Summary
Oct4 is widely considered the most important among the four Yamanaka reprogramming factors. Here, we show that the combination of Sox2, Klf4, and cMyc (SKM) suffices for reprogramming mouse somatic cells to induced pluripotent stem cells (iPSCs). Simultaneous induction of Sox2 and cMyc in fibroblasts triggers immediate retroviral silencing, which explains the discrepancy with previous studies that attempted but failed to generate iPSCs without Oct4 using retroviral vectors. SKM induction could partially activate the pluripotency network, even in Oct4-knockout fibroblasts. Importantly, reprogramming in the absence of exogenous Oct4 results in greatly improved developmental potential of iPSCs, determined by their ability to give rise to all-iPSC mice in the tetraploid complementation assay. Our data suggest that overexpression of Oct4 during reprogramming leads to off-target gene activation during reprogramming and epigenetic aberrations in resulting iPSCs and thereby bear major implications for further development and application of iPSC technology.
Keywords: reprogramming, induced pluripotent stem cells, Sox2, Klf4, Oct4, Myc, developmental potential, tetraploid complementation, retroviral silencing, transcription factor, Yamanaka cocktail, seesaw model of pluripotency
Graphical Abstract
Highlights
-
•
SKM can induce pluripotency in somatic cells in the absence of exogenous Oct4
-
•
SM coexpression activates the retroviral silencing machinery in somatic cells
-
•
Oct4 overexpression drives massive off-target gene activation during reprogramming
-
•
OSKM, but not SKM, iPSCs show abnormal imprinting and differentiation patterns
Velychko et al. showed that Oct4 overexpression during reprogramming causes epigenetic changes deteriorating the quality of iPSCs. SKM reprogramming generates iPSCs with equivalent developmental potential to ESCs, as determined by their ability to generate all-iPSC mice through tetraploid embryo complementation.
Introduction
Since the breakthrough discovery of transcription factor (TF)-driven reprogramming by Shinya Yamanaka (Takahashi and Yamanaka, 2006), multiple studies have addressed the function of each component of the reprogramming cocktail (Li et al., 2010, Malik et al., 2019, Soufi et al., 2012, Sridharan et al., 2009, Tsankov et al., 2015). Subsequent work from Yamanaka’s laboratory showed that, of the four TFs in the reprogramming cocktail—Oct4, Sox2, Klf4, and cMyc (OSKM)—only cMyc could be omitted while still permitting generation of induced pluripotent stem cells (iPSCs) (Nakagawa et al., 2008). Moreover, although Sox2, Kfl4, and cMyc could be replaced by other members of their TF families, Oct4 could not be substituted by either Oct1 or Oct6. Numerous attempts have been made to replace the components of the reprogramming cocktail to understand the molecular role of each factor, as well as potentially improve efficiency and safety of the reprogramming technology (Radzisheuskaya and Silva, 2014).
Although many studies have shown the possibility of substitutions for Oct4 in the reprogramming cocktail (Table S1), Oct4 remains a commonly used molecule for generating iPSCs. In fact, we and others have demonstrated that exogenous expression of Oct4 alone is sufficient for reprogramming cell types that inherently express the other reprogramming factors (Kim et al., 2009a, Kim et al., 2009b, Tsai et al., 2011, Wu et al., 2011). Most of the substitutes for Oct4 that could successfully reprogram to iPSCs work through the direct activation of endogenous Oct4 gene (Buganim et al., 2012, Gao et al., 2013b, Heng et al., 2010, Shi et al., 2008). In vivo, Oct4 serves as a master regulator, playing an integral role in maintaining pluripotency (Niwa et al., 2000, Pesce and Schöler, 2001) and establishing the inner cell mass during development (Nichols et al., 1998).
Intriguing recent studies described Oct4-independent reprogramming cocktails, where Oct4 was replaced with GATA factors (Montserrat et al., 2013, Shu et al., 2013, Shu et al., 2015). Shu et al. hypothesized that Oct4 and Sox2 could orchestrate cellular dedifferentiation during reprogramming by counteracting each other’s effects in the specification of meso-endoderm and neuroectoderm, respectively. The report demonstrated that endoderm lineage specifiers, such as Gata3, Gata4, and Gata6, promoted even higher-efficiency reprogramming compared to Oct4. This study introduced the “seesaw model” of pluripotency, which describes it as a fine balance between the induction of opposing developmental lineages (Shu and Deng, 2013).
Our group previously determined that Oct4 is not critical for establishing totipotency during mouse development; moreover, following nuclear transfer, Oct4-null oocytes can still efficiently reprogram somatic nuclei to pluripotency (Wu et al., 2013). The present work was sparked by the surprising observation that a reprogramming cassette that did not contain Oct4 was able to efficiently generate iPSCs, contrary to the long-standing dogma.
Results
SKM Is Sufficient for Reprogramming Mouse Somatic Cells to Pluripotency
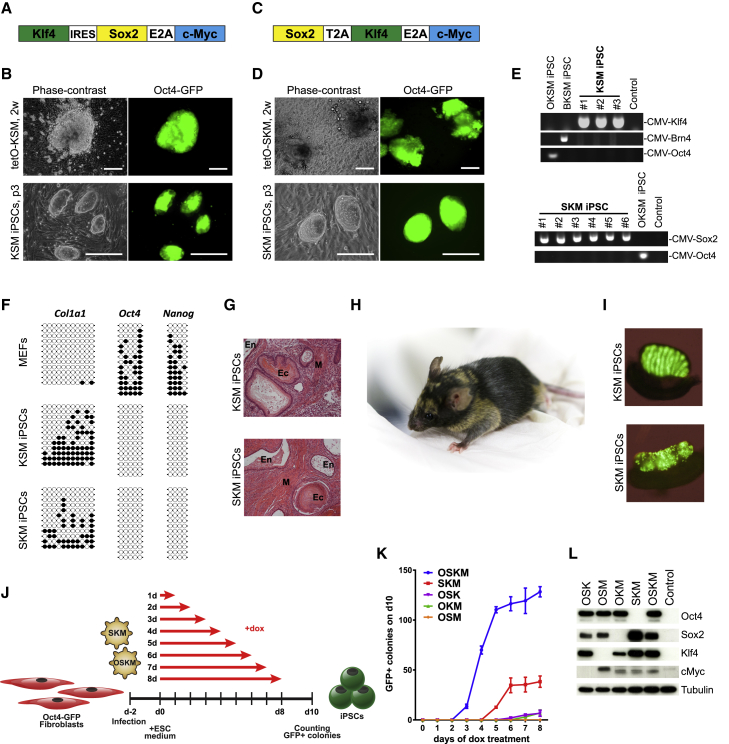
Initially, we removed Oct4 from the widely used tetO-OKSM polycistronic reprogramming cassette (Sommer et al., 2009) to generate a negative control to compare the reprogramming ability of different POU factors (Figure 1A). To our surprise, induction of KSM in Oct4-GFP reporter mouse embryonic fibroblasts (MEFs) (OG2) could still generate GFP+ colonies (Figure 1B). We then deleted Oct4 also from the tetO-OSKM vector (Carey et al., 2009, Carey et al., 2010, Carey et al., 2011; Figure 1C). The SKM cassette also gave rise to GFP+ and alkaline phosphatase-positive colonies, as early as 5 days post-induction (dpi) (Figures 1D and S1B). PCR genotyping confirmed lentiviral integration of SKM/KSM cassettes in all iPSC lines, whereas neither Oct4 transgene nor another POU factor, Brn4, were detected (Figure 1E).
Figure 1.
Sox2, Klf4, and cMyc Can Reprogram to Pluripotency in the Absence of Exogenous POU Factor
(A and C) Scheme of KSM and SKM polycistronic vectors derived from OKSM (A) and OSKM (C) reprogramming cassettes.
(B and D) Generation of iPSCs with tetO-KSM (B) and SKM (D) vectors, respectively. Phase-contrast and fluorescence microscopy images of primary colonies and passaged clonal lines of KSM and SKM iPSCs (scale bars represent 250 μm).
(E) PCR genotyping verifying tetO-KSM and tetO-SKM transgenes in KSM and SKM iPSC lines, respectively, while confirming the absence of Oct4 or Brn4 integration.
(F) Bisulfite sequencing analysis of DNA methylation in Oct4, Nanog, and Col1a1 promoters in MEFs and KSM and SKM iPSC lines.
(G) H&E staining of teratoma sections with representation of three germ layers (ectoderm: keratinizing epithelium; mesoderm: smooth muscles; endoderm: cuboidal and respiratory epithelium).
(H) An adult chimeric mouse generated from SKM iPSC line.
(I) Bright-field and GFP merged images of the gonads from E13.5 KSM and SKM iPSC chimeric embryos.
(J) Schematic representation of the time course reprogramming experiment.
(K) Time course reprogramming experiment of Oct4-GFP MEFs using polycistronic vectors. 103 transduced MEFs were plated on feeders and induced with dox for the indicated number of days. GFP+ colonies were counted on 10 dpi. Error bars represent SD; n = 3.
(L) Western blot analysis of MEFs after transduction of polycistronic vectors, 1 dpi.
The KSM/SKM (hereafter referred to as SKM) iPSCs displayed morphology characteristic of embryonic stem cells (ESCs) and could be expanded for at least 15 passages (Figures 1B and 1D). They stained positive for the pluripotency-specific markers SSEA1 and Nanog (Figure S1A). Methylation analysis of bisulfite-treated DNA revealed that the Oct4 and Nanog promoters were hypomethylated (Figure 1F), indicating epigenetic activation of the pluripotency genes. In contrast, the Col1a1 promoter was hypermethylated in the reprogrammed cell lines, indicating silencing of the somatic gene. The SKM iPSCs gave rise to all three germ layers in teratoma formation assays (Figure 1G) and contributed to the development of viable chimeric mice (Figure 1H), including the germline (Figure 1I).
SKM Reprogramming Is Independent of Expression Cassette or Starting Cell Type
To assess the efficiency and kinetics of SKM versus OSKM reprogramming, we performed a time course reprogramming experiment. OG2 MEFs were transduced with the OSKM, SKM, OSK, OKM, or OSM polycistronic vectors and induced with doxycycline (dox) for 1–8 days (Figure 1J). SKM generated GFP+ colonies after at least 5 days of induction, which is delayed by 2 days compared to OSKM (Figure 1K). The SKM reprogramming efficiency after 6–8 days of induction was approximately 30% of that for OSKM. Surprisingly, the removal of Oct4 from the OSKM cassette was the least detrimental, while removal of Klf4 led to the biggest drop in reprogramming efficiency. Western blot analysis confirmed comparable factor expression and the absence of the factor eliminated from each cassette (Figure 1L). The use of MEFs with Gof18;Rosa26-rtTA background gave a very similar result (Figure S1B).
We ruled out the possibility that the tet-inducible promoter or the reverse tetracycline-controlled transactivator (rtTA) is responsible for reprogramming in the absence of Oct4 by demonstrating that EF1α-SKM/KSM could generate GFP+ colonies in the absence of rtTA (Figure S1C). We also cloned the KSM cassette into the non-integrating, episomal vector to attempt virus-free reprogramming (Okita et al., 2011). Lipofection of episomal KSM into MEFs generated GFP+ colonies that were expanded into stable iPSC lines (Figure S1D) that lost the vector by passage 5 (Figure S1E). We confirmed the pluripotency of integration-free KSM-iPSCs by immunostaining and teratoma assays (Figures S1F and S1G).
To address the question whether SKM reprogramming depends on a specific starting cell population, we transduced presorted Thy− and Thy+ subpopulations of MEFs and found that both could be reprogrammed, although SKM induction in Thy+ cells gave rise to more GFP+ colonies (Figures S1H–S1J). We also demonstrated that adult lung fibroblasts (Figure S1K), immortalized adult tail tip fibroblasts (Figure S1L), and cortical astrocytes (Figures S1M–S1P) could be reprogrammed in the absence of exogenous Oct4. Overall, the efficiency of SKM reprogramming appeared to correlate with the rate of cell division, but not the origin of the cells.
Reprogramming in the Absence of Oct4 Relies on High Cell Proliferation Rate
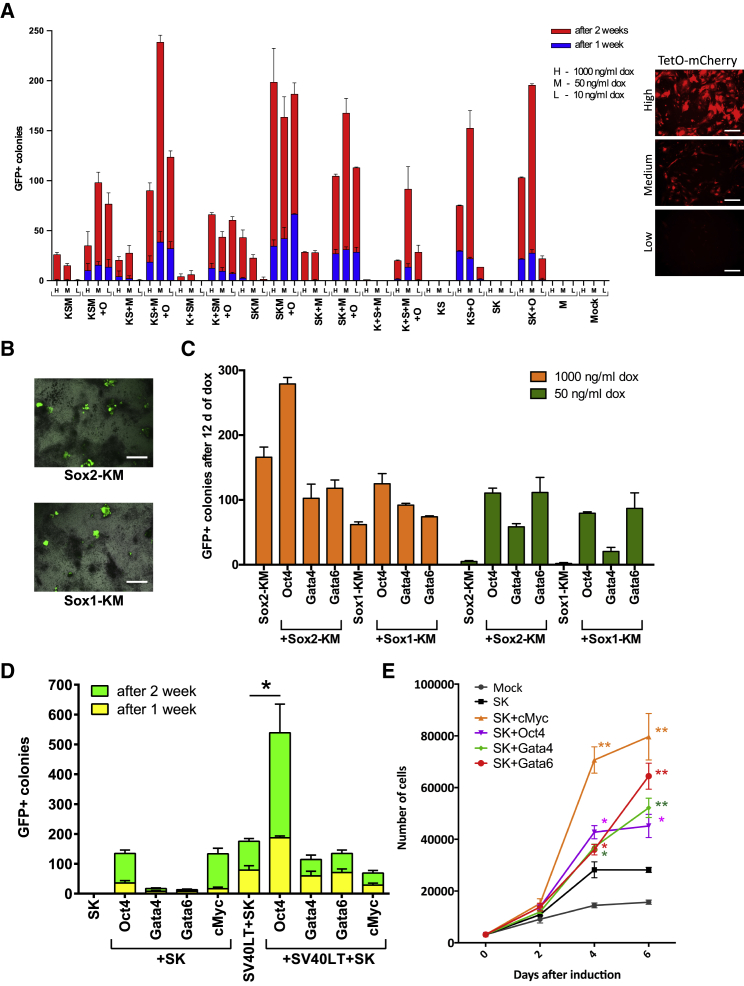
To further understand the components driving SKM reprogramming, we dissected the reprogramming cassettes. We found that neither KS nor SK can reprogram alone, but each could generate GFP+ colonies when combined with Oct4 or cMyc (Figure 2A). We used three different concentrations of dox to induce different levels of reprogramming factor expression. Although even the lowest expression level (10 ng/mL of dox) was sufficient for OSKM reprogramming, reprogramming in the absence of Oct4 required higher levels of expression (50 or 1,000 ng/mL dox). Polycistronic expression of the factors appeared to be beneficial, but not crucial, for SKM reprogramming, as SK+M, KS+M, K+SM, and even S+K+M could generate GFP+ colonies, albeit with very low efficiency in the case of monocistronic induction. Klf4-IRES-Sox2 (KS)+M could generate iPSCs with efficiency comparable to KSM, excluding the possibility of a gain-of-function effect from polyproteins that may arise from uncleaved 2A peptides (Velychko et al., 2019).
Figure 2.
Highly Proliferative Cells Can Be Reprogrammed by Sox2 and Klf4 Alone
(A) Reprogramming Oct4-GFP MEFs with dissected KSM and SKM cassettes. Different levels of transgene expression were achieved by induction with three concentrations of dox: 1 μg/mL (H); 50 ng/mL (M); and 10 ng/mL (L; see the tetO-mCherry panel, right). GFP+ colonies were counted after 7 and 14 dpi. Error bars represent SD; n = 3; scale bars represent 250 μm.
(B) Bright-field and Oct4-GFP merged overview images of Sox1-KM and Sox2-KM iPSCs after 14 dpi (scale bars represent 1 mm).
(C) Reprogramming of Oct4-GFP MEFs with Sox2-KM or Sox1-KM polycistronic cassettes in combination with Oct4, Gata4, or Gata6. Different level of transgene expression was achieved by induction with 1 μg/mL or 50 ng/mL of dox for 12 days. GFP+ colonies were counted on D14. Error bars represent SD; n = 3.
(D) Reprogramming of SV40LT-immortalized Oct4-GFP MEFs by ectopic expression of Sox2-Klf4 bicistronic cassette in combination with Oct4, cMyc, Gata4, or Gata6. Error bars represent SD; n = 3. Statistical significance was calculated with Student’s t test.
(E) Cell proliferation assay. 2,000 MEFs were transduced with indicated constructs in 96-well plates. The cells were harvested and counted after 0, 2, 4, and 6 dpi. SK sample was used as control for calculation of statistical significance. Error bars represent SD; n = 3. Statistical significance was calculated with Student’s t test.
It was reported that the endoderm-specific GATA factors could substitute for Oct4 in reprogramming to pluripotency, while ectoderm-specific Sox1 or Sox3 could replace Sox2 (Shu et al., 2013, Shu et al., 2015). We replaced Sox2 with Sox1 and observed only a 50% decrease in reprogramming efficiency for both the SKM and OSKM cocktails (Figures 2B and 2C). Adding Gata4 or Gata6 did not significantly increase the reprogramming efficiency for either Sox2-KM or Sox1-KM at a high level of transgene expression but increased the number of GFP+ colonies at lower level of induction (Figure 2C).
We next tested whether Gata4 or Gata6 could reprogram in combination with SK only. Indeed, as reported by Shu et al. (2013), SK+Gata4/6 could generate iPSCs; however, the efficiency of reprogramming was 10 times lower than for SK+O or SK+M (Figures S2A and S2B). Moreover, SK+GATA did not generate any GFP+ colonies when induced with a lower dox concentration (Figure S2A).
Given that GATA factors, Oct4, and cMyc share the ability to activate proliferation genes, we next questioned whether SK alone could reprogram highly proliferative cells. We introduced a tet-inducible SV40 Large T antigen (SV40LT) lentiviral vector into MEFs, allowing us to reversibly immortalize fibroblasts during the reprogramming process (Anastassiadis et al., 2010). The immortalization of the starting MEFs allowed for reprogramming with SK only and rendered the addition of cMyc, Gata4, or Gata6 ineffective (Figures 2D, S2C, and S2D). SK iPSCs (Figure S2E) could be passaged at least 12 times while preserving the normal karyotype (Figure S2F). We verified their genomic integrations by PCR genotyping and their pluripotency by immunostaining, teratoma formation, and chimera contribution, including the germline (Figures S2G–S2K).
Cell proliferation assay through the course of reprogramming showed that the addition of Gata4 or Gata6 significantly increased the proliferation of fibroblasts, resulting in 2-fold increase in cell number after 6 dpi compared to SK alone (Figure 2E). Thus, we conclude that GATA factors when combined with SK could reprogram fibroblasts by increasing the proliferation rate in a manner similar to cMyc and SV40LT.
SKM iPSCs Exhibit Dramatically Enhanced Developmental Potential
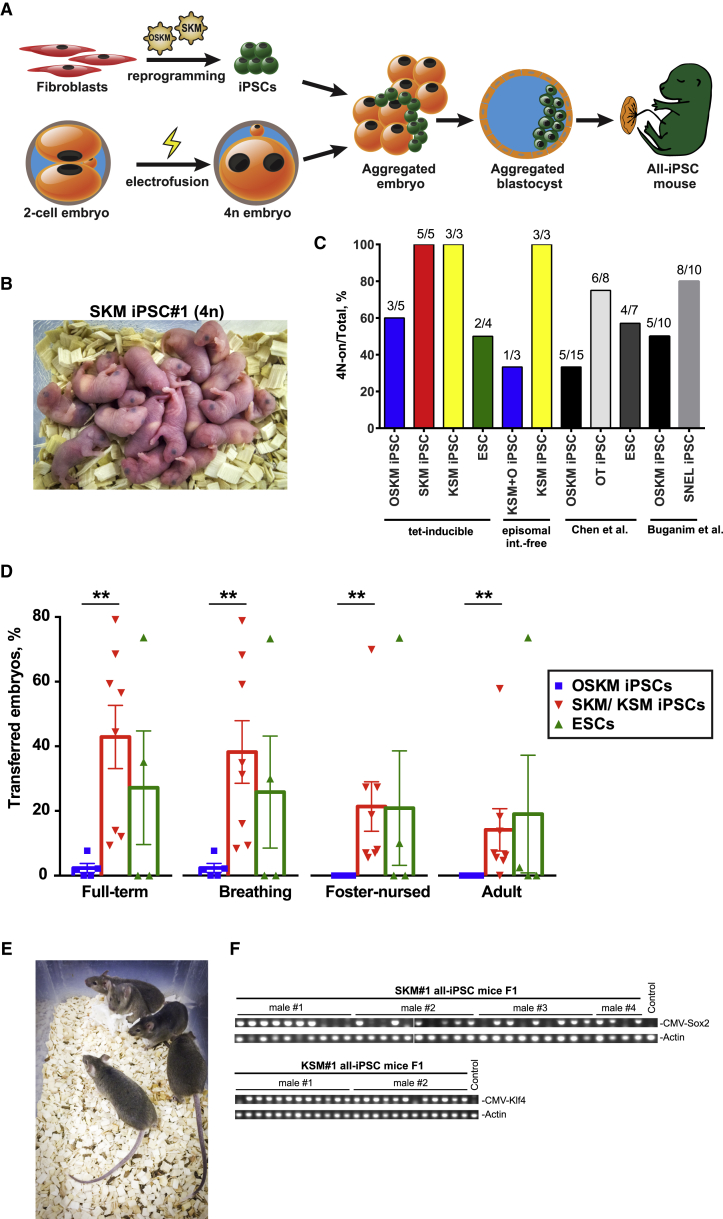
We next evaluated the developmental potential of SKM iPSCs by performing the most stringent test for pluripotency, tetraploid (4N) complementation assay (Figure 3A; Nagy et al., 1990). In this method, the tested iPSCs are aggregated with 4N embryos that can form extra-embryonic tissues but fail to develop viable inner cell mass. Development of such aggregates results in a fetus consisting almost exclusively of iPSCs (all-iPSC mice). We tested 13 clonal tet-inducible iPSC lines derived from Gof18-Rosa26-rtTA MEFs (5 SKM, 3 KSM, and 5 OSKM lines) of the same passage that were generated and cultured in parallel. All the tested lines were preselected for minimal transgene leakage to avoid possible harmful developmental effects (Figure S3A). Additionally, we generated control ESC lines from F1 embryos derived by mating SKM#1 all-iPSC mice to achieve the closest matching genotype. All SKM iPSC lines were capable of generating full-term all-iPSC pups (4N-on; Figures 3B and 3C; Table S2). In contrast, only 3 of 5 OSKM lines were 4N-on (Figure 3C), corroborating published reports (Amlani et al., 2018, Buganim et al., 2014, Chen et al., 2015). Astonishingly, 44.1% ± 9.1% of SKM iPSC- and 27.1% ± 17.5% of ESC-aggregated embryos gave rise to fully developed pups, compared to only 2.3% ± 1.4% for OSKM iPSCs (Figure 3D). The 4N complementation efficiency for OSKM lines was comparable to those of other reports (Figure S3B), whereas the efficiency of SKM iPSCs was much higher than that of OSKM iPSCs or iPSCs generated by alternative cocktails—Oct4+Tet1 (OT) or Sall4+Nanog+Esrrb+Lin28 (SNEL)—which were reported to improve iPSC quality (Buganim et al., 2014, Chen et al., 2015; Figures 3D and S3B). Notably, none of the OSKM all-iPSC pups survived foster nursing (Figure S3B), while 49.5% of SKM all-iPSC pups survived for at least 2 days of foster nursing and 34% survived until adulthood (Figure 3D). All adult SKM all-iPSC mice were healthy and fertile (Figures 3E and 3F). Additionally, we tested 6 integration-free iPSC lines generated by episomal vectors (Figures S1D–S1F). The results were largely consistent: all tested episomal KSM (epiKSM) iPSC lines were 4N-on, while only 1 of 3 epiKSM+O lines was 4N-on, albeit of high quality (Figure 3C; Table S2). The higher variability of episomal comparing to tet-inducible lines is expected, due to the stochastic nature of episomal disappearance and, therefore, lack of factor expression control (Okita et al., 2013). Nevertheless, the rare appearance of a high-quality epiKSM+O line suggests that certain conditions, such as lower expression, fast vector disappearance, or a certain O:KSM ratio, could allow for high-quality iPSCs, even in the presence of Oct4.
Figure 3.
Omitting Oct4 in the Reprogramming Cocktail Increases the Quality of iPSCs
(A) Schematic representation of tetraploid complementation experiment.
(B) All-iPSC pups generated by tetraploid complementation assay with SKM#1 iPSC line. 23 aggregates were transferred to 2 pseudopregnant CD-1 (white) females.
(C) The total ratios of 4N-on versus 4N-off iPSC and ESC lines generated in this study compared to published data.
(D) Percentage of 4N-aggregated embryos derived from tetO-OSKM, SKM/KSM iPSCs or ESCs that gave rise to full-term pups, pups that initiated breathing, pups that survived foster-nursing for at least 48 h, and those survived to adulthood (at least 3 months). Bars are representing the mean between all tested lines. The statistical significance was determined by Mann-Whitney test. Error bars represent SEM.
(E) 6-month-old all-iPSC mice generated by tetraploid complementation assay with SKM#1 iPSC line.
(F) PCR genotyping of F1 offspring of SKM and KSM all-iPSC mice for SKM or KSM transgenes, respectively.
Taken together, the SKM cocktail generates iPSCs with exceptionally high developmental potential, nearly 20-fold higher than that of OSKM, suggesting that exogenous Oct4 detrimentally affects the quality of iPSCs.
Co-expression of Sox2 and cMyc Causes Immediate Retroviral Silencing in Fibroblasts
We hypothesized that early silencing of Moloney murine leukemia virus (MMLV)-based vectors could account for the contradictory data presented in numerous publications reporting the indispensability of Oct4 in the Yamanaka cocktail (Table S1). It is commonly accepted that silencing of retroviral transgenes is a distinct characteristic of pluripotent, but not somatic, cells (Hochedlinger and Plath, 2009).
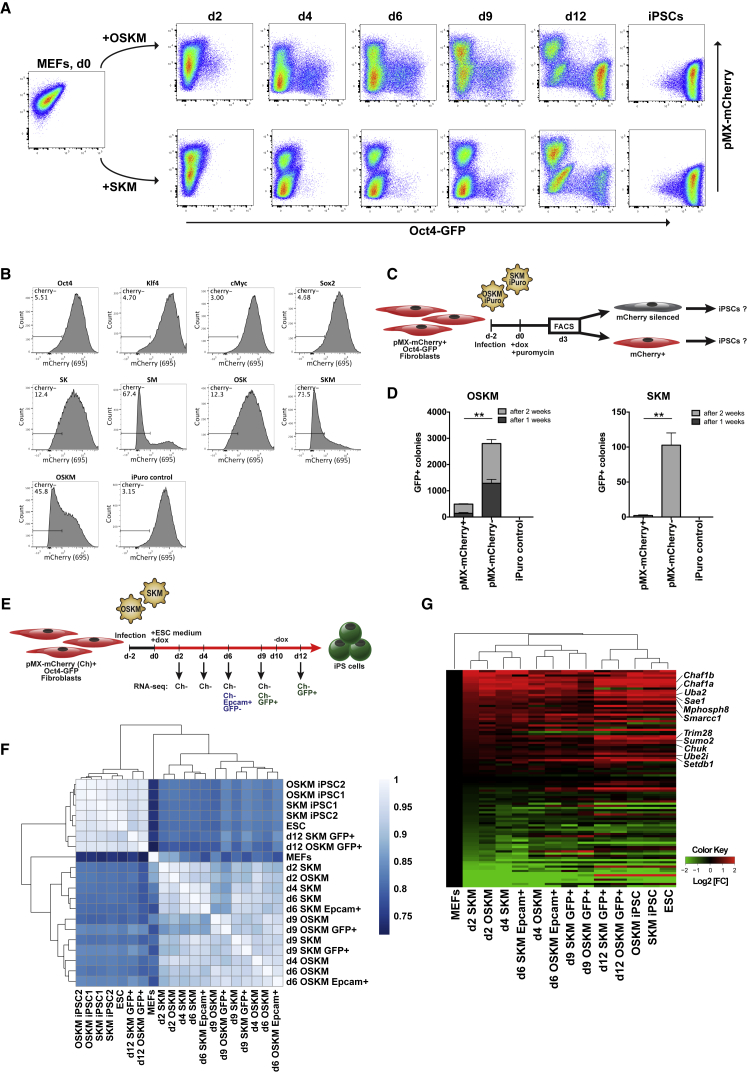
We transduced OG2 MEFs with a retroviral pMX vector expressing mCherry to track the retroviral silencing during reprogramming. Surprisingly, the morphological changes of SKM- and OSKM-induced fibroblasts coincided with the loss of mCherry signal starting as early as 2 dpi (Figures 4A, S4A, and S4B). Later, mCherry− cells formed dome-shaped colonies, within which Oct4-GFP+ cells emerged (Figures 4A and S4B). We quantified the results of time course fluorescence-activated cell sorting (FACS) analysis during OSKM and SKM reprogramming (Figure S4C). Although half of the fibroblasts had already lost mCherry expression after 2 dpi, a third of the mCherry+ cells still expressed fibroblast-specific Thy1, but the majority became double-negative by 4 dpi. These data suggest that retroviral silencing occurs very early during lentivirus-driven reprogramming—even before the loss of somatic cell identity.
Figure 4.
Co-expression of Sox2 and cMyc Leads to Immediate Retroviral Silencing in MEFs
(A) Time course FACS of pMX-mCherry+ Oct4-GFP MEFs during OSKM and SKM reprogramming.
(B) FACS analysis of pMX-mCherry+ MEFs expressing Oct4, Sox2, Klf4, cMyc, and combinations of the factors cloned into tet-inducible polycistronic vector with IRES-Puro on D3 of induction and puromycin selection.
(C) Experimental design to follow the fate of cells that did or did not undergo retrovirus silencing by 3 dpi with OSKM or SKM.
(D) Reprogramming of pMX-mCherry+ Oct4-GFP MEFs with OSKM or SKM. The cells were sorted for mCherry after 3 dpi and plated on a feeder layer. GFP+ colonies were counted after 7 and 14 dpi. Error bars represent SD; n = 3. Statistical significance was calculated with Student’s t test.
(E) Experimental design of time course SKM versus OSKM reprogramming RNA-seq experiment.
(F) Heatmap showing the Spearman correlation coefficient between time course reprogramming samples based on RNA-seq data.
(G) Heatmap depicting the relative fold change (FC) of gene expression after induction with OSKM or SKM. The selected genes were reported as top 100 hits in systemic siRNA screen for reactivation of retrovirus in pluripotent stem cells (Yang et al., 2015). Hierarchical clustering was based on Euclidean distance.
If such immediate silencing occurred during retrovirus-mediated reprogramming, it would preclude conversion of fibroblasts into iPSCs. Indeed, when pMX-mCherry+ cells were infected with retroviral Oct4, Sox2, Klf4, and cMyc, some groups of cells lost mCherry expression and reprogramming was halted (Figure S4D). GFP+ cells could only arise from within mCherry+ clusters that kept expressing the transgenes. Thus, reprogramming with MMLV-based vectors selects for the cells that do not undergo early retroviral silencing.
We dissected the tetO-OSKM cassette to identify the exact factors driving early retrovirus silencing. We cloned the reprogramming factors into a tet-inducible vector carrying IRES-Puro, allowing for the selection of infected cells. Although none of the factors could trigger silencing when induced alone, simultaneous expression of Sox2 and cMyc (SM) was sufficient to silence the retroviral transgene in most of the cells by 3 dpi (Figure 4B).
We tracked the reprogramming fate of the cells that did or did not undergo early retroviral silencing. We sorted the cells on d3 after SKM or OSKM induction and selection and plated the cells on a feeder layer to assess their reprogramming fate (Figure 4C). Overall, the mCherry− populations gave rise to a significantly higher number of GFP+ colonies than the mCherry+ populations for both reprogramming conditions. Although in the case of OSKM reprogramming, the same number of mCherry− cells gave 4 times more GFP+ colonies than the mCherry+ cells, in the case of SKM, the mCherry− fraction gave nearly 50 times more colonies than the mCherry+ fraction (Figure 4D). We therefore confirmed that retroviral silencing occurs early during the reprogramming, which is particularly relevant for SKM induction. Altogether, these results suggest the infeasibility of retrovirus-driven SKM reprogramming (Figure S1C).
Retrovirus Silencing Machinery Is Established Already after 48 h of Reprogramming
To gain deeper insight into the mechanism of SKM reprogramming, we performed RNA sequencing (RNA-seq) over the course of reprogramming (Figure 4E). The pMX-mCherry+ OG2 MEFs were transduced with polycistronic SKM or OSKM; samples were collected after 2, 4, 6, 9, and 12 dpi. Only mCherry− cells were used for RNA isolation to reduce heterogeneity and to eliminate contamination by untransduced cells. Additionally, we sequenced subpopulations of reprogramming intermediates that underwent mesenchymal to epithelial transition (MET) on d6 (mCherry−/GFP−/Epcam+) or activated endogenous Oct4 (mCherry−/GFP+) on d9 and d12 (2 days after dox withdrawal; Figure S4E). Global hierarchical clustering with Spearman correlation as a distance metric revealed that iPSCs as well as d12 samples clustered close to ESCs and far from starting MEFs, while d2–d9 samples clustered in between (Figure 4F).
Previously, Yang et al. (2015) carried out a genome-wide small interfering RNA (siRNA) screen that uncovered the components mediating retrovirus silencing in pluripotent stem cells. We analyzed the expression of the top 100 genes implicated in retroviral silencing in our samples. Remarkably, by d2 of SKM or OSKM induction, the expression of this gene subset already resembled that of ESCs (Figure 4G). Among the genes upregulated on d2 were key drivers of retrovirus silencing: chromatin remodeling by histone chaperones (Chaf1a/b and Smarcc1), sumoylation factors (Sumo2, Ube2i, Sae1, Uba2, and Ube2j2), and chromatin modifiers (Trim28, Setdb1, and Mphosph8). Therefore, we conclude that the retrovirus repressor machinery of pluripotent cells can be established in fibroblasts already around 2 days of SKM or OSKM induction.
Oct4 Is Not Required for Initiation of Pluripotency Program in Somatic Cells
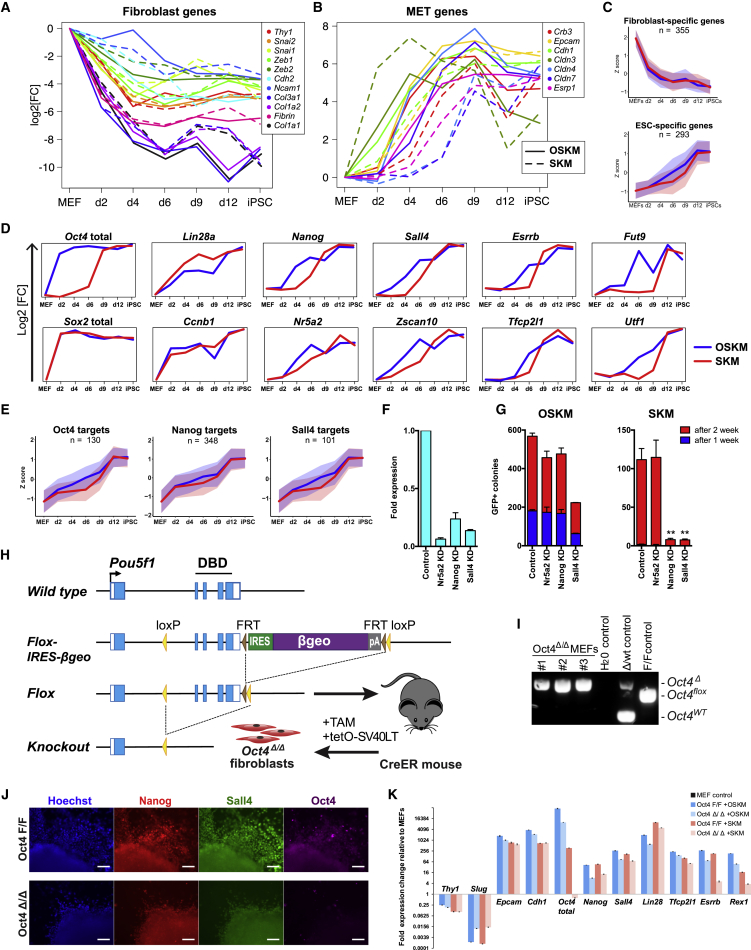
To compare the molecular roadmaps of SKM and OSKM reprogramming, we analyzed gene expression relevant for established reprogramming stages: fibroblast identity loss; MET; and maturation of pluripotency (Li et al., 2010, Polo et al., 2012, Stadtfeld et al., 2008). RNA-seq data showed that downregulation of fibroblast-specific genes (Snai1, Thy1, Ncam1, etc.) starts as early as d2 of OSKM or SKM expression and advances synchronously for both conditions (Figures 5A and 5C). Loss of fibroblast identity, therefore, appears to be independent of exogenous Oct4 expression. MET occurs on d2–d4 for OSKM and d2–d6 for SKM reprogramming, as evident from upregulation of epithelial markers (e.g., Cdh1, Epcam, and Esrp1; Figure 5B). The partially delayed MET in the absence of Oct4 is in accordance with our FACS data showing fewer Epcam+ cells on 6 dpi for SKM versus OSKM (Figure S4E).
Figure 5.
SKM Can Activate Pluripotency Program in Oct4-KO MEFs
(A and B) Time course expression plots of indicated fibroblast (A) and MET (B) genes during OSKM and SKM reprogramming. Only Epcam+ and GFP+ sorted samples are shown for d6 and d9, respectively.
(C) Time course expression of MEF- and ESC-specific genes (Chronis et al., 2017). Only differentially expressed genes (DEGs) with FC ≥ 4 were plotted.
(D) Time course expression plots of indicated genes during OSKM and SKM reprogramming.
(E) Time course expression of Oct4, Nanog (ChIP-Atlas), and Sall4 (Lim et al., 2008) targets in ESCs. Only top peaks (macs score ≥ 200, on Chip-Atlas) within 5 kb of DEGs (FC ≥ 4 in iPSCs versus MEFs) were plotted.
(F) qPCR gene expression analysis of Nr5a2, Nanog, and Sall4 after short hairpin RNA (shRNA)-driven KD during reprogramming after 2 dpi with OSKM. ActB was used as a reference gene. Error bars represent SD; n = 3.
(G) Reprogramming of Oct4-GFP MEFs, expressing Nr5a2, Nanog, Sall4, or control shRNAs with OSKM or SKM. GFP+ colonies were counted after 7 and 14 dpi. Error bars represent SD; n = 3. Statistical significance was calculated with Student’s t test.
(H) Strategy for generating Pou5f1 (Oct4)-KO MEFs.
(I) PCR genotyping confirming homozygous deletion of Oct4 in 3 MEF lines.
(J) Immunofluorescence imaging of Oct4F/F and Oct4Δ/Δ MEFs reprogrammed with SKM, on 7 dpi.
(K) qPCR gene expression analysis of Epcam+ Oct4F/F and Oct4Δ/Δ MEFs reprogrammed with OSKM or SKM on 7 dpi. Error bars represent SD; n = 3.
The maturation stage of reprogramming, marked by upregulation of pluripotency-specific genes, is largely delayed for SKM reprogramming (Figure 5C). Expression plots confirm the absence of Oct4 in SKM and similar levels of Sox2 in SKM versus OSKM samples (Figure 5D). Although some genes (e.g., Lin28a, Nr5a2, and Ccnb1) are upregulated with comparable kinetics for both conditions, other genes are delayed in the absence of Oct4 (e.g., Nanog, Tfcp2l1, and Esrrb). Activation of Oct4 direct targets (Figure 5E), such as Fut9 (SSEA1) and Utf1 (Figure 5D), are among the most delayed in SKM versus OSKM reprogramming, while Nanog and Sall4 targets show less delayed kinetics (Figure 5E).
To understand how endogenous Oct4 is activated during SKM reprogramming, we performed gene knockdown (KD) experiments with 3 candidate factors, Nr5a2, Nanog, and Sall4 (Figure 5F), which showed relatively early upregulation according to the RNA-seq data. All 3 factors were previously reported as Oct4 substitutes in reprogramming (Buganim et al., 2012, Heng et al., 2010). Nr5a2 KD did not affect the efficiency of SKM reprogramming (Figure 5G). Nanog or Sall4 KD, on the other hand, dramatically reduced the number of SKM iPSC colonies but had substantially weaker effects on OSKM reprogramming. Time course immunostaining confirmed that SKM activates Nanog (Figure S5A) prior to Oct4 during reprogramming.
The gene profiling combined with KD experiments suggests that Nanog and Sall4 are the first pluripotency regulators to be activated during SKM reprogramming, although endogenous Oct4 is upregulated in a secondary activation wave. To further understand the role of endogenous Oct4 in SKM reprogramming, we generated Oct4-knockout (KO) fibroblasts (Figure 5H). The MEFs derived from Oct4flox/flox; Rosa26-CreERT2 mice were treated with tamoxifen and immortalized with tetO-SV40LT. Clonal MEFs were picked to ensure homozygous deletion of the Oct4 alleles (Figure 5I). Immunostaining showed that induction of SKM could activate Nanog and Sall4 in the complete absence of Oct4 (Figure 5J). qPCR analysis of Epcam+ cells after 7 dpi showed that SKM can downregulate fibroblast-specific genes (Thy1 and Slug) and activate MET (Epcam and Cdh1) and the pluripotency genes (Nanog, Sall4, Lin28, and Tfcp2l1) in Oct4-KO MEFs (Figure 5K). However, some late pluripotency genes (e.g., Esrrb and Rex1) were only partially upregulated, and Oct4-KO SKM pre-iPSCs could not survive dox withdrawal, suggesting that Oct4 is still needed for finalizing establishment of and for maintaining pluripotency.
Oct4 Overexpression Diverts Cells from a Direct Route to Pluripotency
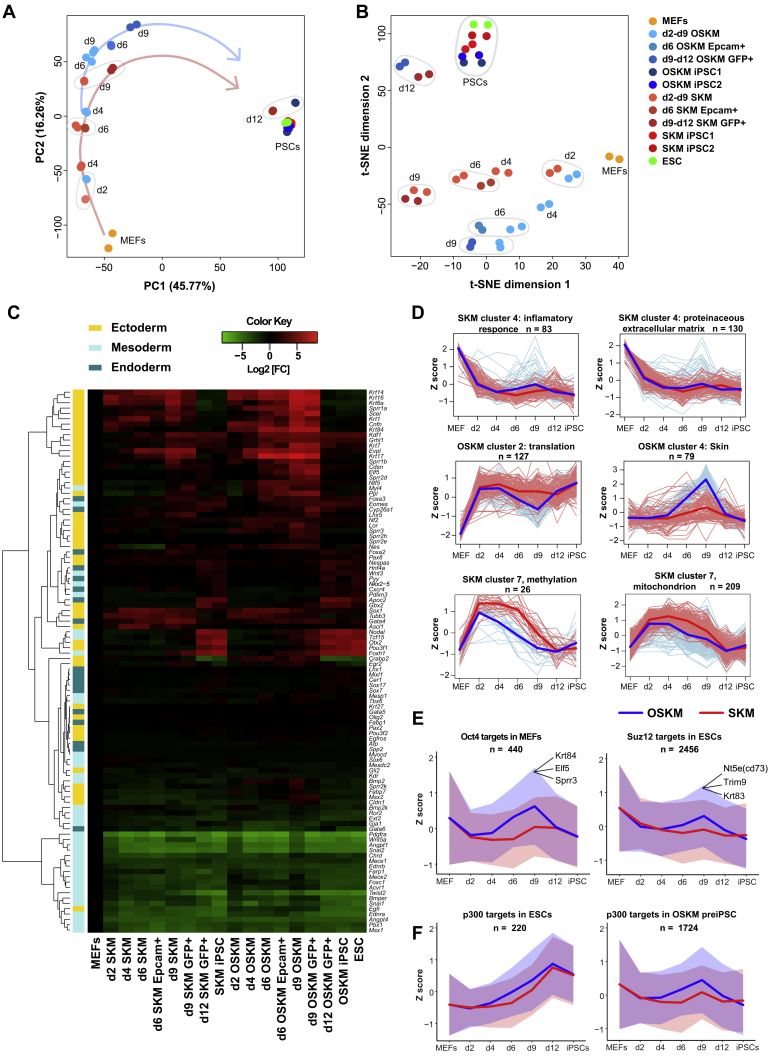
To compare the global gene expression changes occurring during OSKM and SKM reprogramming, we performed two dimensionality reduction methods: principle-component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE) analysis (Figures 6A, 6B, and S6A). As reported previously (Polo et al., 2012), PCA shows that OSKM-reprogrammed cells transiently divert from a direct route to pluripotency on d6, d9, and d12 in the PC2 dimension or even partially re-acquire MEF identity in PC3 dimension (Figures 6A and S6A). In contrast, SKM reprogramming occurs along a more direct PC2 trajectory and diverts further from MEFs in PC3. t-SNE analysis shows an even clearer distinction between SKM and OSKM samples, starting already on 4 dpi.
Figure 6.
Exogenous Oct4 Diverts the Cells from Direct Route to Pluripotency
(A and B) PCA (A) and t-SNE (B) analysis of global gene expression in time course samples during OSKM and SKM reprogramming.
(C) Heatmap depicting the relative FC of gene expression during the course of SKM and OSKM reprogramming based on RNA-seq. Hierarchical clustering was based on Euclidean distance.
(D) Time course plots of representative GO terms of gene cluster characterized by specific kinetics (see Figure S5A). Gene set enrichment analysis was performed by DAVID (enrichment > 2; p < 0.01; false discovery rate [FDR] < 0.1).
(E and F) Time course average expression of Oct4 off targets in MEFs (Chen et al., 2016), Suz12 targets in ESCs (Pasini et al., 2010; E), and p300 targets in ESCs or preiPSCs (Chronis et al., 2017; F). Line graph depicts the average Z score values of expression; shading depicts SD.
It was reported that SKM induction in fibroblasts leads to elevated expression of ectodermal genes, which can be counterbalanced by Oct4 or GATA factors, resulting in pluripotency induction (Shu et al., 2013). However, other studies suggested that OSKM reprogramming also leads to transient elevation of ectodermal genes (González and Huangfu, 2016, Mikkelsen et al., 2008). We compared the effects of SKM with OSKM reprogramming on the expression of ecto-, meso-, and endodermal genes. The mesodermal genes are quickly downregulated after OSKM or SKM induction (e.g., Snai1, Snai2, and Chrd), which coincides with loss of fibroblast identity (Figure 6C). Endodermal genes remain largely unaffected throughout reprogramming; however, in contrast to the report by Shu et al. (2013), many ectodermal genes are transiently upregulated during both SKM and OSKM reprogramming. Moreover, OSKM causes much stronger ectodermal induction (e.g., Krt1, Evpl, and Nes), with some ectodermal genes upregulated only in the presence of exogenous Oct4 (e.g., Lor, Elf5, Krt84, and Sprr3; Figure 6C). Importantly, sorted d6 Epcam+ and d9 Oct4-GFP+ samples show even sharper difference in ectodermal upregulation in OSKM versus SKM reprogramming.
We performed soft clustering to subdivide differentially expressed genes (DEGs) into groups that have similar expression kinetics during reprogramming (Figure S6A). SKM cluster 4, enriched in somatic genes, exhibits rapid downregulation for both conditions. A few genes in this subset, however, are transiently reactivated during the later stages of OSKM reprogramming (Figure 6D). The opposite happens for OSKM cluster 2, enriched in metabolic genes, which shows a sharp upregulation followed by a transient decline on 4–9 dpi for OSKM, but not SKM. These clusters are likely responsible for the partial return of a fibroblast identity in OSKM samples in PC3 dimension (Figure S6A). OSKM cluster 4 contains 1,260 genes, enriched in terms related to epidermis development, which were strongly transiently upregulated on 4–9 dpi with OSKM but remained largely unaffected during SKM reprogramming (Figures 6D and S6C). These changes coincide with the departure of OSKM samples from a direct trajectory to pluripotency in the PC2 plane (Figure 6A). Gene set enrichment analysis of d9 GFP+ OSKM versus SKM samples showed GO terms related to epidermis development among the top enriched (Figures S6D and S6E). On the other hand, the GO terms related to RNA processing, translation, DNA replication, and telomerase maintenance were enriched in d9 GFP+ SKM samples (Figure S6D). Many of OSKM cluster 4 genes are direct Oct4 targets when induced in MEFs but are not bound by Oct4 in ESCs; among those are the above-mentioned keratinocyte-related genes Elf5, Krt84, and Sprr3 (Figures 6C and 6E).
Interestingly, OSKM drives a transient upregulation of genes bound by Suz12 in ESCs (Figure 6E). Suz12, a component of the polycomb repression complex 2 (PRC2), is recruited to poised enhancers in ESCs, preventing them from being transcribed even when bound by TFs (Chen et al., 2008). Among the genes co-bound by Oct4 and Suz12 in ESCs is Nt5e (CD73), an immune-response-related cell-surface antigen used by multiple studies as a marker for partially reprogrammed cells (Hussein et al., 2014, Zunder et al., 2015). We found that Nt5e is strongly upregulated during OSKM, but not SKM, reprogramming. Active ESC enhancers marked by p300, on the other hand, show gradual upregulation during the course of both OSKM and SKM reprogramming, with faster activation in the presence of Oct4, coinciding with faster reprogramming kinetics (Figure 6F). However, OSKM induction transiently recruits p300 and upregulates off-target “pre-iPSC” genes to a higher degree than SKM (Figure 6F).
Overall, our time course gene expression data reveal that the reprogramming trajectories of OSKM and SKM differ predominantly because of profound off-target gene activation driven by ectopically expressed Oct4.
Omitting Exogenous Oct4 Leads to More Faithful Epigenetic Reprogramming
Previous reports have compared iPSCs versus ESCs in 4N complementation assays and found that iPSCs had impaired developmental potential due to loss of imprinting (LOI), karyotypic instability, aberrant gene expression, and mitochondrial defect (Buganim et al., 2014, Stadtfeld et al., 2010, Stadtfeld et al., 2012, Wu et al., 2014, Zhong et al., 2019). We tested OSKM and SKM iPSCs for those abnormalities. G-band karyotyping showed that the majority of PSC lines had a normal karyotype (Figure S7A; Table S2), with the exception of OSKM iPSC#5, KSM iPSC#3, and ESC#3 lines, which had trisomy of chromosomes 14, 1, and 19, respectively (Table S2). Additionally, karyotyping of OSKM- and SKM-sorted GFP+ bulk iPSCs on the second passage showed no significant difference in the rate of chromosomal aberration between the two reprogramming cocktails (Figure S7B). Electron microscopy of 4N-off and 4N-on lines shows no difference in their subcellular organization; the mitochondria of 4N-off OSKM iPSCs exhibit normal cristae structure indistinguishable from SKM iPSCs or ESCs (Figure S7C).
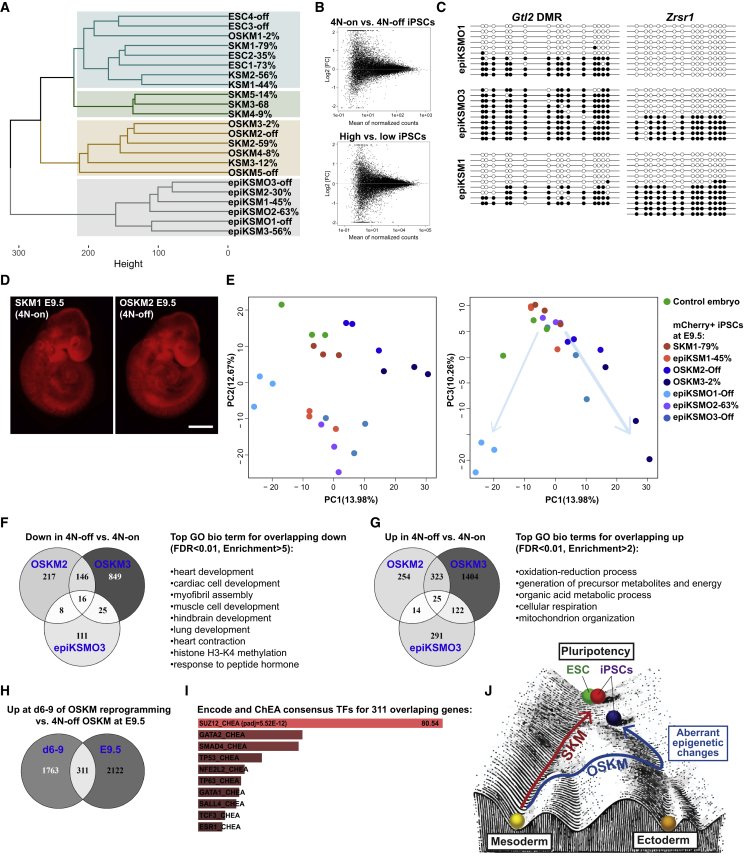
We compared the gene expression in all tested OSKM and SKM iPSCs, including those generated by the episomal method, as well as ESC lines—altogether 23 lines. Although some high-quality tet-inducible SKM lines clustered the closest to 4N-on ESCs, other lines clustered more randomly. Particularly, all the episomal iPSCs cluster further away from ESCs comparing to lentiviral iPSCs (Figure 7A). DE analysis of 15 4N-on versus 4 4N-off iPSCs or 8 highest versus 6 lowest quality iPSC lines by DESeq2 showed no significantly up- or downregulated genes (Figure 7B). We therefore hypothesized that the key might lie in the epigenetic marks, which affect the differentiation of iPSCs.
Figure 7.
OSKM, but Not SKM, Reprogramming Leads to Loss of Imprinting (LOI) and Alterations of iPSC Differentiation during Embryo Development
(A) Hierarchical clustering analysis of the iPSC and ESC lines based on global gene expression. Clustering was based on Euclidean distance.
(B) MA plots displaying differentially expressed (DE) genes in 4 4N-off OSKM lines versus 15 4N-on iPSC lines and 6 low- versus 8 high-quality iPSCs (4N complementation efficiency of ≤2% and ≥30%, respectively). No DEG with p adj < 0.05 were found. The analysis was performed using DESeq2 package. Genes with at least 5 reads in any sample were considered.
(C) Bisulfite sequencing analysis of DNA methylation in Gtl2 and Zrsr1 in representative iPSC lines validating the COBRA results from Figure S7D.
(D) E9.5 embryos generated by 4N complementation with mCherry-labeled SKM and OSKM iPSCs (scale = 500 μm).
(E) PCA analysis of bulk RNA-seq data of mCherry+ cells from E9.5 embryos generated by 4N-complementation with high- or low-quality iPSCs.
(F and G) Venn diagrams showing the overlap between down- and upregulated DEGs in 3 differentiated 4N-off OSKM iPSCs at E9.5 (≥5 reads in any sample; FC > 1.2; p < 0.05 by DESeq2) and the top gene ontology (GO) term enriched in DEGs overlapping in at least 2 of 3 lines.
(H) Venn diagrams showing the overlap between upregulated DEGs from (F) and transiently upregulated DEGs for d6 Epcam+ or d9 Oct4-GFP+ OSKM versus SKM samples (≥5 reads in any sample; FC > 2; p < 0.05 by DESeq2).
(I) Gene set enrichment analysis of overlapping genes from (H). The analysis was performed using Enrichr web server. The bars represent combined Enrichr scores.
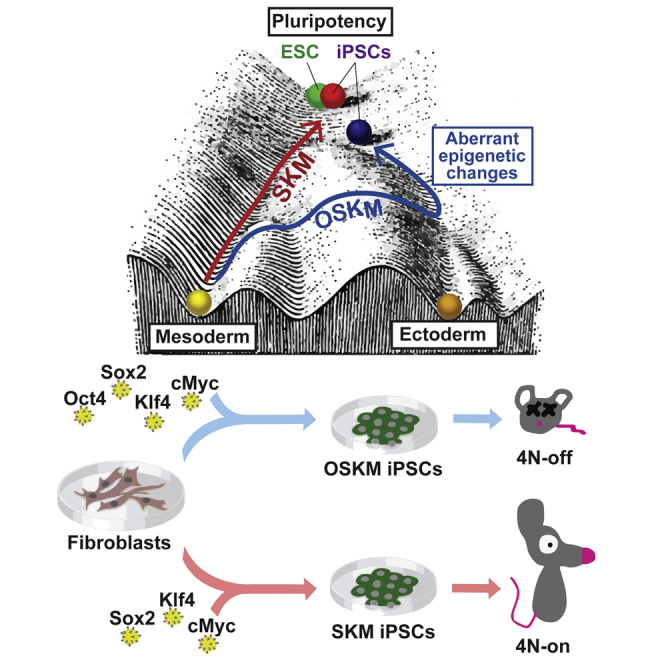
(J) Adapted Waddington’s epigenetic landscape model showing an epigenetic landscape with trajectories representing reprogramming from fibroblasts to pluripotency driven by OSKM or SKM cocktails.
Indeed, the analysis of imprinted loci methylation revealed that 3 of 4 4N-off lines showed LOI in Zrsr1, Peg10, or Gtl2 loci, as determined by combined bisulfite restriction analysis (COBRA) (Figure S7D) and validated by bisulfite sequencing (Figure 7C). On the other hand, all SKM and 4N-on OSKM iPSCs maintain normal imprinting. LOI of all of these loci was previously shown to occur during reprogramming, and aberrations of Zrsr1 and Gtl2 were specifically linked to impaired developmental potential of iPSCs (Carey et al., 2011, Chang et al., 2014, Takikawa et al., 2013).
To track the developmental fate of 4N-off OSKM iPSCs, we marked 3 high-quality and 4 low-quality lines (including integration-free ones) with constitutive mCherry expression. We used these iPSCs to generate 4N aggregates and examined the embryos at embryonic day 9.5 (E9.5). All the tested lines could generate viable embryos with no obvious phenotype at this stage (Figure 7D), and the vast majority of the cells were derived from iPSCs (Figure S7E). We sorted mCherry+ iPSC-derived differentiated cells and performed RNA-seq for 3 embryos per iPSC line, as well as 3 control embryos (altogether 24 embryos).
PCA plots showed that all 3 tested high-quality iPSCs (SKM1, epiKSM1, and epiKSMO2) clustered close to control embryos, although the low-quality lines clustered separately, shifting from the control lines on PC1 and PC3 dimensions (Figure 7E). Moreover, 3 of 4 of these lines (OSKM2, OSKM3, and epiKSMO3) deviate similarly along both dimensions and have many overlapping DEGs (Figures 7F and 7G), although epiKSMO1 embryos clustered separately (Figures 7F and S7F). The GO analysis of overlapping downregulated genes showed the strongest enrichment for genes related to heart development (Figure 7F) but also hindbrain and lung development and histone methylation genes. The upregulated DEGs were enriched for oxidative metabolism (Figure 7G). We compared the lists of the upregulated genes during embryo development with those transiently upregulated on d6 (Epcam+) or d9 (Oct4-GFP+) during OSKM, but not SKM, reprogramming and found 311 overlapping genes. Chromatin immunoprecipitation (ChIP) enrichment analysis (ChEA) of these genes showed the most significant enrichment for Suz12 targets (p adj = 5.52 × 10−12), suggesting misregulation of PRC2 targets.
Taken together, forceful expression of Oct4 does not significantly interfere with karyotype maintenance, mitochondria structure, or gene expression in iPSCs. However, it leads to epigenetic defects, such as LOI and misregulation of polycomb targets, resulting in a compromised differentiation potential.
Discussion
Since the introduction of iPSC technology in 2006, Oct4 has been considered a uniquely essential factor for reprogramming to pluripotency. In the current study, we found that SKM can reprogram mouse somatic cells with efficiency of 30% of that for OSKM, albeit with delayed kinetics. Reprogramming in the absence of exogenous Oct4 requires high transgene expression, a non-retroviral reprogramming vector, and a high rate of cellular proliferation.
Silencing of retroviral transgenes is a prominent feature of pluripotent stem cells, but not of other cell types (Niwa et al., 1983, Stewart et al., 1982, Yang et al., 2015). Retroviral suppression is thought to occur in the final stage of reprogramming and to be a reliable feature distinguishing fully from partially reprogrammed iPSCs (Bar-Nur et al., 2015, Hochedlinger and Plath, 2009). Here, we report that the simultaneous expression of Sox2 and cMyc leads to immediate retroviral silencing (Figures 4 and S4). The gene profiling analysis revealed that fibroblasts activate the key components of retrovirus silencing machinery as early as 2 dpi, demonstrating an inadequacy of using retroviral silencing as a marker for mature iPSCs.
During embryonic development, Oct4 is required for specification of the primitive endoderm and mesoderm (Frum et al., 2013, Zeineddine et al., 2006) and Sox2 for the neuroectoderm lineage (Ferri et al., 2004). Nonetheless, an abundance of published data demonstrate that the two factors act cooperatively to regulate their target genes to induce or maintain pluripotency (Chen et al., 2014, Chronis et al., 2017, Malik et al., 2019, Reményi et al., 2003, Sridharan et al., 2009). Additionally, Oct4 overexpression can rescue pluripotency in Sox2-KO ESCs (Masui et al., 2007) and the enhanced transcriptional activity of Oct4 can compensate for the omission of Sox2 during reprogramming (Marthaler et al., 2016, Wang et al., 2011), suggesting an overlap rather than opposition between the two factors. Our work suggests that Sox2 and Klf4 do not need a counterbalance by meso-endodermal factors, as was proposed by the seesaw model (Shu et al., 2013), but rather they require an additional factor to boost cell proliferation.
GATA factors are known to regulate proliferation of different cell types. For example, Gata3 controls the proliferation of T cells by upregulating cMyc (Wang et al., 2013), Gata4 regulates proliferation of cardiomyocytes by directly activating cyclin D2 and Cdk4 (Rojas et al., 2008), and Gata6 promotes growth of hair follicle progenitor cells by directly activating N-Myc (Wang et al., 2017). We showed that only Oct4, but not cMyc or GATA factors, could enhance the SK reprogramming of highly proliferative SV40LT-immortalized cells and confirmed that GATA factors increase the proliferation rate of SK-induced cells (Figures 2D and 2E). Our findings, along with those of published reports, challenge the premise that GATA factors have exclusively endoderm-inducing function and suggest that their proliferation-boosting properties could explain their role in reprogramming.
Our data show that the activation of endogenous Oct4 and its targets is delayed during SKM reprogramming, whereas the activation of Nanog and its targets occurs more synchronously in the presence or absence of exogenous Oct4 (Figures 5D and 5E). We analyzed published ChIP-seq data (Chronis et al., 2017) and found that, regardless of the presence of Oct4, Sox2 and Klf4 co-occupied the Nanog −5 super-enhancer (SE) already on 2 dpi (Figure S5B). The occupancy at −5 SE locus was lower in Sox2- or Klf4-only samples, suggesting a DNA-binding co-dependency of these two factors. The Oct4 distal enhancer, on the other hand, was occupied by Sox2 and Klf4 only in the presence of Oct4 at 48 h, suggesting that endogenous Oct4 cannot be directly activated by SK (Figure S5C).
Nanog, along with Oct4 and Sox2, is a central regulator of the naive pluripotency network (Boyer et al., 2005, Loh et al., 2006, Wang et al., 2006) and plays a crucial role in the final stage of reprogramming to iPSCs (Silva et al., 2009). We showed that depletion of either Nanog or Sall4 did not strongly affect OSKM reprogramming, although it nearly abolished SKM reprogramming (Figures 5F and 5G). Nanog directly interacts with Sall4 and co-occupies many of the target sites in ESCs, including the Oct4 locus (Zhang et al., 2006). Importantly, SKM induction could downregulate somatic genes and activate MET as well as some pluripotency genes, including Nanog, Sall4, and Lin28, even in Oct4-KO MEFs. Our study indicates that the establishment of pluripotency during SKM reprogramming occurs through direct activation of Nanog, which, in cooperation with Sall4, activates Oct4 and the rest of the pluripotency network (Figure S5D).
Our time course RNA-seq revealed a transient detour from a direct reprogramming trajectory with a prominent upregulation of epidermis-related genes in OSKM intermediates (Figures 6 and S6). An overlap of our RNA-seq data with published ChIP-seq datasets showed that Oct4 directly targets many of the misregulated genes when overexpressed in fibroblasts, but not in ESCs. Moreover, OSKM, but not SKM, reprogramming leads to misregulation of poised genes bound by PRC2 in ESCs and activation of the enhancers specific to “pre-iPSCs,” but not ESCs. In summary, OSKM reprogramming suffers from transient off-target gene upregulation driven by Oct4 overexpression, which can be prevented by omitting Oct4 from the reprogramming cocktail.
The relatively poor developmental potential of iPSCs compared to ESCs is a well-known problem that poses a major hurdle for clinical applications of iPSC technology. In agreement with other reports, half of our OSKM iPSCs generated by either lentiviral or episomal methods cannot give rise to all-iPSC mice. Furthermore, the pups generated from 4N-on OSKM lines have drastically reduced survival rates (Figures 3 and S3; Table S2). Thus, pluripotency of the majority of OSKM iPSCs is compromised. In stark contrast, all 11 tested clonal SKM iPSC lines were capable of generating all-iPSC mice, many of which survived to adulthood. LOI is the most widely reported problem associated with a poor developmental outcome of iPSCs (Carey et al., 2011, Chang et al., 2014). We found that the majority of 4N-off OSKM lines had LOI in either Dlk1-Dio3 or Zrsr1 locus, whereas 4N-on OSKM and all tested SKM iPSCs maintained normal imprinting (Figures 7 and S7). Interestingly, PRC2 can facilitate methylation of imprinted loci and the microRNA (miRNA) expressed from some imprinted loci regulate PRC2, forming a feedback loop (Liu et al., 2010). We found that many transiently upregulated PRC2 targets during OSKM reprogramming were also upregulated during iPSC differentiation within all-iPSC embryos.
A lot of attention has been brought to the reprogramming cocktail components, as even the design of the polycistronic reprogramming cassette can have a dramatic effect on the quality of iPSCs (Carey et al., 2011). The oncogenic potential of cMyc was found to be partially responsible for the poor quality of OSKM iPSCs, as OSK iPSCs showed an increased number of transferred 4N blastocysts that gave rise to full-term pups from 2.6% to 6.5% (Buganim et al., 2014). A few groups have reported that alternative reprogramming cocktails could generate iPSCs with improved developmental potential (Figures 3C and S3B). For example, SNEL could generate iPSCs at a very low efficiency (0.005%) but with a significantly enhanced developmental potential (from 2.6% to 12.8%; Buganim et al., 2014). Here, we report that omitting Oct4 from the reprogramming cocktail allows for the efficient generation of mouse iPSCs (4% total; 30% of OSKM) and increases the average developmental potential of iPSCs from 2.3% to 44.1%—the highest 4N complementation efficiency reported to date.
Oct4, Sox2, and Klf4 are widely considered to be pioneering factors, but their genomic engagement is highly dependent on cofactors and cellular context. Our work suggests that off-target gene activation by Oct4 can potentially have a detrimental effect on the quality of iPSCs. Although we did not address directly whether the mere presence of Oct4 in the beginning of reprogramming or rather its persistent overexpression throughout the process is detrimental, our data suggest that later is the case. Ultimately, this work warrants further development of reprogramming strategies to counterbalance epigenetic aberrations arising from forced expression of reprogramming factors.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat anti-Epcam | eBioscience | 17-5791-82; RRID:AB_2716944 |
| Rat anti-Thy-1.2 | BioLegend | 105318; RRID: AB_492888 |
| Mouse anti-SSEA1 | Millipore | MC-480; RRID:AB_177627 |
| Mouse anti-Oct4 | Santa Cruz Biotechnology | sc-5279; RRID: AB_628051 |
| Goat anti-Sox2 | Santa Cruz Biotechnology | sc-17320; RRID:AB_2286684 |
| Rabbit anti-Klf4 | Santa Cruz Biotechnology | sc-20691; RRID: AB_669567 |
| Rabbit anti-cMyc | Santa Cruz Biotechnology | sc-764; RRID: AB_631276 |
| Rat anti-Nanog | eBioscience | eBioMLC-51; RRID:AB_763613 |
| Mouse anti-Sall4 | Santa Cruz Biotechnology | sc-101147; RRID: AB_1129262 |
| Chemicals, Oligos, and Recombinant Proteins | ||
| FuGENE6 | Promega | E269A |
| Doxycycline hyclate | Sigma | D9891-5G |
| PD0325901 | Cayman Chemical | 13034 |
| CHIR99021 | Tocris | 4423 |
| Human LIF | Prepared in-house | N/A |
| Oligos | Sigma | See Table S3 |
| Critical Commercial Assays | ||
| GoTag green master mix | Promega | M3005 |
| SYBR Green PCR Master | Applied Biosystems | ABS-4367659 |
| EZ DNA Methylation Kit | Zymo Research | D5001 |
| QIAamp DNA mini Kit | QIAGEN | 51306 |
| NucleoSpin RNA | Macherey-Nagel | 740955.250 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6 × C3H mice | Bred in house | N/A |
| CD1 mice | Bred in house | N/A |
| Rosa25-rtTA, Gof18 mice | This study | N/A |
| Oct4flox/flox; Rosa26- Cre ERT2 mice | This study | N/A |
| Deposited Data | ||
| RNA-seq | This study | GEO: GSE137001 |
| Software and Algorithms | ||
| R | R Core Team | https://www.R-project.org |
| DESeq2 | (Love et al., 2014) | http://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| mFUZZ | (Kumar and E Futschik, 2007) | http://bioconductor.org/packages/release/bioc/html/Mfuzz.html |
| CLC 11 | QIAGEN Bioinformatics | https://qiagenbioinformatics.com |
| Venny2.1 | Oliveros, J.C. | https://bioinfogp.cnb.csic.es/tools/venny |
| Enrichr | (Kuleshov et al., 2016) | http://amp.pharm.mssm.edu/Enrichr/ |
| Geneontology | (Ashburner et al., 2000) | http://geneontology.org |
| DAVID | (Huang et al., 2009) | https://david.ncifcrf.gov |
Lead Contact and Materials Availability
Requests for further information should be directed to and will be fulfilled by Sergiy Velychko (sergii.velychko@mpi-muenster.mpg.de). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental Model and Subject Details
Mice
All mice used were bred and housed at the mouse facility Max Planck Institute (MPI) in Muenster, and animal handling was in accordance with MPI animal protection guidelines. A protocol for animal handling and maintenance for this study was approved by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen under the supervision of a certified veterinarian in charge of the MPI animal facility. Mouse embryonic fibroblasts (MEFs) were isolated from male embryos as described previously (Takahashi and Yamanaka, 2006).
Method Details
Cell culture
MEFs and HEK293T cells were cultured in low-glucose DMEM supplemented with 10% FBS (Ch), 1% Glutamax, 1% penicillin-streptomycin, 1% nonessential amino acids (NEAA), 0.5% β-mercaptoethanol (all from Life Technologies). Mouse embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) were grown on irradiated C3H MEFs in high-glucose DMEM medium (Life Technologies) supplemented with 15% KSR, 2% FCS, 1% Glutamax, 1% NEAA, 1% penicillin-streptomycin, 1% β-mercaptoethanol, and 20 ng/ml human recombinant LIF (purified in-house). The cells were routinely tested for Mycoplasma contamination.
Vector construction
pHAGE2-tetO-OKSM (STEMCCA) lentivirus reprogramming vector was kindly provided by Konrad Hochedlinger. All other lentiviral vectors used in this study were cloned into the same pHAGE2 backbone. OSKM was cloned from Col1a 4F2A targeting construct, Addgene #25794 (Carey et al., 2010) using BamHI and SbfI restriction sites. Mouse Oct4, Sox2, Klf4, cMyc, Gata4, Gata6, and mCherry were amplified by PCR and pasted into pHAGE2-tetO using NotI and ClaI restriction sites. KSM, SKM, KS, SK, and SM were cloned using NotI, ClaI, and available restriction sites (Eco47III in Klf4 and RsrII in Sox2). Bluescript-SV40 Large T antigen (SV40LT) construct was kindly provided by Konstantinos Anastassiadis and cloned into pHAGE2 using available NotI and ClaI restriction sites.
For generation of tetO-ires-Puro vectors, NotI-ClaI-ires-Puro-BstBI was amplified by PCR and inserted by NotI and BstBI (compatible with ClaI, destroyed). The rest of the constructs were inserted into tetO-ires-Puro by restriction with NotI, ClaI, and ligation.
For generation of constitutively expressed pHAGE2-EF1alpha vectors, EF1alpha was PCR amplified and inserted into pHAGE2-tetO constructs with SpeI and NotI, replacing tetO-miniCMV promoter. Retroviral vector pMX-SK was generated by blunt-end ligation of NotI and ClaI‒digested SK fragment.
To generate pCXLE constructs, the NotI-ClaI site was inserted by annealing and ligation oligos to EcoRI-digested pCXLE-GFP (addgene #27082) (Okita et al., 2011) KSM or Oct4 were inserted using NotI and ClaI digestion and ligation.
shRNA vectors targeting Nr5a2, Nanog, and Sall4 were generated by ligation of annealed oligos into AgeI and EcoRI sites of pLKO-puro (Addgene #8453) (Stewart et al., 2003). The sequences were Nr5a2 5′-GCAAGTGTCTCAATTTAAA-3′ (Heng et al., 2010), Nanog 5′-CTTGCTTACAAGGGTCTGCTA-3′, Sall4 5′- GCAACCTGAAGGTACACTA-3′ (Tanimura et al., 2013).
Virus production
Fully confluent HEK293T cells were split 1 to 5 on 100-mm dishes. The following day, cells were transfected with 3 μg of lentiviral vector, 2 μg of PAX2, and 1 μg of VSV packaging plasmid using Fugene 6 (Roche) according to the manufacturer’s instructions. In the case of retrovirus, HEK293T cells were split 1 to 7 and transfected with 2 μg of pMX vector and 2 μg of pCL-Eco. Cells were incubated at 37°C with 5% CO2. The virus-containing supernatant was collected and filtered (0.45 μm, Millex-HV, Millipore) 48 h and 72 h after infection. Viral stocks for each experiment were generated simultaneously to ensure equal titers. Virus stocks were titrated by Q-PCR and adjusted accordingly for infections.
Q-PCR analysis
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN). cDNA synthesis was performed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Transcript levels were determined using iTaq SYBR Green Supermix with ROX (Bio-Rad). Gene expression was normalized to the housekeeping genes Hprt2 and Rlp37A, and calculated using the delta Ct algorithm. Primers are listed in Table S3.
Reprogramming
OG2 MEFs or heterozygous ROSA26-rtTA/GOF18 MEFs of passage 3 or 4 were split on 12-well plates (3x104 per well). Simultaneously, the cells were infected with virus-containing supernatant supplemented with 6 μg/ml protamine sulfate (Sigma-Aldrich). 30 μl of non-concentrated tetO-SKM virus and 30 μl of rtTA virus (in the case of OG2 MEFs) were added to 1 mL of fresh medium. The volumes of the rest of the viruses were adjusted accordingly to Q-PCR titration. The cells were washed and medium was replaced with dox-containing (1 μg/ml unless otherwise mentioned) ESC medium (high-glucose DMEM with LIF) 48 hours after infection. Alternatively, the infected cells were passaged on inactivated C3H feeders and next day induced with Dox. The number of GFP+ colonies were counted 1 and 2 weeks after Dox induction. Dox was removed after 12 days of induction (2 days before the second evaluation) to count only transgene-independent iPSC colonies.
For the RNAi experiment, OG2 MEFs were infected with pLKO.puro shRNA viruses and selected with (8 μg/ml) puromycin. After 48 hours, the cells were split and infected with rtTA and tetO-OSKM or tetO-SKM viruses as described above. The selection with (4 μg/ml) puromycin was continued during the whole reprogramming experiment to eliminate the cells that silence shRNA transgenes. Statistical significance was calculated using Student’s t test.
For episomal reprogramming MEFs were plated on 6-well plates (105 per well) and transfected with 2 μg of pCXLE-KSM, or 1 μg of pCXLE-KSM and 1 μg of pCXLE-Oct4 using Fugene 6 (Roche). The transfection was repeated after 48h; the medium was changed to ESC medium after another 48h. Only GFP+ epiKSMO iPSCs that appeared within 1 week were picked to avoid epiKSM iPSCs.
Characterization of iPSCs
For immunostaining, the cells were fixed with 4% PFA, permeabilized, and stained for stage-specific embryonic antigen 1 (SSEA-1, Millipore, MC-480, 1:200), Nanog (eBioscience, eBioMLC-51, 1:1000), Oct4 (Santa Cruz, N-19, sc-8628, 1:500) and Sall4 (Santa Cruz, EE-30, sc-101147, 1:500). Naphtol and fast red were used for alkaline phosphatase (AP) staining.
For bisulfate genomic sequencing, 1 μg of genomic DNA was bisulfate converted using EZ DNA methylation kit (Zymo Research) according to the manufacturer’s protocol. The promoter regions of Oct4, Nanog, and Col1a1 were PCR amplified using HotStart Taq (QIAGEN) using primers listed in Table S3. The PCR products were cloned using TOPO-TA kit (Invitrogen) and sequenced. The data were plotted using QUMA.
Combined bisulphite restriction analysis (COBRA) was used to determine DNA methylation status at imprinting loci. Bisulfite conversion was carried out on 2 ug of isolated genomic DNA using the EZ DNA methylation kit (Zymo research) according to the manufacturer’s protocol. The bisulfite converted DNA was amplified by PCR with primers previously described (Zaitoun et al., 2010). PCR products were then purified using the Nucleospin gel and PCR clean-up (MACHEREY-NAGEL) according to the manufacturer’s protocol. The purified PCR products were digested with appropriated restriction enzymes, and digested patterns were analyzed on 3% agarose gel.
To make chimera embryos, 8-cell embryos were flushed from (C57BL/6 x C3H) F1 females x CD1 males at 2.5 days postcoitum (dpc) and placed in M2 medium (Hogan et al., 1986). 8 to 10 trypsinized iPSC cells were transferred into microdrops of potassium simplex optimized medium (KSOM) under mineral oil; each clump was placed in a depression in the microdrop. Meanwhile, batches of 30-40 embryos were briefly incubated in acidified Tyrode’s solution (Hogan et al., 1986) until dissolution of their zona pellucida. A single embryo was placed on the clump and cultured overnight at 37C, 5% CO2. After 24 hours of culture, chimeric embryos were transferred into 2.5-dpc pseudopregnant recipients for further development.
For the teratoma formation assay, 5 × 106 cells were injected subcutaneously into the flank of SCID mice. After 4‒5 weeks, the teratoma that had developed was excised, fixed in 4% paraformaldehyde, and subjected to histological examination with H&E staining.
Tetraploid (4N) complementation was performed as described before (Russell et al., 2015). Briefly, two-cell–stage embryos were fused with electrical pulses. The fused embryos (tetraploid, 4N) were cultured for 24 hr to the 4-cell stage and aggregated with fifteen trypsin-digested iPSCs. The aggregates were cultured at 37C, 5% CO2 for 24 h and subsequently transferred into one uterine horn of a 2.5 dpc pseudopregnant recipient CD-1 mice for full term development.
Mice were housed in the animal facility of the Max Planck Institute for Molecular Biomedicine in Münster in compliance with the GV-SOLAS guidelines and all animal experiments were approved by the State Office for Nature, Environment and Consumer Protection North Rhine-Westphalia (LANUV).
RNA-sequencing and data analysis
Time-course reprogramming RNA-seq was done for two biological replicates of every condition. ESC and iPSC lines were passaged twice in 2iL medium without feeders for RNA isolation. For embryo sequencing, three E9.5 embryos from at least two independent pregnancies were used for each iPSC lines. The embryos of abnormal size or appearance were discarded. Control embryos were trypsinised and FACS-sored similarly to 4N complementation-derived embryos. NEBNExt ultra Directional RNA Kit with polyA enrichment was used for library preparation. Sequencing was done by HiSeq 3000/4000 (Illumina) with paired-end 75bp or single end 150 bp reads. Reads were aligned to the mm10 reference genome using CLC Genomics Workbench 11.0. DESeq2 R package were used for the differential expression analysis. mFUZZ package was used for time-course gene clustering (Kumar and E Futschik, 2007). Gplots and pheatmap R packages were used for plotting the histograms.
Generating Oct4 knockout fibroblasts
For Oct4 conditional knockout, male Acr-EGFP ESCs (Adachi et al., 2018) were targeted with the construct containing floxed Pou5f1 exon 2-5 together with a FRT-IRES-βgeo-pA cassette and transiently transfected with a FLPe expression vector to remove the FRT cassette. Chimeric mice were generated by aggregation of Oct4WT/flox ESCs with zona pellucida-free eight-cell-stage embryos. Oct4flox/flox; Rosa26-CreERT2 MEFs were conditionally immortalized with lentiviral dox-inducible SV40LT and simultaneously treated with tamoxifen. The clonal lines were genotyped with a mix of following primers: forward 5′-GCTCCAACAACCTGCTCCTCTTCCGCC-3′, 5′-GGATGCTGTGAGCCAAGG-3′, and reverse 5′-GCTTTCTCCAACCGCAGGCTCTCT-3′.
Data and Code Availability
RNA-seq data are available from Gene Expression Omnibus under accession number GEO: GSE137001.
Acknowledgments
We thank Konrad Hochedlinger for providing the tetO-BKSM and tetO-OKSM constructs and Konstantinos Anastassiadis for the Bluescript-SV40LT vector. We are grateful to Martina Sinn for histology, Ingrid Gelker and Maryna Samus for cell culture, Martin Stehling for FACS, Dagmar Zeuschner for electron microscopy, Dong Han for providing Oct4 flox/flox MEFs, and Jonghun Kim and Dong Wook Han for G-band karyotyping. We also thank Janina Fuss, Kurt Stüber, and Anika Witten for RNA-seq; Areti Malapetsas for editing the manuscript; and Oleksandr Dovgusha, Clemens Hug, Eva Kutejova, and Joschka Hey for providing the scripts for RNA-seq analysis. The work was funded by the Max Planck Society and the Max Planck Center grant, as well as the ERC (advanced grant proposal 669168).
Author Contributions
S.V. and H.R.S. conceived the study. S.V. performed most of the experiments; assembled, analyzed, and interpreted data; and wrote the manuscript. K.A. and G.W. generated Oct4flox/flox mouse. K.A. advised on the project and analyzed published ChIP-seq data. K.-P.K. performed DNA methylation assays. Y.H. performed qPCRs. C.M.M. wrote the manuscript. G.W. performed all the mouse experiments, generated ESC lines, and assembled and interpreted the data. H.R.S. supervised the project, interpreted data, and wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: November 7, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2019.10.002.
Contributor Information
Guangming Wu, Email: wu_guangming@grmh-gdl.cn.
Hans R. Schöler, Email: office@mpi-muenster.mpg.de.
Supporting Citations
The following references appear in the Supplemental Information: Eguchi et al., 2016, Fritz et al., 2015, Gao et al., 2013a, Hou et al., 2013, Jerabek et al., 2017, Redmer et al., 2011, Tan et al., 2015, Tapia et al., 2012.
Supplemental Information
References
- Adachi K., Kopp W., Wu G., Heising S., Greber B., Stehling M., Araúzo-Bravo M.J., Boerno S.T., Timmermann B., Vingron M. Esrrb unlocks silenced enhancers for reprogramming to naive pluripotency. Cell Stem Cell. 2018;23:266–275. doi: 10.1016/j.stem.2018.05.020. [DOI] [PubMed] [Google Scholar]
- Amlani B., Liu Y., Chen T., Ee L.-S., Lopez P., Heguy A., Apostolou E., Kim S.Y., Stadtfeld M. Nascent induced pluripotent stem cells efficiently generate entirely iPSC-derived mice while expressing differentiation-associated genes. Cell Rep. 2018;22:876–884. doi: 10.1016/j.celrep.2017.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K., Rostovskaya M., Lubitz S., Weidlich S., Stewart A.F. Precise conditional immortalization of mouse cells using tetracycline-regulated SV40 large T-antigen. Genesis. 2010;48:220–232. doi: 10.1002/dvg.20605. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O., Verheul C., Sommer A.G., Brumbaugh J., Schwarz B.A., Lipchina I., Huebner A.J., Mostoslavsky G., Hochedlinger K. Lineage conversion induced by pluripotency factors involves transient passage through an iPSC stage. Nat. Biotechnol. 2015;33:761–768. doi: 10.1038/nbt.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D.A., Cheng A.W., Itskovich E., Markoulaki S., Ganz K., Klemm S.L., van Oudenaarden A., Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Markoulaki S., van Wietmarschen N., Hoke H., Wu T., Ganz K., Akhtar-Zaidi B., He Y., Abraham B.J., Porubsky D. The developmental potential of iPSCs is greatly influenced by reprogramming factor selection. Cell Stem Cell. 2014;15:295–309. doi: 10.1016/j.stem.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M., Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Beard C., Hanna J., Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat. Methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J.H., Faddah D.A., Buganim Y., Kim J., Ganz K., Steine E.J., Cassady J.P., Creyghton M.P. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Chang G., Gao S., Hou X., Xu Z., Liu Y., Kang L., Tao Y., Liu W., Huang B., Kou X. High-throughput sequencing reveals the disruption of methylation of imprinted gene in induced pluripotent stem cells. Cell Res. 2014;24:293–306. doi: 10.1038/cr.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang Z., Li L., Chen B.-C., Revyakin A., Hajj B., Legant W., Dahan M., Lionnet T., Betzig E. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Gao Y., Huang H., Xu K., Chen X., Jiang Y., Li H., Gao S., Tao Y., Wang H. The combination of Tet1 with Oct4 generates high-quality mouse-induced pluripotent stem cells. Stem Cells. 2015;33:686–698. doi: 10.1002/stem.1879. [DOI] [PubMed] [Google Scholar]
- Chen J., Chen X., Li M., Liu X., Gao Y., Kou X., Zhao Y., Zheng W., Zhang X., Huo Y. Hierarchical Oct4 binding in concert with primed epigenetic rearrangements during somatic cell reprogramming. Cell Rep. 2016;14:1540–1554. doi: 10.1016/j.celrep.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Chronis C., Fiziev P., Papp B., Butz S., Bonora G., Sabri S., Ernst J., Plath K. Cooperative binding of transcription factors orchestrates reprogramming. Cell. 2017;168:442–459.e20. doi: 10.1016/j.cell.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A., Wleklinski M.J., Spurgat M.C., Heiderscheit E.A., Kropornicka A.S., Vu C.K., Bhimsaria D., Swanson S.A., Stewart R., Ramanathan P. Reprogramming cell fate with a genome-scale library of artificial transcription factors. Proc. Natl. Acad. Sci. USA. 2016;113:E8257–E8266. doi: 10.1073/pnas.1611142114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri A.L., Cavallaro M., Braida D., Di Cristofano A., Canta A., Vezzani A., Ottolenghi S., Pandolfi P.P., Sala M., DeBiasi S., Nicolis S.K. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- Fritz A.L., Adil M.M., Mao S.R., Schaffer D.V. CAMP and EPAC signaling functionally replace OCT4 during induced pluripotent stem cell reprogramming. Mol. Ther. 2015;23:952–963. doi: 10.1038/mt.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frum T., Halbisen M.A., Wang C., Amiri H., Robson P., Ralston A. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev. Cell. 2013;25:610–622. doi: 10.1016/j.devcel.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yang J., Tsang J.C.H., Ooi J., Wu D., Liu P. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Reports. 2013;1:183–197. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Chen J., Li K., Wu T., Huang B., Liu W., Kou X., Zhang Y., Huang H., Jiang Y. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- González F., Huangfu D. Mechanisms underlying the formation of induced pluripotent stem cells. Wiley Interdiscip. Rev. Dev. Biol. 2016;5:39–65. doi: 10.1002/wdev.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng J.C.D., Feng B., Han J., Jiang J., Kraus P., Ng J.H., Orlov Y.L., Huss M., Yang L., Lufkin T. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B., Costantini F., Lacy E. Cold Spring Harbor Laboratory Press; 1986. Manipulating the Mouse Embryo: A Laboratory Manual. [Google Scholar]
- Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hussein S.M.I., Puri M.C., Tonge P.D., Benevento M., Corso A.J., Clancy J.L., Mosbergen R., Li M., Lee D.-S., Cloonan N. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- Jerabek S., Ng C.K., Wu G., Arauzo-Bravo M.J., Kim K.P., Esch D., Malik V., Chen Y., Velychko S., MacCarthy C.M. Changing POU dimerization preferences converts Oct6 into a pluripotency inducer. EMBO Rep. 2017;18:319–333. doi: 10.15252/embr.201642958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.B., Sebastiano V., Wu G., Araúzo-Bravo M.J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kim J.B., Greber B., Araúzo-Bravo M.J., Meyer J., Park K.I., Zaehres H., Schöler H.R. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar L., E Futschik M. Mfuzz: a software package for soft clustering of microarray data. Bioinformation. 2007;2:5–7. doi: 10.6026/97320630002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Lim C.Y., Tam W.L., Zhang J., Ang H.S., Jia H., Lipovich L., Ng H.H., Wei C.L., Sung W.K., Robson P. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell. 2008;3:543–554. doi: 10.1016/j.stem.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Liu L., Luo G.Z., Yang W., Zhao X., Zheng Q., Lv Z., Li W., Wu H.J., Wang L., Wang X.J., Zhou Q. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J. Biol. Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y.-H., Wu Q., Chew J.-L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik V., Glaser L.V., Zimmer D., Velychko S., Weng M., Holzner M., Arend M., Chen Y., Srivastava Y., Veerapandian V. Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat. Commun. 2019;10:3477. doi: 10.1038/s41467-019-11054-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthaler A.G., Adachi K., Tiemann U., Wu G., Sabour D., Velychko S., Kleiter I., Schöler H.R., Tapia N. Enhanced OCT4 transcriptional activity substitutes for exogenous SOX2 in cellular reprogramming. Sci. Rep. 2016;6:19415. doi: 10.1038/srep19415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A.A. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montserrat N., Nivet E., Sancho-Martinez I., Hishida T., Kumar S., Miquel L., Cortina C., Hishida Y., Xia Y., Esteban C.R., Izpisua Belmonte J.C. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Nagy A., Gócza E., Diaz E.M., Prideaux V.R., Iványi E., Markkula M., Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa O., Yokota Y., Ishida H., Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Pasini D., Cloos P.A.C., Walfridsson J., Olsson L., Bukowski J.P., Johansen J.V., Bak M., Tommerup N., Rappsilber J., Helin K. JARID2 regulates binding of the polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Pesce M., Schöler H.R. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzisheuskaya A., Silva J.C. Do all roads lead to Oct4? The emerging concepts of induced pluripotency. Trends Cell Biol. 2014;24:275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmer T., Diecke S., Grigoryan T., Quiroga-Negreira A., Birchmeier W., Besser D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reményi A., Lins K., Nissen L.J.L.J., Reinbold R., Schöler H.R., Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A., Kong S.W., Agarwal P., Gilliss B., Pu W.T., Black B.L. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol. Cell. Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R., Ilg M., Lin Q., Wu G., Lechel A., Bergmann W., Eiseler T., Linta L., Kumar P P., Klingenstein M. A dynamic role of TBX3 in the pluripotency circuitry. Stem Cell Reports. 2015;5:1155–1170. doi: 10.1016/j.stemcr.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Do J.T., Desponts C., Hahm H.S., Schöler H.R., Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- Shu J., Deng H. Lineage specifiers: new players in the induction of pluripotency. Genomics Proteomics Bioinformatics. 2013;11:259–263. doi: 10.1016/j.gpb.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Wu C., Wu Y., Li Z., Shao S., Zhao W., Tang X., Yang H., Shen L., Zuo X. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu J., Zhang K., Zhang M., Yao A., Shao S., Du F., Yang C., Chen W., Wu C., Yang W. GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res. 2015;25:169–180. doi: 10.1038/cr.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T.W., Guo G., van Oosten A.L., Barrandon O., Wray J., Yamanaka S., Chambers I., Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A., Donahue G., Zaret K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Maherali N., Breault D.T., Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Ferrari F., Choi J., Walsh R.M., Chen T., Ooi S.S., Kim S.Y., Bestor T.H., Shioda T. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44:398–405. doi: 10.1038/ng.1110. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.L., Stuhlmann H., Jähner D., Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S.A., Dykxhoorn D.M., Palliser D., Mizuno H., Yu E.Y., An D.S., Sabatini D.M., Chen I.S., Hahn W.C., Sharp P.A. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takikawa S., Ray C., Wang X., Shamis Y., Wu T.Y., Li X. Genomic imprinting is variably lost during reprogramming of mouse iPS cells. Stem Cell Res. (Amst.) 2013;11:861–873. doi: 10.1016/j.scr.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F., Qian C., Tang K., Abd-Allah S.M., Jing N. Inhibition of transforming growth factor β (TGF-β) signaling can substitute for Oct4 protein in reprogramming and maintain pluripotency. J. Biol. Chem. 2015;290:4500–4511. doi: 10.1074/jbc.M114.609016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura N., Saito M., Ebisuya M., Nishida E., Ishikawa F. Stemness-related factor Sall4 interacts with transcription factors Oct-3/4 and Sox2 and occupies Oct-Sox elements in mouse embryonic stem cells. J. Biol. Chem. 2013;288:5027–5038. doi: 10.1074/jbc.M112.411173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia N., Reinhardt P., Duemmler A., Wu G., Araúzo-Bravo M.J., Esch D., Greber B., Cojocaru V., Rascon C.A., Tazaki A. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 2012;3:1279. doi: 10.1038/ncomms2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.-Y., Bouwman B.A., Ang Y.-S., Kim S.J., Lee D.-F., Lemischka I.R., Rendl M. Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells. 2011;29:964–971. doi: 10.1002/stem.649. [DOI] [PubMed] [Google Scholar]
- Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., Meissner A. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velychko S., Kang K., Kim S.M., Kwak T.H., Kim K.-P., Park C., Hong K., Chung C., Hyun J.K., MacCarthy C.M. Fusion of reprogramming factors alters the trajectory of somatic lineage conversion. Cell Rep. 2019;27:30–39.e4. doi: 10.1016/j.celrep.2019.03.023. [DOI] [PubMed] [Google Scholar]
- Wang J., Rao S., Chu J., Shen X., Levasseur D.N., Theunissen T.W., Orkin S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen J., Hu J.-L., Wei X.-X., Qin D., Gao J., Zhang L., Jiang J., Li J.-S., Liu J. Reprogramming of mouse and human somatic cells by high-performance engineered factors. EMBO Rep. 2011;12:373–378. doi: 10.1038/embor.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Misumi I., Gu A.D., Curtis T.A., Su L., Whitmire J.K., Wan Y.Y. GATA-3 controls the maintenance and proliferation of T cells downstream of TCR and cytokine signaling. Nat. Immunol. 2013;14:714–722. doi: 10.1038/ni.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A.B., Zhang Y.V., Tumbar T. Gata6 promotes hair follicle progenitor cell renewal by genome maintenance during proliferation. EMBO J. 2017;36:61–78. doi: 10.15252/embj.201694572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Wang H., He J., Kang L., Jiang Y., Liu J., Zhang Y., Kou Z., Liu L., Zhang X., Gao S. Reprogramming of trophoblast stem cells into pluripotent stem cells by Oct4. Stem Cells. 2011;29:755–763. doi: 10.1002/stem.617. [DOI] [PubMed] [Google Scholar]
- Wu G., Han D., Gong Y., Sebastiano V., Gentile L., Singhal N., Adachi K., Fischedick G., Ortmeier C., Sinn M. Establishment of totipotency does not depend on Oct4A. Nat. Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Liu Y., Wen D., Tseng Z., Tahmasian M., Zhong M., Rafii S., Stadtfeld M., Hochedlinger K., Xiao A. Histone variant H2A.X deposition pattern serves as a functional epigenetic mark for distinguishing the developmental potentials of iPSCs. Cell Stem Cell. 2014;15:281–294. doi: 10.1016/j.stem.2014.06.004. [DOI] [PubMed] [Google Scholar]
- Yang B.X.X., El Farran C.A., Guo H.C.C., Yu T., Fang H.T.T., Wang H.F.F., Schlesinger S., Seah Y.F.S.F.S., Goh G.Y.L.Y.L., Neo S.P.P. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitoun I., Downs K.M., Rosa G.J.M., Khatib H. Upregulation of imprinted genes in mice: an insight into the intensity of gene expression and the evolution of genomic imprinting. Epigenetics. 2010;5:149–158. doi: 10.4161/epi.5.2.11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeineddine D., Papadimou E., Chebli K., Gineste M., Liu J., Grey C., Thurig S., Behfar A., Wallace V.A., Skerjanc I.S., Pucéat M. Oct-3/4 dose dependently regulates specification of embryonic stem cells toward a cardiac lineage and early heart development. Dev. Cell. 2006;11:535–546. doi: 10.1016/j.devcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tam W.-L., Tong G.Q., Wu Q., Chan H.-Y., Soh B.-S., Lou Y., Yang J., Ma Y., Chai L. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat. Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- Zhong X., Cui P., Cai Y., Wang L., He X., Long P., Lu K., Yan R., Zhang Y., Pan X. Mitochondrial dynamics is critical for the full pluripotency and embryonic developmental potential of pluripotent stem cells. Cell Metab. 2019;29:979–992.e4. doi: 10.1016/j.cmet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- Zunder E.R., Lujan E., Goltsev Y., Wernig M., Nolan G.P. A continuous molecular roadmap to iPSC reprogramming through progression analysis of single-cell mass cytometry. Cell Stem Cell. 2015;16:323–337. doi: 10.1016/j.stem.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are available from Gene Expression Omnibus under accession number GEO: GSE137001.