Abstract
A 4-bp deletion (c.230_233delATAG) of the ABCB1 gene, frequently found in various dog breeds, results in intolerance to certain drugs routinely used in veterinary medicine, including many chemotherapeutic agents and macrocyclic lactones. The use of rapid and reliable genetic testing is fundamental for early detection of the mutation and prevention of undesirable toxicoses. We developed and compared 2 genotyping tests: PCR–high-resolution melting (PCR-HRM) and PCR–restriction-fragment length polymorphism (PCR-RFLP) to identify the 4-bp deletion in the ABCB1 gene of canine breeds. Amplified PCR products were sequenced in order to confirm different genotypes. Both techniques were efficient in discriminating homozygous wild-type, homozygous mutated, and heterozygous ABCB1 genotypes, and proved to be reproducible and economical methods. The HRM analysis, a sensitive and specific method for the molecular detection of genetic disorders, does not require labeled probes, processing, or separations after PCR.
Keywords: ABCB1 gene, dogs, ivermectin susceptibility, PCR-HRM, PCR-RFLP
P-glycoprotein (P-gp) is an ABCB1 (formerly MDR1) gene product belonging to the ATP-binding cassette superfamily (ABC superfamily) and is among the best characterized drug transport proteins.4 P-gp constitutes an integral part of the blood-brain barrier and is responsible for limiting the passage of different drug compounds into the brain and releasing them back to the bloodstream.4,5,8 ABCB1 has been studied as a candidate gene to assess ivermectin sensitivity given that it was confirmed that affected dogs were homozygous for a 4-bp deletion ABCB1 gene (c.230_233delATAG). This frameshift mutation leads to the emergence of a stop codon, resulting in a truncated P-gp that contains only ~10% of the amino acids in the non-mutated gene product, and displays a complete loss of function.4,8 Dogs with the ABCB1 genetic mutation have a P-gp deficiency and are extremely susceptible to toxicosis from many veterinary drugs in common use, such as ivermectin and derivatives, as well as loperamide, acepromazine, butorphanol, vincristine, and vinblastine, among others.4,5
ABCB1-related drug hypersensitivity is inherited as an autosomal recessive trait. Dogs that are homozygous for the deletion will experience adverse neurologic effects with the administration of a single dose of ivermectin (≥ 120 µg/kg), whereas carrier animals may experience neurotoxicity with a daily dose > 120 µg/kg (e.g., protocols for the treatment of demodectic mange).4 The 4-bp deletion in the ABCB1 gene has been documented in many herding breeds as well as in some mixed breeds. Affected breeds include Australian Shepherd (all sizes), Collie, Border Collie, English Shepherd, Longhaired Whippet, McNab, Old English Sheepdog, Shetland Sheepdog, and Silken Windhound. 6
Several screening tests have been described for genotyping the 4-bp deletion in the ABCB1 gene.1-5,7-9 Genotyping by DNA sequencing requires equipment available only in specialized laboratories. Separation of PCR products by polyacrylamide gel electrophoresis2,7,8 presents the complication of discriminating between wild-type and mutant genotypes in which lengths differ by only 4 bp. An allele-specific PCR for the different genotypes was designed,1 but this technique could be more difficult to perform and less reproducible than other methods. Isothermal amplification was also used to detect the polymorphism.9 Fluorogenic 5’-nuclease assays were established to detect the 4-bp deletion in the ABCB1 gene in dogs.3,7 Our aim was to develop conclusive new methods to identify the 4-bp deletion in ABCB1 genotypes in various canine breeds using PCR–high-resolution melting (PCR-HRM) and PCR–restriction-fragment length polymorphism (PCR-RFLP) techniques.
Blood samples were collected under standard procedure protocol according to our institutional animal ethics committee (CICUAL: Comité institucional de cuidado y uso de animales de experimentación), and with consent of their respective owners. Samples (n = 29) were taken from 28 dogs of different pure breeds and 1 mixed-breed. These dogs were phenotypically healthy animals from private owners and breeders. Dogs had not been tested previously for the presence of the mutant ABCB1 gene. DNA was isolated from blood samples (illustra blood genomic prep mini spin kit; GE Healthcare UK, Little Chalfont, UK). Approximately 70 ng of DNA were used for PCR amplification.
No restriction endonuclease could recognize the mutated sequence, therefore degenerate primers were designed on the basis of published messenger RNA sequences for the P-gp–encoding gene ABCB1 (Gene Bank accessions AF045016 and AJ419568) using dCAPS Finder v.2.0 (http://helix.wustl.edu/dcaps/dcaps.html). The degenerate reverse primer created a recognition site for the restriction endonuclease PvuII in the mutant allele (Table 1). PCR reactions were performed in a final volume of 25 µL containing: 1× PCR buffer, 3 mM MgCl2, 10 mM of each primer, 0.2 mM dNTPs, and 0.2 U Taq polymerase (Invitrogen Life Technologies, Saõ Paulo, Brazil). PCR was carried out under the following cycling program: 5 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 55°C, and 45 s at 72°C, with a final extension at 72°C for 7 min. PCR products were digested with 1 U of PvuII (New England Biolabs, Beverly, MA) at 37°C for 3 h in a 20-µL final volume. Genotype identification was carried out following the restriction fragment pattern obtained from the electrophoresis of the PCR-digested products in stained 2.2% agarose gel (GelRed nucleic acid gel stain; Biotium, Hayward, CA; Fig. 1). The wild-type allele was 134 bp, and the mutant allele had bands of 109 and 21 bp (21 bp band not visible in Fig. 1).
Table 1.
Primers used for amplification by PCR-RFLP and PCR-HRM of the ABCB1 gene.
| Method/Primer | Sequence* (5′–3′) | Size (bp) |
|---|---|---|
| PCR-RFLP | ||
| MDR1 For | CGCTATTCAAATTGGCTTGATAGG | 134/130 |
| MDR1 Rev | GAAATTCCTGCATTTGCACAGC | |
| PCR-HRM | ||
| MDR1 For | CGCTATTCAAATTGGCTTGATAGG | 134/130 |
| MDR1 Rev | GAAATTCCTGCATTTGCAAAGC | |
PCR-HRM = PCR–high-resolution melting; PCR-RFLP = PCR–restriction-fragment length polymorphism.
Underlined letters in the sequence of primers are mismatched to the canine ABCB1 sequence.
Figure 1.
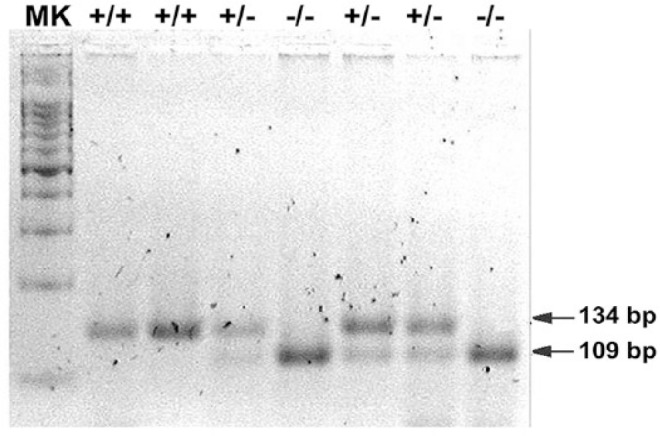
Canine ABCB1 genotypes of wild-type (+/+), heterozygous (+/–), and homozygous mutant (–/–) determined by PCR–restriction-fragment length polymorphism using agarose gel electrophoresis. Lane MK = 100-bp DNA ladder (Promega).
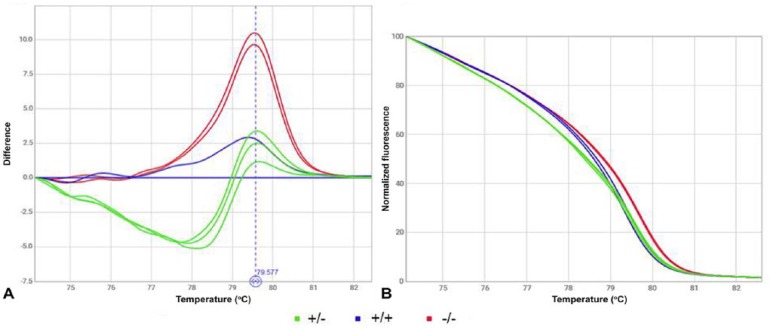
HRM analysis is a post-PCR analysis method used to identify genetic variations in nucleic acid sequences. This simple, fast method is based on PCR melting (dissociation) curve techniques and is enabled by the availability of improved double-stranded DNA (dsDNA) binding dyes along with next-generation real-time PCR instrumentation and analysis software. HRM analysis can differentiate DNA sequences based on their composition, length, GC content, or strand complementarity. In PCR-HRM reactions, non-degenerate primers (Table 1) for the same region used in the PCR-RFLP were checked (Primer Express v.2.0; Applied Biosystems, Foster City, CA). Reactions were performed with 10 µL of MeltDoctor HRM master mix (Applied Biosystems), 5 µM of each primer, and 30 ng of DNA in a 20-µL final volume. Amplification was carried out (StepOne real-time PCR system; Applied Biosystems) with a 40-cycle program of 95°C for 15 s, 60°C for 1 min, followed by a continuous HRM curve analysis of 0.3°C increments from 60–95°C (Fig. 2).
Figure 2.
High-resolution melting profile of the resolved ABCB1 genotypes corresponding to samples of Figure 1. Homozygous +/+ (blue lines), heterozygous +/– (green lines), and homozygous –/– (red lines). A. Fluorescence difference plot normalized to a genotype. B. Normalized melting profile.
Two homozygous wild-type genotypes (Beagle) and 2 homozygous mutant type genotypes (Collie) detected by both techniques were analyzed by DNA sequencing. Amplified PCR products for sequencing were purified (GFX PCR DNA and gel band purification kit; Amersham Biosciences, Piscataway, NJ). Forward primer was used for sequencing homozygous wild-type and homozygous mutated ABCB1 genotypes (3100 DNA sequencer; Applied Biosystems). These sequences were then comparatively analyzed for polymorphisms using BioEdit sequence analysis software (http://www.mbio.ncsu.edu/BioEdit/BioEdit.html).
We tested all of the samples by the 2 methods and results were unequivocal (Table 2), hence both methods were reliable. Three animals (2 Collies and 1 Rough Collie) were homozygous for the ABCB1 mutation. The remaining 6 Collies analyzed were heterozygous for the mutation, and none of the other animals tested had the mutation.
Table 2.
Results of ABCB1 genotyping by dog breed.
| Breed | No. tested | Genotype* |
|---|---|---|
| Argentine Dogo | 4 | +/+ |
| Beagle | 2 | +/+ |
| Bloodhound | 1 | +/+ |
| Border Collie | 1 | +/+ |
| Boxer | 2 | +/+ |
| Collie | 6 | +/– |
| Collie | 2 | –/– |
| Rough Collie | 1 | –/– |
| German Shepherd | 2 | +/+ |
| Golden Retriever | 2 | +/+ |
| Kuvasz | 1 | +/+ |
| Labrador Retriever | 2 | +/+ |
| Malinois | 2 | +/+ |
| Mixed-breed | 1 | +/+ |
Genotypes of the ABCB1 gene: wild-type (+/+), heterozygous (+/–), and homozygous mutant (–/–).
Although both techniques proved to be efficient in genotypic differentiation, PCR-HRM analysis, unlike conventional methods, is a closed-tube method that eliminates processing between amplification and analysis, thus preventing carryover contamination with PCR products. Also, it requires a small amount of DNA. The HRM assay is easier and more cost-effective than probe-based assays or the fluorogenic 5’-nuclease assay. The PCR-HRM assay method resulted in a fast, accurate (sensitivity, specificity), and precise (repeatability, reproducibility) tool to detect the ABCB1 mutation. The PCR-HRM method can also reveal the presence of additional mutations in the product by showing a variation in expected HRM profiles. This could be a disadvantage as it would be necessary to characterize the sample by direct sequencing.
Although PCR-based genotyping methods are simple and reliable, the post-PCR handling necessary to determine the genotypes delays the results and may add a source of contamination. PCR-RFLP is a more strenuous and time-consuming technique, but it allows laboratories with less technology to unequivocally differentiate the genotypes. Both the PCR-HRM and PCR-RFLP techniques proved to be specific and reliable methods for establishing the ABCB1 genotypes in dogs.
Acknowledgments
We thank all of the dogs’ owners for the use of their dogs’ samples in our study.
Footnotes
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: This work was supported by the Argentine University of Buenos Aires (grant V405).
ORCID iD: Graciela Marrube  https://orcid.org/0000-0001-7464-1503
https://orcid.org/0000-0001-7464-1503
References
- 1. Baars C, et al. Allele-specific polymerase chain reaction diagnostic test for the functional MDR1 polymorphism in dogs. Vet J 2008;177:394–397. [DOI] [PubMed] [Google Scholar]
- 2. Geyer J, et al. Development of a PCR-based diagnostic test detecting a nt230(del4) MDR1 mutation in dogs: verification in a moxidectin-sensitive Australian Shepherd. J Vet Pharmacol Ther 2005;28:95–99. [DOI] [PubMed] [Google Scholar]
- 3. Klintzsch S, et al. Detection of the nt230[del4] MDR1 mutation in dogs by a fluorogenic 5’ nuclease TaqMan allelic discrimination method. Vet J 2010;185:272–277. [DOI] [PubMed] [Google Scholar]
- 4. Mealey KL. Canine ABCB1 and macrocyclic lactones: heartworm prevention and pharmacogenetics. Vet Parasitol 2008;158:215–222. [DOI] [PubMed] [Google Scholar]
- 5. Mealey KL, et al. Ivermectin sensitivity in collies is associated with a deletion mutation of the mdr1 gene. Pharmacogenetics 2001;11:727–733. [DOI] [PubMed] [Google Scholar]
- 6. Mealey KL, Meurs KM. Breed distribution of the ABCB1-1Delta (multidrug sensitivity) polymorphism among dogs undergoing ABCB1 genotyping. J Am Vet Med Assoc 2008;233:921–924. [DOI] [PubMed] [Google Scholar]
- 7. Mizukami K, et al. Rapid genotyping assays for the 4-base pair deletion of canine MDR1/ABCB1 gene and low frequency of the mutant allele in Border Collie dogs. J Vet Diagn Invest 2012;24:127–134. [DOI] [PubMed] [Google Scholar]
- 8. Roulet A, et al. MDR1-deficient genotype in Collie dogs hypersensitive to the P-glycoprotein substrate ivermectin. Eur J Pharmacol 2003;460:85–91. [DOI] [PubMed] [Google Scholar]
- 9. Stiedl CP, Weber K. Fast and simple detection methods for the 4-base pair deletion of canine MDR1/ABCB1 gene by PCR and isothermal amplification. J Vet Diagn Invest 2017;29:176–180. [DOI] [PubMed] [Google Scholar]