Abstract
Objective:
To conduct the first support vector machine (SVM)-based study comparing the diagnostic accuracy of T1-weighted magnetic resonance imaging (T1-MRI), F-fluorodeoxyglucose-positron emission tomography (FDG-PET) and regional cerebral blood flow single-photon emission computed tomography (rCBF-SPECT) in Alzheimer’s disease (AD).
Method:
Brain T1-MRI, FDG-PET and rCBF-SPECT scans were acquired from a sample of mild AD patients (n=20) and healthy elderly controls (n=18). SVM-based diagnostic accuracy indices were calculated using whole-brain information and leave-one-out cross-validation.
Results:
The accuracy obtained using PET and SPECT data were similar. PET accuracy was 68∼71% and area under curve (AUC) 0.77∼0.81; SPECT accuracy was 68∼74% and AUC 0.75∼0.79, and both had better performance than analysis with T1-MRI data (accuracy of 58%, AUC 0.67). The addition of PET or SPECT to MRI produced higher accuracy indices (68∼74%; AUC: 0.74∼0.82) than T1-MRI alone, but these were not clearly superior to the isolated neurofunctional modalities.
Conclusion:
In line with previous evidence, FDG-PET and rCBF-SPECT more accurately identified patients with AD than T1-MRI, and the addition of either PET or SPECT to T1-MRI data yielded increased accuracy. The comparable SPECT and PET performances, directly demonstrated for the first time in the present study, support the view that rCBF-SPECT still has a role to play in AD diagnosis.
Keywords: Alzheimer’s disease, support vector machine, MRI, FDG-PET, SPECT
Introduction
Alzheimer’s disease (AD) is characterized by structural and metabolic changes in the brain. The most consistent evidence of AD-related neuronal degeneration includes grey matter (GM) atrophy in the temporal lobes and medial parietal cortex and hypometabolism in the temporoparietal cortex.1,2
AD is mainly diagnosed by detecting characteristic patterns of cognitive decline. Its onset is typically insidious–cognitive decline generally begins years before reaching the threshold of clinical significance and functional impairment. Neuropsychological testing can help identify and quantify such cognitive deficits, thus improving diagnostic certainty. The exclusion of other diseases, which requires blood tests and neuroimaging investigations, is also a fundamental part of the diagnostic process.1,2
T1-weighted magnetic resonance imaging (T1-MRI) is widely used both to rule out non-AD pathological processes (such as brain tumors and infarcts) and to characterize a typical AD-related pattern of atrophy in the temporal lobe and medial parietal cortex. Functional neuroimaging modalities, including 18F-fluorodeoxyglucose-positron emission tomography (FDG-PET) and regional cerebral blood flow single-photon emission computed tomography (rCBF-SPECT) frequently reveal the typical pattern of hypometabolism/hypoperfusion in temporoparietal regions and can be useful in cases of diagnostic uncertainty. While FDG-PET has greater sensitivity and specificity to detect AD-related hypofunctional brain patterns, the rCBF-SPECT method is more readily available and less expensive in clinical settings.3,4 These functional modalities should not be used as the only imaging measures during the diagnostic workup because they cannot provide the structural data necessary to exclude non-AD diagnoses that contribute to cognitive decline.1,2 Although AD studies using mean neuroimaging indices have brought major advances in the characterization of group-wise brain abnormalities,5,6 the diagnosis of individual cases based solely on neuroimaging data is still not possible, given that brain findings can vary greatly between patients.
Diagnosis of early AD can be challenging. The task of summarizing the complex, highly dimensional neuroimaging data into a single label – the presence or absence of disease – has been addressed with the use of machine learning-based pattern recognition techniques in different neuropsychiatric disorders.7 The support vector machine (SVM) is a multivariate machine learning approach for classifying individuals through pattern recognition analysis. The aim of SVM is to find the best hyperplane that separates all data points of one group from those of another group.8,9 SVM can potentially be applied to multimodal neuroimaging data to improve the accuracy of AD diagnosis.10,11
Both FDG-PET and rCBF-SPECT can be combined with neurostructural data to provide complementary information about brain changes associated with AD.1 A recent systematic review found that although FDG-PET can help distinguish between different types of dementia, it is at best only slightly better than rCBF-SPECT in differentiating AD patients from healthy controls.12 Nevertheless, evidence from studies comparing rCBF-SPECT and FDG-PET is still “limited and inadequate” and none of the studies included in the above-mentioned review performed SVM analyses.12
SVM has been used in AD studies investigating combinations of T1-MRI + FDG-PET,13-16 but we are not aware of another SVM study that has investigated SVM performance in discriminating AD from controls with a combination of T1-MRI, FDG-PET and rCBF-SPECT scans acquired from the same sample. Thus, it is still unknown whether a combination of T1-MRI plus FDG-PET is superior to T1-MRI plus rCBF-SPECT.
Intensity normalization is a standard step in studies involving FDG-PET and SPECT, whether for total tracer uptake in the brain or for uptake in a single reference brain region. This procedure minimizes the effects of inter-individual differences in whole-brain signal, allowing regional patterns of change in metabolic activity to be identified.17,18 The cerebellum is often used as reference because it is known to be spared in AD,13,18 and both cerebellar and global uptake normalization have been used in SVM studies on AD.13,19 Based on the hypothesis that the reference region is unaffected by the disease under investigation18 and on previous semi-quantitative FDG-PET and rCBF-SPECT studies comparing AD patients and controls,17,20 we expected cerebellar normalization to improve diagnostic accuracy. However, no extant SVM study has directly compared the diagnostic accuracy of cerebellar and global normalization when distinguishing AD from controls.
The aim of the present study is to evaluate the diagnostic accuracy of SVM in distinguishing AD patients from healthy controls using neurostructural (T1-MRI) and neurofunctional imaging data (both FDG-PET and rCBF-SPECT). Our intention was to measure SVM performance in conjunction with neuroimaging data (both in isolation and in combination) and characterize the optimal combination of imaging modalities, as well as to test whether normalizing radiotracer uptake in the cerebellum (for PET and SPECT data) would improve the diagnostic performance of the classifier.
Methods
Study population
Twenty patients with mild AD and a group of 18 healthy elderly controls were enrolled. The investigation was approved by the ethics committees of the institutions where the study was conducted. Informed consent was obtained from all participants.
Patients were recruited on their first visit to the memory outpatient clinics of the Neurology and Psychiatry departments of the Universidade de São Paulo Medical School. All patients fulfilled the DSM-III-R21 criteria for mild dementia and NINCDS/ADRDA22 criteria for probable AD. The Clinical Dementia Rating (CDR) scale23 was used to assess clinical severity, and only patients with CDR equal to 0.5 or 1 (i.e., mild dementia) were included. Patients with an auditory deficit or uncorrected visual impairment that could have interfered with their cognitive evaluation were excluded, as were those undergoing pharmacological AD treatment with cholinesterase inhibitors. Healthy controls were recruited from activity centers and groups for elderly individuals in the community.
Exclusion criteria for both the AD and control groups were: less than four years of education, age below 60 or above 90 years, psychotropic drug use, diabetes mellitus, the presence of systemic disorders associated with cognitive impairment, or brain lesions incidentally detected in MRI. Family history of AD and information about current and previous medical, neurological and psychiatric illness was obtained during interviews with the participants and their families. Neurologic or psychiatric disorders prior to the onset of AD were also exclusion criteria.
Patients with AD underwent an extensive neuropsychological assessment including the following tests: the Mattis Dementia Rating Scale (DRS),24 the Trail-Making Test parts A and B, the Hopper Visual Organization Test, the Rey Auditory Verbal Learning Test, the Wechsler Memory Scale: logic memory and visual reproduction subtests, the Rey-Osterrieth complex figure test, the Boston Naming Test and the Wisconsin Card Sorting Test. Instrumental activities of daily living were quantified using the Functional Activities Questionnaire.25
Image acquisition and preprocessing
Details on image acquisition and preprocessing are presented as online-only supplementary material (367.7KB, pdf) .
Classification using support vector machine (SVM)
We employed a whole-brain approach involving a mask to exclude voxels outside the brain. This resulted in feature vectors of 219727 voxels for all modalities (each feature corresponds to a brain voxel).
We applied SVM8,9,26,27 using LIBSVM software28 (available in the PRoNTo toolbox)11 to classify controls (class 1) and AD patients (class 2). Considering the large number of features, we used a linear kernel, since there was no need to map them to an even higher dimensional feature space to separate the examples. Due to the comparative nature of our investigation, the soft-margin SVM penalty parameter C was fixed to 1, so no grid search was performed to find the optimal parameter when classifying.
Given that the AD group tended to be older and have fewer years of education than the control group (Table 1), the neuroimaging data was corrected for the effects of age and education. This was achieved through linear algebra operations involving matrix transformation to remove confounding effects from kernels using a residual matrix.29,30
Table 1. Demographic characteristics and neuropsychological performance of both groups, and instrumental activities of daily living of the AD group.
| AD group | Control group | p-value* | |
|---|---|---|---|
| Age | 75.5 (4.0) | 72.7 (4.2) | 0.06 |
| Male/female | 9/11 | 7/11 | 0.70 |
| Education (years) | 7.3 (3.9) | 10.4 (4.8) | 0.05 |
| MMSE | 21.3 (2.8) | 28.1 (1.3) | < 0.01 |
| DRS | 109.30 (12.98) | 134.44 (6.92) | < 0.001 |
| Trail-A | 112.40 (60.84) | 48.11 (16.82) | < 0.001 |
| Trail-B† | 263.62 (75.04) | 132.67 (44.25) | < 0.001 |
| HVOT | 69.25 (10.80) | 58.83 (9.40) | 0.004 |
| RAVLT | 19.60 (6.19) | 36.28 (5.79) | < 0.001 |
| WMS-R-LM | 14.30 (8.32) | 33.56 (7.68) | < 0.001 |
| WMS-R-VR | 12.25 (4.61) | 27.39 (7.45) | < 0.001 |
| ROCF† | 19.76 (10.10) | 26.69 (7.38) | 0.002 |
| BNT | 32.85 (9.21) | 44.39 (7.06) | < 0.001 |
| WCST† | 22.31 (11.86) | 29.29 (9.24) | 0.046 |
| FAQ | 10.00 (6.19) | N/A | - |
Data presented as mean (standard deviation), unless otherwise specified.
AD = Alzheimer’s disease.
p-value for Mann-Whitney tests (continuous variables) or chi-square test (gender). Wilcoxon rank-sum test for neuropsychological performance variables and FAQ.
AD = Alzheimer’s disease; BNT = Boston naming test; DRS = Mattis Dementia Rating Scale (total points)24; FAQ = Functional Activities Questionnaire25; HVOT = Hopper Visual Organization Test (t score, with higher scores reflecting worse performance); MMSE = Mini-Mental State Examination; N/A = not available; RAVLT = Rey Auditory Verbal Learning Test; ROCF = Rey-Osterrieth complex figure test, copy subtest; SD = standard deviation; Trail-A and Trail-B = Trail Making Test parts A and B (time in seconds); WCST = Wisconsin Card Sorting Test, number of correct responses; WMS-R-LM = Wechsler Memory Scale logic memory subtest, immediate recall; WMS-R-VR = Wechsler Memory Scale visual reproduction subtest, immediate recall.
Missing data: there were missing data for Trail-B (four patients), ROCF (one patient) and WCST (seven patients and one control).
During the training phase, the SVM algorithm finds a hyperplane that separates the examples in the input space, thus maximizing the margin of separation between the classes. Support vectors are data points that lie closest to the separating hyperplane. Once the decision function is determined from the training data, it can be used to predict the class label of a new test example.31
To evaluate the generalization ability of the model, the dataset was partitioned into a “training set” and a “testing set.” By repeatedly repartitioning the data this way, we obtained an estimate of the generalization error of the model. We used a leave-one-out cross-validation approach32,33 in which a single example (i.e., one AD patient or healthy control) was left out of testing in each iteration.
Accuracy was obtained through the number of true positives (patients correctly classified) and true negatives (healthy controls correctly classified). Since the number of examples in each class differed, we calculated a balanced accuracy measure, which takes the number of samples in each class into account, giving equal weight to the accuracies obtained in test samples. We also compared the different models using receiver operating characteristic (ROC) curves, plotting the true positive rate against the false positive along different threshold values. The area under the curve (AUC) is a summary measure describing a classifier’s performance across all decision thresholds.
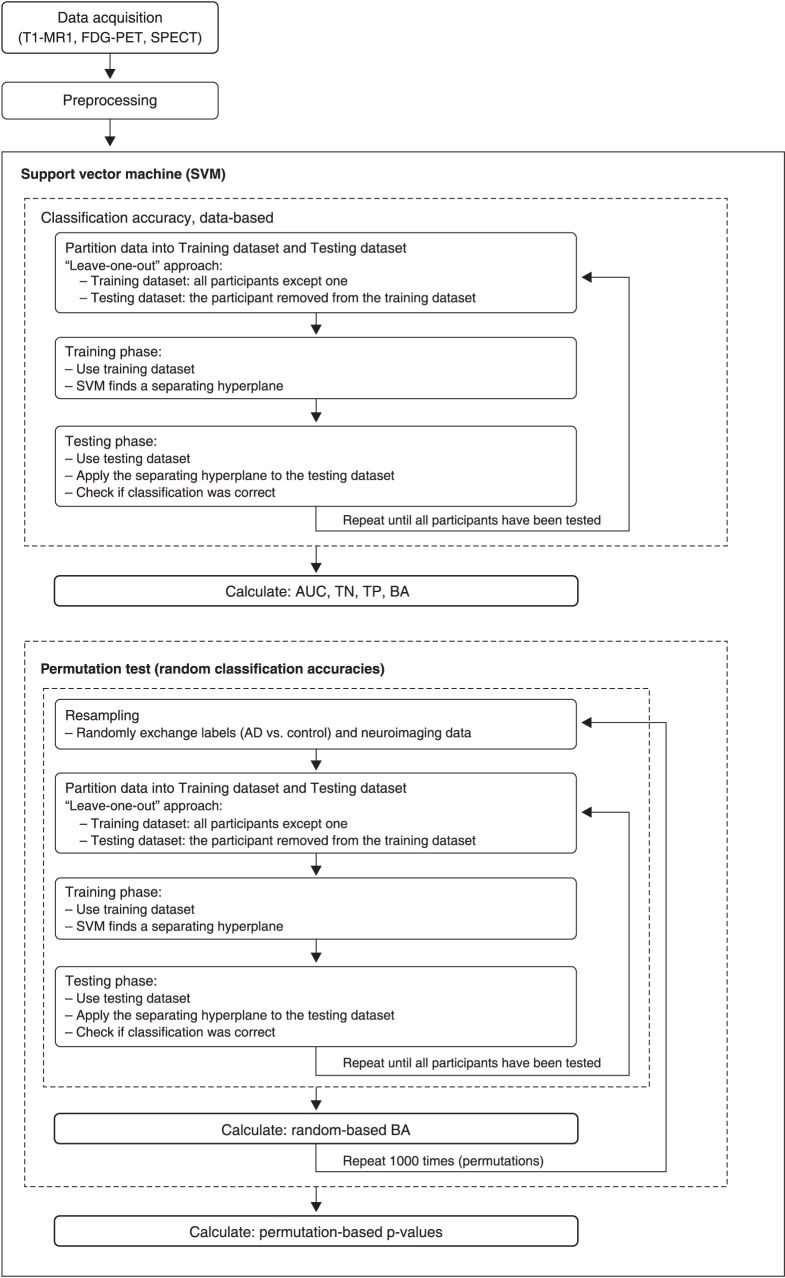
Statistical significance was tested using permutations, a non-parametric approach by which the frequency distribution under the null hypothesis is obtained by calculating many possible combinations under rearrangements of the labels (i.e., “AD patients” vs. “controls”) across the examples. We randomly exchanged the labels associated with the examples and repeated the complete procedure (leave-one-out training and test) 1,000 times. Since permuted labels destroy the correlation between examples and labels, the classification accuracy should be close to chance (around 50%). Figure 1 schematically describes the flow chart of the SVM analysis.
Figure 1. Flow chart of SVM analysis. AD = Alzheimer’s disease; AUC = area under curve; BA = balanced accuracy; FDG-PET = 18F-fluorodeoxyglucose-positron emission tomography; SPECT = single-photon emission computed tomography; SVM = support vector machine; T1-MRI = T1-weighted magnetic resonance imaging; TN = true negative; TP = true positive.
Results
The AD group (11 female) had a mean age of 75.5±4.0 years and their mean educational level was 7.3±3.9 years. The mean Mini-Mental State Examination (MMSE) score was 21.3±2.3, and the mean time of AD was 22.5±11.0 months. All patients presented a typically slow and gradual symptom onset, with clinically relevant cognitive changes beginning 22.5±11 months before study enrollment. The AD group’s mean Functional Activities Questionnaire25 score for instrumental activities of daily living was 10±6.2 points. All AD patients were right-handed. Five patients had a family history of dementia, but without a dominant pattern of inheritance.
Healthy controls (11 female, mean age 72.7±4.2 years, mean education 10.4±4.8 years) presented no memory complaints, their physical and neurological examination was normal and their CDR score was 0 (no deficit). They underwent a comprehensive neuropsychological evaluation (including MMSE) to objectively rule out cognitive deficits,34 and a self-reporting questionnaire35 was used to exclude depression and other mental disorders (all scores were less than 8). All controls except one were right-handed. Volunteers with a family history of AD or any other form of dementia were excluded.
The demographic characteristics of the two groups are presented in Table 1. There was a trend towards greater age and fewer years of formal education in the AD group. Table 1 also presents the neuropsychological assessment data for both groups, as well as the Functional Activities Questionnaire25 scores for the AD group (instrumental activities of daily living). As expected, the neuropsychological performance of the AD group was worse than controls. Further details about the enrolment requirements and sample characteristics have been described in Buchpiguel et al.36
Single modality
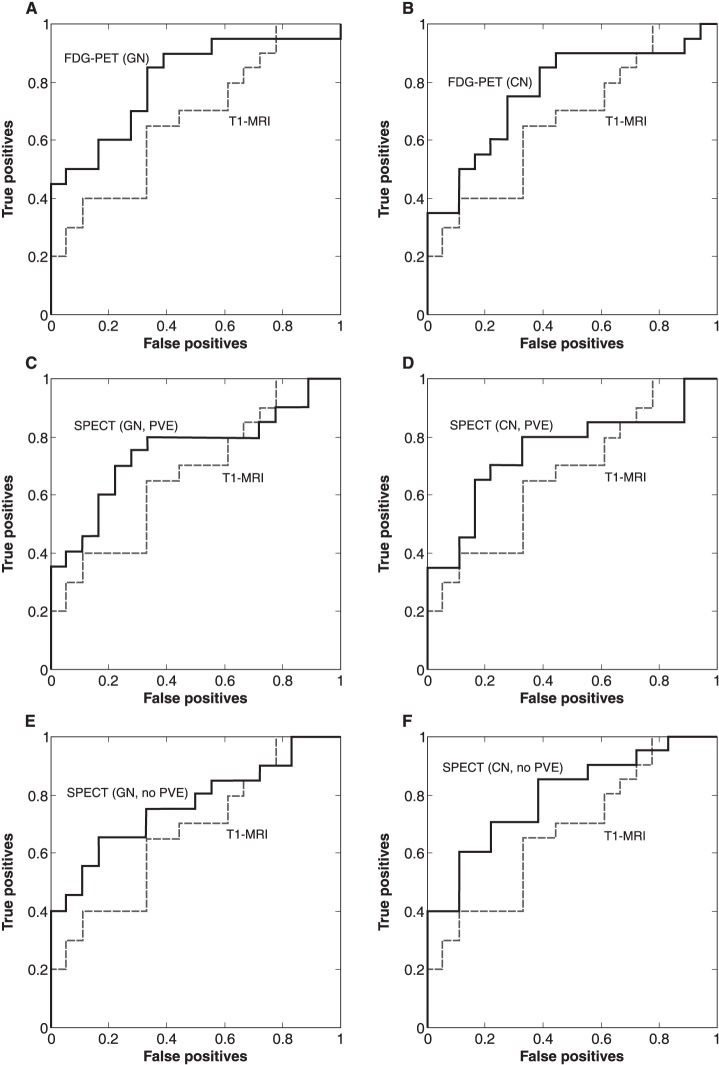
Table 2 presents the results of analyses performed for each modality individually, while Figure 2 presents the ROC curve for each of the functional imaging modalities compared with the T1-MRI data. The best accuracy in distinguishing AD patients from controls was obtained using neurofunctional data. Classification using rCBF-SPECT resulted in an AUC of 0.75∼0.79 and an accuracy of 68∼74%, while FDG-PET data resulted in an AUC of 0.77∼0.81 and an accuracy of 68∼71%. T1-MRI presented the worst performance, with 58% accuracy and an AUC of 0.67. Classification accuracy using neurofunctional data reached statistically significant values, although this did not occur when using only MRI (Table 2). The different preprocessing pipelines for neurofunctional data did not yield clear differences in diagnostic performance.
Table 2. Classification accuracy for each image and combined image modalities.
| Modality | AUC | TP (%) | TN (%) | BA (%) | p-value* |
|---|---|---|---|---|---|
| T1 -MRI | 0.67 | 50.00 | 66.67 | 58.33 | 0.1379 |
| FDG-PET | |||||
| GN | 0.81 | 70.00 | 66.67 | 68.33 | 0.0242 |
| CN | 0.77 | 70.00 | 72.22 | 71.11 | 0.0157 |
| rCBF-SPECT | |||||
| GN | |||||
| PVE | 0.75 | 60.00 | 77.78 | 68.89 | 0.0210 |
| No PVE | 0.76 | 70.00 | 66.67 | 68.33 | 0.0214 |
| CN | |||||
| PVE | 0.75 | 60.00 | 83.33 | 71.67 | 0.0071 |
| No PVE | 0.79 | 70.00 | 77.78 | 73.89 | 0.0043 |
| T1-MRI + FDG-PET (GN) | 0.81 | 75.00 | 66.67 | 70.83 | 0.0198 |
| T1-MRI + FDG-PET (CN) | 0.82 | 70.00 | 72.22 | 71.11 | 0.0014 |
| T1-MRI + rCBF-SPECT (GN, PVE) | 0.78 | 75.00 | 72.22 | 73.61 | 0.0008 |
| T1-MRI + rCBF-SPECT (CN, PVE) | 0.74 | 70.00 | 66.67 | 68.33 | 0.0253 |
| T1-MRI + rCBF-SPECT (GN, no PVE) | 0.78 | 80.00 | 66.67 | 73.33 | 0.0011 |
| T1-MRI + rCBF-SPECT (CN, no PVE) | 0.76 | 75.00 | 72.22 | 73.61 | 0.0003 |
AUC = area under curve; BA = balanced accuracy; CN = cerebellar normalization; FDG-PET = 18F-fluorodeoxyglucose-positron emission tomography; GN = global normalization; PVE = partial volume effect correction; rCBF-SPECT = regional cerebral blood flow single-photon emission computed tomography; T1-MRI = T1-weighted magnetic resonance imaging; TN = true negative; TP = true positive.
Non-parametric statistical significance.
Figure 2. Receiver operating characteristic (ROC) curves of single modality classification. CN = cerebellar normalization; FDG-PET = 18F-fluorodeoxyglucose-positron emission tomography; GN = global normalization; PVE = partial volume effect correction; SPECT = single-photon emission computed tomography; T1-MRI = T1-weighted magnetic resonance imaging.
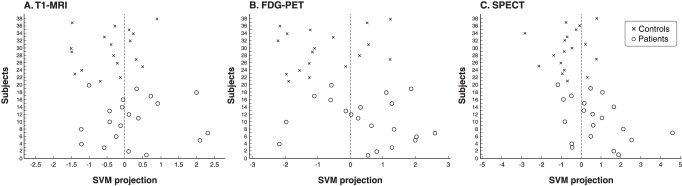
Figure 3 is a graphical representation of predictions for each single modality classification (T1-MRI, FDG-PET and rCBF-SPECT). Since multiple analyses were performed for FDG-PET and rCBF-SPECT, this figure presents only the classifications that resulted in the best accuracies for each modality (i.e., rCBF-SPECT without correction for PVE and both FDG-PET and rCBF-SPECT with cerebellar normalization). Subjects classified as healthy controls are plotted to the left of the dotted vertical line (i.e., with negative SVM projections) while subjects to the right of the vertical line presented positive SVM projections and were thus classified as having AD. Classification using only T1-MRI data resulted in more incorrect labeling than either FDG-PET or rCBF-SPECT alone.
Figure 3. SVM class prediction of individual subjects for each imaging modality. y-axis: subject identification number. x-axis: linear projection for each subject. Subjects receiving negative values (i.e., data points to the left of the vertical dotted line) were classified as controls while those receiving positive values (i.e., to the right of the line) were classified as having Alzheimer’s disease (AD). Misclassifications can be observed as circles to the left of the separating plane (AD patients incorrectly classified as controls, i.e., false negatives) and crosses to the right of it (controls incorrectly classified as AD patients, i.e., false positives). FDG-PET = 18F-fluorodeoxyglucose-positron emission tomography; SPECT = single-photon emission computed tomography; SVM = support vector machine; T1-MRI = T1-weighted magnetic resonance imaging.
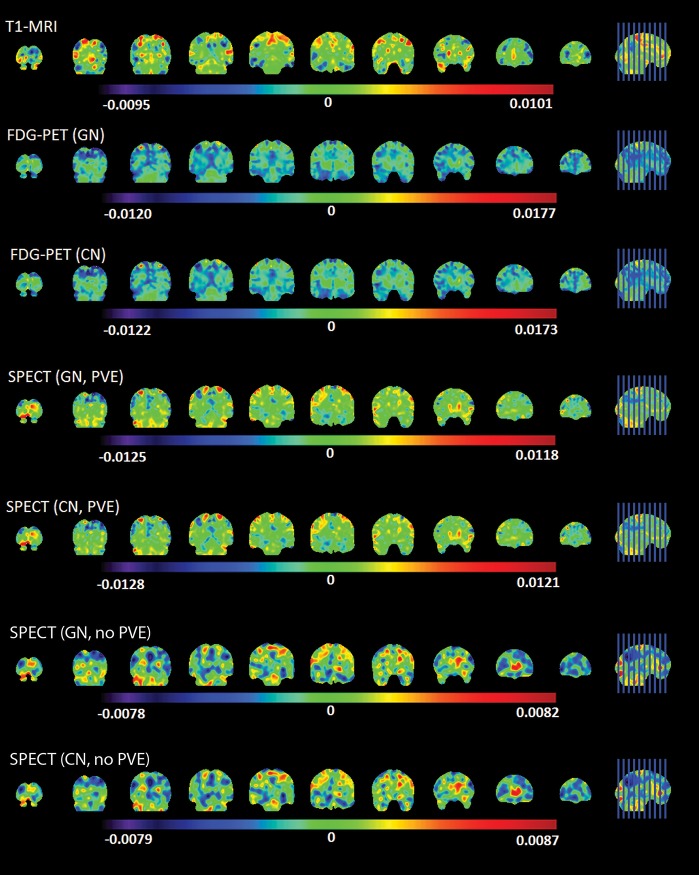
Figure 4 presents the three discriminant maps showing the relative weight of brain voxels resulting from the application of linear SVM to classify AD patients vs. controls using each imaging modality (T1-MRI, FDG-PET and rCBF-SPECT). It is clear in this figure that classifications based on each modality produce different global patterns of anatomical distribution of voxel weights.
Figure 4. Patterns of anatomical distribution of voxel weights resulting from single modality classifications. Positive weights mean a higher relative measure in that voxel for AD patients than controls. Conversely, a negative weight means a higher relative measure for controls. The measures are: GM volume for T1-MRI, regional brain metabolism for FDG-PET and regional cerebral blood flow for rCBF-SPECT. CN = cerebellar normalization; FDG-PET = 18F-fluorodeoxyglucose-positron emission tomography; GN = global normalization; PVE = partial volume effect correction; SPECT = single-photon emission computed tomography; T1-MRI = T1-weighted magnetic resonance imaging.
Combining modalities
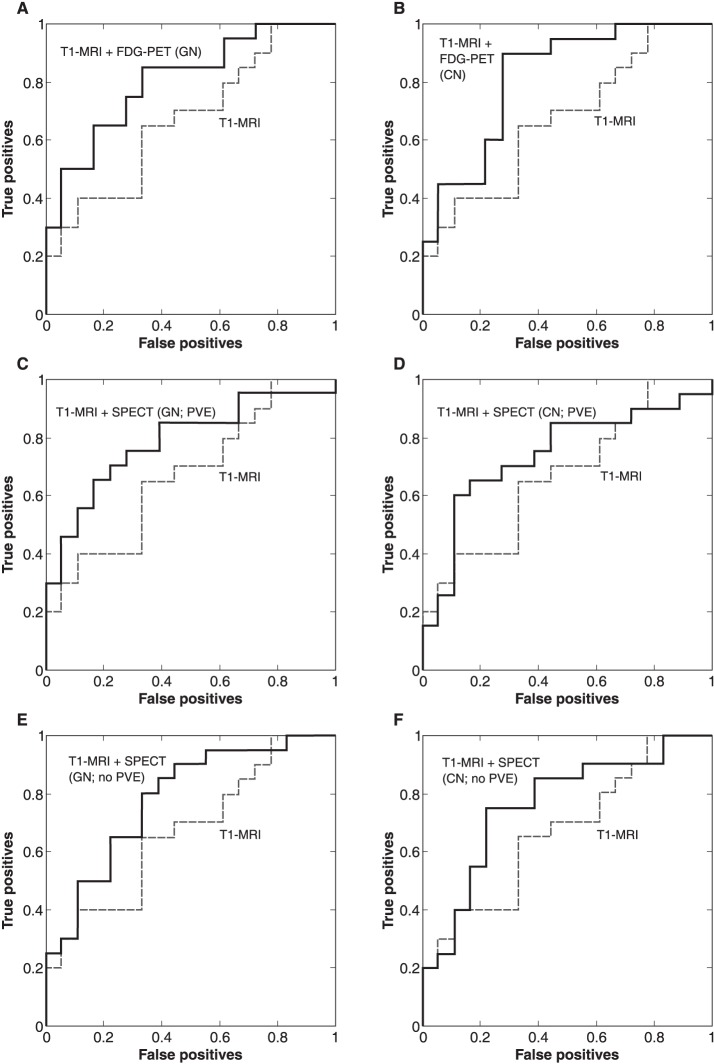
Combining neurofunctional data (FDG-PET or rCBF-SPECT) with T1-MRI yielded better classification accuracies (68∼74% vs. 58%) and improved AUC (0.74∼0.82 vs. 0.67) than neuroanatomical data alone. However, the combination of modalities did not present a clear improvement over isolated neurofunctional data. Table 2 presents the results of analyses with a combination of modalities and Figure 5 shows the ROC curves for each combination of modalities.
Figure 5. Receiver operating characteristic (ROC) curves of classifications using a combination of imaging modalities. CN = cerebellar normalization; FDG-PET = 18F-fluorodeoxyglucose-positron emission tomography; GN = global normalization; PVE = partial volume effect correction; SPECT = single-photon emission computed tomography; T1-MRI = T1-weighted magnetic resonance imaging.
Discussion
To the best of our knowledge, this is the first SVM-based neuroimaging study to conduct comparative diagnostic accuracy assessments of brain T1-MRI, FDG-PET and rCBF-SPECT data acquired from the same AD patients and controls.
The lowest diagnostic accuracy resulted from using only T1-MRI data. One possibility is that nonspecific findings of brain atrophy – variably detected in cognitively preserved elderly subjects – may complicate the discrimination when SVM methods are applied. Moreover, in SVM investigations with a modest sample size (as in our case), variations in a single MRI dataset (for instance due to movement during scanning) may more significantly affect diagnostic performance.37 The higher accuracy indices of neurofunctional data relative to MRI-based data is in accordance with previous T1-MRI and FDG-PET studies,13,14 confirming the idea that brain metabolic markers more accurately differentiate AD-related pathological changes from the effects of healthy aging on the brain.14 Moreover, multimodal neuroimaging studies have suggested that metabolic changes precede atrophy in AD.38 Accordingly, using imaging modalities sensitive to hypometabolism, such as FDG-PET and rCBF-SPECT, resulted in better accuracy than classification with T1-MRI, especially considering that the sample consisted of patients with mild dementia.
Previous studies have used a number of different classification techniques with variable results,39-42 but, to our knowledge, ours is the first to demonstrate that a multivariate analysis resulted in rCBF-SPECT accuracy indices comparable to those of FDG-PET. Our rCBF-SPECT findings agree with the results of other AD SPECT studies.43,44 Thus, this imaging technique may still have a role in AD diagnosis, especially given its broader availability in certain regions than FDG-PET.
In the present study, combining neurofunctional and neuroanatomical data yielded higher accuracy indices than T1-MRI images alone, which agrees with previous studies.13,14 Although adding neurofunctional data to neuroanatomical data yielded better results, the accuracy resulting from the combination of modalities was not clearly superior to that of neurofunctional modalities alone, which also agrees with previous PET and MRI studies.13,14 This may be due to the whole brain approach, since it has been previously shown that using data extracted only from disease-related regions of interest can result in increased MRI-T1+FDG-PET classification accuracy compared to single modality FDG-PET.13,45
Previous SVM-based investigations using FDG-PET or rCBF-SPECT data did not compare the diagnostic accuracy of cerebellar vs. global normalization for distinguishing AD from controls. In this study the accuracy indices were similar for the two tracer normalization procedures. This suggests that although the normalization procedure can yield different results in studies comparing mean functional data between groups of healthy controls and AD,46 the normalization approach is not as critical when classifying individuals using pattern recognition analysis.
There are a number of strengths in the present study that should be highlighted, such as the careful selection and clinical assessment of controls and patients in early-stage AD. All three image modalities were performed with all subjects, allowing for direct comparison of the classifier performance between modalities and combinations of modalities. Moreover, we employed state-of-the-art methods to correct functional neuroimaging data for PVE.
However, the relatively modest sample size is an important limitation, and this probably influenced our relatively low accuracy figures compared to studies with larger samples,37 particularly regarding T1-based MRI data. However, the novelty of our study lies not in reporting how well neuroimaging-based diagnostic performance indices can discriminate AD from controls using SVM, but rather in its direct comparisons of neuroimaging modalities, particularly since both the PET and SPECT data were acquired from the same AD patients and controls. Indeed, the SVM-based evaluation of such a modestly sized but carefully screened sample has still provided sufficient power to produce high diagnostic accuracy scores when either of these neurofunctional modalities was included in the analyses. Another limitation of our study is that we estimated neither the intelligence coefficient of the AD patients and controls nor used a scale to document the presence, severity and clinical impact of neuropsychiatric symptoms, which are very common in AD.47 Nevertheless, the lack of this information is a common limitation in the field, shared by other SVM studies on AD.13-16 It is important to point out that neurologic or psychiatric disorders prior to the onset of AD were exclusion criteria and that the development of neuropsychiatric symptoms has been considered a core feature of AD.47 Therefore, we believe that our study required no data correction for the presence of these symptoms. A third limitation worth mentioning is that the T1-MRI data was acquired using a 1.5 Tesla scanner, and it is possible that higher accuracies would emerge if a 3T scanner had been used. Finally, although the SVM analysis conducted herein allowed the accuracy of a machine learning-based approach to be tested with no need for a priori definitions of anatomical hypotheses, such a whole-brain multivariate approach limits the interpretability of anatomical results. Therefore, although the attribution of voxel weights was available, we refrain from drawing any conclusions about the involvement of specific brain regions. We intend to address this issue in additional investigations involving our multimodal imaging data, using an approach more suitable for extracting anatomical information from SVM analyses.48
A growing number of studies are employing machine learning approaches to deal with the large quantities of data generated by neuroimaging investigations, and future studies should provide new insights and possibilities. For instance, the application of SVM techniques to neurofunctional and neurostructural data has already shown the potential to predict future cognitive decline and conversion from mild cognitive impairment to AD.49 Another possibility is using longitudinal neuroimaging data (i.e. rate of change) as an input for the classifier.50,51 One additional alternative is to apply a probabilistic framework that allows the likelihood of diagnosis to be determined so that the concept of disease as a spectrum from normality to overt AD can be reflected in the analytical approach.52 Another interesting idea is to apply machine-learning techniques to combine neuroimaging data with other types of biomarkers (e.g., CSF Aβ42, t-tau and p-tau; APOE genotype) to further improve accuracy.52,53 Finally, it should be pointed out that although SVM is a popular multivariate machine learning approach, other similar methods have been applied to AD neuroimaging studies, such as the naive-Bayes classifier, the Gaussian process and neural network classifications.52,54-56
In conclusion, SVM performance in distinguishing AD from controls was better with neurofunctional data (FDG-PET and rCBF-SPECT) than with T1-MRI. The combination of neurofunctional and volumetric data also yielded higher accuracies than T1-MR alone. Classification using SPECT presented similar results to those obtained with FDG-PET.
Disclosure
The authors report no conflicts of interest.
Acknowledgements
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP; grants 2004/05551-6 and 2012/50329-6). JMR is supported by the Efficacy and Mechanism Evaluation (EME) Programme of the Medical Research Council (MRC) and National Institute for Health Research (NIHR, United Kingdom) partnership. GFB is supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and FAPESP (grant 2012/50329-6). CAB is supported by FAPESP (grant 2004/05551-6).
References
- 1.Ferreira LK, Busatto GF. Neuroimaging in Alzheimer's disease: current role in clinical practice and potential future applications. Clinics (São Paulo). 2011;66:19–24. doi: 10.1590/S1807-59322011001300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks DJ. Positron emission tomography and single-photon emission computed tomography in central nervous system drug development. NeuroRx. 2005;2:226–36. doi: 10.1602/neurorx.2.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuda H. Role of neuroimaging in Alzheimer's disease, with emphasis on brain perfusion SPECT. J Nucl Med. 2007;48:1289–300. doi: 10.2967/jnumed.106.037218. [DOI] [PubMed] [Google Scholar]
- 5.Busatto GF, Diniz BS, Zanetti MV. Voxel-based morphometry in Alzheimer's disease. Expert Rev Neurother. 2008;8:1691–702. doi: 10.1586/14737175.8.11.1691. [DOI] [PubMed] [Google Scholar]
- 6.Frisoni GB, Pievani M, Testa C, Sabattoli F, Bresciani L, Bonetti M, et al. The topography of grey matter involvement in early and late onset Alzheimer's disease. Brain. 2007;130:720–30. doi: 10.1093/brain/awl377. [DOI] [PubMed] [Google Scholar]
- 7.Orrù G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: A critical review. Neurosci Biobehav Rev. 2012;36:1140–52. doi: 10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Boser BE, Guyon IM, Vapnik VN. A training algorithm for margin optimal classifiers [Internet].; Proceedings of the Fifth Annual Workshop on Computational Learning Theory; 1992. [cited 2017 May 22]. svms.org/training/BOGV92.pdf. [Google Scholar]
- 9.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–97. [Google Scholar]
- 10.Dyrba M, Ewers M, Wegrzyn M, Kilimann I, Plant C, Oswald A, et al. Combining DTI and MRI for the automated detection of Alzheimer’s disease using a large European multicenter dataset. In: Yap PT, Liu T, Shen D, Westin CF, Shen L, editors. Multimodal brain image analysis. Berlin: Springer; 2012. pp. 18–28. [Google Scholar]
- 11.Schrouff J, Rosa MJ, Rondina JM, Marquand AF, Chu C, Ashburner J, et al. PRoNTo: pattern recognition for neuroimaging toolbox. Neuroinformatics. 2013;11:319–37. doi: 10.1007/s12021-013-9178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davison CM, O'Brien JT. A comparison of FDG-PET and blood flow SPECT in the diagnosis of neurodegenerative dementias: a systematic review. Int J Geriatr Psychiatry. 2014;29:551–61. doi: 10.1002/gps.4036. [DOI] [PubMed] [Google Scholar]
- 13.Dukart J, Mueller K, Horstmann A, Barthel H, Möller HE, Villringer A, et al. Combined evaluation of FDG-PET and MRI improves detection and differentiation of dementia. PLoS One. 2011;6:e18111. doi: 10.1371/journal.pone.0018111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dukart J, Mueller K, Barthel H, Villringer A, Sabri O, Schroeter ML, et al. Meta-analysis based SVM classification enables accurate detection of Alzheimer’s disease across different clinical centers using FDG-PET and MRI. Psychiatry Res. 2013;212:230–6. doi: 10.1016/j.pscychresns.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Jie B, Zhang D, Cheng B, Shen D. Alzheimer's Disease Neuroimaging Initiative. Manifold regularized multitask feature learning for multimodality disease classification. Hum Brain Mapp. 2015;36:489–507. doi: 10.1002/hbm.22642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu F, Wee CY, Chen H, Shen D. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer's disease and mild cognitive impairment identification. Neuroimage. 2014;84:466–75. doi: 10.1016/j.neuroimage.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dukart J, Mueller K, Horstmann A, Vogt B, Frisch S, Barthel H, et al. Differential effects of global and cerebellar normalization on detection and differentiation of dementia in FDG-PET studies. Neuroimage. 2010;49:1490–5. doi: 10.1016/j.neuroimage.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Yakushev I, Landvogt C, Buchholz HG, Fellgiebel A, Hammers A, Scheurich A, et al. Choice of reference area in studies of Alzheimer's disease using positron emission tomography with fluorodeoxyglucose-F18. Psychiatry Res. 2008;164:143–53. doi: 10.1016/j.pscychresns.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Horn JF, Habert MO, Kas A, Malek Z, Maksud P, Lacomblez L, et al. Differential automatic diagnosis between Alzheimer's disease and frontotemporal dementia based on perfusion SPECT images. Artif Intell Med. 2009;47:147–58. doi: 10.1016/j.artmed.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen JM, Lakatos A, van Erp TG, Kruggel F, Keator DB, Fallon JT, et al. Empirical derivation of the reference region for computing diagnostic sensitive 18 fluorodeoxyglucose ratios in Alzheimer's disease based on the ADNI sample. Biochim Biophys Acta. 2012;1822:457–66. doi: 10.1016/j.bbadis.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Psychiatric Association . Diagnostic and Statistical Manual of Mental Disorders, Third Revised Edition (DSM-III-R). Washington: American Psychiatric Association; 1987. [Google Scholar]
- 22.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA work group under the auspices of department of health and human services task force on Alzheimer's disease. Neurology. 1984;34:939–44. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 23.Morris JC. The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology. 1993;43:2412–4. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 24.Mattis S. Dementia rating scale - professional manual. Florida: Psychological Assessment Resources; 1988. [Google Scholar]
- 25.Pfeffer RI, Kurosaki TT, Harrah CH, Jr, Chance JM, Filos S. Measurement of functional activities in older adults in the community. J Gerontol. 1982;37:323–9. doi: 10.1093/geronj/37.3.323. [DOI] [PubMed] [Google Scholar]
- 26.Davatzikos C, Shen D, Gur RC, Wu X, Liu D, Fan Y, et al. Whole-brain morphometric study of schizophrenia revealing a spatially complex set of focal abnormalities. Arch Gen Psychiatry. 2005;62:1218–27. doi: 10.1001/archpsyc.62.11.1218. [DOI] [PubMed] [Google Scholar]
- 27.Magnin B, Mesrob L, Kinkingnéhun S, Pélégrini-Issac M, Colliot O, Sarazin M, et al. Support vector machine-based classification of Alzheimer's disease from whole-brain anatomical MRI. Neuroradiology. 2009;51:73. doi: 10.1007/s00234-008-0463-x. [DOI] [PubMed] [Google Scholar]
- 28.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. J ACM TIST. 2011;2:27. [Google Scholar]
- 29.Chu C, Ni Y, Tan G, Saunders CJ, Ashburner J. Kernel regression for fMRI pattern prediction. Neuroimage. 2011;56:662–73. doi: 10.1016/j.neuroimage.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rondina JM, Squarzoni P, Souza-Duran FL, Tamashiro-Duran JH, Scazufca M, Menezes PR, et al. Framingham coronary heart disease risk score can be predicted from structural brain images in elderly subjects. Front Aging Neurosci. 2014;6:300. doi: 10.3389/fnagi.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byun H, Lee SW. Applications of support vector machines for pattern recognition: a survey. In: Lee SW, Verri AL, editors. Pattern recognition with support vector machines; 2002. First International Workshop. Niagara Falls, Canada: 2002 Aug 10. pp. 213–36. [Google Scholar]
- 32.Fu WJ, Carroll RJ, Wang S. Estimating misclassification error with small samples via bootstrap cross-validation. Bioinformatics. 2005;21:1979–86. doi: 10.1093/bioinformatics/bti294. [DOI] [PubMed] [Google Scholar]
- 33.Rondina JM, Hahn T, de Oliveira L, Marquand AF, Dresler T, Leitner T, et al. SCoRS--a method based on stability for feature selection and mapping inneuroimaging [corrected]. IEEE Trans Med Imaging. 2014;33:85–98. doi: 10.1109/TMI.2013.2281398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nitrini R, Caramelli P, Porto CS, Charchat-Fichman H, Formigoni AP, Carthery-Goulart MT, et al. Brief cognitive battery in the diagnosis of mild Alzheimer's disease in subjects with medium and high levels of education. Dement Neuropsychol. 2007;1:32–6. doi: 10.1590/S1980-57642008DN10100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harding TW, de Arango MV, Baltazar J, Climent CE, Ibrahim HH, Ladrido-Ignacio L, et al. Mental disorders in primary health care: a study of their frequency and diagnosis in four developing countries. Psychol Med. 1980;10:231–41. doi: 10.1017/s0033291700043993. [DOI] [PubMed] [Google Scholar]
- 36.Buchpiguel CA, Smid J, Duran FL, Bottino C, Ono CR, Leite CC, et al. Brain MRI, SPECT and PET in early Alzheimer’s disease: a minor mismatch between volumetric and functional findings. Curr Mol Imaging. 2014;3:1–9. [Google Scholar]
- 37.Kloppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, et al. A plea for confidence intervals and consideration of generalizability in diagnostic studies. Brain. 2009;132:e102. doi: 10.1093/brain/awn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jack CR, Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9:119–28. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herholz K, Schopphoff H, Schmidt M, Mielke R, Eschner W, Scheidhauer K, et al. Direct comparison of spatially normalized PET and SPECT scans in Alzheimer's disease. J Nucl Med. 2002;43:21–6. [PubMed] [Google Scholar]
- 40.Morinaga A, Ono K, Ikeda T, Ikeda Y, Shima K, Noguchi-Shinohara M, et al. A comparison of the diagnostic sensitivity of MRI, CBF-SPECT, FDG-PET and cerebrospinal fluid biomarkers for detecting Alzheimer's disease in a memory clinic. Dement Geriatr Cogn Disord. 2010;30:285–92. doi: 10.1159/000320265. [DOI] [PubMed] [Google Scholar]
- 41.Nihashi T, Yatsuya H, Hayasaka K, Kato R, Kawatsu S, Arahata Y, et al. Direct comparison study between FDG-PET and IMP-SPECT for diagnosing Alzheimer's disease using 3D-SSP analysis in the same patients. Radiat Med. 2007;25:255–62. doi: 10.1007/s11604-007-0132-8. [DOI] [PubMed] [Google Scholar]
- 42.Silverman DH. Brain 18F-FDG PET in the diagnosis of neurodegenerative dementias: comparison with perfusion SPECT and with clinical evaluations lacking nuclear imaging. J Nucl Med. 2004;45:594–607. [PubMed] [Google Scholar]
- 43.López MM, Ramírez J, Górriz JM, Alvarez I, Salas-Gonzalez D, Segovia F, et al. SVM-based CAD system for early detection of the Alzheimer's disease using kernel PCA and LDA. Neurosci Lett. 2009;464:233–8. doi: 10.1016/j.neulet.2009.08.061. [DOI] [PubMed] [Google Scholar]
- 44.Padilla P, Górriz JM, Ramírez J, Lang EW, Chaves R, Segovia F, et al. Analysis of SPECT brain images for the diagnosis of Alzheimer's disease based on NMF for feature extraction. Neurosci Lett. 2010;479:192–6. doi: 10.1016/j.neulet.2010.05.047. [DOI] [PubMed] [Google Scholar]
- 45.Yun HJ, Kwak K, Lee JM. Alzheimer's Disease Neuroimaging Initiative. Multimodal discrimination of Alzheimer’s disease based on regional cortical atrophy and hypometabolism. PLoS One. 2015;10:e0129250. doi: 10.1371/journal.pone.0129250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duran FL, Zampieri FG, Bottino CC, Buchpiguel CA, Busatto GF. Voxel-based investigations of regional cerebral blood flow abnormalities in Alzheimer's disease using a single-detector SPECT system. Clinics (São Paulo). 2007;62:377–84. doi: 10.1590/s1807-59322007000400002. [DOI] [PubMed] [Google Scholar]
- 47.Lyketsos CG, Carrillo MC, Ryan JM, Khachaturian AS, Trzepacz P, Amatniek J, et al. Neuropsychiatric symptoms in Alzheimer's disease. Alzheimers Dement. 2011;7:532–9. doi: 10.1016/j.jalz.2011.05.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Castro E, Gómez-Verdejo V, Martínez-Ramón M, Kiehl KA, Calhoun VD. A multiple kernel learning approach to perform classification of groups from complex-valued fMRI data analysis: application to schizophrenia. Neuroimage. 2014;87:1–17. doi: 10.1016/j.neuroimage.2013.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang D, Shen D. Alzheimer's Disease Neuroimaging Initiative. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012;7:e33182. doi: 10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ardekani BA, Bermudez E, Mubeen AM, Bachman AH. Alzheimer's Disease Neuroimaging Initiative. Prediction of incipient Alzheimer's disease dementia in patients with mild cognitive impairment. J Alzheimers Dis. 2017;55:269–81. doi: 10.3233/JAD-160594. [DOI] [PubMed] [Google Scholar]
- 51.Farzan A, Mashohor S, Ramli AR, Mahmud R. Boosting diagnosis accuracy of Alzheimer's disease using high dimensional recognition of longitudinal brain atrophy patterns. Behav Brain Res. 2015;290:124–30. doi: 10.1016/j.bbr.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S, et al. Accurate multimodal probabilistic prediction of conversion to Alzheimer's disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–45. doi: 10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D, Wang Y, Zhou L, Yuan H, Shen D. Alzheimer's Disease Neuroimaging Initiative. Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;55:856–67. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Challis E, Hurley P, Serra L, Bozzali M, Oliver S, Cercignani M. Gaussian process classification of Alzheimer's disease and mild cognitive impairment from resting-state fMRI. Neuroimage. 2015;112:232–43. doi: 10.1016/j.neuroimage.2015.02.037. [DOI] [PubMed] [Google Scholar]
- 55.Dyrba M, Ewers M, Wegrzyn M, Kilimann I, Plant C, Oswald A, et al. Robust automated detection of microstructural white matter degeneration in Alzheimer's disease using machine learning classification of multicenter DTI data. PLoS One. 2013;8:e64925. doi: 10.1371/journal.pone.0064925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Wang S, Phillips P, Yang J, Yuan TF. Three-dimensional eigenbrain for the detection of subjects and brain regions related with Alzheimer's disease. J Alzheimers Dis. 2016;50:1163–79. doi: 10.3233/JAD-150988. [DOI] [PubMed] [Google Scholar]