus strain. Arch Virol 2011;156:1567–1574. [DOI] [PubMed] [Google Scholar]
- 11. Martella V, et al. Genetic heterogeneity of porcine enteric caliciviruses identified from diarrhoeic piglets. Virus Genes 2008;36:365–373. [DOI] [PubMed] [Google Scholar]
- 12. Martínez MA, et al. Molecular detection of porcine enteric caliciviruses in Venezuelan farms. Vet Microbiol 2006;116:77–84. [DOI] [PubMed] [Google Scholar]
- 13. Miller LC, et al. Evaluation of two real-time polymerase chain reaction assays for porcine epidemic diarrhea virus (PEDV) to assess PEDV transmission in growing pigs. J Vet Diagn Invest 2015;28:20–29. [DOI] [PubMed] [Google Scholar]
- 14. Palmquist JM, et al. Detection of porcine teschovirus and enterovirus type II by reverse transcription-polymerase chain reaction. J Vet Diagn Invest 2002;14:476–480. [DOI] [PubMed] [Google Scholar]
- 15. Reuter G, et al. Enteric caliciviruses in domestic pigs in Hungary. Arch Virol 2006;152:611–614. [DOI] [PubMed] [Google Scholar]
- 16. Song DS, et al. Multiplex reverse transcription-PCR for rapid differential detection of porcine epidemic diarrhea virus, transmissible gastroenteritis virus, and porcine group A rotavirus. J Vet Diagn Invest 2006;18:278–281. [DOI] [PubMed] [Google Scholar]
- 17. Sozzi E, et al. Molecular characterization and phylogenetic analysis of VP1 of porcine enteric picornaviruses isolates in Italy. Transbound Emerg Dis 2010;57:434–442. [DOI] [PubMed] [Google Scholar]
- 18. Sun RQ, et al. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis 2012;18:161–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang C, et al. Rapid and real-time detection of porcine sapelovirus by reverse transcription loop-mediated isothermal amplification assay. J Virol Methods 2014;203:5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang W, et al. The first Chinese porcine sapovirus strain that contributed to an outbreak of gastroenteritis in piglets. J Virol 2008;82:8239–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]


