Summary
Radiotherapy and chemotherapy disrupt bone vasculature, but the underlying causes and mechanisms enabling vessel regeneration after bone marrow (BM) transplantation remain poorly understood. Here, we show that loss of hematopoietic cells per se, in response to irradiation and other treatments, triggers vessel dilation, permeability, and endothelial cell (EC) proliferation. We further identify a small subpopulation of Apelin-expressing (Apln+) ECs, representing 0.003% of BM cells, that is critical for physiological homeostasis and transplant-induced BM regeneration. Genetic ablation of Apln+ ECs or Apln-CreER-mediated deletion of Kitl and Vegfr2 disrupt hematopoietic stem cell (HSC) maintenance and contributions to regeneration. Consistently, the fraction of Apln+ ECs increases substantially after irradiation and promotes normalization of the bone vasculature in response to VEGF-A, which is provided by transplanted hematopoietic stem and progenitor cells (HSPCs). Together, these findings reveal critical functional roles for HSPCs in maintaining vascular integrity and for Apln+ ECs in hematopoiesis, suggesting potential targets for improving BM transplantation.
Keywords: irradiation, bone marrow transplantation, hematopoietic stem cell, stem cell niche, vessel regeneration, Apln (Apelin), VEGF, Esm1, VEGFR, endothelial cell heterogeneity
Graphical Abstract
Highlights
-
•
Loss of hematopoietic cells phenocopies irradiation-induced vascular defects
-
•
Identification and characterization of Apln+ ECs in adult BM
-
•
Apln+ ECs regulate HSC maintenance and steady-state hematopoiesis
-
•
Apln+ ECs expand, respond to HSPCs, and drive post-transplantation recovery
Chen and colleagues identify an Apln+ subpopulation of bone marrow endothelial cells (ECs), which are distinct from other sinusoidal ECs. Apln+ ECs regulate HSC maintenance, hematopoiesis, hematopoietic reconstitution, and vascular regeneration after irradiation. Crosstalk between Apln+ ECs and donor-cell-derived VEGF-A can be manipulated to improve bone marrow transplantation efficiency.
Introduction
Irradiation or chemotherapy followed by allogenic stem cell transplantation is a major therapeutic approach for the treatment of leukemia and other malignant and non-malignant diseases (Copelan, 2006, Ferrara et al., 2009, Kondo et al., 2003, Morgan et al., 2017, Rafii and Lyden, 2003, Thomas et al., 1975). High transplantation efficiency is beneficial for both patients and donors (Ballen et al., 2013, Copelan, 2006, Ratajczak and Suszynska, 2016). Hematopoietic stem cell (HSC) engraftment, proliferation, and differentiation are critically regulated by niche microenvironments, which involve endothelial cells (ECs) and vessel-associated mesenchymal cells (Comazzetto et al., 2019, Crane et al., 2017, Ding et al., 2012, Doan et al., 2013a, Greenbaum et al., 2013, Himburg et al., 2018, Hoggatt et al., 2016, Mendelson and Frenette, 2014, Rafii et al., 2016, Ramasamy et al., 2016, Severe et al., 2019, Sugiyama et al., 2006, Zhao et al., 2014). Whereas it is proposed that lethal irradiation triggers acute loss of ECs, vessel dilation or swelling and changes in permeability have been also reported (Bowers et al., 2018, Doan et al., 2013b, Hassanshahi et al., 2017, Himburg et al., 2016, Hooper et al., 2009, Li et al., 2008, Slayton et al., 2007, Zhou et al., 2015). Thus, our understanding of irradiation and chemotherapy-induced vascular damage and the factors promoting regeneration remain insufficient.
In development but also in the adult skeletal system, ECs are heterogeneous and form different subpopulations (Baryawno et al., 2019, Itkin et al., 2016, Kusumbe et al., 2014, Langen et al., 2017, Rafii et al., 2016, Tikhonova et al., 2019). It is unclear whether all capillary ECs are equally important for hematopoiesis and HSC maintenance. Apelin, encoded by gene Apln, is a secreted peptide whose function is reported to be redundant with Elabela/Apela (Helker et al., 2015, Ho et al., 2017, Kim, 2014, Kuba et al., 2019). Apelin is primarily secreted from a subset of capillary ECs, namely, Apln+ ECs, which can be genetically targeted by Apln-CreER knockin mice (Chen et al., 2016, Liu et al., 2015, Tian et al., 2013). In the growing retinal vasculature, Apln expression is enriched in tip ECs at the distal end of vessel sprouts (del Toro et al., 2010). Apln expression also marks highly proliferative ECs in many developing organs (Langen et al., 2017, Liu et al., 2015, Pitulescu et al., 2017), whereas Apln+ ECs largely disappear in the adult vasculature consistent with its quiescent, non-proliferative status (Liu et al., 2015).
Here, we have combined inducible mouse genetics, flow cytometry, RNA sequencing (RNA-seq), and advanced imaging approaches to show that adult Apln+ ECs are critical for the maintenance of steady-state hematopoiesis as well as vascular regeneration and hematopoietic reconstitution after bone marrow (BM) transplantation.
Results
Irradiation-Induced Changes in Bone ECs
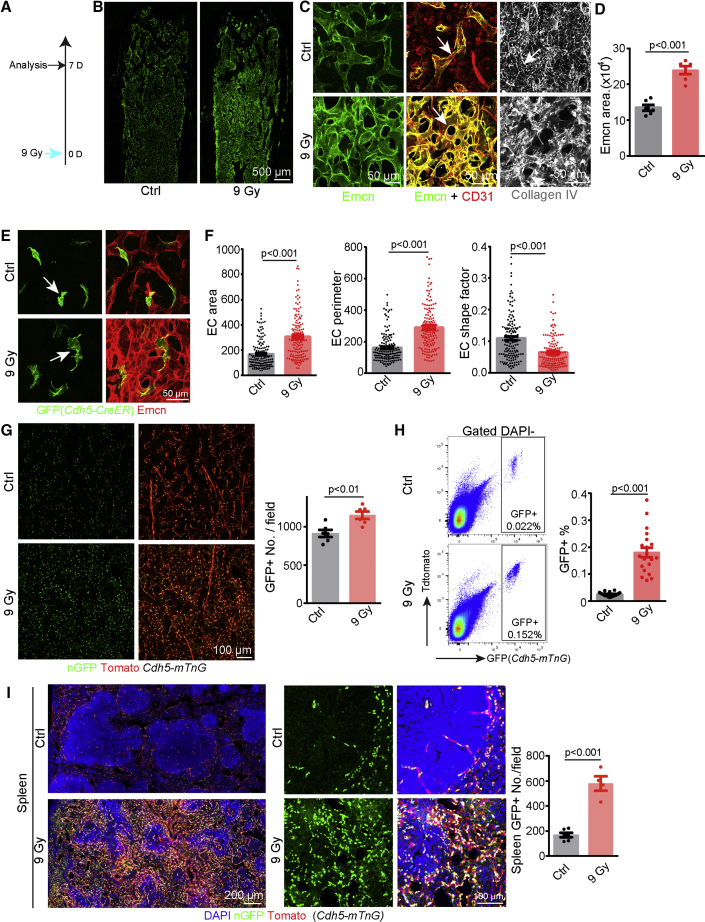
Making use of advanced bone processing and imaging protocols, we found that lethal total body irradiation (9 Gy) of adult mice results in profound alterations of the long bone vasculature at 7 days post-irradiation, including the disruption of columnar capillaries in the metaphysis, dilation of sinusoidal capillaries in the diaphysis, and expansion of the vascular area in bone, visualized by immunostaining of the sialoglycoprotein Endomucin (Emcn) (Figures 1A–1D and S1A–S1C). Irradiation also causes an increase in vessel permeability, as indicated by enhanced tracer extravasation (Figure S1C). Whereas CD31+ Emcn– hematopoietic cells are largely absent after irradiation, endothelial CD31 expression is upregulated, and vessel-associated collagen IV+ reticular fibers are disrupted (Figure 1C). At 1 day after irradiation, BM vascular morphology is already changed with higher expression of Emcn relative to 3 h post-irradiation (Figure S1D). At 4 days post-irradiation, alterations in vascular morphology, such as vessel dilation, are more profound, indicating dynamic changes over several days (Figure S1D). To reveal the morphological changes of ECs at single-cell level, we treated Cdh5(PAC)-CreERT2 Rosa26-mTmG double-transgenic mice (Table S1) with low doses of tamoxifen. The analysis of rare recombined and therefore isolated GFP+ cells indicates substantial changes in EC size and shape complexity at single-cell resolution (Figures 1E and 1F). Next, we checked the density of ECs after irradiation, which was aided by Cdh5(PAC)-mtdTomato-nGFP (Cdh5-mTnG) reporter mice in which membrane-anchored tdTomato fluorescent protein labels the surface and histone H2B-coupled GFP (nGFP) the nuclei of ECs (Jeong et al., 2017) (Table S1). Confirming the specificity of this reporter, H2B-GFP signal co-localizes with nuclear ERG, a transcription factor and EC marker, in postnatal day (P) 6 heart endothelium but not in the EC-derived mesenchymal cell in heart valves (Figure S1E). Further proving that Cdh5-mTnG reporter marks genuine ECs but not EC-derived cell populations, GFP signal in bone decorates Emcn+ and VEGFR2+ (vascular endothelial growth factor receptor 2+) ECs without labeling CD31+ Emcn–, B220+, and lineage committed hematopoietic cells (Figures S1F–S1J). Lethal irradiation of adult Cdh5-mTnG mice revealed a significant increase in EC density and percentage both by analysis of bone sections and flow cytometry (Figures 1G and 1H). Moreover, a higher number and ratio of GFP+EdU+ signals are detected in Cdh5-mTnG bone sections after irradiation (Figure S1K). In contrast, active caspase 3 immunostaining and annexin V binding, which indicate apoptosis, are not increased in bone ECs at 3 or 24 h post-irradiation (Figures S1L and S1M). These results show that irradiation disrupts the normal organization of bone capillaries and leads to increases in EC density and vascular permeability.
Figure 1.
Irradiation-Induced Changes in the Vasculature of BM and Spleen
(A) Schematic representation of protocol for lethal irradiation analysis.
(B) Tile scan overview images of Emcn-stained vessels in adult femur after irradiation.
(C) Emcn, CD31, and collagen IV immunostaining of control and irradiated BM, as indicated. Arrows mark Emcn+ CD31+ vessels in middle panel and collagen IV+ reticular fiber on the right.
(D) Quantification of Emcn+ area in imaging field (n = 6 per group).
(E) Morphology of individual ECs (arrows) in control and irradiated bones of Cdh5-CreER R26-mTmG mice. Low dosage of tamoxifen was injected 6 days after irradiation.
(F) Quantification of area, perimeter, and shape factor from control (n = 147 from 3 mice) and 9 Gy (n = 140 from 4 mice) single ECs. Shape factor is a numerical indication of how similar a 2D shape is to a perfect circle, which has a shape factor of 1.
(G) Nuclear GFP+ (nGFP+) ECs in control and irradiated Cdh5-mTnG diaphysis. Graph shows quantitation of GFP+ cells (n = 6 in each group).
(H) FACS plot of GFP+ cells from control and irradiated Cdh5-mTnG mice. Graph show frequency of GFP+ cells (n = 20 in each group).
(I) Tile scan overview images and selected maximum intensity projections of spleen vessels in Cdh5-mTnG mice. Quantification of GFP+ nuclei in each image field (Ctrl n = 6; 9 Gy n = 4) is shown.
Error bars, mean ± SEM. p values, two-tailed unpaired Student’s t test.
See also Figure S1.
To investigate whether irradiation disrupts the vasculature in other organs, we analyzed different vascular networks in Cdh5-mTnG mice. Strikingly, no overt changes in vessel morphology and EC density were visible in retina, heart, skin, and small intestinal villi, which have no extramedullary hematopoietic function (Figure S1N–S1Q). In spleen, a major adult extramedullary organ, lethal irradiation disrupts the white pulp and leads to significant higher density of splenic vessels (Figure 1I). These findings support that ECs are far more resistant to irradiation than hematopoietic cells and suggest that vascular alterations after irradiation are particularly strong in organs with hematopoietic function. Very similar to irradiation, the chemotherapy drug 5-fluoracil (5-FU) (Noach et al., 2003) induces profound changes in the bone vasculature including expansion of the Emcn+ area and dilation of sinusoidal vessels, disruption of collagen IV+ reticular fibers, CD31 upregulation in ECs, morphological alterations of ECs at the single-cell level, and increased EC density (Figures S2A–S2H).
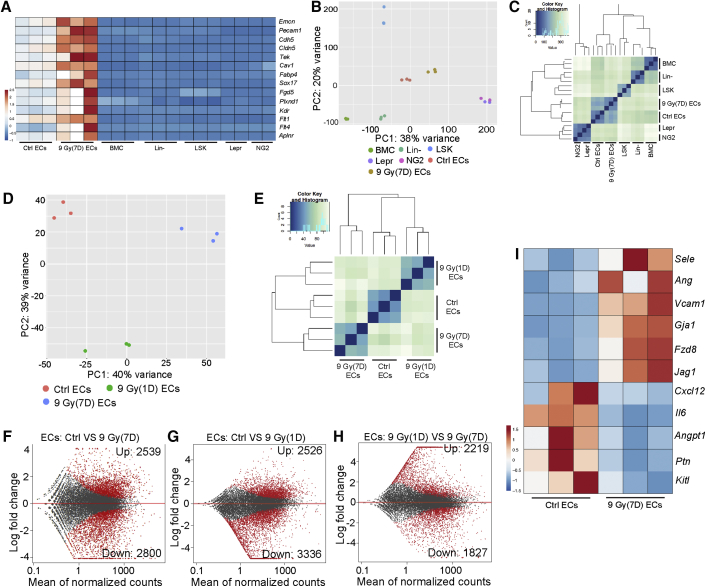
To identify molecular changes in the bone endothelium after irradiation, we used fluorescence-activated cell sorting (FACS) to isolate GFP+ ECs from adult (8–12 weeks old) Cdh5-mTnG long bone for RNA-seq analysis. These FACS-sorted GFP+ cells show significant enrichment of core EC markers, confirming their endothelial identity (Figure 2A). Principal-component analysis (PCA) divides normal (control) GFP+ ECs and irradiated GFP+ ECs (7 days post-irradiation) into two separate clusters, both of which are distinct from other BM cell groups (Figure 2B). According to unsupervised clustering, normal ECs and irradiated ECs are more similar to hematopoietic cells than to bone mesenchymal cell populations (Figure 2C). Furthermore, we found that the endothelial transcriptome is significantly changed only 1 day after irradiation (Figures 2D and 2E). EC transcriptomes at 1 day and 7 days post-irradiation are also significantly different from each other, suggesting dynamic changes consistent with the structural alterations in the bone vasculature at different time points after the treatment (Figures 2D, 2E, and S1B–S1D). At 7 days after irradiation, GFP+ bone ECs show profound changes in their transcription profile with more than 2,000 upregulated genes and downregulated genes at p < 0.05 significance level relative to control GFP+ ECs (Figures 2F, 2G, and S2I). More than 4,000 differentially expressed genes were also detected between 1 and 7 days after irradiation (Figure 2H). Several molecular signals associated with vascular niche function are significantly changed in ECs exposed to irradiation (Figure 2I), whereas genes related to DNA damage and senescence are not significantly altered (Figures S2J and S2K). These data indicate that irradiation substantially changes the transcription profile of bone ECs, including genes associated with EC niche function.
Figure 2.
Irradiation Induces Dynamic Changes in the Transcription Profile of BM ECs
(A) Heatmap of selected EC-enriched genes in BM cells, Lin– cells, LSK cells, ECs, irradiated ECs (9 Gy, 7 days), Lepr+, and NG2+ cells.
(B) PCA plot of control and irradiated ECs together with other BM cell types. Data for Lepr+ and NG2+ cells were previously published (Asada et al., 2017).
(C) Sample distance analysis of different BM cell types, as indicated.
(D and E) PCA plot (D) and sample distance analysis (E) of ctrl ECs and ECs at 1 day or 7 days after irradiation.
(F–H) MA plot showing differentially expressed genes (red dots) between Ctrl ECs and ECs at 7 days after irradiation (9 Gy) (F), Ctrl ECs and ECs at 1 day after irradiation (G), and ECs at 1 and 7 days after irradiation (H), as indicated.
(I) Heatmap of selected HSC niche-related genes after irradiation.
See also Figure S2.
Loss of Hematopoietic Cells Phenocopies Irradiation-Induced Vascular Defects
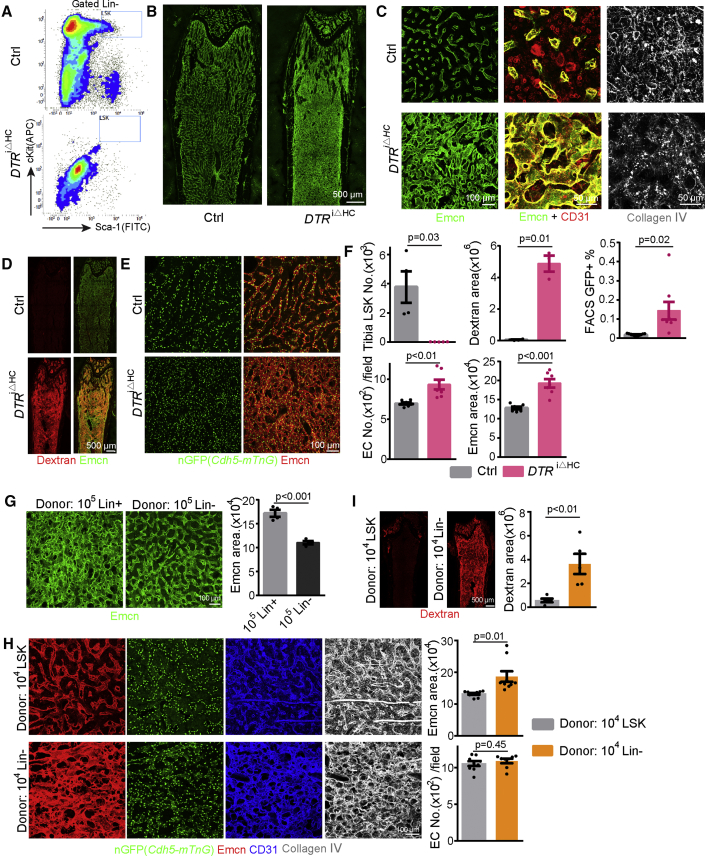
Irradiation and 5-FU treatment target multiple cell types (Maxwell et al., 2003, Yahyapour et al., 2018). As BM hematopoietic cells are largely eliminated after irradiation and chemotherapy, we reasoned that this loss might be a key contributor to the observed vascular defects. To test this hypothesis, we generated Vav1-Cre Rosa26-DTR mice (DTRiΔHC) expressing diphtheria toxin receptor in hematopoietic cells (Table S1). The Vav1-Cre line used in these experiments does not induce recombination in ECs in adult femur (Figures S3A–S3D). Moreover, diphtheria toxin injection into Vav1-Cre Cre-negative mice has no effect on hematopoietic cells and bone vessels (Figure S3E). In contrast, diphtheria toxin injection in DTRiΔHC animals results in significant deletion of hematopoietic cells including Lin– Sca1+ cKit+ (LSK) multipotent progenitors (Figures 3A, 3F, S3F, and S3G). This hematopoietic cell ablation model, which is independent of irradiation and chemotherapy, leads to expansion of the Emcn+ area, dilation of sinusoidal vessels, higher EC density, morphological alterations of BM ECs, disruption of reticular fibers, and increased permeability (Figures 3B–3F), phenocopying the vascular alterations after irradiation and 5-FU treatment. Furthermore, RNA-seq analysis of sorted DTRiΔHC ECs reveals similar changes in niche-related gene transcripts (Figures S3H–S3J). These data indicate that hematopoietic cells maintain physiological properties of the bone vasculature and suggest the existence of reciprocal interactions between hematopoietic cells and ECs.
Figure 3.
Diphtheria Toxin-Mediated Hematopoietic Cell Ablation Mimics Irradiation-Induced Vascular Defects
(A) FACS plot of LSK cells 4 days after diphtheria toxin injection in DTRiΔHC and control mice.
(B) Tile scan overview images of Emcn-stained vessels in DTA-treated control and DTRiΔHC long bone.
(C) High-magnification images showing Emcn, CD31, and collagen IV immunostaining in DTRiΔHC and control femur.
(D) Tile scan images showing extravasation of fluorescent Dextran in DTRiΔHC and control long bone.
(E) Cdh5-mTnG–controlled GFP+ EC nuclei and Emcn staining in DTRiΔHC and control diaphysis.
(F) Quantification of LSK cell number (Ctrl = 4, DTRiΔHC= 5), Emcn+ area (Ctrl = 7, DTRiΔHC= 7), Dextran+ area (Ctrl = 4, DTRiΔHC= 3), and GFP+ cells per image (Ctrl = 7, DTRiΔHC= 7, frequency of GFP+ cells by FACS (Ctrl = 11, DTRiΔHC= 8).
(G) Representative images and quantification of Emcn+ area at 16 days after irradiation and transplantation of 4 × 105 Lin– or Lin+ cells (n = 4 in each group)
(H) Emcn, GFP+ EC nuclei, CD31, and collagen IV at 16 days after irradiation and transplantation of 2 × 104 Lin– or LSK cells. Quantification of Emcn area (LSK = 10, Lin– = 9) and GFP+ EC nuclei in each image field (LSK = 9, Lin– = 8).
(I) Tile scan overview images showing Dextran extravasation at 16 days after irradiation and transplantation of 2 × 104 Lin– or LSK cells. Quantification of Dextran+ area (LSK = 5, Lin– = 5).
Error bars, mean ± SEM. p values, two-tailed unpaired Student’s t test. See also Figure S3.
Transplantation of the same number of different hematopoietic cell subpopulations after irradiation showed that non-committed, lineage-negative cells (Lin–: Ter119– CD5– CD11b– CD45R– Ly-6G/C–) induce vessel regeneration more efficiently than lineage committed cells (Lin+) (Figures 3G and S3K). Multipotent progenitor LSK cells are even more efficient in this assay and induce vessel regeneration within 16 days even when a lower number of cells was transplanted (Figures 3H, 3I, and S3L). Following the transplantation of LSK cells, recovering BM vessels show improved vessel integrity and a reduction of the Emcn+ area, indicating reduced vessel dilation (Figures 3H and 3I). BM regeneration after irradiation and transplantation does not involve the differentiation of donor-derived LSK cells into ECs in the recipient vasculature (Figures S3N and S3O). Mathematical regression analysis indicates that the number of BM nuclear cells (BMNCs) in a broad range (2 × 106 to 8 × 106 cells) is not proportional to the extent of vessel regeneration (Figures S3M, S7L, and S7Q). These data argue that LSK cells, which are enriched in hematopoietic stem and progenitor cells (HSPCs), have specific properties that promote the recovery of the recipient vasculature after irradiation more efficiently than other hematopoietic cell subsets.
Identification of Apln+ ECs in Adult BM
Whereas different EC subpopulations have been proposed to provide vascular niches for HSPCs in BM, it is currently unknown whether the recovery of the bone vasculature after irradiation or 5-FU treatment is mediated by all or only certain ECs. Bmx-CreERT2 transgenic mice enable efficient and irreversible genetic labeling of arterial ECs (Ehling et al., 2013, Xu et al., 2018). To check whether arterial cells contribute to bone vessel regeneration, we generated Bmx-CreERT2 Rosa26-mTmG (Bmx-mTmG) double-transgenic mice in which tamoxifen injection irreversibly labels arterial ECs and their descendants by GFP expression (Table S1). Irradiation and 5-FU treatment did not induce significant morphological changes of arteries (Figures S4A, S4B, and S2F). Bmx-CreERT2 mice mediated lineage tracing indicates no apparent contribution of arterial ECs to regenerating sinusoidal vessels after irradiation and BM transplantation (Figure S4C) (Ehling et al., 2013, Xu et al., 2018).
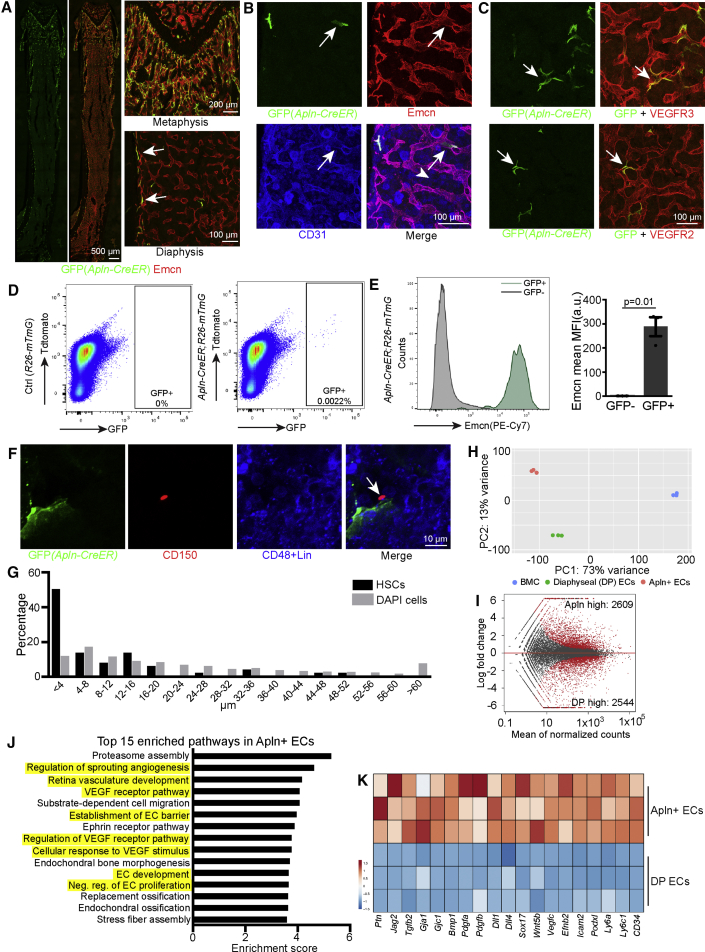
Apelin (Apln) is a peptide ligand secreted by a subpopulation of angiogenic ECs in different growing and regenerating organs (Tian et al., 2013). To analyze the distribution of Apln+ ECs in adult bone, we generated adult Apln-CreER Rosa26-mTmG (Apln-mTmG) mice and tracked the GFP-labeled ECs shortly (2 days) after tamoxifen injection (Table S1). Apln-CreER-labeled cells were predominantly found in the metaphysis, endosteum, and the transition area between the metaphysis and diaphysis (Figure 4A). GFP+ cells express Emcn, CD31, VEGFR2, VEGFR3, and VE-cadherin confirming their endothelial identity (Figures 4B, 4C, and S4H). Apln+ ECs express Sca-1 but are, unlike arterial ECs, not covered by αSMA+ vascular smooth muscle cells (vSMCs) (Figure S4I). FACS-isolated Apln-CreER-labeled ECs represent about 0.0032% ± 0.0003% of total BM cells and 12.4% ± 1.3% of bone ECs (Figures 4D and 4E). Strikingly, Apln+ EC are closely associated with a subset of CD150+ CD48– Lin– HSCs that are located in the metaphysis and the transition zone between the metaphyseal and diaphyseal vasculature (Figures 4F and 4G). To understand whether Apln+ ECs are similar to other BM ECs, we isolated Apln-CreER-labeled (GFP+) cells from Apln-mTmG mice by FACS for RNA-seq analysis. These cells were compared to ECs from the Cdh5-mTnG diaphysis (Figure S4D). Confirming their endothelial identity, both sorted cell populations show significantly higher expression of EC markers relative to other BM cells, but they also show substantial differences in gene expression and form separate clusters in principal components analysis (Figures 4H and S4E). While 2,609 genes are significantly enriched in Apln+ ECs, 2,544 genes show significantly higher expression in the diaphyseal endothelium (Figures 4I and S4F). Based on Gene Ontology (GO) biological process analysis, most of the 15 top enriched pathways in Apln+ ECs are related to angiogenesis and, in particular, vascular endothelial growth factor (VEGF) signaling (Figure 4J). In contrast, pathways enriched in diaphyseal ECs relate to leukocyte adhesion, tethering and rolling, which is consistent with the known function of sinusoidal vessels in the trafficking of hematopoietic cells (Figure S4G). Accordingly, numerous mRNAs associated with HSC niche function of the endothelium, such as Ptn (encoding Pleiotrophin), gap-junction proteins, and Notch ligands are substantially higher in Apln+ ECs (Figure 4K) (Guo et al., 2017, Himburg et al., 2010, Taniguchi Ishikawa et al., 2012, Tikhonova et al., 2019), suggesting that Apln+ ECs may potentially play a role in hematopoiesis. In addition, Apln+ ECs have special molecular properties and show expression of certain arterial markers, such as Dll4 (Delta-like 4), Efnb2 (ephrin-B2), Ly6a (Sca-1), Gja1 (connexin 43), Cav1 (caveolin-1), and the transcription factor Sox17, even though they are not typical arterial cells based on their localization and the lack of αSMA+ cell coverage (Figures 4K, S4E, and S4I). These data indicate that Apln+ ECs in adult bone represent a specialized endothelial subpopulation.
Figure 4.
Identification of Apln+ ECs in Adult BM
(A) Confocal tile scan images (left) showing distribution of GFP+ cells in Apln-mTmG mice. High magnifications (right) show enlarged regions of the metaphysis and diaphysis. Arrows mark GFP+ cells in endosteum.
(B and C) Emcn and CD31 (B) and VEGFR2 and VEGFR3 (C) staining in adult Apln-mTmG femur. Arrows mark GFP+ ECs.
(D) FACS plots of GFP+ cells isolated from Apln-mTmG mice.
(E) Histogram of Emcn expression in GFP– and GFP+ cells. Quantification of mean fluorescence intensity (MFI) in GFP– and GFP+ cells (n = 3 per group). Error bars, mean ± SEM. p values, two-tailed unpaired Students’ t test.
(F) CD150+ Lin– CD48– HSC (arrow) and GFP+ EC in the metaphysis and metaphysis-diaphysis transition area of Apln-mTmG mice.
(G) Distribution of distance between DAPI+ cells (n = 3,872) or CD150+ Lin– CD48– HSCs (n = 52) and GFP+ ECs cells in the metaphysis and metaphysis-diaphysis transition area of Apln-mTmG mice.
(H) PCA plot of total BM cells (BMC), Apln+ ECs (from whole bone), and diaphyseal ECs (DP ECs).
(I) MA plot showing differentially expressed genes between Apln+ ECs and DP ECs.
(J) Top 15 enriched pathways in GO analysis in Apln+ ECs relative to DP ECs.
(K) Heatmap of selected genes in Apln+ ECs.
See also Figure S4.
Apln+ ECs Regulate Steady-State Hematopoiesis
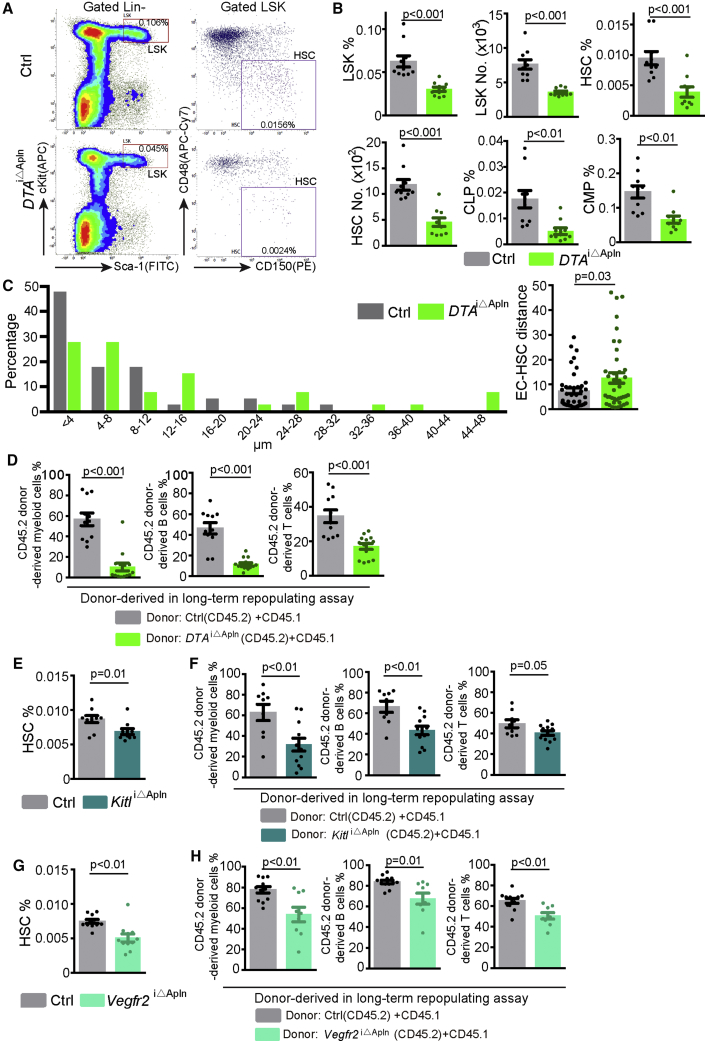
To investigate the function of Apln+ ECs, we generated Apln-CreER Rosa26-DTA (DTAiΔApln) mice enabling tamoxifen-induced elimination of Apln+ ECs (Figure S4M). This procedure damages metaphyseal vessels in direct proximity of the growth plate, results in slight dilation of sinusoidal vessels in the diaphysis, and increases the permeability of the bone vasculature (Figures S4N and S4O). Notably, vascular defects in the DTAiΔApln model are morphologically distinct from the damage seen after myeloablative treatments, namely, irradiation, 5-FU, or the DTRiΔHC model (Figures S4N, 1B, 1C, S2B, 3B, and 3C). Based on flow cytometric analysis, the ablation of Apln+ ECs leads to profound reductions in the percentage and absolute number of LSK cells and HSCs (Figures 5A and 5B). The remaining CD150+ CD48– Lin– HSCs are found in larger distance from BM vessels in DTAiΔApln mice (Figure 5C). Depletion of long-term HSCs in DTAiΔApln bone was confirmed by CD45.1/CD45.2 long-term competitive repopulation experiments (Figures 5D and S5A). We also detected a significant decrease of common lymphoid progenitors and common myeloid progenitors (Figure 5B), indicating that Apln+ ECs are required for the maintenance of both multipotent and oligopotent HSPCs. The number of BMNCs is not significantly altered, whereas the percentage of B lymphocytes decreases significantly in DTAiΔApln bone (Figure S5B). In contrast, the percentages of CD45– Ter119– CD31– CD51+ CD140a+ and of CD45– Ter119– CD31– Lepr+ CD140b+ perivascular cells are not changed significantly (Figure S5C). Peripheral blood (PB) count results show a significant decrease of lymphocyte and increase in the percentage of granulocytes in DTAiΔApln mice relative to control (Figure S5D). FACS analysis indicates a significantly higher percentage of myeloid cells in DTAiΔApln PB, whereas the percentage of total CD45+ leukocytes is not significantly changed (Figure S5E). Moreover, certain proinflammatory cytokines, in particular, interleukin (IL)-1α and IL-6, are elevated in blood plasma of DTAiΔApln mice (Figure S5F). The percentage change of myeloid cells and B lymphocyte are also seen in liver but not spleen of DTAiΔApln mice (Figures S5G and S5H). These results indicate that Apln+ ECs help to maintain a normal, non-inflammatory milieu and play a critical role in steady-state hematopoiesis.
Figure 5.
Apln+ ECs Regulate HSC Maintenance and Steady-State Hematopoiesis
(A) FACS plot of LSK cells and HSCs in control and DTAiΔApln mice. Numbers in boxes represent the percentage of the cell population among all BM cells.
(B) Quantification of the number and percentage of LSK cells, HSCs, and oligopotent progenitor cells in control (n = 10) and DTAiΔApln (n = 10) mice. CLP, common lymphoid progenitor; CMP, common myeloid progenitor.
(C) Quantitative distribution of distance between CD150+CD48-Lin– HSCs and vessels in whole BM of control (n = 40 HSC from 4 mice) and DTAiΔApln mice (n = 40 HSC from 5 mice).
(D) Long-term competitive repopulation assay showing donor-derived (CD45.2, control = 11 and DTAiΔApln = 14) myeloid cells, B cells, and T cells.
(E) Percentage of BM HSCs in control (n = 10) and KitliΔApln mice (n = 10).
(F) Long-term competitive repopulation assay showing donor-derived (CD45.2, control = 9 and KitliΔApln = 12) myeloid cells, B cells, and T cells.
(G) Percentage of BM HSCs in control (n = 11) and Vegfr2iΔApln mice (n = 11).
(H) Long-term competitive repopulating assay showing donor-derived (CD45.2, control = 12 and Vegfr2iΔApln = 9) myeloid cells, B cells, and T cells.
Error bars, mean ± SEM. p values, two-tailed unpaired Student’s t test. See also Figure S5.
Stem cell factor (SCF, encoded by the gene Kitl in mice) is an essential regulator of HSC self-renewal and function (Asada et al., 2017, Comazzetto et al., 2019, Ding et al., 2012, Xu et al., 2018). Apln-CreER-mediated inactivation of the Kitl gene causes a significantly lower percentage of HSCs in the BM of the resulting KitliΔApln mutants relative to littermate controls (Figure 5E). This reduction of HSCs in KitliΔApln mice is confirmed by CD45.1/CD45.2 long-term competitive repopulating assay (Figure 5F), whereas the PB count in KitliΔApln mice is normal (Figure S5I). In Apln+ ECs, genetic inactivation of VEGF receptor 2 (Vegfr2iΔApln), which is the major receptor of VEGF-A, also causes a significant reduction in HSC percentage (Figures 5G, 5H, and S5J). These results indicate that Apln+ ECs and signaling processes in these cells are necessary for HSC maintenance.
Apln-CreER mice have been generated by a knockin of tamoxifen-inducible CreER into the X-linked Apln locus (Tian et al., 2013), which disrupts the function of the gene globally so that Apln-CreER hemizygous male mice (T/y) are null mutants lacking the ability to generate apelin peptide. Excluding the possibility that the defects in Apln-CreER males are caused by lost Apln function, hematopoiesis and hematopoietic reconstitution are comparable for Apln-CreER hemizygous (T/y) and littermate wild-type (+/y) male mice in the absence of tamoxifen treatment (Figures S5K and S5L). Together, these results indicate that a small subpopulation of Apln+ ECs but not apelin expression is required for steady-state hematopoiesis and HSC maintenance.
Role of Apln+ ECs in BM Transplantation and Vascular Regeneration
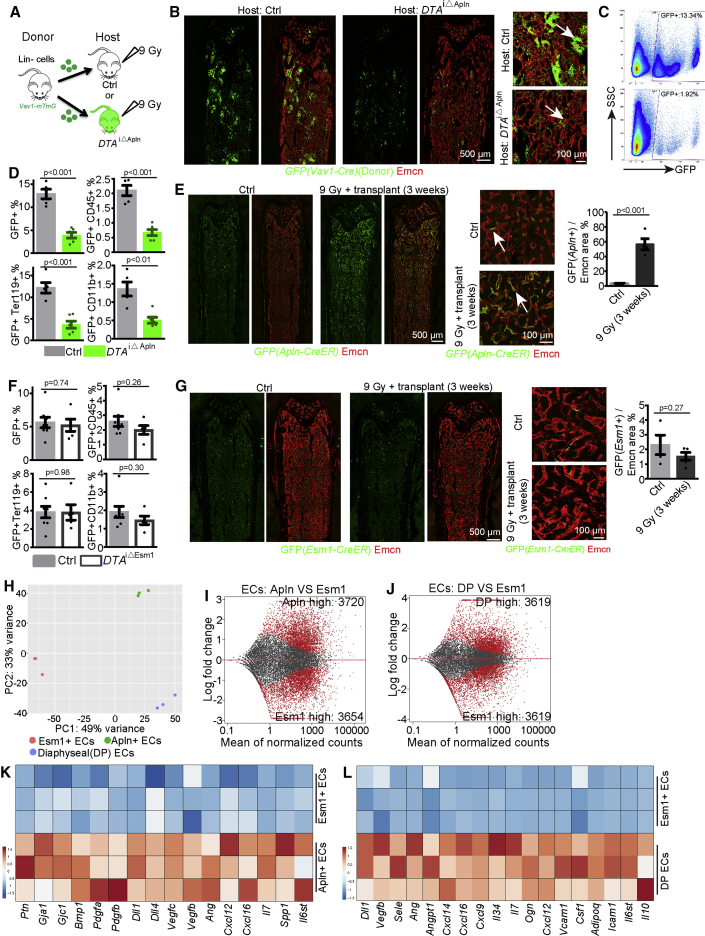
To investigate potential functional roles of Apln+ ECs in hematopoietic reconstitution, we transplanted BM cells into DTAiΔApln and littermate control recipients. Transplantation of genetically labeled (GFP+) BM cells from Vav1-Cre Rosa26-mTmG donors and immunostaining of bone sections and quantitative FACS suggested limited engraftment and expansion of transplanted hematopoietic cells in BM of DTAiΔApln recipients at 5 days after lethal irradiation, indicating that Apln+ ECs are necessary for efficient transplantation (Figures 6A–6D). To understand how Apln+ ECs contribute to hematopoietic reconstitution, we performed lineage tracing experiments. Adult Apln-mTmG were treated with 4-hydroxytamoxifen (4-OHT) at 1 day before lethal irradiation and transplantation, whereas control Apln-mTmG mice received the same dosage of 4-OHT without irradiation. In these latter animals, the distribution of Apln-CreER-labeled ECs is not substantially changed between 1 and 22 days after 4-OHT administration (Figures 6E and 4A). In contrast, irradiated Apln-mTmG mice show a profound increase of GFP+ cells in the diaphyseal vasculature, indicating that the regeneration of bone vessels involves the expansion of Apln+ ECs into the diaphyseal (i.e., sinusoidal) vascular network (Figure 6E). Irradiation without transplantation can trigger similar expansion of Apln+ ECs, indicating that this process does not require allogenic BM transplantation (Figure S6D). The descendant cells generated by the expansion of Apln+ ECs are reduced but do not regress completely after stable engraftment of the transplanted hematopoietic cells (Figure S6E). The progeny of Apln+ ECs includes cells that are covered by αSMA+ vSMCs, suggesting adult Apln+ ECs have the potential to generate arterial ECs in response to irradiation stress (Figure S6F). Together, these results indicate that Apln+ ECs contribute to allogenic transplantation and actively participate in the remodeling of the BM vasculature after irradiation.
Figure 6.
Apln+ ECs Are Necessary for BM Transplantation
(A) Transplantation of cells from Vav1-Cre R26-mTmG mice into Ctrl and DTAiΔApln hosts.
(B) Tile scan overview images and selected maximum intensity projections of control and DTAiΔApln bone 5 days after irradiation and transplantation with 3 × 105Vav1-Cre R26-mTmG Lin– cells. Arrows indicate donor-derived GFP+ cells.
(C) FACS plots of GFP+ cells from control and DTAiΔApln host BM.
(D) Quantification of GFP+, GFP+CD45+, GFP+Ter119+, and GFP+CD11b+ cells in control (n = 5) and DTAiΔApln (n = 6) host BM.
(E) Tile scan overview images and selected maximum intensity projections showing lineage tracing of GFP+ cells in Apln-mTmG long bone with or without irradiation and transplantation. Arrows mark GFP+ Emcn+ ECs. Quantification of GFP+ area relative to Emcn+ area in diaphysis (n = 4 in each group).
(F) Quantitative analysis of transplantation efficiency in DTAiΔEsm1 mice at 5 days after irradiation and transplantation of 105 Lin– cells from Vav1-mTmG mice (Ctrl = 8; DTAiΔEsm1 = 5).
(G) Lineage tracing of GFP+ cells in Esm1-CreER R26-mTmG mice with the same protocol as in (E). Quantification of GFP+ area relative to Emcn+ area in diaphysis (Ctrl = 4; 9 Gy = 5).
(H) PCA plot of Apln+ ECs, Esm1+ ECs, and total DP ECs under steady state.
(I and J) MA plot showing genes with differential expression between Esm1+ ECs and Apln+ ECs (I) and between Esm1+ ECs and total DP ECs (J).
(K and L) Heatmap showing expression of selected angiocrine or niche-related genes in Apln+ ECs relative to Esm1+ ECs (K), and in Esm1+ ECs relative to total DP ECs (L).
Error bars, mean ± SEM. p values, two-tailed unpaired Student’s t test. See also Figure S6.
We also investigated another subpopulation of BM ECs, namely, Esm1+ ECs, which were genetically labeled in Esm1-CreER R26-mTmG mice (Figure S6G) (Pitulescu et al., 2017, Rocha et al., 2014). Esm1+ ECs and Apln+ ECs show similar labeling percentages in the diaphysis under steady-state conditions (Figure S6G). In contrast to Apln+ ECs, genetic ablation experiments with Esm1-CreER R26-DTA mice (DTAiΔEsm1) (Figures S6H and S6L) fail to influence the percentage of HSCs, BM hematopoietic cell composition, PB composition under steady-state conditions (Figures S6H–S6K), and transplantation efficiency (Figures 6F and S6L–S6M). Moreover, Esm1+ ECs are not preferentially located in the metaphysis and endosteum and fail to expand after irradiation (Figure 6G). We used FACS to isolate Esm1+ ECs (Figure S6N) for RNA-seq analysis. These cells show significant enrichment of core EC markers relative to hematopoietic cells and mesenchymal cells (Figure S6O) but are distinct from Apln+ ECs and total diaphyseal ECs (Figures 6H–6J and S6P). Remarkably, Esm1+ ECs show low expression of a broad range of niche factors and angiocrine factors (Figures 6K and 6L), further arguing that this subpopulation of ECs is not critical for hematopoiesis and transplantation. These results show that not all BM ECs are equally important for HSCs maintenance, hematopoiesis, and transplantation. Moreover, the phenotype in DTAiΔApln mutants is most probably induced by specific ablation of Apln+ ECs and not by side effects of EC death or unspecific toxicity of diphtheria toxin.
Crosstalk between Apln+ ECs and HSPC during BM Regeneration
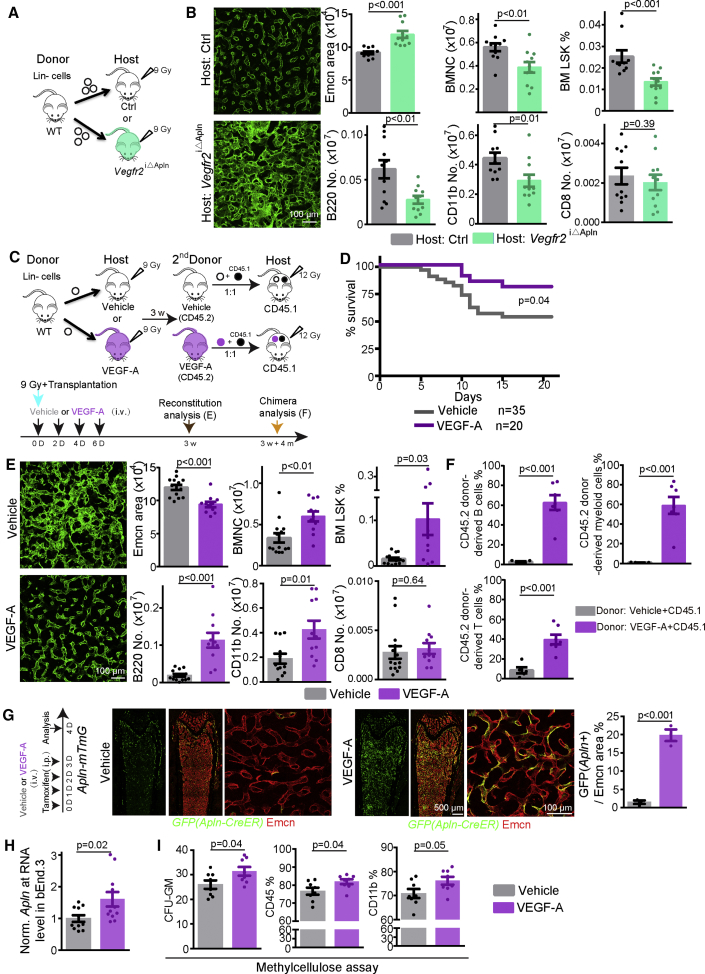
The results above indicate that HSPC/LSK cells and Apln+ ECs play critical roles after irradiation and transplantation (Figures 3G–3I and 6A–6E), which raises the question whether the activity of these cell populations is coupled during hematopoietic reconstitution and bone vessel regeneration. Analysis of RNA-seq results of LSK cells and Lin– HSPCs in combination with previously published data for Lepr+ and NG2/Cspg4+ perivascular niche cells (Asada et al., 2017) indicates that LSK cells are a major source of VEGF-A (encoded by the gene Vegfa) in BM (Figure S7A). Higher expression of VEGF-A protein in LSK cells is confirmed by ELISA (Figure S7B). Irradiation significantly disrupts autocrine expression of Vegfa by ECs, which, together with the loss of LSK cells, results in profoundly reduced Vegfa expression in total BM cells (Figures S7A–S7C). Previous work has shown that donor-cell-derived VEGF-A is necessary for transplantation (Gerber et al., 2002). This finding is confirmed by our own transplantation experiments with BM cells from Vav1-Cre Vegfa (VegfaΔHC) double-heterozygous mice into lethally irradiated recipients (Figures S7F–S7H) (Ferrara et al., 1996). However, it was unclear whether hematopoietic cells or ECs are the cellular target of VEGF-A, as both possibilities are favored by different reports (Gerber et al., 2002, Hooper et al., 2009). To address this question, we have combined inactivation the Kdr (Vegfr2) gene, which encodes the main receptor for VEGF-A, in different cells types. In BM transplantation experiments, donor cells from hematopoietic cell-specific Vav1-Cre Vegfr2 (floxed/floxed) (Vegfr2ΔHC) mutants and control animals do not lead to significant differences in transplantation efficiency or recipient BMNC number (Figures S7I–S7J). However, Vegfr2ΔHC BM donor transplantation results in a significant increase of CD11b+ myeloid cells relative to control BM (Figure S7J), indicating a role of VEGFR2 in hematopoietic cell differentiation.
Expression of Vegfr2 is strongly increased in irradiated ECs (Figure S7D), and RNA-seq shows that VEGF signaling pathway components are enriched in Apln+ ECs (Figures 4J and S7E). To test whether Apln+ ECs might respond to VEGF-A from transplanted BM cells, we generated tamoxifen-induced adult Apln-CreER Vegfr2 (floxed/floxed) (Vegfr2iΔApln) mutants. Transplantation of Lin– HSPCs into lethally irradiated Vegfr2iΔApln animals results in delayed bone vessel normalization, lower abundance of BMNCs, LSK cells, myeloid cells, and B cells, but not T cells, relative to littermate control recipients (Figures 7A, 7B, and S7K–S7M). In an independent approach, we performed transplantation experiments with pan-endothelial Cdh5-CreERT2 Vegfr2 (floxed/floxed) (Vegfr2iΔEC) recipients (Figure S7N). Relative to littermate control recipients, lethally irradiated Vegfr2iΔEC mutants show impaired vascular regeneration, lower percentage of LSK cells, and fewer myeloid cells and B cells at 2.5 weeks after transplantation (Figure S7N).
Figure 7.
Crosstalk of HSPC with Apln+ ECs in BM Vascular Regeneration and Hematopoietic Reconstitution
(A) Diagram depicting the transplantation of wild-type Lin– cells into Ctrl and Vegfr2iΔApln host mice.
(B) Bone vessels at 2.5 weeks after transplantation in control or Vegfr2iΔApln host mice. Quantification of Emcn+ area, percentage of LSK cells, number of BMNCs, B220+, CD11b+, and CD8+ cells in control (n = 10–11) and Vegfr2iΔApln (n = 11) host mice.
(C) Scheme depicting VEGF-A treatment in combination with transplantation of wild-type Lin– cells.
(D) Survival curve of irradiated mice transplanted with 104 Lin– cells and intravenous injection of PBS (vehicle; n = 35) or recombinant VEGF-A (n = 20).
(E) Bone vessels at 3 weeks after irradiation and treatment with vehicle (PBS + Lin– cells) or VEGF-A (VEGF-A + Lin– cells). Quantification of Emcn+ area, percentage of LSK cells, number of BMNCs, B220+, CD11b+, and CD8+ cells in vehicle (n = 12–14) and VEGF-A (n = 11)-treated mice.
(F) Secondary long-term competitive repopulating assay with donor-derived CD45.2 cells from 1st transplant of vehicle or VEGF-A-treated recipients. Graphs show percentage of CD45.2 donor-derived myeloid cells, B cells, and T cells (vehicle = 5, VEGF-A = 7).
(G) Confocal tile scan overview and high-magnification images of GFP and Emcn signals in the Apln-mTmG diaphysis after infusion of vehicle or VEGF-A. Quantification of GFP+ cell relative to Emcn+ (vehicle = 3, VEGF-A = 3).
(H) Apln transcripts in cultured bEnd.3 cells. Actb was used as control (vehicle = 11, VEGF-A = 11).
(I) Quantification of CFU-GM number, CD45% and CD11b% in methylcellulose assays (700 Lin– HSPC were seeded). Vehicle (n = 8) or VEGF-A (4 μg/ml; n = 8) were added to standard culture medium.
Error bars, mean ± SEM. p values, two-tailed unpaired Student’s t test. See also Figure S7.
Conversely, combining the transplantation of a limited number of 104 Lin– BM cells after lethal irradiation with intravenous administration of recombinant VEGF-A improves recipient survival, normalization of the BM vasculature, repopulation efficiency in primary transplantations, and significantly increases HSC frequency relative to vehicle-treated controls (Figures 7C–7F and S7O–S7R). To understand how VEGF-A improves transplantation efficiency, we administrated VEGF-A to Apln-mTmG mice together with tamoxifen. In these experiments, VEGF-A infusion significantly increases the percentage of Apln+ ECs in diaphysis (Figure 7G). VEGF-A also significantly increases the transcription of Apln in cultured ECs in vitro (Figure 7H). Thus, VEGF-A can change the molecular properties of ECs in adult BM and endothelial Apln expression. Consistent with previous research (Gerber et al., 2002), VEGF-A has also direct effects on Lin– HSPCs and promotes the formation of leukocytes in methylcellulose assay in vitro (Figures 7I and S7S).
Discussion
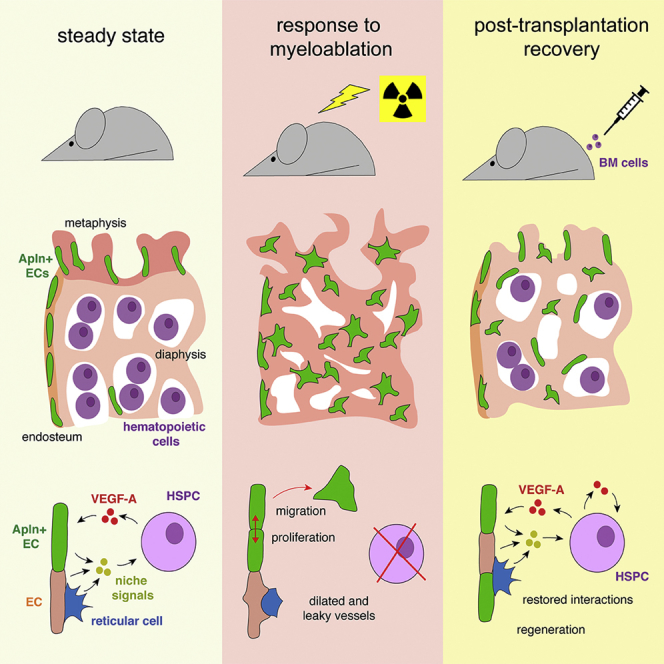
In this study, we provide new insight into the nature of irradiation-induced damage to the bone vasculature and the dynamic changes in BM vessels during regeneration and repopulation. Irradiation leads to vessel dilation, increased EC density, and vascular permeability, which are accompanied by changes in EC morphology and gene expression in bone. These vascular defects are phenocopied by a genetic model of hematopoietic cell ablation and are efficiently rescued by transplantation of multipotent LSK cells, which are a major source of VEGF-A. Furthermore, our work identifies a subpopulation of bone ECs, namely, Apln+ ECs, as important regulators of HSC function and BM regeneration after transplantation. While previous work has already established that ECs and associated mesenchymal cells provide niches controlling HSC self-renewal and differentiation (Asada et al., 2017, Comazzetto et al., 2019, Ding et al., 2012, Méndez-Ferrer et al., 2010, Sugiyama et al., 2006, Tikhonova et al., 2019, Xu et al., 2018), we now assign critical functional properties to a small subset of Apln+ ECs.
Our previous work has shown that neonatal Apln+ ECs are enriched in CD31hi Emcnhi (type H and E) ECs, mediate postnatal osteogenesis, and generate other EC subtypes during development (Langen et al., 2017). The current study extends the function of Apln+ ECs from osteogenesis to adult hematopoiesis and BM transplantation. Adult Apln+ ECs express niche factors, such as connexin gap-junction proteins, Notch ligands, and pleiotrophin (PTN), that have been previously associated with HSPC function and BM reconstitution (Figure 4K). Expression of PTN, a secreted and matrix-binding growth factor, is increased in BM ECs after myelosuppressive irradiation and EC-specific inactivation of the murine Ptn gene impairs HSC regeneration (Himburg et al., 2010, Himburg et al., 2018). These results argue that adult Apln+ ECs are an important source of niche factors.
Recent single-cell RNA-seq profiling classifies BM ECs into two groups of cells defined as Stab2high ECs and Ly6ahigh ECs, the latter includes 11% of total ECs (Tikhonova et al., 2019). Our own data show that Apln+ ECs represent approximately 12% of total ECs, and Apln+ ECs share a strikingly similar transcription profile with Ly6ahigh ECs (Figure S4J), suggesting that these two subsets may be identical or overlapping entities. While other endothelial subpopulations, namely, Bmx+ arterial ECs and Esm1+ capillary ECs, show no appreciable expansion after irradiation, ECs derived from the Apln+ population increase substantially in number and expand into the endothelium of the regenerating BM cavity. Hematopoietic reconstitution and vascular regeneration after irradiation are promoted by VEGF-A provided by transplanted BM cells and, in particular, by HSPCs and LSK cells, consistent with previous reports addressing the crucial role of the VEGF pathway in BM regeneration (Gerber et al., 2002, Hooper et al., 2009). Thus, regeneration of the vasculature during hematopoietic reconstitution involves reciprocal interactions between the transplanted BM cells and expanding, niche factor-providing Apln+ ECs.
Apln+ ECs may play an anti-inflammatory role required to maintain a balanced BM microenvironment. Apln+ ECs show enriched expression of anti-inflammatory factors such as Mmp, Tgfb2, Nos3, and Ptgis (Aguirre et al., 2017, Bougaki et al., 2010, Jin et al., 2014, Manicone and McGuire, 2008, Dorris and Peebles, 2012, Knudsen et al., 1986). Accordingly, ablation of Apln+ ECs leads to an increase of PB myeloid cells, which is a hallmark of numerous conditions with compromised BM function including aging, inflammation, or compromised stromal support (Kusumbe et al., 2016, Mann et al., 2018, Pietras, 2017, Tikhonova et al., 2019).
Based on the findings presented above, we propose that Apln+ ECs in the adult skeletal system represent a population of specialized ECs, which are characterized by progenitor properties, are a source of niche factors, control multipotent, and oligopotent hematopoietic progenitors, and promote BM regeneration after myeloablative treatment. Our results provide insights into the cellular and molecular mechanisms controlling BM transplantation and highlight Apln+ ECs as a potential therapeutic target in this process.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER(RRID) |
|---|---|---|
| Antibodies | ||
| Endomucin | Santa Cruz | RRID:AB_2100037 |
| GFP-Alexa488 | Invitrogen | RRID:AB_221477 |
| CD31 | R&D | RRID:AB_2161028 |
| Collagen IV | AbD Serotec | RRID:AB_2082660 |
| ERG | Abcam | RRID:AB_10864794 |
| PDGFR-beta | eBioscience | RRID:AB_467493 |
| VEGFR2 | R&D | RRID:AB_355500 |
| VEGFR3 | R&D | RRID:AB_355563 |
| BCAM | R&D | RRID:AB_2811217 |
| Caveolin-1 | Cell signaling | RRID:AB_2072166 |
| Lineage-biotin cocktail | Miltenyi Biotec | RRID:AB_1103214 |
| CD150-Alexa647 | Biolegend | RRID:AB_2239178 |
| CD48-biotin | eBioscience | RRID:AB_466470 |
| Caspase3 | Cell Signaling | RRID:AB_2341188 |
| B220-biotin | BD pharmingen | RRID:AB_10053179 |
| RFP (Tomato) | MBL | RRID:AB_591279 |
| Ki67 | Abcam | RRID:AB_443209 |
| VE-cadherin | R&D | RRID:AB_2077789 |
| Sca-1 | Biolegend | RRID:AB_756190 |
| αSMA | eBioscience | RRID:AB_2574362 |
| Linegae-biotin cocktail | Miltenyi Biotec | RRID:AB_1103214 |
| Sca1-FITC | eBioscience | RRID:AB_465334 |
| cKit-APC | BD pharmingen | RRID:AB_398536 |
| Ter119-APC | eBioscience | RRID:AB_469474 |
| CD45-FITC | eBioscience | RRID:AB_465049 |
| CD45R(B220)-APC | Invitrogen | RRID:AB_10392558 |
| Ly-6G-APC | eBioscience | RRID:AB_469475 |
| CD11b-APC | Biolegend | RRID:AB_312795 |
| CD3-APC | eBioscience | RRID:AB_469313 |
| CD4-APC | eBioscience | RRID:AB_469323 |
| CD8-APC | eBioscience | RRID:AB_469335 |
| CD150-PE | Biolegend | RRID:AB_313683 |
| CD48-APC-Cy7 | BD pharmingen | RRID:AB_10644381 |
| CD34-eFluor450 | eBioscience | RRID:AB_2043837 |
| CD127-PE | eBioscience | RRID:AB_465845 |
| CD16/CD32 PE-Cy7 | eBioscience | RRID:AB_469598 |
| Ter-119 Pacific Blue | Biolegend | RRID:AB_2251160 |
| CD45- Pacific Blue | Biolegend | RRID:AB_493535 |
| CD51-PE | eBioscience | RRID:AB_465704 |
| CD140a-APC | eBioscience | RRID:AB_529482 |
| CD140b-APC | eBioscience | RRID:AB_1548743 |
| Lepr-biotin | R&D | RRID:AB_2296953 |
| CD31-Alexa488 | R&D | RRID:AB_10972784 |
| CD31-APC | R&D | RRID:AB_10971931 |
| Cdh5-APC | eBioscience | RRID:AB_10598508 |
| CD31-Pe/Cy7 | Biolegend | RRID:AB_2572182 |
| Endomucin | Santa Cruz | RRID:AB_2100037 |
| CD45.1-FITC | invitrogen | RRID:AB_2539698 |
| CD45.2 Pacific Blue | Biolegend | RRID:AB_492872 |
| CD45 PE-Cy7 | eBioscience | RRID:AB_2734986 |
| Annexin V-APC | BD pharmingen | 550474 |
| Anti-biotin Microbeads | Miltenyi Biotec | RRID:AB_2811216 |
| Biological Samples | ||
| Mouse blood | This paper | N/A |
| Mouse bone marrow cells | This paper | N/A |
| Mouse spleen, retina, heart, skin, small intestine | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human recombinant VEGF-A | Reliatech | 300-076 |
| Dextran Texas Red | ThermoFisher Scientific | D1830 |
| MethoCult™ medium | Stem cell technologies | GF M3434 |
| EdU | Thermofisher scientific | A10044, |
| Tamoxifen | Sigma | T5648 |
| 4OH-tamoxifen | Sigma | H7904 |
| Peanut oil | Sigma | P2144 |
| Vybrant DyeCycle Violet stain |
Invitrogen |
V35003 |
| Critical Commercial Assays | ||
| Legendplex mouse inflammation panel | Biolegend | 740446 |
| Mouse VEGF-A ELISA Kit | Cloud-Clone Corp | SEA143Mu |
| SMART-Seq v4 Ultra Low Input RNA kit for Sequencing | Takara | 634894 |
| Nextera XT DNA Library Preparation Kit | Illumina | 15031942 |
| Click-iT EdU Alexa-647 imaging Kit | Invitrogen | C10340 |
| RNeasy plus Micro Kit | QIAGEN | 74034 |
| iScript™ cDNA Synthesis Kit | BioRad | 170-8891 |
| Deposited Data | ||
| RNA sequencing | This paper | GSE115422 |
| Experimental Models: Cell Lines | ||
| bEnd.3 | ATCC | ATCC CRL-2299 |
| Experimental Models: Organisms/Strains | ||
| Cdh5 membrane-Tdtomato H2B-EGFP(Cdh5-mTnG) | Our lab | PMID: 28959057 |
| Rosa26-mT/mG | The Jackson Laboratory | RRID:IMSR_JAX:007576 |
| Cdh5(PAC)-CreERT2 | Our lab | RRID:MGI:3848984 |
| Apln-CreERT2 | Bin Z lab | PMID: 23797856 |
| Bmx-CreERT2 | Our lab | PMID: 23785053 |
| Vav1-Cre | The Jackson Laboratory | RRID:IMSR_JAX:008610 |
| VEGFatm2Gne | Ferrara N lab | PMID: 10021335 |
| Kdrtm1Wag | Nagy A lab | PMID: 14550787 |
| Rosa26-DTR | The Jackson Laboratory | RRID:IMSR_JAX:007900 |
| Rosa26-DTAtm1(DTA)Mrc | The Jackson Laboratory | RRID:IMSR_JAX:010527) |
| Kitl | The Jackson Laboratory | RRID:IMSR_JAX:017861 |
| Esm1-CreERT2 | Our lab | PMID: 25057127 |
| Software and Algorithms | ||
| Fiji | NIH | ImageJ 1.51 s |
| Volocity | PerkinElmer | 6.3 |
| Photochop | Adobe | CS6 |
| Illustrator | Adobe | CS6 |
| Graphpad Prism | GraphPad | 7 |
| Microsoft Excel | Microsoft | 15.40 |
| FACS Diva | BD | 8.0.1 |
| FACSuite | BD | 1.0.5.3841 |
| Legendplex | Biolegend | 8.0 |
| FlowJo | BD | 10.4.2 |
| TopHat | John Hopkins Univ. | version 2.1.1 |
| HTSeq-count | PMID: 25260700 | version 0.6.1 |
| DESeq2 | Bioconductor | version 3.9 |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ralf H. Adams (ralf.adams@mpi-muenster.mpg.de). This study did not generate new unique reagents.
Experimental Model and Subject Details
8-15 week-old mice were used as young adults and C57BL/6 males were used as wild-type mice for all analysis unless stated otherwise. Genetically modified mouse models used in this study are summarized in Table S1. Littermates with appropriate genotypes were used as controls of genetically modified mutants whenever possible (see further details below).
Cdh5(PAC)-mtdTomato-nGFP (Cdh5-mTnG) reporter mice were previously described and used for labeling of EC nuclei and FACS-sorting (Jeong et al., 2017).
Vav1-Cre (de Boer et al., 2003) transgenic mice were interbred with Rosa26-DTR (Buch et al., 2005) animals to generate Vav1-Cre+/T Rosa26-DTR+/lox (DTRiΔHC) mice. Vav1-Cre+/+ Rosa26-DTR+/lox littermates were used as controls. 25 μg/kg diphtheria toxin (Sigma, D0564) was administrated to DTR iΔHC and control mice for 2 consecutive days and animals were analyzed 4 days after first injection. DTR iΔHC mutants were interbred with Cdh5-mTnG animals to generate Vav1-Cre+/T Rosa26-DTR+/lox Cdh5-mTnG+/T triple transgenic mice for the visualization of EC nuclei and flow cytometry analysis. Vav1-Cre transgenic mice were interbred with conditional (floxed) Vegfa mutants (Gerber et al., 1999) mice to generate Vav1-Cre+/T Vegfa+/lox (VegfaΔHC) animals and Vav1-Cre+/+ Vegfa+/lox controls. Vav1-Cre transgenic mice were also interbred with conditional (floxed) Kdr (Vegfr2) (Haigh et al., 2003) mutants to generate Vav1-Cre+/T Kdrlox/lox (Vegfr2ΔHC) mice and Vav1-Cre+/+ Kdrlox/lox controls.
In the Rosa26-mTmG reporter (Muzumdar et al., 2007) background, Cre activity leads to an irreversible switch from constitutive membrane-anchored tdTomato protein expression to membrane-anchored GFP. Cdh5(PAC)-CreERT2 (Wang et al., 2010) transgenic mice were interbred with Rosa26-mTmG reporter animals to generate Cdh5-CreERT2 Rosa26-mTmG mice enabling genetic labeling of all ECs. For the morphological analysis of ECs at single cell level, 10 μg tamoxifen (Sigma, T5648) were injected to Cdh5-CreERT2 Rosa26-mTmG mice 6 days after irradiation or 5-FU treatment. Bmx-CreERT2 (Ehling et al., 2013) transgenic mice were interbred with the Rosa26-mTmG strain to generate Bmx-CreERT2 Rosa26-mTmG for genetic labeling of arterial ECs. 500 μg tamoxifen were administered on 2 consecutive days to Bmx-CreERT2 Rosa26-mTmG mice for analysis in irradiation experiments with or without transplantation.
Apln-CreERT2 (T/y) hemizygous males were used for all experiments with this strain to avoid mosaic Cre recombinase expression due to X-inactivation in (T/x) heterozygous females (Tian et al., 2013). Apln-CreERT2 transgenic animals were bred into the Rosa26-mTmG background to generate Apln-CreERT2 Rosa26-mTmG (Apln-mTmG) mice for genetic labeling, lineage tracing and flow cytometry analysis of Apln+ ECs in bone. 1mg tamoxifen was injected intraperitoneally to Apln-mTmG mice on three consecutive days for the labeling and flow cytometric analysis of Apln+ ECs. 1mg 4-hydroxytamoxifen (4-OHT; Sigma, H7904) was injected intraperitoneally at 1 day before irradiation for lineage tracing of Apln+ ECs and their descendants after transplantation. Apln-CreERT2 transgenic mice were interbred with Rosa26-DTA (Wu et al., 2006) animals to generate Apln-CreERT2T/y Rosa26-DTAlox/lox (DTAiΔApln) males and Apln-CreERT2+/y Rosa26-DTAlox/lox controls. Two injections of 1mg tamoxifen were used to induce CreERT2 activity and the ablation of Apln+ ECs (Figure S4M). Apln-CreERT2 transgenic mice were interbred with Kdr conditional (floxed) animals to generate Apln-CreERT2T/y Kdrlox/lox (Vegfr2iΔApln) mutants and Apln-CreERT2+/y Kdrlox/lox controls. 2 mg tamoxifen were injected on 5 consecutive days to Vegfr2iΔApln and control mice for transplantation experiments. Apln-CreERT2 transgenic mice were interbred with Kitltm2.1Sjm (encoding SCF) conditional (floxed) animals to generate Apln-CreERT2T/y Kitllox/lox (KitliΔApln) mutants and Apln-CreERT2+/y Kitllox/lox controls. 2mg tamoxifen were injected on 5 consecutive days to KitliΔApln and control mice for analysis.
Cdh5(PAC)-CreERT2 transgenic mice were interbred with Kdr conditional (floxed) animals to generate Cdh5(PAC)-CreERT2+/T Kdrlox/lox (Vegfr2iΔEC) mutants and Cdh5(PAC)-CreERT2+/+ Kdrlox/lox controls. 1mg tamoxifen was injected on 5 consecutive days to Vegfr2 iΔEC and control mice for transplantation experiments. Esm1-CreERT2 transgenic mice were interbred with Rosa26-mTmG and Rosa26-DTA animals to generate Esm1-CreERT2+/T Rosa26-mTmG and Esm1-CreERT2+/T Rosa26-DTA (DTAiΔEsm1) mice. Esm1-Cre mice were treated with exactly the same tamoxifen injection protocol as Apln-Cre animals (Figures S6H and S6L).
All animals were routinely genotyped using respective PCR protocols. Protocols and primer sequences can be provided upon request. All the animals were housed in the animal facility at the Max Planck Institute for Molecular Biomedicine or the Institute for Nutritional Sciences and Shanghai Institute of Biochemistry and Cell Biology, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences. All experiments were performed according to the institutional guidelines and laws, following the protocols approved by local and national animal ethics committees.
Method Details
Cryosectioning, immunohistochemistry and permeability assays
Adult bones were dissected and immediately placed in ice-cold 4% paraformaldehyde (PFA-PBS) solution and fixed under gentle agitation overnight. Bone samples were placed in 0.5M EDTA (Ph 8.0) for at least 2 days, dehydrated in 20% sucrose-1% polyvinylpyrrolidone PBS solution for at least 2 days, and embedded in PBS containing 15% sucrose, 8% gelatin and 1% polyvinylpyrrolidone for storage at −80°C and cryosectioning on a Leica CM3050 cryostat using low profile blades. For immunostaining, bone sections were rehydrated in PBS, permeabilized for 15 min in 0.5% Triton X-100 PBS solution and blocked for 30 min in PBS containing 1% BSA, 2% donkey serum, 0.3% Trion X100 PBS (blocking buffer) at room temperature. Sections were probed with primary antibodies diluted in blocking buffer at 4°C overnight. After incubation, sections were washed three times with PBS and incubated with appropriate Alexa Fluor-conjugated secondary antibodies (1:100 to 1:400, Invitrogen) diluted in blocking buffer at room temperature for 2 hours. Nuclei were stained with DAPI during secondary antibody incubation. After that, sections were washed three times with PBS, mounted with Fluoromount-G (0100-01, Southern Biotech) and kept in 4°C for imaging.
For the analysis of vascular permeability in bone, 1mg Dextran Texas Red (ThermoFisher Scientific, D1830, 70000MW) was injected into the tail vein. Mice were sacrificed 15 minutes after injection. Femurs were dissected and directly placed in ice-cold 4% PFA for overnight fixation. Further processing of femur samples was exactly the same as for immunostainings. Texas Red signal was excited with a 594nm laser during confocal microscope without signal enhancement.
Flow cytometry
Bones were dissected and crushed by pestle for more than 3 times before cells were collected in 2% FCS-PBS solution. The tissue was immersed in 6ml dissociation solution (2% FCS-PBS solution with approximate 145U/ml type 4 GIBCO collagenase) and incubated at 37°C for 30 minutes. Samples were filtered using 70 μm Nylon cell strainer to get single cell suspensions. Reporter mice expressing fluorescent proteins were directly used for flow cytometry. Primary antibodies were diluted in 2% FCS-PBS solution and incubated with cells on ice for 40 minutes. Cells were washed 3 times by 2% FCS-PBS solution and incubated with secondary antibodies for 40 minutes. Cells were washed 3 further times again and used for flow cytometry. Cells were resuspended in PBS/2%FCS supplemented with 1 μg/ml DAPI to allow exclusion of nonviable cells when required. Cell sorting was performed on a FACSAriaIIu cell sorter (BD Biosciences, San Jose, CA) using an 85 μm nozzle. Cell analysis was performed using BD FACS Verse.
Irradiation, transplantation, lineage separation and VEGF-A treatment
Mice were exposed to a lethal dose of Gamma irradiation (137Cs, GammaCell 40, Best Theratronics) of 9 Gy and typically analyzed 7 days after irradiation (shown as “9 Gy” in the results), unless another experimental regime is mentioned in the legend. Bone marrow cells (or Lin-/LSK cells) were transplanted 4 to 6 hours after irradiation. For competitive long-term repopulating assays, CD45.1 host mice were lethally irradiated (12 Gy) and transplanted with 5 × 105 donor-derived (CD45.2 background) BM cells together with 5 × 105 host-derived (CD45.1 background) bone marrow cells. Host mice were sacrificed about 16 weeks after transplantation to determine chimerism in bone marrow and peripheral blood using FACS analysis.
For separation of lineage-positive cells, bone marrow cells were collected and filtered as described for flow cytometry analysis. Biotinylated-lineage cocktail antibodies (Miltenyi Biotec, 130-092-613) were mixed with cells at 4°C for 20 minutes. Antibody-conjugated BM cells were washed 2 times and then incubated with anti-biotin antibody-conjugated magnetic microbeads (Miltenyi Biotec, 130-105-637) at 4°C for 30 minutes. The cells were washed 2 times and separated with LS column (MiltenyiBiotec, 130-042-401). Lineage negative or positive cells were counted and transplanted as outlined in the relevant results.
In VEGF-A treatment experiments, 104 lineage-negative cells were transplanted together with 2 μg VEGF-A (Reliatech, 300-076) 4 to 6 hours after lethal irradiation. In the following days, 2 μg VEGF-A or vehicle (PBS) were intravenously injected into recipients as indicated in Figure 7C. 3 weeks after irradiation, vehicle and VEGF-A treated mice were sacrificed for analysis or for secondary transplantation (long-term repopulating assays). Before intravenous injection, VEGF-A activity was validated in culture ECs.
Blood count, blood plasma cytokine analysis and bone marrow cell number counting
Peripheral blood was extracted from the submandicular vein, collected in EDTA-K tube and analyzed with a Scil Vet ABC plus according to the manufacturer’s instructions. To analyze peripheral blood cells using FACS, whole blood was centrifuged at 1000 g for 10 minutes and blood plasma was separated. Blood cells layers were resuspended for FACS analysis. The remaining blood plasma was used to calculate cytokine levels by using the Biolegend Legendplex mouse inflammation panel (13-plex; Biolegend, 740446). The experimental process and quantification analysis were performed following the manufacturer’s protocol. Data analysis was performed using software provided by Biolegend. Manual gating was used to define beads A and B, while an automatic gating strategy was used to gate individual cytokine in APC-PE plot. Cytokine levels lower than detection level were excluded from analysis.
For counting of BM nucleated cells, BM cells were processed to generate single cell suspensions, as described for flow cytometry analysis. Red blood cells were removed using RBC lysis buffer (ebioscience, 00-4300-54). Total cell numbers were determined using cell counting plates or a Luna 2 automated cell counter (Logos Biosystems). The number of different cell types in BM was calculated based on BMNC counting and flow cytometry analysis with appropriate markers.
ELISA
To collect bone marrow supernatant for ELISA analysis, 1 femur was dissected, flushed into 1ml PBS solution and immediately kept on ice. Solutions were centrifuged at 300 g for 5 minutes. Supernatant was stored at −80°C. To collect BM cellular protein for ELISA analysis, Lin- and Lin+ cells were separated. After counting the number of cells, Lin- or Lin+ cells were centrifuged and immersed in NP40 solution with proteinase inhibitor cocktail. The cell-NP40 solution was rotated at 4°C for 30 minutes and then centrifuged at 15000 g for 30 minutes. The remaining supernatant was collected and stored at −80°C. ELISA for VEGF-A was performed with a kit following the manufacturer’s instructions (Cloud-Clone Corp, SEA143Mu).
In vitro methycellulose and bEnd.3 assays
Approximate 700 Lin- cells from wild-type mice were magnetically separated and cultured in MethoCult™ medium (GF M3434, Stem cell technologies) in a 35mm dish. 4μg/ml VEGF-A or vehicle (PBS) were added to the MethoCult™ medium for in vitro culture. Lin- HSPCs from the same batch were equally distributed, then exposed to vehicle (PBS) or VEGF-A for half an hour before seeding. Cells and medium were incubated in 37°C 5%CO2 cell incubator. 7 days after incubation, CFU number was counted. After counting, cells were washed 4 times with 2% FCS-PBS solution and filtered with 70 μm Nylon cell strainer to get single cell solution for FACS analysis. The FACS staining protocol was the same as for BM cells (see above).
bEnd.3 endothelial cells were cultured in base medium (including DMEM, Glutamine and P/S) plus 5% FCS in 37°C incubator with 5% CO2. Cells were passaged into 6 well-plates and, at 70%–80% confluency, the culture medium was changed to base medium plus 1μg/ml VEGF-A or base medium plus PBS vehicle without any serum. The cells were collected into RLT lysis buffer for RNA extraction at 24 hours after a final culture medium change.
EdU incorporation
EdU (A10044, Thermofisher scientific) was dissolved in PBS. 40mg/kg EdU-PBS was injected to Cdh5-mTnG mice at 7 consecutive days after irradiation. Bone samples were processed as described above. Click-iT EdU Alexa-647 imaging Kit (C10340, Invitrogen) was used to visualize EdU signals. Single optical slices instead of z stacked images were used for data analysis. Co-localization of GFP and EdU signal were analyzed and recorded in Volocity (PerkinElmer).
RNA extraction and quantitative PCR
RNA was extracted using RNeasy plus Micro Kit (74034, QIAGEN) and cDNA was generated with iScript™ cDNA Synthesis Kit (#170-8891, BioRad). Quantitative PCR with reverse transcription was performed with a Bio-rad CFX96 real-time PCR system using FAM-conjugated Taqman probes for Vegfa (Mm00437306_m1) and Apln (Mm00443562_m1). Gene expression levels were normalized to the endogenous VIC-conjugated Actb probe (4352341E) as control.
RNA sequencing and data analysis
Total RNAs were extracted using RNeasy Plus Micro kit (QIAGEN) according to the manufacturer’s instructions. Quality and quantity of RNA samples were analyzed with a Bioanalyzer and RNA 6000 pico kit (Agilent). Double strand cDNA was synthesized using SMART-Seq v4 Ultra Low Input RNA kit for Sequencing (Takara) and sequencing libraries were constructed with Nextera XT DNA Library Preparation Kit (Illumina) according to the manufacturer’s instructions. TruSeq Stranded Total RNA kit (Illumina) was used for the library preparation of total BMC and Lin- samples. The resulting sequencing library was sequenced with 2 × 75 bp paired-end reads on MiSeq or NextSeq 500 sequencer (Illumina). Sequenced reads were aligned to the mouse (mm10) reference genome with TopHat (version 2.1.1), and the aligned reads were used for the transcript quantification by using HTSeq-count (version 0.6.1). DESeq2 was used to identify differentially expressed genes across the samples.
Raw data for Lepr+ and NG2+ cells were derived from previous work (Asada et al., 2017) (GEO accession number: GSE89811).
Confocal imaging and image processing
Stained bone sections were imaged with laser scanning confocal microscopes (Leica SP5 or SP8, Zeiss LSM780) after immunohistochemistry. Quantitative analysis of mutant phenotypes was done with the same microscope and identical imaging acquisition setting for mutant and control samples. Overview images of bone were automatically generated using the tile-scan function of SP5 and LSM780 associated software and the resulting pictures combine multiple individual maximum intensity images. Maximum projections were generated by Volocity or Fiji. Volocity (PerkinElmer), Fiji (open source; http://fiji.sc/), Photoshop and Illustrator (Adobe) softwares were used for image processing in compliance with Cell Stem Cell’s guide for digital images. In general, original images were loaded into Volocity and brightness-contrast modifications were applied to the whole image. Images exported from Volocity were rotated and cropped in Fiji. Quantification of cell number, length, and area was performed in Volocity and Fiji. Shape factor is a numerical indication of how similar a 2D shape is to a perfect circle. A shape factor of 1 indicates a perfect sphere.
Quantification and Statistical Analysis
No statistical methods were used to predetermine sample size. Mice that died before the completion of experimental protocols were excluded from analysis, which was a pre-established criterion before the experiment. No randomization and blinding were used. Samples were tested using two-tailed Student’s t test. P value less than 0.05 was considered to be statistically significant. Statistical data were drawn from normally distributed group. Before the Student’s t-Test, samples from different groups were tested using F-test to identify the variances between groups. F value less than 0.05 indicated samples have significantly different variances. All results are represented as mean ± s.e.m. Number of animals or cells represents biological replicates.
Data and Code Availability
RNA-seq raw data supporting this work have been deposited in GEO, under accession number GSE115422. All original data are available upon reasonable request to Lead Contact.
Acknowledgments
Funding was provided by the Max Planck Society, the University of Münster, the Deutsche Forschungsgemeinschaft cluster of excellence “Cells in Motion”, the European Research Council (AdG 339409 AngioBone; AdG 786672 PROVEC), and the Chinese Academy of Science. Y.L. was supported by Christiane Nüsslein-Volhard Stiftung.
Author Contributions
Q.C., Y.L., and R.H.A. designed the study, interpreted results, and wrote the manuscript. Q.C. and Y.L. performed most experiments. H.-W.J. analyzed RNA-seq data and wrote part of the manuscript. M.S. performed FACS experiments. V.V.D. performed part of the FACS and immunostaining experiments. B.Z. provided critical mouse models.
Declaration of Interests
The authors declare no competing interests.
Published: November 21, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2019.10.006.
Supplemental Information
References
- Aguirre A., Blázquez-Prieto J., Amado-Rodriguez L., López-Alonso I., Batalla-Solís E., González-López A., Sánchez-Pérez M., Mayoral-Garcia C., Gutiérrez-Fernández A., Albaiceta G.M. Matrix metalloproteinase-14 triggers an anti-inflammatory proteolytic cascade in endotoxemia. J. Mol. Med. (Berl.) 2017;95:487–497. doi: 10.1007/s00109-017-1510-z. [DOI] [PubMed] [Google Scholar]
- Asada N., Kunisaki Y., Pierce H., Wang Z., Fernandez N.F., Birbrair A., Ma’ayan A., Frenette P.S. Differential cytokine contributions of perivascular haematopoietic stem cell niches. Nat. Cell Biol. 2017;19:214–223. doi: 10.1038/ncb3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballen K.K., Gluckman E., Broxmeyer H.E. Umbilical cord blood transplantation: the first 25 years and beyond. Blood. 2013;122:491–498. doi: 10.1182/blood-2013-02-453175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno N., Przybylski D., Kowalczyk M.S., Kfoury Y., Severe N., Gustafsson K., Kokkaliaris K.D., Mercier F., Tabaka M., Hofree M. A Cellular Taxonomy of the Bone Marrow Stroma in Homeostasis and Leukemia. Cell. 2019;177:1915–1932. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougaki M., Searles R.J., Kida K., Yu J., Buys E.S., Ichinose F. Nos3 protects against systemic inflammation and myocardial dysfunction in murine polymicrobial sepsis. Shock. 2010;34:281–290. doi: 10.1097/SHK.0b013e3181cdc327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers E., Slaughter A., Frenette P.S., Kuick R., Pello O.M., Lucas D. Granulocyte-derived TNFα promotes vascular and hematopoietic regeneration in the bone marrow. Nat. Med. 2018;24:95–102. doi: 10.1038/nm.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T., Heppner F.L., Tertilt C., Heinen T.J., Kremer M., Wunderlich F.T., Jung S., Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Chen Q., Zhang H., Liu Y., Adams S., Eilken H., Stehling M., Corada M., Dejana E., Zhou B., Adams R.H. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat. Commun. 2016;7:12422. doi: 10.1038/ncomms12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comazzetto S., Murphy M.M., Berto S., Jeffery E., Zhao Z., Morrison S.J. Restricted Hematopoietic Progenitors and Erythropoiesis Require SCF from Leptin Receptor+ Niche Cells in the Bone Marrow. Cell Stem Cell. 2019;24:477–486. doi: 10.1016/j.stem.2018.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copelan E.A. Hematopoietic stem-cell transplantation. N. Engl. J. Med. 2006;354:1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- Crane G.M., Jeffery E., Morrison S.J. Adult haematopoietic stem cell niches. Nat. Rev. Immunol. 2017;17:573–590. doi: 10.1038/nri.2017.53. [DOI] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- del Toro R., Prahst C., Mathivet T., Siegfried G., Kaminker J.S., Larrivee B., Breant C., Duarte A., Takakura N., Fukamizu A. Identification and functional analysis of endothelial tip cell-enriched genes. Blood. 2010;116:4025–4033. doi: 10.1182/blood-2010-02-270819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., Morrison S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan P.L., Himburg H.A., Helms K., Russell J.L., Fixsen E., Quarmyne M., Harris J.R., Deoliviera D., Sullivan J.M., Chao N.J. Epidermal growth factor regulates hematopoietic regeneration after radiation injury. Nat. Med. 2013;19:295–304. doi: 10.1038/nm.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan P.L., Russell J.L., Himburg H.A., Helms K., Harris J.R., Lucas J., Holshausen K.C., Meadows S.K., Daher P., Jeffords L.B. Tie2(+) bone marrow endothelial cells regulate hematopoietic stem cell regeneration following radiation injury. Stem Cells. 2013;31:327–337. doi: 10.1002/stem.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris S.L., Peebles R.S., Jr. PGI2 as a regulator of inflammatory diseases. Mediators Inflamm. 2012;2012:926968. doi: 10.1155/2012/926968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling M., Adams S., Benedito R., Adams R.H. Notch controls retinal blood vessel maturation and quiescence. Development. 2013;140:3051–3061. doi: 10.1242/dev.093351. [DOI] [PubMed] [Google Scholar]
- Ferrara N., Carver-Moore K., Chen H., Dowd M., Lu L., O’Shea K.S., Powell-Braxton L., Hillan K.J., Moore M.W. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- Ferrara J.L., Levine J.E., Reddy P., Holler E. Graft-versus-host disease. Lancet. 2009;373:1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber H.P., Hillan K.J., Ryan A.M., Kowalski J., Keller G.A., Rangell L., Wright B.D., Radtke F., Aguet M., Ferrara N. VEGF is required for growth and survival in neonatal mice. Development. 1999;126:1149–1159. doi: 10.1242/dev.126.6.1149. [DOI] [PubMed] [Google Scholar]
- Gerber H.P., Malik A.K., Solar G.P., Sherman D., Liang X.H., Meng G., Hong K., Marsters J.C., Ferrara N. VEGF regulates haematopoietic stem cell survival by an internal autocrine loop mechanism. Nature. 2002;417:954–958. doi: 10.1038/nature00821. [DOI] [PubMed] [Google Scholar]
- Greenbaum A., Hsu Y.M., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., Link D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Poulos M.G., Palikuqi B., Badwe C.R., Lis R., Kunar B., Ding B.S., Rabbany S.Y., Shido K., Butler J.M., Rafii S. Endothelial jagged-2 sustains hematopoietic stem and progenitor reconstitution after myelosuppression. J. Clin. Invest. 2017;127:4242–4256. doi: 10.1172/JCI92309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigh J.J., Morelli P.I., Gerhardt H., Haigh K., Tsien J., Damert A., Miquerol L., Muhlner U., Klein R., Ferrara N. Cortical and retinal defects caused by dosage-dependent reductions in VEGF-A paracrine signaling. Dev. Biol. 2003;262:225–241. doi: 10.1016/s0012-1606(03)00356-7. [DOI] [PubMed] [Google Scholar]
- Hassanshahi M., Hassanshahi A., Khabbazi S., Su Y.W., Xian C.J. Bone marrow sinusoidal endothelium: damage and potential regeneration following cancer radiotherapy or chemotherapy. Angiogenesis. 2017;20:427–442. doi: 10.1007/s10456-017-9577-2. [DOI] [PubMed] [Google Scholar]
- Helker C.S., Schuermann A., Pollmann C., Chng S.C., Kiefer F., Reversade B., Herzog W. The hormonal peptide Elabela guides angioblasts to the midline during vasculogenesis. eLife. 2015;4 doi: 10.7554/eLife.06726. Published online May 27, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg H.A., Muramoto G.G., Daher P., Meadows S.K., Russell J.L., Doan P., Chi J.T., Salter A.B., Lento W.E., Reya T. Pleiotrophin regulates the expansion and regeneration of hematopoietic stem cells. Nat. Med. 2010;16:475–482. doi: 10.1038/nm.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg H.A., Sasine J., Yan X., Kan J., Dressman H., Chute J.P. A Molecular Profile of the Endothelial Cell Response to Ionizing Radiation. Radiat. Res. 2016;186:141–152. doi: 10.1667/RR14444.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himburg H.A., Termini C.M., Schlussel L., Kan J., Li M., Zhao L., Fang T., Sasine J.P., Chang V.Y., Chute J.P. Distinct Bone Marrow Sources of Pleiotrophin Control Hematopoietic Stem Cell Maintenance and Regeneration. Cell Stem Cell. 2018;23:370–381. doi: 10.1016/j.stem.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L., van Dijk M., Chye S.T.J., Messerschmidt D.M., Chng S.C., Ong S., Yi L.K., Boussata S., Goh G.H., Afink G.B. ELABELA deficiency promotes preeclampsia and cardiovascular malformations in mice. Science. 2017;357:707–713. doi: 10.1126/science.aam6607. [DOI] [PubMed] [Google Scholar]
- Hoggatt J., Kfoury Y., Scadden D.T. Hematopoietic Stem Cell Niche in Health and Disease. Annu. Rev. Pathol. 2016;11:555–581. doi: 10.1146/annurev-pathol-012615-044414. [DOI] [PubMed] [Google Scholar]
- Hooper A.T., Butler J.M., Nolan D.J., Kranz A., Iida K., Kobayashi M., Kopp H.G., Shido K., Petit I., Yanger K. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin T., Gur-Cohen S., Spencer J.A., Schajnovitz A., Ramasamy S.K., Kusumbe A.P., Ledergor G., Jung Y., Milo I., Poulos M.G. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016;532:323–328. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.W., Hernández-Rodríguez B., Kim J., Kim K.P., Enriquez-Gasca R., Yoon J., Adams S., Schöler H.R., Vaquerizas J.M., Adams R.H. Transcriptional regulation of endothelial cell behavior during sprouting angiogenesis. Nat. Commun. 2017;8:726. doi: 10.1038/s41467-017-00738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Wi H.J., Choi M.H., Hong S.T., Bae Y.M. Regulation of anti-inflammatory cytokines IL-10 and TGF-β in mouse dendritic cells through treatment with Clonorchis sinensis crude antigen. Exp. Mol. Med. 2014;46:e74. doi: 10.1038/emm.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Apelin-APJ signaling: a potential therapeutic target for pulmonary arterial hypertension. Mol. Cells. 2014;37:196–201. doi: 10.14348/molcells.2014.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen P.J., Dinarello C.A., Strom T.B. Prostaglandins posttranscriptionally inhibit monocyte expression of interleukin 1 activity by increasing intracellular cyclic adenosine monophosphate. J. Immunol. 1986;137:3189–3194. [PubMed] [Google Scholar]
- Kondo M., Wagers A.J., Manz M.G., Prohaska S.S., Scherer D.C., Beilhack G.F., Shizuru J.A., Weissman I.L. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- Kuba K., Sato T., Imai Y., Yamaguchi T. Apelin and Elabela/Toddler; double ligands for APJ/Apelin receptor in heart development, physiology, and pathology. Peptides. 2019;111:62–70. doi: 10.1016/j.peptides.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., Adams R.H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507:323–328. doi: 10.1038/nature13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., Itkin T., Mäe M.A., Langen U.H., Betsholtz C., Lapidot T., Adams R.H. Age-dependent modulation of vascular niches for haematopoietic stem cells. Nature. 2016;532:380–384. doi: 10.1038/nature17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langen U.H., Pitulescu M.E., Kim J.M., Enriquez-Gasca R., Sivaraj K.K., Kusumbe A.P., Singh A., Di Russo J., Bixel M.G., Zhou B. Cell-matrix signals specify bone endothelial cells during developmental osteogenesis. Nat. Cell Biol. 2017;19:189–201. doi: 10.1038/ncb3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.M., Hu Z., Jorgenson M.L., Wingard J.R., Slayton W.B. Bone marrow sinusoidal endothelial cells undergo nonapoptotic cell death and are replaced by proliferating sinusoidal cells in situ to maintain the vascular niche following lethal irradiation. Exp. Hematol. 2008;36:1143–1156. doi: 10.1016/j.exphem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Liu Q., Hu T., He L., Huang X., Tian X., Zhang H., He L., Pu W., Zhang L., Sun H. Genetic targeting of sprouting angiogenesis using Apln-CreER. Nat. Commun. 2015;6:6020. doi: 10.1038/ncomms7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicone A.M., McGuire J.K. Matrix metalloproteinases as modulators of inflammation. Semin. Cell Dev. Biol. 2008;19:34–41. doi: 10.1016/j.semcdb.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M., Mehta A., de Boer C.G., Kowalczyk M.S., Lee K., Haldeman P., Rogel N., Knecht A.R., Farouq D., Regev A. Heterogeneous Responses of Hematopoietic Stem Cells to Inflammatory Stimuli Are Altered with Age. Cell Rep. 2018;25:2992–3005. doi: 10.1016/j.celrep.2018.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell P.J., Longley D.B., Latif T., Boyer J., Allen W., Lynch M., McDermott U., Harkin D.P., Allegra C.J., Johnston P.G. Identification of 5-fluorouracil-inducible target genes using cDNA microarray profiling. Cancer Res. 2003;63:4602–4606. [PubMed] [Google Scholar]
- Mendelson A., Frenette P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 2014;20:833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R.A., Gray D., Lomova A., Kohn D.B. Hematopoietic Stem Cell Gene Therapy: Progress and Lessons Learned. Cell Stem Cell. 2017;21:574–590. doi: 10.1016/j.stem.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Noach E.J., Ausema A., van Os R., Akkerman I., Koopal S., Weersing E., Dontje B., Vellenga E., de Haan G. Chemotherapy prior to autologous bone marrow transplantation impairs long-term engraftment in mice. Exp. Hematol. 2003;31:528–534. doi: 10.1016/s0301-472x(03)00068-7. [DOI] [PubMed] [Google Scholar]
- Pietras E.M. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 2017;130:1693–1698. doi: 10.1182/blood-2017-06-780882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu M.E., Schmidt I., Giaimo B.D., Antoine T., Berkenfeld F., Ferrante F., Park H., Ehling M., Biljes D., Rocha S.F. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat. Cell Biol. 2017;19:915–927. doi: 10.1038/ncb3555. [DOI] [PubMed] [Google Scholar]
- Rafii S., Lyden D. Therapeutic stem and progenitor cell transplantation for organ vascularization and regeneration. Nat. Med. 2003;9:702–712. doi: 10.1038/nm0603-702. [DOI] [PubMed] [Google Scholar]
- Rafii S., Butler J.M., Ding B.S. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S.K., Kusumbe A.P., Itkin T., Gur-Cohen S., Lapidot T., Adams R.H. Regulation of Hematopoiesis and Osteogenesis by Blood Vessel-Derived Signals. Annu. Rev. Cell Dev. Biol. 2016;32:649–675. doi: 10.1146/annurev-cellbio-111315-124936. [DOI] [PubMed] [Google Scholar]
- Ratajczak M.Z., Suszynska M. Emerging Strategies to Enhance Homing and Engraftment of Hematopoietic Stem Cells. Stem Cell Rev Rep. 2016;12:121–128. doi: 10.1007/s12015-015-9625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha S.F., Schiller M., Jing D., Li H., Butz S., Vestweber D., Biljes D., Drexler H.C., Nieminen-Kelhä M., Vajkoczy P. Esm1 modulates endothelial tip cell behavior and vascular permeability by enhancing VEGF bioavailability. Circ. Res. 2014;115:581–590. doi: 10.1161/CIRCRESAHA.115.304718. [DOI] [PubMed] [Google Scholar]
- Severe N., Karabacak N.M., Gustafsson K., Baryawno N., Courties G., Kfoury Y., Kokkaliaris K.D., Rhee C., Lee D., Scadden E.W. Stress-Induced Changes in Bone Marrow Stromal Cell Populations Revealed through Single-Cell Protein Expression Mapping. Cell Stem Cell. 2019;25:570–583. doi: 10.1016/j.stem.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slayton W.B., Li X.M., Butler J., Guthrie S.M., Jorgensen M.L., Wingard J.R., Scott E.W. The role of the donor in the repair of the marrow vascular niche following hematopoietic stem cell transplant. Stem Cells. 2007;25:2945–2955. doi: 10.1634/stemcells.2007-0158. [DOI] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Taniguchi Ishikawa E., Gonzalez-Nieto D., Ghiaur G., Dunn S.K., Ficker A.M., Murali B., Madhu M., Gutstein D.E., Fishman G.I., Barrio L.C., Cancelas J.A. Connexin-43 prevents hematopoietic stem cell senescence through transfer of reactive oxygen species to bone marrow stromal cells. Proc. Natl. Acad. Sci. USA. 2012;109:9071–9076. doi: 10.1073/pnas.1120358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas E., Storb R., Clift R.A., Fefer A., Johnson F.L., Neiman P.E., Lerner K.G., Glucksberg H., Buckner C.D. Bone-marrow transplantation (first of two parts) N. Engl. J. Med. 1975;292:832–843. doi: 10.1056/NEJM197504172921605. [DOI] [PubMed] [Google Scholar]
- Tian X., Hu T., Zhang H., He L., Huang X., Liu Q., Yu W., He L., Yang Z., Zhang Z. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res. 2013;23:1075–1090. doi: 10.1038/cr.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova A.N., Dolgalev I., Hu H., Sivaraj K.K., Hoxha E., Cuesta-Dominguez A., Pinho S., Akhmetzyanova I., Gao J., Witkowski M. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569:222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nakayama M., Pitulescu M.E., Schmidt T.S., Bochenek M.L., Sakakibara A., Adams S., Davy A., Deutsch U., Lüthi U. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465:483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- Wu S., Wu Y., Capecchi M.R. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Xu C., Gao X., Wei Q., Nakahara F., Zimmerman S.E., Mar J., Frenette P.S. Stem cell factor is selectively secreted by arterial endothelial cells in bone marrow. Nat. Commun. 2018;9:2449. doi: 10.1038/s41467-018-04726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahyapour R., Salajegheh A., Safari A., Amini P., Rezaeyan A., Amraee A., Najafi M. Radiation-induced Non-targeted Effect and Carcinogenesis; Implications in Clinical Radiotherapy. J. Biomed. Phys. Eng. 2018;8:435–446. [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Perry J.M., Marshall H., Venkatraman A., Qian P., He X.C., Ahamed J., Li L. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med. 2014;20:1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- Zhou B.O., Ding L., Morrison S.J. Hematopoietic stem and progenitor cells regulate the regeneration of their niche by secreting Angiopoietin-1. eLife. 2015;4:e05521. doi: 10.7554/eLife.05521. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq raw data supporting this work have been deposited in GEO, under accession number GSE115422. All original data are available upon reasonable request to Lead Contact.